Abstract
Low bone mineral density (BMD) is a major risk factor for osteoporotic fracture. Studies of BMD in families and twins have shown that this trait is under strong genetic control. To identify regions of the genome that contain quantitative trait loci (QTL) for BMD, we performed independent genomewide screens, using two complementary study designs. We analyzed unselected nonidentical twin pairs (1,094 pedigrees) and highly selected, extremely discordant or concordant (EDAC) sib pairs (254 pedigrees). Nonparametric multipoint linkage (NPL) analyses were undertaken for lumbar spine and total-hip BMD in both cohorts and for whole-body BMD in the unselected twin pairs. The maximum evidence of linkage in the unselected twins (spine BMD, LOD 2.7) and the EDAC pedigrees (spine BMD, LOD 2.1) was observed at chromosome 3p21 (76 cM and 69 cM, respectively). These combined data indicate the presence, in this region, of a gene that regulates BMD. Furthermore, evidence of linkage in the twin cohort (whole-body BMD; LOD 2.4) at chromosome 1p36 (17 cM) supports previous findings of suggestive linkage to BMD in the region. Weaker evidence of linkage (LOD 1.0–2.3) in either cohort, but not both, indicates the locality of additional QTLs. These studies validate the use, in linkage analysis, of large cohorts of unselected twins phenotyped for multiple traits, and they highlight the importance of conducting genome scans in replicate populations as a prelude to positional cloning and gene discovery.
Introduction
Osteoporosis is a disease characterized by reduced bone strength and the development of fractures, particularly of the spine, hip, and wrist. These fractures usually occur with minimal trauma and may cause prolonged suffering, incapacity, and premature mortality. The fractures may be due to failure to achieve an adequate bone mass in early adulthood, loss of bone mass with accompanying bone architecture changes with age, or both (Rodan et al. 1996).
Bone mass and susceptibility to osteoporotic fracture can be assessed by measurement of bone mineral density (BMD). A low BMD, estimated by dual-energy x-ray absorptiometry (DEXA), is known to be one of the strongest risk factors for osteoporotic fracture. Extensive studies of BMD in twins have shown that at both axial and appendicular sites, BMD is highly heritable (H2=0.50–0.90) (Hunter et al. 2001; Recker and Deng 2002). In addition, estimates of heritability in pre- and postmenopausal women are similar, suggesting that environmental influences may not diminish the relevance of genetic control of bone mass in older women (Hunter et al. 2001). Further substantiation that osteoporosis may have a genetic basis comes from the observation that there is familial increase in risk of fracture. Keen et al. (1999) demonstrated that the risk of wrist fracture may be increased as much as fourfold for a woman who has a first-degree relative who has also had this type of fracture. Moreover, it is now recognized that individual genes that regulate BMD are likely to exert different effects at different body sites and that different genes may have general (whole-body), site-specific, and sex-specific effects (MacGregor et al. 1997).
On the basis of this evidence for the genetic regulation of BMD, genome screens have been conducted in cohorts of various ethnic groups. The data from previous linkage studies in humans have highlighted a number of regions that may contain QTLs for BMD (Devoto et al. 1998; Niu et al. 1999; Koller et al. 2000) or femoral structure (Koller et al. 2001). In relation to BMD, Devoto et al. (1998) performed whole-genome scans in seven large pedigrees with recurrent low bone density and obtained maximum evidence of linkage, by parametric analysis, to 11q (LOD 2.08). They also provided evidence of linkage, using nonparametric analysis, to 1p36 (LOD 2.29) and 2p24-23 (LOD 2.25). Niu et al. (1999) performed whole-genome scans in 96 Chinese families. Utilizing data from forearm densitometry, they reported evidence of linkage for BMD as a QTL at 2p24-23 (LOD 2.15) and 13q14-21 (LOD 1.67). Johnson et al. (1997) reported linkage for high bone mass to 11q12-13 in a single large pedigree. In another whole-genome scan of healthy white and African American sibs (Koller et al. 2000), linkage was shown for lumbar spine BMD in the white subset, at 1q21-23, 6p12-11, 11q12-13, and 22q12-13 (LODs of 3.64, 2.13, 1.65, and 1.82, respectively); for femoral neck BMD, at 5q33-35 (LOD 2.03); and for trochanter BMD, at 14q31-32 (LOD 1.99). Recently, Karasik et al. (2002) reported suggestive linkage from multipoint analysis of trochanter BMD to 21q, with lower LOD scores for Ward’s BMD at 8q24 and spine BMD to 14q21.3.
In the present study, female twins, unselected for BMD, were examined. This is a population in which the BMD and genotype frequencies for bone-related traits would be expected to be representative of white women in the United Kingdom. Large numbers of pairs (>1,000) are necessary to provide adequate statistical power when using the twin-study design, but there is the advantage that multiple related phenotypes can be studied simultaneously. In addition, we studied a second cohort made up of white women from pedigrees containing an extremely discordant or concordant (EDAC) sibling pair. The EDAC approach provides increased statistical power because of the selection process (Carey and Williamson 1991; Risch and Zhang 1996; Dolan and Boomsma 1998). Genomewide screens of these two complementary cohorts were used to locate evidence of QTLs for BMD in what is, to our knowledge, the largest linkage study reported to date.
Material and Methods
Subjects and Clinical Assessment
Unselected twin pairs
Twins were identified from the St Thomas’ UK adult twin registry and were invited to participate in the study. Female twin pairs were 18–80 years of age and were measured for an extensive range of clinical phenotypes related to cardiovascular disease, obesity, diabetes, and osteoporosis. Here, we report only the data relevant to the study of BMD. Both twins attended the clinic together for the collection of clinical data, which included age, height, and weight. Measurement of anterior-posterior projection of lumbar spine (L1–4), total-hip (femoral neck, trochanter, and intertrochanter), and whole-body BMD was made using DEXA (QDR 2000W, Hologic), as described elsewhere (Hunter et al. 2001). General medical, gynecological, and lifestyle questionnaires were completed at interview.
EDAC
Probands were identified from bone-densitometry databases at participating centers or were recruited from newspaper and press advertisements in the United Kingdom, Belgium, New Zealand, and Australia. Probands were required to be white women, to be 25–83 years of age, and to have a lumbar spine, femoral neck, or total-hip BMD Z score <−1.5. The use of BMD Z scores, which define the number of standard deviations above or below mean of an age-matched control population, provides a correction of BMD for age in the linkage analysis. The exclusion criteria applied to these subjects were myeloma, malignancy with skeletal involvement, prolonged use of steroids (>5mg/day for >6 mo), anorexia, premature menopause (at <40 years of age), unstable thyroid disease, primary hyperparathyroidism, antiepileptic therapy, history of osteomalacia, amenorrhea lasting >6 mo, and rheumatoid arthritis. Each individual who qualified as a proband and had at least one sister willing to participate was invited to enroll in the study. Exclusion criteria were also applied to participating sibs. Female sibs then underwent lumbar spine and total-hip DEXA (Hologic). Clinical data (including age, height, and weight) were recorded, and subjects completed medical, gynecological, and lifestyle questionnaires. Two hundred fifty-four families were identified that contained an EDAC pair comprising a proband and at least one sister with spine, hip, or femoral neck BMD Z <−1.0 or >1.0. These thresholds for BMD represent the upper and lower 16% of the age-matched BMD distribution. This design was selected to provide the most efficient use of genotyping resources. The EDAC pairs and any available additional sibs were used for the genome screen. The final group used in the analysis comprised 587 individuals.
Ethics approval
All subjects from both cohorts provided written informed consent, and the institutional ethics committees of participating institutions approved the experimental protocols.
Genotyping
Genotyping was performed on DNA extracted from venous blood. Microsatellite marker–based genotyping was undertaken using standard ABI Prism (Applied Biosystems) fluorescence-based genotyping methodologies (Reed 1994; Pritchard 1995). Genome scans included the analysis of as many as 737 markers on each of the twins. Specifically, marker loci were amplified in 10-μl single-plex PCRs in 384-well microtiter plates. Amplification products were pooled and precipitated and then combined with loading buffer, formamide, and an internal size standard (GeneScan-500, Applied Biosystems); products were then separated by size and were detected using ABI Prism 377 automated sequencers (Applied Biosystems).
For the EDAC families, the genome scans were performed using HuSNP GeneChips (Affymetrix), as well as microsatellite markers. The HuSNP GeneChip is a valid alternative to a microsatellite genome scan with 10-cM marker spacing (Wilson et al. 2000; Thompson and Reed 2001). However, a disadvantage is the inflexibility and nonuniformity of the genomic distribution of the marker loci assayed. For the present study, 33 microsatellite markers were also assayed, to achieve more-uniform coverage across the genome. In all, as many as 1,008 biallelic and microsatellite markers were included in this analysis.
Genotypes from both analyses were processed and maintained within Phenobase (Sequenom), a proprietary database of clinical and genetic information. Consistencies between genotypes and family relationships and/or twin zygosity were routinely investigated, and this included analysis of identity-by-state relationships in families. Discrepant pairs and MZ twins identified by this analysis were excluded from further analyses. Random duplicate genotyping was routinely undertaken throughout the study and indicated a genotyping error rate of <1% for both microsatellite and biallelic-marker genotyping.
Statistical Analysis
Nonparametric multipoint linkage (NPL) analysis (MAPMAKER/SIBS, v. 2.0; Kruglyak and Lander 1995) was performed separately on the twin and EDAC data sets. Marker loci map positions were determined from the Généthon linkage maps. The positions of marker loci that were not included in these maps was interpolated from alternative physical and genetic maps (Center for Medical Genetics, Marshfield Research Foundation; UCSC Genome Bioinformatics) which comprised both the marker in question and the flanking Généthon marker loci (Dib et al. 1996). Those for which a position could not be accurately determined were excluded from the analysis. Lumbar spine, total-hip, and whole-body BMD (g/cm2) or lumbar spine and total-hip BMD Z scores were analyzed as quantitative traits in unselected twin pairs or EDAC pedigrees, respectively. Prior to commencement of linkage analysis, the BMD Z score was recalculated using published normal-range data for the lumbar spine site (Hologic 1999) and the NHANES III reference data for the total hip (Looker et al. 1995), to ensure consistency across the data set. No age correction of BMD data was necessary for the twin pairs. Some BMD results (<3%) obtained by use of Lunar instruments were corrected for the difference between the two types of densitometers, using the published correction formula (Steiger 1995; Hanson 1996). Simulated P values, calculated on a chromosomewide and genomewide basis, were derived using a permutation approach from a minimum of 500 random reassignments of the phenotype data, whereas genotypes were held constant (proprietary software, Sequenom).
All twin pairs were from independent families, and no additional sibs were considered in the analysis of the twin cohort. The majority of EDAC families had only a single sib pair contributing to the analysis (table 1). However, in some families, genome scanning was performed for as many as seven female sibs, and weighting for multiple pairwise comparisons was used in the statistical analyses to minimize bias. Analysis of clinical data was performed using Statistica (Statsoft, v. 6.0). The Mann-Whitney U test was used to perform the statistical analysis of clinical data because of the difference in data distribution in the two cohorts. Clinical data were examined for deviation from normal distribution by use of the χ2 test and are presented as median, interquartile range, and number of individuals.
Table 1.
Composition of Families Used for Two Whole-Genome Screens
| Variable | Twins | EDAC |
| No. of families | 1,094 | 254 |
| Total no. of individuals | 2,188 | 587 |
| No. of possible sib pairs | 1,094 | 444 |
| No. of sibs in family: | ||
| 2 | 1,094 | 198 |
| 3 | 0 | 39 |
| 4 | 0 | 13 |
| 5 | 0 | 3 |
| 7 | 0 | 1 |
Results
The median age of the twin cohort was significantly less than that of the EDAC cohort (P<.001) (table 2). The median height of unselected twins was greater than that of the EDAC cohort (P<.001); however, the median BMIs of the two groups were not different (P>.05).
Table 2.
Clinical Data for Unselected Twins and Subjects from EDAC Pedigrees
|
Median (Interquartile Range),No. of Individuals among |
||
| Characteristic | Unselected Twins | EDAC Pedigrees |
| Age (years) | 48.1 (15.6), 2,188 | 59.0 (16.1), 587a |
| Height (m) | 1.63 (.09), 2,185 | 1.59 (.09), 535a |
| Weight (kg) | 64.2 (14.3 ), 2,184 | 61.0 (14.0), 534a |
| BMI (kg/m2) | 24.2 (5.3), 2,184 | 24.2 (5.4), 534 |
| BMD (g/cm2): | ||
| Spine | 1.005 (.186), 2,167 | .795 (.210), 584a |
| Total hip | .925 (.172), 2,164 | .757 (.186), 562a |
| Whole body | 1.141 (.145), 2,168 | N/A |
P<.001.
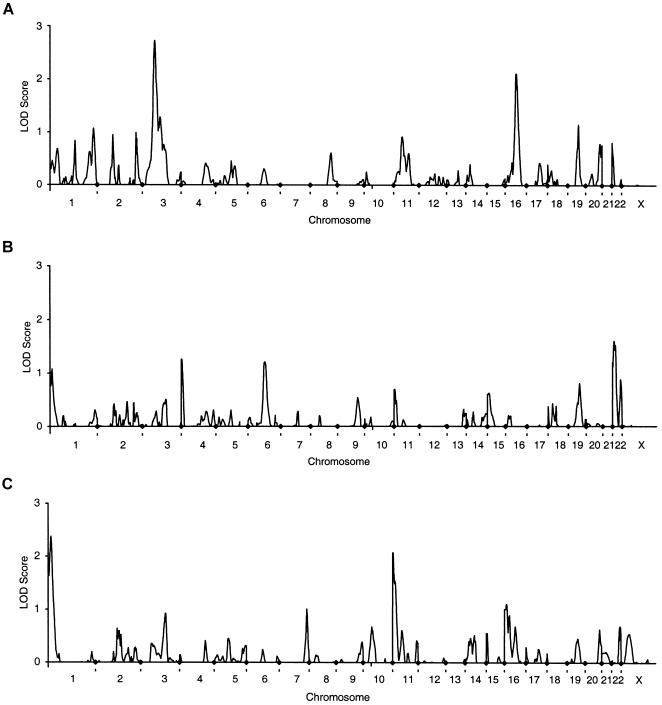
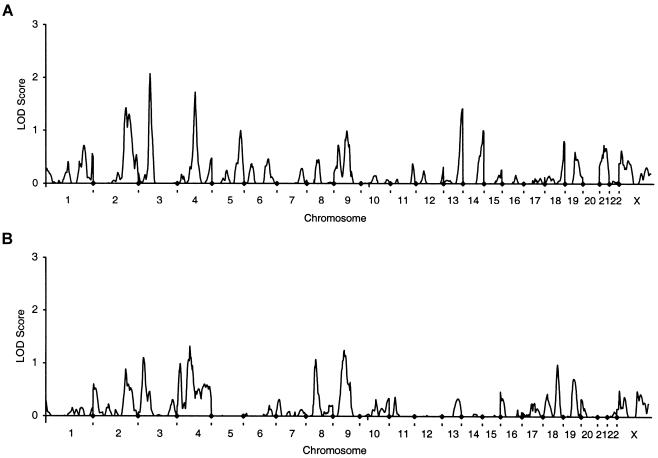
The strongest evidence of linkage that was found consistently in both cohorts was to spine BMD on chromosome 3 at 69–76 cM (figs. 1 and 2). Neither of these peaks achieved genomewide significance; however, both achieved low P values when assessed in the context of chromosomewide significance (tables 3 and 4). The approximate support interval for the chromosome 3 linkages (flanked by markers D3S1298 and D3S1285) extend between 64 cM and 91 cM in the unselected twins and between 62 cM and 82 cM in the EDAC group. The mean ± SD information content (Kruglyak and Lander 1995) in this interval is 0.50±0.04 for the unselected twins and 0.60±0.07 for the EDAC cohort.
Figure 1.
Genomewide scans for QTL that regulate BMD in 1,094 twin pairs. A, lumbar spine. B, total hip. C, whole body. Equivalent LOD scores were calculated from the NPL Z scores output by MAPMAKER/SIBS, using the relationship Z2/4.6052 = 1 LOD.
Figure 2.
Genomewide scans for QTL that regulate BMD in 254 pedigrees containing an EDAC pair. A, lumbar spine. B, total hip.
Table 3.
Genomic Location of Maximum NPL for BMD Phenotypes in Unselected Twins
| Chromosome | Position(cM)a | Phenotype | No. ofIndividuals | MaximumNPL Z | MaximumLOD | Pempb |
| 1 | 13 | Total hip | 2,139 | 2.23 | 1.08 | NS |
| 1 | 17 | Whole body | 2,158 | 3.31 | 2.38 | .030 |
| 1 | 270 | Lumbar spine | 2,161 | 2.22 | 1.07 | NS |
| 3 | 76 | Lumbar spine | 2,161 | 3.54 | 2.72 | .008 |
| 3 | 109 | Lumbar spine | 2,161 | 2.43 | 1.29 | NS |
| 4 | 2 | Total hip | 2,139 | 2.41 | 1.26 | NS |
| 6 | 105 | Total hip | 2,139 | 2.36 | 1.21 | NS |
| 11 | 2 | Whole body | 2,158 | 3.09 | 2.08 | .014 |
| 16 | 13 | Whole body | 2,158 | 2.26 | 1.11 | NS |
| 16 | 67 | Lumbar spine | 2,161 | 3.12 | 2.11 | .008 |
| 19 | 66 | Lumbar spine | 2,161 | 2.30 | 1.15 | NS |
| 22 | 10 | Total hip | 2,139 | 2.72 | 1.60 | .038 |
Distance from the p telomere.
Pemp = chromosomewide empirically determined P value. NS=P>.05.
Table 4.
Genomic Location of Maximum NPL for BMD Phenotypes in Families Containing an EDAC Pair[Note]
| Chromosome | Position(cM) | Phenotype | No. ofIndividuals | MaximumNPL Z | MaximumLOD | Pemp |
| 2 | 203 | Lumbar spine | 583 | 2.56 | 1.42 | NS |
| 2 | 221 | Lumbar spine | 583 | 2.45 | 1.30 | NS |
| 3 | 32 | Total hip | 543 | 2.25 | 1.10 | NS |
| 3 | 69 | Lumbar spine | 583 | 3.09 | 2.07 | .014 |
| 4 | 83 | Total hip | 543 | 2.47 | 1.32 | NS |
| 4 | 115 | Lumbar spine | 583 | 2.81 | 1.72 | .032 |
| 8 | 62 | Total hip | 543 | 2.23 | 1.08 | NS |
| 9 | 69 | Total hip | 543 | 2.40 | 1.25 | NS |
| 13 | 110 | Lumbar spine | 583 | 2.55 | 1.41 | NS |
Note.— Column headings are as defined in table 3 footnotes.
The data from the unselected twins highlighted 11 additional genomic regions with a LOD >1 for the three phenotypes analyzed (table 3. These included evidence of linkage to spine BMD on chromosome 1 at 270 cM, chromosome 3 at 109 cM, chromosome 16 at 67 cM and chromosome 19 at 66 cM. There was also weak evidence of linkage to total-hip BMD in four regions (chromosome 1 at 13 cM, chromosome 4 at 2 cM, chromosome 6 at 105 cM, chromosome 22 at 10 cM), and these were different from those linked to spine BMD. There was suggestive linkage to whole-body BMD on chromosome 1 at 17 cM, and there was weaker evidence of linkage on chromosome 11 at 2 cM and on chromosome 16 at 13 cM. The only evidence of linkage to more than one phenotype in the same chromosomal region was for total-hip BMD and whole-body BMD on chromosome 1. The approximate support interval extends 31 cM from the p telomere and is flanked by D1S199. The mean ± SD information content in this interval is 0.52±0.07.
Analysis of the EDAC pedigree data defined eight more genomic locations for the possible presence of genes that regulate BMD (table 4). Weak evidence for linkage to spine BMD was shown on chromosome 2 at 203 cM and 221 cM, on chromosome 4 at 115 cM, and on chromosome 13 at 110 cM. Total-hip BMD in this cohort showed weak evidence of linkage to chromosome 3 at 32 cM, on chromosome 4 at 83 cM, chromosome 8 at 62 cM, and on chromosome 9 at 69 cM. There was no concordance between peaks for spine and hip BMD in this data set. It may be that the evidence of linkage of total-hip BMD to chromosome 3 at 32 cM reflects the presence of the same gene(s) defined in both cohorts by spine BMD linkages, which are ∼40 cM away. However, concordance of peaks is normally restricted to genetic distances <20 cM.
Empirical P values derived using simulations are given in tables 3 and 4. These are for guidance only, because of the small number of permutations applied. Although seven of the peaks achieved a P value <.05 when considered in the context of chromosomewide significance, only the peaks at 76 cM on chromosome 3 and 67 cM on chromosome 16 had a P value <.01.
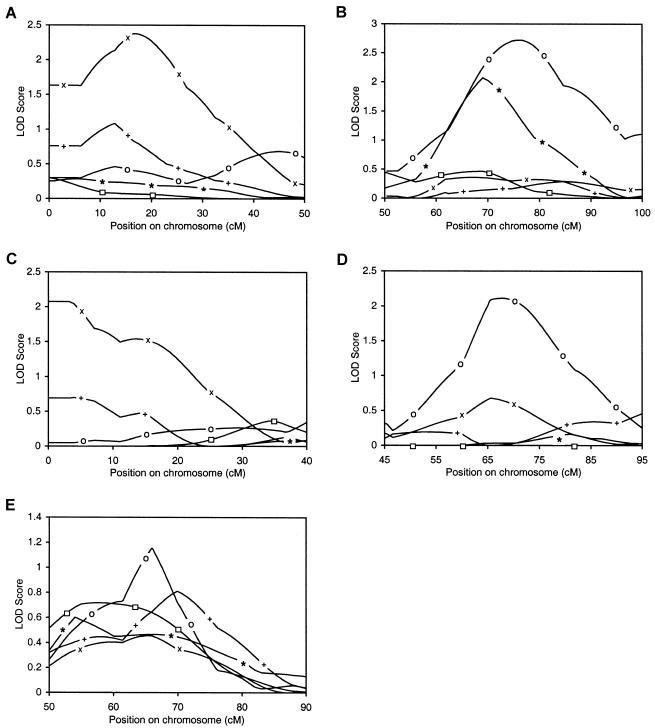
In contrast to peaks on chromosome 1 at 17 cM (fig. 3A) and chromosome 3 at 76 cM (fig. 3B), which are supported by evidence from the analysis of other phenotypic data within the same cohort or in the other cohort; chromosome 11 at 2–15 cM (fig. 3C) and chromosome 16 at 67 cM (fig. 3D) have less evidence of replication. Several other peaks did have weak support from other phenotypes, and these included chromosome 2 at 203 cM and 221 cM, chromosome 9 at 69 cM, chromosome 16 at 13 cM, and chromosome 19 at 66 cM, but these data did not reach the threshold of LOD =1. Furthermore, given the phenotypic correlation between BMD measurements at different body sites, some similarity between the multipoint linkage plots for each of the phenotypes might be expected (e.g., correlation coefficients for spine BMD and total-hip BMD and for whole-body BMD and total-hip BMD in unselected twins are 0.71 and 0.75, respectively). Of these latter peaks, only the chromosome 19 peak is replicated for phenotypes from both cohorts (fig. 3E). In the unselected twins, for spine, hip, and whole-body BMD, very weak evidence of linkage to chromosome 19 was maximal between 65 cM and 70 cM (LOD = 1.15, 0.81, 0.46, respectively). Linkage data from the EDAC pedigrees for spine and hip BMD was maximal at 54–58 cM (LOD = 0.6 and 0.72, respectively). Although in this instance the magnitudes of the linkages are not great, the apparent consistency in both cohorts is intriguing.
Figure 3.
Comparison of linkage data for lumbar spine (o), total-hip (+) and whole-body (×) BMD in 1,094 unselected twin pairs and for lumbar spine (*) and total-hip (□) BMD in 254 pedigrees containing an EDAC pair at 1p36 (D1S468 to D1S199) (A), 3p22-14 (D3S1277 to D3S1566) (B), 11pter-14 (D11S4046 to D11S904) (C), 16p12-q23 (D16S3068 to D16S515) (D), and 19q13 (D19S414 to D19S571) (E).
As might be expected from the highly polymorphic nature of dinucleotide-repeat polymorphisms, the average information content was higher in the microsatellite screen than in the predominantly biallelic-marker–based screen (mean ± SD 0.64±0.12 and 0.47±0.14, respectively).
In the unselected twins, the BMD data for lumbar spine and total-hip sites were normally distributed (P>.05). The distribution of whole-body BMD in the unselected twins was leptokurtic (P<.01). Spine BMD data for the EDAC cohort were not normally distributed (P=.01) and showed positive skewness and negative kurtosis, as might be expected from the selection criteria. Data on total-hip BMD in the EDAC cohort was not significantly different (P>.05) from a normal distribution. This may be a result of the bias toward selection on the basis of spine-BMD criteria in the cohort. As would be expected from the age of the individuals and the selection process, the median spine and total-hip BMD values were lower in the EDAC families than in the unselected twins (P<.001 for each measure; table 2). The probands for the EDAC pedigrees had very low BMD at both spine and hip sites (median, interquartile range [number of individuals] 0.723, 0.144 [253] and 0.704, 0.139 [232]). Siblings that qualified as concordant from these pedigrees also had low BMD for both spine and hip (0.819, 0.166 [220] and 0.753, 0.166 [220], respectively). Data (median, interquartile range [number of individuals]) for the discordant siblings were 1.064, 0.172 (69) for spine BMD and 0.944, 0.163 (69) for hip BMD. There was a predominance of pedigrees selected on the basis of spine-BMD data in the present study. Thirty-four percent of the total number of pedigrees was selected on the basis of spine BMD in the proband and the concordant sibling, with a further 16% selected among discordant siblings. This compares with only 11% and 2%, respectively, when both siblings were selected on the basis of hip BMD data.
Discussion
In the present study, maximal evidence of linkage to spine BMD on chromosome 3 (62–91 cM, 3p21.33-3p14.1) is observed in two independent study populations. The conventional paradigm for genomewide linkage analysis and positional cloning has recently come under criticism when applied to the study of complex multifactorial diseases, such as diabetes, asthma, cardiovascular disease, and osteoporosis (Altmuller et al. 2001). However, much of this criticism relates to the inability of follow-up studies to replicate findings and, ultimately, to deliver disease genes. Independent replication of linkage is a fundamental prerequisite for the commencement of positional cloning studies, and, despite the failure of either peak on chromosome 3 to reach the accepted threshold for genomewide significance, these data must be seen as evidence for the presence of a gene controlling BMD. Our data demonstrate that for studies of structural such traits as BMD, replication is possible when large studies are conducted among subjects with comparable ethnic backgrounds.
Since neither peak on chromosome 3 reached genomewide significance, the issue of combining the data for further analysis could be considered. However, the two genome screens have very different study designs, and each study is subject to a different ascertainment bias. Furthermore, the marker sets are distinct, so a combined data set would contain a very high rate of missing data; genotypes for markers typed in each study would be missing for individuals in the other. This creates substantial computational complexity. Combining the data and performing a reanalysis would therefore yield a result difficult to interpret, so we have not attempted to do this.
The approximate support interval for this chromosome 3 peak encompasses a broad and relatively gene-dense region. There are ∼350–400 genes reported in this area, and a substantial number of these are expressed in bone. These include the following: parathyroid receptor 1 (PTHR1), which has a role in skeletal development (Zhang et al. 1998) and calcium homeostasis (Kovacs et al. 2001); tetranectin (TNA), a gene that may function in mineralization during embryogenesis (Wewer et al. 1994); and macrophage stimulating 1 (MST1), a gene that produces a protein that may be capable of stimulating osteoclastic resorption (Kurihara et al. 1998). Linkage of hip and spine BMD to this region of chromosome 3 has been reported elsewhere (Duncan et al. 1999) by researchers who examined microsatellite markers surrounding 23 candidate genes, including PTHR1. They examined just two microsatellite markers in this region and found suggestive evidence of linkage of hip BMD to D3S1289 (69.1 cM; LOD 2.7–3.5) with weaker linkage for lumbar spine BMD to D3S3559 (63.3 cM; LOD 1.3–1.6). Thus, studies in three independent cohorts of similar ethnic background now provide evidence of linkage for BMD to this region of the genome. We report linkage to spine BMD in both cohorts, and we found no convincing evidence of linkage for the total-hip phenotype to this region. One possibility for this apparent discrepancy may be a predominance of pedigrees that are informative for spine BMD in our cohorts. The overall power of both studies is modest, and larger studies will likely be required to accurately discern whether there are site-specific or general effects on BMD.
In the light of these combined data, polymorphisms within the regulatory elements of the PTHR1 gene continue to be strongly implicated for a role in the regulation of BMD, and it may be appropriate to begin positional candidate or positional cloning studies in this region. However, given the broad support interval and the large number of genes therein, one should be cautious in speculating about the potential known or novel genes responsible.
Evidence of linkage was also observed with hip BMD at 1p36 within a 31-cM support interval, a region also thought to contain 350–400 genes. This peak in our data also provides replication of published data (Devoto et al. 1998). Since the first identification of this region as the possible location of a QTL regulating BMD, a fine mapping study of the 1p36 region has been completed (Devoto et al. 2001). Tumor necrosis factor receptor superfamily, member 1B (TNFRSF1B) has emerged as one of the leading candidates. In a recent preliminary report, Spotila et al. (2001) have shown significant association of spine BMD to an M196R polymorphism in this gene. Albagha et al. (2002) have provided further support for a role of TNFRSF1B in the regulation of BMD. They reported an association between BMD and genetic variation within the 3′ UTR of the gene, but it must be noted that the effect in that study was not mediated by the M196R polymorphism. Another candidate, procollagen-lysine, 2-oxoglutarate 5-dioxygenase (PLOD), which encodes an enzyme that catalyzes the hydroxylation of lysyl residues in collagenlike peptides, failed to show association with BMD (Spotila et al. 2001). There are a substantial number of other genes in this region with known expression in bone. Further work will be required to determine if TNFRSF1B is responsible for the observed linkage in both studies.
Among our results, weak evidence of linkage of whole-body BMD to 11p does not appear to be supported by data from other phenotypes in our cohort or the existing literature. The LRP5 locus is on chromosome 11 and has been reported to have a role in controlling BMD variability (Little et al. 2002), but it is at 11q13, >40 cM away from this peak. Similarly, weak evidence of linkage of spine BMD to chromosome 16 also remains unsupported. Among the peaks that achieved a LOD of 1–2.3, the chromosome 2q peak for spine BMD in the EDAC pedigrees and the peak for spine BMD in unselected twins on 19q are probably the most interesting. Hocking et al. (2001) have reported suggestive linkage in families with Paget’s disease to 2q36, a region within the approximate support interval of this linkage peak. This region contains a number of important candidate genes, including insulinlike growth factor binding proteins 2 and 5 (IGFBP2 and IGFBP5) and Indian hedgehog (IHH) genes (National Center for Biotechnology Information). The chromosome 19 support interval encompasses both transforming growth factor–β1 (TGFβ1) and apolipoprotein E (ApoE) genes, among others. Genetic variation in both these genes has been shown to be associated with BMD (Shiraki et al. 1997; Grainger et al. 1999; Salamone et al. 2000; Keen et al. 2001). Whether other genes in this region also contribute to the variance in BMD remains to be determined.
Numerous other regions of the genome have been highlighted from genomewide screens of humans and provide possible locations for other genes that may participate in the regulation of BMD, but they are not supported by our data. These include 1q21-23q, 11q12-13, and 14q21.3, among others (Johnson et al. 1997; Koller et al. 2000; Karasik et al. 2002). Consideration of this issue could include the possibility of false negatives in our data and of false positives in these other published reports. Other potential sources of difference include ethnic background, differences in exposure to environmental factors that might modulate the effects of the genes (e.g., calcium intake or sunlight exposure) and variations in study design (including selection criteria, menopausal status, sample size, and linkage marker selection).
We chose three sites for the study of BMD, since they represent an economical design for consideration of the two main clinically relevant sites for osteoporotic fracture (spine and hip) plus an overall assessment of gene effects (whole body). Although other sites (e.g., the forearm) might have been examined, the data were not available for all sib pairs, and the clinical relevance of such measurements is less clear. Similarly, we do not present analysis of the interrelated BMD hip regions (e.g., femoral neck, trochanter, intertrochanter, Ward’s), which are compromised by virtue of the high phenotypic correlation within the anatomical site.
With respect to the distribution of spine and hip BMD in the EDAC cohort, one might expect the distribution to be strongly bimodal. This was not the case. Although families entered the study by virtue of the presence of an EDAC pair, many pedigrees included additional sibs who did not conform to the criteria (table 1). These were included in the analysis, since they present additional information on identity-by-descent in the family and contribute to the linkage analysis, albeit with less power than highly selected individuals. Moreover, even members of the EDAC group show within-individual variation in BMD at various body sites. An individual who had a measurement of BMD Z <−1.5 at the spine might still have a total-hip BMD of Z=0.2; thus, the entire distribution of BMD values is represented in the cohort. Since we sought to localize genes with major effect on BMD, whether site specific or not, we have not restricted the recruitment criteria to a single clinical site (e.g., lumbar spine). Instead, we have included spine, hip, and whole-body phenotypes in an attempt to localize any gene providing substantial regulation of BMD. We chose to accept a stringent criterion for the proband (lowest 6.7% of the age-matched distribution) but a less stringent criterion for the other sibling (highest or lowest 15.9% of the age-matched distribution). Our goal was to enhance the power of the study by selecting subjects with evidence of a familial trend for low BMD while maintaining a balance with the practicalities of recruiting sufficient pedigrees to empower the study.
Although this is a large study in comparison with some of the genome screens published for BMD to date, the power of the study remains relatively modest, and the ability to detect genes of small effect is poor. For example, Fulker et al (1994) have shown that a sample size of 200 sib pairs would provide adequate power (80%–90%, at a significance level of .01) to detect QTL only if they account for ⩾25% of trait heritability. By comparison, Risch and Zhang (1996) give sample-size requirements to detect a QTL with the use of extremely discordant pairs. They estimate that the number of pairs required to detect linkage (e.g., LOD 3.0) for a trait of heritability 0.3, for an additive model, is ∼150 pairs. Although our EDAC cohort is approximately double this size, its statistical power appears to be adequate only for the detection of those genes that explain a relatively large amount of the variance in the trait. The analysis of the unselected twin cohort includes many more individuals than that of the EDAC cohort. However, because the twins are unselected, the power of that cohort to detect genes of minor effect is also small. Dolan et al. (1999) have estimated that, for a trait with heritability similar to that of BMD, 5,000 unselected twins are likely to be comparable to a sample of 250 EDAC pairs drawn from 15% of the distribution. Thus, despite the size of the cohort, the power of the unselected-twin study is probably similar to that of the EDAC cohort.
Two distinct, but complementary, approaches were used in the present study to define the location of QTLs for BMD in white women: one unselected and one highly selected. Large numbers of unselected DZ twins are available in the general population, and many countries maintain twin registers, making these an ideal resource for large-scale linkage studies. Such schemes do not generally exist for sibling pairs, who are also more difficult to recruit than twins. Unselected twins might be expected to be a good model for linkage studies of complex diseases in the population at large, because twins have been shown to be comparable to general population singletons for a range of chronic age-related phenotypes, including BMD and other bone parameters (Andrew et al. 2001). Furthermore, extensively phenotyped twins represent an economical study design for the simultaneous completion of multipoint linkage studies of several complex diseases for the cost of a single genome screen. In contrast, the use of highly specific selection criteria, as required for an affected sibling pair or EDAC design, effectively prohibits the cohort from being used in the study of other diseases. For example, because BMD was the selection criterion for this study, there were insufficient data to study the important clinical endpoint of vertebral fracture. Other data, such as bone- and calcium-metabolism–related biochemistry, were also lacking in the EDAC cohort, but analysis of these phenotypes will be possible for the unselected twins in the future. Selected study designs are generally regarded as a more cost-effective design than similarly sized unselected cohorts, for the detection of linkage for a single phenotype. Certainly, the perception of economy in the use of the EDAC design appears to be supported by our data. We analyzed many more unselected twins than EDAC family members.
For many complex diseases, age is crucially important in disease expression. Because of the precisely shared age—and, usually, a shared upbringing—for twins, a number of common important environmental influences, such as sunlight exposure, childhood exercise, and calcium intake, are also controlled (MacGregor et al. 2000). Little reliable information on shared upbringing is available to compare between siblings in our EDAC cohort, and the higher median age of this group suggests a substantially greater period of influence of environmental factors that might affect BMD. Such factors may not act equally on both sibs, and the increased environmental variance may be one cause of the lower number and magnitude of linkage peaks observed for this cohort, compared with the unselected twins. However, it must also be noted that some of the linkage peaks observed in one cohort and not the other could be due to differences in marker number or distribution, which vary across the genome in the marker sets we used. Therefore, evidence of linkage in only one cohort is not a sufficient reason to support rejection of a region from consideration for further study.
In conclusion, the present article details what is, to our knowledge, the largest genome screen for the study of linkage for BMD reported to date. The number and magnitude of the linkage peaks—and the concordance of peaks from the analysis of unselected twins with those from our EDAC genome screen and the literature—provide evidence for the value of our approach. This is particularly well illustrated by replication of the linkage of spine BMD to 3p21 and total-hip and whole-body BMD to 1p36. We have identified two genomic regions for which there is now strong evidence of the presence of genes that regulate BMD. Replication of these findings is provided within our own data, as well as in the literature. Although the approximate support intervals for these regions remain relatively broad, these data provide an important advance toward identifying appropriate genomic regions for the commencement of positional-cloning or positional-candidate studies.
Acknowledgments
The authors acknowledge N. Gilchrist, J.Y. Reginster, I. Fogelman, and I. Smith, who contributed patients to the study. We thank N. Morrison, F. Dudbridge, T. Andrew, and R. Middelberg for advice and assistance. P. Major, R. Keen, G. Blake, G. Surdulescu, R. Retallack, E. Jamieson, and the staffs of Sequenom and Gemini Genomics contributed to patient recruitment, phenotyping, and genotyping. The Chronic Disease Research Foundation funded part of the EDAC recruitment. The Twin Research unit received funding from the Wellcome Trust, the Arthritis Research Campaign, and the British Heart Foundation.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation http://research.marshfieldclinic.org/genetics/ (for marker order and genetic distance between markers)
- Généthon, http://www.cephb.fr/bio/ceph-genethon-map.html (for marker order and genetic distance between markers)
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/ (for identification of candidate genes in loci of interest)
- UCSC Genome Bioinformatics, http://genome.cse.ucsc.edu/ (for marker order and genetic distance between markers)
References
- Albagha OM, Tasker PN, McGuigan FE, Reid DM, Ralston SH (2002) Linkage disequilibrium between polymorphisms in the human TNFRSF1B gene and their association with bone mass in perimenopausal women. Hum Mol Genet 11:2289–2295 [DOI] [PubMed] [Google Scholar]
- Altmuller J, Palmer LJ, Fischer G, Scherb H, Wjst M (2001) Genomewide scans of complex human diseases: true linkage is hard to find. Am J Hum Genet 69:936–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew T, Hart DJ, Snieder H, de Lange M, Spector TD, MacGregor AJ (2001) Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res 4:464–477 [DOI] [PubMed] [Google Scholar]
- Carey G, Williamson J (1991) Linkage analysis of quantitative traits: increased power by using selected samples. Am J Hum Genet 49:786–796 [PMC free article] [PubMed] [Google Scholar]
- Devoto M, Shimoya K, Caminis J, Ott J, Tenenhouse A, Whyte MP, Sereda L, Hall S, Considine E, Williams CJ, Tromp G, Kuivaniemi H, Ala-Kokko L, Prockop DJ, Spotila LD (1998) First-stage autosomal genome screen in extended pedigrees suggests genes predisposing to low bone mineral density on chromosomes 1p, 2p and 4q. Eur J Hum Genet 6:151–157 [DOI] [PubMed] [Google Scholar]
- Devoto M, Specchia C, Li HH, Caminis J, Tenenhouse A, Rodriguez H, Spotila LD (2001) Variance component linkage analysis indicates a QTL for femoral neck bone mineral density on chromosome 1p36. Hum Mol Genet 10:2447–2452 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Dolan CV, Boomsma DI (1998) Optimal selection of sib pairs from random samples for linkage analysis of a QTL using the EDAC test. Behav Genet 28:197–206 [DOI] [PubMed] [Google Scholar]
- Dolan CV, Boomsma DI, Neale MC (1999) A simulation study of the effects of assignment of prior identity-by-descent probabilities to unselected sib pairs, in covariance-structure modeling of a quantitative-trait locus. Am J Hum Genet 64:268–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan EL, Brown MA, Sinsheimer J, Bell J, Carr AJ, Wordsworth BP, Wass JA (1999) Suggestive linkage of the parathyroid receptor type 1 to osteoporosis. J Bone Miner Res 14:1993–1999 [DOI] [PubMed] [Google Scholar]
- Fulker DW, Cardon LR (1994) A sib-pair approach to interval mapping of quantitative trait loci. Am J Hum Genet 54:1092–1103 [PMC free article] [PubMed] [Google Scholar]
- Grainger DJ, Heathcote K, Chiano M, Snieder H, Kemp PR, Metcalfe JC, Carter ND, Spector TD (1999) Genetic control of the circulating concentration of transforming growth factor type beta1. Hum Mol Genet 8:93–97 [DOI] [PubMed] [Google Scholar]
- Hanson J (1997) Standardization of femur BMD. J Bone Miner Res 12:1316–1317 [DOI] [PubMed] [Google Scholar]
- Hocking LJ, Herbert CA, Nicholls RK, Williams F, Bennett ST, Cundy T, Nicholson GC, Wuyts W, Van Hul W, Ralston SH (2001) Genomewide search in familial Paget disease of bone shows evidence of genetic heterogeneity with candidate loci on chromosomes 2q36, 10p13, and 5q35. Am J Hum Genet 69:1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hologic (1999) Bone density reference data. In: Favus MJ (ed) Primer on the metabolic bone diseases and disorders of mineral metabolism. 4th ed. Lippincott Williams & Wilkins, Philadelphia, pp 483–484 [Google Scholar]
- Hunter DJ, de Lange M, Andrew T, Snieder H, MacGregor AJ, Spector TD (2001) Genetic variation in bone mineral density and calcaneal ultrasound: a study of the influence of menopause using female twins. Osteoporos Int 12:406–411 [DOI] [PubMed] [Google Scholar]
- Johnson ML, Gong G, Kimberling W, Recker SM, Kimmel DB, Recker RB (1997) Linkage of a gene causing high bone mass to human chromosome 11 (11q12-13). Am J Hum Genet 60:1326–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasik D, Myers RH, Cupples LA, Hannan MT, Gagnon DR, Herbert A, Kiel DP (2002) Genome screen for quantitative trait loci contributing to normal variation in bone mineral density: the Framingham Study. J Bone Miner Res 17:1718–1727 [DOI] [PubMed] [Google Scholar]
- Keen RW, Hart DJ, Arden NK, Doyle DV, Spector TD (1999) Family history of appendicular fracture and risk of osteoporosis: a population-based study. Osteoporos Int 10:161–166 [DOI] [PubMed] [Google Scholar]
- Keen RW, Snieder H, Molloy H, Daniels J, Chiano M, Gibson F, Fairbairn L, Smith P, MacGregor AJ, Gewert D, Spector TD (2001) Evidence of association and linkage disequilibrium between a novel polymorphism in the transforming growth factor beta 1 gene and hip bone mineral density: a study of female twins. Rheumatology (Oxford) 40:48–54 [DOI] [PubMed] [Google Scholar]
- Koller DL, Econs MJ, Morin PA, Christian JC, Hui SL, Parry P, Curran ME, Rodriguez LA, Conneally PM, Joslyn G, Peacock M, Johnston CC, Foroud T (2000) Genome screen for QTLs contributing to normal variation in bone mineral density and osteoporosis. J Clin Endocrinol Metab 85:3116–3120 [DOI] [PubMed] [Google Scholar]
- Koller DL, Liu G, Econs MJ, Hui SL, Morin PA, Joslyn G, Rodriguez LA, Conneally PM, Christian JC, Johnston CC Jr, Foroud T, Peacock M (2001) Genome screen for quantitative trait loci underlying normal variation in femoral structure. J Bone Miner Res 16:985–991 [DOI] [PubMed] [Google Scholar]
- Kovacs CS, Chafe LL, Fudge NJ, Friel JK, Manley NR (2001) PTH regulates fetal blood calcium and skeletal mineralization independently of PTHrP. Endocrinology 142:4983–4993 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES (1995) Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet 57:439–454 [PMC free article] [PubMed] [Google Scholar]
- Kurihara N, Tatsumi J, Arai F, Iwama A, Suda T (1998) Macrophage-stimulating protein (MSP) and its receptor, RON, stimulate human osteoclast activity but not proliferation: effect of MSP distinct from that of hepatocyte growth factor. Exp Hematol 26:1080–1085 [PubMed] [Google Scholar]
- Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, et al (2002) A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet 70:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looker AC, Johnston CC Jr, Wahner HW, Dunn WL, Calvo MS, Harris TB, Heyse SP, Lindsay RL (1995) Prevalence of low femoral bone density in older U.S. women from NHANES III. J Bone Miner Res 10:796–802 [DOI] [PubMed] [Google Scholar]
- MacGregor AJ, Snieder H, Keen RW, Spector TD (1997) Do different sets of genes influence trabecular and cortical bone density at different anatomical sites? J Bone Miner Res Suppl 12:S492 [Google Scholar]
- MacGregor AJ, Snieder H, Schork NJ, Spector TD (2000) Twins: novel uses to study complex traits and genetic diseases. Trends Genet 16:131–134 [DOI] [PubMed] [Google Scholar]
- Niu T, Chen C, Cordell H, Yang J, Wang B, Wang Z, Fang Z, Schork NJ, Rosen CJ, Xu X (1999) A genome-wide scan for loci linked to forearm bone mineral density. Hum Genet 104:226–33 [DOI] [PubMed] [Google Scholar]
- Pritchard LE, Kawaguchi Y, Reed PW, Copeman JB, Davies JL, Barnett AH, Bain SC, Todd JA (1995) Analysis of the CD3 gene region and type 1 diabetes: application of fluorescence-based technology to linkage disequilibrium mapping. Hum Mol Genet 4:197–202 [DOI] [PubMed] [Google Scholar]
- Recker RR, Deng HW (2002) Role of genetics in osteoporosis. Endocrine 17:55–66 [DOI] [PubMed] [Google Scholar]
- Reed PW, Davies JL, Copeman JB, Bennett ST, Palmer SM, Pritchard LE, Gough SC, Kawaguchi Y, Cordell HJ, Balfour KM, Jenkins SC, Powell EE, Vignal A, Todd JA (1994) Chromosome-specific microsatellite sets for fluorescence-based, semi-automated genome mapping. Nat Genet 7:390–395 [DOI] [PubMed] [Google Scholar]
- Risch NJ, Zhang HP (1996) Mapping quantitative trait loci with extreme discordant sib pairs: sampling considerations. Am J Hum Genet 58:836–843 [PMC free article] [PubMed] [Google Scholar]
- Rodan GA, Raisz LG, Bilezikian JP (1996) Pathophysiology of osteoporosis. In: Bilezikian JP, Raisz LG, Rodan LG (eds) Principles of bone biology. 1st ed. Academic Press, San Diego, pp 979–990 [Google Scholar]
- Salamone LM, Cauley JA, Zmuda J, Pasagian-Macaulay A, Epstein RS, Ferrell RE, Black DM, Kuller LH (2000) Apolipoprotein E gene polymorphism and bone loss: estrogen status modifies the influence of apolipoprotein E on bone loss. J Bone Miner Res 15:308–314 [DOI] [PubMed] [Google Scholar]
- Shiraki M, Shiraki Y, Aoki C, Hosoi T, Inoue S, Kaneki M, Ouchi Y (1997) Association of bone mineral density with apolipoprotein E phenotype. J Bone Miner Res 12:1438–1445 [DOI] [PubMed] [Google Scholar]
- Spotila LD, Rodriguez H, Koch M, Stoltzfus S, Epley W, Tenenhouse HS, Tenenhouse A (2001) Linkage disequilibrium mapping at chromosome 1p36.2 for bone mineral density. J Bone Miner Res Suppl 16:S167 [Google Scholar]
- Steiger P (1995) Standardization of spine BMD measurements. J Bone Miner Res 10:1602–1603 [DOI] [PubMed] [Google Scholar]
- Thompson D, Reed PW (2001) Comparison of SNP and microsatellite marker based genome scanning for linkage. SNP 2000: Third International Meeting on Single Nucleotide Polymorphism and Complex Genome analysis. Hum Mutat 17:327–347 [Google Scholar]
- Wewer UM, Ibaraki K, Schjorring P, Durkin ME, Young MF, Albrechtsen R (1994) A potential role for tetranectin in mineralization during osteogenesis. J Cell Biol 127:1767–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SG, Reed PW, Bailey M, Thompson E, Pask R, Kleyn P, GENOS Study Collaborators (2000) HuSNP GeneChips for linkage mapping in sib pairs concordant for low bone mineral density. Calcif Tissue Int 67:A484 [Google Scholar]
- Zhang P, Jobert AS, Couvineau A, Silve C (1998) A homozygous inactivating mutation in the parathyroid hormone/parathyroid hormone-related peptide receptor causing Blomstrand chondrodysplasia. J Clin Endocrinol Metab 83:3365–3368 [DOI] [PubMed] [Google Scholar]