Abstract
Genetic variants in a gene on 6p22.3, dysbindin, have been shown recently to be associated with schizophrenia (Straub et al. 2002a). There is no doubt that replication in other independent samples would enhance the significance of this finding considerably. Since the gene is located in the center of the linkage peak on chromosome 6p that we reported earlier, we decided to test six of the most positive DNA polymorphisms in a sib-pair sample and in an independently ascertained sample of triads comprising 203 families, including the families for which we detected linkage on chromosome 6p. Evidence for association was observed in the two samples separately as well as in the combined sample (P=.00068 for SNP rs760761). Multilocus haplotype analysis increased the significance further to .00002 for a two-locus haplotype and to .00001 for a three-locus haplotype. Estimation of frequencies for six-locus haplotypes revealed one common haplotype with a frequency of 73.4% in transmitted, and only 57.6% in nontransmitted, parental haplotypes. All other six-locus haplotypes occurring at a frequency of >1% were less often transmitted than nontransmitted. Our results represent a first successful replication of linkage disequilibrium in psychiatric genetics detected in a region with previous evidence of linkage and will encourage the search for causes of schizophrenia by the genetic approach.
Schizophrenia is a complex genetic disorder affecting ∼1% of the population worldwide. A number of linkage studies have been performed to map the underlying susceptibility genes, but most of these studies were not supported by subsequent attempts to replicate them in independent samples. Following initial reports (Moises et al. 1995; Schwab et al. 1995; Straub et al. 1995; Wang et al. 1995), chromosome 6p (locus SCZD3 [MIM 600511]) has been studied by several groups, yielding some positive results (Schizophrenia Linkage Collaborative Group for Chromosomes 3 1996; Brzustowicz et al. 1997; Turecki et al. 1997; Hwu et al. 2000) but some negative results as well. Most of these studies were reviewed in the Chromosome 6 Workshop Report (Nurnberger and Foroud 1999).
There is some consensus that the classical linkage approach to complex disorders may not be as straightforward as for monogenic disorders with Mendelian inheritance. In particular, linkage analysis in complex disorders may not be capable of mapping a potential susceptibility gene to a defined position, which would allow successful application of the classical positional cloning approach. However, a positive linkage finding in complex disorders could serve as a starting point for identification of candidate genes in an even larger region, by using the now-available information on sequence variants and genes that allows association/linkage disequilibrium studies.
Straub et al. (2002a) recently reported association of schizophrenia with genetic variants in a candidate gene on 6p22.3, the gene for the dystrobrevin-binding protein 1, dysbindin, which is located within the linkage peak on chromosome 6p, as reported elsewhere (Straub et al. 1995). Dysbindin binds to α- and β-dystrobrevin in muscle and brain. In addition to its presence in muscles, it has been detected in axon terminals in the cerebellum and hippocampus of the adult mouse brain (Benson et al. 2001), and a possible function in glutaminergic synapses has been discussed (Benson et al. 2001). Straub et al. (2002a) obtained a high degree of statistical significance for association of schizophrenia with several DNA polymorphisms between exon 5 and exon 1 in the dysbindin gene (P=.008–.0001 for three marker haplotypes) in the 270 Irish high-density pedigrees in which linkage had been reported earlier (Straub et al. 1995). No mutation affecting function has been detected yet (Straub et al. 2002a).
We reported elsewhere support for potential susceptibility genes on 6p by multipoint affected sib-pair linkage analysis in a sample of 54 sib-pair families from Germany and Israel (Schwab et al. 1995). Two broad peaks were observed in the region of linkage obtained in this study, one distal between markers D6S260 and D6S285, spanning a distance of ∼3.1 Mb, and a second one between marker D6S461 (distal to the HLA region) and the HLA region. Therefore, we decided to test six of the most positive SNPs reported by Straub et al. (2002a) for association in a sib-pair sample that included the original families demonstrating linkage, as well as in a sample of independently ascertained triads. The used SNP markers (names, distances, and polymorphic site are given in table 1) span a chromosomal interval of 36.2 kb between exon 5 and exon 1 in the dysbindin gene.
Table 1.
Allele Frequencies and P Values for Six SNP Markers Located within the Dysbindin Gene
|
Triads |
Sib Pairs |
Combined |
||||||||||||
| Frequency |
Frequency |
Frequency |
||||||||||||
| SNP (NCBI No.) | Distance(kb) | Polymorphisma | Pseudocontrolsb | Parentsc | T/Ud | P | Pseudocontrolsb | Parentsc | T/Ud | P | Pseudocontrolsb | Parentsc | T/Ud | P |
| P1635e (rs3213207) | … | A/G | .160 | .132 | 1.65 | .0741 | .206 | .162 | 1.72 | .0540 | .176 | .143 | 1.69 | .0052 |
| P1325 (rs1011313) | 5.3 | C/T | .128 | .097 | 2.25 | .0163 | .121 | .107 | 1.63 | .1725 | .126 | .101 | 1.87 | .0092 |
| P1765 (rs2619528) | 16.4 | C/T | .256 | .223 | 1.46 | .0845 | .287 | .232 | 1.53 | .0945 | .267 | .226 | 1.50 | .0140 |
| P1320 (rs760761) | 1.3 | C/T | .273 | .229 | 1.62 | .0260 | .312 | .246 | 1.71 | .0210 | .287 | .235 | 1.67 | .0007 |
| P1763 (rs2619522) | 2.5 | A/C | .250 | .211 | 1.60 | .0415 | .242 | .207 | 1.32 | .2850 | .247 | .209 | 1.44 | .0300 |
| P1578 (rs1018381) | 3.4 | C/T | .099 | .089 | 1.29 | .4233 | .069 | .065 | 1.00 | 1.000 | .089 | .081 | 1.14 | .6900 |
Second allele is the rare allele.
Frequency of the rare alleles in all nontransmitted parental alleles.
Frequency of the rare allele in all parental alleles.
Transmission disequilibrium (T = number of transmissions of allele “1”; U = number of transmissions of allele “2” from heterozygous parents to their affected children).
SNP, as described by Straub et al. (2002a).
The sample containing the affected sib-pair families was an extended sample of the 54 families used in our earlier linkage report (Schwab et al. 1995). For the present study, nine families had to be removed because we had no permanent lymphoblast cell lines available, and the supply of DNA was finished. The sample was increased to 78 families by adding 33 newly ascertained sib-pair families. Linkage was still present in this extended sample, since multipoint-linkage analysis with the GENEHUNTER program (Kruglyak et al. 1996), using the six SNP markers, produced a nonparametric LOD score of 2.55 (P=.0055). The second independently ascertained sample consisted of 125 “triad” families (affected offspring with parents). For each of these triads, a history of psychiatric disorder (at least one additional first- or second-degree member with a psychiatric disorder) was available. Diagnosis was based on the DSM-IV criteria and comprised the core phenotypes schizophrenia and schizoaffective disorder, mainly schizophrenic (on the basis of the RDC criteria). The sib-pair sample included six families from Israel (four of non-Ashkenazi, one of Ashkenazi, and one of Arab origin), which contributed to the reported linkage results (Schwab et al. 1995), and one family from Hungary. The triad families included nine families from Hungary. The families from Hungary were ascertained in the area of Pecs, an area where Germans founded large settlements in the last centuries. The families from Germany were collected in the southern (Bavaria) and southwestern (Mainz, Bonn) parts of Germany and should present a typical German population formed by mixing Celts, Romans, and Germanic tribes ∼1,500–2,000 years ago.
The six SNPs were analyzed by template-directed dye-terminator incorporation followed by detection of fluorescence polarization in a Tecan Ulta 384 reader, as described by Chen et al. (1999). An AcycloPrime-FP SNP Detection Kit (Perkin Elmer) was used for allele-specific incorporation of fluorescence-labeled Acyclo Terminators. Sequences of oligonucleotides for PCR and primer extension were the same as described by Straub et al. (2002a). Genotypes that could not be unambiguously assigned were cross-validated by alternative methods, such as SSCP analysis (for P1578 and P1765), denaturing high-performance liquid chromatography (for P1325 and P1320), and PCR-RFLP (for P1635, the polymorphic site represented a recognition site for BsrI). Availability of parental genotype in all cases facilitated error checking by examination of Mendelian inheritance. To detect markers likely to contain a larger number of undetected genotyping errors, the distribution of genotypes in pseudocontrols (created by combining the parental alleles not transmitted to the first child) was tested for deviation from Hardy-Weinberg equilibrium by an exact test (Weir 1996). For both samples and all markers, the resulting P values were >.05.
The frequency of the rare allele in pseudocontrols is given for each polymorphism in table 1. This approach is the preferred method of estimating population allele frequencies from a sample of families ascertained through an affected child. Similar data do not exist from the study of Straub et al. (2002a). It should be noted that apparent differences in identities of the common and rare nucleotides that appear to exist between the report of Straub et al. (2002a) and the present report are due to labeling errors. These errors have been corrected in an erratum (Straub et al. 2002b). According to the authors, these labeling errors had no effect on the statistical results or their interpretation. For the purpose of comparison with the frequencies calculated by Straub et al. (2002a), table 1 also contains the frequency of the rare allele estimated in the parents of our samples (independent of transmission). The rare allele occurs for all six SNP markers in both samples at a higher frequency than in the sample of Irish families reported by Straub et al. (2002a). For markers P1325 and P1578, the values reported by Straub et al. (2002a) were within the CI (.08 – .124 and .062 – .103, respectively) and, therefore, were not significantly different. For the other marker, the level of statistical significance of the difference could not be estimated, since no information about the precision of estimates was available for the Straub study (Straub et al. 2002a).
The more frequent variant of each marker was always preferentially transmitted, as shown in table 1, by the ratio of number of transmissions of allele 1 to the number of transmissions of allele 2 (T/U), with one exception: marker P1578 in the sib-pair sample. Under a multiplicative model, the transmission ratio T/U (table 1) is an estimator of genotype relative risk (Altshuler et al. 2000). Highest estimated relative risk ratio was detected for marker P1325 in the triads (T/U=2.25). To assess the significance of the observed transmission disequilibrium in the sample of triads, the transmission/disequilibrium test (TDT) was used as originally proposed by Spielman et al. (1993). Since the TDT is not a valid test for the null hypothesis of no association in the presence of linkage when applied to a sample of families with more than one affected offspring, P values for the sib-pair sample were calculated by an extension of the method of Zhao et al. (2000). In brief, P values were calculated on the basis of simulated samples. Each simulated sample is obtained either by randomly assigning to all offspring, with equal chance, the observed genotypes or by randomly assigning to all offspring the nontransmitted genotypes at all sites. The test statistic is recomputed for each simulated sample, and the fraction of simulations resulting in a test statistic greater than or equal to the observed statistic gives the empirical P value. One polymorphism (P1320) was significantly associated with schizophrenia in both samples (sib pairs: P=.021; triads: P=.026). Since the more frequent allele showed excess transmission in the triads as well as in the sib pairs, evidence of association increased when both samples were combined. In the combined sample, the P value reached .00068 (estimated on the basis of R=200,000 simulated samples) for P1320. For four additional markers, a P value of <.05 was observed. Since the six families from Israel contributed to the linkage finding, the entire sample was used for the present association study. However, since these families may represent a different ethnic background, we estimated transmission of alleles without them in the sib-pair and in the combined sample. Lower P values were obtained for all markers, in particular for marker P1765 (sib pairs: P=.007; combined sample: P=.0014) and marker P1320 (sib pairs: P=.005; combined sample: P=.0002). This suggests that these families do not contribute to the association finding. The low number of families precluded testing this sample separately for association.
For haplotype analysis, we calculated first the standardized linkage disequilibrium coefficient D′ (Lewontin 1988) for each pair of markers on the basis of estimates for the frequencies of nontransmitted parental two-locus haplotypes. In accordance with the data reported by Straub et al. (2002a), we observed the smallest D′ for marker pair P1325–P1320. For the remaining pairs of markers, D′ values ranged between .55 and 1.00 (data not shown) and indicated very strong linkage disequilibrium between the markers.
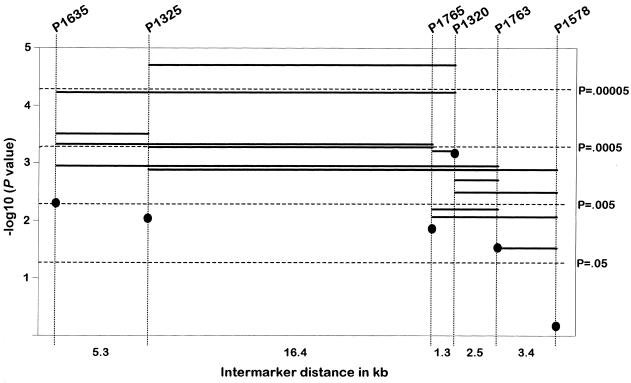
All combinations of up to five markers were tested in the sib-pair sample, the sample of triads, and the combined sample for association with schizophrenia by the extension of Zhao et al. (2000), as described above. The smallest P values for haplotypes with two markers were observed for P1325–P1320 (triads: P=.00083 [R=200,000]; sib pairs: P=.00925 [R=20,000]; sib pairs without Israeli families: P=.0012; combined: P=.00002 [R=200,000]; combined sample without Israeli families: none of the 200,000 replicated samples resulted in a test statistic greater than or equal to the observed statistic). The SD of an empirical P value based on 200,000 replications is ∼2×10-5 when the true P value is <10-4. Therefore, the true P value corresponding to an empirical P value of .00002 is almost certainly <.0001. Even if we take into account that a total of 186 tests was performed, an empirical P value of .00002 remains significant after a very crude and conservative correction for the variation of empirical P values and for multiple testing. For the combined sample, all P values corresponding to haplotypes consisting of adjacent markers are depicted graphically in figure 1. Lowest P values are obtained for haplotypes containing the marker P1325.
Figure 1.
Association of single markers (filled circles), 2-, 3-, 4-, and 5-locus haplotypes (represented by lines between the markers) in the combined sample of 203 families with schizophrenia.
Six-locus haplotype frequencies were calculated by expressing the likelihood of the whole family as a function of parameters (haplotype frequencies) but allowing for different frequencies of haplotypes being transmitted or being nontransmitted to the first (or second) affected offspring in the family. The maximum-likelihood estimates of haplotypes with a frequency ⩾1% are shown in table 2. Several points are evident from this table. First, >97% of the distribution is concentrated in only seven different haplotypes. Second, the frequency estimates obtained for the sample of triads and the sib-pair sample are very similar. Interchanging the first child with the second affected child in the sib-pair sample showed nearly no effect on the resulting frequency estimates. Third, there is one very common haplotype, which is composed of the common allele at each locus. This haplotype is the only haplotype that occurs at a larger frequency in transmitted than in nontransmitted haplotypes. Although this observation is completely consistent with the pattern of single-marker results obtained for our samples (table 1), it is in contrast to the results reported by Straub et al. (2002a) for the Irish families in which the rare allele was preferentially transmitted for each of the six markers. These differences appeared to persist after correction of errors in allele-frequency identity by Straub et al. (2002b). It suggests that genetic variants conferring risk of schizophrenia are inherited in the Irish families on a different haplotype. Thus, assumption of different mutation events is required to explain the differences between the association results reported by Straub et al. (2002a) and those in this report. Identification of the variants conferring risk for the disorder may clarify this point. Association of different mutations with different haplotypes is well known for monogenic disorders like cystic fibrosis (Sereth et al. 1993), Wilson disease (Thomas et al. 1994, 1995), and breast cancer (Meindl 2002). Association reflecting linkage disequilibrium with common alleles also has been reported recently for association of variants in the ADAM33 gene with asthma and bronchial hyperresponsiveness (Van Eerdewegh et al. 2002).
Table 2.
Haplotype Frequencies for the Six-Marker Haplotype
|
Frequency in |
||||||
| Sib Pairs |
||||||
| Triads |
FirstAffected |
SecondAffected |
||||
| Haplotypea | Tb | NTc | Tb | NTc | Tb | NTc |
| 1 1 1 1 1 1 | .738 | .594 | .731 | .566 | .736 | .560 |
| 1 1 1 2 1 1 | .016 | .021 | .019 | .039 | .013 | .045 |
| 1 1 2 2 2 2 | .071 | .086 | .051 | .065 | .045 | .071 |
| 1 2 1 1 1 1 | .061 | .103 | .083 | .098 | .077 | .104 |
| 1 2 1 2 1 1 | .012 | .013 | ||||
| 2 1 2 1 2 1 | .012 | .013 | .032 | .038 | ||
| 2 1 2 2 2 1 | .090 |
.143 |
.090 |
.175 |
.115 |
.149 |
| Sum | .988 | .959 | .987 | .974 | .986 | .980 |
Order of markers: P1635, P1325, P1765, P1320, P1763, P1578; allele coding: 1 = common allele, 2 = rare (polymorphisms as in table 1).
Frequency in transmitted haplotypes.
Frequency in nontransmitted haplotypes.
In conclusion, the intention of our study was to evaluate the association reported by Straub et al. (2002a) in a family sample that showed evidence for linkage in the same region and can be considered, with the exception of six Israeli families, as having an ethnic background similar to the Irish families. We detected strong evidence for association of the same polymorphisms in the dysbindin gene with schizophrenia, but for different alleles, suggesting a population-specific susceptibility allele in the Irish families. In contrast to case-control studies, the family-based design used in our study protects against an increased rate of false-positive associations due to population stratification, it further facilitates haplotype analysis by providing more information on phase, and it aids in detecting typing errors by the possibility to test for Mendelian inheritance. Despite the strong evidence we provide for the existence of susceptibility alleles in this region, this statistical evidence has to be confirmed by identification of the alleles conferring susceptibility for the disease.
Acknowledgments
We thank all patients and their family members; without their cooperation, this work would not have been possible. We gratefully acknowledge information provided by R. E. Straub and information and supply of oligonucleotide primers by B. P. Riley and K. S. Kendler (prior publication). For contribution of clinical information and blood samples, we are grateful to M. Gross, T. G. Schulze, D. Müller, and P. Falkai and to C. Hanses, V. Guttenthaler, and A. Lenzen for technical assistance. The study was supported by Deutsche Forschungsgemeinschaft Sonderforschungsbereich SFB400 (to W.M. and D.B.W.), the German-Israeli Foundation for Scientific Research (to B.L. and D.B.W.), and Nationales Genomforschungsnetz, Germany (to D.B.W.). M.K. is supported by grant KN 378/1-1 (Project D1 of FOR 423) from the Deutsche Forschungsgemeinschaft.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- National Center for Biotechnology Information (NCBI), Single Nucleotide Polymorphism, http://www.ncbi.nlm.nih.gov/SNP/ (for reference identification numbers for SNPs)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SCZD3 [MIM 600511])
References
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES (2000) The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26:76–80 [DOI] [PubMed] [Google Scholar]
- Benson MA, Newey SE, Martin-Rendon E, Hawkes R, Blake DJ (2001) Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J Biol Chem 276:24232–24241 [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Honer WG, Chow EW, Hogan J, Hodgkinson K, Bassett AS (1997) Use of a quantitative trait to map a locus associated with severity of positive symptoms in familial schizophrenia to chromosome 6p. Am J Hum Genet 61:1388–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Levine L, Kwok PY (1999) Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res 9:492–498 [PMC free article] [PubMed] [Google Scholar]
- Hwu HG, Lin MW, Lee PC, Lee SF, Ou-Yang WC, Liu CM (2000) Evaluation of linkage of markers on chromosome 6p with schizophrenia in Taiwanese families. Am J Med Genet 96:74–78 [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC (1988) On measures of gametic disequilibrium. Genetics 120:849–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl A (2002) Comprehensive analysis of 989 patients with breast or ovarian cancer provides BRCA1 and BRCA2 mutation profiles and frequencies for the German population. Int J Cancer 97:472–480 [DOI] [PubMed] [Google Scholar]
- Moises HW, Yang L, Kristbjarnarson H, Wiese C, Byerley W, Macciardi F, Arolt V, et al (1995) An international two-stage genome-wide search for schizophrenia susceptibility genes. Nat Genet 11:321–324 [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Foroud T (1999) Chromosome 6 workshop report. Am J Med Genet 88:233–238 [DOI] [PubMed] [Google Scholar]
- Schizophrenia Linkage Collaborative Group for Chromosomes 3, 6 and 8 (1996) Additional support for schizophrenia linkage on chromosomes 6 and 8: a multicenter study. Am J Med Genet 67:580–594 [DOI] [PubMed] [Google Scholar]
- Schwab SG, Albus M, Hallmayer J, Honig S, Borrmann M, Lichtermann D, Ebstein RP, Ackenheil M, Lerer B, Risch N, Maier W, Wildenauer DB (1995) Evaluation of a susceptibility gene for schizophrenia on chromosome 6p by multipoint affected sib-pair linkage analysis. Nat Genet 11:325–327 [DOI] [PubMed] [Google Scholar]
- Sereth H, Shoshani T, Bashan N, Kerem BS (1993) Extended haplotype analysis of cystic fibrosis mutations and its implications for the selective advantage hypothesis. Hum Genet 92:289–295 [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–516 [PMC free article] [PubMed] [Google Scholar]
- Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, Cesare AJ, Gibberman A, Wang X, O'Neill FA, Walsh D, Kendler KS (2002a) Genetic Variation in the 6p22.3 Gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet 71:337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2002b) Erratum. Am J Hum Genet 71:1007 [Google Scholar]
- Straub RE, MacLean CJ, O'Neill FA, Burke J, Murphy B, Duke F, Shinkwin R, Webb BT, Zhang J, Walsh D, Kendler KS (1995) A potential vulnerability locus for schizophrenia on chromosome 6p24–22: evidence for genetic heterogeneity. Nat Genet 11:287–293 [DOI] [PubMed] [Google Scholar]
- Thomas GR, Bull PC, Roberts EA, Walshe JM, Cox DW (1994) Haplotype studies in Wilson disease. Am J Hum Genet 54:71–78 [PMC free article] [PubMed] [Google Scholar]
- Thomas GR, Roberts EA, Walshe JM, Cox DW (1995) Haplotypes and mutations in Wilson disease. Am J Hum Genet 56:1315–1319 [PMC free article] [PubMed] [Google Scholar]
- Turecki G, Rouleau GA, Joober R, Mari J, Morgan K (1997) Schizophrenia and chromosome 6p. Am J Med Genet 74:195–198 [PubMed] [Google Scholar]
- Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, Torrey D, et al (2002) Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature 418:426–430 [DOI] [PubMed] [Google Scholar]
- Wang S, Sun CE, Walczak CA, Ziegle JS, Kipps BR, Goldin LR, Diehl SR (1995) Evidence for a susceptibility locus for schizophrenia on chromosome 6pter–p22. Nat Genet 10:41–46 [DOI] [PubMed] [Google Scholar]
- Weir BS (1996) Genetic data analysis II. Sinauer Associates, Sunderland, MA [Google Scholar]
- Zhao H, Zhang S, Merikangas KR, Trixler M, Wildenauer DB, Sun F, Kidd KK (2000) Transmission/disequilibrium tests using multiple tightly linked markers. Am J Hum Genet 67:936–946 [DOI] [PMC free article] [PubMed] [Google Scholar]