Abstract
Skeletal muscle makes up approximately 40% of the total body mass, providing structural support and enabling the body to maintain posture, to control motor movements and to store energy. It therefore plays a vital role in whole body metabolism. Skeletal muscle displays remarkable plasticity and is able to alter its size, structure and function in response to various stimuli; an essential quality for healthy living across the lifespan. Exercise is an important stimulator of extracellular and intracellular stress signals that promote positive adaptations in skeletal muscle. These adaptations are controlled by changes in gene transcription and protein translation, with many of these molecules identified as potential therapeutic targets to pharmacologically improve muscle quality in patient groups too ill to exercise. MicroRNAs (miRNAs) are recently identified regulators of numerous gene networks and pathways and mainly exert their effect by binding to their target messenger RNAs (mRNAs), resulting in mRNA degradation or preventing protein translation. The role of exercise as a regulatory stimulus of skeletal muscle miRNAs is now starting to be investigated. This review highlights our current understanding of the regulation of skeletal muscle miRNAs with exercise and disease as well as how they may control skeletal muscle health.
Keywords: miRNA, skeletal muscle, exercise, disease, ageing
Introduction
Maintaining skeletal muscle metabolism, size and contractile function are prerequisites for whole body health throughout life. Skeletal muscle is highly sensitive to extracellular and intracellular signals elicited by contractions from endurance and resistance exercise. These signals are the catalyst for numerous physiological adaptations including enhanced substrate metabolism, mitochondrial biogenesis, angiogenesis, muscle growth and regeneration (Hawke, 2005; Hawley et al., 2006; Léger et al., 2006; Russell, 2010). In contrast, a lack of exercise and muscle contraction, as seen in numerous neuromuscular, musculoskeletal and chronic diseases, as well as in limb immobilization following trauma, sedentary lifestyles or with ageing, negatively impacts on skeletal muscle metabolism, size and contractile function. These negative responses play a role in the on-set and progression of secondary diseases, such as diabetes and cardiovascular disease, increase the severity of chronic diseases and limit the availability of treatment options.
Extracellular and intracellular signals, activated by exercise or disease and inactivity, influence transcriptional and translational regulation of genes encoding proteins that control skeletal muscle metabolism, growth, regeneration and contraction (Dela et al., 1994; Russell et al., 2003, 2005; Short et al., 2003; Wadley et al., 2007). The control of these transcription and translation processes is regulated by transcription factor activation (Keller et al., 2001; McGee et al., 2006), histone modification (McGee et al., 2009) and DNA methylation (Nakajima et al., 2010; Barres et al., 2012). However, the discovery of microRNAs (miRNAs) (Lee et al., 1993; Reinhart et al., 2000) has revealed another level of complexity in transcriptional and translational regulation (Bartel, 2004). Our understanding of how exercise and disease regulate miRNA expression and activity as well as their messenger RNA (mRNA) targets implicated in skeletal muscle health is rudimentary. Exploring this field will advance our knowledge of the mechanisms behind skeletal muscle health and disease and potentially reveal novel therapeutic targets that may be used as a means to improve health outcomes for people suffering from muscular disorders.
The regulation of mRNAs by microRNAs
MiRNAs play an imperative role in the maintenance of healthy cellular function. The primary role of miRNAs is to specifically inhibit protein expression (Olsen and Ambros, 1999; Lee et al., 2004; Wightman et al., 2004; Humphreys et al., 2005; Pillai et al., 2005; Huili et al., 2010) and this can be achieved either by degrading specific mRNA species or by repressing protein translation. Overall, mRNA degradation accounts for the majority of miRNA activity (Huili et al., 2010). The precise mechanism of miRNA targeting and activity still remains to be fully understood. However, miRNA activity appears to be largely dependent on its binding capacity to the target mRNA molecule (Brennecke et al., 2005; Hu and Bruno, 2011). Most miRNA binding sites are located in the 3′ untranslated region (UTR) of the target mRNA species and exist in multiple copies (Hu and Bruno, 2011). There are 2 known binding types for miRNAs (Brennecke et al., 2005). The first is perfect Watson-Crick complementary binding between the 5′ end of the miRNA and the 3′ UTR of the target mRNA; although some miRNA can target the 5′ UTR. This region of the miRNA is called the “seed” region, occurring at base position 2–8 on the 5′ end of the miRNA. This perfect binding within the seed region is sufficient to suppress mRNA activity on its own. The second type of binding is imperfect binding between the seed region and the 3′ UTR but with compensatory binding at the 3′ end of the mRNA molecule. Figure 1 depicts the possible types of miRNA/mRNA interactions along with the effect of miRNA binding on mRNA degradation and translational repression.
Figure 1.
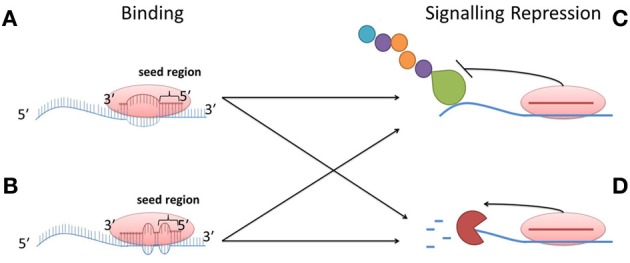
Schematic representation of possible miRNA—mRNA interactions. (A) Perfect Watson-Crick complementary binding. (B) Imperfect binding, with compensatory binding at the 3′ end of the mRNA molecule. Once bound to the mRNA molecule, the miRNA can either (C) inhibit protein translation, or (D) induce mRNA degradation.
Understanding how miRNAs target and bind mRNAs has led to the development of a number of different algorithms and bioinformatics websites such as miRWalk and TargetScan (Lewis et al., 2005; Dweep et al., 2011). These softwares are commonly used to predict specific mRNA/miRNA interactions. However, miRNA binding rules are complex and are not completely understood, resulting in a lack of consensus in the literature. Foremost, establishing direct cause-and-effect links between miRNAs and mRNA targets is key to understanding the underlying molecular mechanisms behind health and disease and thus the development of effective and targeted therapies.
Regulation of microRNA biogenesis machinery
MiRNA biogenesis is a complex process requiring co-ordination of primary miRNA (pri-miRNA) transcription in the nucleus. The pri-miRNA is cleaved by the RNase-III type endonuclease Drosha associated with Pasha (also known as DGCR8) into a precursor molecule referred to as the pre-miRNA. The pre-miRNA is then exported into the cytoplasm by exportin 5 (XPO5) and cleaved a second time by Dicer (Lee et al., 2003; Lund et al., 2004). This gives rise to a duplex strand that is unwound by RNA helicases. The miRNA strands are separated and incorporated into the RNA-induced silencing complex (RISC), providing specificity for the RISC to identify and bind to the 3′ or 5′ UTR of the target mRNAs (Khvorova et al., 2003; Bartel, 2004; Lee et al., 2009). There has been very little investigation into the exercise-induced regulation of the miRNA biogenesis machinery. Work from our group has shown that in the 3 h following a single bout of endurance exercise in untrained males there is an increase in XPO5 mRNA as well as Drosha and Dicer mRNA (Russell et al., 2013). XPO5 and Drosha mRNA are also increased in old but not young subjects within 6 h following a single bout of resistance exercise (Drummond et al., 2008b). Interestingly, this change occurred in parallel with a decrease in XPO5 protein levels in both groups. In addition to its role in exporting pre-miRNA from the nucleus to the cytoplasm, XPO5 also stabilizes pre-miRNAs (Lund et al., 2004), suggesting that XPO5 increases the pool of pre-miRNAs. The upregulation of XPO5 in skeletal muscle following exercise may be an adaptive response to aid processing of new pre-miRNAs that regulate exercise-induced adaptations in muscle. However, further work is required to elucidate the regulation and expression of miRNA biogenesis machinery in muscle and to understand how this affects the miRNA pool.
Regulation of microRNAs in healthy muscle and exercise
Many miRNAs can be highly and specifically enriched in certain tissues (Sood et al., 2006). Skeletal muscle enriched miRNAs, referred to as myomiRs, include miR-1, miR-133a, miR-133b, miR-206, miR-208, miR-208b, miR-486 and miR-499 (McCarthy and Esser, 2007; Callis et al., 2008). The transcriptional regulation of muscle enriched miRNAs is under the control of myogenic regulatory factors (MRFs), such as MyoD, myogenin, Myf5 and MRF4 (Rao et al., 2006; Rosenberg et al., 2006), that tightly control skeletal muscle regeneration (Rudnicki and Jaenisch, 1995; Tajbakhsh et al., 1996; Sabourin and Rudnicki, 2000). These miRNAs can be arranged in polycistronic clusters and transcribed together or in parallel with protein-coding genes (Sweetman et al., 2008). MiRNAs expressed in skeletal muscle are modulated during multiple biological processes involved in skeletal muscle growth, development and maintenance, including atrophy and hypertrophy (McCarthy and Esser, 2007; McCarthy et al., 2007, 2009).
Skeletal muscle atrophy and hypertrophy models have been used to characterize the role and regulation of miRNAs potentially involved in maintaining skeletal muscle mass (see Table 1). Following 7-days of hypertrophy-inducing functional overload of the mouse plantaris muscle, expression of miR-1 and miR-133a was decreased by 50% when compared to control muscle (McCarthy and Esser, 2007). MiR-1 and miR-133a were proposed to contribute to muscle hypertrophy by the removal of their transcriptional inhibitory effect on growth factors such as IGF-1. In support of this hypothesis a regulatory feedback loop was demonstrated in vitro where IGF-1 downregulated miR-1 via the Akt/FoxO3a pathway (Elia et al., 2009). It was also shown that FoxO3a increased levels of miR-1 resulting in reduced IGF-1 protein levels.
Table 1.
Regulation of miRNAs by exercise and disuse.
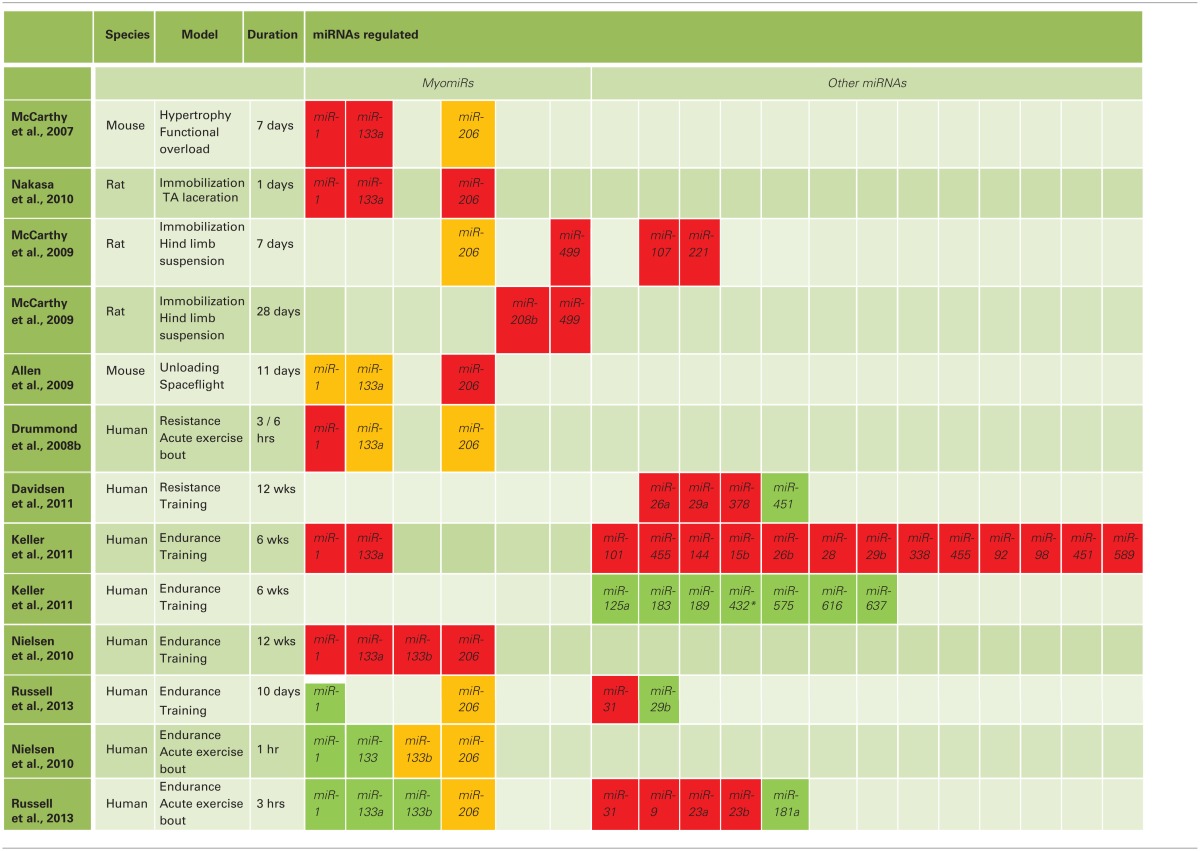
Red, downregulated; Green, upregulated; Orange, no change.
Unloading of skeletal muscle by immobilization, hind limb suspension (HS) or exposure to microgravity during space flights decreases muscle mass (Allen et al., 2009; McCarthy et al., 2009). Muscle immobilization in rats, induced by the laceration of the tibialis anterior, is associated with a decrease in miR-1, miR-133a and miR-206 levels 1 day post-intervention (Nakasa et al., 2010). MiR-107, miR-221, miR-499 and miR-208b were all downregulated following 7 days of rat HS (McCarthy et al., 2009). Eleven days of spaceflight decreased miR-206 expression (Allen et al., 2009). This decrease was paralleled by an upregulation of FoxO1, atrogin-1 and myostatin mRNAs; all regulators of muscle atrophy (Bodine et al., 2001; Kim et al., 2012). MiR-206 promotes differentiation of C2C12 myoblasts (Kim et al., 2006) and skeletal muscle regeneration following injury in mice (Liu et al., 2012). Whether miR-206 plays a direct or indirect role in repressing the atrophy genes is unknown. However, atrogin-1 degrades MyoD (Tintignac et al., 2005), which in turn positively regulates miR-206 (Chen et al., 2006); however, the existence of a miR-206/MyoD/atrogin-1 regulatory loop has not been investigated.
Exercise plays an important role in maintaining muscle health throughout the lifespan, with resistance exercise a potent anabolic stimulus enhancing muscle protein synthesis and muscle growth (Fry, 2004; Léger et al., 2006; Kumar et al., 2009; Phillips, 2009; Koopman et al., 2011). Few studies have investigated the changes in skeletal muscle miRNA species following resistance exercise in humans. MiR-1 expression is reduced 3 and 6 h following an single bout of resistance exercise, while no changes were observed in miR-133a and miR-206 levels (Drummond et al., 2008b). Following a 12-week resistance-training program aimed at inducing muscle hypertrophy, a difference in miRNA regulation was observed in skeletal muscle of subjects defined as “high responders” vs. “low responders” to the resistance exercise training; “low responders” having little or no muscle hypertrophy following the training intervention (Davidsen et al., 2011). The training protocol resulted in an increase in skeletal muscle miR-451 expression and a decrease in miR-26a, miR-29a and miR-378 expression in the “low responder” group only. Low muscle hypertrophy response to resistance exercise training in healthy young subjects is referred to as anabolic resistance (Baar and Esser, 1999; Terzis et al., 2008); a phenomenon also linked to age-related muscle wasting or sarcopenia in the elderly. Whether miR-451, miR-26a, miR-29a and miR-378 contributes to an attenuated hypertrophy response in young healthy subjects and the mechanisms they control now requires experimental validation.
Endurance exercise is another modulator of skeletal muscle miRNA expression. Following 12 weeks of endurance training, expression of the myomiRs miR-1, miR-133a, miR-133b and miR-206 were all significantly down regulated. These miRNAs returned to pre-training baseline levels 2 weeks after the cessation of training (Nielsen et al., 2010). In contrast, 10 days of endurance training increased miR-1, concomitantly with an increase in miR-29b and a decrease in miR-31 (Russell et al., 2013). With respect to a single bout of endurance exercise, miR-1 and miR-133a levels increased in the untrained state, however this acute response was not observed in the trained state (Nielsen et al., 2010). In addition, we observed that in the 3 h period following a single bout of endurance exercise, miR-1, -133a, -133-b and miR-181a were all increased. In contrast miR-9, -23a, -23b and -31 were decreased (Russell et al., 2013). We also demonstrated in vitro, via a reporter assay, that miR-31 directly interacts with HDAC4 (Russell et al., 2013), a component of the MAPK pathway (Symons et al., 2009), as well as with NRF1, which is involved in mitochondrial biogenesis and metabolism. These studies demonstrate that myomiR expression is sensitive to acute and chronic endurance exercise, as well as inactivity. However, their precise targets and the molecular processes regulated remain to be established. Other studies found no correlation between miRNA expression and components of the signaling pathways involved in skeletal muscle adaptation to endurance exercise, such as the MAPK pathway (Kramer and Goodyear, 2007) or the TGF-β pathway (Schabort et al., 2009), suggesting that the individual myomiRs may not regulate these targets in response to endurance exercise. However, multiple miRNAs may need to work together to regulate several key proteins involved in pathway signaling. Following 6 weeks of supervised endurance training in young sedentary males ~800 gene transcripts were regulated and referred to as the training-responsive transcriptome (TRT) (Timmons et al., 2010). Three DNA sequences identified as runt-related transcription factor 1 (RUNX1), sex determining region Y box-9 (SOX9), and paired box gene-3 (PAX3) transcription factor binding sites were overexpressed in the TRT post-training and bioinformatics analyses confirmed RUNX1, SOX9, and PAX3 as potential modulators of muscle aerobic adaptation. MiRNA screening of these subjects also identified 14 miRNAs that were decreased and 7 that were increased in skeletal muscle (Keller et al., 2011). Of the 14 miRNAs that were decreased miR-92, -98, -101 and 104 were predicted to target RUNX1, SOX9 and PAX3. This suggests that the down regulation of these 4 miRNAs during endurance training may permit aerobic adaptation to occur.
Regulation of microRNAs in myopathies
MiRNAs are essential regulators of skeletal muscle health and their implication in the onset and progression of myopathies and chronic diseases associated with muscle wasting and dysfunction is of high interest. Eisenberg and colleagues observed 185 miRNAs to be commonly dysregulated across ten human primary muscular disorders (Eisenberg et al., 2007). Of these, miR-146b, miR-155, miR-214, miR-221 and miR-222 were consistently increased in almost all of the disease conditions and samples tested. Myotonic dystrophy type 1 (DM1) is the most frequently inherited neuromuscular disorder in adults. MiR-206, a regulator of muscle regeneration (Liu et al., 2012), was specifically augmented in DM1 patients when compared to healthy controls (Gambardella et al., 2010). Additionally miR-1 and miR-335 are upregulated and miR-29b, miR-29c and miR-33 downregulated in DM1 patients, when compared to control subjects suspected of a neuromuscular disorder but not presenting any pathological features (Perbellini et al., 2011). Furthermore, the cellular localization of miR-1, miR-133b and miR-206 appears disrupted in DM1 muscle. Similarly, 11 miRNAs, including the muscle enriched miRNA miR-208, are dysregulated in muscle samples from patients with myotonic dystrophy type 2 (DM2) (Greco et al., 2012).
Duchenne muscular dystrophy (DMD) is the most common and severe form of muscular dystrophy characterized by the absence of the structural membrane protein dystrophin. The muscle-enriched miR-206 is downregulated in mdx mice, a well-established animal model for DMD (McCarthy et al., 2007; Yuasa et al., 2008) and miR-206 loss-of-function accelerates the dystrophic phenotype (Liu et al., 2012). In addition to miR-206, another 11 miRNAs were found to be dysregulated in both DMD patients and mdx mice (Greco et al., 2009). In mice, these dysregulations could be rescued following therapeutic intervention, such as HDAC inhibition or restoration of nitric oxide (NO) signaling; treatments reported to ameliorate the mdx phenotype (Colussi et al., 2008). In DMD samples, miR-31 and miR-486 were also identified as regulators of muscle regeneration (Greco et al., 2009). In human DMD myoblasts, miR-31 inhibition increases dystrophin content. MiR-31 modulation is therefore proposed as a possible therapeutic strategy to ameliorate the DMD phenotype (Cacchiarelli et al., 2011). Interestingly, miR-486 expression was not altered in muscle from patients with Becker muscular dystrophy who expresses a partially functional dystrophin protein (Eisenberg et al., 2007). MiR-486 is proposed to play an important regulatory role in the PTEN (phosphatase and tensin homolog deleted on chromosome 10)/Akt pathway in dystrophin deficient (Alexander et al., 2011) and normal muscle (Small et al., 2010). The sapje mutant zebrafish is a model presenting a more severe dystrophic phenotype than the mdx mouse (Bassett and Currie, 2004). MiR-199a-5p is elevated in both sapje zebrafish and human DMD samples when compared to their respective control samples (Alexander et al., 2013). It was demonstrated that miR-199a-5p inhibits the expression of several components of the Wnt signaling pathway, a pathway that regulates satellite cell maintenance and differentiation (Polesskaya et al., 2003; Le Grand et al., 2009).
Chronic diseases associated with muscle wasting are associated with miRNA dysregulation. MiR-1 downregulation is observed in patients with chronic obstructive pulmonary disease (COPD) and is associated with a downregulation of the MRTF-SRF axis (Lewis et al., 2012); an important transcriptional complex regulating muscle gene expression (Cen et al., 2004; Charvet et al., 2006; Miano et al., 2007). Amyotrophic lateral sclerosis (ALS), a severe motor neuron disorder, is characterized by progressive degeneration of upper and lower motor neurons, a decline in strength, severe muscle atrophy, respiratory insufficiency (Pasinelli and Brown, 2006) and mitochondrial dysfunction (Menzies et al., 2002). We have recently identified an increase in miR-23a in skeletal muscle of ALS patients when compared to healthy controls (Russell et al., 2012). It was established in vitro that miR-23a negatively regulates peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) (Russell et al., 2012), a key activator of mitochondrial biogenesis and function. Therapeutic inhibition of miR-23a may rescue PGC-1α activity and ameliorate the ALS phenotype, however this remains to be established. Sixteen miRNAs were dysregulated in patients with laminopathies, a class of myopathies presenting mutations in the lamin A/C gene (Sylvius et al., 2011). Of these, miR-100, miR-192 and miR-135, were directly involved in C2C12 myoblast proliferation and differentiation. In muscle from children suffering from dermatomyositis, an upregulation of 33 miRNAs was observed (Eisenberg et al., 2007). However, miR-126 was specifically downregulated in patients in the early stage of the disease when compared to healthy controls (Kim et al., 2012). MiR-126 is proposed to play a specific role in the early but not in the late stage of juvenile dermatomyositis by promoting the expression of the vascular cell adhesion molecule 1 (VCAM-1), a protein normally expressed in developing but not in mature healthy muscle fibers.
MiRNAs are responsible for the regulation of numerous gene networks and pathways in muscle. Consequently, they are important modulators of skeletal muscle health and many miRNAs are dysregulated in specific muscle disease conditions. Some of these miRNAs play a direct role in muscle cell proliferation or differentiation; however, whether the changes observed in miRNA levels actively contribute or are a consequence of the disease development remains mostly unknown. Identifying the miRNAs dysregulated and understanding their role in muscle diseases is therefore a crucial step in the development of targeted therapeutic strategies.
Age-associated regulation of microRNAs in skeletal muscle
Ageing is a condition associated with changes in skeletal muscle size and function as well as the regulation of miRNAs. Ageing studies using C. elegans identified changes in miRNAs associated with lifespan and cellular senescence (Boehm and Slack, 2005; Ibáñez-Ventoso et al., 2006; Yamakuchi and Lowenstein, 2009; De Lencastre et al., 2010). Of particular interest, let-7 miRNA is decreased with age in C. elegans (Ibáñez-Ventoso et al., 2006; De Lencastre et al., 2010). In contrast, observations in older humans have identified that two miRNAs from the let-7 family of miRNAs, let-7b and let-7e, are elevated in skeletal muscle when compared to young subjects (Drummond et al., 2008a, 2011). The sequence of C. elegans (cel)-let-7 varies slightly from human (has)-let-7b and hsa-let-7e and this difference may be sufficient to confer different mRNA targets for these three miRNAs. Caution must be taken when comparing the expression of miRNAs between different species. MiRNAs have historically been named in the same order as they have been discovered. However, occasional discrepancies exist between species; therefore, a direct comparison between correspondingly named miRNAs cannot always be made. In addition, the mRNAs targeted by a same miRNA can differ between species, although their biological function can be conserved. The function of the let-7 family of miRNAs is similar in both C. elegans and humans. The primary role of the let-7 miRNAs appears to be anti-proliferative, as observed in human cancer cells (Johnson et al., 2007; Nishino et al., 2008; Dong et al., 2010; Zhao et al., 2010; Lee et al., 2011) and in mouse neuronal stem cells (Nishino et al., 2008). The elevation of let-7 miRNAs may be responsible for the impaired ability to activate and proliferate satellite cells in the elderly skeletal muscle, therefore contributing to the attenuated skeletal muscle regenerative capacity in the elderly (Carlson et al., 2009). Accordingly, bioinformatics analysis identified cell cycle regulation and cellular growth and proliferation as the most highly ranked cellular processes likely to be regulated by the 2 let-7 miRNAs in humans. In old mice with muscle atrophy, 57 miRNAs were differentially regulated in the quadriceps when compared to young mice (Hamrick et al., 2010). Of the dysregulated miRNAs, several were predicted to target genes involved in myogenesis, including Mef2, SRF, cardiotrophin 1, myogenin and the cell cycle regulator type IIA activin receptor. Figure 2 summarizes the known and potential roles of miRNAs in myogenesis in the elderly. In order to elucidate which of these miRNAs are important in the age-related muscle wasting, their mRNA targets and specific roles in muscle regeneration and protein synthesis need to be established.
Figure 2.
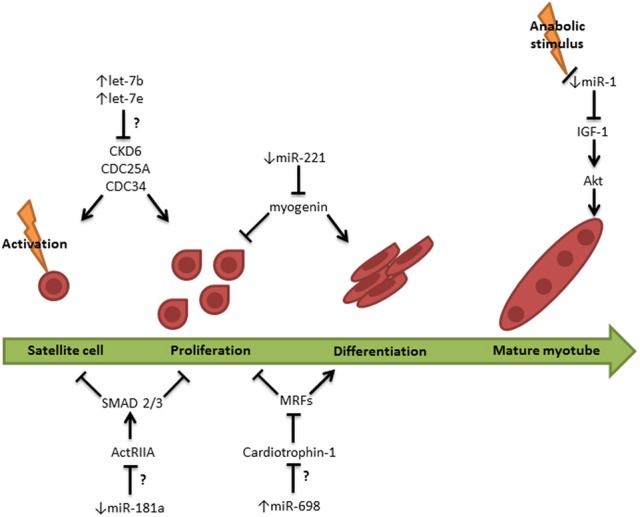
The potential role of miRNAs in the attenuated myogenic process in the elderly. Let-7b and let-7e may contribute to the inhibition of satellite cell activation and proliferation via downregulating cell cycle regulators CKD6, CDC25A, and CDC34. In addition miR-181a may inhibit ActRIIA (activin type IIa receptor) and as a consequence permit activation of the proliferation inhibitors SMAD2/3. The combination of miR-221 and miR-698 further inhibits proliferation by down regulating various MRFs and driving terminal differentiation of the myocytes. Anabolic stimulus is known to reduce miR-1. Failure to downregulate miR-1 in elderly skeletal muscle following anabolic stimulation may contribute to anabolic resistance in mature myotubes via inhibition of IGF-1/Akt signaling., ↑ = stimulatory pathway; or, T = inhibitory pathway; ↑, upregulated miRNA; ↓, downregulated miRNA; ?, cause-and-effect relationship not established.
The most natural way to promote muscle growth is by ingesting good quality protein and engaging in resistance exercise training which is able to stimulate muscle protein synthesis (Yarasheski et al., 1993; Hasten et al., 2000; Drummond et al., 2008a; Katsanos et al., 2008; Kumar et al., 2009; Symons et al., 2009). However, elderly subjects demonstrate an impaired protein synthetic response to resistance exercise (Cuthbertson et al., 2005; Kumar et al., 2009). To date, only one study has investigated the expression of miRNAs in young and old subjects following a protein-stimulating intervention protocol (Drummond et al., 2008b). MiR-1, miR-133a and miR-206 were measured in the muscle of young and old subjects following an acute bout of resistance exercise and ingestion of essential amino acids (EAA). Only miR-1 expression was reduced in the young, but not the old subjects, 3 and 6 h post-exercise and EAA ingestion. A regulatory role for miR-1 as an IGF-1 inhibitor has previously been established (McCarthy and Esser, 2007; Elia et al., 2009). Failure to downregulate miR-1 in elderly subjects following an acute bout of resistance exercise and EAA ingestion may be partially responsible for the attenuated muscle protein synthesis in response to anabolic stimuli. However, more miRNA targets need to be investigated to fully appreciate the role of miRNAs in age-related muscle wasting and to understand the potential mechanisms attenuating protein synthesis following resistance exercise.
MicroRNAs as biomarkers of disease and exercise-induced adaptation
A key feature of miRNAs is their resistance to ribonucleases (RNases) and therefore their presence and potential stability in blood (Chen et al., 2008; Gilad et al., 2008; Turchinovich et al., 2011). Indeed, miRNAs exist within exosomes, lipoprotein and ribonucleoprotein complexes, which protect them from RNases digestion (Valadi et al., 2007; Zhang et al., 2010; Vickers et al., 2011). MiRNAs can be highly expressed in specific tissues, (Sood et al., 2006) although their role in circulation is not yet clear. Circulating miRNAs can originate from tissues with hematopoietic cells an abundant source of circulating miRNAs (Kosaka et al., 2010; Pritchard et al., 2012).
The existence and stability of miRNAs in circulation has led to the search for miRNA biomarkers for various diseases such as cancer, type 2 diabetes, hepatic diseases and coronary diseases (Chen et al., 2008; Mitchell et al., 2008; D'Alessandra et al., 2010; Huang et al., 2010; Pigati et al., 2010; Brase et al., 2011; Freedman et al., 2012). Aberrant expression of specific miRNAs in the circulation may be reflective of disease burden in a specific tissue. It follows that circulating miRNAs may be beneficial as biomarkers for skeletal muscle disease or skeletal muscle adaptation to exercise. To date, only one study has looked at the circulating miRNAs dysregulated in a muscle disorder. Roberts et al. identified an increase in 57 circulating miRNAs in the mdx mouse when compared to wild-type controls (Roberts et al., 2013), including miR-1, miR-133a, and miR-206. The same study demonstrated that miR-1 levels were elevated in the serum of wild-type mice 15 min after a cardiotoxin injection in the tibialis anterior muscle, suggesting that high levels of circulating miR-1 are associated with muscle degeneration and injury.
Changes in plasma and whole blood miRNA profiles have been observed following acute and chronic exercise and in association with training-induced changes in muscle performance (Baggish et al., 2011; Uhlemann et al., 2012; Aoi et al., 2013; Bye et al., 2013; Sawada et al., 2013; Tonevitsky et al., 2013). Table 2 summarizes the circulating miRNAs regulated by exercise, training and fitness level. Circulating miR-133a increases immediately following a marathon race and a single bout of resistance exercise (lateral pulldown, leg press, butterfly) (Uhlemann et al., 2012) but not after a single bout of cycling or treadmill exercise (Baggish et al., 2011; Uhlemann et al., 2012) in trained subjects. In contrast, circulating miR-133a was too lowly expressed at rest and following cycling exercise to be reliably measured (Aoi et al., 2013). Immediately following an acute bout of resistance exercise (bench press and bilateral leg press) in untrained subjects, no changes in circulating miRNA expression was observed (Sawada et al., 2013). Circulating miR-21 and miR-222 are increased following a single bout of cycling exercise and after 90 days of rowing training (Baggish et al., 2011). However, circulating miR-21 and miR-222 appear highly expressed in individuals with low VO2max when compared to individuals with high VO2max (Bye et al., 2013). These discrepancies may be explained by differences in the exercise protocols used. However, it is becoming apparent that various RNA extraction protocols introduce differences in the expression levels of the measured miRNAs (McAlexander et al., 2013). In addition, hemolysis is a major source of plasma miRNAs (Pritchard et al., 2012) and caution needs to be taken during blood draw and plasma separation to avoid miRNA contamination from red blood cells. The expression of plasma miRNAs derived from hematopoietic cells correlates strongly with hematopoietic cell number (Pritchard et al., 2012). Therefore, changes in circulating miRNA levels within subjects may reflect exercise-induced changes in blood cell numbers (Tanimura et al., 2009; Connes et al., 2013; Tonevitsky et al., 2013) rather than muscle-specific adaptations. Tonevitsky et al. completed a miRNA array in whole blood of trained individuals following a single bout of treadmill exercise. MiR-21-5p, miR-24-2-5p, miR-27a-5p, miR-181a-5p and miR-181b-5p were all regulated immediately post-exercise and during recovery (Tonevitsky et al., 2013). Bioinformatics analysis predicted these miRNAs to target exercise-responsive processes including immune function, apoptosis, membrane trafficking and transcriptional regulation. However, these relationships have not been experimentally validated.
Table 2.
Regulation of circulating miRNAs by exercise, training, and fitness level.
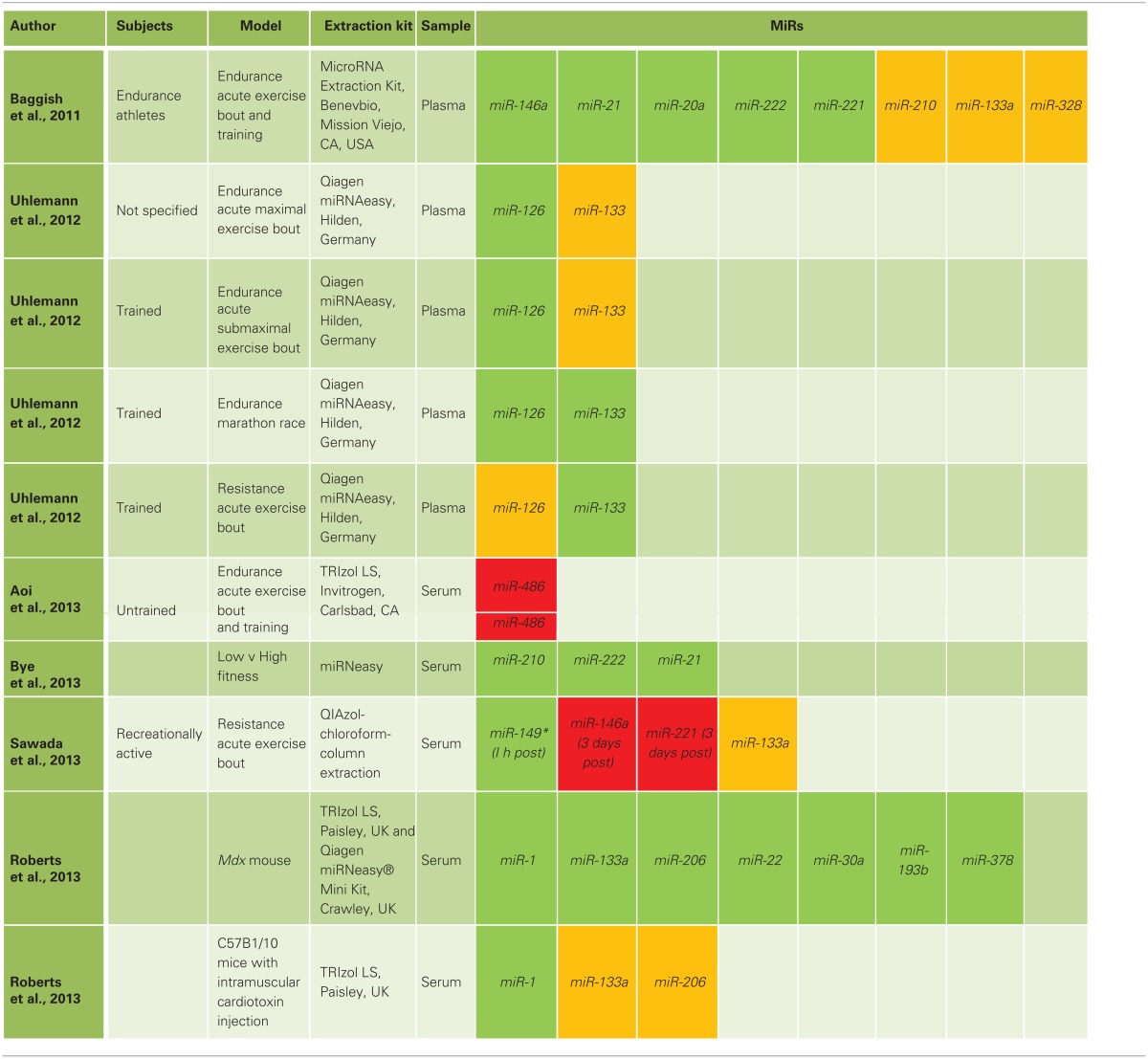
Circulating miRNAs regulated by acute exercise were measured immediately post-exercise. Red, downregulated; Green, upregulated; Orange, no change.
An ideal miRNA biomarker candidate for muscle disease or adaptation to exercise should not be expressed by hematopoietic cells but rather be predominantly expressed in the tissue of interest, such as miR-133a (Callis et al., 2008) and miR-210; the latter described as a hypoximiR (Devlin et al., 2011). More work is required to determine whether circulating miRNAs can serve as stable blood-based biomarkers for underlying skeletal muscle diseases and exercise-induced muscle adaptations.
Concluding remarks
MiRNAs are positive regulators of myogenesis. Their expression levels change following a single bout of exercise and exercise training and following nutritional interventions. A dysregulation of various miRNAs occurs in myopathies, in chronic diseases associated with muscle atrophy as well as with ageing. These observations suggest that skeletal muscle miRNAs play an important role in muscle adaptation and maladaptation. The identification of circulating miRNAs and their regulation following exercise and in disease suggests that they may be useful biomarkers of health and adaptation to treatment interventions. These observations also imply that miRNAs might be amenable to therapeutic intervention. However, at present we have little knowledge relating to how changes in skeletal muscle or circulating miRNAs influence, either directly or indirectly, changes in skeletal muscle regeneration, size, function, metabolism and consequently whole body health. Establishing the causal roles of skeletal muscle miRNAs in vivo is now required to significantly advance this exciting field.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Séverine Lamon is supported by an Alfred Deakin postdoctoral fellowship from Deakin University.
References
- Alexander M. S., Casar J. C., Motohashi N., Myers J. A., Eisenberg I., Gonzalez R. T., et al. (2011). Regulation of DMD pathology by an ankyrin-encoded miRNA. Skelet. Muscle 1, 27 10.1186/2044-5040-1-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M. S., Kawahara G., Motohashi N., Casar J. C., Eisenberg I., Myers J. A., et al. (2013). MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. Cell Death Differ. 20, 1194–1208 10.1038/cdd.2013.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen D. L., Bandstra E. R., Harrison B. C., Thorng S., Stodieck L. S., Kostenuik P. J., et al. (2009). Effects of spaceflight on murine skeletal muscle gene expression. J. Appl. Physiol. 106, 582–595 10.1152/japplphysiol.90780.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi W., Ichikawa H., Mune K., Tanimura Y., Mizushima K., Naito Y., et al. (2013). Muscle-enriched microRNA miR-486 decreases in circulation in response to exercise in young men. Front. Physiol. 4:80 10.3389/fphys.2013.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baar K., Esser K. (1999). Phosphorylation of p70S6k correlates with increased skeletal muscle mass following resistance exercise. Am. J. Physiol. Cell Physiol. 276, C120–C127 [DOI] [PubMed] [Google Scholar]
- Baggish A. L., Hale A., Weiner R. B., Lewis G. D., Systrom D., Wang F., et al. (2011). Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J. Physiol. 589, 3983–3994 10.1113/jphysiol.2011.213363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres R., Yan J., Egan B., Treebak J. T., Rasmussen M., Fritz T., et al. (2012). Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 15, 405–411 10.1016/j.cmet.2012.01.001 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bassett D., Currie P. D. (2004). Identification of a zebrafish model of muscular dystrophy. Clin. Exp. Pharmacol. Physiol. 31, 537–540 10.1111/j.1440-1681.2004.04030.x [DOI] [PubMed] [Google Scholar]
- Bodine S. C., Latres E., Baumhueter S., Lai V. K. M., Nonez L., Clarke B. A., et al. (2001). Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704 10.1126/science.1065874 [DOI] [PubMed] [Google Scholar]
- Boehm M., Slack F. (2005). A developmental timing microRNA and its target regulate life span in C. elegans. Science 310, 1954–1957 10.1126/science.1115596 [DOI] [PubMed] [Google Scholar]
- Brase J. C., Johannes M., Schlomm T., Fälth M., Haese A., Steuber T., et al. (2011). Circulating miRNAs are correlated with tumor progression in prostate cancer. Int. J. Cancer 128, 608–616 10.1002/ijc.25376 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R. B., Cohen S. M. (2005). Principles of MicroRNA–target recognition. PLoS Biol. 3:e85 10.1371/journal.pbio.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bye A., Røsjø H., Aspenes S. T., Condorelli G., Omland T., Wisløff U. (2013). Circulating MicroRNAs and aerobic fitness – The HUNT-Study. PLoS ONE 8:e57496 10.1371/journal.pone.0057496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacchiarelli D., Incitti T., Martone J., Cesana M., Cazzella V., Santini T., et al. (2011). miR-31 modulates dystrophin expression: new implications for Duchenne muscular dystrophy therapy. EMBO Rep. 12, 136–141 10.1038/embor.2010.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis T. E., Deng Z., Chen J.-F., Wang D.-Z. (2008). Muscling Through the microRNA World. Exp. Biol. Med. 233, 131–138 10.3181/0709-MR-237 [DOI] [PubMed] [Google Scholar]
- Carlson M. E., Suetta C., Conboy M. J., Aagaard P., Mackey A., Kjaer M., et al. (2009). Molecular aging and rejuvenation of human muscle stem cells. EMBO Mol. Med. 1, 381–391 10.1002/emmm.200900045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen B., Selvaraj A., Prywes R. (2004). Myocardin/MKL family of SRF coactivators: key regulators of immediate early and muscle specific gene expression. J. Cell. Biochem. 93, 74–82 10.1002/jcb.20199 [DOI] [PubMed] [Google Scholar]
- Charvet C., Houbron C., Parlakian A., Giordani J., Lahoute C., Bertrand A., et al. (2006). New role for serum response factor in postnatal skeletal muscle growth and regeneration via the interleukin 4 and insulin-like growth factor 1 pathways. Mol. Cell. Biol. 26, 6664–6674 10.1128/MCB.00138-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., et al. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 10.1038/ng1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., et al. (2008). Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18, 997–1006 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- Colussi C., Mozzetta C., Gurtner A., Illi B., Rosati J., Straino S., et al. (2008). HDAC2 blockade by nitric oxide and histone deacetylase inhibitors reveals a common target in Duchenne muscular dystrophy treatment. Proc. Natl. Acad. Sci. U.S.A. 105, 19183–19187 10.1073/pnas.0805514105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connes P., Simmonds M. J., Brun J.-F., Baskurt O. K. (2013). Exercise hemorheology: classical data, recent findings and unresolved issues. Clin. Hemorheol. Microcirc. 53, 187–199 [DOI] [PubMed] [Google Scholar]
- Cuthbertson D., Smith K., Babraj J., Leese G., Waddell T., Atherton P., et al. (2005). Anabolic signaling deficits underlie amino acid resistance of wasting, aging muscle. FASEB J. 19, 422–424 [DOI] [PubMed] [Google Scholar]
- D'Alessandra Y., Devanna P., Limana F., Straino S., Di Carlo A., Brambilla P. G., et al. (2010). Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur. Heart J. 31, 2765–2773 10.1093/eurheartj/ehq167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsen P. K., Gallagher I. J., Hartman J. W., Tarnopolsky M. A., Dela F., Helge J. W., et al. (2011). High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J. Appl. Physiol. 110, 309–317 10.1152/japplphysiol.00901.2010 [DOI] [PubMed] [Google Scholar]
- De Lencastre A., Pincus Z., Zhou K., Kato M., Lee S. S., Slack F. J. (2010). MicroRNAs both promote and antagonize longevity in C. elegans. Curr. Biol. 20, 2159–2168 10.1016/j.cub.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dela F., Ploug T., Handberg A., Petersen L. N., Larsen J. J., Mikines K. J., et al. (1994). Physical training increases muscle GLUT4 protein and mRNA in patients with NIDDM. Diabetes 43, 862–865 10.2337/diabetes.43.7.862 [DOI] [PubMed] [Google Scholar]
- Devlin C., Greco S., Martelli F., Ivan M. (2011). miR-210: more than a silent player in hypoxia. IUBMB Life 63, 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q., Meng P., Wang T., Qin W., Qin W., Wang F., et al. (2010). MicroRNA Let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS ONE 5:e10147 10.1371/journal.pone.0010147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond M. J., Dreyer H. C., Pennings B., Fry C. S., Dhanani S., Dillon E. L., et al. (2008a). Skeletal muscle protein anabolic response to resistance exercise and essential amino acids is delayed with aging. J. Appl. Physiol. 104, 1452–1461 10.1152/japplphysiol.00021.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond M. J., McCarthy J. J., Fry C. S., Esser K. A., Rasmussen B. B. (2008b). Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am. J. Physiol. Endocrinol. Metab. 58, E1333–E1340 10.1152/ajpendo.90562.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond M. J., McCarthy J. J., Sinha M., Spratt H. M., Volpi E., Esser K. A., et al. (2011). Aging and microRNA expression in human skeletal muscle: a microarray and bioinformatics analysis. Physiol. Genomics 43, 595–603 10.1152/physiolgenomics.00148.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep H., Sticht C., Pandey P., Gretz N. (2011). miRWalk – database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 44, 839–847 10.1016/j.jbi.2011.05.002 [DOI] [PubMed] [Google Scholar]
- Eisenberg I., Eran A., Nishino I., Moggio M., Lamperti C., Amato A. A., et al. (2007). Distinctive patterns of microRNA expression in primary muscular disorders. Proc. Natl. Acad. Sci. U.S.A. 104, 17016–17021 10.1073/pnas.0708115104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia L., Contu R., Quintavalle M., Varrone F., Chimenti C., Russo M. A., et al. (2009). Reciprocal regulation of MicroRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation 120, 2377–2385 10.1161/CIRCULATIONAHA.109.879429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman J., Ercan B., Morin K., Liu C.-T., Tamer L., Ayaz L., et al. (2012). The distribution of circulating microRNA and their relation to coronary disease. F1000 Res. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A. C. (2004). The role of resistance exercise intensity on muscle fibre adaptations. Sports Med. 34, 663–679 10.2165/00007256-200434100-00004 [DOI] [PubMed] [Google Scholar]
- Gambardella S., Rinaldi F., Lepore S. M., Viola A., Loro E., Angelini C., et al. (2010). Overexpression of microRNA-206 in the skeletal muscle from myotonic dystrophy type 1 patients. J. Transl. Med. 8, 48 10.1186/1479-5876-8-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad S., Meiri E., Yogev Y., Benjamin S., Lebanony D., Yerushalmi N., et al. (2008). Serum MicroRNAs are promising novel biomarkers. PLoS ONE 3:e3148 10.1371/journal.pone.0003148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco S., Perfetti A., Fasanaro P., Cardani R., Capogrossi M. C., Meola G., et al. (2012). Deregulated microRNAs in myotonic dystrophy type 2. PLoS ONE 7:e39732 10.1371/journal.pone.0039732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco S., Simone M. D., Colussi C., Zaccagnini G., Fasanaro P., Pescatori M., et al. (2009). Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J. 23, 3335–3346 10.1096/fj.08-128579 [DOI] [PubMed] [Google Scholar]
- Hamrick M. W., Herberg S., Arounleut P., He H.-Z., Shiver A., Qi R.-Q., et al. (2010). The adipokine leptin increases skeletal muscle mass and significantly alters skeletal muscle miRNA expression profile in aged mice. Biochem. Biophys. Res. Commun. 400, 379–383 10.1016/j.bbrc.2010.08.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasten D. L., Pak-Loduca J., Obert K. A., Yarasheski K. E. (2000). Resistance exercise acutely increases MHC and mixed muscle protein synthesis rates in 78–84 and 23–32 yr olds. Am. J. Physiol. Endocrinol. Metab. 278, E620–E626 [DOI] [PubMed] [Google Scholar]
- Hawke T. J. (2005). Muscle stem cells and exercise training. Exerc. Sport Sci. Rev. 33, 63–68 10.1097/00003677-200504000-00002 [DOI] [PubMed] [Google Scholar]
- Hawley J. A., Hargreaves M., Zierath J. R. (2006). Signalling mechanisms in skeletal muscle: role in substrate selection and muscle adaptation. Essays Biochem. 42, 1–12 10.1042/bse0420001 [DOI] [PubMed] [Google Scholar]
- Hu Z., Bruno A. E. (2011). The Influence of 3′UTRs on MicroRNA function inferred from human SNP data. Comp. Funct. Genomics 2011:910769 10.1155/2011/910769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Huang D., Ni S., Peng Z., Sheng W., Du X. (2010). Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer 127, 118–126 10.1002/ijc.25007 [DOI] [PubMed] [Google Scholar]
- Huili G., Ingolia N. T., Weissman J. S., Bartel D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 10.1038/nature09267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys D. T., Westman B. J., Martin D. I. K., Preiss T. (2005). MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. (English). Proc. Natl. Acad. Sci. U.S.A. 102, 16961–16966 10.1073/pnas.0506482102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez-Ventoso C., Yang M., Guo S., Robins H., Padgett R. W., Driscoll M. (2006). Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell 5, 235–246 10.1111/j.1474-9726.2006.00210.x [DOI] [PubMed] [Google Scholar]
- Johnson C. D., Esquela-Kerscher A., Stefani G., Byrom M., Kelnar K., Ovcharenko D., et al. (2007). The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 67, 7713–7722 10.1158/0008-5472.CAN-07-1083 [DOI] [PubMed] [Google Scholar]
- Katsanos C. S., Chinkes D. L., Paddon-Jones D., Zhang X.-J., Aarsland A., Wolfe R. R. (2008). Whey protein ingestion in elderly persons results in greater muscle protein accrual than ingestion of its constituent essential amino acid content. Nutr. Res. 28, 651–658 10.1016/j.nutres.2008.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C., Steensberg A., Pilegaard H., Osada T., Saltin B., Pedersen B. K., et al. (2001). Transcriptional activation of the IL-6 gene in human contracting skeletal muscle: influence of muscle glycogen content. FASEB J. 15, 2748–2750 [DOI] [PubMed] [Google Scholar]
- Keller P., Vollaard N. B., Gustafsson T., Gallagher I. J., Sundberg C. J., Rankinen T., et al. (2011). A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J. Appl. Physiol. 110, 46–59 10.1152/japplphysiol.00634.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Reynolds A., Jayasena S. D. (2003). Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209 10.1016/S0092-8674(03)00801-8 [DOI] [PubMed] [Google Scholar]
- Kim E., Cook-Mills J., Morgan G., Sredni S. T., Pachman L. M. (2012). Increased expression of vascular cell adhesion molecule 1 in muscle biopsy samples from juvenile dermatomyositis patients with short duration of untreated disease is regulated by miR-126. Arthritis Rheum. 64, 3809–3817 10.1002/art.34606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. K., Lee Y. S., Sivaprasad U., Malhotra A., Dutta A. (2006). Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 174, 677–687 10.1083/jcb.200603008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman R., Gleeson B., Gijsen A. P., Groen B., Senden J. M. G., Rennie M. J., et al. (2011). Post-exercise protein synthesis rates are only marginally higher in type I compared with type II muscle fibres following resistance-type exercise. Eur. J. Appl. Physiol. 111, 1871–1878 10.1007/s00421-010-1808-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N., Iguchi H., Ochiya T. (2010). Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 101, 2087–2092 10.1111/j.1349-7006.2010.01650.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer H. F., Goodyear L. J. (2007). Exercise, MAPK, and NF-kappaB signaling in skeletal muscle. J. Appl. Physiol. 103, 388–395 10.1152/japplphysiol.00085.2007 [DOI] [PubMed] [Google Scholar]
- Kumar V., Selby A., Rankin D., Patel R., Atherton P., Hildebrandt W., et al. (2009). Age-related differences in the dose-response relationship of muscle protein synthesis to resistance exercise in young and old men. J. Physiol. 587, 211–217 10.1113/jphysiol.2008.164483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Ajay S. S., Yook J. I., Kim H. S., Hong S. H., Kim N. H., et al. (2009). New class of microRNA targets containing simultaneous 5'-UTR and 3'-UTR interaction sites. Genome Res. 19, 1175–1183 10.1101/gr.089367.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Lee S.-T., Chu K., Oh H.-J., Im W.-S., Lim J.-Y., Kim S.-K., et al. (2011). Let-7 microRNA inhibits the proliferation of human glioblastoma cells. J. Neurooncol. 102, 19–24 10.1007/s11060-010-0286-6 [DOI] [PubMed] [Google Scholar]
- Lee S. W., Dai G., Hu Z., Wang X., Du J., Mitch W. E. (2004). Regulation of muscle protein degradation: coordinated control of apoptotic and Ubiquitin-Proteasome systems by Phosphatidylinositol 3 kinase. J. Am. Soc. Nephrol. 15, 1537–1545 10.1097/01.ASN.0000127211.86206.E1 [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., et al. (2003). The nuclear RNase III drosha initiates microRNA processing. Nature 425, 415–419 10.1038/nature01957 [DOI] [PubMed] [Google Scholar]
- Léger B., Cartoni R., Praz M., Lamon S., Dériaz O., Crettenand A., et al. (2006). Akt signalling through GSK-3beta, mTOR and Foxo1 is involved in human skeletal muscle hypertrophy and atrophy. J. Physiol. 576, 923–933 10.1113/jphysiol.2006.116715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grand F., Jones A. E., Seale V., Scimè A., Rudnicki M. A. (2009). Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell 4, 535–547 10.1016/j.stem.2009.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A., Riddoch-Contreras J., Natanek S. A., Donaldson A., Man W. D., Moxham J., et al. (2012). Downregulation of the serum response factor/miR-1 axis in the quadriceps of patients with COPD. Thorax 67, 26–34 10.1136/thoraxjnl-2011-200309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are MicroRNA targets. Cell 120, 15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Liu N., Williams A. H., Maxeiner J. M., Bezprozvannaya S., Shelton J. M., Richardson J. A., et al. (2012). microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J. Clin. Invest. 122, 2054–2065 10.1172/JCI62656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Güttinger S., Calado A., Dahlberg J. E., Kutay U. (2004). Nuclear export of MicroRNA precursors. Science 303, 95–98 10.1126/science.1090599 [DOI] [PubMed] [Google Scholar]
- McAlexander M. A., Phillips M. J., Witwer K. W. (2013). Comparison of methods for miRNA extraction from plasma and quantitative recovery of RNA from plasma and cerebrospinal fluid. Front. Genet. 4:83 10.3389/fgene.2013.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J. J., Esser K. A. (2007). MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J. Appl. Physiol. 102, 306–313 10.1152/japplphysiol.00932.2006 [DOI] [PubMed] [Google Scholar]
- McCarthy J. J., Esser K. A., Andrade F. H. (2007). MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. Am. J. Physiol. Cell Physiol. 62, C451–C457 10.1152/ajpcell.00077.2007 [DOI] [PubMed] [Google Scholar]
- McCarthy J. J., Esser K. A., Peterson C. A., Dupont-Versteegden E. E. (2009). Evidence of MyomiR network regulation of β-myosin heavy chain gene expression during skeletal muscle atrophy. Physiol. Genomics 39, 219–226 10.1152/physiolgenomics.00042.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee S. L., Fairlie E., Garnham A. P., Hargreaves M. (2009). Exercise-induced histone modifications in human skeletal muscle. J. Physiol. 587, 5951–5958 10.1113/jphysiol.2009.181065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee S. L., Sparling D., Olson A. L., Hargreaves M. (2006). Exercise increases MEF2- and GEF DNA-binding activity in human skeletal muscle. FASEB J. 20, 348–349 [DOI] [PubMed] [Google Scholar]
- Menzies F. M., Ince P. G., Shaw P. J. (2002). Mitochondrial involvement in amyotrophic lateral sclerosis. Neurochem. Int. 40, 543–551 10.1016/S0197-0186(01)00125-5 [DOI] [PubMed] [Google Scholar]
- Miano J. M., Long X., Fujiwara K. (2007). Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Cell Physiol. 292, C70–C81 10.1152/ajpcell.00386.2006 [DOI] [PubMed] [Google Scholar]
- Mitchell P. S., Parkin R. K., Kroh E. M., Fritz B. R., Wyman S. K., Pogosova-Agadjanyan E. L., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Takeoka M., Mori M., Hashimoto S., Sakurai A., Nose H., et al. (2010). Exercise effects on methylation of ASC gene. Int. J. Sports Med. 31, 671–675 10.1055/s-0029-1246140 [DOI] [PubMed] [Google Scholar]
- Nakasa T., Ishikawa M., Shi M., Shibuya H., Adachi N., Ochi M. (2010). Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J. Cell. Mol. Med. 14, 2495–2505 10.1111/j.1582-4934.2009.00898.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S., Scheele C., Yfanti C., Akerstrom T., Nielsen A. R., Pedersen B. K., et al. (2010). Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J. Physiol. 588, 4029–4037 10.1113/jphysiol.2010.189860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J., Kim I., Chada K., Morrison S. J. (2008). Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell 135, 227–239 10.1016/j.cell.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen P. H., Ambros V. (1999). The lin-4 regulatory RNA controls developmental timing in caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216, 671–680 10.1006/dbio.1999.9523 [DOI] [PubMed] [Google Scholar]
- Pasinelli P., Brown R. H. (2006). Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat. Rev. Neurosci. 7, 710–723 10.1038/nrn1971 [DOI] [PubMed] [Google Scholar]
- Perbellini R., Greco S., Sarra-Ferraris G., Cardani R., Capogrossi M. C., Meola G., et al. (2011). Dysregulation and cellular mislocalization of specific miRNAs in myotonic dystrophy type 1. Neuromuscul. Disord. 21, 81–88 10.1016/j.nmd.2010.11.012 [DOI] [PubMed] [Google Scholar]
- Phillips S. M. (2009). Physiologic and molecular bases of muscle hypertrophy and atrophy: impact of resistance exercise on human skeletal muscle (protein and exercise dose effects). Appl. Physiol. Nutr. Metab. 34, 403–410 10.1139/H09-042 [DOI] [PubMed] [Google Scholar]
- Pigati L., Yaddanapudi S. C. S., Iyengar R., Dong-Ja K., Hearn S. A., Danforth D., et al. (2010). Selective release of MicroRNA species from normal and malignant mammary epithelial cells. PLoS ONE 5:e13515 10.1371/journal.pone.0013515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R. S., Bhattacharyya S. N., Artus C. G., Zoller T., Cougot N., Basyuk E., et al. (2005). Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309, 1573–1576 10.1126/science.1115079 [DOI] [PubMed] [Google Scholar]
- Polesskaya A., Seale P., Rudnicki M. A. (2003). Wnt signaling induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration. Cell 113, 841–852 10.1016/S0092-8674(03)00437-9 [DOI] [PubMed] [Google Scholar]
- Pritchard C. C., Kroh E., Wood B., Arroyo J. D., Dougherty K. J., Miyaji M. M., et al. (2012). Blood cell origin of circulating micrornas: a cautionary note for cancer biomarker studies. Cancer Prev. Res. 5, 492–497 10.1158/1940-6207.CAPR-11-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P. K., Kumar R. M., Farkhondeh M., Baskerville S., Lodish H. F. (2006). Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. U.S.A. 103, 8721–8726 10.1073/pnas.0602831103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis Elegans. Nature 403, 901–906 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Roberts T. C., Godfrey C., McClorey G., Vader P., Briggs D., Gardiner C., et al. (2013). Extracellular microRNAs are dynamic non-vesicular biomarkers of muscle turnover. Nucleic Acids Res. [Epub ahead of print]. 10.1093/nar/gkt724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M. I., Georges S. A., Asawachaicharn A., Analau E., Tapscott S. J. (2006). MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J. Cell Biol. 175, 77–85 10.1083/jcb.200603039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki M. A., Jaenisch R. (1995). The MyoD family of transcription factors and skeletal myogenesis. Bioessays 17, 203–209 10.1002/bies.950170306 [DOI] [PubMed] [Google Scholar]
- Russell A. P. (2010). Molecular regulation of skeletal muscle mass. Clin. Exp. Pharmacol. Physiol. 37, 378–384 10.1111/j.1440-1681.2009.05265.x [DOI] [PubMed] [Google Scholar]
- Russell A. P., Feilchenfeldt J., Schreiber S., Praz M., Crettenand A., Gobelet C., et al. (2003). Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-gamma coactivator-1 and peroxisome proliferator-activated receptor-alpha in skeletal muscle. Diabetes 52, 2874–2881 10.2337/diabetes.52.12.2874 [DOI] [PubMed] [Google Scholar]
- Russell A. P., Hesselink M. K., Lo S. K., Schrauwen P. (2005). Regulation of metabolic transcriptional co-activators and transcription factors with acute exercise. FASEB J. 19, 986–988 10.1096/fj.04-3168fje [DOI] [PubMed] [Google Scholar]
- Russell A. P., Lamon S., Boon H., Wada S., Guller I., Brown E. L., et al. (2013). Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short term endurance training. J. Physiol. 591, 4637–4653 10.1113/jphysiol.2013.255695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A. P., Wada S., Vergani L., Hock M. B., Lamon S., Leger B., et al. (2012). Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiol. Dis. 49C, 107–117 [DOI] [PubMed] [Google Scholar]
- Sabourin L. A., Rudnicki M. A. (2000). The molecular regulation of myogenesis. Clin. Genet. 57, 16–25 10.1034/j.1399-0004.2000.570103.x [DOI] [PubMed] [Google Scholar]
- Sawada S., Kon M., Wada S., Ushida T., Suzuki K., Akimoto T. (2013). Profiling of Circulating MicroRNAs after a bout of acute resistance exercise in humans. PLoS ONE 8:e70823 10.1371/journal.pone.0070823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schabort E. J., Van Der Merwe M., Loos B., Moore F. P., Niesler C. U. (2009). TGF-beta's delay skeletal muscle progenitor cell differentiation in an isoform-independent manner. Exp. Cell Res. 315, 373–384 10.1016/j.yexcr.2008.10.037 [DOI] [PubMed] [Google Scholar]
- Short K. R., Vittone J. L., Bigelow M. L., Proctor D. N., Rizza R. A., Coenen-Schimke J. M., et al. (2003). Impact of aerobic exercise training on age-related changes in insulin sensitivity and muscle oxidative capacity. Diabetes 52, 1888–1896 10.2337/diabetes.52.8.1888 [DOI] [PubMed] [Google Scholar]
- Small E. M., O'Rourke J. R., Moresi V., Sutherland L. B., McAnally J., Gerard R. D., et al. (2010). Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. U.S.A. 107, 4218–4223 10.1073/pnas.1000300107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood P., Krek A., Zavolan M., Macino G., Rajewsky N. (2006). Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. U.S.A. 103, 2746–2751 10.1073/pnas.0511045103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman D., Goljanek K., Rathjen T., Oustanina S., Braun T., Dalmay T., et al. (2008). Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev. Biol. 321, 491–499 10.1016/j.ydbio.2008.06.019 [DOI] [PubMed] [Google Scholar]
- Sylvius N., Bonne G., Straatman K., Reddy T., Gant T. W., Shackleton S. (2011). MicroRNA expression profiling in patients with lamin A/C-associated muscular dystrophy. FASEB J. 25, 3966–3978 10.1096/fj.11-182915 [DOI] [PubMed] [Google Scholar]
- Symons T. B., Sheffield-Moore M., Wolfe R. R., Paddon-Jones D. (2009). A moderate serving of high-quality protein maximally stimulates skeletal muscle protein synthesis in young and elderly subjects. J. Am. Diet. Assoc. 109, 1582–1586 10.1016/j.jada.2009.06.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S., Bober E., Babinet C., Pournin S., Arnold H., Buckingham M. (1996). Gene targeting the myf-5 locus with nlacZ reveals expression of this myogenic factor in mature skeletal muscle fibres as well as early embryonic muscle. Dev. Dyn. 206, 291–300 [DOI] [PubMed] [Google Scholar]
- Tanimura Y., Kon M., Shimizu K., Kimura F., Kono I., Ajisaka R. (2009). Effect of 6-day intense Kendo training on lymphocyte counts and its expression of CD95. Eur. J. Appl. Physiol. 107, 227–233 10.1007/s00421-009-1119-1 [DOI] [PubMed] [Google Scholar]
- Terzis G., Georgiadis G., Stratakos G., Vogiatzis I., Kavouras S., Manta P., et al. (2008). Resistance exercise-induced increase in muscle mass correlates with p70S6 kinase phosphorylation in human subjects. Eur. J. Appl. Physiol. 102, 145–152 10.1007/s00421-007-0564-y [DOI] [PubMed] [Google Scholar]
- Timmons J. A., Knudsen S., Rankinen T., Koch L. G., Sarzynski M., Jensen T., et al. (2010). Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J. Appl. Physiol. 108, 1487–1496 10.1152/japplphysiol.01295.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintignac L. A., Lagirand J., Batonnet S., Sirri V., Leibovitch M. P., Leibovitch S. A. (2005). Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J. Biol. Chem. 280, 2847–2856 10.1074/jbc.M411346200 [DOI] [PubMed] [Google Scholar]
- Tonevitsky A., Maltseva D., Abbasi A., Samatov T., Sakharov D., Shkurnikov M., et al. (2013). Dynamically regulated miRNA-mRNA networks revealed by exercise. BMC Physiol. 13:9 10.1186/1472-6793-13-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A., Weiz L., Langheinz A., Burwinkel B. (2011). Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39, 7223–7233 10.1093/nar/gkr254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlemann M., Möbius-Winkler S., Fikenzer S., Adam J., Redlich M., Möhlenkamp S., et al. (2012). Circulating microRNA-126 increases after different forms of endurance exercise in healthy adults. Eur. J. Prev. Cardiol. [Epub ahead of print]. 10.1177/2047487312467902 [DOI] [PubMed] [Google Scholar]
- Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J. J., Lotvall J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- Vickers K. C., Palmisano B. T., Shoucri B. M., Shamburek R. D., Remaley A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 13, 423–433 10.1038/ncb2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadley G. D., Konstantopoulos N., Macaulay L., Howlett K. F., Garnham A., Hargreaves M., et al. (2007). Increased insulin-stimulated Akt pSer473 and cytosolic SHP2 protein abundance in human skeletal muscle following acute exercise and short-term training. J. Appl. Physiol. 102, 1624–1631 10.1152/japplphysiol.00821.2006 [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. (2004). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 116, 855–862 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- Yamakuchi M., Lowenstein C. J. (2009). MiR-34, SIRT1, and p53: the feedback loop. Cell Cycle 8, 712–715 10.4161/cc.8.5.7753 [DOI] [PubMed] [Google Scholar]
- Yarasheski K. E., Zachwieja J. J., Bier D. M. (1993). Acute effects of resistance exercise on muscle protein synthesis rate in young and elderly men and women. Am. J. Physiol. Endocrinol. Metab. 265, E210–E214 [DOI] [PubMed] [Google Scholar]
- Yuasa K., Hagiwara Y., Ando M., Nakamura A., Takeda S., Hijikata T. (2008). MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Struct. Funct. 33, 163–169 10.1247/csf.08022 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu D., Chen X., Li J., Li L., Bian Z., et al. (2010). Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell 39, 133–144 10.1016/j.molcel.2010.06.010 [DOI] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S., Lang M.-F., Yang S., Li W., et al. (2010). MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 1876–1881 10.1073/pnas.0908750107 [DOI] [PMC free article] [PubMed] [Google Scholar]


