SUMMARY
Epigenetic mechanisms have been proposed to play crucial roles in mammalian development, but their precise functions are only partially understood. To investigate epigenetic regulation of embryonic development, we differentiated human embryonic stem cells into mesendoderm, neural progenitor cells, trophoblast-like cells, and mesenchymal stem cells, and systematically characterized DNA methylation, chromatin modifications, and the transcriptome in each lineage. We found that promoters that are active in early developmental stages tend to be CG rich and mainly engage H3K27me3 upon silencing in non-expressing lineages. By contrast, promoters for genes expressed preferentially at later stages are often CG poor and primarily employ DNA methylation upon repression. Interestingly, the early developmental regulatory genes are often located in large genomic domains that are generally devoid of DNA methylation in most lineages, which we termed DNA methylation valleys (DMVs). Our results suggest that distinct epigenetic mechanisms regulate early and late stages of ES cell differentiation.
Introduction
Embryonic development is a complex process that remains to be understood despite knowledge of the complete genome sequences of many species and rapid advances in genomic technologies. A fundamental question is how the unique gene expression pattern in each cell type is established and maintained during embryogenesis. It is well accepted that the gene expression program encoded in the genome is executed by transcription factors that bind to cis-regulatory sequences and modulate gene expression in response to environmental cues (Young, 2011). Growing evidence now shows that maintenance of such cellular memory depends on epigenetic marks such as DNA methylation and chromatin modifications (Bird, 2002; Kouzarides, 2007).
DNA methylation at promoters has been shown to silence gene expression and thus has been proposed to be necessary for lineage-specific expression of developmental regulatory genes, genomic imprinting and X chromosome inactivation (Bird, 2002). Indeed, the DNA methyltransferases DNMT1 or DNMT3a/3b double knockout mice exhibit severe defects in embryogenesis and die before mid-gestation, supporting an essential role for DNA methylation in embryonic development (Li et al., 1992; Okano et al., 1999). On the other hand, mouse embryonic stem cells (mESCs) lacking all three DNMTs can survive and self-renew, and can even begin to differentiate to some germ layers (Jackson et al., 2004; Tsumura et al., 2006), raising the possibility that DNA methylation is dispensable for at least initial lineage-specification in early embryos. Thus, the role of DNA methylation in animal development needs to be more precisely defined. Like DNA methylation, chromatin modifications have also been shown to play a key role in animal development. Enzymes responsible for methylation of histone H3 at lysine 4, 9 and 27, in particular, are essential for embryogenesis (Kouzarides, 2007) (Vastenhouw and Schier, 2012). Additionally, depletion of the histone acetyltransferase p300 or CBP also leads to early embryonic lethality (Yao et al., 1998). While both DNA methylation and chromatin modifications are critical for mammalian development, the exact role of each epigenetic mark in the maintenance of lineage-specific gene expression patterns remains to be defined.
In humans, studying the epigenetic mechanisms regulating early embryonic development often requires access to embryonic cell types that are currently difficult or impractical to obtain. Human embryonic stem cells (hESCs) (Thomson et al., 1998) can be differentiated into a variety of precursor cell types, providing an in vitro model system for studying early human developmental decisions. We have established protocols for differentiation of hESCs to various cell states including trophoblast-like cells (TBL)(Xu et al., 2002), mesendoderm (ME) (Yu et al., 2011), neural progenitor cells (NPCs)(Chambers et al., 2009; Chen et al., 2011), and mesenchymal stem cells (MSCs) (Vodyanik et al., 2010). The first three states represent developmental events that mirror critical developmental decisions in the embryo (the decision to become embryonic or extraembryonic, the decision to become mesendoderm or ectoderm, the decision to become surface ectoderm or neuroectoderm, respectively). MSCs are fibroblastoid cells that are capable of expansion and multi-lineage differentiation to bone, cartilage, adipose, muscle and connective tissues (Vodyanik et al., 2010). The specific hESC derivatives chosen thus reflect key lineages in the human embryo and also represent those lineages that currently can be produced in sufficient quantity and purity for epigenomic studies. These lineages will complement other cells from more mature sources, many of which have had their epigenomes well characterized (Hawkins et al., 2010; Lister et al., 2009; Zhu et al., 2013). Importantly, epigenomic analysis of these cell types allows for investigation of chromatin and transcriptional changes that drive the initial developmental fate decisions.
Here we used high throughput approaches to examine the differentiation of hESCs into four cell types, by generating in-depth maps of transcriptomes, a large panel of histone modifications, and base-resolution maps of DNA methylation for each cell type. Our study provided a full view of the dynamic epigenomic changes accompanying cellular differentiation and lineage specification. As outlined below, an integrative analysis of these datasets provided us with substantial insights into the role of DNA methylation and chromatin modifications in animal development.
Results
Generation of comprehensive epigenome reference maps for hESCs and four hESC derived lineages
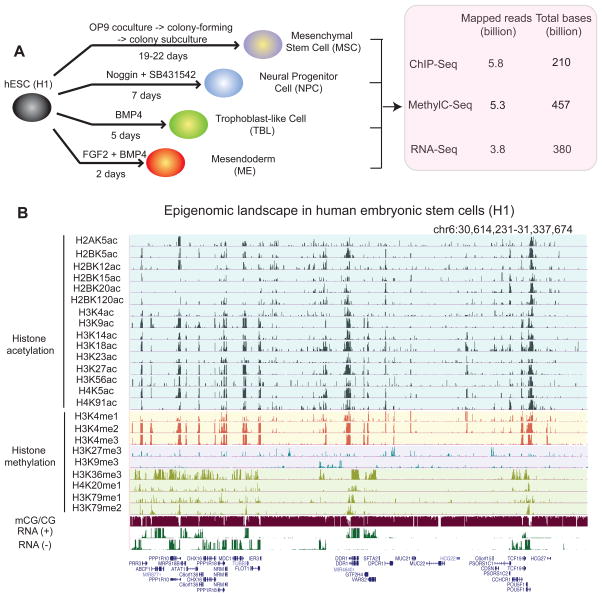
We differentiated the hESC line H1 to mesendoderm (ME), trophoblast-like cells (TBL), neural progenitor cells (NPCs), and mesenchymal stem cells (MSCs) (Figure 1A) (Supplementary Methods). ME, TBL, and NPC differentiation occurred quickly (2 days, 5 days, and 7 days respectively) compared to that of MSC (19–22 days). The expression of various marker genes in these cells was confirmed using immunofluorescence and FACS, and the purity of each cell population ranged from 93% to 99% (Figure S1A–C). ME, NPCs, and MSCs possess further differentiation potentials as shown in Figure S1D–E (for ME and NPCs) and our previous study (for MSCs)(Vodyanik et al., 2010). On the other hand, the nature of TBL is still currently under debate (Bernardo et al., 2011; Xu et al., 2002). As a control for terminally differentiated cells, we also cultured and analyzed IMR90, a primary human fetal lung fibroblast cell line. For each cell type, we mapped DNA methylation at base resolution using MethylC-Seq (Lister et al., 2009) (20–35x total genome coverage, or 10–17.5x coverage per strand). We also mapped the genomic locations of 13–24 chromatin modifications by ChIP-Seq. Additionally, we performed paired-end (100bp x 2) RNA-Seq experiments, generating more than 150 million uniquely mapped reads for every cell type (Figure 1A–B). At least two biological replicates were carried out for each analysis and the data were publicly released as part of the NIH Roadmap Epigenome Project (http://www.epigenomebrowser.org/). Selected data are also available at http://epigenome.ucsd.edu/differentiation.
Figure 1. Generation of comprehensive epigenome reference maps for hESCs and four hESC derived lineages.
(A) Schematic of hESC differentiation procedures and a summary of the epigenomic datasets produced in this study. (B) A snapshot of the UCSC genome browser shows the DNA methylation level (mCG/CG), RNA-Seq reads (+, Watson strand; -, Crick strand), and ChIP-Seq reads (RPKM) of 24 chromatin marks in H1. See also Figure S1.
Identification of differentially expressed genes in hESC-derived cells
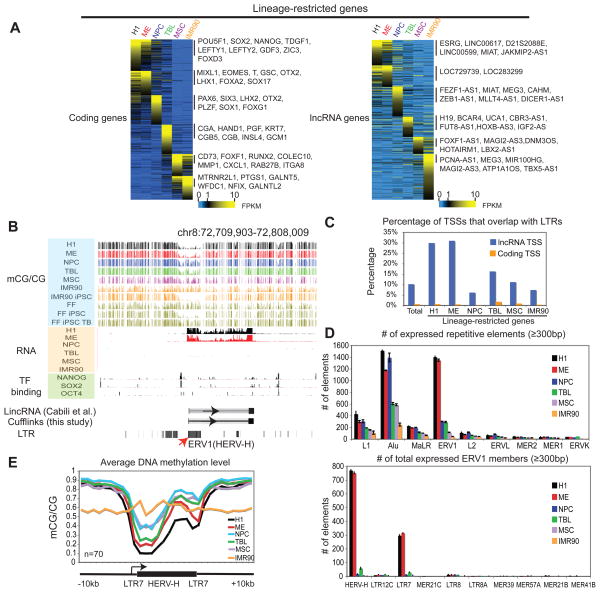
We first asked how the genome is differentially transcribed when hESCs are differentiated into each cell type. To do so, we examined the expression of 19,056 RefSeq coding genes (33,797 isoforms), among which 76.6% (14,595) were expressed in at least one cell type (Figure S2A). Using an entropy-based method (Barrera et al., 2008; Schug et al., 2005) (Figure S2B), we identified 2,408 genes that showed cell type-specific expression (Figure 2A and Figure S2A). For convenience, we use “lineage-restricted genes” to reflect both H1-specific and differentiated cell-specific genes. As expected, known lineage markers were highly expressed in the corresponding cell types (Figure 2A). It is worth noting that in line with a previous report (Yu et al., 2011), the ME cells also express high levels of the hESC regulators NANOG, POU5F1, and a reduced but significant level of SOX2. We then investigated a cohort of long non-coding RNA (lncRNA) genes and detected significant levels of transcripts for 2,175 known and 281 novel lncRNA genes in at least one cell type (Figure 2A and Figure S2A). Using the same entropy-based approach, we found 930 lncRNA genes defined as lineage restricted (Figure S2C), which constitute 37.9% of total expressed lncRNA genes. By contrast, only 16.5% of expressed coding genes are characterized as lineage restricted (Figure S2D). The above analysis defined a large number of coding and non-coding genes that are differentially expressed in H1 and its derived cells. The lists of all lineage-restricted genes are included in Table S1.
Figure 2. Identification of lineage-restricted transcripts in H1 and the H1-derived cells.
(A) Heatmaps showing the expression levels of lineage-restricted coding genes (left) and lncRNA genes (right). Genes are organized by the lineage in which their expression is enriched. Note that certain genes (such as SOX2) can be expressed in more than one cell type. (B) The levels of DNA methylation, RNA, as well as the binding of NANOG, SOX2, and POU5F1, are shown around an annotated lincRNA gene with the promoter overlapping a HERV-H element. (C) The percentages of TSSs that overlap with LTRs are shown for coding genes (yellow) and lncRNA genes (blue) for all genes (total) or lineage-restricted genes. (D) The numbers of expressed (FPKM≥1), mappable repetitive elements are shown in each cell type for various repeat classes (top) or subclasses of ERV1 (bottom). Data are represented as mean +/− standard deviation based on two replicates of RNA-Seq. (E) The average DNA methylation level in each cell type is shown for a subset of H1-specific HERV-H elements. See also Figure S2.
Intriguingly, the promoters of several lncRNA genes highly expressed in H1 overlap with the long terminal repeat (LTR)-containing retrotransposons (Figure 2B). This appears to be a general phenomenon as we observed that significant percentages of transcription start sites (TSSs) of lncRNA genes directly fall into LTRs (Figure 2C). The percentages are notably higher for H1 and ME enriched lncRNA genes (30% and 31%, respectively), which are in contrast to those of coding genes (< 2%). By quantifying the transcription levels of all major classes of mappable repetitive elements, we found that the ERV1 (class I endogenous retrovirus) elements are preferentially expressed in H1 and ME, but not in other cell types (Figure 2D, top). Strikingly, such lineage-specific expression occurs almost exclusively at the ERV1 subfamily HERV-H and its flanking LTR elements LTR7 (Figure 2D, bottom). Together, HERV-H and LTR7 account for more than 43% of LTRs that are present at H1 and ME specific lncRNA gene promoters. A gene ontology analysis of coding genes near H1-specific HERV-H/LTR7 sites revealed an enrichment of POU5F1 targeted genes (p-value = 4E-15), consistent with a previous study showing that NANOG and POU5F1 preferentially bind to repetitive elements(Kunarso et al., 2010). We did not find significant enrichment of LTR subclasses for other lineage-restricted lncRNA genes. Repetitive elements are known to be regulated by DNA methylation and H3K9me3 in ESCs (Leung and Lorincz, 2012). We do not find significant enrichment of H3K9me3 around most HERV-H elements (data not shown). By contrast, a subset of the H1-specific HERV-H elements (n=70) show hypomethylation in H1 and ME, but gain DNA methylation in other H1-derived cells (Figure 2B and 2E). Notably, the overall low level of DNA methylation in IMR90 reflects its globally hypomethylated genome, likely due to the presence of partially methylated domains (PMDs) (Figure S2E–F) (Lister et al., 2009). Additionally, by examining published methylomes (Lister et al., 2011), we found that DNA methylation at these regions was depleted upon reprogramming of IMR90 to iPSCs and was then reestablished when IMR90-derived iPSCs were differentiated to trophoblast-like lineage (Figure 2B). Together, these data suggest that many non-coding RNA genes may be transcriptionally regulated by endogenous retroviral sequences. Of particular interests, the expression of HERV-H/LTR7 is closely correlated with the state of pluripotency and may be regulated by DNA methylation.
Dynamic DNA methylation and chromatin modifications at promoters of lineage-restricted transcripts
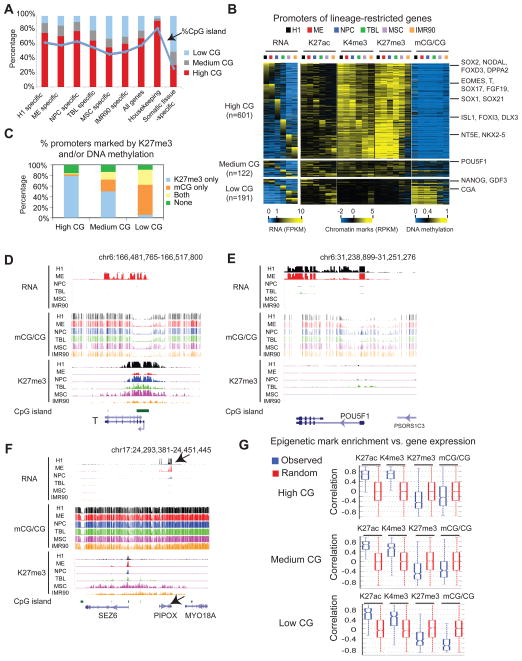
Previous studies have shown that the promoters for somatic tissue specific genes are often CG poor and lack CpG islands (CGIs), in contrast to those for housekeeping genes which are CG rich and predominantly contain CGIs (Barrera et al., 2008; Schug et al., 2005). Therefore, we asked if early lineage-restricted promoters also demonstrate similar features as tissue specific promoters. We first identified promoters for each lineage-restricted gene, and excluded those with ambiguous active promoters (Supplementary Methods). Next, we divided the promoters into three groups based on CG density (high, medium, and low) (Figure S3A). Surprisingly, genes preferentially expressed in early embryonic lineages H1, ME, and NPC tend to be CG rich and contain CGIs (Figure 3A). The percentages of CGI-containing promoters decreased for genes enriched in MSCs and IMR90, which are at relatively late development stages. By contrast, a much lower percentage of promoters (23%) contain CGIs for somatic tissue-specific genes identified from 18 human tissues (Zhu et al., 2008) (Figure 3A). We further verified this using an independent set of somatic tissue-specific genes (35%) (Chang et al., 2011). These data suggest the promoters used for lineage specification in early stages of cell differentiation have distinct sequence features compared to those in more mature cell types.
Figure 3. Epigenetic regulation of promoters for lineage-restricted genes.
(A) Bar graphs showing the percentages of promoters in the high, medium and low CG classes for genes that are enriched in each cell type, all RefSeq genes, housekeeping genes, and somatic tissue-specific genes identified in (Zhu et al., 2008). The percentages of promoters that contain CGIs are also shown (blue line). (B) Heatmaps showing the average levels of RNA, H3K27ac, H3K4me3, H3K27me3 and DNA methylation for promoters of lineage-restricted genes. Histone modifications, TSS +/− 2kb; DNA methylation, TSS +/− 200bp; promoter CG density: TSS +/− 500bp. (C) Bar graphs showing the percentages of promoters that are marked by DNA methylation or K27me3 in at least one cell type. (D–F) The levels of RNA, DNA methylation, and K27me3 are shown for the locus containing T (D), POU5F1 (E), or PIPOX (F). PIPOX (black arrow) is a low CG promoter-containing gene located in a K27me3 domain in MSCs and IMR90 where it is also repressed. (G) The distribution of Pearson correlation coefficients between gene expression level and the levels of various histone modifications or DNA methylation at promoters. See also Figure S3.
DNA methylation machinery has been shown to be a mechanism of gene silencing during cell differentiation (Bird, 2002). In addition, the Polycomb protein complex, which deposits H3K27me3 at target genes, can also repress developmental genes (Boyer et al., 2006; Lee et al., 2006). We set to determine which promoters are subject to regulation by DNA methylation, or H3K27me3, or both. A detailed analysis showed that promoters with high CG density tend to be enriched for H3K27me3, while those with low CG density are preferentially marked by DNA methylation (Figure 3B–C). This is exemplified by the promoters of the ME marker T (high CG, with a CGI) and the hESC marker POU5F1 (medium CG, no CGIs) (Figure 3D–E). Notably, while both H3K27me3 and DNA methylation are largely anti-correlated with gene expression, high CG promoters are often marked by reduced but significant enrichment of H3K27me3 even when they are active (Figure 3B and Figure 3D). It has been shown that the PRC2 complex can be directly recruited by CG rich sequences (Mendenhall et al., 2010). Consistent with this model, our data indicate that the sequence of a promoter could contribute to the epigenetic mechanisms that affect its regulation.
Notably, the majority of developmental regulatory genes, including SOX2, NODAL, EOMES, T, SOX17, and SOX1, belong to the high CG group and are marked by H3K27me3 (Figure 3B). DNA methylation, on the other hand, marks a relatively small number of lineage-restricted genes, including NANOG and POU5F1. A gene ontology analysis also showed that lineage-restricted genes with high CG promoters are enriched for developmental genes, embryonic morphogenesis and pattern specification, while those with low CG promoters contain genes that function in plasma membrane, disulfite bond and protein kinase cascade. As controls, somatic tissue-specific promoters are largely CG poor, often showing high level of DNA methylation; housekeeping gene promoters are predominantly CG rich, showing neither DNA methylation nor H3K27me3 in these cells (Figure S3B). Interestingly, some CG-poor promoters are also marked by low levels of H3K27me3. These promoters are largely observed in the expanded H3K27me3 domains (Figure 3B and Figure 3F, black arrow), a broad pattern of enrichment for H3K27me3 (Hawkins et al., 2010) (Zhu et al., 2013) that frequently occurs in MSCs and IMR90, but less so in H1 and other H1-derived cells (Figure S3C and data not shown). These observations suggest that the expansion of H3K27me3 may be a mechanism to lock low CG promoters in a repressed state in later development stages. Consistently, H3K27me3 shows similar negative correlations with gene expression in all three classes (Figure 3G). By contrast, DNA methylation shows the strongest negative correlation with gene expression for low CG genes (see Figure S3D for the analysis of additional histone modifications). Together, our data suggest that while H3K27me3 may play a widespread role in regulating key factors of cellular differentiation, DNA methylation is involved in modulation of many somatic tissue-specific genes and a limited number of, albeit critical, developmental regulators.
Dynamic DNA methylation and chromatin modifications at enhancers reflect lineage-restricted gene expression
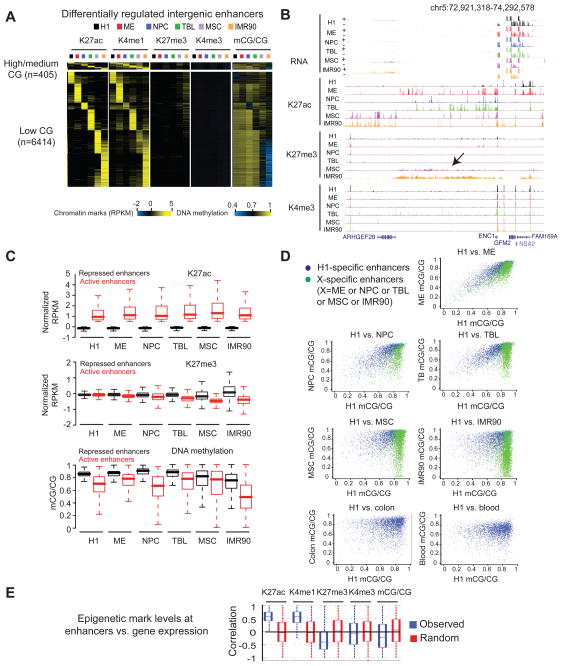
Enhancers are distal regulatory elements that mediate tissue and developmental stage-specific gene expression (Ong and Corces, 2011). To examine the potential role of DNA methylation and chromatin modifications at enhancers, we first identified a total of 103,982 putative enhancer sites in the six cell types (Table S2), using an enhancer prediction method described recently (Rajagopal et al., 2013) (Supplementary Methods). By examining the level of H3K27ac, a marker for active enhancers (Creyghton et al., 2010; Rada-Iglesias et al., 2011), we classified 32,423 enhancers as lineage restricted using the entropy-based analysis (Figure S4A) (Table S2 and Supplementary Methods). We validated these enhancers using several approaches, by showing that they extensively overlap with the binding sites of transcriptional regulators or DNase I hypersensitive sites (John A. Stamatoyannopoulos, unpublished data)(Figure S4B); they show evolutional conservation in sequences (Figure S4C); they are enriched for motifs of transcription factors known to function in each lineage (Figure S4D and Table S3); and their neighboring genes demonstrate functional enrichment that is related to their lineage identities (Figure S4E). Finally, we constructed 8 GFP reporters containing various lineage-specific enhancers and injected them in zebrafish embryos. A high percentage of these enhancers (50%) demonstrated activity in vivo in specific lineages regardless of their positions relative to the reporter gene (Figure S4F). Together, these data suggest that we have identified a set of lineage-restricted enhancers of high quality in hESCs and hESC-derived cells.
We subsequently examined the dynamic epigenetic modifications at lineage-restricted enhancers. As these modifications at intragenic enhancers can be confounded by the activity of their hosting genes, we focused on intergenic lineage-restricted enhancers (n = 6,819) for this analysis (enhancers present in PMDs in IMR90 were also excluded). Most enhancers are CG poor (94%), and appear to be depleted of H3K27me3 (Figure 4A). However, weak enrichment of H3K27me3 is observed at a subset of enhancers in MSCs and IMR90. These enhancers are largely active in H1, ME, NPCs and TBL, but not in MSCs and IMR90, as indicated by the levels of H3K27ac. A closer examination revealed that these enhancers are preferentially present in the H3K27me3 domains specific to MSCs and IMR90 (see Figure 4B for an example). In IMR90 and MSCs, repressed enhancers are marked by higher level of H3K27me3 compared to active enhancers (Figure 4C). By contrast, this is less evident for enhancers in H1 and other H1-derived cells. These results are consistent with the mode that the H3K27me3 domains that arise in differentiated cells may function to repress enhancers that are active in other lineages (Hawkins et al., 2010; Zhu et al., 2013).
Figure 4. Epigenetic regulation of lineage-restricted enhancers.
(A) Heatmaps showing the average levels of H3K27ac, H3K4me1, H3K4me3, H3K27me3, and DNA methylation around the centers of lineage-restricted enhancers. Histone modifications, enhancer center +/− 2kb; DNA methylation, enhancer center +/− 500bp; CG density, enhancer center +/− 500bp. (B) The epigenetic landscape at an intergenic locus showing a low level of H3K27me3 and absence of H3K27ac in MSC and IMR90. (C) Boxplots showing the levels of H3K27ac (top), H3K27me3 (middle) and DNA methylation (bottom) at active and repressed enhancers in each cell type. (D) Scatterplots showing the levels of DNA methylation in each cell type at H1-specific enhancers (blue) and differentiated cell-specific enhancers (green). In the last two panels, colon- and blood-specific enhancer information (green dots) is not available in (Berman et al., 2012; Li et al., 2010). (E) Boxplots showing the distribution of Pearson correlation coefficients between the levels of various histone modifications or DNA methylation at enhancers and the expression level of their potential target genes. See also Figure S4.
Our data also showed that the presence of DNA methylation negatively correlates with the activity of enhancers (Figure 4C). Interestingly, while some H1-specific enhancers acquire DNA methylation in MSCs and IMR90, this is less evident in ME, NPCs and TBL (Figure 4A and 4D). These data are in line with a recent study showing that inactive regulatory elements tend to progressively gain DNA methylation over time during cell differentiation (Bock et al., 2012). By contrast, differentiated cell-specific enhancers appear highly methylated in lineages where they are inactive. We do not observe significant differences between H1-specific and differentiated cell-specific enhancers in their proximity to the nearest TSSs (data not shown). Notably, some H1-specific enhancers remain hypo-methylated even in MSCs, IMR90, and two human tissues: peripheral blood mononuclear cells (Li et al., 2010) and the colon (mucosa) (Berman et al., 2012)(Figure 4D). The functions of these hypo-methylated enhancers remain to be explored. Together, these data indicate that H3K27me3 is preferentially enriched at a subset of enhancers in later stage of cellular differentiation. By contrast, DNA methylation is widely present at enhancers of all stages and negatively correlates with their activity.
We further examined if the presence of DNA methylation or H3K27me3 may correlate with the expression of genes that are potentially regulated by enhancers. To do so, we identified candidate target genes of lineage-restricted enhancers using correlative analyses (Ernst et al., 2011) (Table S4 and Supplementary Methods). At enhancers, histone acetylation is generally positively correlated with the expression of enhancer-targeted genes (Figure 4E and Figure S4G). H3K27me3 and DNA methylation, by contrast, show inverse relationship with gene expression of their potential target genes. The analysis results for expanded histone marks are included in Figure S4G.
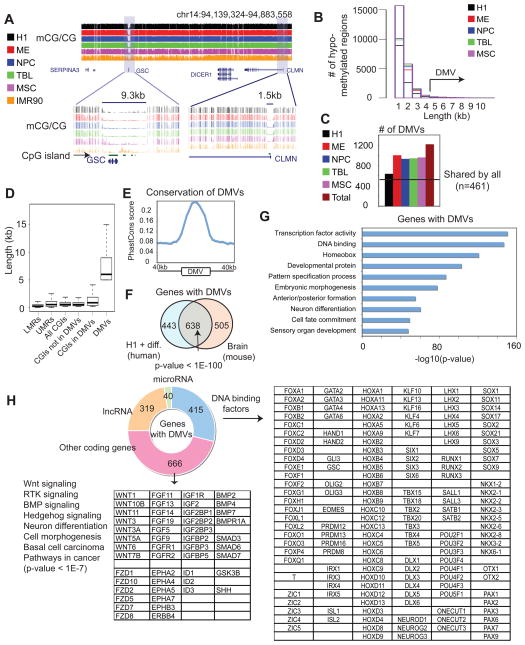
Identification of DNA Methylation Valleys (DMVs)
Previously, low methylation regions (LMRs) and unmethylated regions (UMRs) have been suggested to function as cis elements (Stadler et al., 2011). Applying the same approach by Stadler et al., we defined 5,323 to 31,158 UMRs and 32,744 to 74,541 LMRs in H1 and its derived lineages (Table S5). Indeed, over 85% of UMRs and 42% of LMRs are present in either enhancers or promoters. Surprisingly, while LMR and UMRs are generally short (median lengths 252bp and 532bp, respectively), a number of loci show a much wider depletion of DNA methylation. Interestingly, they often appear near genes for transcription factors and developmental regulators. For example, a 9.3kb hypomethylated region is observed at GSC, a transcription factor specifically expressed in ME (Figure 5A). This unmethylated region covers the entire gene body and regions beyond, in contrast to a typical UMR (CLMN, Figure 5A). We sought to investigate if such broad DNA methylation depletion around developmental genes is a general phenomenon. By examining all continuous hypomethylated regions in H1 and the H1 derived cells (Figure S5A and Figure 5B), we identified those that are at least 5kb long, which constitute less than 3.2% of all hypomethylated regions in any cell type. We named these regions DNA Methylation Valleys (DMVs). IMR90 was excluded from this part of our study due to the presence of PMDs in these cells (Lister et al., 2009)(Figure S2F), which would confound the analyses. Genome wide, we identified 639, 1004, 933, 944, and 962 DMVs in H1, ME, NPC, TBL, and MSC, respectively, among which 461 are shared by all cell types (Figure 5C, see Table S6 for the full lists). Together these regions occupy 1,220 distinct genomic loci. Strikingly, nearly every DMV (99.7%) contains at least one known (89.9%) or putative promoter (9.8%, as indicated by the presence of H3K4me3). The majority of DMVs (93.8%, n = 1,144) contain at least one CGI. Interestingly, while 51.8% DMVs contain one or less CGI, 23.7% (289) DMVs contain at least three CGIs (Figure S5B). These DMVs range in size from 5kb to 68kb, which are much larger than the CGIs in these regions (Figure 5D). About 67% of DMVs contain at least half non-CGI sequences even when we used a much larger CGI list (n = 63,956) (Irizarry et al., 2009) instead of the UCSC CGI list (n = 27,639). We then asked if DMVs are conserved across species. Indeed, DMVs show high level of sequence conservation (Figure 5E). Additionally, we searched for DMVs in mice using a brain methylome that we recently obtained (Xie et al., 2012). Strikingly, a large number of genes with DMVs in humans (638, or 59%, p-value < 1E-100) are also present in DMVs in mice (Figure 5F). Finally, many DMVs (>40%) found in H1 and its derivatives were also observed to be such in adult tissues (Berman et al., 2012; Li et al., 2010) (Figure S5C), suggesting that DMVs are not artifacts of cell culture. The different numbers of DMVs in various cell types may be in part attributed to variations in sequencing depth and methylome coverage of promoters (Figure S5D).
Figure 5. Genes within DNA Methylation Valleys (DMVs) are strongly enriched for transcription factors and developmental genes.
(A) DNA methylation levels for a DMV (GSC) and a nearby typical UMR (CLMN) are shown. (B) Histograms showing the distribution of the lengths of hypomethylated regions in various cell types. (C) The numbers of DMVs found in various cell types. The horizontal line indicates the number of DMVs shared by all cell types. (D) The distribution of lengths of various genomic elements as indicated. (E) The average conservation level (PhastCons scores) around DMVs. (F) A Venn diagram showing the overlap of genes with DMVs in humans (H1 and its derived cells) and in mice (frontal cortex). (G) Gene ontology analysis results for DMV genes in H1 and the H1-derived cells. (H) A breakdown of the types of DMV genes in H1 and the H1-derived cells, with examples shown in the tables. See also Figure S5.
Intriguingly, DMVs contain a unique set of genes. In total, 1,086 coding genes are found in the 1,220 DMVs (Table S7). The majority (91.5%) of their promoters are CG rich (Figure S5E). No significant differences in gene sizes are found for DMV genes with CGIs compared to non-DMV genes with CGIs (data not shown). Strikingly, a GO analysis showed that these genes are strongly enriched for functional groups in transcription factors, homeobox family, developmental protein, and embryonic morphogenesis (Figure 5G). In fact, 38.4% (415) of coding genes in DMVs encode DNA binding proteins (Figure 5H). These genes include hESC and lineage markers such as SOX2, POU5F1, ZIC3 (hESC); EOMES, T, GSC (ME); GLI3, SIX3, LHX3, PAX6 (NPC); GATA2, GATA6 (TBL); and RUNX1 (MSC). This list also includes transcription factor families that are located in clusters (such as HOX), as well as those that reside in different locations (such as FOX, ZIC, GATA, KLF, SIX, TBX, LHX, DLX). In addition, genes in DMVs are strongly enriched for those encoding components of development signaling pathways, including WNT, receptor tyrosine kinase (RTK), BMP, and Hedgehog (Figure 5H). Furthermore, there are 319 lncRNA genes with promoters that overlap with DMVs, including 22 novel lncRNA genes identified in this study (Figure 5H and Table S7). Finally, we found 40 microRNA genes in DMVs (Figure 5H and Table S7), 12 (30%) of which are known to be hESC-specific (such as mir-302/367) (Suh et al., 2004), or within 10kb of lineage-restricted genes that we identified (data not shown). Taken together, our data have revealed a unique class of genomic regions that show wide depletion of DNA methylation, and are strongly associated with transcription factor genes and developmental genes.
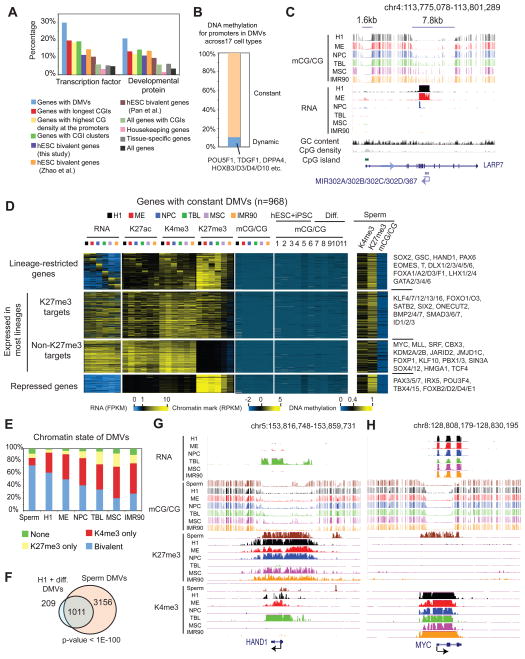
The majority of DMVs remain largely unmethylated upon cell differentiation
Previously, bivalent genes marked by H3K4me3 and H3K27me3 were shown to be highly enriched for developmental genes (Bernstein et al., 2006). Interestingly, DMV genes appear to be more enriched for transcription factors and developmental genes compared to bivalent genes in hESCs as defined in this study or previous studies (Pan et al., 2007; Zhao et al., 2007) (Figure 6A and Figure S6A). Additionally, genes in DMVs are not simply genes with long CGIs, high promoter CG density, or CGI clusters (Supplementary Method) (Figure 6A and Figure S6B). We then asked whether DMVs undergo dynamic epigenetic regulation upon H1 differentiation. We examined the DNA methylation levels in H1, the H1-derived cells, and a panel of published methylomes (see Figure 6D and its legend for the list) (Berman et al., 2012; Li et al., 2010; Lister et al., 2011). Interestingly, most of the promoters in DMVs (89.5%, n=968) remain hypomethylated in all cell types (Figure 6B and Figure S6C). The other 113 promoters demonstrate methylation level at or above 0.4 in at least one cell type (Figure 6B and Figure S6C), including those at several HOX genes as shown previously (Bock et al., 2012; Laurent et al., 2010), and genes that have low CG promoters include POU5F1 (Figure 3E), DPPA4 (not shown), and the hESC-specific microRNA gene cluster mir-302/367 (Figure 6C). Notably, the expression of the mir-302/367 cluster can reprogram somatic cells to pluripotent cells (Anokye-Danso et al., 2011). The activity of mir-302/367 may be regulated by DNA methylation as indicated by the hypermethylation of the associated DMV upon differentiation (Figure 6C). Therefore, a small subset of DMVs, including those at the HOX genes and a number of CG poor promoters, shows dynamic DNA methylation during cell differentiation.
Figure 6. DMVs largely remain hypomethylated in sperm and many terminally differentiated cell types.
(A) Percentages of genes that belong to various gene ontology groups are shown as bar graphs for coding genes in DMVs (n = 1,081), genes with longest CGIs (n = 1,081), genes with the highest promoter CG densities (n=1,081), genes with CGI clusters (n = 1,019), hESC bivalent genes as defined in this study (n=2,401) or in previous studies (Zhao et al., 2007, n=1,797 after gene symbol conversion; Pan et al., 2007, n=3,301 after gene symbol conversion), all RefSeq genes, housekeeping genes (n = 3,140) and somatic tissue-specific genes (n = 885) as defined in (Zhu et al., 2008). (B) A bar graph showing the percentages of promoters in DMVs that demonstrate dynamic DNA methylation (mCG/CG ≥ 0.4 in any cell types) or constant DNA methylation (mCG/CG < 0.4 in any cell types). (C) The levels of DNA methylation and RNA are shown near mir-302A/302B/302C/302D/367. A transcript, likely the hosting transcript for this microRNA gene cluster, is observed mainly in H1 and ME (only - strand RNA reads are shown for simplicity). (D) Heatmaps showing RNA, H3K27ac, H3K4me3, H3K27me3 and DNA methylation levels for promoters of genes with DMVs within various categories. The levels of DNA methylation in additional 11 cell types and sperm, as well as the levels of H3K4me3 and H3K27me3 in sperm, are also shown. 1, hESC H9; 2–4, foreskin fibroblast (FF)-derived iPSC lines (19.11,6.9,19.7); 5, adipose-derived stem (ADS) cell iPSCs; 6, FF iPSC-derived trophoblast-like cells; 7, ADS; 8, ADS-derived adipocytes; 9, FF (Lister et al., 2011); 10, PMBC (blood) (Li et al., 2010); 11, colon tissue (Berman et al., 2012). (E) The chromatin state (presence of H3K4me3 and/or H3K27me3) of DMVs is shown for various cell types. (F) The overlap of DMVs is shown between those in H1 and its derived cells, and those in sperm. (G–H) The epigenetic landscape is shown for the DMV associated with the gene HAND1 (G) or MYC (H). See also Figure S6.
Next, we examined DMVs that remain hypomethylated upon cell differentiation. Among all 968 coding genes that are located in these DMVs, 259 are defined as aforementioned lineage-restricted genes. Most promoters of these genes are CG rich and are marked differentially by H3K27me3 in various lineages, while lacking DNA methylation in general (Figure 6D). Additionally, 134 genes are repressed in all 6 cell types and are also predominantly marked by H3K27me3, including HOXC5/C12/D3/D4, FOXB2/D2/D4/E1, and PAX3/5/7 (Figure 6D). We then examined genes with DMVs that are expressed in most lineages (≥4) in the current study, including those that are marked by H3K27me3 in at least one of the 6 cell types, and those that are not marked by H3K27me3 in any cell types (Figure 6D). The first group shows somewhat weak lineage-restricted expression. The second group is active in all 6 cell types. Gene ontology analysis shows that this group is not enriched for housekeeping genes, but instead is still strongly enriched for transcription regulators, such as MYC, MLL, SRF, and CBX3, and several histone demethylase genes KDM2A/2B, JARID2 and JMJD1C. Together, DMV genes appear to be largely marked by H3K4me3 and/or H3K27me3 (Figure 6D–E). Interestingly, this is also true in sperm as we examined datasets from published studies (Hammoud et al., 2009; Molaro et al., 2011) (Figure 6D–E), Consistent with the notion that many bivalent developmental genes become monovalent upon cell differentiation (Bernstein et al., 2006), a larger portion of DMVs bear only either H3K4me3 or H3K27me3 in differentiated cells compared to that in sperm or H1 (Figure 6E). Interestingly, the sperm genome contains more DMVs than those in other cell types (n=4,167), and most DMVs in H1 and the H1-derived cells (82.9%) are also present in sperm (Figure 6F). These observations are exemplified at two loci near HAND1 (Figure 6G) and MYC (Figure 6H). Therefore, we conclude that the majority of genes in DMVs remain hypomethylated upon H1 differentiation, and are pre-marked by H3K27me3 and/or H3K4me3 in sperm.
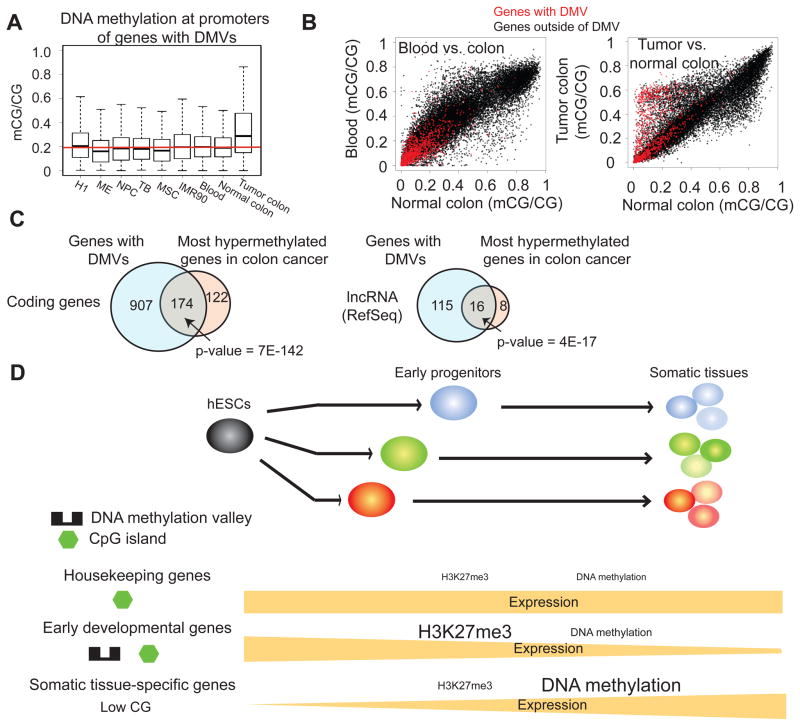
Genes with DMVs are hypermethylated in cancer
As promoters with DMVs are preferentially hypomethylated in most cells that we examined, we sought to examine if this is also true in cancer. Notably, DMV genes are enriched for genes involved in cancer pathways (Figure 5H), tumor suppressor genes (n=120, p-value = 2E-20) and oncogenes (n=72, p-value=5E-14) (Cancer Gene Database, Memorial Sloan-Kettering Cancer Center) (Table S7). Interestingly, by examining base-resolution methylomes for normal and tumor colon tissues (Berman et al., 2012), we found that promoters in DMVs gain significant levels of DNA methylation in the tumor tissue (Figure 7A). Genome wide, 54.0% of DMVs (n=659) overlap with the “methylation prone elements” in colon cancer (Berman et al., 2012). Conversely, 28.9% of methylation prone elements (n=1,493) overlap with DMVs. As the majority of methylation prone elements (71%) are in non-promoter regions (Berman et al., 2012) but DMVs are present almost exclusively at promoters, we focused on the promoter regions for the following analysis. Strikingly, promoters that gain most DNA methylation in the tumor sample (ΔmCG/CG ≥ 0.4) strongly overlap with DMVs identified in H1 and the H1-derived cells (Figure 7B–C). This is true for promoters of both coding genes and lncRNA genes. Similar results were obtained using two additional hypermethylated gene lists in breast cancer and colorectal cancer (Figure S7A). As a control, promoters with DMVs remain hypomethylated in blood cells (Figure 7B). Importantly, most hypermethylated tumor suppressor genes in colon cancer are also DMV genes (16/22, p-value = 1E-17). Unexpectedly, 12 oncogenes are also hypermethylated in colon cancer, among which 9 are DMV genes (p-value = 2E-11). Previously, it was shown that many hypermethylated genes in cancer are Polycomb targets (Bracken and Helin, 2009). Consistently, 87.2% (575/659) of hypermethylated DMVs, compared to 42% (236/561) of non-hypermethylated DMVs, are marked by K27me3 in H1. Taken together, these data suggest that while DMV genes are preferentially maintained DNA methylation free in normal cells, they are prone to hypermethylation in cancer.
Figure 7. DMVs are preferentially methylated in cancer.
(A) Boxplots showing the distribution of the DNA methylation levels at promoters in DMVs for various cell types. (B) Scatterplots showing the DNA methylation levels at promoters between colon and blood (left), and normal and tumor colon (right). Red, promoters with DMVs; black, all other promoters in the genome. (C) Venn diagrams showing the overlaps between genes of which the promoters are hypermethylated in colon cancer (ΔmCG≥0.4, at least 10 CGs covered) and genes with DMVs, for coding genes (left) and lncRNA genes (right). (D) A model for three classes of promoters with distinct sequence features and epigenetic regulation mechanisms in cell differentiation. See the main text for details and also Figure S7.
Discussion
It has long been recognized that epigenetic mechanisms play a critical role in mammalian development, but precisely how DNA methylation and chromatin modifications contribute to development has not yet been clearly elucidated. In this study, we focused on hESCs as a model and generated by far the most comprehensive reference epigenome maps of a multi-lineage differentiation system in humans. Importantly, we demonstrated that the majority of genes differentially expressed in early progenitors are CG rich and appear to employ H3K27me3-mediated repression in non-expressing cells. Conversely, genes differentially expressed in later stages are largely CG poor and preferentially show DNA methylation-mediated gene silencing (Figure 7D). Surprisingly, we found over 1,200 loci, termed DNA Methylation Valleys, that largely remain unmethylated in most cell types that we examined. These regions are uniquely enriched for transcription factor and developmental regulatory genes. Interestingly, DMVs frequently gain abnormal DNA methylation in cancer, suggesting that alterations in DNA methylation machinery might be an important epigenetic mechanism aiding tumorigenesis. Our analysis also confirmed dynamic changes of DNA methylation and chromatin marks at enhancers correlate with gene expression, suggesting that a potential role of epigenetic modulators in regulating enhancer activities.
Distinct epigenetic mechanisms at lineage-restricted genes expressed at early and late stages of ES cell differentiation
Previous studies have shown that somatic tissue-specific promoters tend to be CG poor (Barrera et al., 2008; Schug et al., 2005). However, we found that a large number of CG-rich promoters appear to drive lineage-specific expression in hESC-derived early precursor cells. In line with previous studies, these CG-rich promoters tend to employ Polycomb, but not DNA methylation, for repression (Meissner et al., 2008; Mendenhall et al., 2010; Mohn et al., 2008). By contrast, dynamic DNA methylation is frequently observed at the late stage lineage-restricted promoters, which are characterized by CG-poor sequences. Similar results were obtained when we analyzed two published time-course datasets for single lineage hESC differentiation to trophoblast (Xu et al., 2002) (Figure S7B) or cardiovascular cells (Paige et al., 2012)(Figure S7C). Together, these data add to the notion that low and high CG promoters are regulated by distinct epigenetic regulatory mechanisms (Meissner et al., 2008), and further suggest a temporal relationship of DNA methylation and Polycomb in regulating cell type specific genes.
DMVs are a special class of genomic loci subject to exquisite epigenetic control
Interestingly, many genes encoding for key regulators of embryonic development reside in hypomethylated domains, or DMVs. Importantly, these DMVs are also preferentially hypomethylated in sperm, raising the possibility that these DMVs may be established even earlier. Why are developmental regulatory genes preferentially located in DMVs? One possibility is that DNA methylation at these regions may be incompatible with maintenance of the pluripotency or multipotency of these cells. We noticed that many DMV genes demonstrate a bivalent state (H3K4me3 and H3K27me3), which is linked to poised transcription that may enable developmental genes to be more flexibly modulated(Bernstein et al., 2006). DNA methylation, on the other hand, may be required for more stable silencing of genes in terminally differentiated cells. Another possibility is that the genetic programs regulating embryonic development may actually evolve separately from, or prior to, the evolution of DNA methylation machinery. Supporting this hypothesis, DNA methylation is either absent (such as in Drosophila and C. elegans) or varies considerably in its pattern relative to gene activity in invertebrates(Feng et al., 2010; Zemach et al., 2010). On the other hand, the Polycomb family of factors regulates key developmental regulatory genes in both invertebrates and vertebrates in a more conserved manner. Several mechanisms of DNA hypomethylation at DMVs can be envisioned. DMVs may be recognized by proteins, such as the Tet family, that actively remove DNA methylation (Wu and Zhang, 2011). Alternatively, DMVs may be associated with histone modifications or histone variants, such as H3K4me3 or H2A.Z, that are incompatible to DNA methylation (Cedar and Bergman, 2009). Future experiments are needed to determine which of the above mechanisms could be responsible for DMV formation in the mammalian genome.
Experimental Procedures
hESC differentiation
H1 cells were differentiated according to previously established protocols to mesendoderm (Yu et al., 2011), trophoblast-like cells (Xu et al., 2002), neural progenitor cells (Chambers et al., 2009; Chen et al., 2011), and mesenchymal stem cells (Vodyanik et al., 2010). Details of the differentiation methods can be found in Supplemental Methods.
MethylC-Seq library generation and sequencing
Genomic DNA from H1 and the H1-derived cells was extracted and sonicated. Sequencing library was constructed using NEBNext DNA Sample Prep Reagent Set 1 (NEB). Methylated adapters were used in place of the standard genomic DNA adapters from Illumina. Ligation products were purified, bisulfite treated, PCR amplified, and sequenced using HiSeq2000 (Illumina).
ChIP-Seq library generation and sequencing
H1 and the H1-derived cells were processed following a ChIP protocol as previously described (Hawkins et al., 2010). ChIP libraries were prepared and sequenced using the Illumina instrument as per manufacturer’s instructions.
RNA-Seq library generation and sequencing
Total RNA from H1 and the H1-derived cells was extracted and sequencing libraries were constructed using the TruSeq RNA Sample Prep Kit (Illumina) according to manufacturer’s instructions with modifications to confer strand-specificity (see Supplementary Methods for details).
Accession numbers
All data have been deposited to the Sequence Read Archive (SRA), accession SRP000941.
Data analyses
Details of bioinformatic analyses can be found in Supplemental Information.
Supplementary Material
Highlights.
Epigenome was mapped in-depth for hESCs and four hESC derived cell types
Lineage restricted genes and regulatory sequences were identified in these cell types
Distinct mechanisms regulate lineage-restricted genes at early and late stages
Developmental genes tend to reside in large genomic domains devoid of DNA methylation
Acknowledgments
This work is supported by the NIH Epigenome Roadmap Project (U01 ES017166) and also in part by NSFC 91019016 and NIH R01 HG001696 (MQZ), NIH P01 GM081629 (JAT) and CIRM RN2-00905-1 (BR). NCC is funded by grants from the NIH/NHLBI. HY is funded by grants from the American Heart Association (12POST12050080). We thank Dr. Tomek Swigut and Dr. Joanna Wysocka for sharing the zebrafish enhancer reporter vector. We thank Dr. John Stamatoyannopoulos for generating and providing access to the DNase-Seq datasets. We also thank members of the Ren lab for helpful comments of the manuscript. BR, JAT, JRE, WW, and MQZ designed and supervised the research. ZH, JZ, PY, NEP, KS, JAB, and IS performed/supervised the H1 differentiation experiments. WX, RDH, DL, AYL, AK, SK, and CY, and SK performed ChIP-Seq experiments. RL and JN performed MethylC-Seq experiments. MAU, YL, and YZ performed RNA-Seq experiments. HY and NCC performed/supervised the enhancer-reporter assay in zebrafish. WX, MS, NR, PR, JWW, ST, TW, SAS, YZ, RL, HC, LEE, UW, AK, ZX, WYC, and RS analyzed data. WX, BR, and RS prepared the manuscript. Despite Cell Journal’s policy to limit the max number of co-corresponding authors, BR, JAT, JRE, WW and MQZ are equally responsible for the analysis results.
Footnotes
Note Added in Proof
While this manuscript was in revision, the following related papers describing hESC-specific expression of the HERV-H retrotransposable elements appeared: Kelley, D.R., and Rinn, J.L. (2012). Transposable elements reveal a stem cell specific class of long noncoding RNAs. Genome Biol 13, R107; Santoni, F.A., Guerra, J., and Luban, J. (2012). HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology 9, 111.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera LO, Li Z, Smith AD, Arden KC, Cavenee WK, Zhang MQ, Green RD, Ren B. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 2008;18:46–59. doi: 10.1101/gr.6654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC, et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9:144–155. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bock C, Beerman I, Lien WH, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A. DNA Methylation Dynamics during In Vivo Differentiation of Blood and Skin Stem Cells. Mol Cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nature reviews Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Cheng WC, Chen CR, Shu WY, Tsai ML, Huang CL, Hsu IC. Identification of human housekeeping genes and tissue-selective genes by microarray meta-analysis. PLoS One. 2011;6:e22859. doi: 10.1371/journal.pone.0022859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Wu H, Feinberg AP. A species-generalized probabilistic model-based definition of CpG islands. Mamm Genome. 2009;20:674–680. doi: 10.1007/s00335-009-9222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Krassowska A, Gilbert N, Chevassut T, Forrester L, Ansell J, Ramsahoye B. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol. 2004;24:8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DC, Lorincz MC. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem Sci. 2012;37:127–133. doi: 10.1016/j.tibs.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhu J, Tian G, Li N, Li Q, Ye M, Zheng H, Yu J, Wu H, Sun J, et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, Smith AD. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–1041. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal N, Xie W, Li Y, Wagner U, Wang W, Stamatoyannopoulos J, Ernst J, Kellis M, Ren B. RFECS: A Random-Forest Based Algorithm for Enhancer Identification from Chromatin State. PLoS Comput Biol. 2013;9:e1002968. doi: 10.1371/journal.pcbi.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ., Jr Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Schier AF. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol. 2012;24:374–386. doi: 10.1016/j.ceb.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodyanik MA, Yu J, Zhang X, Tian S, Stewart R, Thomson JA, Slukvin II. A mesoderm-derived precursor for mesenchymal stem and endothelial cells. Cell Stem Cell. 2010;7:718–729. doi: 10.1016/j.stem.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL, Ren B. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Pan G, Yu J, Thomson JA. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 2011;8:326–334. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, et al. Genome-wide Chromatin State Transitions Associated with Developmental and Environmental Cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, He F, Hu S, Yu J. On the nature of human housekeeping genes. Trends Genet. 2008;24:481–484. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.