Abstract
The major challenge in cancer therapy is to efficiently translocate drug molecules into cancer tumors without doing any damage to healthy tissues. Since there exist pH gradients between tumor and normal tissues, pH-sensitive materials may have great potential to overcome such challenge. Here, we report one new type of pH-responsive drug delivery system where pH-sensitive polymers are introduced to control the cellular uptake of nanoparticles under different pH environments through dissipative particle dynamics simulations. Interestingly, the behavior of cellular uptake of nanoparticles here exhibits “smart” pH-responsive properties: for lower and higher pH, the nanoparticles can be taken up by cell membranes, while for pH in middle range, the endocytosis is blocked. Further, it is found that receptor-ligand interactions as well as surface charge property of nanoparticles and membranes can also have important impacts on the endocytosis. The present study may give some significant insights into future stimulus-responsive medical materials design.
Efficient delivery of nanoparticles into cell interiors is of great importance in biomedicine1,2. For example, how to efficiently transport drug molecules into cancer tumors without doing any damage to healthy tissues is still a huge challenge in cancer therapy3. Recently, the pH-responsive materials which can exhibit a sharp change in physicochemical properties under different pH environments have shown a great potential to realize such functions4,5,6, because some solid tumor tissues form a mildly acidic environment (pH ~ 6.5), lower than that in the normal tissues and blood (pH ~ 7.4)4,5,6.
However, there is still little knowledge about the cellular uptake of nanoparticles, especially for how nanoparticles translocate across cell membranes under different external pH. Although some previous experimental studies have shown that the change of pH may be utilized to transport nanoparticles into targeted tissues7,8,9,10, it is rather difficult to systematically probe and visualize the endocytosis process under various conditions, due to available experimental technologies. Furthermore, the in vivo and in vitro efficiencies are still low because there exist lots of barriers for tumor drug delivery11. It is therefore essential to have a thorough understanding of the detailed molecular mechanism of the translocation process under different pH environments. Moreover, since the physicochemical properties of nanoparticle and the ligands on its surface may have important impacts on the cellular uptake of nanomaterials12,13,14,15,16,17,18, can we design any new types of stimulus materials to make the best use of the variable properties of nanomaterials and well control the endocytosis process under different environments?
In this study, we undergo the first computational study to design such type of pH-sensitive nanomaterials (i.e., nanoparticle-polymers complexes) by using dissipative particle dynamics (DPD) simulations19,20. Differing from the main methods in experiments where nanoparticle (or/and its covalent ligands) may show different charge properties under different pH environments5,6,7,8,9, the key idea here is to use external pH-sensitive polymers to well control the cellular uptake of nanoparticles. As we will show below, the uptake behaviors here can show multiply pH-responsive, which may have some advantages over previous designed materials in real applications. Further, the effect of properties of nanoparticles, ligands and membranes on the cellular uptake will also be studied.
Results
Idea of pH-sensitive nanoparticle-polymers complex design
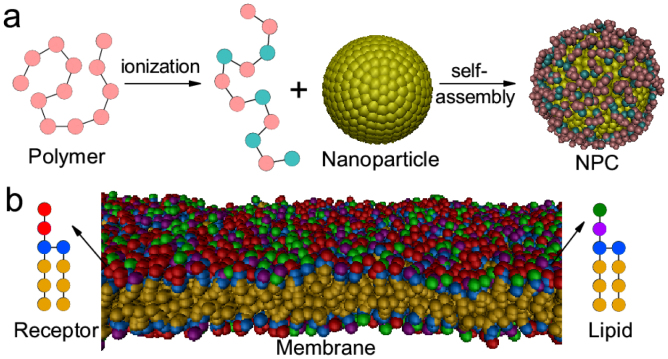
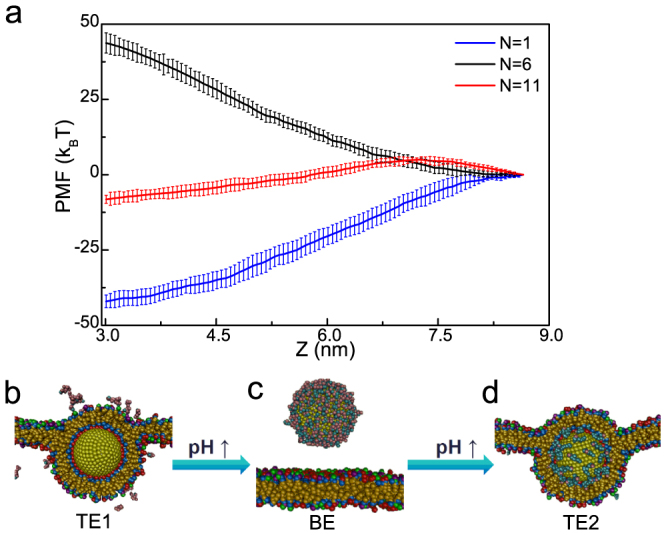
Figure 1 shows the coarse-grained models of different components in our simulations. The nanoparticle-polymers complex (NPC) is composed of one nanoparticle and some pH-sensitive polymers (Fig. 1a), and can be spontaneously formed due to electrostatic interactions when placing the nanoparticle in the polymer solutions (see Supplementary Fig. S1a). The nanoparticle is fabricated by arranging hydrophilic DPD beads (P) on a fcc lattice with lattice constant α = 0.40 nm into a desired geometry shape and volume, and all beads comprising a nanoparticle move as a rigid body21,22. The surface beads on the particle are treated as ligand ones17,23. Each surface bead carries a charge of +e (the surface charge density is set as σP = 2.5 e/nm2 unless otherwise stated), and its ionization degree is kept 100% in the simulations (some chemical group like 1,8-Bis(dimethylamino)naphthalene24 may be used to reach such demand when pH is not very large). The pH-sensitive polymer is composed of twelve hydrophilic beads (C), and some beads may carry a charge of −e, which depends on the system's pH value (for real applications, many chemical groups like carboxyl group can be utilized in the pH-sensitive polymer design25). For the simplicity, we use the relationship between the fraction (θ) of charged polymer segments and external pH as follows26,27: 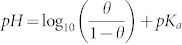 , where pKa is the acidity constant of the polymer. Note that the charged monomers are randomly generated, but with a fixed charge number N per polymer (N = 12θ in a fixed polymer length of 12) during the simulations27. By doing some simple transformation, we have
, where pKa is the acidity constant of the polymer. Note that the charged monomers are randomly generated, but with a fixed charge number N per polymer (N = 12θ in a fixed polymer length of 12) during the simulations27. By doing some simple transformation, we have 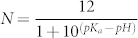 . From this equation, we can easily find that N increases with the increase of pH.
. From this equation, we can easily find that N increases with the increase of pH.
Figure 1. Schematic illustration of the models in the simulations.
(a) Snapshot of the nanoparticle-polymers complex (NPC, i.e., the nanoparticle with pH-sensitive polymers assembling onto its surface); (b) Snapshot of membranes and architecture of lipids and receptors. Green bead represents lipid head containing +e, purple bead is lipid head containing –e, while blue bead stands for lipid head with no charge; the orange bead represents lipid tail, the red bead stands for receptor head bead, yellow bead is particle bead, and the polymers are formed of the cyan beads (with charge –e) and pink beads.
Each amphiphilic lipid consists of a headgroup containing four connected hydrophilic beads (H) and two tails with respective three hydrophobic beads (T) (Fig. 1b)28,29,30,31. The first head bead carries a charge of +e, while the second head bead carries a charge of −e; the remaining two beads are uncharged32. Particularly, when modeling the negatively charged lipids, non-charged hydrophilic bead is used to take place of the first positive charged bead in lipid molecule. The receptor molecule has the same conformation of lipid molecules17,23,33, but its first two head beads (R) are uncharged and can interact with the ligand bead (L) via soft Lennard-Jones (LJ) potentials22. When lipids and receptors are immersed in the water, they can form a stable membrane (see Supplementary Fig. S1b for the density profile of all types of beads). Additionally, the percent of the receptors in the membrane is set to be 50%17,23,33, and the nanoparticle radius is fixed as 4 nm.
Endocytosis of nanoparticle-polymers complex under different external pH
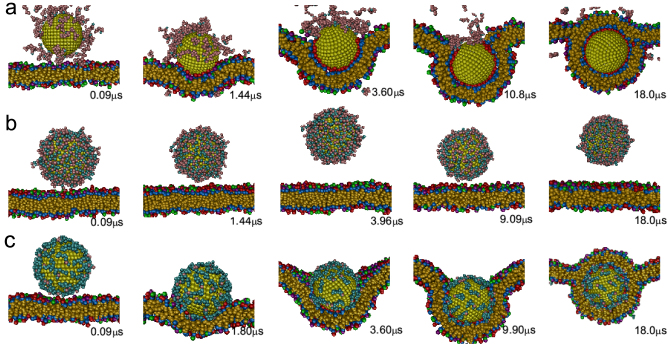
As shown in Fig. 2, there exist big differences in the endocytosis process under different pH values — the endocytosis of nanoparticle will be blocked by the associated polymers on its surface under middle-range pH, whereas the nanoparticle can be fully engulfed by the membrane under low and high pH.
Figure 2. Time sequence of the snapshots of interactions between the nanoparticle and membranes in the presence of pH-sensitive polymers under different ionized degrees.
(a) N = 1 (i.e., pH = pKa − 1.04); (b) N = 6 (i.e., pH = pKa); (c) N = 11 (i.e., pH = pKa + 1.04).
When pH is low (pH ≤ pKa − 0.48, i.e., N ≤ 3), the number of ionized monomers is small. As a result, the adsorption of polymers on the particle surface is so weak that the polymers cannot prevent the strong attraction between the receptors on the membrane and ligands on the nanoparticle surface. Therefore, the polymers will detach from the nanoparticle surface with the increase of the engulfment degree. Finally, the particle can be totally engulfed by the membrane, with no polymers on its surface (Fig. 2a).
When pH is in middle range (pKa − 0.30 ≤ pH ≤ pKa + 0.30, i.e., 4 ≤ N ≤ 8), there are several monomers ionized in each polymer, so the polymers can absorb on the nanoparticle surface stably. Additionally, since the electrostatic repulsion between adsorbed polymers is not sufficiently strong, the number of adsorbed polymers can be very large (see Supplementary Fig. S2). As a result, the nanoparticle is totally surrounded by the polymers. Given that the receptor-ligand interaction is short-ranged, the receptors on the membrane cannot interact with the ligands on the particle surface. Therefore, in this situation the NPC will just weakly attach to or get away from the membrane (Fig. 2b).
When pH is high (pH ≥ pKa + 0.48, i.e., N ≥ 9), the number of ionized monomers becomes large, which makes the polymers strongly absorbed onto the nanoparticle surface. However, because of the strong electrostatic repulsion between charged polymer monomers themselves, the number of adsorbed polymers is small. As a result, the polymers cannot totally prevent the nanoparticle from receptors so that the nanoparticle can also attach onto the membrane and then be engulfed by cell membranes. However, differing from the former case of low pH, there still exist some polymers on the nanoparticle surface after it is totally engulfed (Fig. 2c). This is because in this case the electrostatic interaction becomes strong (which is comparable to the receptor-ligand interaction) so that the polymers are very hard to detach from the nanoparticle surface.
To obtain further insight of the three different endocytosis processes, we calculate the potential of mean force (PMF) of them along the membrane-normal direction by using steered molecular dynamics (SMD)34,35. Figure 3a shows the PMF as a function of the distance (Z) of particle from the central plane of the bilayer when N = 1, 6, and 11, corresponding to three different engulfment modes shown in Figs. 3b–d, respectively. For N = 1, as the nanoparticle approaches the membrane, the PMF monotonously decreases and there is no energy barrier in the PMF curve in this case, indicating that the endocytosis of nanoparticle is spontaneous. While for N = 6, the PMF monotonously increases with the decrease of the distance (Z) between nanoparticle and membranes, so the engulfment of particles is impossible in this case. When N is 11, the PMF firstly increases with the decrease of Z, which is due to small deformation of membranes and weak repulsive interactions between polymers and membranes. Nevertheless, when the NPC is closer to the membrane, the PMF decreases because of the strong attractive receptor-ligand interactions. The energy barrier in this case is just about 5 kBT, and could be overcome only by thermal motions, therefore the NPC may also be engulfed by membranes.
Figure 3. Comparison of the three different interaction modes.
(a) Potential of mean force (PMF) of nanoparticles as a function of its distance (Z) from the central plane of the lipid bilayer in three different modes. (b)–(d) show the snapshots of final equilibrium of interactions between NPC and membranes under different external pH: (b) N = 1, (c) N = 6, and (d) N = 11. Error bars are obtained by taking the standard derivation of ten independent simulations. In order to distinguish the three modes, we name them as total engulfment type 1 (TE1), blocked engulfment (BE), and total engulfment type 2 (TE2), respectively.
Effect of receptor-ligand interaction as well as charge property of nanoparticle and lipid membrane on the endocytosis
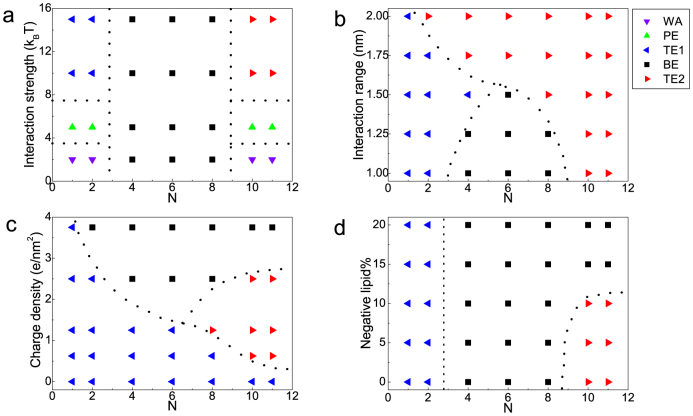
To deeply understand the mechanism of the endocytosis of nanoparticles in the presence of pH-sensitive polymers, we further study other factors on the nanoparticle internalization. As discussed above, there may exist three different phases (i.e., TE1, BE, and TE2) in the interactions between nanoparticles and membranes. Additionally, another two phases including weak absorption (WA) and partially engulfment (PE) can also occur when the receptor-ligand interaction is not strong enough according to previous studies17,23. As a result, here there are five different phases. Note that the difference between TE1 and TE2 is whether there exist polymers on the nanoparticle surface.
As shown in Fig. 4a, when the receptor-ligand interaction strength is weak, there will be no obvious engulfment of the NPC. With the increase of the receptor-ligand interaction strength, the NPC will just be partially engulfed by membranes and when the strength is strong enough, the NPC can be totally engulfed under both low and high pH conditions. However, when N is in the range of 4–8 (i.e., pH is in [pKa − 0.3, pKa + 0.3]), the engulfment will be blocked because the ligand can not interact with receptors, even for stronger receptor-ligand interaction. Therefore, the increase of interaction strength in this case has little effect on the nanoparticle internalization.
Figure 4. Phase diagrams describing the equilibrium state of the particle-membrane systems.
(a) the receptor-ligand interaction strength verse the ionized degree N, where rcut = 1.0 nm and σP = 2.5 e/nm2; (b) the receptor-ligand interaction range verse the ionized degree N, where 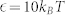 and σP = 2.5 e/nm2; (c) the surface charge density verse the ionized degree N, where rcut = 1.0 nm and
and σP = 2.5 e/nm2; (c) the surface charge density verse the ionized degree N, where rcut = 1.0 nm and 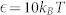 . In (a)–(c), the membrane is neutrally charged. (d) Negative lipid fraction (i.e., the number of negative lipids/the total number of lipids and receptors) verse the ionized degree N, where
. In (a)–(c), the membrane is neutrally charged. (d) Negative lipid fraction (i.e., the number of negative lipids/the total number of lipids and receptors) verse the ionized degree N, where 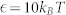 , σP = 2.5 e/nm2, and rcut = 1.0 nm.
, σP = 2.5 e/nm2, and rcut = 1.0 nm.
Besides the interaction strength, the range of the receptor-ligand interaction is also a characteristic quantity36,37, which may have significant effects on the cellular internalization of nanoparticles. As shown in Fig. 4b, with the increase of the interaction range, the change of the phases in low and high pH is not obvious except that the phase (N = 2, rcut = 2.0 nm) changes from TE1 to TE2 (the reason is that when the interaction range is large, the ligand on the nanoparticle surface can still interact with receptors on the membrane even if some polymers attach to the nanoparticle). Importantly, the NPC can be engulfed by membranes when the ionized number is in the range of 4–8 because the polymers on the particle fail to protect the “longer” range interactions between ligands and receptors. Here the final equilibrium is also TE2, based on the same reason discussed above. In this sense, the receptor-ligand interaction range may be more important for cellular uptake of nanoparticles than the interaction strength.
Further, we examine the effect of surface charge density of nanoparticle on the internalization. As shown in Fig. 4c, when the surface charge density is zero, the polymers do not attach onto the nanoparticle due to lack of electrostatic attractive interactions. As a result, the engulfment of nanoparticle will not be affected by polymers, and the nanoparticle can be totally engulfed with no polymers on its surface (TE1). With the increase of surface charge density, the phases in high pH case will begin to change from TE1 to TE2, because in that case the polymers carrying more charges will not detach from the nanoparticle surface during the engulfment. As the charge density increases, more polymers will absorb onto the nanoparticle surface so that the engulfment of nanoparticle will be blocked. Even if the ionized number N is small (e.g., N = 2, σP = 3.75 e/nm2), the block could also happen when the charge density is large enough. In addition, in the case of high pH and high surface charge density, since the electrostatic repulsion between highly charged polymers become less important (compared to the electrostatic attraction between nanoparticle and polymers), more polymers can attach onto the nanoparticle surface to prevent the interaction between nanoparticles and membranes. As a result, under that condition (e.g., N = 10, σP = 3.75 e/nm2), the engulfment will also be blocked.
Since there are more anionic molecules (e.g., sialic acid) abundant on the surface of cancer cells as compared to normal cells38, it is very important to study the effect of membrane charge property on the endocytosis. For the sake of the simplicity, here we just use the negatively charged lipids to model the anionic molecules in the membrane39. When N ≤ 2, the zeta potential(ζ)40,41 of NPC is above 40 mV (see Supplementary Fig. S3), so there will exist the attractive electrostatic interaction between the NPC and negative membranes, which can help the cellular uptake process. Thus the final state under low pH will still be total engulfment. With the increase of N, ζ decreases largely (when N = 4, ζ is about 5 mV, and when N ≥ 6, ζ is below −30 mV, see Supplementary Fig. S3). As a result, the attractive electrostatic interaction between the NPC and membranes becomes very weak when N = 4 so that it will not affect the endocytosis process. While when N ≥ 6, the electrostatic interaction becomes repulsive, which will greatly prevent the endocytosis, so the final state in middle pH still is blocked engulfment. Generally, as shown in Fig. 4d, the negative membrane has little impact on the phase behaviors under low and middle pH. On the contrary, since the NPC under high pH is negative-charged, when the membrane carries more negative charges, the repulsive electrostatic interactions will become very strong, which can induce the failure of the adsorption of the NPC onto cell membranes. Therefore, the endocytosis will be blocked in this situation (Fig. 4d).
Discussion
In the present study, we report one new type of pH-responsive drug delivery system with the addition of pH-sensitive polymers for controlling the cellular uptake of nanoparticles under different pH environments. Importantly, as summarized in Figs. 3b–d, the endocytosis process shows triple-pH-responsive, i.e., under lower and higher pH conditions, the particles can be engulfed by cell membranes, while in the middle range of pH, the endocytosis process is blocked. This triple-pH-responsive property is quite different from those reported by previous studies7,8,9,10 and may have some advantages over them in multi-pH-responsive process. For example, the designed NPC with triple-pH-sensitive property may have the capability to distinguish the multi tissues of various pH values. It is known that there may exist pH gradient between different tumor types42 (e.g., pH in sarcomas is about 7.0 while in astrocytomas it is about 6.5). In this case, we can choose some type of polymers (e.g., PAMAM-PEG-mannose43) with pKa of about 7.0. When the environmental pH is about 6.5 (N ~ 2 – 3) or 7.5(N ~ 9 – 10), the nanoparticle (e.g., quantum dots) can be well translocated into cell interiors, whereas in the case of pH of about 7.0 (N ~ 6), fewer nanoparticles are taken up by cells. Therefore, there exist big differences of fluorescence intensity, which can distinguish not only the normal tissues from cancer tumors but also different tumors.
Furthermore, the NPC can be also used in drug delivery for cancer therapy, where the polymer with pKa of about 7.5 could be chosen (e.g., PLGA-b-PBLG25). When pH is about 6.5 (N ~ 1, typical cancer cell environment), the cellular uptake of particles can be very high, whereas the uptake will become low under pH of about 7.5 (N ~ 6) for normal cell, which can make the best use of drugs. Moreover, since here the uptake is low in normal cells, this may also decrease its potential toxicity44. Thus we believe that the high targeted efficiency as well as the low nanotoxicity will make it a good type of drug carriers for cancer therapy.
Finally, we point out the feasibility and application of our reported drug delivery system in real experiments. To some extent, the present drug delivery system may be just suitable for in vitro experiment. While for in vivo experiment, since there exist other charged macromolecules45 which could also affect the cellular uptake, it becomes very complicated in that situation and is beyond the present system. Nevertheless, if we treat the charged polymers in our system as external charged macromolecules (e.g., proteins), the present study can give some useful insights into another important question in drug delivery — how do external macromolecules affect the cellular uptake of nanomaterials? Recently, an experimental study has illustrated that nanoparticles may lose its targeting ability when placed in a complex biological environment46. Here we may give a quantitative theoretical explanation for it, i.e., the adsorption of external macromolecules on nanoparticle surface can suppress the specific interactions between receptors and ligands. Generally, our results show that the cellular uptake of nanoparticles can be well controlled by the properties of nanoparticles, pH-sensitive polymers, cell membranes, and external environment. Therefore, our study can help better understand the physical mechanism of receptor-mediated endocytosis of pH-sensitive nanomaterials, and may provide significant ideas for engineering new types of stimulus-responsive materials in biomedicine.
Methods
We briefly summarize the simulation method used in this work and the details can be found in Supplementary Information. Dissipative particle dynamics (DPD) is a coarse-grained simulation technique with hydrodynamic interaction19. The dynamics of the elementary units which are so-called DPD beads, is governed by Newton's equation of motion. Typically, there are three types of pairwise forces in DPD, i.e., the conservative force, dissipative force, and random force. Here, in order to include electrostatic interaction between charged beads, the Coulomb force is incorporated into our DPD simulations20,27. Further, a modified LJ potential is introduced to mimic the receptor-ligand interactions22. Additionally, we also use a harmonic bond to ensure the integrality of lipids and polymers, and a three-body potential to depict the rigidity of lipid tails28,47. All simulations are performed in the NVT ensembles using the velocity-Verlet integration algorithm. The size of the simulation box is 65rc × 65rc × 40rc with the number density of 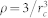 . The integration time step Δt = 36 ps and each simulation time is at least 18.0 μs.
. The integration time step Δt = 36 ps and each simulation time is at least 18.0 μs.
Author Contributions
H.M.D. and Y.Q.M. conceived and designed the simulations. H.M.D. and Y.Q.M. performed the simulations. H.M.D. and Y.Q.M. analysed the data and co-wrote the paper. All authors discussed the results and commented on the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
This work is supported by the National Natural Science Foundation of China (Nos. 91027040 and 31061160496), the National Basic Research Program of China (No.2012CB821500), the Scientific Research Foundation of Graduate School of Nanjing University (No. 2013CL01), and the College Postgraduate Research and Innovation Project of Jiangsu province (No. CXZZ13_0036). We are grateful to the High Performance Computing Center (HPCC) of Nanjing University for doing the numerical calculations in this paper on its IBM Blade cluster system.
References
- Lammers T., Aime S., Hennink W. E., Storm G. & Kiessling F. Theranostic nanomedicine. Acc. Chem. Res. 44, 1029–1038 (2011). [DOI] [PubMed] [Google Scholar]
- Chou L. Y. T., Ming K. & Chan W. C. W. Strategies for the intracellular delivery of nanoparticles. Chem. Soc. Rev. 40, 233–245 (2011). [DOI] [PubMed] [Google Scholar]
- Ferrari M. Cancer nanotechnology: Opportunities and challenges. Nat. Rev. Cancer 5, 161–171 (2005). [DOI] [PubMed] [Google Scholar]
- Gil E. S. & Hudson S. M. Stimuli-reponsive polymers and their bioconjugates. Prog. Polym. Sci. 29, 1173–1222 (2004). [Google Scholar]
- Fleige E., Quadir M. A. & Haag R. Stimuli-responsive polymeric nanocarriers for the controlled transport of active compounds: Concepts and applications. Adv. Drug Deliver. Rev. 64, 866–884 (2012). [DOI] [PubMed] [Google Scholar]
- Schmaljohann D. Thermo- and pH-responsive polymers in drug delivery. Adv. Drug Deliver. Rev. 56, 1655–1670 (2006). [DOI] [PubMed] [Google Scholar]
- Kim D., Lee E. S., Oh K. T., Gao Z. G. & Bae Y. H. Doxorubicin-loaded polymeric micelle overcomes multidrug resistance of cancer by double-targeting folate receptor and early endosomal pH. Small 4, 2043–2050 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. Z. et al. Sheddable ternary nanoparticles for tumor acidity-targeted siRNA delivery. ACS Nano 6, 771–781 (2012). [DOI] [PubMed] [Google Scholar]
- Su J., Chen F., Cryns V. L. & Messersmith P. B. Catechol polymers for pH-responsive, targeted drug delivery to cancer cells. J. Am. Chem. Soc. 133, 11850–11853 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo R. et al. Multistage pH-responsive liposomes for mitochondrial-targeted anticancer drug delivery. Adv. Mater. 24, 3659–3665 (2012). [DOI] [PubMed] [Google Scholar]
- Grossman J. H. & McNeil S. E. Nanotechnology in cancer medicine. Phys. Today 65, 38–42 (2012). [Google Scholar]
- Nel A. E. et al. Understanding biophysicochemical interactions at the nano-Bio interface. Nat. Mater. 8, 543–557 (2009). [DOI] [PubMed] [Google Scholar]
- Mitragotri S. & Lahann J. Physical approaches to biomaterial design. Nat. Mater. 8, 15–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. L., Li J., Lykotrafitis G., Bao G. & Suresh S. Size-dependent endocytosis of nanoparticles. Adv. Mater. 21, 419–424 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H. J., Shi W. D. & Freund L. B. Mechanics of receptor-mediated endocytosis. Proc. Natl. Acad. Sci. U. S. A. 102, 9469–9474 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chithrani B. D., Ghazani A. A. & Chan W. C. W. Determining the size and shape denendence of gold nanoparticle uptake into mammalian cells. Nano Lett. 6, 662–668 (2006). [DOI] [PubMed] [Google Scholar]
- Ding H. M. & Ma Y. Q. Role of physicochemical properties of coating ligands in receptor-mediated endocytosis of nanoparticles. Biomaterials 33, 5798–5802 (2012). [DOI] [PubMed] [Google Scholar]
- Verma A. & Stellacci F. Effect of surface properties on nanoparticle-cell interactions. Small 6, 12–21 (2010). [DOI] [PubMed] [Google Scholar]
- Groot R. D. & Warren P. B. Dissipative particle dynamics: Bridging the gap between atomistic and mesoscopic simulations. J. Chem. Phys. 107, 4423–4435 (1997). [Google Scholar]
- Groot R. D. Electrostatic interactions in dissipative particle dynamics — simulation of polyelectrolytes and anionic surfactants. J. Chem. Phys. 118, 11265 (2003). [Google Scholar]
- Lin X. B., Wang C. L., Wang M., Fang K. & Gu N. Computer simulation of the effects of nanoparticles' adsorption on the properties of supported lipid bilayer. J. Phys. Chem. C 116, 17960–17968 (2012). [Google Scholar]
- Yang K. & Ma Y. Q. Computer simulation of the translocation of nanoparticles with different shapes across a lipid bilayer. Nat. Nanotechnol. 5, 579–583 (2010). [DOI] [PubMed] [Google Scholar]
- Vácha R., Veracoechea J. M. & Frenkel D. Receptor-mediated endocytosis of nanoparticles of various shapes. Nano Lett. 11, 5391–5395 (2011). [DOI] [PubMed] [Google Scholar]
- Alder R. W., Bowman P. S., Steele W. R. S. & Winterman D. R. The remarkable basicity of 1,8-bis(dimethylamino)naphthalene. Chem. Commun. (London) 723–724 (1968). [Google Scholar]
- Yang Y. N. et al. pH-dependent self-assembly of amphiphilic poly(L-glutamic acid)-block-poly-(lactic-co-glycolic acid) copolymers. Polymer 51, 2676–2682 (2010). [Google Scholar]
- Tagliazucchi M. & Szleifer I. Stimuli-responsive polymers grafted to nanopores and other nano-curved surfaces: structure, chemical equilibrium and transport. Soft Matter 8, 7292–7305 (2012). [Google Scholar]
- Melchor M. G., Mayoral E., Velazquez M. E. & Alejandre J. Electrostatic interactions in dissipative particle dynamics using the Ewald sums. J. Chem. Phys. 125, 224107 (2006). [DOI] [PubMed] [Google Scholar]
- Ding H. M., Tian W. D. & Ma Y. Q. Designing nanoparticle translocation through membranes by computer simulations. ACS Nano 6, 1230–1238 (2012). [DOI] [PubMed] [Google Scholar]
- Li Y. F., Li X. J., Li Z. H. & Gao H. J. Surface-structure-regulated penetration of nanoparticles across a cell membrane. Nanoscale 4, 3768–3775 (2012). [DOI] [PubMed] [Google Scholar]
- Yue T. T. & Zhang X. R. Cooperative effect in receptor-mediated endocytosis of multiple nanoparticles. ACS Nano 6, 3196–3205 (2012). [DOI] [PubMed] [Google Scholar]
- Pogodin S., Werner M., Sommer J. & Baulin V. A. Nanoparticle-induced permeability of lipid membranes. ACS Nano 6, 10555–10561 (2012). [DOI] [PubMed] [Google Scholar]
- Izvekov S. & Voth G. A. A multiscale coarse-graining method for biomolecular systems. J. Phys. Chem. B 109, 2469–2473 (2005). [DOI] [PubMed] [Google Scholar]
- Shi X. H., Bussche A. V. D., Hurt R. H., Kane A. B. & Gao H. J. Cell entry of one-dimensional nanomaterials occurs by tip recognition and rotation. Nat. Nanotechnol. 6, 714–719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H. M. & Ma Y. Q. Interactions between Janus particles and membranes. Nanoscale 4, 1116–1122 (2012). [DOI] [PubMed] [Google Scholar]
- Park S. & Schulten K. Calculating potential of mean force from steered molecular dynamics simulations. J. Chem. Phys. 120, 5946–5961 (2004). [DOI] [PubMed] [Google Scholar]
- Bongrand P. Ligand-receptor interactions. Rep. Prog. Phys. 62, 921–968 (1999). [Google Scholar]
- Merkel R., Nassoy P., Leung A., Ritchie K. & Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 397, 50–53 (1999). [DOI] [PubMed] [Google Scholar]
- Riedl S., Zweytick D. & Lohner K. Membrane-active host defense peptides — challenges and perspectives for the development of novel anticancer drugs. Chem. Phys. Lipids 164, 766–781 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. L., Ding H. M. & Ma Y. Q. Translocation of polyarginines and conjugated nanoparticles across asymmetric membranes. Soft Matter 9, 1281–1286 (2013). [Google Scholar]
- Diehl A. & Levin Y. Smoluchowski equation and the colloidal charge reversal. J. Chem. Phys. 125, 054902 (2006). [DOI] [PubMed] [Google Scholar]
- Wang Z. Y. & Ma Y. Q. Insights from Monte Carlo simulations on charge inversion of planar electric double layers in mixtures of asymmetric electrolytes. J. Chem. Phys. 133, 064704 (2010). [DOI] [PubMed] [Google Scholar]
- Griffiths J. R. Are cancer cells acidic. Br. J. Cancer 64, 425–427 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood K. C., Little S. R., Langer R. & Hammond P. T. A family of hierarchically self-assembling linear-dendritic hybrid polymers for highly efficient targeted gene delivery. Angew. Chem. Int. Ed. 44, 6704–6708 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang Y. J. et al. Tuning the autophagy-inducing activity of lanthanide-based nanocrystals through specific surface-coating peptides. Nat. Mater. 11, 817–826 (2012). [DOI] [PubMed] [Google Scholar]
- Lynch I. & Dawson K. A. Protein-nanoparticle interactions. Nano Today 3, 40–47 (2008). [Google Scholar]
- Salvati A. et al. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 8, 137–143 (2013). [DOI] [PubMed] [Google Scholar]
- Alexeev A., Uspal W. E. & Balazs A. C. Harnessing Janus nanoparticles to create controllable pores in membranes. ACS Nano 2, 1117–1122 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information