Abstract
Sirt5, localized in the mitochondria, is a member of sirtuin family of NAD+-dependent deacetylases. Sirt5 was shown to deacetylate and activate carbamoyl phosphate synthase 1. Most recently, Sirt5 was reported to be the predominant protein desuccinylase and demalonylase in the mitochondria because the ablation of Sirt5 enhanced the global succinylation and malonylation of mitochondrial proteins, including many metabolic enzymes. In order to determine the physiological role of Sirt5 in metabolic homeostasis, we generated a germline Sirt5 deficient (Sirt5−/−) mouse model and performed a thorough metabolic characterization of this mouse line. Although a global protein hypersuccinylation and elevated serum ammonia during fasting were observed in our Sirt5−/− mouse model, Sirt5 deficiency did not lead to any overt metabolic abnormalities under either chow or high fat diet conditions. These observations suggest that Sirt5 is likely to be dispensable for the metabolic homeostasis under the basal conditions.
Sirt5 is one of the seven members of the sirtuin family of NAD+-dependent deacetylases. Together with Sirt3 and Sirt4, Sirt5 localizes in the mitochondria1,2. Activation of Sirt1, the founding member of the sirtuin family, has many beneficial effects on energy and metabolic homeostasis, improving health span3,4,5. The role of mitochondrial sirtuins as metabolic sensors in different metabolic pathways is only beginning to be appreciated.
Sirt3 is thought to be the predominant mitochondrial protein deacetylase based on the fact that the absence of Sirt3 in mice, but not Sirt4 or Sirt5, enhances global acetylation of mitochondrial proteins6. Sirt3 regulates the acetylation level and enzymatic activity of a vast set of metabolic enzymes such as long-chain acyl-CoA dehydrogenase, glutamate dehydrogenase (GDH), isocitrate dehydrogenase 2, ornithine transcarbamoylase7,8. In line with the important molecular function of Sirt3, germline Sirt3−/− mice are sensitized to high fat diet (HFD)-induced obesity, hyperlipidemia, insulin resistance, and steatohepatitis9. Despite the fact that liver and skeletal muscle are the most important tissues determining whole body metabolism, specific deletion of Sirt3 in liver or skeletal muscle has almost no effect on the global metabolic homeostasis10, pointing to the importance of other tissues in Sirt3 signaling.
Sirt4 lacks deacetylase activity but instead demonstrates an ADP-ribosyl transferase activity11,12. The ability of Sirt4 to ADP-ribosylate and inactivate GDH indicates the involvement of Sirt4 in glutamine metabolism12. Indeed, Sirt4 expression is suppressed by mammalian target of rapamycin complex 1 (mTORC1), thus activating GDH and promoting glutamine metabolism and cell proliferation in human cancer13,14.
Sirt5 was shown to deacetylate and activate carbamoyl phosphate synthase 1 (CPS1)15, the rate-limiting enzyme catalyzing the first step of the urea cycle for ammonia disposal. The activation of CPS1 in Sirt5−/− mice is impaired during fasting with a consequent accumulation of ammonia in the blood. Sirt5 is furthermore reported to exhibit potent desuccinylase and demalonylase activities16,17. Sirt5 knockout mice show global protein hypersuccinylation and hypermalonylation including hypersuccinylated CPS1, indicating Sirt5 is the major protein desuccinylase and demalonylase in mitochondria. The ability of Sirt5 to remove acyl modifications, including acetylation, succinylation, and malonylation, on lysine residues establishes Sirt5 as a deacylase rather than as a deacetylase. Of note in this context, Sirt6 is recently discovered to efficiently remove long-chain fatty acyls, such as myristoyl groups, from lysine residues18, further supporting the emerging recognition of sirtuins as deacylases.
Malonylation and succinylation were detected in multiple metabolic enzymes in mitochondria16,17. A very recent succinylome study further revealed that Sirt5 desuccinylates a mast set of metabolic enzymes in mitochondria that are involved in amino acid degradation, the tricarboxylic acid (TCA) cycle, and fatty acid metabolism19. All these molecular evidences point toward possible important role of Sirt5 in metabolic homeostasis. Since the physiological role of Sirt5 in metabolic homeostasis at the level of whole body has not been reported, we generated a Sirt5 deficient mouse model and performed a thorough metabolic characterization of this mouse line using a standardized phenotyping protocol20,21. Surprisingly, under the conditions that we studied, deletion of Sirt5 leads only to minor metabolic consequences. The absence of a phenotype under basal conditions hence warrants further study to define the biological role of Sirt5 in challenged conditions where Sirt5 function may be solicited.
Results
Generation and validation of a Sirt5 deficient mouse model
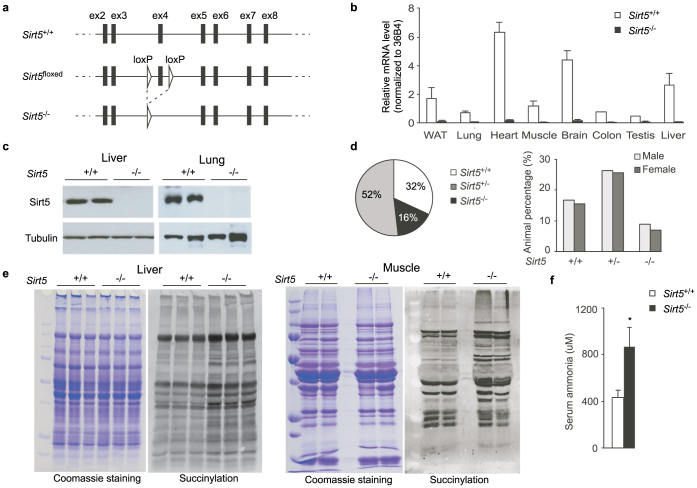
To investigate the impact of Sirt5 on whole body metabolism, we generated a mouse line in which the exon 4 of the Sirt5 locus was flanked with LoxP sites using a standard gene targeting strategy22 (Fig. 1a). The resulting Sirt5floxed mice were bred with CMV-Cre transgenic mice23 to generate germline Sirt5−/− mice. mRNA analysis demonstrated that the Sirt5 gene is successfully inactivated in different tissues in the Sirt5−/− mice (Fig. 1b). Protein analysis further supported a complete disruption of Sirt5 expression in the knockout animals (Fig. 1c). To generate experimental cohorts, Sirt5+/− mice were first backcrossed with C57BL/6J mice to reach 97% background purity. Male and female Sirt5+/− mice were then bred to generate Sirt5+/+ and Sirt5−/− littermates. Of note, the Sirt5−/− mice were not born at the expected Mendelian ratio, although the ratio of male and female Sirt5−/− mice is close to 1:1 (Fig. 1d). The proportion of Sirt5−/− mice was about 60% of what was expected, suggesting that there was prenatal loss of approximately 40% of Sirt5−/− offspring. However, the surviving Sirt5−/− mice appeared generally normal.
Figure 1. Generation and validation of a Sirt5 deficient mouse model.
(a). Targeting strategy for disruption of the mouse Sirt5 gene. LoxP sites were inserted to flank exon 4 of the Sirt5 gene. The resulting Sirt5floxed mice were bred with CMV-Cre transgenic mice to generate Sirt5−/− mice. (b). Sirt5 mRNA levels in selected tissues of Sirt5+/+ and Sirt5−/− mice. Three pairs of 8-week-old wild-wt(wt) and knockout male mice were used for the validation. (c). Sirt5 protein levels in liver and lung of Sirt5+/+ and Sirt5−/− mice. Two pairs of 8-week-old wt and knockout male mice were used for protein analysis. (d). The birth and sex ratio of Sirt5+/+, Sirt5+/−, Sirt5−/− mice. 328 mice were genotyped and analyzed. (e). Global protein succinylation in liver and gastrocnemius muscle. 3 pairs of 12-week-old wt and knockout male mice were fasted for 24 hours and sacrificed for protein analysis. (f). Serum ammonia levels. 12-week-old Sirt5+/+ and Sirt5−/− male mice were fasted for 24 hours and sacrificed. Blood was collected by cardiac puncture. Serum was immediately frozen in liquid nitrogen and used for measurement of ammonia levels. N = 4. * P< 0.05.
We first evaluated global protein succinylation in two major metabolic organs, i.e. the liver and gastrocnemius muscle, of Sirt5+/+ and Sirt5−/− mice. In line with the previous reports demonstrating the robust desuccinylase activity of Sirt516,17,19, various proteins were hypersuccinylated in Sirt5−/− liver and skeletal muscle (Fig. 1e). We further measured serum ammonia levels in Sirt5+/+ and Sirt5−/− littermates after 24-hour of fasting. In agreement with published data showing the involvement of Sirt5 in the control of the urea cycle15, there was a marked increase in blood ammonia level in Sirt5−/− mice during fasting (Fig. 1f).
Metabolic characterization of chow fed Sirt5 deficient mice
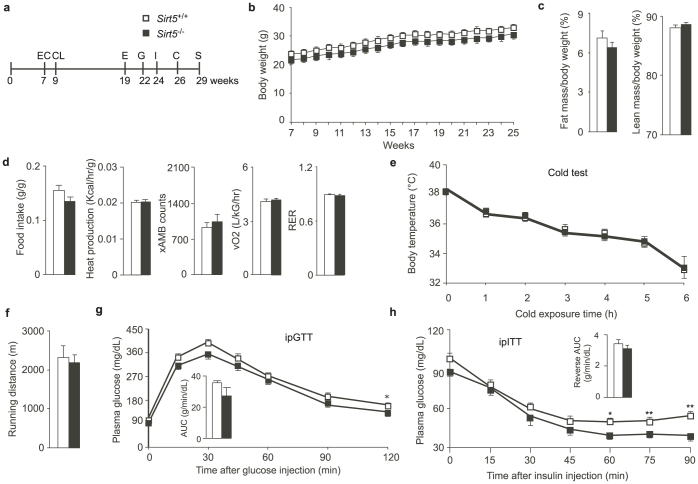
We then subjected chow fed Sirt5−/− male mice to a standardized phenotyping protocol (Fig. 2a). Although the curve of weight gain of the Sirt5−/− mice was continuously lower than that of Sirt5+/+ littermates, the difference was not significant (Fig. 2b). The body composition was similar between Sirt5+/+ and Sirt5−/− littermates (Fig. 2c). Furthermore, during indirect calorimetry using the comprehensive lab animal monitoring system (CLAMS), food intake, body heat production, spontaneous locomotor activity, oxygen consumption, respiratory exchange ratio (RER) were comparable between Sirt5+/+ and Sirt5−/− mice (Fig. 2d). The mice were then subjected to different physiological challenges. The cold test revealed a similar tolerance of Sirt5+/+ and Sirt5−/− mice to the cold (Fig. 2e). In an endurance run, the distance run to exhaustion of Sirt5−/− mice was also similar to that of wt controls (Fig. 2f). Sirt5−/− mice demonstrated a slight trend towards improved glucose tolerance compared to wt littermates during an ipGTT test, although only the 2 hr time point reached statistical significance (Fig. 2g). In line with the results of the ipGTT, Sirt5−/− animals are more sensitive to insulin than Sirt5+/+ mice, an effect that was particularly noted during the later phases of the ipITT test (Fig. 2h).
Figure 2. Metabolic phenotyping of chow fed Sirt5−/− mice.
(a). Experimental schedule of the clinical phenotyping protocol. EC: EchoMRI measurement to non-invasively monitor body fat and lean mass; CL: energy expenditure by indirect calorimetry (CLAMS system); E: endurance run test; G: intra-peritoneal glucose tolerance test (ipGTT); I: intra-peritoneal insulin tolerance test (ipITT); C: cold test; S: sacrifice. N = 9 for all the tests, except the endurance running (N = 6) because some mice were reluctant to run in the test. (b). Body weight evolution of Sirt5+/+ and Sirt5−/− male mice between week 7 to 25 of age. (c). Fat mass and lean mass as determined by EchoMRI. (d). Mice were monitored in the CLAMS system for 48 h. The data obtained in the last 24 h (6 h light, 12 h dark, and 6 h light) were used for the calculation. Food intake and heat production were normalized to body weight. xAMB counts indicated the horizontal movement of the mice. (e). Cold tolerance test. The mice were exposed to 4 °C for 6 h. Mice had free access to water but no food or bedding was provided. (f). Endurance run test. The mice were habituated to the treadmill by running for 10 min at speed of 9 m/min for four consecutive days. On the fifth day, the mice ran until they were exhausted. The speed started at 9 m/min. Every 10 min, the speed was increased 1 m/min. (g). ipGTT. The mice were fasted overnight and then an intraperitoneal (i.p.) injection with 2 g/kg glucose was performed. Plasma glucose was monitored at the indicated intervals in the next 2 hours. The bar graph in the inset represents the incremental area under the curve (AUC). (h). ipITT. The mice were fasted overnight and then i.p. injected with 0.5 U/kg insulin. The plasma glucose was monitored in the next 90 minutes. The bar graph in the inset represents the reverse AUC. * P< 0.05, ** P < 0.01.
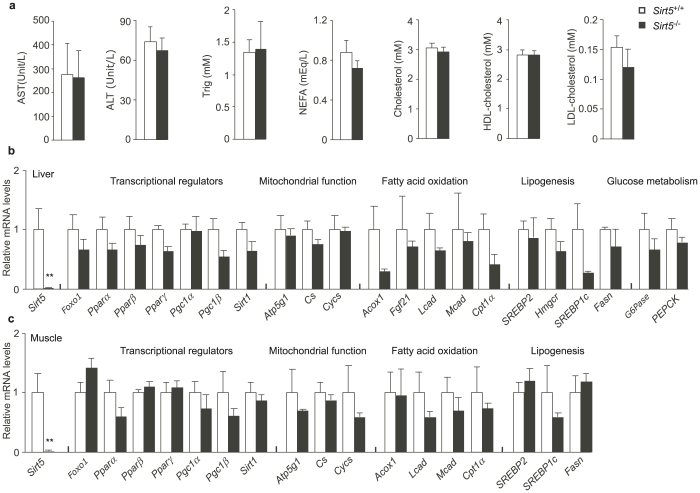
We also measured the levels of major metabolites in the plasma of Sirt5+/+ and Sirt5−/− mice. The aspartate transaminase (AST), alanine transaminase (ALT), triglycerides, non-essential fatty acids (NEFA), cholesterol, high-density lipoprotein (HDL)-cholesterol, and low density lipoprotein (LDL)-cholesterol were comparable in the plasma of chow-fed Sirt5+/+ and Sirt5−/− mice (Fig. 3a). At the molecular levels, we examined the liver and skeletal muscle for the expression of a vast set of metabolic genes involved in transcriptional regulation of metabolism (Foxo1, Ppars, Pgc1s, Sirt1), mitochondrial function (Atp5g1, Cs, Cycs), fatty acid oxidation (Acox1, Lcad, Mcad, Cpt1α), lipogenesis (Fasn, SREBP1c), and glucose metabolism (PEPCK, G6Pase) (Fig. 3b and 3c). The deletion of Sirt5 had, however, no significant impact on the expression of genes involved in the major metabolic pathways.
Figure 3. Metabolite and transcription analysis of chow-fed Sirt5−/− mice after sacrifice.
The 12-week-old wt and knockout male mice were fasted for 24 hours and sacrificed. N = 4–6. (a). AST, ALT, Triglycerides (Trig), NEFA, cholesterol, HDL-cholesterol, and LDL-cholesterol in plasma. (b) and (c). The expression profile of a selected set of metabolic genes in the liver and gastrocnemius muscle. mRNA level of each gene was normalized to the housekeeping gene 36B4 and further normalized to the expression level in Sirt5+/+ mice. ** P < 0.01.
Metabolic characterization of Sirt5 deficient mice fed a HFD
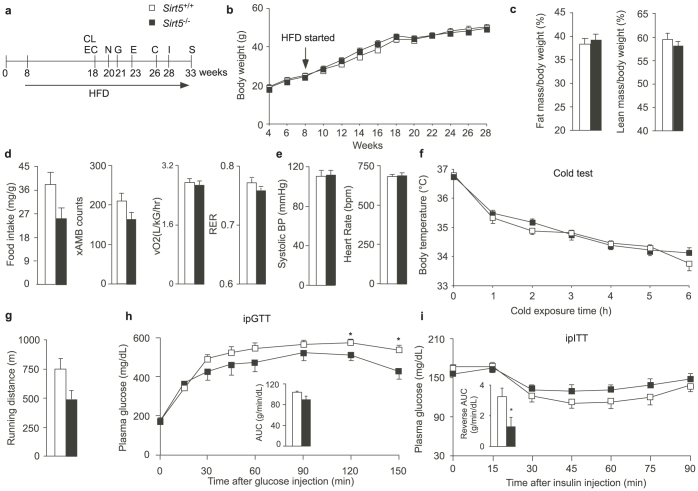
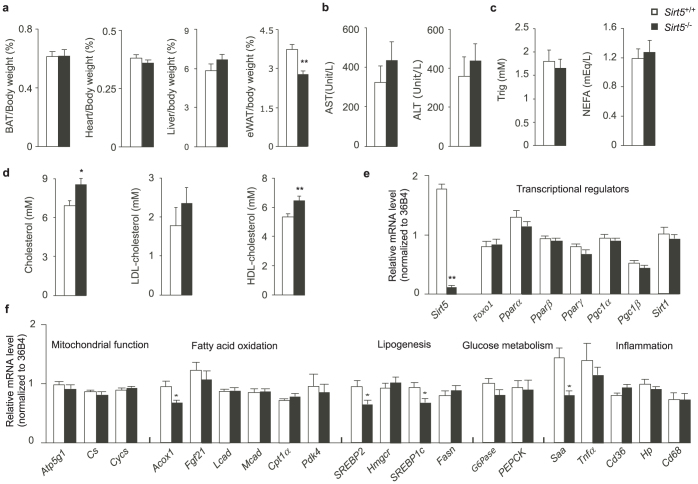
Since no striking metabolic phenotypes were observed in Sirt5−/− mice fed chow diet, we challenged male Sirt5+/+ and Sirt5−/− littermates with HFD for 10 weeks and then reevaluated their metabolic profiles using a similar phenotyping protocol as described in Fig. 2a (Fig. 4a). No significant difference in weight gain was observed between Sirt5+/+ and Sirt5−/− mice (Fig. 4b). Consistently, body composition (Fig. 4c), food intake, spontaneous locomotor activity, oxygen consumption, and RER (Fig. 4d) were similar between the two genotypes. In addition, both the systolic blood pressure and heart rate were indistinguishable between Sirt5+/+ and Sirt5−/− mice (Fig. 4e). Also on HFD, the tolerance of Sirt5+/+ and Sirt5−/− mice to cold was comparable (Fig. 4f). Although during an endurance run, the distance run to exhaustion of Sirt5−/− mice tended to be less than that of control mice, the difference was not significant (Fig. 4g). Consistent to what was observed under chow diet, Sirt5−/− and Sirt5+/+ littermates on HFD showed only a slight difference in glucose tolerance during the later phases of an ipGTT (Fig. 4h). Both genotypes showed equal insulin sensitivity during the ipITT test under HFD, although the reverse AUC indicated a difference between Sirt5+/+ and Sirt5−/− mice (Fig. 4i).Upon sacrifice, we weighed the metabolic organs and determined the levels of plasma metabolites in HFD-fed Sirt5+/+ and Sirt5−/− mice. The weights and gross morphology of Sirt5−/− brown adipose tissue (BAT), heart, and liver were similar to those of wt mice (Fig. 5a). The epididymal white adipose tissue (eWAT) of Sirt5−/− mice weighed less than Sirt5+/+ littermates. The levels of AST, ALT (Fig. 5b), triglycerides, and NEFA (Fig. 5c) were comparable in the plasma of mice with the two genotypes. Plasma cholesterol was higher in Sirt5−/− mice, probably accounted for by the increased level of HDL-cholesterol (Fig. 5d).
Figure 4. Metabolic phenotyping of Sirt5−/− mice fed a HFD.
(a). Experimental schedule of the clinical phenotyping protocol. For abbreviations see legend of Fig. 2a; N: non-invasive blood pressure measurement. N = 8 for all the tests, except the endurance running (N = 5), because some mice were reluctant to run in the test. (b). The body weight evolution of Sirt5+/+ and Sirt5−/− male mice during the period of week 4 to 28. HFD started at the week 8. (c). Fat mass and lean mass. (d). CLAMS results include food intake, spontaneous locomotor activity, oxygen consumption, and RER. (e). The blood pressure and heart rate were measured for 4 days. 15 measurements/mouse were performed each day. The data obtained in the last two days were used for calculation. (f). Cold tolerance test. (g). Endurance run test. (h). ipGTT. (i). ipITT. * P< 0.05.
Figure 5. Metabolic analysis of HFD-treated Sirt5−/− mice after sacrifice.
The 33-week-old wt and knockout male mice after 25-week of HFD treatment were fasted for 6 hours and sacrificed. N = 8. (a). The weight of BAT, heart, liver, and eWAT. All the data were normalized to body weight. (b). Plasma AST and ALT. (c). Triglycerides and NEFA in plasma. (d). Cholesterol, LDL-cholesterol, and HDL-cholesterol in plasma. (e) and (f). The expression profile of a selected set of metabolic and inflammation genes in the liver. * P< 0.05, ** P < 0.01.
At the molecular level, we again examined the livers of HFD fed mice for the expression of metabolic genes. Similar to the results in chow fed mice, the deletion of the Sirt5 gene had no striking impact on the expression of genes involved in the major metabolic pathways (Fig. 5e, f). Besides reduced expression levels of Acox1, SREBP1c and SREBP2, most of the mRNAs were unchanged in the livers of HFD fed Sirt5−/− mice.
Discussion
We report here the generation and metabolic characterization of whole body Sirt5 knockout mice. The germline deletion of Sirt5 caused mice to be born at an abnormal Mendelian ratio, with the number of live-born Sirt5−/− offspring being reduced by 40%. This partial loss of Sirt5−/− offspring pointed towards a potential important role of Sirt5 in embryogenesis or early development.
The surviving Sirt5−/− mice were characterized by a global protein hypersuccinylation in both liver and skeletal muscle. The Sirt5−/− mice showed elevated levels of blood ammonia during fasting, but otherwise were metabolically similar to their wt littermates under basal conditions. The general metabolic and physiological parameters of the two genotypes (weight gain, body composition, spontaneous activity, oxygen consumption, response to cold exposure, and endurance) were comparable between Sirt5+/+ and Sirt5−/− mice. Sirt5−/− mice tended to have a slightly improved glucose tolerance and a trend towards insulin sensitization, although the changes were rather minimal. Under HFD, the absence of Sirt5 did not protect or sensitize mice to the development of HFD-induced obesity, hypertension, and insulin resistance. Of note, the obese Sirt5−/− mice may tolerate glucose slightly better than Sirt5+/+ mice and showed increased serum cholesterol levels. At the molecular level, Sirt5 deficiency did not lead to any overt expression change of the genes involved in the major metabolic pathways, except for the reduced expression of Acox1, SREBP1c, and SREBP2 under HFD.
The fact that multiple and high-level of acylation (acetylation, succinylation, and malonylation) occurs on various mitochondrial proteins favors the possibility of mitochondrial sirtuins being involved in the sensing and removal of inadvertent acyl modifications on proteins16,17,19,24,25. This detoxifying and regulatory mechanism may become particularly important under extreme stress conditions, as was demonstrated for Sirt3. Initially Sirt3−/− mice were reported to be metabolically normal under basal conditions6. Similarly, skeletal muscle and liver-specific Sirt3 deficiency did not induce any phenotypic abnormalities under standard chow and HFD conditions10. However, when exposed to extreme stress, such as seen after fasting followed by cold exposure, 48 hours of fasting, or after consumption of a HFD for almost one year, the Sirt3−/− mice developed evident metabolic abnormalities, including defective adaptive thermogenesis, dysfunction of β-oxidation, diet-induced obesity, or insulin resistance9,26,27. It is unclear what causes these differences between the various Sirt3−/− mice strains, but different study conditions (e.g. stress) and the contribution of tissues beyond the muscle and liver to the more pronounced phenotypes observed in germline Sirt3−/− mice could explain these discrepancies. Notably in this context is also the fact that the substantial deacetylation of CPS1 by Sirt5 was observed after 48 hours of fasting15. Unfortunately, we are unable to test such stringent stress conditions because of ethical concerns in Switzerland.
In summary, we report here the generation and phenotypic characterization of a germline Sirt5−/− mouse line. Strikingly, no overt phenotype was observed in Sirt5−/− mice fed chow or HFD, indicating that Sirt5 may be dispensable for basal homeostasis, under these conditions. Further studies are, however, warranted to explore a role of Sirt5 in particular challenged and stressed conditions. Given the emerging role of protein succinylation in immune response28 and the genetic link between a single-nucleotide polymorphism (SNP) in Sirt5 and brain aging29, such studies should also focus on the role of Sirt5 in immune response and neurodegeneration, or carcinogenesis.
Methods
Generation and maintenance of the Sirt5−/− mice
Sirt5floxed mice were generated using standard gene targeting procedures22, using 129SV embryonic stem (ES) cells. These animals were crossed with CMV-Cre transgenic mice23 to generate germline Sirt5−/− mice, which were subsequently backcrossed for 5 generations with C57BL/6J mice. All mice were maintained in a temperature-controlled (23°C) facility with a 12 hr light/dark cycle and were given free access to food and water. Regular chow diet (2018) and high-fat diet (TD.06414) were obtained from Harlan.
Clinical phenotyping of the Sirt5−/− mice
All animal work in the manuscript was performed according to the validated standard operating procedures (SOPs)20,21, as defined and validated by the Eumorphia program (see: http://empress.har.mrc.ac.uk/). Animal experiments were approved by the veterinary ethics committee of the canton of Vaud - Switzerland (Permit ID 2163). We subjected our mice to non-invasive monitoring of body fat and lean mass by EchoMRI. Indirect calorimetry to monitor food consumption, body temperature, O2 consumption, CO2 production, and animal activity was measured using CLAMS system (Columbus Instruments). The blood pressure and heart rate was measured by a computerized tail-cuff system (Visitech Systems) in conscious animals.
For the cold test, the body temperature was recorded using a thermometer (Bioseb) with a rectal probe (Physitemp Instruments). A variable speed belt treadmill enclosed in a plexiglass chamber with a stimulus device consisting of a shock grid attached to the rear of the belt (Panlab) was used for the endurance test. Exhaustion was assumed when mice received more than 50 shocks in a 2.5 min interval30. ipGTT and ipITT were performed in animals that were fasted overnight. Glucose quantification was done with the Glucose RTU (BioMérieux Inc). Plasma metabolites were determined by enzymatic assays (Roche). Serum ammonia was measured using ammonia assay kit (Sigma AA0100) based on reductive amination using L-glutamate dehydrogenase.
The fasting experiments were performed under the Cornell Institutional Animal Care and Use Committee (IACUC) protocol #2011-0098. Food was depleted for 24 hours during the fasting, but animals could access water freely.
Quantitative RT-PCR
qRT-PCR was performed as described31. Briefly, RNA was extracted from tissues using TRIzol reagent (Invitrogen). cDNA was generated using QuantiTect Rev (Qiagen). The real-time PCR measurement of individual cDNAs was performed using SYBR green dye in the LightCycler System (Roche Diagnostics). A list of primers is included in the Supplementary Table S1 online.
Western blot analysis
Tissue samples were lysed in lysis buffer (50 mM Tris pH 7.9, 150 mM NaCl, 1 mM EDTA, 1% NP40, 10 mM sodium butyrate, 5 mM nicotinamide, protease inhibitor tablet). Liver and muscle samples were used to extract intact mitochondria32, which were lysed in lysis buffer and subjected to western blot. Protein extracts were separated by SDS-PAGE and transferred onto nitrocellulose membranes. Anti-Sirt5 was purchased from Abcam. Anti-Tubulin was purchased from Santa Cruz. Anti-succinyllysine antibody was purchased from PTM Biolabs.
Statistical analyses
Statistical analyses were performed with non-parametric Student's t-test. Data are expressed as mean ± SEM. Statistical significance is displayed as * (p < 0.05) or ** (p < 0.01).
Author Contributions
J.Y. directed and performed most experiments. S.S. measured the serum ammonia level, succinylation in the muscle, and coordinated the collaboration at Cornell. L.N. participated in the metabolic phenotyping of the mouse models. NM measured serum metabolites and analyzed gene expression in the chow-fed mice. B.H. measured the succinylation in the liver. R.W. and H.L. directed the collaboration work at Cornell. K.S. and J.A. directed all the work. J.Y., K.S. and J.A. wrote the manuscript. All authors reviewed the manuscript.
Supplementary Material
supplementary table s1
Acknowledgments
JA is the Nestlé Chair in Energy Metabolism. The work in the Auwerx lab is supported by the EPFL, the ERC (2008-AdG-23138), the Velux Stiftung, and the SNSF. The work in the Lin and Weiss labs is supported by NIH R01CA163255. JY was supported by a FEBS fellowship. The authors thank the members of the Auwerx lab for kind help and inspiring discussions, Pei Xin Lim, Tim Pierpont, and John Stupinski from the Weiss lab for mouse strain maintenance and tissue collection.
References
- Pirinen E., Lo Sasso G. & Auwerx J. Mitochondrial sirtuins and metabolic homeostasis. Best Pract Res Clin Endocrinol Metab 26, 759–770 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L. & Mostoslavsky R. Fine tuning our cellular factories: sirtuins in mitochondrial biology. Cell Metab 13, 621–626 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. & Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Ann N Y Acad Sci 1173 Suppl 1, E10–19 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkiadaki A. & Guarente L. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat Rev Endocrinol 8, 287–296 (2012). [DOI] [PubMed] [Google Scholar]
- Houtkooper R. H., Pirinen E. & Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 13, 225–238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard D. B. et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 27, 8807–8814 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J. C., He W. & Verdin E. Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J Biol Chem 287, 42436–42443 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack M. N. & Finkel T. Mitochondrial metabolism, sirtuins, and aging. Cold Spring Harb Perspect Biol 4, a013102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey M. D. et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell 44, 177–190 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Marcos P. J. et al. Muscle or liver-specific Sirt3 deficiency induces hyperacetylation of mitochondrial proteins without affecting global metabolic homeostasis. Sci Rep 2, 425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 130, 1095–1107 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis M. C. et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126, 941–954 (2006). [DOI] [PubMed] [Google Scholar]
- Jeong S. M. et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell 23, 450–463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csibi A. et al. The mTORC1 Pathway Stimulates Glutamine Metabolism and Cell Proliferation by Repressing SIRT4. Cell 153, 840–854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Lomb D. J., Haigis M. C. & Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137, 560–570 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C. et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics 10, M111 012658 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J. et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. et al. SIRT5-Mediated Lysine Desuccinylation Impacts Diverse Metabolic Pathways. Mol Cell 50, 919–930 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champy M. F. et al. Mouse functional genomics requires standardization of mouse handling and housing conditions. Mamm Genome 15, 768–783 (2004). [DOI] [PubMed] [Google Scholar]
- Champy M. F. et al. Genetic background determines metabolic phenotypes in the mouse. Mamm Genome 19, 318–331 (2008). [DOI] [PubMed] [Google Scholar]
- Argmann C. A., Chambon P. & Auwerx J. Mouse phenogenomics: the fast track to "systems metabolism". Cell Metab 2, 349–360 (2005). [DOI] [PubMed] [Google Scholar]
- Schwenk F., Baron U. & Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 23, 5080–5081 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. C. et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23, 607–618 (2006). [DOI] [PubMed] [Google Scholar]
- Choudhary C. et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 (2009). [DOI] [PubMed] [Google Scholar]
- Hirschey M. D. et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows W. C. et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell 41, 139–149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill G. M. et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 496, 238–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorioso C., Oh S., Douillard G. G. & Sibille E. Brain molecular aging, promotion of neurological disease and modulation by sirtuin 5 longevity gene polymorphism. Neurobiol Dis 41, 279–290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H. et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell 147, 827–839 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige J. N. et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab 8, 347–358 (2008). [DOI] [PubMed] [Google Scholar]
- Frezza C., Cipolat S. & Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc 2, 287–295 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary table s1