Abstract
Sotos syndrome is a childhood overgrowth syndrome characterized by a distinctive facial appearance, height and head circumference >97th percentile, advanced bone age, and developmental delay. Weaver syndrome is characterized by the same criteria but has its own distinctive facial gestalt. Recently, a 2.2-Mb chromosome 5q35 microdeletion, encompassing NSD1, was reported as the major cause of Sotos syndrome, with intragenic NSD1 mutations identified in a minority of cases. We evaluated 75 patients with childhood overgrowth, for intragenic mutations and large deletions of NSD1. The series was phenotypically scored into four groups, prior to the molecular analyses: the phenotype in group 1 (n=37) was typical of Sotos syndrome; the phenotype in group 2 (n=13) was Sotos-like but with some atypical features; patients in group 3 (n=7) had Weaver syndrome, and patients in group 4 (n=18) had an overgrowth condition that was neither Sotos nor Weaver syndrome. We detected three deletions and 32 mutations (13 frameshift, 8 nonsense, 2 splice-site, and 9 missense) that are likely to impair NSD1 functions. The truncating mutations were spread throughout NSD1, but there was evidence of clustering of missense mutations in highly conserved functional domains between exons 13 and 23. There was a strong correlation between presence of an NSD1 alteration and clinical phenotype, in that 28 of 37 (76%) patients in group 1 had NSD1 mutations or deletions, whereas none of the patients in group 4 had abnormalities of NSD1. Three patients with Weaver syndrome had NSD1 mutations, all between amino acids 2142 and 2184. We conclude that intragenic mutations of NSD1 are the major cause of Sotos syndrome and account for some Weaver syndrome cases but rarely occur in other childhood overgrowth phenotypes.
Introduction
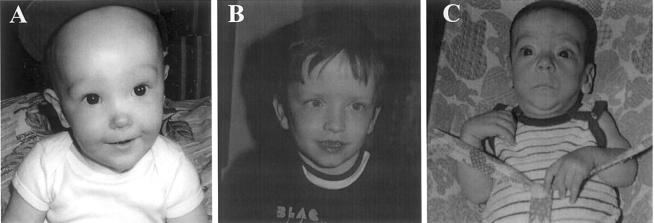
Sotos syndrome (MIM 117550), previously known as “cerebral gigantism,” was initially described in 1964, with hundreds of cases subsequently reported (Sotos et al. 1964; Cole and Hughes 1994). The cardinal features are pre- and postnatal accelerated somatic growth, characteristic facial appearance (i.e., macrocephaly, prominent jaw, and a high hairline with sparse hair growth), advanced bone age, and developmental delay (Cole and Hughes 1994). Additional features that may be present include neonatal hypotonia, seizures, scoliosis, strabismus, congenital heart defects, and cancer (Kaneko et al. 1987; Hersh et al. 1992; Cole and Hughes 1994; Noreau et al. 1998; Opitz et al. 1998; Sweeney et al. 2002). To date, the diagnosis of Sotos syndrome has been based primarily on the presence of the characteristic facial appearance in a large child (fig. 1A). However, although the combination of craniofacial features is distinctive, individual components are nonspecific, and diagnosis can be very difficult for the inexperienced clinician. Furthermore, phenotypic overlap with other childhood overgrowth conditions increases the challenge of accurate clinical diagnosis (Cole and Hughes 1994; Cole 1998).
Figure 1.
Typical facial phenotype in overgrowth groups 1, 2, and 3. A, Group 1 (classic facial gestalt of Sotos syndrome). B, Group 2 (patients with similarities to Sotos syndrome but with some atypical characteristics; primarily the facial features were not classical). C, Group 3 (Weaver syndrome). Patients in Group 4 did not have a distinctive facial phenotype but were characterized by not having the facial phenotypes present in the other three groups.
The condition showing greatest phenotypic overlap with Sotos syndrome is Weaver syndrome (MIM 277590). This condition was originally described in 1974, and the cardinal features are accelerated growth, distinctive facies, advanced bone age, and developmental delay (Weaver et al. 1974; Cole et al. 1992). The facial appearance is somewhat similar to that in Sotos syndrome, but experienced dysmorphologists believe they are distinct (fig. 1C). Additional features occurring in Weaver syndrome include a hoarse low-pitched cry, metaphyseal flaring of the femurs, deep-set nails, prominent finger pads, and camptodactyly (Cole et al. 1992; Opitz et al. 1998; Proud et al. 1998). There has been considerable debate as to whether Sotos and Weaver syndromes are representative of locus or allelic heterogeneity (Cole 1998; Opitz et al. 1998).
Both Sotos and Weaver syndromes usually occur sporadically, although occasional families exhibiting autosomal dominant inheritance of these conditions have been reported (reviewed by Cole and Hughes 1990; Opitz et al. 1998; Proud et al. 1998). The absence of multiple-case families has hampered efforts to identify causative gene(s). However, recently a child with Sotos syndrome and a t(5;8)(q35;q24.1) translocation was reported (Imaizumi et al. 2002), and the gene disrupted by the 5q35 breakpoint was identified as NSD1 (nuclear receptor SET-domain–containing protein) (Kurotaki et al. 2002).
The functions of NSD1 have not been fully elucidated, but it is thought to act as a transcriptional intermediary factor capable of both negatively and positively influencing transcription, depending on the cellular context (Huang et al. 1998; Kurotaki et al. 2001). NSD1 contains multiple functional domains, including the SET (SU[VAR]3-9,E[Z],trithorax) domain, which was initially identified in Drosophila genes involved in chromatin-mediated regulation during development (reviewed by Jenuwein 2001). NSD1 also contains a SET-associated Cys-rich (SAC) domain adjacent to the SET domain. The combination of SAC and SET domains is present in proteins that function as histone-methyltransferases (HMTases), and both are required for HMTase activity (Rea et al. 2000). Thus, it is possible that NSD1 is involved in histone modification and the regulation and maintenance of chromatin states. NSD1 also contains five plant homeodomain (PHD) domains. The PHD domain is a zinc finger–like motif that predominantly occurs in proteins that function at the chromatin level (Aasland et al. 1995). The consensus motif is C4HC3, and NSD1 PHD-I and PHD-IV conform to this. However, NSD1 PHD domains II, III, and V contain a His in place of the final Cys and are thus known as “PHD-H2 fingers” (Huang et al. 1998). NSD1 contains two PWWP (proline-tryptophan-tryptophan-proline) domains. The role of the PWWP motif has not been established, but it is found in regulatory factors and de novo methyltransferases and is thought to be involved in protein-protein interactions (Stec et al. 2000). Finally, NSD1 contains two distinct nuclear receptor (NR) interaction domains, NID−L and NID+L, which are found in NR corepressors and coactivators, respectively (Kurotaki et al. 2001).
Analysis of NSD1 in 42 Japanese patients with sporadic Sotos syndrome revealed 19 with a 2.2-Mb microdeletion of 5q35 encompassing NSD1 and 4 with intragenic NSD1 mutations, predicted to inactivate the protein (Kurotaki et al. 2002). We have evaluated a phenotypically characterized series of patients with overgrowth syndrome for deletions and mutations of NSD1, to investigate the phenotypic and molecular spectra of NSD1 aberrations in childhood overgrowth.
Subjects and Methods
Patients
The research was approved by the London Multicentre Research Ethics Committee, and consent was obtained from all patients and/or parents. DNA was extracted by standard methods. Samples were obtained from two sources: 48 patients were ascertained as part of a previous study and were phenotypically scored independently by three clinical geneticists (Cole and Hughes 1994). Extensive clinical details and serial photographs of these cases have been published elsewhere (Cole and Hughes 1990, 1991, 1994, 1995; Cole et al. 1992, 1995). Twenty-seven patients were ascertained as part of the ongoing Childhood Overgrowth Study. Patients eligible for this study must fulfill four of the following criteria: (a) height >97th percentile, (b) occipito-frontal circumference >97th percentile, (c) bone age >90th percentile, (d) dysmorphic facial features, (e) developmental delay, and (f) congenital anomaly or malformations. The 27 newly ascertained patients were phenotypically scored by three clinical geneticists (T.R.P.C., H.E.H., and I.K.T.). All patients were assigned to one of four groups: patients in group 1 had classical Sotos syndrome (fig. 1A); patients in group 2 had a disease with similarities to Sotos syndrome but with some atypical characteristics, primarily concerning the facial features, which were not fully consistent with the gestalt of Sotos syndrome (fig. 1B); patients in group 3 had Weaver syndrome (fig. 1C); and patients in group 4 had an overgrowth phenotype but did not have either Sotos syndrome or Weaver syndrome. Within group 4, there was one patient considered to have Marshall-Smith syndrome (MIM 602535) and four with a diagnosis of autosomal dominant macrocephaly (MIM 153470; MIM 605309). The remaining group 4 patients did not have a specific recognized overgrowth syndrome. Patients with a confirmed molecular diagnosis of an overgrowth condition, such as Beckwith-Wiedemann syndrome (MIM 130650) or Simpson-Golabi Behmel syndrome (MIM 312870), were excluded. In cases where all three scores were not concordant, the majority score was accepted. No patient was scored into three different categories, and no patient was scored into both groups 1 and 4 or groups 3 and 4 by the assessors. There were high levels of concordant scoring for patients in groups 1, 3, and 4. For group 2, the level of concordant scoring was lower, with several patients scored as either group 1 or 4 by one assessor. Parental DNA was obtained wherever possible. The numbers of patients and parental samples analyzed in each group are shown in table 1.
Table 1.
Phenotypic Delineation of Patients with Childhood Overgrowth, Indicating Numbers of Affected Individuals and Parents Analyzed and the Number of Patients with NSD1 Deletions and Mutations
|
No. of Patients with |
||||||
| Group | Phenotype | No. of Patients Analyzed | NSD1Deletions | NSD1Mutations | Both ParentsAnalyzed | One ParentAnalyzed |
| 1 | Classic Sotos syndrome | 37 | 2 | 26 | 19 | 6 |
| 2 | Sotos-like, but with atypical features | 13 | 1 | 3 | 6 | 0 |
| 3 | Weaver syndrome | 7 | 0 | 3 | 5 | 2 |
| 4 | Childhood overgrowth, but not Sotos or Weaver syndrome | 18 | 0 | 0 | 4 | 11 |
NSD1 Microdeletion Analyses
To identify microdeletions encompassing NSD1, we analyzed polymorphic microsatellite markers within and surrounding NSD1. We developed new microsatellite markers through use of the UCSC Human Genome Project Working Draft sequence (UCSC Genome Bioinformatics Web site). We searched the 2-Mb interval encompassing NSD1 for dinucleotide, trinucleotide, and tetranucleotide repeat elements. Flanking amplifying primers were designed using Primer3 software (Primer3 Web site). All markers were amplified using a touchdown protocol, cycling from 68°C to 50°C, with the exception of SOT19 and SOT20, for which the PCR was performed at a single annealing temperature of 55°C. Of the 22 markers designed, 9 were highly informative and worked reliably in the analyses. The forward primer for each marker was end-labeled with γ[32P]-ATP, and the PCR products were electrophoresed on denaturing polyacrylamide gels and were exposed to x-ray film. The positions of the markers relative to NSD1 were calculated using the UCSC Working Draft sequence. Two markers, SOT3 and SOT17, are intragenic within intron 2 and intron 17 of NSD1, respectively. All the markers are well within the 2.2-Mb interval reported as commonly deleted in patients with Sotos syndrome (Kurotaki et al. 2002). All patients (n=75) and parents (n=87) were analyzed at all markers. The physical locations relative to NSD1, primer sequences, and sizes of the nine markers are given in table 2.
Table 2.
Primer Sequences for Chromosome 5q35 Microsatellite Markers and Positions Relative to NSD1
|
Primer Sequence(5′→3′) |
||||
| Marker | Position relativeto NSD1a(kb) | Forward | Reverse | Size(bp) |
| SOT12 | 645 cen | GATAAACCACAACCCCAACC | ACGTAGCTAGGCACCACCAT | 172 |
| SOT11 | 391 cen | CAGGCTCGTTCATTCACAAA | AGGCAAAATTTCCTCCCATC | 203 |
| SOT10 | 383 cen | GACTGCAAGGAGCTTGAACC | GGGCCACCATACACTTGTTC | 148 |
| SOT1 | 181 cen | GGGAAAGTTGACAGGATTTTGA | GCAAATAGGGCATCTGCAAG | 226 |
| SOT4 | 26 cen | GGTCCTCCACACATTCTGCT | ACATGCCCTATGACCTGGAA | 201 |
| SOT3 | Intragenic | GCACCGTTTTACAGTCCTACTT | CTGCAGTGAGCCAAGACCAT | 216 |
| SOT17 | Intragenic | GGCATTGTTCCTGGATGAGA | GGAGATGGATGTTGCAGTGA | 201 |
| SOT19 | 284 tel | CCCCTTTGTATGGGGTCTTT | CCTGGGTGACACAGTGAGACT | 201 |
| SOT20 | 290 tel | CCAGTTCCATCCAAGACACA | CATTTGATCCAGAAATCCCACT | 267 |
The position relative to the first base of NSD1 is given for markers centromeric (cen) to NSD1. The position relative to the last base of NSD1 is given for markers telomeric (tel) to NSD1. Marker positions are based on the UCSC Human Genome Project Working Draft.
Patients who were homozygous at both intragenic markers and at all intragenic polymorphisms detected in the mutation screen were additionally screened for a whole gene deletion, through use of a method of multiplex PCR amplification of short fluorescent fragments, using MLH1 as a control (Charbonnier et al. 2002). 6-Fam–labeled primers for NSD1 exons 6, 8, and 21 and MLH1 exons 18 and 4 were simultaneously PCR amplified. The resulting product was electrophoresed on an ABI 3100 sequencer (ABI Perkin Elmer) and analyzed with GENOTYPER software. All experiments were repeated six times. A consistent, reproducible 0.5 reduction (i.e., a reduction by one half) in peak height of all three NSD1 exons, compared with MLH1 exons, was taken as indicative of a deletion of one copy of NSD1 in that individual.
NSD1 Mutation Analyses
NSD1 contains 23 exons, the first of which is noncoding. Primers were designed to amplify the remaining 22 exons and intron-exon boundaries of NSD1, using conformation-sensitive gel electrophoresis (CSGE) (Ganguly et al. 1993). In our laboratory, we estimate the sensitivity of CSGE to detect small deletions/insertions and point mutations to be ⩾90%. Primer sequences were designed using the NSD1 genomic sequence (GenBank accession number AF395588) and Primer 3 software. Exons >400 bp were amplified using overlapping primer pairs. The gene was screened in 40 fragments. The primer sequences and sizes for the NSD1 mutation screen are shown in table 3. All fragments were amplified using a touchdown 68°C–50°C protocol. Genomic DNA from patients showing mobility shifts on CSGE was bidirectionally sequenced using the BigDyeTerminator Cycle Sequencing Kit and a 3100 automated sequencer (ABI Perkin Elmer). All 75 patients were screened through all 40 fragments. In mutation-positive patients, parental DNA (if available) was analyzed by direct sequencing for the specific mutation identified in the proband. Mutations were considered pathogenic if they were likely to result in premature truncation of the protein or in exon skipping (small insertions and deletions, nonsense and splice-site mutations). To decide whether missense alterations were likely to be pathogenic, we considered whether they occurred de novo, whether they were present in 200 control individuals, and whether they were at conserved residues in the mouse orthologue and human paralogues. For comparison of NSD1 missense mutations with mouse nsd1 and human NSD2 and NSD3, the BLAST program was used (NCBI BLAST Home Page).
Table 3.
Primer Pairs Used to Amplify the NSD1 Coding Sequence, and Sizes of PCR Products
|
Primer Sequence(5′→3′) |
|||
| Exon | Forward | Reverse | Size(bp) |
| 2A | AGAGTCGAGTCAGATGGCCTA | GATCCATCAGCAGACCCATT | 355 |
| 2B | GTGGAACATCCCAAAATGCT | TCTGTGACTGGCTGTTCTGG | 367 |
| 2C | TGGCTTTCTGCACTTTGAGA | GAAGGGCTGCTTTTTCATTG | 317 |
| 2D | GCCATTCTTGCCATTAGCTC | TTTCCCTTTAAGTGGCCTGT | 323 |
| 3 | TGCTTTTTCAGAAGGCTAATAGG | TCATTCACAAAATGTTCCAAGG | 332 |
| 4 | GCAATGATGTGGCTGTTCTC | TCCAATCTGGGAAACAGAGC | 364 |
| 5A | TCTGATTTCATCTCCCTTTTCC | GGCTTTTCCTTCTCATCTGC | 315 |
| 5B | ATGCCATTTGAAGACTGCAC | TCCACAGGAAGAAAACAGAAAA | 360 |
| 5C | TCCACAGGAAGAAAACAGAAAA | TATGGGATCCAGGTCACTGC | 323 |
| 5D | GGAAAAGCGAAGTGATTCCA | TCTGACTGGGGTTTGTGAAC | 348 |
| 5E | GGGTTGTACTAAGAGTGCAGAGC | TTAGAAATGCTGGCCAAAGG | 348 |
| 5F | TATGGCAGAACCCCCAGTTA | CGCTGCTCCTTCGTCTTACT | 381 |
| 5G | TGGAACATCAAAGCCATCAA | CGCCAGATAATGCAGAGTCA | 332 |
| 5H | GGCTCCACACACAATTCAGA | CTCCCTGCAGTACAGCATCA | 334 |
| 5I | ATGCTTTTTCAGCCCAAATG | CTGGGCCTTTTCCGTTTT | 312 |
| 5J | GATGTGCATTTCGATAGCAAG | GCTCTGTCAGTGGTTCCTCA | 343 |
| 5K | GGTCTTACTTCCTAGTGACC | TATCACATTTAGATGTCCTTAC | 255 |
| 6 | ATGTGGTTTCCCATCTGGTT | TGACATTGAAGATAAAATTGCAG | 300 |
| 7 | AACAATTTTGGCCTGTGGAC | TCAAATACTGAGACCCCAACC | 345 |
| 8 | TTGTGCCCAGTTTCTAAATCA | TGCAAAACAGCCTTTCATGT | 327 |
| 9 | TGGCAGCTGACAATTCAGAC | CTCACTGGTCGGGCTTACAC | 268 |
| 10 | CCCGTTTTCCTAATCCACAA | CCTCTGGCGTGAAAAGTAGC | 310 |
| 11 | AGGGGGTCAAATGGAAGAGA | GATGGAGTGGGTTTCCCTTT | 273 |
| 12 | TCACCTCCTTTTCTGCCACT | CCCAGTGTTGCCACAAAATA | 337 |
| 13 | TGGGTTCAGACGATGTCAAA | TCTGTTGCCAATTAAACTGAGG | 383 |
| 14 | TCCATCATCTTAGTGGTCATTCC | TCCAGTGGCAATATGATGAAA | 411 |
| 15 | TGGATGTACACATACATGACTTGC | AAGAGGGGAGGAGTACCATGA | 342 |
| 16 | ATTTTCCTAATGCCTTGCAG | GGCAGTTTCAAAATGGAGCA | 364 |
| 17 | TCTCCAACTTAAAGGGGAAAAA | AGAGTGGGAAGAGCCAGCTA | 273 |
| 18 | GGACGTGAATTGTCTTCTGCT | TCAAGCAACTGCAAAGAGGA | 381 |
| 19 | CTGCTGCTGACAGTGGTAGG | TATGGCTGGGACAACACAAA | 386 |
| 20 | AAATTTTAATCCACAGCAGAGGTC | GTGGTGATGGTTGCACAAAA | 356 |
| 21 | TCTTGGGAGTTGGTATCCTTTG | AACACTGTTAGGGAGGGAGGA | 257 |
| 22 | GAATGAGGCTCAGAGAGGGTA | AAATGGCATGAGACCCTGAG | 360 |
| 23A | GGAAGGTCATCATCCACACC | GATTGCTCTGCCAGGTGAGT | 356 |
| 23B | TGGGGAGATCCGTGAGTATG | CTGGTCACTGGAGAGGGTTT | 369 |
| 23C | AATCCCAATCCTTGGTTTCC | GGTCTGGACCACAGCTGATA | 380 |
| 23D | CCTACTGACAAACCCCATGC | CTGCTGCTTTCCCAGATGTC | 346 |
| 23E | AGCAAAGGTCTGGGGCATA | CAGGGACTTTGCTCTGTGGT | 395 |
| 23F | TTTTAGGTCTCTCGGGAAGG | GGGGCAGCTTGTTTGTTC | 331 |
Results
NSD1 Microdeletion Analyses
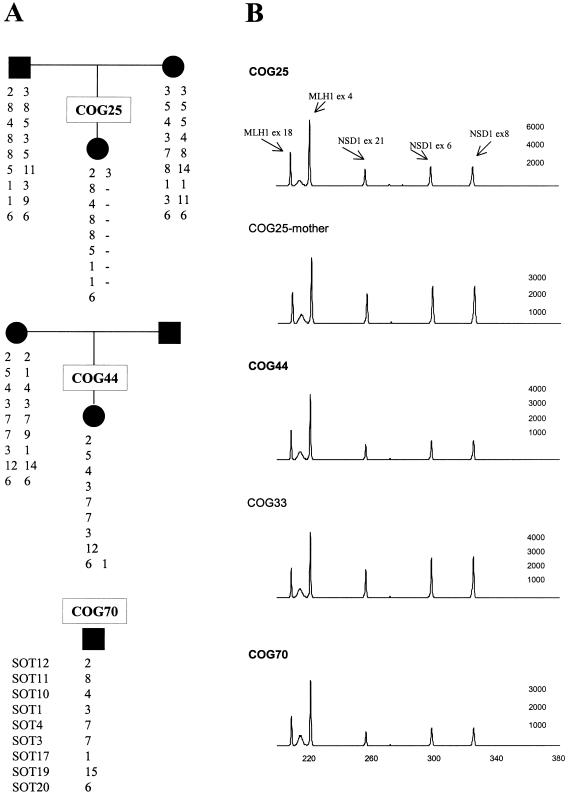
We analyzed DNA from 75 patients and 87 parents at nine microsatellite markers in the 1.5-Mb interval encompassing NSD1, including two intragenic markers (SOT3 and SOT17). In 69 patients, a microdeletion encompassing NSD1 was excluded by the presence of two alleles of differing sizes at one or both intragenic markers and/or by heterozygosity for NSD1 mutations or polymorphisms detected by CSGE. Three patients (COG25, COG44, and COG70) are highly likely to have hemizygous deletions resulting in the loss of one whole copy of NSD1 (and surrounding genes), since they carry a single allele at a minimum of eight of the nine markers analyzed (fig. 2A). For COG25, DNA from both parents was available and confirmed a maternal deletion telomeric to SOT12 in the affected child (fig. 2A). In addition, the multiplex PCR demonstrated a consistent 0.5 reduction in peak height of NSD1 exons (fig. 2B). COG70 has a single allele at all nine SOT markers and is homozygous for all 15 intragenic polymorphisms. Moreover, the multiplex PCR demonstrated a 0.5 reduction in NSD1 exon peak height in six separate experiments. Thus, although we do not have parental DNA or cells for FISH analysis, our results very strongly suggest that COG70 has a deletion of one copy of NSD1. Similarly, COG44 has a single allele at eight consecutive markers, is homozygous for all intragenic polymorphisms, and consistently shows a 0.5 reduction in peak height of NSD1 exons in the multiplex PCR (fig. 2). Only maternal DNA was available and was consistent with transmission of a maternal allele at all SOT markers (fig. 2A). Thus, it is very likely that COG44 has a deletion of the paternal copy of NSD1. Three patients (COG7, COG23, and COG50) were homozygous at the two intragenic markers and were homozygous for the common alleles at the 15 NSD1 polymorphisms, but they did not show reduction of NSD1 exon peak height in the multiplex PCR and are thus very unlikely to have a whole gene deletion.
Figure 2.
NSD1 deletions in three patients with childhood overgrowth—COG25, COG44, and COG70. A, Marker allele haplotypes for nine microsatellite markers spanning NSD1, demonstrating maternal deletion in COG25, probable paternal deletion in COG44, and deletion of unknown origin in COG70. B, Fluorescent multiplex PCR demonstrating a 0.5 reduction of NSD1 exons relative to MLH1, in COG25, COG44, and COG70, and a normal ratio in two control samples: (1) the mother of COG25 and (2) COG33 (who has an intragenic NSD1 mutation). The Y-axis shows fluorescence (in arbitrary units), and the X-axis indicates the size (in bp).
NSD1 Mutation Analyses
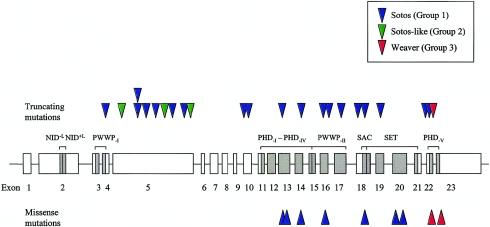
Genomic DNA from all 75 patients was screened for intragenic NSD1 mutations by CSGE. In total, 32 mutations were identified (table 4). Twenty-one mutations were predicted to result in premature truncation of the protein because of frameshift or nonsense alterations. Two splice-site mutations were identified and are also likely to be pathogenic, although the precise effect of these mutations was not determined. The truncating mutations were spread fairly evenly through the gene, between exons 4 and 23 (fig. 3). One mutation (R604X) was identified in two separate patients, COG22 and COG54. Analysis of parental samples for COG22 demonstrated that the mutation had occurred de novo and, hence, these two identical mutations have arisen separately. The mutation is at a CpG dinucleotide, which is known to be particularly susceptible to mutation (Cooper and Krawczak 1989). All other mutations were identified only once. Samples from both parents were available for eight patients with truncating NSD1 mutations, and all were wild type, indicating the mutations had occurred de novo in the affected child.
Table 4.
Pathogenic NSD1 Mutations Identified in 75 Patients with Childhood Overgrowth
| Mutation | ProteinChangea | Exon | Patient | PhenotypicGroup | Results of MutationAnalysis ofParental Samplesb |
| Deletion: | |||||
| 1171delC | Q391fsX418 | 4 | COG14 | 1 | No mutation in parents |
| 1727delA | N576fsX598 | 5 | COG57 | 1 | … |
| 2576delAT | H859fsX873 | 5 | COG69 | 2 | … |
| 3383delCT | S1128fsX1129 | 5 | COG67 | 1 | … |
| 4883delT | M1628fsX1641 | 13 | COG45 | 1 | No mutation in mother |
| 6001delC | L2001fsX2001 | 19 | COG77 | 1 | … |
| 6302delA | K2101fsX2149 | 22 | COG29 | 1 | No mutation in parents |
| Insertion: | |||||
| 2807-8insA | Y936fsX936 | 5 | COG75 | 1 | … |
| 3549-50insT | E1184fsX1184 | 5 | COG73 | 2 | … |
| 5008-9insG | A1670fsX1672 | 14 | COG66 | 1 | … |
| 5744-5insT | M1915fsX1919 | 18 | COG30 | 1 | No mutation in parents |
| 6431-2ins17 | A2144fsX2155 | 22 | COG17 | 1 | No mutation in parents |
| 6450-1insC | K2151fsX2165 | 22 | COG6 | 3 | No mutation in mother |
| Nonsense: | |||||
| 1492C→T | R498X | 5 | COG51 | 2 | … |
| 1810C→T | R604X | 5 | COG22 | 1 | No mutation in parents |
| 1810C→T | R604X | 5 | COG54 | 1 | … |
| 2323C→T | Q775X | 5 | COG74 | 1 | No mutation in parents |
| 4411C→T | R1471X | 10 | COG33 | 1 | No mutation in mother |
| 4417C→T | R1473X | 10 | COG21 | 1 | No mutation in parents |
| 5332C→T | R1778X | 16 | COG63 | 1 | … |
| 5861G→A | W1954X | 18 | COG28 | 1 | No mutation in parents |
| Splice-site: | |||||
| IVS15-1G→C | Int15 | COG72 | 1 | … | |
| IVS16-2delA | Int16 | COG35 | 1 | No mutation in mother | |
| Missense: | |||||
| 4847A→T | H1616L | 13 | COG24 | 1 | No mutation in parents |
| 4910T→C | L1637P | 13 | COG31 | 1 | No mutation in parents |
| 5022C→G | C1674W | 14 | COG52 | 1 | … |
| 5375G→T | G1792V | 16 | COG76 | 1 | No mutation in parents |
| 5773T→C | C1925R | 18 | COG34 | 1 | No mutation in mother |
| 6014G→A | R2005Q | 20 | COG5 | 1 | No mutation in parents |
| 6050G→A | R2017Q | 20 | COG27 | 1 | No mutation in parents |
| 6429C→G | H2143E | 22 | COG15 | 3 | No mutation in parents |
| 6548G→C | C2183S | 23 | COG62 | 3 | No mutation in parents |
Frameshift mutations are designated according to the following example: Q391fsX418 refers to a frameshift mutation in which Q391 is the first amino acid altered, with termination of the ORF at residue 418.
If the result of a parental analysis is not given, the sample was not available for analysis.
Figure 3.
Schematic representation of NSD1, showing the distribution of intragenic mutations identified in the present study and the associated phenotype. Exons of NSD1 are shown as boxes with the exon number underneath. Functional domains of NSD1 are shown as shaded boxes, with the domain name above. Mutations are shown as triangles, with truncating mutations (insertions, deletions, nonsense, and splice-site) above the gene and missense mutations below the gene. One mutation (R604X) was identified twice and is depicted as two triangles, one above the other.
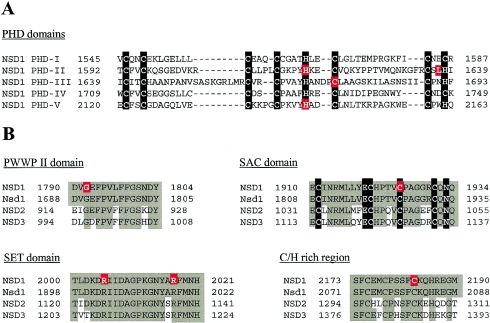
We identified nine missense mutations, and these were clustered in the C-terminal half of NSD1, between exons 13 and 23 (fig. 3). For seven patients with missense mutations, analysis of parental samples indicated the mutations had occurred de novo and, hence, are highly likely to be pathogenic (table 4). Samples from both parents were not available for the remaining missense alterations; however, these mutations (C1925R and C1674W) were not present in the other 74 patients or 200 control individuals. Further evidence supporting a pathogenic role for the missense alterations was obtained from analysis of their position within NSD1. All occur at residues within functional domains and are conserved not only in mouse Nsd1 but also in the two known human paralogues of NSD1, NSD2 (also known as MMSET [Chesi et al. 1998] and WHSC1 [Stec et al. 1998]) and NSD3 (Angrand et al. 2001). Four missense mutations (H1616L, L1637P, C1674W, and H2143E) occur within the PHD domains, and three of these occur at consensus Cys/His residues and would be expected to have critical effects on substrate binding by these domains (fig. 4A). Two further missense mutations also occur at Cys residues: C2183S, in a Cys/His-rich region adjacent to PHD-V, which may correspond to another zinc finger–like motif and is present in mouse Nsd1 and in human NSD2 and NSD3 (fig. 4B); and C1925R, which is in the consensus motif of the SAC domain (fig. 4B). The R2005Q and R2017Q mutations occur within the highly conserved SET domain (fig. 4B), and G1792V is in the PWWP-II domain (fig. 4B).
Figure 4.
NSD1 missense mutations in patients with childhood overgrowth. Residues altered by missense mutations are shown in red boxes. Consensus residues in the PHD and SAC domains are shown in black boxes. Conserved residues in NSD1, mouse Nsd1, NSD2, and NSD3 are shown in shaded boxes. A, The alignment of five PHD domains in NSD1 and the position of the missense mutations identified in the present study. B, Missense mutations in PWWPII, SAC, SET, and C/H-rich domains and in corresponding regions in mouse Nsd1, NSD2, and NSD3.
We identified 15 polymorphisms (table 5). These were considered nonpathogenic, since they were identified in multiple individuals, both affected and unaffected, and were present in patients with a separate pathogenic NSD1 mutation. Moreover, several are intronic or result in synonymous amino acid changes.
Table 5.
Polymorphisms in the NSD1 Gene
| Location andNucleotide Change | ProteinChange |
| Exon 5: | |
| 1749G→A | E583E |
| 1792T→C | L599L |
| 1840T→G | V614L |
| 2071G→A | A691T |
| 2176T→C | S726P |
| 3106G→C | A1036P |
| 3271C→A | L1091I |
| 3705T→C | N1235N |
| Intron 10: | |
| IVS10-9delT | … |
| Intron 17: | |
| IVS17-22G→A | … |
| Intron 18: | |
| 1VS18-132G→A | … |
| Exon 23: | |
| 6750G→A | M2250I |
| 6782T→C | M2261T |
| 6829C→T | L2277L |
| 6903G→C | G2301G |
Correlation of NSD1 Aberrations and Phenotype
There was a strong correlation between the presence of NSD1 mutation and phenotype (table 1; fig. 3). Twenty-six of 32 mutations and two of three deletions were identified in group 1 (classic Sotos) patients. Thus, we identified a pathogenic NSD1 aberration in 28 of 37 group 1 patients, a frequency of 76%. Of 13 group 2 (Sotos-like) patients, 3 harbored NSD1 mutations, and 1 group 2 patient had an NSD1 deletion. The overall frequency of NSD1 alterations in the group 2 patients was thus 4 of 13 patients (30%). In contrast, none of the 18 group 4 (non-Sotos, non-Weaver overgrowth) patients harbored either NSD1 mutations or deletions. Of 7 patients with Weaver syndrome, 3 (42%) harbored NSD1 mutations (fig. 3). Intriguingly, these were all within a 40–amino acid region between 2142 and 2184 in exons 22 and 23 and were the most C-terminal mutations identified (fig. 3).
Discussion
Recently, deletions and mutations in NSD1 were reported in a series of Japanese patients with Sotos syndrome (Kurotaki et al. 2002). To evaluate the contribution of NSD1 aberrations to childhood overgrowth syndromes, we have undertaken an analysis of NSD1 in a phenotypically characterized series of childhood overgrowth syndrome cases from Britain. Our results demonstrate that 76% of patients with Sotos syndrome harbor deleterious alterations of NSD1. Thus, functional abrogation of one copy of NSD1 is the major—and possibly the only—cause of Sotos syndrome. We did not identify mutations or deletions in nine patients with classic Sotos syndrome. Although it is possible that mutations of another gene can lead to Sotos syndrome in a minority of cases, it is equally plausible that the patients with unexplained Sotos syndrome in our series have underlying aberrations in NSD1 that were not detected, either because of lack of sensitivity of the screening technique or because they result in alterations that are not detectable by our methods, such as genomic rearrangements of NSD1 or regulatory mutations.
The frequency (76%) of pathogenic NSD1 aberrations in our series of patients with Sotos syndrome is very similar to the 77% reported by Kurotaki et al. (2002). However, the spectrum of NSD1 aberrations is rather different in the two series. In our series, whole gene deletions occurred in only 8% (3/37) of patients with Sotos syndrome, compared with 66% (20/30) of patients in the Japanese study. Furthermore, Kurotaki et al. (2002) reported a common 2.2-Mb deletion in 19 patients. Although we have not defined the precise breakpoints in our three patients with Sotos syndrome who have deletions, the centromeric breakpoint in COG25 must be telomeric to SOT12, and the telomeric breakpoint in COG44 must be centromeric to SOT21. This positions these breakpoints well within the 2.2-Mb interval reported, and, thus, these two patients are very unlikely to carry the common deletion found in the Japanese patients with Sotos syndrome. Conversely, we detected intragenic mutations in 70% (26/37) of our patients with Sotos syndrome, whereas the Japanese group identified NSD1 mutations in only 10% (4/38). Clinical evaluation of our three patients with 5q35 microdeletions suggest they have more-severe learning difficulties, a coarser facial gestalt, and less pronounced overgrowth than is typical for patients with intragenic NSD1 mutations.
The reasons for the disparity in NSD1 pathogenetic spectra in the Japanese and British populations are not clear. It could simply reflect the particular patients analyzed in these initial studies, and, as more cases are screened, the frequencies of mutations and deletions in the different populations may become more comparable. Indeed, it is quite likely that the difference in NSD1 mutation frequency is explicable by a relative enrichment of true Sotos syndrome in our group 1, since many of the 75 patients we analyzed were initially referred with a potential diagnosis of Sotos syndrome (i.e., group 1) but were rescored as group 2, 3, or 4 by clinical geneticists with expertise in overgrowth conditions.
The disparity in the incidence of NSD1 deletions in the two series is more difficult to resolve. We cannot be missing many deletions, since 69 of 75 patients had NSD1 mutations and/or polymorphisms that were identified by a heteroduplex assay (CSGE) and, hence, were only detectable if two copies of the gene were present. A possible explanation is a specific Japanese overgrowth phenotype, caused by a 5q35 microdeletion, which is very uncommon in Britain. A separate “Japanese Sotos” phenotype has been postulated in the past, to account for the higher incidence of congenital heart disease in Japanese patients with Sotos syndrome (Kaneko et al. 1987). However, the Japanese NSD1 deletions occurred in patients with sporadic Sotos syndrome and, thus, seem unlikely to be caused by a single Japanese founder.
Whatever the reason(s) for these apparent differences, the very high frequency of mutations in our study clearly indicates that intragenic NSD1 mutations are the major cause of Sotos syndrome in the British population. We therefore believe that Sotos syndrome should be considered a single-gene disorder analogous to neurofibromatosis type 1, in which the majority of cases are due to NF1 mutations, with a minor proportion (5%–10%) attributable to chromosomal aberrations that result in deletion of one copy of the gene (Lopez-Correa et al. 2000). We do not consider Sotos syndrome a genomic disorder, comparable to syndromes such as Williams or Smith-Magenis, in which the predominant pathogenetic mechanism is a recurrent chromosomal microdeletion (Stankiewicz and Lupski 2002).
Given that Sotos syndrome is generally caused by hemizygous mutations in an autosomal nonimprinted gene, it is of interest that it does not behave as an autosomal dominant trait and that familial cases are so rare. The mental impairment in Sotos syndrome is usually in the mild/moderate range and is not sufficiently severe to account for the extreme paucity of familial Sotos cases. All of the 15 mutations in which we could assess inheritance occurred de novo, and the 5q35 microsatellite analyses confirmed paternity in all cases. Furthermore, of the 37 group 1 patients with classic Sotos syndrome, only one, COG76, has a family history of the disease, in that her daughter also has Sotos syndrome. Unfortunately, DNA from the daughter was not available for analysis, but she is very likely to carry the G1762V mutation that has occurred de novo in her mother.
The most likely reason for the lack of familial cases is an underlying defect in fertility associated with NSD1 mutations. Delayed menarche, oligomenorrhea, and increases in the rate of spontaneous abortions and stillbirths have been reported in patients with Sotos syndrome (Opitz et al. 1998). Since the diagnostic criteria for the syndrome were reported only relatively recently, it is only now that a generation of children with diagnosed Sotos syndrome are reaching reproductive age, and, thus, the incidence and nature of fertility defects may become clearer in the near future. It is possible that rare cases of familial Sotos syndrome may be explicable by NSD1 mutations that result in Sotos syndrome but not in reproductive impairment. It is noteworthy, in this regard, that the only patient in our series who has a family history of Sotos syndrome has a missense mutation in the PWWP-II domain, and functions of other NSD1 domains may be intact in carriers of this mutation. However, further investigation of patients with familial Sotos syndrome and of the functions of NSD1 are required before conclusions about the postulated reproductive impairment in Sotos syndrome can be drawn.
We analyzed seven patients with Weaver syndrome and identified three NSD1 mutations. Weaver syndrome is a childhood overgrowth syndrome that shares many phenotypic features with Sotos syndrome but is much less common. Identification of NSD1 mutations in patients with Weaver syndrome demonstrates that the two conditions are allelic. However, we have investigated only a small number of patients with Weaver syndrome, and NSD1 mutations were not identified in over half of them, all of whom have phenotypes typical of the syndrome. It remains possible, therefore, that a separate, second Weaver syndrome gene exists. Additional patients with Weaver syndrome need to be screened, to more accurately assess the frequency of NSD1 mutations in this condition.
Our results revealed a strong correlation between clinical phenotype and presence of a deleterious NSD1 mutation or deletion. Most striking was the absence of any mutations or deletions in the 18 group 4 patients with a childhood overgrowth syndrome that was definitely not either Sotos syndrome or Weaver syndrome. The group 4 patients are likely to have a heterogeneous group of conditions, and the majority of patients in this group were felt to have an overgrowth condition that did not correspond to any recognized syndrome. Four of 13 group 2 (Sotos-like, but with some atypical features) patients harbored pathogenic NSD1 alterations. The individual phenotypic scores for three of these were available, and, for each, one clinician (who had actually met the patient) scored the patient as group 1, whereas the other two (who only reviewed photographs) scored the patient as group 2. The fourth mutation-positive group 2 patient was part of the original study that established the diagnostic criteria of Sotos syndrome (Cole and Hughes 1994). This patient was initially scored as group 1 on the basis of the facial gestalt alone but was rescored as group 2 because his bone age was not advanced. Thus, our results suggest that NSD1 aberrations are rather specific for Sotos and Weaver syndromes, and the frequency in other phenotypes is likely to be very low. However, given the challenges of accurate clinical diagnosis of Sotos and Weaver syndromes, a low threshold for diagnostic NSD1 screening in overgrowth conditions would be prudent, until the associated range of phenotypes becomes clearer.
Our data provide preliminary indications of possible genotype-phenotype associations. The majority of known NSD1 functional domains are in exons 11–23, and all missense mutations we identified were clustered in these domains, within highly conserved regions in mouse Nsd1 and human NSD2 and NSD3. It is possible that missense mutations outside these domains do not cause Sotos syndrome. Perhaps the most intriguing finding is the clustering of the three Weaver syndrome mutations within a 40–amino acid region in exons 22 and 23. This region is in the PHD-V domain and the adjacent Cys/His-rich region, and all are 5′ to the SET domain, which may retain some function. It is possible that the position of mutations in the patients with Weaver syndrome is not coincidental and reflects a genotype-phenotype correlation, whereby mutations within the C-terminal region of the gene, after the SET domain, give rise to a Weaver phenotype. However, a mutation within this 40–amino acid region was also identified in a patient with classic Sotos syndrome, indicating that the correlation, if substantiated, cannot be completely specific.
Both Sotos and Weaver syndromes are associated with a wide spectrum of additional abnormalities, including congenital heart disease, scoliosis, and cancer. Investigation of mutational spectra in relation to the phenotype of individual patients may reveal further genotype-phenotype associations and may also be helpful in clarifying the complex functions and interactions of NSD1. The reason why functional abrogation of NSD1, a bifunctional transcriptional regulator with a postulated role in histone and chromatin modification, results in overgrowth and the wide phenotypic spectrum of Sotos and Weaver syndromes remains to be elucidated.
Acknowledgments
We are extremely grateful to all the families with childhood overgrowth conditions that participated in this research, as well as to the clinicians that recruited them to the study, including Amanda Collins, Fiona Stewart, Lucy Raymond, Evan Reid, and Daniela Pilz. We are grateful to Andy Futreal, for the identification of marker SOT3, and to Mike Stratton and Janet Shipley, for helpful discussions. This research was supported by the Institute of Cancer Research (United Kingdom).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the NSD1 genomic sequence [accession number AF395588])
- NCBI BLAST Home Page, http://www.ncbi.nlm.nih.gov/blast/ (for comparison of human NSD1 [GenBank accession number AF395588; RefSeq protein sequence NP_071900] with mouse Nsd1 [GenBank accession number AF419220, RefSeq protein sequence NP_032765] and human NSD2 [GenBank accession number AF083389, RefSeq protein sequence NP_579890] and human NSD3 [GenBank accession number AF332469, RefSeq protein sequence NP_075447])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Sotos syndrome [MIM 117550], Weaver Syndrome [MIM 277590], Marshall Smith syndrome [MIM 602535], autosomal dominant macrocephaly [MIM 605309, MIM 153470], Beckwith-Wiedemann syndrome [MIM 130650], Simpson-Golabi Behmel syndrome [MIM 312870], Neurofibromatosis type 1 [MIM 162200], Williams syndrome [MIM 194050], Smith-Magenis syndrome [MIM 182290])
- Primer3, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi (for designing microsatellite markers amplifying repetitive elements in the vicinity of NSD1 and NSD1 mutation screening primers)
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for identification and positioning of repetitive sequence elements flanking NSD1)
References
- Aasland R, Gibson TJ, Stewart AF (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 20:56–59 [DOI] [PubMed] [Google Scholar]
- Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P (2001) NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics 74:79–88 [DOI] [PubMed] [Google Scholar]
- Charbonnier F, Olschwang S, Wang Q, Boisson C, Martin C, Buisine M-P, Puisieux A, Frebourg T (2002) MSH2 in contrast to MLH1 and MSH6 is frequently inactivated by exonic and promoter rearrangements in hereditary nonpolyposis colorectal cancer. Cancer Res 62:848–853 [PubMed] [Google Scholar]
- Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL (1998) The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 92:3025–3034 [PubMed] [Google Scholar]
- Cole T (1998) Growing interest in overgrowth. Arch Dis Child 78:200–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TRP, Dennis NR, Hughes HE (1992) Weaver syndrome. J Med Genet 29:332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1995) Weaver syndrome: seven new cases and a review of the literature. In: Donnai D, Winter RM (eds) Congenital malformation syndromes. Chapman & Hall, London, pp 267–280 [Google Scholar]
- Cole TRP, Hughes HE (1990) Sotos syndrome. J Med Genet 27:571–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1991) Autosomal dominant macrocephaly: benign familial macrocephaly or a new syndrome? Am J Med Genet 41:115–124 [DOI] [PubMed] [Google Scholar]
- ——— (1994) Sotos syndrome: a study of the diagnostic criteria and natural history. J Med Genet 31:20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1995) Sotos syndrome. In: Donnai D, Winter RM (eds) Congenital malformation syndromes. Chapman & Hall, London, pp 254–266 [Google Scholar]
- Cooper DN, Krawczak M (1989) Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet 83:181–188 [DOI] [PubMed] [Google Scholar]
- Ganguly A, Rock MJ, Prockop DJ (1993) Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci USA 90:10325–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh JH, Cole TRP, Bloom AS, Bertolone SJ, Hughes HE (1992) Risk of malignancy in Sotos syndrome. J Pediatr 120:572–574 [DOI] [PubMed] [Google Scholar]
- Huang N, vom Baur E, Garnier J-M, Lerouge T, Vonesch J-L, Lutz Y, Chambon P, Losson R (1998) Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J 17:3398–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi K, Kimura J, Matsuo M, Kurosawa K, Masuno M, Niikawa N, Kuroki Y (2002) Sotos syndrome associated with a de novo balanced reciprocal translocation t(5;8)(q35;q24.1). Am J Med Genet 107:58–60 [DOI] [PubMed] [Google Scholar]
- Jenuwein T (2001) Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol 11:266–273 [DOI] [PubMed] [Google Scholar]
- Kaneko H, Tsukahara M, Tachibana H, Kurashige H, Kuwano A, Kajii T (1987) Congenital heart defects in Sotos sequence. Am J Med Genet 26:569–576 [DOI] [PubMed] [Google Scholar]
- Kurotaki N, Harada N, Yosiura K, Sugano S, Niikawa N, Matsumoto N (2001) Molecular characterisation of NSD1, a human homologue of the mouse Nsd1 gene. Gene 279:197–204 [DOI] [PubMed] [Google Scholar]
- Kurotaki N, Imaizumi K, Harada N, Masuno M, Kondoh T, Nagai T, Ohashi H, Naritomi K, Tsukahara M, Makita Y, Sugimoto T, Sonoda T, Hasegawa T, Chinen Y, Tomita H, Kinoshita A, Mizuguchi T, Yoshiura K, Ohta T, Tatsuya K, Fukushima Y, Niikawa N, Matsumoto N (2002) Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet 30:365–366 [DOI] [PubMed] [Google Scholar]
- Lopez Correa C, Brems H, Lazaro C, Marynen P, Legius E (2000) Unequal meiotic crossover: a frequent cause of NF1 microdeletions. Am J Hum Genet 66:1969–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noreau DR, Al-Ata J, Jutras L, Teebi AS (1998) Congenital heart defects in Sotos syndrome. Am J Med Genet 79:327–328 [DOI] [PubMed] [Google Scholar]
- Opitz JM, Weaver DW, Reynolds JF (1998) The syndromes of Sotos and Weaver: reports and review. Am J Med Genet 79:294–304 [DOI] [PubMed] [Google Scholar]
- Proud VK, Braddock SR, Cook L, Weaver DD (1998) Weaver syndrome: autosomal dominant inheritance of the disorder. Am J Med Genet 79:305–310 [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methytransferases. Nature 406:593–599 [DOI] [PubMed] [Google Scholar]
- Sotos JF, Dodge PR, Muirhead D, Crawford JD, Talbot NB (1964) Cerebral gigantism in childhood. N Engl J Med 271:109–116 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 [DOI] [PubMed] [Google Scholar]
- Stec I, Nagl SB, van Ommen GJB, den Dunnen JT (2000) The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett 473:1–5 [DOI] [PubMed] [Google Scholar]
- Stec I, Wright TJ, van Ommen G-J, de Boer PAJ, van Haeringen A, Moorman AFM, Altherr MR, den Dunnen JT (1998) WHSC1, a 90kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet 7:1071–1082 [DOI] [PubMed] [Google Scholar]
- Sweeney E, Fryer A, Donnai D (2002) Sotos syndrome: two cases with severe scoliosis. Clin Dysmorphol 11:121–124 [DOI] [PubMed] [Google Scholar]
- Weaver DA, Graham CB, Thomas IT, Smith DW (1974) A new overgrowth syndrome with accelerated skeletal maturation, unusual facies and camptodactyly. J Pediatr 44:547–552 [DOI] [PubMed] [Google Scholar]