Abstract
Scapuloperoneal spinal muscular atrophy (SPSMA) and hereditary motor and sensory neuropathy type IIC (HMSN IIC, also known as HMSN2C or Charcot-Marie-Tooth disease type 2C (CMT2C)) are phenotypically heterogeneous disorders involving topographically distinct nerves and muscles. We originally described a large New England family of French-Canadian origin with SPSMA and an American family of English and Scottish descent with CMT2C1,2. We mapped SPSMA and CMT2C risk loci to 12q24.1–q24.31 with an overlapping region between the two diseases3,4. Further analysis reduced the CMT2C risk locus to a 4-Mb region5. Here we report that SPSMA and CMT2C are allelic disorders caused by mutations in the gene encoding the transient receptor potential cation channel, subfamily V, member 4 (TRPV4). Functional analysis revealed that increased calcium channel activity is a distinct property of both SPSMA- and CMT2C-causing mutant proteins. Our findings link mutations in TRPV4 to altered calcium homeostasis and peripheral neuropathies, implying a pathogenic mechanism and possible options for therapy for these disorders.
SPSMA is characterized by progressive scapuloperoneal atrophy and weakness, laryngeal palsy, congenital absence of muscles and developmental abnormalities of the bones1. Pathological studies of biopsy and autopsy samples from an individual with SPSMA revealed severe muscle fiber–type grouping and atrophy (Fig. 1a), extensive fatty replacement, increased endomysial fibrosis, marked variability of fiber size, and many fibers with multiple internal nuclei, fiber splitting and multiple nuclear bags (Fig. 1b and Supplementary Fig. 1a). Both type 1 and type 2 fibers showed atrophy as demonstrated by ATPase staining (Fig. 1c). These pathological changes were particularly severe in the gastrocnemius muscle. However, brain and spinal cord sections revealed normal numbers of motor neurons in the motor cortex and spinal cord. There was no gliosis in the anterior horns. The lateral corticospinal tracts were well preserved (Fig. 1d and Supplementary Fig. 1b). These findings, together with previous clinical and neurophysiological data1, support a diagnosis of spinal neurogenic amyotrophy due to peripheral motor neuropathy.
Figure 1.
Pathology of an individual with SPSMA. (a) Phosphotungstic acid hematoxylin (PTAH) staining of muscle biopsy showing severe muscle fiber–type grouping and atrophy (arrow). (b) Hematoxylin and eosin (H&E) staining of muscle autopsy sample showing severe muscle fiber–type grouping and atrophy (arrow). (c) ATPase staining (pH 9.4) of muscle autopsy samples showing both small type 1 and type 2 fibers and fiber type grouping. (d) Luxol fast blue/H&E staining of spinal cord sections showing a normal number of anterior horn cells. A representative motor neuron in the anterior horn is indicated by an arrow.
Because the entire 4-Mb CMT2C-linked region is included in the 14-Mb SPSMA-linked region and SPSMA and CMT2C share some common clinical features, including characteristic vocal cord paresis1–5, we considered that SPSMA and CMT2C may be clinical variants of the same genetic entity. We thus focused on identifying the genetic defects underlying these two related disorders.
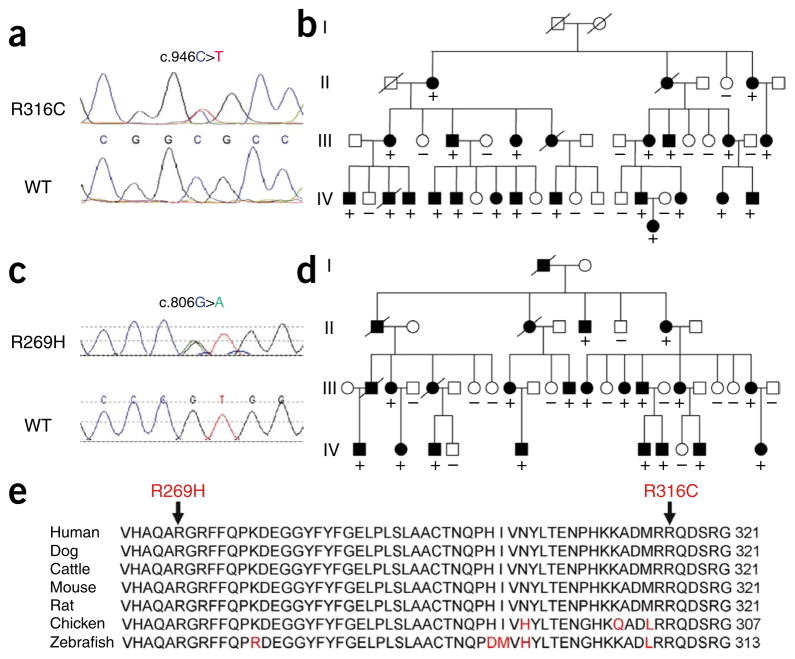
We first excluded mutations in HSPB8, which is linked to distal hereditary motor neuropathy (dHMN) typeIIA6 and CMT2L7 near the CMT2C locus. We also excluded mutations in UBE3B, USP30 and LHX5 (LIM homeobox 5), which share homology to either UBA1 (previously called UBE1) or FHL1, as mutations in UBA1 and FHL1 are linked to infantile spinal muscular atrophy8 and scapuloperoneal myopathy9, respectively. We then sequenced a total of 62 genes, including all the 56 known and predicted genes in the minimum 4-Mb CMT2C region, using DNA samples from individuals with SPSMA. We identified a heterozygous mutation in the TRPV4 gene10–12. The mutation, C946T, occurs in exon 6 and is predicted to result in an amino acid substitution of arginine by cysteine at codon 316 (R316C) (Fig. 2a). This C946T mutation co-segregated with the disease in the large SPSMA pedigree (Fig. 2b). The mutation causing this substitution was not present in the SNP database or in over 600 control samples. The other genetic variants we found either were polymorphisms and/or did not co-segregate with the disease. Thus, in the SPSMA-affected family, the R316C-causing substitution in TRPV4 was the only genetic defect we found in the coding exons of all the genes within the minimum region shared by both the SPSMA and CMT2C loci.
Figure 2.
TRPV4 mutations in SPSMA and CMT2C pedigrees. (a) A heterozygous mutation, C946T, resulting in R316C, was identified in exon 6 of TRPV4 in the SPSMA family. The wild-type (WT) sequence is shown in the lower panel. (b) All the affected members whose DNA samples were available for sequencing analysis had the R316C substitution. (c) A heterozygous mutation, G806A (leading to the R269H amino acid substitution) in exon 5 of TRPV4 was identified in the CMT2C family. The WT sequence is shown in the lower panel. (d) All the affected members whose DNA samples were available for sequencing analysis had this R269H substitution. (e) Evolutionary conservation of amino acids in the mutated region of TRPV4 in different species. Comparison of human (Homo sapiens) TRPV4 and its orthologs in dog (Canis lupus familiaris), cattle (Bos taurus), mouse (Mus musculus), rat (Rattus norvegicus), chicken (Gallus gallus) and zebrafish (Danio rerio). Amino acids identical to those in human TRPV4 are in black letters; nonidentical ones are denoted in red letters. The positions of the C-terminal amino acids are shown on the right. The mutated amino acids are indicated by arrows on the top. (+) indicates TRPV4 mutation; (−) indicates wild type.
We then extended our genetic analysis of TRPV4 to the original CMT2C-affected family2,4. Previous sequence analysis of 45 genes in the CMT2C region did not reveal any pathogenic mutation. However, analysis of TRPV4 revealed a heterozygous mutation, G806A, leading to R269H, in exon 5 of the TRPV4 gene in this large CMT2C pedigree (Fig. 2c). This R269H substitution co-segregated with the disease in this family (Fig. 2d). The G806A mutation was not present in the SNP database, and we did not detect it in over 700 control samples. Both Arg316 and Arg269 of TRPV4 are conserved among human, rat, mouse, chicken, stickleback and zebrafish (Fig. 2e).
Notably, both R269H and R316C occur in the ankyrin repeat–containing region of the cytoplasmic N terminus, which usually mediates protein-protein interactions (Supplementary Fig. 2)13,14. TRPV4 splice variants affecting this region have shown defects in oligomerization, leading to accumulation of TRPV4 monomers in the endoplasmic reticulum15. Because these variants are unable to be targeted to the plasma membrane, they are functionally inactive15. To examine whether the mutants have defects in subcellular trafficking, we analyzed the subcellular distribution of human wild-type TRPV4 (wtTRPV4) and mutant TRPV4 with the R269H or R316C substitutions in transiently transfected cells from the HEK293 line. We found that exogenous TRPV4 was present on the plasma membrane. Both mutants had a similar pattern of subcellular localization to wtTRPV4 in the transfected cells (Fig. 3), suggesting that these two alterations might not interfere with channel assembly and intracellular trafficking. This notion was supported by cell surface biotinylation assay, which did not show a significant difference in the level of TRPV4 at the plasma membrane between the wild-type and mutant proteins (data not shown).
Figure 3.
Localization of wild-type and mutant TRPV4 on the plasma membrane. (a–l) Confocal microscopy was performed using HEK293 cells transfected with plasmids pIRES2-ZsGreen1 containing wtTRPV4 (a–d), R269H (e–h) or R316C (i–l). Cells expressing exogenous TRPV4 were labeled by green fluorescent protein (GFP) (c, g, k). TRPV4 is shown by blue (a, e, i) and cadherin by red (b, f, j). Merged images are shown on the right panels (d, h, l). Arrows indicate TRPV4 signals on the plasma membrane. Arrowheads indicate representative cells without significant expression of GFP and TRPV4 (f, h).
The TRPV4 channel responds to a large variety of stimuli16. To test the potential effects of the mutations of interest on calcium channel activity, we analyzed the calcium channel activity of transiently transfected HEK293 cells using internal Fura-2 fluorescence ratio (activity at 340 nm/activity at 380 nm) as an indicator of the intra-cellular Ca2+ levels, which depend on Ca2+ influx. We examined the calcium levels of the transfected cells in response to various stimuli, including the TRPV4-specific agonist 4α-phorbol 12,13-didecanoate (4αPDD) (Fig. 4), osmotic cell swelling (Fig. 5), moderate heat (37 °C) (Supplementary Fig. 3) and endogenous agonist arachi-donic acid (Supplementary Fig. 4). We observed a consistent pattern of channel response to different stimuli. First, in nonstimulus conditions, cells transfected with either wild-type or mutant proteins showed increased basal intracellular calcium levels when compared to the nontransfected cells. Second, the presence of the TRPV4 mutant protein resulted in substantially higher basal calcium levels than wtTRPV4 protein, suggesting an increased constitutive activity for the mutants. Finally, the maximum and net increase in calcium levels in cells transfected with mutant TRPV4 were markedly higher than those in cells transfected with wtTRPV4 when activated by all the stimuli tested, suggesting that the mutations confer an increased channel activity (Figs. 4 and 5 and Supplementary Figs. 3 and 4). The increased calcium channel activity could be inhibited by the TRPV antagonist ruthenium red (data not shown).
Figure 4.
Effect of mutations on TRPV4 activity when stimulated with 4αPDD. (a–c) Effect of stimulation with 4αPDD (2 μM) on internal fluorescence ratio in wtTRPV4- (a), R269H- (b) and R316C-transfected (c) HEK293 cells. (d) Application of 4αPDD induced an increase in intracellular calcium ([Ca2+]i). Average increases, basal and maximum values are given. For each condition, n > 11 in at least three independent recordings. *, significant differences when compared with wtTRPV4 (two-tailed Student’s t test, P < 0.001). Error bars, means ± s.e.m.
Figure 5.
Effect of mutations on TRPV4 activity when stimulated with hypotonic solution. (a–c) Effect of stimulation with a hypotonic stimulus (HTS) (200 mOsm) on internal fluorescence ratio in wtTRPV4- (a), R269H- (b) and R316C-transfected (c) HEK293 cells. (d) Application of HTS induced an increase in intracellular calcium ([Ca2+]i). Average increases, basal and maximum values are given. For each condition, n > 15 in at least three independent recordings. *, significant differences when compared with wtTRPV4 (two-tailed Student’s t test, P < 0.005). Error bars, means ± s.e.m.
We then examined the effect of the CMT2C-linked substitution R269H on the electrophysiological properties of TRPV4. Whole-cell patch-clamp recordings were obtained from transiently transfected HEK293 cells. Although nontransfected cells had only small basal currents that did not show TRPV4 rectification (data not shown), cells transfected with either wtTRPV4 or TRPV4 carrying the R269H change showed large, outward-rectifying basal currents (Fig. 6a, b). Addition of the TRPV blocker ruthenium red to the bath markedly reduced the currents (Fig. 6a, b); the ruthenium red–sensitive component was then obtained by offline digital subtraction (Fig. 6c, d). These data show that most of the basal current in the transfected cells was actually mediated by TRPV4 channels (Fig. 6e, f). Consistent with the calcium imaging data, the ruthenium red–sensitive TRPV4 conductance was higher in cells expressing the CMT2C-linked mutant compared to wtTRPV4 (at 100 mV, the normalized conductance of the ruthenium red–sensitive component was 0.354 ± 0.47 nS/pF and 2.105 ± 0.57 nS/pF for cells expressing the wild-type (n = 9) and the mutant channels (n = 6), respectively (P < 0.01; Fig. 6g)). Similar to a previous study12, we also observed that about half of the wild-type (22 out of 47) and mutant (15 out of 27) TRPV4-transfected HEK293 cells did not show rectifying TRPV4 currents.
Figure 6.
Whole-cell recordings of TRPV4 currents from transfected HEK293 cells. (a, b) Voltage clamp recordings of the currents elicited by a slow voltage ramp (from -100 to 100 mV, 600 ms) in control conditions (black traces) and in the presence of 20 μM ruthenium red (red traces) in cells expressing wtTRPV4 (a) or R269H (b) channels. (c, d) Ruthenium red–sensitive current (obtained by digital subtraction) in cells expressing wtTRPV4 (c) and R269H (d). (e) Plot summarizing the amount of current (normalized to capacitance) recorded at +100 mV in control conditions (black bars) and in the presence of ruthenium red (red bars) in cells expressing the wild-type and mutated channels. (f) Plot summarizing the size of the ruthenium red–sensitive current in cells expressing the two channel types. (g) The ruthenium red–sensitive current was transformed into conductance. The conductance of R269H-expressing cells was ~6 times larger (0.35 ± 0.5 in wild type and 2.1 ± 0.6 in the mutant, 9 and 6 cells, respectively, P < 0.01). Error bars, means ± s.e.m.
TRPV4 is a vanilloid receptor–related transient receptor potential channel17, and it plays an important role in neural signaling18. Recently, two missense mutations (resulting in the R616Q and V620I substitutions) in TRPV4 were identified in two families with brachyolmia19. Subsequently, six TRPV4 missense mutations were found in spondylo-metaphyseal dysplasia, Kozlowski type (SMDK), and two TRPV4 mis-sense mutations were found in metatropic dysplasia20. Brachyolmia, SMDK and metatropic dysplasia are autosomal dominant dysplasias of the bones with variable severity. Among eight skeletal dysplasia–linked mutations tested in vitro, seven resulted in increased basal calcium channel activity, suggesting that a gain-of-function mechanism, possibly an increase in calcium channel activity, underlies the skeletal dysplasias19,20. This hypothesis is supported by the observation that overexpression of mouse wild-type Trpv4 in zebrafish causes marked shortening and curvature of the axial skeleton21.
Notably, some individuals in the SPSMA-affected family also presented variable skeletal abnormalities in addition to neuromuscular phenotypes. The skeletal abnormalities included congenital hip dysplasia, scoliosis, smaller hands with clinodactyly, and one arm or leg shorter than the other1. Identification of the R316C substitution in the SPSMA-affected family suggests that the R316C mutant may share a common property with other skeletal dysplasia–linked mutants in triggering skeletal abnormalities, and another distinct property with CMT2C-linked R269H substitutions in triggering neuropathic phenotypes. Thus, the SPSMA-linked R316C mutant may have two different properties that bridge these two clinically distinct groups of disorders (skeletal dysplasias and peripheral neuropathies).
Sensory impairment was found in some individuals with CMT2C but was not obvious in most of the individuals with SPSMA, with the exception of reduced vibratory sense at 256 Hz in the feet in some individuals with SPSMA1,2. However, it has become apparent that mutations in the same gene can cause distinct phenotypes or a spectrum of related phenotypes, as in CMT and hereditary motor neuropathy (HMN). For example, mutations in GARS cause CMT2D and dHMN5 (ref. 22), mutations in HSPB1 cause CMT2F and dHMN2B23, and mutations in HSPB8 result in CMT2L7 and dHMN2A6. These phenomena suggest that other genetic and environmental factors may modulate disease phenotype.
The pathogenic mechanism underlying the mutant TRPV4–mediated peripheral neuropathies is not yet clear. As R316C and R269H mutants have increased basal and maximum calcium channel activities compared to wtTRPV4, a gain-of-function rather than a loss-of-function mechanism is likely to be involved. This notion is supported by a previous observation that mice lacking Trpv4 do not show apparent neuromuscular abnormalities24,25. TRPV4 expression and/or function can be regulated by other auxiliary proteins such as OS-9, WNK4, AQP5 and the AIP4 ubiquitin ligase21,26–29. It has been demonstrated that PACSIN3 strongly inhibits the basal activity of TRPV4 and its activation by cell swelling and heat, but not by 4αPDD. Specific mutations of proline residues near the first ARD in the N terminus of TRPV4 abolish binding of PACSIN3 and render the channel insensitive to PACSIN3-induced inhibition30. These findings highlight the functional importance of the sequence alterations in TRPV4 and the interactions of TRPV4 with other proteins.
TRPV4 is widely expressed in diverse cells, but the R269H and R316C substitutions prominently affect axons of lower motor neurons. We have considered four possible mechanistic relationships in the development of SPSMA and CMT2C. First, motor neuron firing produces membrane potential changes in the axons, which may enhance the gain-of-function properties of these mutants more robustly than in electrically less-active tissues. Second, these mutations may alter the interactions of TRPV4 with protein(s) specifically expressed in the lower motor neurons. This may lead to an increased TRPV4 activity similar to the modulation of TRPV4 activity by PACSIN3 (ref. 30). Third, the pathology may be mediated by local and tissue-specific availability of endogenous or environmental agonists, leading to an increase in the gain of function of the TRPV4 mutants. The deleterious response seen in the axons may be related to their larger surface area, as opposed to the cell body, which is spared. Lastly, the proximal-to-distal gradient of axonal diameter, internodal distances and limb temperature may interplay with the specific properties of each mutant to produce the specific topographic distribution of nerve damage seen as more proximal in SPSMA and more distal in CMT2C. Although our findings open a new avenue for understanding the pathogenesis of peripheral neuropathy, further studies are needed to elucidate the link between specific TRPV4 properties and clinically distinct disorders caused by mutant TRPV4. To account for the three clinically distinct but sometimes overlapping phenotypes associated with TRPV4 mutations, we propose that the collective syndrome be named TRPV4-associated peripheral neuropathy and bony dysplasia, or TRPV4-PNAB.
ONLINE METHODS
Subjects and samples
This study was approved by the local institutional review boards. DNA and other samples were taken after obtaining written informed consent. This study included a large New England family of French-Canadian origin affected with SPSMA and an American family of English and Scottish descent affected with CMT2C described previously1,2.
Genetic analysis
Genomic DNA was extracted from whole peripheral blood, transformed lymphoblastoid cell lines or available tissues by standard methods (Qiagen). Intronic primers covering sequences of interest (such as coding exons) were designed at least 50 bp away from the intron-exon boundaries. When a PCR product was over 500 bp long, multiple overlapping primers were designed with an average of 50 bp overlap. Primers were designed with the help of an Oligo analyzer (IDT, IA) and ExonPrimer software (Institute of Human Genetics). Forty nanograms of genomic DNA were used for PCR amplification with high-fidelity TaKaRa LA Taq (Takara). The amplification protocol consisted of the following steps: incubation at 95 °C for 1 min, 32 cycles of 95 °C (30 s), 58 °C (30 s) and 72 °C (1 min), and a final 5 min extension at 72 °C. Unconsumed dNTPs and primers were digested with Exonuclease I and Shrimp Alkaline Phosphatase (ExoSAP-IT, USB). When nonspecific PCR amplification occurred, the PCR products were separated by 1.5% agarose gel and the specific PCR product was cut out from the gel and purified using QIAquick Gel Extraction Kit (QIAGEN Science). For sequencing of a PCR product, fluorescent dye–labeled single-strand DNA was amplified with Beckman Coulter sequencing reagents (GenomeLab DTCS Quick Start Kit) followed by single-pass bidirectional sequencing with CEQ 8000 Genetic Analysis System (Beckman Coulter) (Northwestern University) or using dye termination chemistry with 3730xl sequencer (Applied Biosystems; Mayo Clinic).
Expression vectors
A full length human cDNA clone (IMAGE: 40125977) was used as a template. Two primers anchored with an XhoI (TRPV4-TP1) and BamHI (TRPV4-TP2) were used to amplify the full-length coding sequence. The amplified fragment was cloned into the plasmid vector pBluescript M13. The TRPV4 sequence was verified by direct sequencing. The R316C mutation was introduced into the plasmid vector by site-directed mutagenesis using a primer containing the R316C mutation (TRPV3-R316C) and R269H was introduced using a primer containing the R269H mutation (TRPV4-R269H). For primer sequences, see Supplementary Table 1. The XhoI-BamHI fragment containing wild-type TRPV4 or the substitution R316CC or R269H was released from the pBluescript M13 vector and cloned into the XhoI and BamHI sites of a dual expression vector, pIRES2-ZsGreen1 (Clontech).
Expression of wild-type and mutant TRPV4
Human embryonic kidney (HEK293) cells were grown on collagen-coated glass coverslips in Dulbecco’s modified Eagle’s medium containing 10% (v/v) human serum, 2 mM L-glutamine, 2 U/ml penicillin and 2 mg/ml streptomycin at 37 °C in a humidity- controlled incubator with 5% CO2. The cells were transiently transfected with expression vectors (wild-type TRPV4, and the substitutions R316C or R269H) using Lipofectamine 2000 (Invitrogen).
Confocal microscopy
HEK293 cells were seeded on collagen-coated cover-slides 24 h before transfection. Twenty-four hours after transfection, the cells were fixed with 3% paraformaldehyde and 0.02% glutaraldehyde for 15 min. Ice-cold methanol was used to permeabilize cells. Rabbit antibody to TRPV4 (1:50, Chemicon) and mouse antibody to cadherin (1:200, Abcam) were used as primary antibodies. Alexa Fluor 555 goat antibody to mouse (1:500, Invitrogen) and Alexa Fluor 633 goat antibody to rabbit (1:250, Invitrogen) were used as secondary antibodies. Digital images were captured and analyzed with Carl Zeiss LSM 510 META laser scanning confocal microscopes.
Calcium imaging and intracellular Ca2+ measurements
Intracellular free calcium concentration was measured using digital video microfluorimetry. Twenty-four hours after transfection, cells were rinsed briefly with HEPES buffer (120 mM NaCl, 5.4 mM KCl, 1.6 mM MgCl2, 1.8 mM CaCl2, 11 mM glucose and 25 mM HEPES, pH 7.2) and loaded with 4 μM Fura-2 AM (Molecular Probes) in HEPES buffer for 30 min at room temperature. Cultures were then rinsed and kept in the dark in HEPES buffer at room temperature for an additional 30 min to allow for complete dye de-esterification. The coverslips were then mounted on the stage of a Nikon Diaphot inverted epifluorescence microscope equipped for digital fluorescence microscopy. After excitation at 340 nm (Ca2+-bound) and 380 nm (Ca2+-free), fluorescence was digitally monitored at 520 nm. Fluorescence at 340 nm compared to 380 nm (F340/F380) ratios were collected before and during treatment with 10 μM arachidonic acid, 2 μM 4-α-phorbol-12,13-didecanoate (4αPDD) or 200 mOsm hypotonic saline (HTS) using MetaFluor software (Universal Imaging Corporation). For temperature activation, cells were incubated in HEPES at 14 °C and stimulated with HEPES at 37 °C. Ruthenium red (20 μM) was used to block the effects of these stimuli. Measurements were calibrated using the Grynkiewicz equation31. Values for Rmin and Rmax (where R is the fluorescence ratio of 340 nm/380 nm) were determined by applying Ca2+-free solution and high Ca2+–containing solution in the presence of 5 μM ionomycin, respectively. The dissociation constant (Kd) of 224 nM for Fura-2 a.m. was used for calculations. Imaging experiments were performed at room temperature (22 °C), unless otherwise stated. A two-tailed unpaired Student’s t test (P < 0.05) was used for statistical analysis.
Electrophysiology
Whole-cell voltage-clamp recordings were performed on HEK293 cells transiently transfected with wild-type or R269H TRPV4 cDNA. For experiments with ruthenium red blockage, cells were incubated with ruthenium red (10 μm) after transfection. Similar GFP-fluorescent cells were selected for experiments. Patch pipettes were pulled from World Precision Instruments (WPI) glass (PG10-165) using a horizontal puller (P97, Sutter) and had a resistance of 2–3 MΩ when filled with internal solution consisting of 140 mM CsCl, 2 mM MgCl2, 10 mM EGTA, 2 mM Na2ATP, 0.1 mM NaGTP and 10 mM HEPES, pH 7.3 (adjusted with CsOH). The series resistance was 4–14 MΩ and was compensated between 65% and 75%. The extracellular solution consisted of 100 mM NaCl, 6 mM KCl, 2 mM MgCl2, 1.5 mM CaCl2, 10 mM glucose and 10 mM HEPES, pH 7.38 (adjusted with NaOH) and ~315 mOsm (adjusted with sucrose). Whole-cell currents were recorded using an Axopatch 200B amplifier (Molecular Devices). Cells were held at −30 mV and currents were elicited by slow voltage ramps (from −100 mV to +100 mV, 0.6 s duration), filtered at 5 KHz and sampled at 10 KHz. Drugs were bath-applied at a rate of ~4 ml/min. The current density was determined by normalizing the current to the cell capacitance measured in voltage clamp using a −5-mV square pulse from a holding potential of −30 mV.
Ruthenium red was prepared in a 20-mM stock solution (in water) and stored at 2–4 °C. Working solutions were prepared freshly daily and were bath applied. After drug application, the dish was discarded even if complete wash-out of the drug was obtained.
Supplementary Material
Acknowledgments
The authors acknowledge the support from the National Institute of Neurological Disorders and Stroke (NS050641), the Les Turner ALS Foundation, the Vena E. Schaff ALS Research Fund, the Harold Post Research Professorship, the Herbert and Florence C. Wenske Foundation, the David C. Asselin MD Memorial Fund, the Help America Foundation, the Les Turner ALS Foundation/Herbert C. Wenske Foundation Professorship and the Epilepsy Foundation. We thank R. J. Miller and A. Belmadani for help with the calcium imaging studies and J. Caliendo for proofreading this manuscript.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
AUTHOR CONTRIBUTIONS
T.S., H.-X.D. and C.J.K. conceived this project. H.-X.D., J.Y., Y.S., Y.W., Y.Y. and H.Z. did the sequencing analysis. F.F., Y.S. and H.-X.D. performed the calcium imaging analysis. H.-J.Y. and M.M. performed the whole-cell patch-clamp recordings. N.S. collected family information and samples. E.T.H.-W. did pathological analysis. C.J.K., R.D., P.J.D. and T.S. did clinical studies. H.-X.D., C.J.K., M.M. and T.S. analyzed the data and wrote the paper.
References
- 1.DeLong R, Siddique T. A large New England kindred with autosomal dominant neurogenic scapuloperoneal amyotrophy with unique features. Arch Neurol. 1992;49:905–908. doi: 10.1001/archneur.1992.00530330027010. [DOI] [PubMed] [Google Scholar]
- 2.Dyck PJ, et al. Hereditary motor and sensory neuropathy with diaphragm and vocal cord paresis. Ann Neurol. 1994;35:608–615. doi: 10.1002/ana.410350515. [DOI] [PubMed] [Google Scholar]
- 3.Isozumi K, et al. Linkage of scapuloperoneal spinal muscular atrophy to chromosome 12q24.1-q24.31. Hum Mol Genet. 1996;5:1377–1382. doi: 10.1093/hmg/5.9.1377. [DOI] [PubMed] [Google Scholar]
- 4.Klein CJ, et al. The gene for HMSN2C maps to 12q23–24: a region of neuromuscular disorders. Neurology. 2003;60:1151–1156. doi: 10.1212/01.wnl.0000055900.30217.ea. [DOI] [PubMed] [Google Scholar]
- 5.McEntagart ME, et al. Confirmation of a hereditary motor and sensory neuropathy IIC locus at chromosome 12q23-q24. Ann Neurol. 2005;57:293–297. doi: 10.1002/ana.20375. erratum 57, 609, 2005. [DOI] [PubMed] [Google Scholar]
- 6.Irobi J, et al. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat Genet. 2004;36:597–601. doi: 10.1038/ng1328. [DOI] [PubMed] [Google Scholar]
- 7.Tang BS, et al. Small heat-shock protein 22 mutated in autosomal dominant Charcot-Marie-Tooth disease type 2L. Hum Genet. 2005;116:222–224. doi: 10.1007/s00439-004-1218-3. [DOI] [PubMed] [Google Scholar]
- 8.Ramser J, et al. Rare missense and synonymous variants in UBE1 are associated with X-linked infantile spinal muscular atrophy. Am J Hum Genet. 2008;82:188–193. doi: 10.1016/j.ajhg.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinzii CM, et al. X-linked dominant scapuloperoneal myopathy is due to a mutation in the gene encoding four-and-a-half-LIM protein 1. Am J Hum Genet. 2008;82:208–213. doi: 10.1016/j.ajhg.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liedtke W, et al. Vanilloid receptor-related osmotically activated channel (VR-OAC), a candidate vertebrate osmoreceptor. Cell. 2000;103:525–535. doi: 10.1016/s0092-8674(00)00143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strotmann R, Harteneck C, Nunnenmacher K, Schultz G, Plant TD. OTRPC4, a nonselective cation channel that confers sensitivity to extracellular osmolarity. Nat Cell Biol. 2000;2:695–702. doi: 10.1038/35036318. [DOI] [PubMed] [Google Scholar]
- 12.Wissenbach U, Bodding M, Freichel M, Flockerzi V. Trp12, a novel Trp related protein from kidney. FEBS Lett. 2000;485:127–134. doi: 10.1016/s0014-5793(00)02212-2. [DOI] [PubMed] [Google Scholar]
- 13.Jin X, Touhey J, Gaudet R. Structure of the N-terminal ankyrin repeat domain of the TRPV2 ion channel. J Biol Chem. 2006;281:25006–25010. doi: 10.1074/jbc.C600153200. [DOI] [PubMed] [Google Scholar]
- 14.Phelps CB, Huang RJ, Lishko PV, Wang RR, Gaudet R. Structural analyses of the ankyrin repeat domain of TRPV6 and related TRPV ion channels. Biochemistry. 2008;47:2476–2484. doi: 10.1021/bi702109w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arniges M, Fernandez-Fernandez JM, Albrecht N, Schaefer M, Valverde MA. Human TRPV4 channel splice variants revealed a key role of ankyrin domains in multimerization and trafficking. J Biol Chem. 2006;281:1580–1586. doi: 10.1074/jbc.M511456200. [DOI] [PubMed] [Google Scholar]
- 16.Nilius B, Owsianik G, Voets T, Peters JA. Transient receptor potential cation channels in disease. Physiol Rev. 2007;87:165–217. doi: 10.1152/physrev.00021.2006. [DOI] [PubMed] [Google Scholar]
- 17.Pedersen SF, Owsianik G, Nilius B. TRP channels: an overview. Cell Calcium. 2005;38:233–252. doi: 10.1016/j.ceca.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 18.Liedtke W. Molecular mechanisms of TRPV4-mediated neural signaling. Ann NY Acad Sci. 2008;1144:42–52. doi: 10.1196/annals.1418.012. [DOI] [PubMed] [Google Scholar]
- 19.Rock MJ, et al. Gain-of-function mutations in TRPV4 cause autosomal dominant brachyolmia. Nat Genet. 2008;40:999–1003. doi: 10.1038/ng.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krakow D, et al. Mutations in the gene encoding the calcium-permeable ion channel TRPV4 produce spondylometaphyseal dysplasia, Kozlowski type and metatropic dysplasia. Am J Hum Genet. 2009;84:307–315. doi: 10.1016/j.ajhg.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, et al. OS-9 regulates the transit and polyubiquitination of TRPV4 in the endoplasmic reticulum. J Biol Chem. 2007;282:36561–36570. doi: 10.1074/jbc.M703903200. [DOI] [PubMed] [Google Scholar]
- 22.Antonellis A, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evgrafov OV, et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- 24.Liedtke W, Friedman JM. Abnormal osmotic regulation in trpv4−/ − mice. Proc Natl Acad Sci USA. 2003;100:13698–13703. doi: 10.1073/pnas.1735416100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki M, Mizuno A, Kodaira K, Imai M. Impaired pressure sensation in mice lacking TRPV4. J Biol Chem. 2003;278:22664–22668. doi: 10.1074/jbc.M302561200. [DOI] [PubMed] [Google Scholar]
- 26.Fu Y, Subramanya A, Rozansky D, Cohen DM. WNK kinases influence TRPV4 channel function and localization. Am J Physiol Renal Physiol. 2006;290:F1305–F1314. doi: 10.1152/ajprenal.00391.2005. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, et al. A role for AQP5 in activation of TRPV4 by hypotonicity: concerted involvement of AQP5 and TRPV4 in regulation of cell volume recovery. J Biol Chem. 2006;281:15485–15495. doi: 10.1074/jbc.M600549200. erratum 283, 3688, 2008. [DOI] [PubMed] [Google Scholar]
- 28.Sidhaye VK, et al. Transient receptor potential vanilloid 4 regulates aquaporin-5 abundance under hypotonic conditions. Proc Natl Acad Sci USA. 2006;103:4747–4752. doi: 10.1073/pnas.0511211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wegierski T, Hill K, Schaefer M, Walz G. The HECT ubiquitin ligase AIP4 regulates the cell surface expression of select TRP channels. EMBO J. 2006;25:5659–5669. doi: 10.1038/sj.emboj.7601429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’hoedt D, et al. Stimulus-specific modulation of the cation channel TRPV4 by PACSIN3. J Biol Chem. 2008;283:6272–6280. doi: 10.1074/jbc.M706386200. [DOI] [PubMed] [Google Scholar]
- 31.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.