Abstract
Proteins that interact with voltage-gated sodium (Nav) channels are important in channel sorting and modulation. In this study, we identified the transcriptional regulator, Sin3B, as a novel binding partner of Nav channels in a yeast two-hybrid screen and confirmed the interaction using pull-down assays, co-immunoprecipitation, and immunofluorescence-colocalization. Because both long (~1100-residue) and short (N-terminal 293 residues) Sin3B variants interacted with Nav channels, binding occurred within the N-terminal region containing two paired-amphipathic helix domains. In Nav channels, Sin3B bound to a 132-residue portion of the cytoplasmic C-terminus. Expression of the short Sin3B variant strongly reduced native sodium current and Nav-channel gating charge in the neuronal cell line N1E-115, without affecting the voltage-dependence of activation. Because the total amount of channel protein was unchanged by Sin3B, binding of Sin3B likely decreases the number of channels in the plasma membrane, suggesting that interaction with Sin3B influences Nav-channel trafficking or stability in the membrane.
Voltage-gated sodium (Nav) channels are essential for action potential generation and electrical excitability in neurons, muscle cells, and neuroendocrine cells. The pore-forming α subunit of Nav channels is part of a macromolecular complex that includes auxiliary β subunits, cytoskeletal proteins, and signaling proteins such as protein kinases1,2. These protein-protein interactions modulate channel function and also serve to anchor the channels at crucial membrane sites, such as nodes of Ranvier and axon initial segments. We set out to identify novel proteins that interact with Nav channels and thus might be components of the channel complex that contribute to channel trafficking and/or function. Because the cytoplasmic C-terminus of the Nav-channel α subunit is a known site of interaction with other proteins, we focused on proteins that bind to this part of the channel. Here, we present evidence that short and long splice variants of the multifunctional transcriptional regulatory protein, Sin3B, interact with C-terminal regions of Nav1.2 and Nav1.6, which are two important CNS sodium channel α subunits whose axonal expression is developmentally regulated3. When expressed in N1E-115 cells, the short isoform of Sin3B strongly reduced endogenous sodium current density and gating charge, but not the overall amount of Nav-channel protein, consistent with an effect on targeting or stabilization of voltage-gated sodium channels in the plasma membrane.
In its gene regulatory role, Sin3B serves as a molecular adapter for multiple repressive transcription factors as well as histone deacetylases (HDAC), which are in turn responsible for gene silencing4. Although it is predominantly a nuclear protein, Sin3B can translocate from nucleus to cytoplasm depending upon interaction with other transcription factors5,6. In addition, there is evidence that Sin3B is not restricted to the nucleus in myoblasts7, and the structurally related protein Sin3A can be found in both nucleus and cytosol in large pyramidal neurons of the frontal cortex8. Therefore, we suggest that Sin3B may be used in both nuclear and non-nuclear contexts as a platform for assembling and/or stabilizing protein complexes, including the Nav-channel complex.
Results
Sin3B interacts with voltage-gated sodium channels
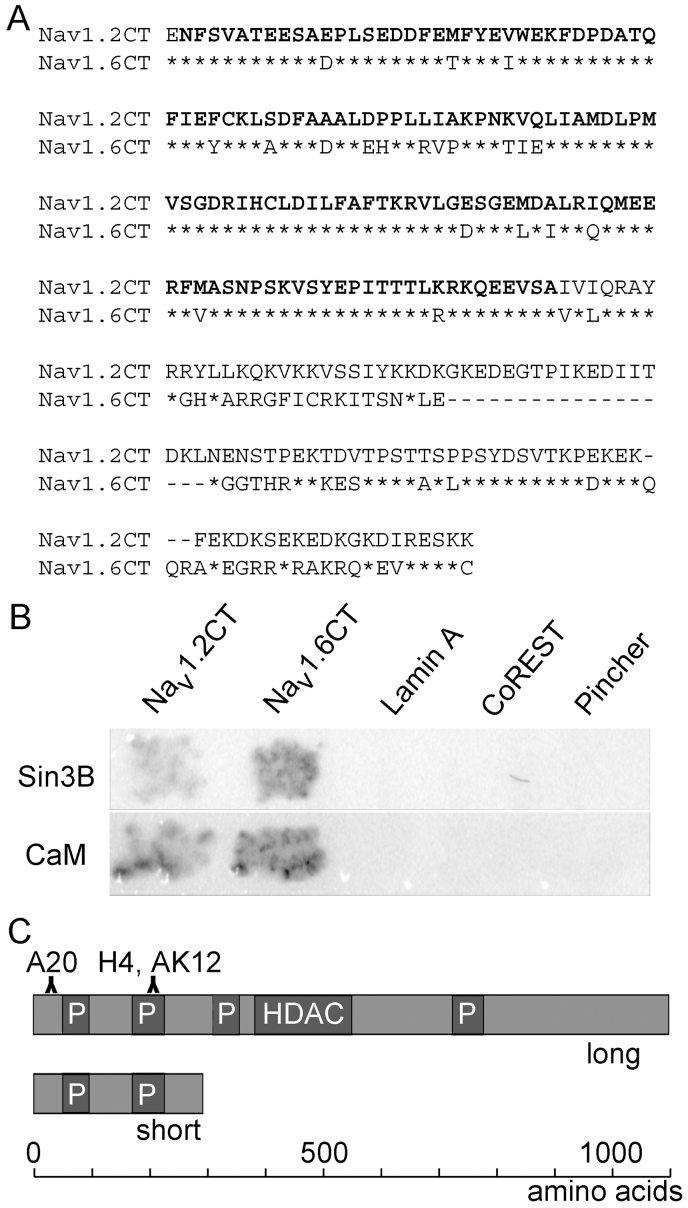
To identify novel binding partners for Nav channels, we conducted a yeast two-hybrid assay using the cytoplasmic C-terminal regions of sodium-channel α subunits Nav1.2 and Nav1.6 (Nav1.2CT and Nav1.6CT; Fig. 1A) as bait constructs. Specificity of identified binding partners for Nav channels was tested by using a set of three unrelated proteins, Lamin A, CoREST and pincher, as negative controls. Lamin A is a component of the nuclear membrane, CoREST is a corepressor protein associated with the silencing transcription factor REST, and pincher is a chaperone protein involved in endocytic trafficking. We identified over 50 different proteins able to interact specifically with the NavCT baits, including proteins previously known to bind to C-termini of Nav channels, like FHF4 (one clone) and calmodulin I, II, and III (>1500 clones). Among the new putative interacting partners, the most repeated protein was a short isoform of Sin3B, identified in four independent clones. Mating assays showed that Sin3B interacted with both Nav1.2CT and Nav1.6CT, but not with the three negative control baits (Fig. 1B; upper panel). The known NavCT interactor calmodulin served as a positive control for the assay (Fig. 1B; lower panel). The four detected Sin3B clones showed minor differences among them; for example, in clones T22–131 and T22–713, sequences started on the second triplet, while clones T22–702 and T22–748 contained 12 and 27 bases upstream of the start codon. However, all of them contained the C-terminus and 3′ UTR characteristic of the short variant, isoform 2 7,9, which is frequently denoted as Sin3B 293. Mature isoform 2 mRNA encodes a protein of 293 amino acids, compared with ~1100 amino acids for long variants (Fig. 1C). Sin3B 293 contains paired amphipathic helix (PAH) domains 1 and 2, common to all Sin3B isoforms, but lacks other domains that are relevant for transcriptional repression activity, such as the histone deacetylase (HDAC)-binding domain9. Although the physiological role of the short isoform of Sin3B is not yet clear, it is expressed in several tissues including brain, heart, and skeletal muscle7.
Figure 1. Sin3B interacts with the C-terminal cytoplasmic tail of Nav channels in yeast.
(A) Sequence comparison of C-terminal fragments of Nav1.2 and Nav1.6 that were used to construct baits for a two-hybrid screen, with identical residues indicated by asterisks. The bold residues indicate the high-homology region (HHR) in the proximal part of the C-terminus. (B) Coexpression of Sin3B (upper panel) and calmodulin (CaM, lower panel) with Nav C-termini in yeast. L40 cells expressing Sin3B or CaM fused with Gal4 activation domain were mated with AMR70 cells expressing Nav1.2CT or Nav1.6CT bait or a control bait (LaminA, CoREST, or Pincher). Mated cells were tested for β-galactosidase activity by incubating with X-gal. Only cells expressing Nav1.2CT and Nav1.6CT turned blue (shown in black in the gray scale image), indicating that both Sin3B and CaM interact specifically with Nav. (C) Schematic illustration of the long and short isoforms of Sin3B protein, indicating the PAH domains (P) and the region where histone deacetylase (HDAC) binds. Locations of the epitopes for the three anti-Sin3B antibodies (A20, H4, and AK12) used in immunoblots are indicated above the long isoform.
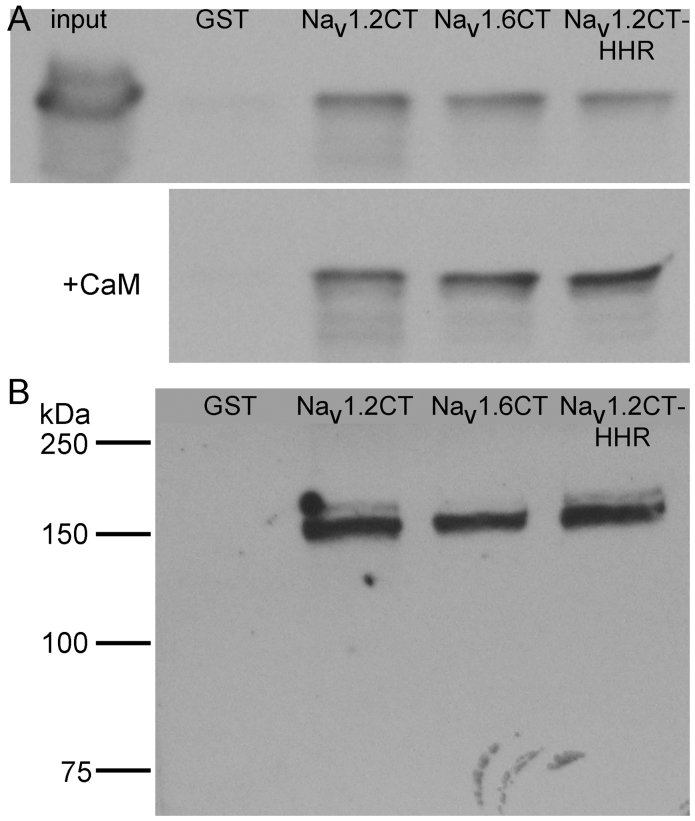
To verify the results of yeast two-hybrid assays indicating interaction between Sin3B and C-termini of Nav channels, we carried out biochemical tests using recombinant and native proteins. First, we expressed GST-Nav1.2CT and GST-Nav1.6CT fusion proteins in bacteria and purified the recombinant proteins using glutathione-sepharose beads. We then incubated the beads containing GST-bait with lysate of bacteria expressing recombinant His6-tagged Sin3B 293, and bound Sin3B was analyzed by immunoblotting with anti-Sin3B or anti-polyhistidine antibody after SDS-PAGE. Figure 2A (upper panel) shows that both GST-Nav1.2CT and GST-Nav1.6CT, but not GST alone, were able to pull down Sin3B 293. As with the two-hybrid assay (Fig. 1B), Sin3B interacted with both Nav1.2CT and Nav1.6CT in the GST pull-down assay. Therefore, the Sin3B binding site is likely located within the high homology region (HHR; see Fig. 1A, bold residues), where the two Nav C-termini are almost identical. To test this, we examined whether Sin3B 293 interacts with GST-Nav1.2CTHHR, which is truncated on the C-terminal end near the point of divergence of the two Nav C-terminal sequences. This fusion protein also effectively bound recombinant His6-tagged Sin3B 293 (Fig. 2A, upper panel), demonstrating that Sin3B interacts with the proximal portion of the C-terminus of Nav channels, nearest the final transmembrane segment of the channel.
Figure 2. Sin3B interacts with the C-terminal cytoplasmic tail of Nav channels in vitro and in vivo.
(A) Recombinant His6-Sin3B 293 was incubated with glutathione-sepharose beads preloaded with Nav C-terminus/GST fusion proteins (GST-Nav1.2CT, GST-Nav1.6CT, or GST-Nav1.2HHR) or GST alone. After washing, supernatant and bound proteins were collected, electrophoresed and blotted with anti-Sin3B antibody A20. The detected bands are at ~35 kDa, near the predicted molecular mass for His6-Sin3B 293. Similar results were observed in immunoblots using anti-polyhistidine antibody. In the upper panel, beads preloaded with the indicated GST-NavCT proteins were incubated only with His6-tagged Sin3B 293. In the bottom panel, beads were first incubated with recombinant calmodulin (CaM) prior to incubation with His6-Sin3B 293. (B) Native Sin3B from HEK293 cells was pulled down by the indicated GST-NavCT fusion proteins, but not by GST alone. Bound proteins were electrophoresed and blotted with anti-Sin3B antibody A20. A single band matching the size of the long isoform of human Sin3B was detected, indicating that the native long isoform of Sin3B binds the C-terminus of Nav.
Calmodulin is known to bind to the C-terminus of Nav channels and modulate channel gating and/or current density10,11,12,13,14,15. Presumably because of its high abundance in neurons16,17, calmodulin was by far the most repeated partner found in our Y2H screen. Therefore, it is important to determine if binding of calmodulin could occlude subsequent interaction of Nav C-termini with Sin3B, because such competition could influence the relevance of Sin3B binding in vivo. However, as shown in Fig. 2A (lower panel), prior incubation with calmodulin did not affect subsequent binding of His6-tagged Sin3B 293 to GST-tagged Nav C-terminal baits. This is consistent with the demonstrated binding of Sin3B within the HHR of Nav1.2, which does not include the calmodulin-binding IQ motif, and it suggests that calmodulin and Sin3B bind independently to non-overlapping sites. This separation of calmodulin and Sin3B binding sites was further confirmed by pull-down of recombinant calmodulin by GST-Nav1.2CT but not by GST-Nav1.2HHR (data not shown), both of which bound Sin3B (Fig. 2A).
We next determined whether the GST-NavCT fusion proteins could interact with native Sin3B in a GST pull-down assay. Glutathione-sepharose beads preincubated with GST, GST-Nav1.2CT, GST-Nav1.6CT, or GST-Nav1.2HHR were mixed with lysate from HEK293 cells, and binding of endogenous Sin3B was detected in immunoblots using anti-Sin3B antibody. As shown in Fig. 2B, all three of the GST-NavCT fusion proteins bound to full-length, native Sin3B from HEK293 cells, whereas GST alone produced no detectable binding. This further confirms the interaction between Sin3B and the C-terminal fragment of Nav channels. Since both full-length Sin3B and Sin3B 293 interact with NavCT, we conclude that the binding site is within the N-terminal fourth of the full protein, which includes PAH domains 1 and 2.
Sin3B is associated with the membrane fraction
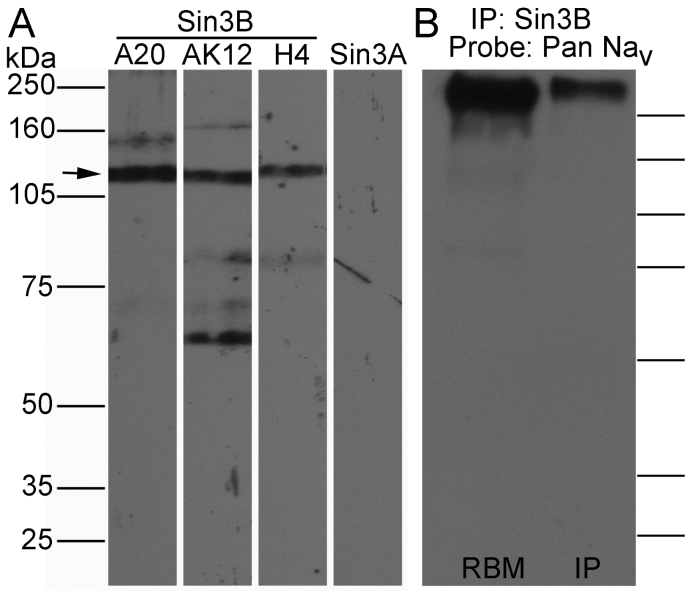
A nuclear protein like Sin3B may seem unlikely to have the opportunity to interact with a membrane protein like Nav channel in intact neurons, despite their demonstrated binding in in vitro assays. However, under certain circumstances transcription factors can shuttle in and out of the nucleus18. Also, although Sin3 proteins are well known members of many transcription complexes and are expected to be mainly nuclear, there is evidence that immunoreactivity for Sin3B and the related protein Sin3A can be found in cytosol of myoblasts and neurons7,8. To determine if Sin3B may be present in the membrane domain, we isolated membrane fractions from adult mouse and rat brain and tested for Sin3A and Sin3B in immunoblots. Three different antibodies for Sin3B revealed the presence of the long isoform of Sin3B in membrane fractions (Fig. 3A), whereas no signals of Sin3A or the short isoform of Sin3B were detected. This suggests that Sin3B can localize to the membrane compartment, where Nav channels reside.
Figure 3. Sin3B is detected in the membrane fraction in brain, and associates with Nav.
(A) Rat brain membrane fractions were probed with anti-Sin3B antibodies A20, AK12, or H4, or with the anti-Sin3A antibody K20. Bands at the appropriate molecular mass (arrow) for long isoforms of Sin3B were detected by all three anti-Sin3B antibodies, but Sin3A was undetectable. (B) Nav channels in a rat brain membrane preparation co-immunoprecipitated with Sin3B, using anti-Sin3B antibody A20. Immunoblot was then probed with pan-specific anti-Nav1 antibody K58/35 (Pan Nav). Left lane: rat brain membrane preparation (RBM). Right lane: material immunoprecipitated (IP) with anti-Sin3B. Presence of the long isoform of Sin3B in both lanes was subsequently confirmed by reprobing with anti-Sin3B antibody (not shown). Molecular weight indicators on the right have the same values as in A.
Next, we tried to immunoprecipitate Nav-channel protein from brain membrane fraction in complex with Sin3B. As shown in Fig. 3B, antibody against Sin3B was able to co-immunoprecipitate Nav channels. Therefore, Sin3B is not only present in membrane fractions, but it also can interact with Nav channels. However, we were not able to show the reverse immunoprecipitation of Sin3B with antibody against Nav channels. This may indicate that a relatively high amount of the total Sin3B associated with the membrane fraction is bound to Nav channels, but the amount of Nav channels bound to Sin3B is small compared to the total amount of Nav channels. Results from immunostaining, presented in the next section, suggest that this interpretation may be correct.
Sin3B and Nav immunostaining colocalize in a subset of neuronal processes in the brain
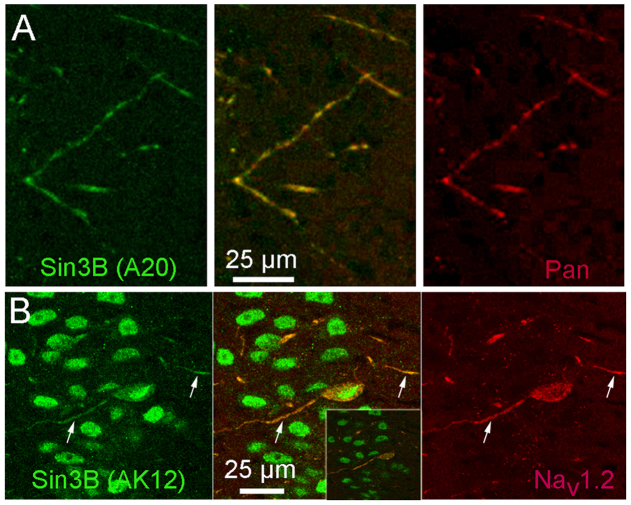
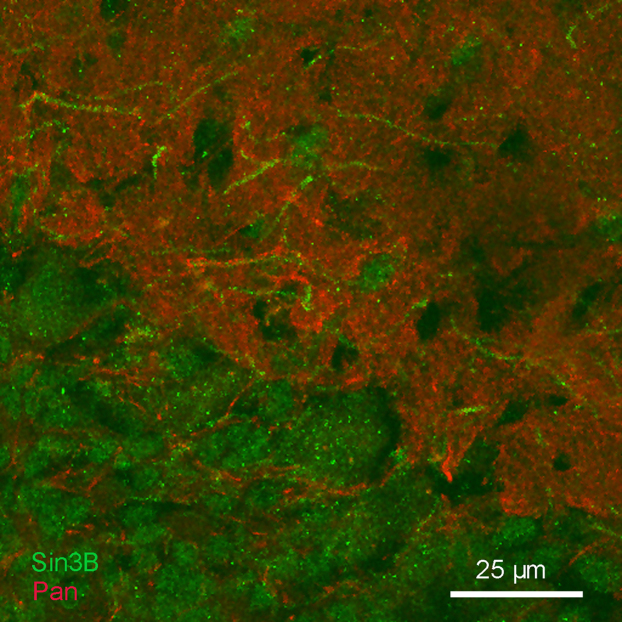
Because the results of immunoblots suggested the presence of Sin3B in the membrane fraction of brain lysate, we examined the distribution of immunostaining with anti-Sin3B antibodies in cryosections of mouse and rat hippocampal region CA1, which we selected because the cell bodies of neurons occupy a compact layer, allowing clear segregation of Sin3B staining in cell nuclei from staining in the surrounding neuropil. As expected, intense Sin3B immunoreactivity was present in nuclei, but in addition, extranuclear Sin3B immunoreactivity was readily detected above background in a sparse subset of neurites in the neuropil (Fig. 4). Similar staining in neuronal processes was observed with all three of the different anti-Sin3B antibodies. We next determined whether extranuclear Sin3B immunostaining coincided with Nav channels in the Sin3B-positive processes, using double-labeling with pan-specific or isoform-specific antibodies against Nav channels. As shown in Fig. 4A, pan-specific anti-Nav-channel staining colocalized closely with extranuclear Sin3B, consistent with our biochemical demonstrations of interaction between the two proteins. Staining with an antibody selective for Nav1.2 (ref. 19) also colocalized with extranuclear Sin3B (Fig. 4B). Therefore, the results suggest that the interaction between Sin3B and Nav channels detected using biochemical methods also occurs in situ in brain neurons. Although extranuclear Sin3B immunostaining was associated with Nav channels, most anti-Nav-channel staining in the brain was not associated with Sin3B immunostaining (Fig. 5), as expected from the fact that extranuclear Sin3B immunoreactivity was observed only in a sparse subset of neuronal processes, whereas Nav channels are ubiquitous. This suggests that our failure to co-immunoprecipitate Sin3B with pan-specific Nav-channel antibody (see above) was likely due to the low proportion of total brain Nav channels associated with Sin3B.
Figure 4. Sin3B immunoreactivity is found outside the nucleus, where it colocalizes with immunostaining for Nav channels A.
Discrete processes in the molecular layer of region CA1 of the hippocampus were labeled with anti-Sin3B (A20, green) as well as with a pan-specific antibody against Nav channels (Pan, red). The middle panel shows superposition of the anti-Nav-channel and anti-Sin3B staining. Images are single confocal optical sections. B. Extranuclear staining with anti-Sin3B (AK12, green) colocalized with immunostaining for Nav1.2 (red). Arrows indicate neuronal processes where the two immunoreactivities coincide. Images are Z-axis projections of a series of 5 consecutive confocal optical sections taken in CA1; a single optical section is shown in the inset, for comparison. Note that immunostaining for Sin3B in panels A and B used two different anti-Sin3B antibodies.
Figure 5. Most immunoreactivity for Nav channels (red; pan-specific antibody against Nav channels) was not associated with extranuclear staining for Sin3B.
Cryosection of rat cerebellum, showing extensive Sin3B immunostaining (green) in cell nuclei and sparse extranuclear Sin3B in a subset of processes.
Sin3B 293 reduces sodium current and surface expression of sodium channels
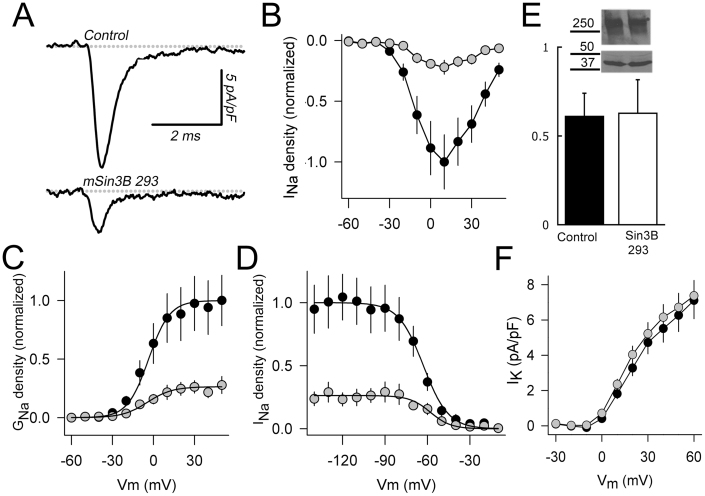
To determine if the interaction with Sin3B has any consequences for Nav-channel function, we overexpressed Sin3B 293 in the N1E-115 mouse neuroblastoma cell line and analyzed native sodium currents using the whole-cell patch-clamp technique 48 h after transfection. We used Sin3B 293 because the short isoform binds to Nav channels (Figs. 1 and 2) but lacks domains required for interaction with transcriptional complexes, which therefore avoids the complications related to effects on gene transcription that might arise from overexpressing full-length Sin3B. Figure 6A shows representative sodium currents recorded from cells transfected with Sin3B 293:DsRed in a 1:1 molar ratio, or DsRed alone (control cells). Sodium current density was conspicuously smaller in cells transfected with Sin3B 293 at all voltages tested (Fig. 6B), but no differences in current kinetics were apparent. Activation curves (Fig. 6C) were fitted with a Boltzmann equation (Methods) to obtain VG1/2 and kG for each cell. Average values of these parameters were nearly identical between control cells (VG1/2 = 3.6 ± 2.5 mV, kG = 7.7 ± 0.9, n = 13) and cells transfected with Sin3B 293 (VG1/2 = 4.4 ± 2.6 mV, kG = 9.0 ± 1.2, n = 8). A significant reduction of ~73% in Gmax was observed in cells expressing Sin3B 293 (p = 0.02). Additionally, inactivation curves were obtained by eliciting INa at −10 mV (test pulse) after applying 200-ms inactivating prepulses at different voltages. Figure 6D shows inactivation curves for control and Sin3B 293-transfected cells normalized by the average Imax obtained for control cells. Again, maximal sodium current density was >70% smaller in cells expressing Sin3B 293. In addition, a small but statistically significant shift of Vh1/2 was observed, from −62.8 ± 1.0 mV in control cells to −58.0 ± 0.8 mV in Sin3B-transfected cells (p = 0.002). In contrast, values for kh remained similar between groups, 8.8 ± 1.0 for control cells (n = 10) and 7.3 ± 0.6 for Sin3B-expressing cells (n = 8). We chose to use Sin3B 293 rather than a long isoform of Sin3B because the former lacks the HDAC-binding domain and is therefore unlikely to act as a repressor of Nav-channel transcription. Nevertheless, we did explore if the reduction of INa density was associated with a global reduction of Nav-channel protein expression. As immature neurons have been reported to have an important intracellular pool of sodium channels, accounting for up 77% of total immunoreactivity20, whole cell lysates, including plasma membrane and intracellular pool, were tested. As shown in Fig. 6E, the total amount of Nav channels was not significantly different in immunoblots from control and Sin3B-expressing cells at 48 h after transfection. Therefore, we ruled out a possible effect of Sin3B 293 on sodium-channel gene transcription as the cause for the observed reduction in sodium current density. Additionally, we recorded potassium currents at several voltages in control cells and cells expressing Sin3B 293 (Fig. 6F). However, both currents and I-V curves showed no differences, indicating that the effect of Sin3B 293 on sodium currents is rather specific.
Figure 6. Sin3B reduces native sodium currents in N1E-115 neuroblastoma cells.
(A) Typical sodium currents recorded under voltage-clamp in transfected N1E-115 cells. Currents were elicited by depolarizing steps from a holding potential of −80 mV to +40 mV. Upper trace: control cell, expressing DsRed alone. Lower trace: cell expressing Sin3B 293 and DsRed. (B) Current-Voltage relationship. Sodium currents were recorded at different test voltages from a holding potential of −80 mV. Control cells (black dots) showed a larger current at all voltages tested than Sin3B-expressing cells (gray dots). (C) Conductance-voltage curves were calculated for each cell, and normalized to the average Gmax of control cells (black dots). A strong reduction of Gmax was observed in Sin3B-expressing cells (gray dots). Analysis of data showed no changes in VG1/2 or kG. (D) Inactivation curves. INa was recorded at −10 mV after inactivating pre-pulses of 200 ms. Peak currents were normalized to the average of INa recorded in control cells (black dots) and plotted as function of the pre-pulse voltage. The data from each cell were fitted with Boltzmann equations to calculate Vh1/2 and kh. On average, a small (5 mV) but statistically significant shift to the right was observed in Sin3B-expressing cells (gray dots). (E) Immunoreactivity for Nav channels from whole cell lysates, from three independent experiments, was measured by densitometry and normalized to the immunoreactivity of actin in control (black) and Sin3B-expressing cells (white). Inset: representative inmmunoblots with Pan Nav (upper panel) and anti-actin (lower panel) antibodies in control N1E-115 cells (lane 1) and cells expressing Sin3B 293 (lane 2). (F) In contrast, potassium currents were unaffected by overexpression of Sin3B at any voltage tested.
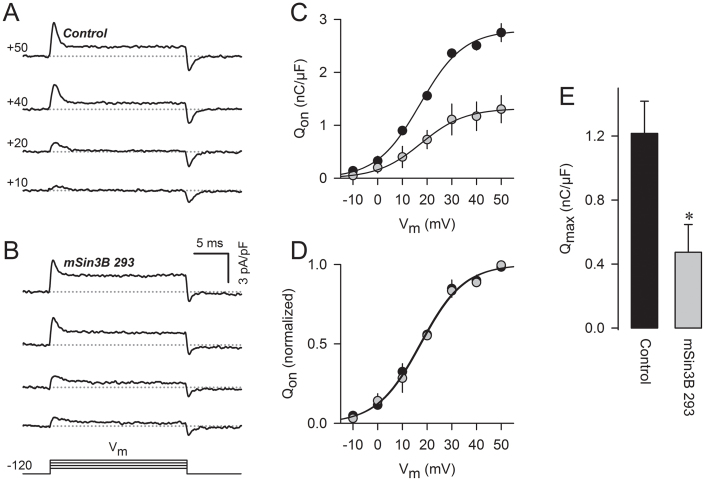
The smaller sodium current observed in N1E-115 cells transfected with Sin3B 293 could reflect reduced expression of Nav channels in the plasma membrane, or a decrease in channel open probability. To examine this issue, we measured the gating charge movement associated with channel activation (Qon). The maximum value of Qon (Qmax) is considered a very reliable index of Nav-channel density in the membrane, that is independent of open probability after activation (reviewed in ref. 21). In these experiments, the holding potential was set at −120 mV during pre-pulses to maximize the fraction of channels available for activation. Examples of gating currents observed at different levels of depolarization are shown in Fig. 7A (control cells) and Fig. 7B (Sin3B 293-transfected cells), and the voltage dependence of Qon obtained from such recordings is summarized in Fig. 7C. Fits of the Boltzmann equation (Methods) to the Qon activation data revealed no difference in VQ1/2 or kQ between control and Sin3B 293-expressing cells, which can be appreciated when the data are normalized with respect to Qmax and superimposed (Fig. 7D). Since the activation curves for both Qon (Fig. 7C) and INa (Fig. 6C) showed no differences in either half-activation voltages or slope factor, it seems improbable that Sin3B is modifying the voltage sensor movement. This strongly suggests that the reduction in Gmax most probably reflects a change in the number of channels at the membrane. In agreement with this interpretation, Qmax was reduced by approximately 60% (P = 0.013) when Sin3B 293 was overexpressed (Fig. 7E), which is similar to the reduction in sodium current induced by Sin3B 293 (Fig. 6). We conclude that interaction with Sin3B leads to a decrease in the number of sodium channels in the plasma membrane of N1E-115 cells. Since Sin3B expression did not affect the total amount of Nav-channel protein (Fig. 6E), this decrease in channel density in the membrane likely represents an effect on trafficking to the membrane and/or on the stability of the channel protein in the plasma membrane.
Figure 7. Sin3B reduces gating charge associated with sodium-channel activation in N1E-115 cells.
(A) Examples of gating currents evoked by the indicated depolarizations in a control cell transfected with DsRed alone. (B) Examples of gating currents in a cell expressing Sin3B 293 and DsRed. (C) Voltage-dependence of Nav-channel gating charge (Qon) measured in control and Sin3B 293-expressing cells. Solid lines are fits of equation 3 (Methods) to the data. (D) Data from panel C were normalized with respect to the observed maximum Qon for each condition. (E) Average values of Qmax for control cells expressing DsRed alone and cells expressing both DsRed and Sin3B 293. Data were obtained from a total of 9 cells for each group. * P = 0.013.
Discussion
We used a yeast two-hybrid system to identify Sin3B as an interacting partner of the C-terminal region of voltage-gated sodium channels. The interaction was confirmed in vitro by pull-down of recombinant proteins as well as of native Sin3B from non-neuronal cells. We were also able to co-immunoprecipitate both proteins from brain tissue and to detect colocalization of Sin3B and Nav-channel immunostaining in a subset of neuronal processes in the brain. Also, we observed a strong reduction of sodium current density in N1E-115 cells overexpressing the short isoform, Sin3B 293, without a change in overall amount of Nav-channel protein detected in immunoblots. Altogether, our results indicate that the reduction of sodium current is due to direct interaction between the two proteins. Therefore, we add Sin3B to the growing list of proteins that bind to the C-terminus of Nav-channel α subunits, including calmodulin, members of the fibroblast growth factor homologous factor family (FHF1B, FHF2, and FHF4), Nedd4-like ubiquitin ligase, and syntrophin (reviewed in ref. 22). We have shown that calmodulin and Sin3B bind independently to the Nav-channel C-terminus, but it seems unlikely that a single channel could bind so many partners at the same time within the same region. More probably, binding to different partners would occur on subpopulations of channels and/or at different stages in the life cycle of Nav channels.
The mechanism by which Sin3B 293 reduces Nav-channel density at the membrane is not yet clear. One possibility is that the binding of Sin3B interferes with the normal trafficking of Nav channels towards the plasma membrane, causing intracellular accumulation of the channels. Alternatively, the presence of Sin3B on the C-terminus could facilitate retrieval of Nav channels from the plasma membrane, either directly or indirectly, so that the balance is shifted toward internalization. For example, it could help to recruit AP2 and/or other components of the clathrin-dependent pit assembly responsible for internalization of NaV1.2 (ref. 23). In addition to the short isoform, full-length Sin3B can also bind to Nav channels, and this interaction could in turn recruit enzymes that modify the channel protein and regulate its interaction with other partner proteins.
While long isoforms of Sin3B have a well-established role as transcription cofactors, the role of the short isoform remains unclear. The short isoform contains PAH1 and PAH2, which are necessary for binding to DNA-binding repressors REST and NCoR, but it lacks other domains that are relevant for transcriptional repression activity, such as the HDAC-binding domain. Therefore, is been proposed that Sin3B 293 could antagonize Sin3B/HDAC-mediated co-repression of genes24. In our experiments, it is unlikely that the down-regulation of sodium current is due to Sin3B 293-mediated repression of Nav-channel gene expression, because the channel protein level was the same in control and transfected cells. On the other hand, binding of REST/Sin3B/HDAC complex to REST-sensitive regulatory elements in Scn2a results in repression/silencing of transcription25. Therefore, competitive binding of Sin3B 293 to REST could remove suppression and upregulate Nav-channel expression. However, although N1E-115 neuroblastoma cells express mRNA for several Nav-channel α-subunits26,27, single cell RT-PCR experiments suggest that Nav1.2 is the predominant isoform in this cell line27. Therefore, REST-dependent suppression of Scn2a expression may already be removed, preventing any further upregulation by overexpression of Sin3B 293.
Our results suggest that Sin3B regulates Nav-channel activity not through effects on gene expression but by direct binding to the channel. However, Sin3B is not the first regulator of transcription that has been proposed to have additional functions outside the cell nucleus. For example, the ubiquitously expressed transcription factor, TFII-I, regulates the activity of plasma membrane calcium channel TRPC3, by competing with the channel for binding to the regulatory protein, phospholipase C18. Another example is CtBP2, which was first identified as a component of transcriptional complexes, but is now known to be a component of ribbon synapses28. The CtBP2 homolog, CtBP1 (also known as BARS), is also associated with ribbon synapses29 and in addition has been implicated in intracellular membrane trafficking, membrane fission, and regulation of the microtubule cytoskeleton (reviewed by ref. 30). Thus, Sin3B may be a member of this growing group of transcription factors with a double life, within and outside of the nucleus. Further investigation is needed to determine under what conditions Sin3B would leave the nucleus and bind to Nav channels in the plasma membrane. In this regard, it is interesting that Sin3A has been reported to be largely excluded from the nuclei of pyramidal neurons in Huntington disease patients8, but it is not yet known if Sin3B behaves similarly. The picture of the interaction of Nav channels with cytosolic proteins is only beginning to emerge. Knowing the interactions between Nav channels and intracellular proteins such as Sin3B will help to better understand the fine-tuning of these functionally significant neuronal channels.
Methods
Plasmids
cDNA fragments encoding the C-terminal regions of Nav1.2 and Nav1.6 (Nav1.2CT and Nav1.6CT) were prepared by PCR from adult mouse brain cDNA (Ambion Inc, Austin, TX). Appropriate restriction sites were added to the PCR primers to clone fragments in frame with the DNA-binding domain of LexA in two-hybrid bait plasmid pSTT91 (ref. 31). The resulting plasmids pSN12CT and pSN16CT contain bases 5278–5964 (XM_980330) and 5293–5934 (AF049617) of Nav1.2 and Nav1.6 coding regions, corresponding to amino acids 1760–1988 and 1765–1978 of the respective α-subunits. The bait plasmids were confirmed not to have endogenous GAL4 activity when expressed in yeast. C-terminal cDNAs were also subcloned into pGEX to produce GST-tagged versions of Nav1.2CT and Nav1.6CT (plasmids pGN12CT and pGN16CT). A shorter version of pGN12CT, pGN12CT-HHR, was produced by introducing an early stop codon just before the calmodulin-binding IQ motif. Negative-control bait plasmids encoding LexA-LaminA, LexA-CoREST and LexA-Pincher were generous gifts from Drs. Rolf Sternglanz, Gail Mandel, and Simon Halegoua, respectively. After identification of cDNA of interacting clones by sequencing, the full-length cDNA for Sin3B 293 was subcloned from plasmid T22–702 into pGEX and pET28 vectors to generate plasmids pGSin3B and pESin3B, encoding GST- and His6-tagged versions of Sin3B 293, respectively. Finally, we also subcloned Sin3B 293 into the mammalian vector pMyc-CMV (plasmid pMycSin3B) for functional assays in N1E-115 cells. Correct orientation and reading frame of all plasmids were verified by sequencing.
Yeast two-hybrid assay (Y2H)
The procedure followed for yeast two-hybrid screens was largely as described by Park and Sternglanz32,33. For the initial screen, yeast cells of strain L40 were co-transfected with a commercial two-hybrid plasmid library (Clontech, catalog #ML4008AH; fusion with GAL4 activation domain) derived from adult mouse brain cDNA, together with the desired bait plasmid. Transfected cells were then plated on medium deficient in histidine, leucine, and tryptophan, on which L40 yeast cells are able to grow only if they contain both bait and prey plasmids and if the LexA/GAL4 activator is formed by interaction of bait and prey proteins. After 3 days at 30°C, white colonies >2 mm in diameter were picked and replated on triple-deficient medium. After 3 more days, colonies were tested for LexA-driven β−galactosidase activity, and the positives were replated and retested two more times. To determine specificity of prey interaction with the bait used in the screen, positive colonies were grown under conditions that promote loss of the bait plasmid, and the resulting prey-only cells were then crossed with mating-proficient AMR70 yeast cells transfected with a negative control bait plasmid, or with the original bait plasmid to reconfirm the original interaction. Colonies were then tested for β-galactosidase activity, as an index of bait-prey interaction.
Glutathione S-transferase (GST) pull-down assay
pGEX, pGN12CT, and pGN16CT were introduced in the E. coli strain BL21, and synthesis of GST, GST-Nav1.2CT, and GST-Nav1.6CT proteins was induced with 1 mM IPTG for 4 h at 37°C. Cells were then lysed by snap freezing and digestion with lysozyme, and DNA was degraded by sonication. Glutathione sepharose beads were incubated with the lysate for 1 h at 4°C and then washed extensively with ice-cold binding buffer (50 mM Tris-HCl, pH 7.5, 120 mM NaCl, 2 mM EGTA, 0.1% triton X-100, 2 mM DTT). BL21 cells transformed with pESin3B were used to produce His6-Sin3B, and then lysed as described above. The His6-Sin3B lysate was incubated with the GST, GST-Nav1.2CT, or GST-Nav1.6CT pre-loaded beads for 3 h at 37°C. Beads were washed extensively with binding buffer and resuspended in 2X Laemmli reducing sample buffer (RSB). Pulled-down proteins were resolved by standard SDS-PAGE, electroblotted, and then subjected to Western blot with anti-polyhistidine or anti-Sin3B antibodies. In another set of experiments, designed to investigate whether binding of calmodulin may interfere with Sin3B-NavCT interaction, beads pre-loaded with GST or GST-fusion proteins were first incubated with 10 μg of recombinant calmodulin, produced in BL21 bacteria, for 1 hr at RT, washed three times in binding buffer, and then incubated with the His6-Sin3B cell lysate.
Pull-down from HEK293 cell lysate
Untransfected HEK293 cells were detached with PBS/1 mM EDTA, pelleted and washed with 50 mM Tris-HCl/120 mM NaCl. Then cells were lysed in binding buffer. Lysate was pre-cleared by spinning at 12000×G for 5 min. Supernatant was transferred to a fresh tube, aliquoted and frozen until use. Protein concentration was determined by Bradford assay. 50 μl of beads pre-loaded with GST or GST-NavCT baits were incubated with 0.5 mg of protein at 37 or 4°C for 3 h. Beads were washed four times with binding buffer, then resuspended in 2X RSB. Pulled-down proteins were electrophoresed and immunoblotted with anti-Sin3B antibodies.
Preparation of brain membranes
Brain membranes were prepared form a freshly dissected adult rat brain. Animal use followed guidelines established by the National Institutes of Health and was approved by the Institutional Animal Care and Use Committee at SUNY Stony Brook. Rats were killed by CO2 inhalation. Immediately after death, brains were dissected out and homogenized in ice-cold 0.3 M sucrose/5 mM sodium phosphate buffer, with 100 mM NaF and protease inhibitors (2 μg/ml aprotinin, 1 μg/ml leupeptin, 2 μg/ml antipain, 10 μg/ml benzamidine, and 0.5 mM PMSF). The homogenate was centrifuged at 3000×G to remove nuclei and debris. The supernatant was centrifuged at 38500×G for 90 minutes to pellet membranes. Membranes were resuspended in sucrose buffer, and protein concentration was determined by Bradford assay.
Immunoprecipitation
Membranes were solubilized in lysis buffer containing 1% Triton X-100, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10 mM EDTA, 10 mM iodoacetamide, 10 mM sodium azide, protease inhibitors and 1 mg/ml BSA. The solubilized fraction was incubated overnight at 4°C with 1 μg of rabbit anti-Sin3B A20 antibody. Immune complexes were incubated with protein-G coupled to sepharose beads for 2 h, and then washed several times in lysis buffer. Finally, the beads were resuspended in 2X RSB and pelleted. Immunoprecipitated proteins were then fractionated by SDS-PAGE and immunoblotted with pan-specific Nav-channel antibody. Then antibodies were stripped out and the blot was probed again with anti-Sin3B (A20).
Western blots, immunostaining, and antibodies
Proteins were size fractioned by standard SDS-PAGE methods in 5%, 9% or 12% polyacrylamide gels and then transferred to Hybond nitrocellulose membranes (Amersham). Blocking and incubation with antibodies were carried out in blotto (20 mM Tris-HCl, pH 8.0, NaCl 150 mM, 4% milk). Primary antibodies were detected with anti-IgG antibodies conjugated to horseradish peroxidase. Immunoreactive bands were then visualized by chemiluminescence. Anti-polyhistidine antibody was purchased from Novagen (EDM biosciences, Germany). Two polyclonal rabbit anti-Sin3B antibodies directed against the N-terminus (A20) and the PAH2 domain (AK12) of human Sin3B and a mouse monoclonal antibody (H4) against the PAH2 domain of mouse Sin3B were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Additionally, we used an antibody raised in rabbit against the N-terminus of Sin3A (K20, Santa Cruz). Antibody K58/35 binds to all voltage-gated sodium channel isoforms (Pan Nav-channel antibody; Sigma). Monoclonal anti-actin antibody was a generous gift form Dr. Manuel Hernandez (Cinvestav, Mexico). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit antibodies were purchased from Santa Cruz Biotechnology. Immunofluorescence staining of brain cryosections was carried out as described previously34, using the following antibodies: anti-Sin3B (A20, AK12, H4; 1:200), Pan Nav (K58/35; 1:3000), anti-Nav1.2 (K69/3; 1:1500; NeuroMab), and anti-Nav1.1 (K74/71; 1:5000; NeuroMab). Images were acquired using an Olympus FV300 confocal microscope.
N1E-115 cell culture and transfection
Neuroblastoma N1E-115 cells were kept in culture media consisting of DMEM supplemented with 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and L-glutamine (4 mM). For patch-clamp experiments, cells were plated at 15600 cells/cm2 on coverslips, and transfected 24 h after plating with pDsRed-N1 alone or in combination with pMycSin3B (molar ratio 1:1), using 0.5 μg of total DNA and the appropriate amount of Fugene HD (Roche). For other experiments, N1E-115 cells were plated at 2500 cell/cm2 in 10 cm plastic dishes and allowed to duplicate twice before transfection. A total amount of 2 μg of DNA was used. For Western blot analysis, cells were harvested in 2X RSB supplemented with 10 mM NaF, 1 mM NaVO4, and protease inhibitors (Roche). All subsequent experiments were performed 48 h after transfection.
Electrophysiology
N1E-115 cells were subjected to voltage-clamp using the whole-cell patch-clamp technique. Briefly, electrical seals of ~10 GΩ were formed between cell membrane and borosilicate glass electrodes. The electrical resistance of electrodes was approximately 2 MΩ, when filled with the internal solution. Composition (all in mM) of internal and external solutions varied according with the experimental goal. For isolated sodium current recording, internal: 130 CsCl, 10 NaCl, 1 CaCl2, 10 Hepes, 10 EGTA, 0.05 NaGTP, 5 MgATP and 5 glucose, 290 mOsm; external: 135 NaCl, 20 TEA-Cl, 2 CaCl2, 1 MgCl2, 10 Hepes, 0.5 CdCl2, 311 mOsm. For isolated potassium current recording, internal: 115 KCl, 30 NaCl, 1 CaCl2, 2 MgCl2 10 EGTA, 2 Na2-ATP2, 10 HEPES, 5 glucose; external: 145 NaCl, 5 KCl, 2 CaCl2, 0.5 CdCl2, 1 MgCl2, 0.006 TTX, 10 HEPES, 10 glucose. For NaV-channel gating currents, internal, 145 Cs-Asp, 10 Cs-EGTA, 2 CaCl2, 10 HEPES, 5 glucose, 5 MgATP, 0.05 GTP-Tris; external: 150 TEA-Cl, 2 CaCl2, 1 MgCl, 0.5 CdCl2, 10 HEPES, 10 glucose; The pH was adjusted to 7.3 in all solutions, and experiments were performed at room temperature (22–24°C). Cell membrane capacitance (Cm) was estimated by subtracting linear charge movements, which were acquired under both cell-attached and whole-cell conditions, as described in ref. 35. The series resistance was electronically cancelled (~60–80%), resulting in time constants for charging Cm of approximately 80 μs. Linear currents were eliminated following determination of Cm, using the capacitance cancellation feature of the amplifier and P/8 leak subtraction protocol. Sodium currents (INa) were filtered at 10 KHz, and digitally sampled every 10 μs. The holding potential was −80 mV, and the test pulses lasted 15 ms. For I-V curves, INa was measured at the peak, normalized by Cm, and plotted as a function of membrane potential during the test pulse (Vm). For each cell, the extrapolated reversal potential (Vrev) was estimated and used to calculate sodium conductance (GNa), according to the modified Ohm's law (GNa = INa/(Vm − Vrev)). There was no significant difference in the average values of Vrev between control and Sin3B 293-expressing cells (58 ± 6 mV and 65 ± 5 mV, respectively; p = 0.5). GNa was plotted as a function of Vm and fitted according to the following Boltzmann equation:
where Gmax represents the maximal conductance, VG1/2 is the potential required to activate 0.5 of Gmax, and kG is a slope factor. To investigate the voltage-dependence of inactivation, a two-pulse protocol was used, in which a test pulse to −10 mV was applied following 200 ms prepulses set at different voltages. Subsequently, each data set (a plot of peak INa density during the test pulse, versus prepulse voltage) was fitted with a Boltzmann equation of the form:
where Imax is the calculated maximal current density, kh is a slope factor, and Vh1/2 is the midpoint potential. Nav-channel gating currents were measured as described in previous studies36. Currents were elicited from a holding potential of −120 mV to remove inactivation. Nine cells of each group were analyzed. To estimate the amount of intramembrane charge movement, the outward component of the resulting traces was integrated and normalized by Cm (Qon). The values of Qon were subsequently plotted as a function of Vm, and fitted according to the following Boltzmann equation:
where Qmax represents the maximum amount of intramembrane charge movement, V1/2 is the voltage required to activate 50% of Qmax, and k is a slope factor.
Statistical analysis
Unless otherwise noted, results are presented as mean ± standard error. Significant differences were determined at the P = 0.05 level, using the Student's t-test for two-tailed unpaired samples.
Author Contributions
A.V. designed and performed experiments, analyzed data, constructed figures, and wrote the manuscript. G.A. designed and performed experiments and analyzed data. G.M. designed experiments, analyzed data, constructed figures, and wrote the manuscript. All authors reviewed the manuscript.
Acknowledgments
Supported by NIH grant R01EY003821 to Gary Matthews, Conacyt grant 56733 to Guillermo Avila, and PAPCA 2013 grant to Ana V. Vega.
References
- Meadows L. S. & Isom L. L. Sodium channels as macromolecular complexes: implications for inherited arrhythmia syndromes. Cardiovasc. Res. 67, 448–458 (2005). [DOI] [PubMed] [Google Scholar]
- Levitan I. B. Signaling protein complexes associated with neuronal ion channels. Nat. Neurosci. 9, 305–310 (2006). [DOI] [PubMed] [Google Scholar]
- Boiko T. et al. Compact myelin dictates the differential targeting of two sodium channel isoforms in the same axon. Neuron 30, 91–104 (2001). [DOI] [PubMed] [Google Scholar]
- Silverstein R. A. & Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 47, 1–17 (2005). [DOI] [PubMed] [Google Scholar]
- Grimes J. A. et al. The co-repressor Sin3A is a functional component of the REST-CoREST repressor complex. J. Biol. Chem. 275, 9461–9467 (2000). [DOI] [PubMed] [Google Scholar]
- Shiio Y. et al. Identification and characterization of SAP25, a novel component of the Sin3 corepressor complex. Mol. Cell Biol. 26, 1386–1397 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q. et al. The winged-helix/forkhead protein myocyte nuclear factor beta (MNF-beta) forms a co-repressor complex with mammalian Sin3B. Biochem. J. 345, 335–343 (2000). [PMC free article] [PubMed] [Google Scholar]
- Boutell J. M. et al. Aberrant interactions of transcriptional repressor proteins with the Huntington's disease gene product, huntingtin. Hum. Mol. Genet. 8, 1647–1655 (1999). [DOI] [PubMed] [Google Scholar]
- Koipally J., Renold A., Kim J. & Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 18, 3090–3100 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M. et al. Novel interaction of the voltage-dependent sodium channel (VDSC) with calmodulin: does VDSC acquire calmodulin-mediated Ca2+-sensitivity? Biochemistry 39, 1316–1323 (2000). [DOI] [PubMed] [Google Scholar]
- Deschênes I. et al. Isoform-specific modulation of voltage-gated Na(+) channels by calmodulin. Circ. Res. 90, E49–57 (2002). [DOI] [PubMed] [Google Scholar]
- Tan H. L. et al. A calcium sensor in the sodium channel modulates cardiac excitability. Nature 415, 442–447 (2002). [DOI] [PubMed] [Google Scholar]
- Herzog R. I., Liu C., Waxman S. G. & Cummins T. R. Calmodulin binds to the C terminus of sodium channels Nav1.4 and Nav1.6 and differentially modulates their functional properties. J. Neurosci. 23, 8261–8270 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K. A. & Caldwell J. H. Modulation of skeletal and cardiac voltage-gated sodium channels by calmodulin. J. Physiol. 565, 349–370 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. S., Hudmon A., Waxman S. G. & Dib-Hajj S. D. Calmodulin regulates current density and frequency-dependent inhibition of sodium channel Nav1.8 in DRG neurons. J. Neurophysiol. 96, 97–108 (2006). [DOI] [PubMed] [Google Scholar]
- DeLorenzo R. J. Role of calmodulin in neurotransmitter release and synaptic function. Ann. N. Y. Acad. Sci. 356, 92–109 (1980). [DOI] [PubMed] [Google Scholar]
- Kortvely E., Palfi A., Bakota L. & Gulya K. Ontogeny of calmodulin gene expression in rat brain. Neuroscience 114, 301–316 (2002). [DOI] [PubMed] [Google Scholar]
- Caraveo G., van Rossum D. B., Patterson R. L., Snyder S. H. & Desiderio S. Action of TFII-I outside the nucleus as an inhibitor of agonist-induced calcium entry. Science 314, 122–125 (2006). [DOI] [PubMed] [Google Scholar]
- Berendt F. J., Park K.-S. & Trimmer J. S. Multisite phosphorylation of voltage-gated sodium channel α subunits from rat brain. J. Proteome Res. 9, 1976–1984 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J., Rossie S. & Catterall W. A. A large intracellular pool of inactive Na channel α subunits in developing rat brain. Proc. Natl. Acad. Sci. USA 82, 4847–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong C. M. Sodium channels and gating currents. Physiol. Reviews 61, 644–683 (1981). [DOI] [PubMed] [Google Scholar]
- Shao D., Okuse K. & Djamgoz M. B. Protein-protein interactions involving voltage-gated sodium channels: Post-translational regulation, intracellular trafficking and functional expression. Int. J. Biochem. Cell Biol. 41, 1471–1481 (2009). [DOI] [PubMed] [Google Scholar]
- Garrido J. J., Fernandez F., Giraud P., Mouret I., Pasqualini E., Fache M.-P. Jullien F. & Dargent B. Identification of an axonal determinant in the C-terminus of the sodium channels NaV1.2. EMBO J. 20, 5950–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romm E., Nielsen J. A., Kim J. G. & Hudson L. D. Myt1 family recruits histone deacetylase to regulate neural transcription. J. Neurochem. 93, 1444–1453 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J. A. et al. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80, 949–957 (1995). [DOI] [PubMed] [Google Scholar]
- Benzinger G. R., Tonkovich G. S. & Hanck D. A. Augmentation of recovery from inactivation by site-3 Na channel toxins. A single-channel and whole-cell study of persistent currents. J. Gen. Physiol. 113, 333–346 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh J. K. & Quandt F. N. Down-regulation of Na channel expression by A23187 in N1E-115 neuroblastoma cells. Brain Res. 706, 343–346 (1996). [DOI] [PubMed] [Google Scholar]
- Schmitz F., Königstorfer A. & Südhof T. C. RIBEYE, a component of synaptic ribbons: a protein's journey through evolution provides insight into synaptic ribbon function. Neuron 28, 857–872 (2000). [DOI] [PubMed] [Google Scholar]
- tom Dieck S. et al. Molecular dissection of the photoreceptor ribbon synapse: physical interaction of Bassoon and RIBEYE is essential for the assembly of the ribbon complex. J. Cell Biol. 168, 825–836 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corda D., Colanzi A. & Luini A. The multiple activities of CtBP/BARS proteins: the Golgi view. Trends Cell Biol. 16, 167–173 (2006). [DOI] [PubMed] [Google Scholar]
- Sutton A. et al. A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol. Cell Biol. 21, 3514–3522 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. & Sternglanz R. Two separate conserved domains of eukaryotic DNA topoisomerase I bind to each other and reconstitute enzymatic activity. Chromosoma 107, 211–215 (1998). [DOI] [PubMed] [Google Scholar]
- Park H. & Sternglanz R. Identification and characterization of the genes for two topoisomerase I-interacting proteins from Saccharomyces cerevisiae. Yeast 15, 35–41 (1999). [DOI] [PubMed] [Google Scholar]
- Vega A. V., Henry D. L. & Matthews G. Reduced expression of Nav1.6 sodium channels and compensation by Nav1.2 channels in mice heterozygous for a null mutation in Scn8a. Neurosci. Lett. 442, 69–73 (2008). [DOI] [PubMed] [Google Scholar]
- Meza U., Avila G., Felix R., Gomora J. C. & Cota G. Long-term regulation of calcium channels in clonal pituitary cells by epidermal growth factor, insulin, and glucocorticoids. J. Gen. Physiol. 104, 1019–1038 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Mondragón R., Vega A. V. & Avila G. Long-term modulation of Na+ and K+ channels by TGF-β1 in neonatal rat cardiac myocytes. Pflugers Arch. 461, 235–247 (2011). [DOI] [PubMed] [Google Scholar]