Summary
Little is known about molecular recognition of acetylated N-termini, despite prevalence of this modification among eukaryotic cytosolic proteins. We report that the family of human DCN-like (DCNL) co-E3s, which promote ligation of the ubiquitin-like protein NEDD8 to cullin targets, recognizes acetylated N-termini of the E2 enzymes UBC12 and UBE2F. Systematic biochemical and biophysical analyses reveal 40- and 10- fold variations in affinities amongst different DCNL-cullin and DCNL-E2 complexes, which contribute to widely ranging efficiencies of different NEDD8 ligation cascades. Structures of DCNL2 and DCNL3 complexes with N-terminally acetylated peptides from UBC12 and UBE2F illuminate a common mechanism by which DCNL proteins recognize N-terminally acetylated E2s, and how selectivity for interactions dependent on N-acetyl-methionine can be established through sidechains recognizing distal residues. Distinct preferences of UBC12 and UBE2F peptides for inhibiting different DCNLs, including the oncogenic DCNL1 protein, suggest it may be possible to develop small molecules blocking specific N-acetyl-methionine-dependent protein interactions.
Introduction
Approximately 50%–90% of eukaryotic cytosolic proteins are co-translationally N-terminally acetylated either on Met, or the resultant N-terminus following processing by Met aminopeptidase (Arnesen, 2011; Kalvik and Arnesen, 2012). Important functions for N-terminal acetylation can be inferred from genetic experiments in which N-terminal acetyltransferase enzymes were deleted from budding yeast either alone or in synthetic lethal screens, or were knocked down in mammalian cells (reviewed in (Arnesen, 2011; Starheim et al., 2012)). Nonetheless, only a few specific functions of N-terminal acetylation have been reported. Examples include roles of N-terminal Met acetylation in tropomyosin-actin complex formation (Coulton et al., 2010; Polevoda et al., 2003; Singer and Shaw, 2003), and in trafficking of certain GTPases (Behnia et al., 2004; Setty et al., 2004). At this point, little is known about potential regulation of and by N-terminal acetylation. However, metabolic changes mediated by expression of the antiapoptotic Bcl-2 family member Bcl-xL influences the extent of cellular protein N-terminal acetylation (Yi et al., 2011). Bcl-xL expression modulates levels of acetyl-CoA, which provides the acetyl group to be transferred to N-termini. Notably, decreased N-terminal acetylation upon Bcl-xL overexpression is thought to play a role in apoptotic resistance (Yi et al., 2011). Although there are presently no known N-terminal deacetylases, N-terminal acetylation can serve to target proteins as substrates for ubiquitination by the yeast ubiquitin E3 ligase Doa10, thereby directing some N-terminally acetylated proteins for proteasomal degradation (Hwang et al., 2010). Protein-protein interactions that sequester acetylated N-termini have been proposed potentially to protect N-terminally acetylated proteins from Doa10-dependent degradation (Hwang et al., 2010; Zhang et al., 2010). Nonetheless, detailed structural mechanisms by which N-terminal acetylation can influence protein activities are largely unknown.
Recently, N-terminal Met acetylation was shown to be critical for a specific protein-protein interaction that enhances ligation of the ubiquitin-like protein (UBL), NEDD8, to a Lys in the WHB subdomain of the CUL1 C-terminal domain (CTD) (Scott et al., 2011). Like other UBLs, NEDD8 is ligated by distinctive E1-E2-E3 cascades. N-terminal acetylation of the E2, UBC12, was shown to play a role in NEDD8 ligation via a “dual E3” mechanism (Scott et al., 2011; Scott et al., 2010). One E3, RBX1, acts as a conventional RING ligase: RBX1’s β-strand recruits the CUL1 substrate, and RBX1’s RING domain binds the labile thioester-linked UBC12~NEDD8 intermediate and promotes NEDD8 ligation (Scott et al., 2010). However, the linker between RBX1’s CUL1 binding site and UBC12-binding RING domain is flexible. The conformation is harnessed by a co-E3, Dcn1 in budding yeast or DCNL1 in human cells, which binds other CUL1 and UBC12 surfaces to juxtapose UBC12’s active site and CUL1’s acceptor Lys for the NEDD8 ligation reaction. Whereas RBX1-mediated NEDD8 ligation is independent of the state of UBC12’s N-terminus, N-terminal acetylation contributes 2 orders of magnitude to the Kd for UBC12 binding to the Dcn1/DCNL1 PONY domain (“Potentiation of Neddylation”, also referred to with a “P” superscript: for example DCNL1P) (Scott et al., 2011). Crystal structures of both yeast and human Dcn1P/DCNL1P complexes with N-terminally acetylated UBC12 peptides revealed that interactions are dominated by burial of UBC12’s N-acetyl-Met in a deep hydrophobic pocket in Dcn1/DCNL1 (Scott et al., 2011).
Lower eukaryotes such as budding yeast have only one NEDD8 E2 (Ubc12), one RBX protein (Rbx1, also called Hrt1), and one Dcn co-E3 (Dcn1). However, typical human cells express two NEDD8 E2s (UBC12 and UBE2F), two RBX proteins (RBX1 and RBX2), five distinct Dcn-like proteins (DCNL 1–5), and numerous cullins (Huang et al., 2009; Kamura et al., 1999; Kipreos et al., 1996; Kurz et al., 2005; Ohta et al., 1999; Seol et al., 1999; Skowyra et al., 1999; Tan et al., 1999). For the best-studied mammalian cullins (CUL1, CUL2, CUL3, CUL4A and B, and CUL5), NEDD8 ligation from the two different E2s involves particular pairings with a cullin’s associated RBX protein: UBC12 is specific for RBX1 and mediates neddylation of RBX1-associated CUL1, 2, 3 and 4. RBX2 is specific for UBE2F, which mediates neddylation of the RBX2 partner, CUL5 (Huang et al., 2009). However, other than the interaction between DCNL1P and N-terminally acetylated UBC12, the extent to which the different DCNL PONY domains can stimulate cullin neddylation by the different NEDD8 E2s, and roles of E2 N-terminal acetylation in this process, have not been explored. This is of interest because cullin-RBX complexes assemble with other subunits to form the largest family of ubiquitin E3s, the cullin-RING ligases (CRLs), with ~300 CRLs encoded by the human genome (Deshaies and Joazeiro, 2009). NEDD8 ligation favors an active conformation for CRL ubiquitin ligase catalytic activity, and is estimated to elicit ~20% of all 26S proteasomal degradation (Bennett et al., 2010; Duda et al., 2008; Saha and Deshaies, 2008; Soucy et al., 2009; Yamoah et al., 2008).
To gain insights into structural mechanisms underlying NEDD8 ligation by mammalian enzymes, we performed systematic biochemical, biophysical, and crystallographic analyses of human CUL-DCNL-E2 interactions using purified recombinant proteins. Our data provide a biophysical rationale for previously described in vivo promiscuity between DCNL and cullin components of human NEDD8 cascades (Bennett et al., 2010; Huang et al., 2011; Kim et al., 2008; Kurz et al., 2008; Meyer-Schaller et al., 2009), and reveal that N-terminal acetylation of NEDD8 E2s is generally important for DCNL activation of NEDD8 ligation to cullins. The data also show how selectivity for protein-protein interactions dependent on Met N-terminal acetylation can be established by subtle differences in recognition of downstream residues among highly homologous structures, and indicate that it may be possible to inhibit specific protein interactions mediated by N-acetyl-Met.
Results
N-terminal acetylation of human UBE2F expressed in eukaryotic cells
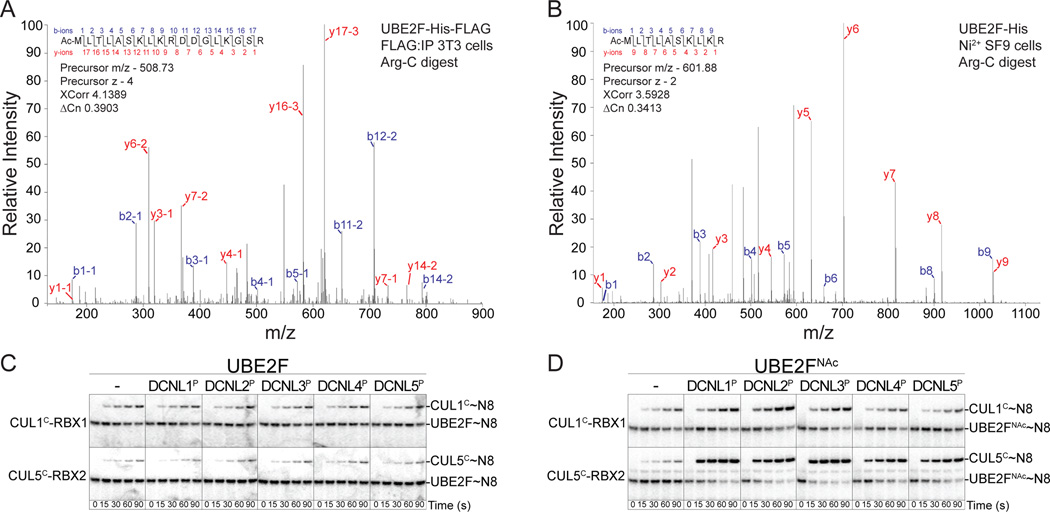
To investigate the modification status of UBE2F’s N-terminus expressed in eukaryotic systems, we purified C-terminally tagged forms of the protein. In NIH 3T3 cells, we retrovirally expressed human UBE2F harboring a C-terminal His6 and FLAG-tag (Huang et al., 2009). Following anti-FLAG immunoprecipitation, LC-MS/MS analysis revealed UBE2F as retaining its N-terminal methionine and being N-terminally acetylated (Fig. 1A). Similar results were obtained for C-terminally His-tagged UBE2F purified after baculovirus-mediated expression in insect cells (Fig. 1B, S1A). N-terminally acetylated UBE2F is referred to hereafter as UBE2FNAc.
Figure 1. Role of N-terminal acetylation of the NEDD8 E2 UBE2F in human DCNL PONY domain-stimulated NEDD8 ligation.
(A) MS/MS spectrum resulting from the N-terminal peptide after Arg-C digestion/desalting of human UBE2F expressed in NIH 3T3 cells following affinity purification via a C-terminal tag. The corresponding XCorr and ACN values are indicated as well as the y (red) and b (blue) ions used to match the peptide sequence.
(B) MS/MS spectrum of the N-terminal peptide from Arg-C digested UBE2F-His purified from insect cells upon baculovirus-mediated expression.
(C) Pulse-chase [32P]~NEDD8 transfer from unacetylated UBE2F to either CUL1CTD-RBX1 (labeled CUL1C-RBX1) or CUL5CTD-RBX2 (labeled CUL5C-RBX2) in the absence or presence of the indicated DCNL PONY domain.
(D) Pulse-chase [32P]~NEDD8 transfer assays as in panel C, except with the N-terminally acetylated E2 UBE2FNAc.
DCNL PONY domains can potentiate NEDD8 ligation from N-terminally acetylated UBE2F
Can N-terminally acetylated UBE2F mediate NEDD8 ligation? In the absence of DCNL1, RBX1 can promote efficient NEDD8 transfer from UBE2F to an associated CUL1 C-terminal domain and RBX2 can promote efficient NEDD8 transfer from UBE2F to an associated CUL5 C-terminal domain (Huang et al., 2009). To assess potential roles of UBE2F N-terminal acetylation, we compared activities of UBE2FNAc with unacetylated UBE2F prepared in E. coli as a SUMO fusion protein (Mossessova and Lima, 2000), which after cleavage with the protease SENP2 yields an unacetylated N-terminal Met (Fig. S1B). Using a pulse-chase assay, we exclusively examined NEDD8 transfer from the E2s to cullin targets (Fig. 1C, D). Briefly, after E1-mediated “pulse” generation of a thioester-linked E2~[32P]-NEDD8 intermediate, a cullin CTD-RBX complex was added, and radiolabeled NEDD8 was “chased” from the E2 to the cullin. In the absence of a DCNL, NEDD8 transfer to the CUL1 or CUL5 CTDs in complex with RBX1 or RBX2, respectively, is insensitive to whether or not UBE2F is N-terminally acetylated, and there was no effect of any DCNL PONY domain on NEDD8 transfer from unacetylated UBE2F (Fig. 1C). However, under the conditions of our assays, the PONY domains from all 5 DCNL family members potently stimulated NEDD8 transfer from UBE2FNAc to RBX2-associated CUL5CTD, and PONY domains from DCNLs 1–3 also clearly stimulated NEDD8 transfer from UBE2FNAc to RBX1-associated CUL1CTD (Fig. 1D).
DCNL-CUL and DCNL-NEDD8 E2 interactions contribute to specificity of NEDD8 ligation pathways in vitro
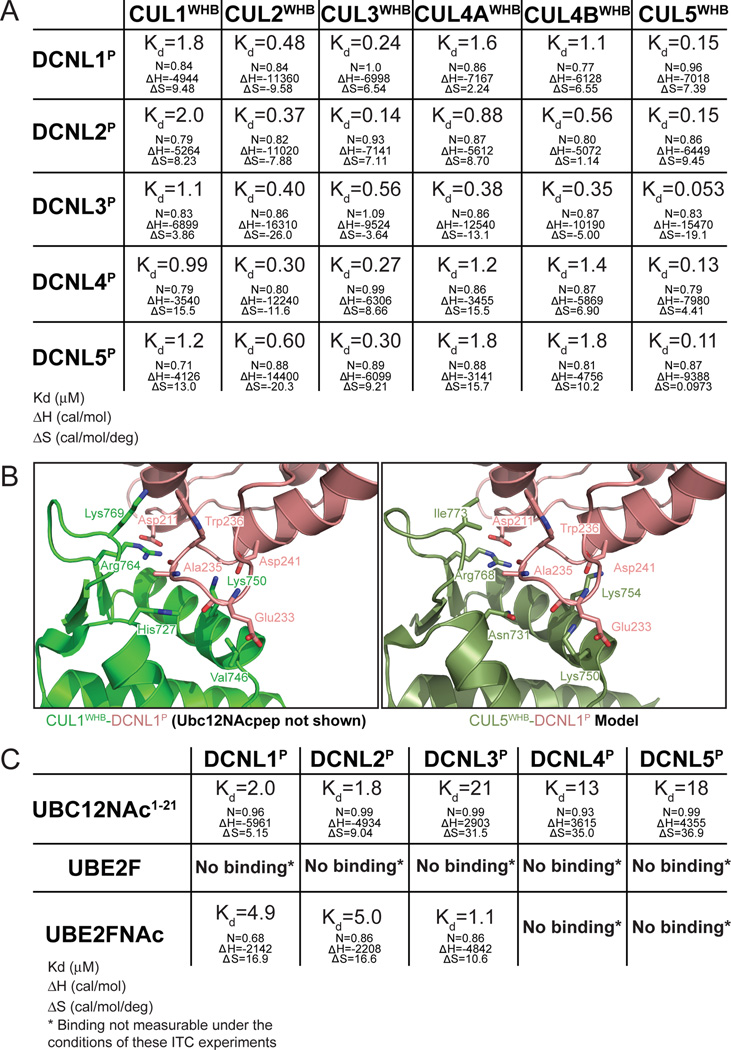
To gain insights into the extent of DCNL activity toward the different cullins and NEDD8 E2s, we systematically characterized DCNL PONY domain interactions. First, we focused on DCNL-cullin complexes. Previous studies localized interactions to the PONY domain from DCNL1 and the WHB subdomain of CUL1 (Kim et al., 2008; Kurz et al., 2008; Scott et al., 2011; Scott et al., 2010). Thus, we focused on these regions, which are conserved among all human family members (Fig. S2) (Kim et al., 2008; Kurz et al., 2008). All 30 pairwise combinations of the 5 DCNL PONY domains and 6 cullin WHB subdomains showed interactions by isothermal titration calorimetry (ITC), albeit with different affinities (Fig. 2A, S2). The observed 40-fold range in Kd values appears largely dictated by the identity of the cullin, rather than by the DCNL family member. As examples, the PONY domains from all 5 DCNL family members bind to the WHB subdomains from CUL2, CUL3, and CUL5 with Kd values in the submicromolar range, whereas interactions with the CUL1, CUL4A and CUL4B WHB subdomains displayed Kd values in the micromolar range. Indeed, the WHB subdomain from CUL5 binds tightly to all the different PONY domains with only a ~3-fold range in Kd (0.05–0.15 µM), and the WHB subdomain from CUL1 binds much more weakly to all the different PONY domains with only a ~2-fold range in Kd (0.99–2.0 µM). Sequence alignments in light of a prior DCNL1P-CUL1WHB crystal structure (Scott et al., 2011; Scott et al., 2010) explain this promiscuity, as key interacting residues are conserved in all 5 DCNL PONY domains and the various cullin WHB subdomains (Kim et al., 2008; Kurz et al., 2008). However, assorted amino acid variations spread throughout the interaction surface likely account for differences in affinities (Fig. S2). Some differences are observed in a docking model in which the CUL5WHB structure (Duda et al., 2008) is superimposed onto CUL1WHB from the complex with DCNL1P (Scott et al., 2011) (Fig. 2B). Among the DCNL-interacting residues least conserved among the cullins are Val746 in CUL1/Lys750 in CUL5, and Lys769 in CUL1/Ile773 in CUL5 (Fig. S2). These may contribute to the more favorable interactions for CUL5, where Lys750 would be predicted to form a salt-bridge with DCNL1 Glu233, which is conserved as Glu or Asp among all 5 DCNLs, and Ile773 would be predicted to pack against a conserved DCNL Trp sidechain (Fig. 2B).
Figure 2. Human DCNL PONY domain interactions with different cullins and N-terminally acetylated NEDD8 E2s.
(A) Thermodynamic parameters determined by ITC for binding between the indicated human DCNL PONY domains and cullin WHB subdomains.
(B) Previously published structure of CUL1WHB (green) - DCNL1P(salmon) (3TDU.pdb, UBC12NAc peptide not shown) complex (Scott et al., 2011) and model of CUL5WHB (olive) (Duda et al., 2008)- DCNL1P(salmon) (Scott et al., 2011), highlighting DCNL-CUL interacting residues.
(C) Thermodynamic parameters determined by ITC for binding between the indicated human DCNL PONY domains and unacetylated and N-terminally acetylated versions of UBE2F, and an N-terminally acetylated peptide corresponding to UBC12.
We also performed ITC experiments to examine interactions between the different DCNL PONY domains with the NEDD8 E2s (Fig. 2C, S2). Because of challenges with producing the large amounts of UBC12NAc in insect cells required for ITC, we examined interactions between the different DCNL PONY domains with a 21-residue peptide corresponding to the N-terminal helix of UBC12NAc (Fig. 2C). The Kd values of ~2 µM obtained for DCNL1P and DCNL2P binding to this UBC12 peptide are similar to those reported previously for DCNL1P binding to full-length UBC12NAc or to an acetylated N-terminal 26-residue peptide (Scott et al., 2011). However, there was striking variation in affinities, with 5–10-fold higher Kd values for interactions between the N-terminally acetylated UBC12 peptide and PONY domains from DCNL3, DCNL4, or DCNL5. By contrast, UBE2FNAc protein preferentially binds DCNL3P with a Kd of ~1 µM, with a 5-fold decrease in binding to DCNL1P and DCNL2P, and Kd values for binding to DCNL4P and DCNL5P were too high for us to measure. We could not measure binding between unacetylated UBE2F and any of the DCNL PONY domains. Thus, N-terminal acetylation is a common component of NEDD8 E2 interactions with DCNL family members.
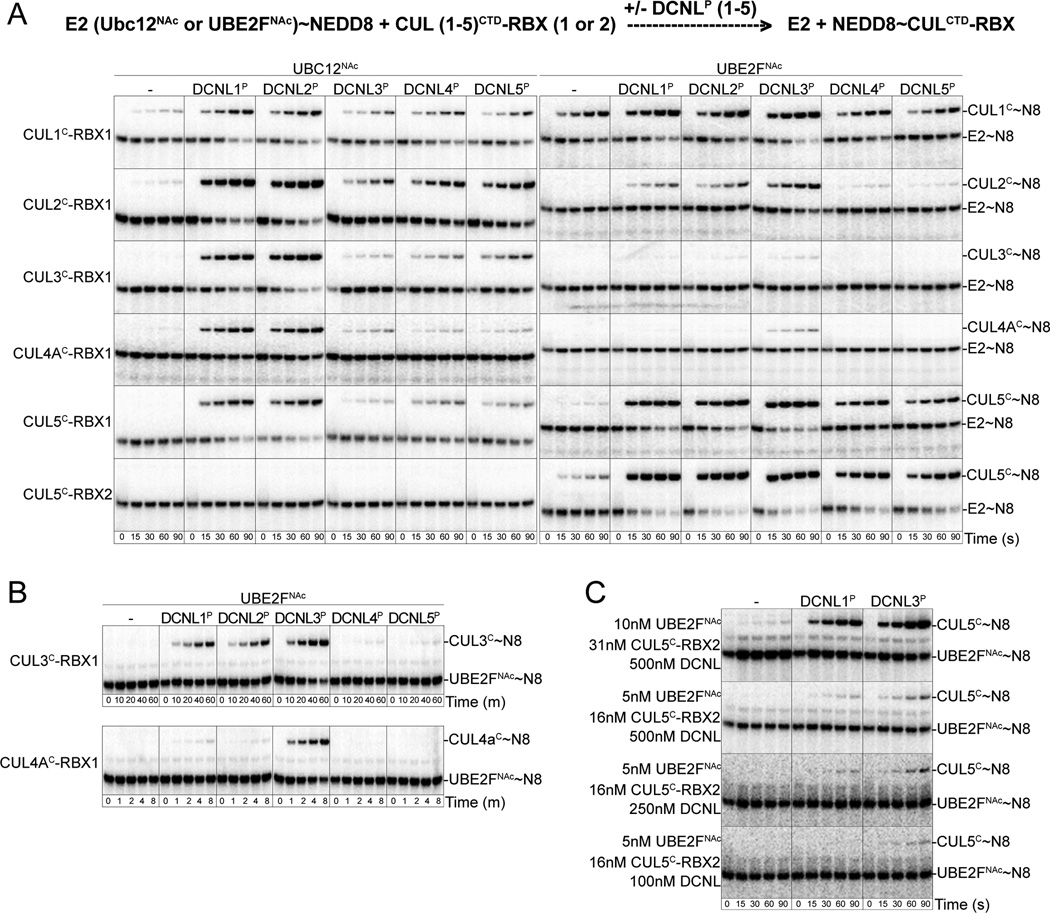
To understand the functional consequences of different DCNL affinities for distinct cullins and NEDD8 E2s, we examined activation of NEDD8 transfer from UBC12NAc and UBE2FNAc to different CULCTD-RBX complexes. All five DCNL PONY domains stimulate NEDD8 transfer from UBC12NAc to all five cullin CTD-RBX1 complexes (Fig. 3A). Even in the presence of a DCNL PONY domain, however, UBC12’s specificity for RBX1 was retained, as NEDD8 was not ligated to CUL5CTD in complex with its native RBX partner, RBX2. Furthermore, all 5 DCNL PONY domains also could stimulate NEDD8 transfer from UBE2FNAc, as observed for CUL2CTD in complex with RBX1 and CUL5CTD in complex with either RBX1 or RBX2.
Figure 3. Variations in DCNL PONY domain activation of NEDD8 transfer from N-terminally acetylated NEDD8 E2s to different cullin C-terminal domain/RBX complexes.
(A) As indicated in the schematic diagram, pulse-chase [32P]~NEDD8 transfer from UBC12NAc (left) or UBE2FNAc (right) to the indicated cullin C-terminal domain-RBX complexes in the absence or presence of the indicated DCNL PONY domain. For comparison, all reactions were carried out under the same conditions.
(B) Extended time courses from (A) for pulse-chase [32P]~NEDD8 transfer from UBE2FNAc to either CUL3CTD-RBX1 or CUL4ACTD-RBX1 in the absence or presence of the indicated DCNL PONY domain.
(C) Pulse-chase reactions with the indicated concentrations of various components, monitoring [32P]~NEDD8 transfer from UBE2FNAc to CUL5CTD-RBX2 in the absence or presence of the PONY domain from DCNL1 or DCNL3.
However, under the conditions of our assays, the different DCNL/cullin/NEDD8 E2 combinations displayed great variation in overall neddylation efficiencies, which generally correlate with a combination of DCNL-independent activity and the DCNLP-cullin and DCNLP-E2 interaction affinities (Fig. 3). For example, in accordance with DCNLP-E2 interaction affinities, DCNL1P and DCNL2P generally most potently stimulate NEDD8 transfer from UBC12NAc, DCNL3P generally most potently stimulates NEDD8 transfer from UBE2FNAc, and DCNL4P and DCNL5P are generally less active toward both E2s (Fig. 2, 3). Similarly, the very high affinity interactions for all 5 DCNLs with the CUL5WHB, taken together with relatively high basal DCNL-independent neddylation activity, explain the high level of UBE2FNAc-mediated NEDD8 modification of CUL5CTD-RBX2 in the presence of all of the DCNL PONY domains. It is only at very low protein concentrations that the neddylation reaction is slowed to the point where DCNL3P promotes slightly more NEDD8 ligation to CUL5CTD from UBE2FNAc (Fig. 3C). At the opposite end of the spectrum, CUL3CTD-RBX1 and CUL4ACTD-RBX1 are very efficiently neddylated by UBC12NAc in the presence of DCNL1P and DCNL2P, and the reactions are generally inefficient with UBE2FNAc (Fig. 3A, B). For CUL3, whose WHB binds with similar affinities to all the DCNL PONY domains, neddylation efficiency is largely determined by DCNLP-E2 interaction strength, with PONY domains from DCNL1, 2, and 3 all activating UBE2FNAc, with DCNL3P displaying the greatest effect on UBE2FNAc-mediated CUL3CTD neddylation (Fig. 3B). The DCNL3P-UBE2FNAc specificity is particularly evident for CUL4A, likely due in part to enhanced affinity of CUL4A’s WHB for DCNL3P.
Peptide inhibition of DCNL-dependent UBE2FNAc-mediated NEDD8 ligation
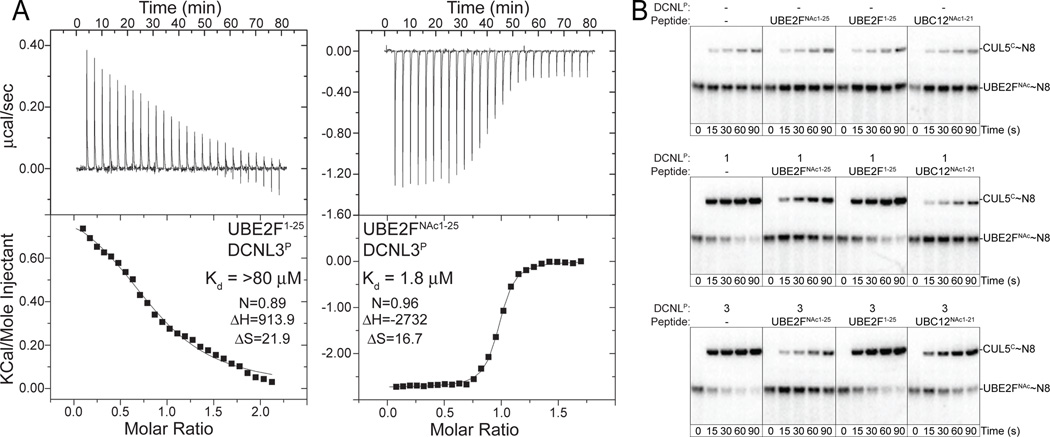
We next probed whether the specificity of DCNL-NEDD8 E2 interactions could be recapitulated with peptides. As with DCNL1P binding to an N-terminally acetylated peptide from UBC12 (Fig. 2C), DCNL3P binds an N-terminally acetylated peptide from UBE2F, and N-terminal acetylation strongly increases UBE2F peptide binding to DCNL3P (Fig. 4A). To test whether a peptide could compete with a full-length NEDD8 E2, we examined the effects of adding peptides to NEDD8 ligation assays. CUL5CTD in complex with its natural partner, RBX2, was used as the target for these experiments because UBE2F has been shown to mediate NEDD8 ligation to CUL5 in cells (Huang et al., 2009), and because multiple DCNL proteins stimulate this reaction in vitro (Fig. 3A, C). Peptides corresponding to both UBC12NAc and UBE2FNAc inhibited DCNL1P-dependent activity (Fig. 4B). The peptide derived from UBC12NAc inhibited this reaction more potently, reducing the level of neddylation to that observed in the absence of DCNL stimulation. In agreement with the observed interaction preferences measured by ITC, opposite specificity was observed for DCNL3P, for which the peptide derived from UBE2FNAc was a more effective inhibitor (Fig. 2C, 4A, 4B). N-terminal acetylation was absolutely required for the UBE2F peptide mediated inhibition. Furthermore, the peptide inhibition is specific for DCNL stimulated NEDD8 transfer, as none of the peptides influenced the basal level of neddylation in the absence of a DCNL PONY domain. Thus, the DCNL-E2 preferences observed by binding and in vitro neddylation reactions were recapitulated by short peptide inhibitors (Fig. 4B).
Figure 4. Peptide inhibition of DCNL activation of NEDD8 ligation depends on N-terminal acetylation.
(A) ITC data for interactions between DCNL3P and unacetylated or N-terminally acetylated peptides from UBE2F. Upper panels show raw power data recorded during titration experiments, and lower panels show fits of standard binding equations after integration of the raw data, using Origin (v. 7.0) software provided from MicroCal.
(B) Pulse-chase monitoring [32P]~NEDD8 transfer from UBE2FNAc to CUL5CTD-RBX2 in the absence or presence of the PONY domain from DCNL1 or DCNL3, and unacetylated or N-terminally acetylated peptides corresponding to UBC12 or UBE2F.
Crystal structures of DCNL2P-UBC12NAc peptide and DCNL3P-UBE2FNAc peptide complexes: a common mode of DCNL PONY domain interactions with N-terminally acetylated NEDD8 E2s
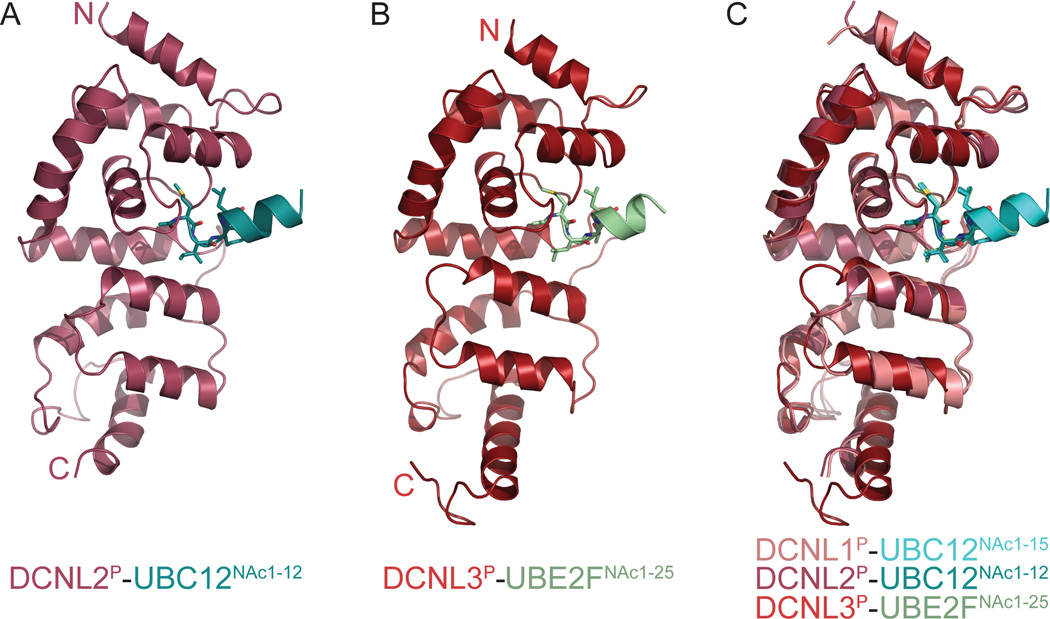
To better understand how the different DCNL co-E3s interact with the different NEDD8 E2s, we undertook a broad campaign to obtain crystal structures of complexes with distinctive partners. We were able to obtain crystals of a complex between DCNL2P and a 12-residue peptide from UBC12NAc (referred to as UBC12NAc1–12) that diffracted to 3.3 Å resolution (Fig. 5A, Table 1, Fig. S3), and of a complex between DCNL3P and a 25-residue peptide from UBE2FNAc (referred to as UBE2FNAc1–25) that diffracted to 2.4 Å resolution (Fig. 5B, Table 1, Fig. S3), and determined their structures using molecular replacement (see Experimental Procedures). Upon comparison with a prior DCNL1P-UBC12NAc1–15 complex (Scott et al., 2011), the structures reveal a common mode of NEDD8 E2 interactions with DCNL PONY domains, with average RMSDs of 0.6 Å between DCNL1P-UBC12NAc1–15 and DCNL2P-UBC12NAc1–12, and of 1.4 Å between DCNL1P- UBC12NAc1–15 and DCNL3P-UBE2FNAc1–25 (Fig. 5C, Table S1).
Figure 5. Overall conserved mode of DCNL PONY domain interactions with acetylated N-terminal helices from NEDD8 E2s.
(A) Cartoon representation of overall structure of DCNL2 PONY domain (raspberry) complex with peptide from UBC12NAc (teal) with N-acetyl-Met1, Ile2, and Leu4 shown in sticks.
(B) Cartoon representation of overall structure of DCNL3 PONY domain (brick) complex with peptide from UBE2FNAc (lime) with N-acetyl-Met1, Leu2, and Leu4 shown in sticks.
(C) Superposition of DCNL2P-UBC12NAc1–12 and DCNL3P-UBE2FNAc1–25 crystal structures with prior structure of DCNL1P (pink)-UBC12NAc1–15 (cyan) (3TDU.pdb) (Scott et al., 2011).
Table 1.
Data collection and refinement statistics
| DCNL2P : NAcUBC121–12 | DCNL3P : NAcUBE2F1–25 | |
|---|---|---|
| ACCESSION CODES | 4GAO.pdb | 4GBA.pdb |
| DATA COLLECTION | ||
| Beamline | APS 24-ID-E | ALS 8.2.2 |
| λ | 0.97915 | 0.97910 |
| Space group | P21 | P21 |
| Complexes in a.u. | 4 | 2 |
| Cell dimensions | ||
| a, b, c (Å) | 48.57, 190.15, 49.07 | 83.47, 44.58, 101.22 |
| α, β, γ (°) | 90.00, 101.75, 90.00 | 90.00, 103.30, 90.00 |
| Resolution (Å) | 3.3 | 2.4 |
| Rmerge (%) | 18.2 (44.9) | 11.0 (47.4) |
| I/σI | 6.2 (1.7) | 15.4 (1.9) |
| Completeness (%) | 98.9 (93.2) | 99.7 (97.9) |
| Redundancy | 3.2 (2.5) | 1.9 (1.8) |
| REFINEMENT | ||
| Resolution (Å) | 3.3 | 2.4 |
|
Reflections Work set/Test set |
11723/643 | 27346/1466 |
| Rwork/Rfree | 25.2/29.6 | 19.1/23.0 |
| No. atoms | ||
| Protein/Ligand | 6027/270 | 3277/159 |
| Solvent | 135 | |
| Bromides | 2 | |
| B-factors (Å2) | ||
| Protein+peptide | 51.3 | 43.9 |
| Solvent | 43.7 | |
| Bromides | 49.7 | |
| Wilson B (Å2) | 55.3 | 36.9 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.009 | 0.008 |
| Bond angles (°) | 0.951 | 1.032 |
| RAMACHANDRAN (%) |
||
| Preferred regions | 97.7 | 96.6 |
| Allowed regions | 2.3 | 3.4 |
| Disallowed regions | 0.0 | 0.0 |
Data for highest resolution shell is shown in parentheses. Rwork = ∑|FoFc|/∑FoRfree is the cross-validation of R-factor, with 5–10% of the total reflections omitted in model refinement.
As observed for previous PONY domain structures (Kurz et al., 2008; Scott et al., 2011; Scott et al., 2010; Sethe Burgie et al., 2011; Yang et al., 2007), DCNL2P and DCNL3P consist entirely of helices (Fig. 5). The N-terminally acetylated peptides from both NEDD8 E2s also form helical structures. The interactions are anchored by the N-acetyl-methionine from a NEDD8 E2 docking in a deep hydrophobic pocket at the center of the DCNL PONY fold.
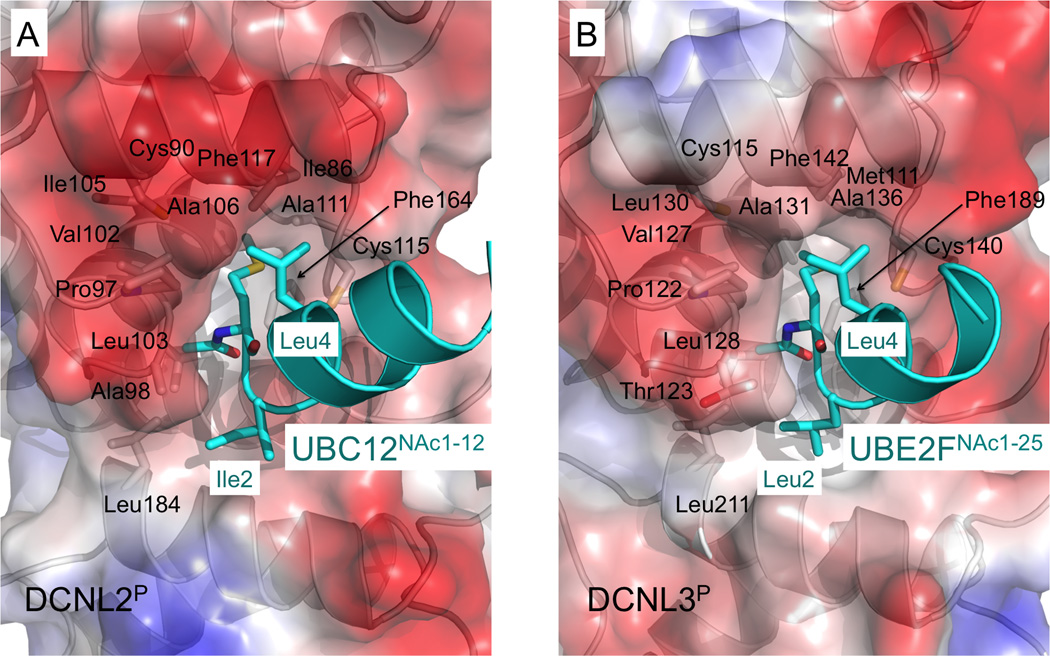
The structures explain the general requirement of NEDD8 E2 N-terminal acetylation in binding to DCNL PONY domains, as an N-terminal positive charge would block burial of N-acetyl-methionine into the DCNL hydrophobic pocket (Fig. 6). In the DCNL2P-UBC12NAc1–12 and DCNL3P-UBE2FNAc1–25 structures, these interactions involve the methyl moiety of the acetyl group from the E2 packing in a hydrophobic pocket consisting of the a-carbon from DCNL2P Ala98/DCNL3P Thr123 and sidechains from DCNL2P Val102, Leu103, and Leu184 and DCNL3P Val127, Leu128, and Leu211, respectively. The E2’s amide makes a hydrogen bond to the backbone carbonyl of a Pro conserved in both DCNL PONY domains (97 in DCNL2P and 122 in DCNL3P). Also, the Met sidechain from UBC12NAc1–12 extends into a deep hydrophobic channel formed by sidechains from DCNL2P’s Ile86, Cys90, Pro97, Val102, Ile105, Ala106, Ala111, Cys115, Phe117, and Phe164. The Met sidechain from UBE2FNAc1–25 is buried in the corresponding hydrophobic groove formed by DCNL3P’s Met111, Cys115, Pro122, Val127, Leu130, Ala131, Ala136, Cys140, Phe142, and Phe189. The hydrophobic NEDD8 E2 residues at positions 2 (Ile in UBC12 and Leu in UBE2F) and 4 (Leu in both UBC12 and UBE2F) further pack against their respective DCNL partners to seal the N-acetyl-Met1 in place. The DCNL2P and DCNL3P residues interacting with N-acetyl-Met correspond to those observed previously from DCNL1P (Scott et al., 2011).
Figure 6. A DCNL hydrophobic pocket surrounds N-acetyl-methionine from a NEDD8 E2.
(A) Close-up view of DCNL2P-UBC12NAc1–12 structure, with DCNL2P surface colored by electrostatic potential, and UBC12NAc1–12 shown in cyan with N-acetyl-Met1, Ile2, and Leu4 shown in sticks.
(B) Close-up view of DCNL3P- UBE2FNAc1–25 structure, with DCNL3P surface colored by electrostatic potential, and UBE2FNAc1-25 shown in cyan with N-acetyl-Met1, Leu2, and Leu4 shown in sticks.
Identification of a DCNL residue influencing selective NEDD8 E2 interactions
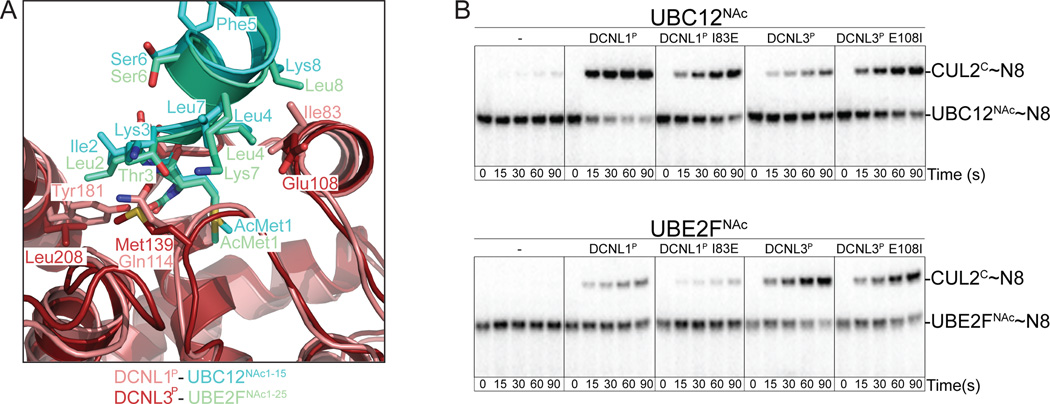
We inspected the crystal structures for possible clues as to the basis for the observed preferences for the different DCNL family members toward UBC12NAc and UBE2FNAc. In addition to the conserved interactions, some DCNL residues are poised to make limited contacts to residues downstream of the N-terminus from the helix of the interacting NEDD8 E2 (Fig. 7A). Of note is a residue that is hydrophobic in DCNL1P (Ile83) and DCNL2P (Val83), but acidic in DCNL3P (Glu108). This is poised to dock in a hydrophobic surface in UBC12NAc, between the sidechains of Leu4, Leu7, and the aliphatic portion of the sidechain from Lys8. For DCNL3P, the aliphatic portion of Glu108 contacts a hydrophobic surface from a slightly different arrangement of sidechains on UBE2FNAc, from Leu4, the aliphatic portion of Lys7, and Leu8. To test for a role in specificity, we swapped the Ile and Glu from DCNL1P and DCNL3P, respectively, and assayed activation of NEDD8 transfer from UBC12NAc and UBE2FNAc to the CUL2CTD-RBX1 complex (Fig. 7B). CUL2 was chosen as a representative target for these experiments because both DCNL1P and DCNL3P potently stimulate NEDD8 ligation from both E2s to CUL2 (Fig. 3A). For both NEDD8 E2s, a Glu substitution in place of DCNL1P’s Ile83 reduces activity. Thus, the Glu alone is not a positive determinant of specificity for UBE2FNAc. However, a DCNL3P mutant with Glu108 replaced by Ile is more active toward UBC12NAc, and shows similar activity as wild-type DCNL3P toward UBE2FNAc. Although we do not know the extent to which UBC12NAc is repelled by Glu108 or attracted by the Ile substitution in DCNL3P, the data imply that DCNL PONY domain residues contacting portions of NEDD8 E2s downstream of the N-acetyl-Met can influence specificity.
Figure 7. Amino acid identity downstream of N-acetyl-methionine influences E2 specificity of DCNL-stimulated NEDD8 ligation.
(A) Close-up views of superimposed DCNL1P-UBC12NAc1–15 (Scott et al., 2011) and DCNL3P-UBE2FNAc1–25 crystal structures highlighting interactions with residues downstream of the N-terminus.
(B) Pulse-chase assays monitoring [32P]~NEDD8 transfer from UBC12NAc (top panel) or UBE2FNAc (lower panel) to CUL2CTD-RBX1 in the absence or presence of the wild-type and indicated mutant versions of the PONY domains from DCNL1 and DCNL3.
Discusssion
Multiple DCNL proteins have been implicated in augmenting NEDD8 ligation to multiple cullins in vivo (Huang et al., 2011; Kim et al., 2008; Meyer-Schaller et al., 2009; Wu et al., 2011). However, the relative biochemical potential and thermodynamic preferences for different DCNL PONY domain, cullin WHB subdomain, and NEDD8 E2 interactions have remained largely uncharacterized. Here we found that all PONY domains can stimulate cullin neddylation from both E2s, with a wide-range of NEDD8 ligation efficiencies for different enzyme combinations (Fig. 3). In vitro NEDD8 ligation efficiency appears to be related to a combination of the innately varying abilities of UBC12 and UBE2F to mediate NEDD8 ligation to different RBX-CUL complexes (Huang et al., 2009), and the wide range of DCNL affinities for the different CUL WHB subdomains and NEDD8 E2s (Fig. 2, 3). Our data indicate that intrinsic RBX-E2 interaction specificity would dominate pathway establishment in vivo, because none of the DCNL PONY domains can overcome the exclusion of UBC12-mediated NEDD8 ligation to CUL5CTD associated with RBX2 (Fig. 3). In vivo, overexpression of UBE2F can compensate for knockdown of UBC12 and promote NEDD8 ligation to CUL1 or CUL2, presumably in complex with RBX1. However, UBE2F knockdown only leads to decreased levels of neddylated CUL5, suggesting that endogenous UBE2F function is restricted to RBX2 (Huang et al., 2009). This specificity is likely a result of the relatively high-level expression of UBC12, which would occupy RBX1, and low-level expression of UBE2F combined with availability of RBX2 (Huang et al., 2009). Thus, RBX2’s tolerance for UBE2F establishes NEDD8 E2 specificity for its only known endogenous cullin partner, CUL5 in vivo (Huang et al., 2009; Kamura et al., 2004). CUL5’s WHB subdomain binds all DCNL PONY domains with high affinity (Fig. 2), suggesting that multiple DCNL proteins could contribute to CUL5 neddylation in cells. Although only the PONY domains from DCNLs 1–3 show appreciable interaction with UBE2FNAc, in the context of the multiple interaction surfaces for DCNL/NEDD8 E2/RBX/CUL complexes, the effective concentrations of any individual component will be much higher, which might overcome the low intrinsic affinities between DCNL4 or DCNL5 and both NEDD8 E2s (Fig. 2, 3).
In a related vein, at least some fraction of DCNL-CUL interactions are also not redundant, as lysates from HeLa cells after siRNA-mediated knockdown of DCNL1 or DCNL3, or from null MEFs or testes of DCNL1 knockout mice, showed reduced levels of the NEDD8-ligated species for multiple cullins (Huang et al., 2011; Kim et al., 2008; Meyer-Schaller et al., 2009). Subcellular localization of neddylation enzymes can also be regulated. For example, UBC12 is largely nuclear, and nuclear localization of DCNL1 is important for its role as a co-NEDD8 E3 (Huang et al., 2011; Wu et al., 2011). Besides the conserved PONY domain, the DCNLs display a range of N-terminal sequences, which can also influence localization (Meyer-Schaller et al., 2009). Membrane localized DCNL3 apparently plays an important role in NEDD8 ligation to CUL3 (Meyer-Schaller et al., 2009). In addition to subcellular partitioning of the different components of NEDD8 ligation pathways, it is also possible that the distinct DCNL N-terminal regions could impart other forms of regulation. Furthermore, different expression levels may also influence whether the different DCNLs function as co-E3s for NEDD8, as our data show that even in the absence of any biological regulation, differences in interaction affinities impact DCNL/NEDD8 E2/CUL partnering at various protein concentrations (Fig. 2, 3). Differences in innate, DCNL-independent neddylation efficiencies for CUL-RBX complexes may also influence sensitivity to DCNL co-E3 activity. In this regard, we note that the CUL3CTD-RBX1 complex shows the lowest level of DCNL-independent neddylation under a range of experimental conditions, with both acetylated and unacetylated UBC12 and UBE2F ((Huang et al., 2009) and Fig. 3). Interestingly, a single point mutation in the DCNL PONY binding region of the CUL3 WHB almost entirely eliminated neddylation of HA-tagged CUL3 expressed in HeLa cells (Meyer-Schaller et al., 2009). Given our data that all DCNL PONY domains can in principle interact with the WHB subdomains from cullins 1–5 (Fig. 2), future studies will be required to deconvolute DCNL-CUL specificities in vivo, including spatio-temporal regulation.
Our data revealed that N-terminal acetylation not only of UBC12, but also of UBE2F is required for DCNL activation (Figs. 1, 3). With two new structures described herein, crystallographic analyses now show the detailed molecular interactions for the three highest affinity DCNL-NEDD8 E2 complexes: DCNL1P-UBC12NAc, DCNL2P-UBC12NAc, and DCNL3P-UBE2FNAc. The structures reveal common mechanisms by which DCNL-NEDD8 E2 complexes depend on acetylation: the acetyl group both neutralizes a positive charge at a NEDD8 E2’s N-terminus, which would obstruct the interaction, and also makes positive interactions upon burial along with the rest of the N-acetyl-Met in a deep pocket within a DCNL PONY domain. Thus, the PONY domain joins a short list of known acetylation-specific interaction modules. Comparing N-acetyl-methionine recognition by PONY domains of the DCNL family to acetyl-lysine recognition by modules such as bromodomains reveals a common feature: N-acetyl-methionine and acetyl-lysine are both inserted into a hydrophobic pocket, in which it would be unfavorable to bury the positive charge masked by acetylation (Fig. S4) (Dhalluin et al., 1999; Owen et al., 2000; Sanchez and Zhou, 2009). Notably, recent development of selective bromodomain inhibitors that occupy the hydrophobic pocket and prevent acetyl-lysine binding (Filippakopoulos et al., 2010; Nicodeme et al., 2010) raises the possibility that N-acetylmethionine binding sites may also be targeted by small molecules. Analysis of the surface of the DCNL1P (Scott et al., 2011), DCNL2P, and DCNL3P PONY domains with the program CASTp (Dundas et al., 2006) indicates surface exposed pockets of 347, 355, and 379 Å3, respectively, when averaged over all copies per asymmetric unit, which would seem plausible for chemical targeting. This is of particular importance for DCNL1, as the gene encoding human DCNL1, which is also called “SCCRO” for Squamous Cell Carcinoma-Related Oncogene, is amplified in the 3q26.3 region in squamous cell carcinoma (SCC). Notably, DCNL1 amplification and overexpression in SCC of mucosal origin is associated with adverse clinical outcome (Estilo et al., 2003; Sarkaria et al., 2006). NIH 3T3 cell lines overexpressing DCNL1 display many attributes associated with transformation, including colony formation on soft agar and oncogenicity in a xenograft assay in nude mice (Sarkaria et al., 2006). Thus, the N-acetyl-methionine binding site in DCNL1 may be an attractive candidate for development of small molecule therapeutics.
This raises the question of whether there is any specificity among perhaps numerous protein-protein interactions dependent on N-acetyl-Met. In some cases, the acetyl group has been proposed to provide a hydrogen bond to establish secondary structure required for protein-protein interactions, without directly mediating contacts to partner proteins (Frye et al., 2010; Greenfield et al.,1994). However, we anticipate that there will be many cases like DCNL-NEDD8 E2 complexes, in which the interaction is driven by direct contacts with the N-acetyl-Met. In this regard, it is informative to compare the highly homologous DCNL1-UBC12NAc, DCNL2-UBC12NAc, and DCNL3-UBE2FNAc complexes. The structures not only reveal subtle differences in the N-acetyl-Met binding pockets, but also differences in the constellation of amino acids contacting proximal residues. This is particularly evident when comparing interactions with the PONY domains from either DCNL1 or DCNL2, which are ~82% identical in sequence, to the more divergent but still more than 40% identical PONY domain of DCNL3 (Table S1, Fig. 6). Encouragingly, we find that short N-terminal peptides from UBC12NAc and UBE2FNAc display distinct preferences for inhibiting DCNL1P- or DCNL3P-activated NEDD8 ligation, respectively (Fig. 4B), and that a single point mutation can significantly enhance the activity of DCNL3P’s PONY domain toward UBC12NAc to eliminate this specificity under our assay conditions (Fig. 7B). Thus, our results suggest the potential for selective manipulation of N-acetyl-methionine-dependent protein-protein interactions.
Experimental Procedures
Mass spectrometry
C-terminally tagged UBE2F was purified from NIH 3T3 and SF9 cells. Peptides generated from ArgC digestion of sample proteins were desalted offline using C18 stage tips. Peptides were eluted from the stage tip, dried down using a speed vac, and resuspended in 10µl 5% formic acid, 5% acetonitrile. Peptide mixtures were separated by in line reverse phase using an 18 cm × 150 µm (ID) column packed with C18 (MAGIC C18 5 µm particle, 200 angstrom pore size) using a 50-minute 8%-26% acetonitrile gradient. MS/MS data was generated using an LTQ-Velos mass spectrometer (Thermo), a data-dependent top10 instrument method, and zoom scan for MS1 assignments. Data was acquired using CID with the normalized collision energy set to 35% with activation times of 10ms. MS/MS triggering thresholds were set to 2000 and a 30s dynamic exclusion was used with an exclusion list size of 500. Resultant MS/MS spectra were searched using Sequest against a concatenated forward and reverse human I PI database (v3.6). Methionine oxidation (+15.99), acetylation (+42.01), and the combined modification (+58.09) were set as dynamic modifications.
Intact protein mass spectra were obtained with RPLC-desalted samples introduced by flow injection, using an ion trap mass spectrometer (Bruker).
Isothermal Titration Calorimetry (ITC)
Measurements were performed using a MicroCal ITC200. For DCNLP:CULWHB experiments, protein samples were buffer matched by desalting over a NAP-5 column (GE Healthcare) into 25 mM Tris, 100 mM NaCl, 1 mM BME, pH 7.6. DCNLP samples were placed in the sample cell at a final concentration of 100 µM at 22 °C. The ligand, CULWHB (1 mM) was constantly injected (2.5 µL). The interval time between each injection was 3 minutes and the duration of each injection was 5 seconds. For DCNLP: UBC12NAc1-21 experiments, UBC12NAc1–21 was dissolved by weight to a final concentration of 10 mM, and DCNLP samples buffer matched in 50 mM HEPES, 125 mM NaCl, 1 mM BME, pH 7.0. DCNLP was placed into the sample cell at a final concentration of 400 µM at 12 °C. The ligand, UBC12NAc1–21 (4 mM) was constantly injected (1.5 µl). The interval time between each injection was 3 minutes and the duration of each injection was 3 seconds. For experiments with UBE2FNAc, protein samples were buffer matched by desalting over a NAP-5 column into 25 mM HEPES, 125 mM NaCl, 1 mM BME, pH 7.0. UBE2FNAc was placed into the sample cell at a final concentration of 100 µM at 16 °C. The ligand, DCNLP (1 mM) was constantly injected (2.5 µl). The interval time between each injection was 3 minutes and the duration of each injection was 5 seconds. Obtained spectra were evaluated using Origin (V 7.0) to determine heats of binding and Kd values. All ITC experiments were performed independently at least two times, with similar results. Values from one experiment are presented.
Crystallography
Crystals were grown by the hanging-drop vapor-diffusion method. Crystals of DCNL2P-UBC12NAc1–12 were grown at 4 °C in 20% PEG2000 MME, 0.1 M NaBr, 3% sorbitol in a 1:1 drop of protein:mother liquor. The crystals were harvested from mother liquor supplemented with 30% of a 50:50 mixture of glycerol:ethylene glycol prior to flash-freezing in liquid nitrogen. Reflection data were collected at NECAT ID-24-E at the Advanced Photon Source. The DCNL2P-UBC12NAc1–12 crystals diffracted weakly and the diffraction spots were not spherical in shape, resulting in a relatively high Rmerge. The data are of similar quality for all resolution shells. Processing the data using different frame combinations, different spot and background sizes, and different programs all gave similar results. The crystals belong to space group P21 with four DCNL2P-UBC12NAc1–12 complexes in the asymmetric unit. Crystals of DCNL3P-UBE2FNAc1–25 were grown at 4 °C in 2.28 M sodium malonate, pH 7.0 in a 2:1 drop of protein:mother liquor. The crystals grew as multiple clusters. Single crystals of the complex were obtained by streak-seeding into 2.05 M sodium malonate, pH 7.0. The crystals were harvested from mother liquor supplemented with 30% glycerol prior to flash-freezing in liquid nitrogen. Reflection data were collected at beamline 8.2.2 at the Advanced Light Source. The DNCL3P-UBE2FNAc1-25 data included numerous ice rings, which presumably contribute to the high Rmerge value. The crystals belong to space group P21 with two DNCL3P-UBE2FNAc1–25 complexes in the asymmetric unit.
Data were processed with HKL2000 (Otwinowski and Minor, 1997). Phases for both structures were obtained by molecular replacement (MR) using PHASER (McCoy et al., 2007) using the following search models: for the DCNL2P-UBC12NAc1–12 structure, four copies of chains A and F from 3TDU.pdb (Scott et al., 2011); for the DCNL3P-UBE2FNAc1-25 structure, two copies of residues 93–269 from a DCNL3P model generated by the Modweb server (Eswar et al., 2006; Sali and Blundell, 1993). For the DCNL2P-UBC12NAc1–12 structure, the peptide was included in the MR. Thus, simulated annealing (SA) composite omit maps were used throughout model building stages to reduce model bias during subsequent fitting. The final model contains only peptide residues and/or their associated sidechains present in the SA Fo-Fc omit maps at 2.5a. For DCNL3P–UBE2FNAc1-25, the peptide was built manually, and general manual rebuilding was performed with COOT (Emsley and Cowtan, 2004). Refinement was performed using Phenix (Adams et al., 2010) and Refmac (Murshudov et al., 1997). For the DCNL2P–UBC12NAc1–12 complex, medium 2-fold NCS restraints were used: DCNL2P protomers A with D, and B with G. Peptides were not included in NCS restraints. Details of refinement are provided in Table 1.
Biochemical assays
DCNLP-mediated co-E3 activity was monitored using pulse-chase assays. For the “pulse”, 10 µM of the indicated versions of UBC12 or UBE2F were charged with [32P]-NEDD8, for 15 minutes at room temperature using 0.1 µM E1, 15 µM [32P]-NEDD8, in 50 mM HEPES, 100 mM NaCl, 1.5 mM ATP, 2.5 mM MgCl2, pH 7.5. Formation of an E2~[32P]-NEDD8 intermediate was quenched with 50 mM EDTA on ice for 5 minutes. Other than in Fig. 3C where additional details are provided, chase reactions involved dilution of the E2~[32P]-NEDD8 thioester conjugate to 40 nM in 100 mM Tris, 100 mM NaCl, 50 mM EDTA, 0.5 mg/ml BSA, pH 6.8. Chase reactions were initiated at 0°C by the addition of 125 nM CULCTD-RBX with or without 500 nM DCNLP. Aliquots were removed at the indicated times and quenched with 2X SDS-PAGE sample buffer. Reaction products were heated at 70°C for 1 minute and separated on 4–12% NuPAGE gels (Invitrogen). Dried gels were exposed to a Storm (GE) Phosphoimager screen.
For peptide inhibition experiments, pulse-chase assays were performed as described above in the absence or presence of 300 µM unacetylated or acetylated UBC12 or UBE2F peptides.
Supplementary Material
Highlights.
-
-
Systematic analyses of DCNL-cullin and DCNL-E2 interactions in NEDD8 ligation
-
-
N-terminal acetylation is required for both human NEDD8 E2s to interact with DCNLs
-
-
Structures show a general mechanism for DCNL binding to N-terminally acetylated E2s
-
-
Selectivity among similar N-acetyl-Met-dependent protein interactions is observed
Acknowledgments
This work was supported by ALSAC, the St. Jude Cancer Center Core grant, NIH R01GM069530 to BAS, Damon Runyon postdoctoral fellowship DRG 2061-10 to JL, R01GM070565 to JWH, and the Howard Hughes Medical Institute. BAS is an Investigator of the Howard Hughes Medical Institute. EJB is funded by New Scholar awards from the Sidney Kimmel Foundation for Cancer Research and the Ellison Medical Foundation. We thank L Borg, D Duda, S Gygi, R Cassell, P Rodrigues, K Kodali, V Pagala, J Peng, I Kurinov, and C Ralston for experimental assistance, reagents, and/or discussions, and DW Miller, S Bozeman, and J Bollinger for administrative/computational support. NECAT is supported by NIH NCRR RR-15301, APS by US DOE W-31-109-ENG-38, and ALS by US DOE DE-AC02-05CH11231.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession codes
Coordinates and structure factor data have been deposited in the RCSB with accession codes 4GAO and 4GBA, for immediate release upon publication.
Author contributions
JKM, DCS, EJB, JL, DK and DJM designed, performed, and/or analyzed the experiments; JKM, DCS and BAS wrote the manuscript with contributions from all authors; JWH and BAS provided guidance, advice, and assistance on all aspects of the work.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesen T. Towards a functional understanding of protein N-terminal acetylation. PLoS Biol. 2011;9:e1001074. doi: 10.1371/journal.pbio.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R, Panic B, Whyte JR, Munro S. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat Cell Biol. 2004;6:405–413. doi: 10.1038/ncb1120. [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulton AT, East DA, Galinska-Rakoczy A, Lehman W, Mulvihill DP. The recruitment of acetylated and unacetylated tropomyosin to distinct actin polymers permits the discrete regulation of specific myosins in fission yeast. Journal of cell science. 2010;123:3235–3243. doi: 10.1242/jcs.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic acids research. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Estilo CL, P OC, Ngai I, Patel SG, Reddy PG, Dao S, Shaha AR, Kraus DH, Boyle JO, Wong RJ, et al. The role of novel oncogenes squamous cell carcinoma-related oncogene and phosphatidylinositol 3-kinase p110alpha in squamous cell carcinoma of the oral tongue. Clin Cancer Res. 2003;9:2300–2306. [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr. Protoc. Protein Sci. Chapter 2. 2006 doi: 10.1002/0471140864.ps0209s50. Unit 2.9. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye J, Klenchin VA, Rayment I. Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diveristy of N-terminal recognition. Biochemistry. 2010;49:4908–4920. doi: 10.1021/bi100349a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield NJ, Stafford WF, Hitchcock-DeGregori SE. The effect of N-terminal acetylation on the structure of an N-terminal tropomyosin peptide and alpha alpha-tropomyosin. Protein Sci. 1994;3:402–410. doi: 10.1002/pro.5560030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Kaufman AJ, Ramanathan Y, Singh B. SCCRO (DCUN1D1) promotes nuclear translocation and assembly of the neddylation E3 complex. J Biol Chem. 2011;286:10297–10304. doi: 10.1074/jbc.M110.203729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvik TV, Arnesen T. Protein N-terminal acetyltransferases in cancer. Oncogene. 2012 doi: 10.1038/onc.2012.82. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AY, Bommelje CC, Lee BE, Yonekawa Y, Choi L, Morris LG, Huang G, Kaufman A, Ryan RJ, Hao B, et al. SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation. J Biol Chem. 2008;283:33211–33220. doi: 10.1074/jbc.M804440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Kurz T, Ozlu N, Rudolf F, O'Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, Peter M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435:1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Schaller N, Chou YC, Sumara I, Martin DD, Kurz T, Katheder N, Hofmann K, Berthiaume LG, Sicheri F, Peter M. The human Dcn1-like protein DCNL3 promotes Cul3 neddylation at membranes. Proc Natl Acad Sci U S A. 2009;106:12365–12370. doi: 10.1073/pnas.0812528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Molecular cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr D Biol Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology, Macromolecular Crystallography, part A. 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B, Cardillo TS, Doyle TC, Bedi GS, Sherman F. Nat3p and Mdm20p are required for function of yeast NatB Nalpha-terminal acetyltransferase and of actin and tropomyosin. J Biol Chem. 2003;278:30686–30697. doi: 10.1074/jbc.M304690200. [DOI] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of molecular biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel. 2009;12:659–665. [PMC free article] [PubMed] [Google Scholar]
- Sarkaria I, P Oc, Talbot SG, Reddy PG, Ngai I, Maghami E, Patel KN, Lee B, Yonekawa Y, Dudas M, et al. Squamous cell carcinoma related oncogene/DCUN1D1 is highly conserved and activated by amplification in squamous cell carcinomas. Cancer Res. 2006;66:9437–9444. doi: 10.1158/0008-5472.CAN-06-2074. [DOI] [PubMed] [Google Scholar]
- Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Monda JK, Grace CR, Duda DM, Kriwacki RW, Kurz T, Schulman BA. A dual E3 mechanism for Rub1 ligation to Cdc53. Mol Cell. 2010;39:784–796. doi: 10.1016/j.molcel.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethe Burgie E, Bingman CA, Makino S, Wesenberg GE, Pan X, Fox BG, Phillips GN., Jr Structural architecture of Galdieria sulphuraria DCN1L. Proteins. 2011;79:1329–1336. doi: 10.1002/prot.22937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, Strochlic TI, Tong AH, Boone C, Burd CG. Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat Cell Biol. 2004;6:414–419. doi: 10.1038/ncb1121. [DOI] [PubMed] [Google Scholar]
- Singer JM, Shaw JM. Mdm20 protein functions with Nat3 protein to acetylate Tpm1 protein and regulate tropomyosin-actin interactions in budding yeast. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7644–7649. doi: 10.1073/pnas.1232343100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Starheim KK, Gevaert K, Arnesen T. Protein N-terminal acetyltransferases: when the start matters. Trends in biochemical sciences. 2012;37:152–161. doi: 10.1016/j.tibs.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- Wu K, Yan H, Fang L, Wang X, Pfleger C, Jiang X, Huang L, Pan ZQ. Mono-ubiquitination drives nuclear export of the human DCN1-like protein hDCNL1. The Journal of biological chemistry. 2011;286:34060–34070. doi: 10.1074/jbc.M111.273045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc Natl Acad Sci U S A. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhou J, Sun L, Wei Z, Gao J, Gong W, Xu RM, Rao Z, Liu Y. Structural basis for the function of DCN-1 in protein Neddylation. J Biol Chem. 2007;282:24490–24494. doi: 10.1074/jbc.C700038200. [DOI] [PubMed] [Google Scholar]
- Yi CH, Pan H, Seebacher J, Jang IH, Hyberts SG, Heffron GJ, Vander Heiden MG, Yang R, Li F, Locasale JW, et al. Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell. 2011;146:607–620. doi: 10.1016/j.cell.2011.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kulkarni K, Hanrahan SJ, Thompson AJ, Barford D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. The EMBO journal. 2010;29:3733–3744. doi: 10.1038/emboj.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.