Abstract
Warfarin is the most commonly used oral anticoagulant worldwide. Warfarin has a narrow therapeutic index, requiring frequent monitoring of the INR to achieve therapeutic anticoagulation. The role of pharmacogenomics in warfarin disposition and response has been well-established in adults, but remains unclear for pediatric patients. In this review, we focus on the important CYP2C9 and VKORC1 variants involved in warfarin response, our current understanding of warfarin disposition and pharmacogenomics, and recent warfarin pharmacogenetic studies in pediatric patients. Finally, we discuss the need for future prospective pediatric studies and the clinical implications of developing pharmacogenetic-based dosing algorithms in children.
Keywords: warfarin, pharmacogenomics, CYP2C9, VKORC1
INTRODUCTION
Since receiving FDA approval in 1954, warfarin has been the most widely used oral anticoagulant worldwide, with 0.5–1.5% of the population receiving the drug annually.[1] Approved indications include prophylaxis and treatment of venous thrombosis and its extension, pulmonary embolism, prophylaxis and treatment of thromboembolic complications associated with atrial fibrillation and/or cardiac valve replacement, and reduction in the risk of death, recurrent myocardial infarction, and thromboembolic events after myocardial infarction.[2] Although the majority of warfarin usage occurs in adults, it is also the mainstay of oral anticoagulation therapy in children and adolescents, most commonly in the setting of primary or secondary deep venous thromboses, indwelling central venous access catheters and congenital heart disease with prosthetic valves and/or endovascular stents.[3]
Despite its utility, warfarin has a narrow therapeutic index that can contribute to its therapeutic failure (i.e. thromboses) or toxicity (i.e. bleeding). A wide variability in warfarin sensitivity among patients makes determination of the optimal warfarin dose a time and effort intensive task. Multiple factors, including age, height, diet, concomitant medications, and indication for warfarin contribute to this variability. Adjustment of warfarin doses to obtain therapeutic levels of anti-coagulation is dictated by frequent measurement of the International Normalized Ratio (INR), and therapeutic INR levels typically range from 2–3.5, with a significant increase in risk of adverse events above or below this range.[4] Therefore, maintaining a therapeutic INR is essential to prevent either untoward bleeding or thrombosis.
Such variability in warfarin response and the increased risk of complications when patients are above or below therapeutic INR highlight a need for a prescribing approach accounting for the wide interpatient variability in dose requirement. Clinicians had noted this variability in dosing for years, but despite clinical and pharmacokinetic studies, the wide range in warfarin dosing requirements was poorly understood.[5] Polymorphisms in the genes encoding vitamin K epoxide reductase complex subunit 1 (VKORC1) and cytochrome P450 2C9 (CYP2C9) were subsequently discovered to account for a substantial amount of variability in warfarin sensitivity.[6–8] The incorporation of genetic information into warfarin dosing has the potential to improve clinical practice substantially. However, limited information is available in children receiving warfarin. Tables I and II provide a summary of a number of influential adult and pediatric warfarin pharmacogenetic studies to date.[7, 9–18]
Table I.
Notable studies in warfarin pharmacogenomics for adults
| Study | Study Type | N | Ethnicities | Ages (years) |
Major Findings |
|---|---|---|---|---|---|
| Aithal et al (Lancet 1999) | Case-control | 36 | not listed | 55–88 | Patients requiring low-dose warfarin are 6x more likely to have a CYP2C9 variant |
| Taube et al (Blood 2000) | Retrospective cohort | 561 | not listed | not listed | Mean dose with variant CYP2C9 alleles 61–86% of dose for wild type |
| Higashi et al (JAMA 2002) | Retrospective cohort | 185 | 91% European 4% Asian 3% African 2% Hispanic |
60 (mean) | CYP2C9 genotype is associated with (1) warfarin maintenance dose, (2) time to stable warfarin dosing, (3) rate of above-range INRs, and (4) bleeding events in patients taking warfarin |
| Rieder et al (NEJM 2005) | Retrospective cohort | 186 (primary) 357 (replication) |
100% European-American | ≥18 | Stratification of VKORC1 haplotypes into low-, intermediate-, and high-dose |
| D’Andrea et al. (Blood 2005) | Retrospective cohort | 147 | 100% Caucasian | 15–84 | Genetic variants of the VKORC1 gene locus modulate the mean daily dose of drug prescribed to acquire the target anticoagulation intensity. |
| Aquilante et al. (CPT 2006) | Cross-sectional | 350 | 91.1% White 7.1% Black 1.4% Hispanic 0.6% Asian/Other |
22–89 | Patients with the VKORC1 A/A and G/A genotypes required 50.4% and 29.4% lower weekly doses of warfarin, respectively, compared to those with the wild-type G/G genotype. Patients carrying 1 CYP2C9 variant allele (*2, *3, or *5) required 23.8% lower weekly doses of warfarin; patients with 2 variant alleles required 35.3% lower weekly doses of warfarin than those with the homozygous wild-type genotype. |
| Schelleman et al. (CPT 2007) | Prospective cohort | 317 | 51.1% African American 48.9% Caucasian |
median 59 | VKORC1 1173C/T and −1639G/A are associated with warfarin maintenance dose requirements among both African Americans and Caucasians. However, these polymorphisms may not be as useful in predicting over-anticoagulation among African Americans. |
| Cooper et al. (Blood 2008) | GWAS | 554 | 100% White | ≥18 | VKORC1 genotype (rs9923231), and CYP2C9 carrier status (either *2 or *3) predicted approximately 41.2% of the total variance in stabilized dose |
| Schwarz et al (NEJM 2008) | Prospective cohort | 297 | 89.2% White 9.8% Black 1% Hispanic |
47–74.8 | VKORC1 has greater impact during initiation, but CYP2C9 also significant in maintenance |
| Gage et al. (CPT 2008) | Prospective cohort | 1015 (derivation) 292 (validation) |
83% Caucasian 15% African-American 2% Hispanic 2% Other/Mixed |
65 (mean) | A pharmacogenetic equation explained 53–54% of the variability in the warfarin dose, while a clinical equation explained only 17–22% of the dose variability (P < 0.001). Development of nonprofit website http://www.WarfarinDosing.org |
| Wadelius et al. (Blood 2009) | Prospective cohort | 1496 | Majority Swedish | 18–92 | VKORC1 SNPs rs2359612 and rs9923231 explained 29.8% and and 29.3% of variance in stable warfarin dose; CYP2C9*2 and *3 explain 11.8% of variance in stable warfarin dose |
| Takeuchi et al. (PLoS Genet 2009) | GWAS | 1053 | 100% Swedish | 18–92 | Detected VKORC1 and CYP2C9, already known to cause 40% of the variability in warfarin dose, and discovered a new gene (CYP4F2) contributing 1%–2% of the variability |
| IWPC (NEJM 2009) | Retrospective cohort | 4043 (derivation) 1009 (validation) |
55.3% White 30.3% Asian 8.9% Black 5.5% Mixed/Missing |
10–90 (12 <19) |
Development and validation of warfarin dosing algorithm including genotype |
| Limdi et al. (Blood 2010) | Retrospective cohort | 4886 | 63.7% White 22.6% Asian 13.7% Black |
10-over 90 (10 ≤19) |
VKORC1 −1639G>A and 1173C>T individually explained the greatest variance in dose in all 3 racial groups. Incorporation of additional VKORC1 SNPs or haplotypes did not further improve dose prediction. |
| Gong et al. (Blood 2011) | Prospective cohort | 167 | 95.2% White 2.4% African-American 1.8% Asian 0.6% other |
≥18 | Use of a dosing algorithm incorporating pharmacogenomics eliminates VKORC1 and CYP2C9 genotype-related differences in attainment of first therapeutic INR |
IWPC = International Warfarin Pharmacogenomics Consortium.
Table II.
Warfarin pharmacogenomics studies in children.
| Study | Study Type | N | Ethnicities | Ages (years) (mean) |
Major Findings |
|---|---|---|---|---|---|
| Nowak et al (2010) | Prospective Cohort | 59 (34 on Warfarin) |
100% White | 1–19 (15) |
Age accounts for 28.3% of dose variability; no role for genotype |
| Kato et al (2011) | Retrospective Cohort | 48 | 100% Japanese | 0.5–19 yo (6.6) |
VKORC1 is a major determinant of required warfarin dose; CYP2C9 could not be evaluated |
| Biss et al (Blood 2012) | Cross-sectional | 120 | 75.8% White 8.3% Indian 5% Black 5% Asian 5.8% Other |
1–18 (11) |
VKORC1, CYP2C9, height, and indication account for 72.4% of dose variability; IWPC algorithm consistently overestimates necessary dose |
| Moreau et al (Blood 2012) | Retrospective Cohort | 118 (83 on warfarin) |
>90% White | 0.25–18 (8.4) |
VKORC1 accounts for 18.2% of dose variability, CYP2C9 2%, and height 48.1% |
| Nguyen et al (Pediatric Cardiology 2012) | Prospective Cohort | 37 (all pts with heart disease) |
73% White 18.9% Black 8.1% Asian |
1.8–18.6 (9.6) |
VKORC1 is a major determinant of required warfarin dose; with CYP2C9 only accounting for 5% of variability; age, height, and goal INR also contribute |
WARFARIN DISPOSITION AND METABOLISM
Warfarin is the most widely-used member of a family of anticoagulants known as Vitamin K antagonists. Vitamin K is an essential cofactor in the gamma-glutamyl-carboxylase-mediated activation of coagulation factors II, VII, IX, and X.[19] VKORC1 is an integral membrane protein which converts vitamin K epoxide to vitamin K, thus activating the Vitamin K dependent coagulation factors II, VII, IX, and X, and allowing the coagulation cascade to continue. Warfarin inhibits VKORC1, reducing the amount of active coagulation factors available and interrupting the coagulation cascade. (Figure 1)[19, 20]
Figure 1. Warfarin consists of 2 enantiomers, R- and S- warfarin, which inhibit VKORC1. S-warfarin is the more active enantiomer.
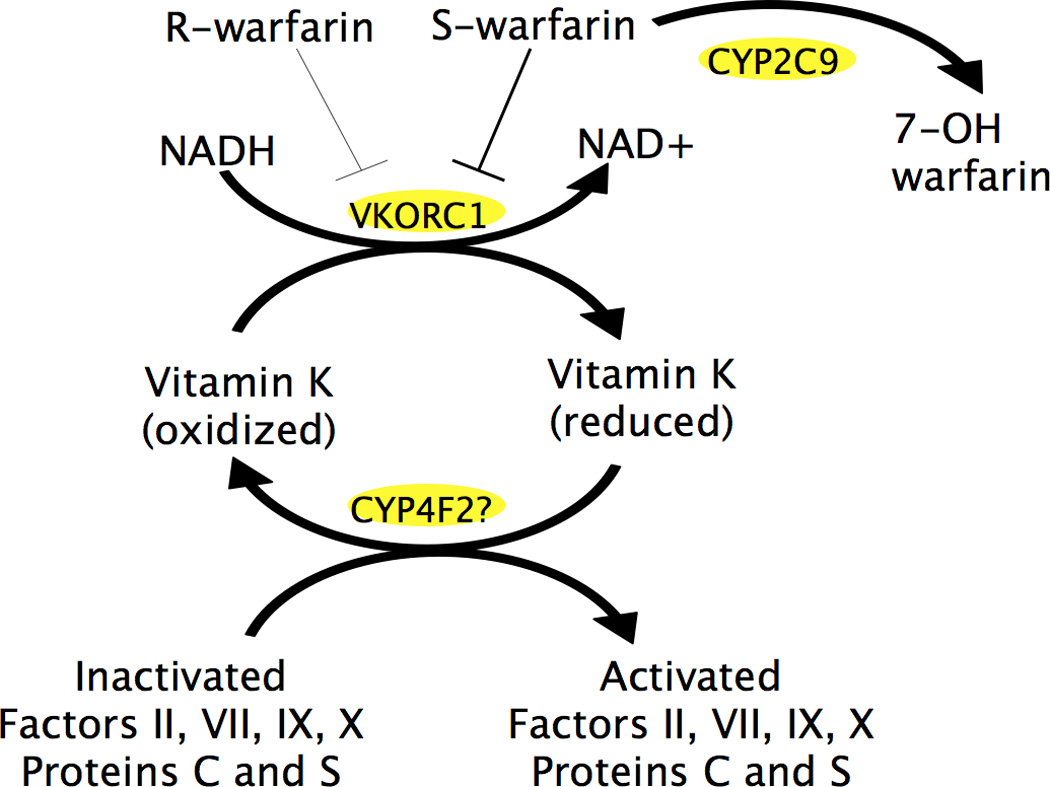
VKORC1 is an integral membrane protein which reduces vitamin K epoxide to vitamin K, thus activating the Vitamin K dependent coagulation factors II, VII, IX, and X, and allowing the coagulation cascade to continue. CYP2C9 is a cytochrome P450 enzyme which metabolizes S-warfarin to its inactive form, 7-OH warfarin. The role of CYP4F2, another cytochrome P450 enzyme, is poorly understood, but is thought to be involved in Vitamin K oxidation.
CYP2C9 is a phase I drug-metabolizing enzyme that is responsible for the metabolism of S-warfarin, the more active enantiomer of the drug, into 6-hydroxy and 7-hydroxy metabolites.[7] CYP2C9 makes up ~20% of total hepatic CYP content contributes to the metabolism of ~15% of clinical drugs, including amiodarone, phenobarbital, and rosuvastatin, along with warfarin.[21]
PHARMACOGENETICS OF WARFARIN RESPONSE
It has been well-established in adults that polymorphisms in CYP2C9 and VKORC1 affect the metabolism and response to warfarin and thereby the dose required to achieve a stable therapeutic INR.[17] The majority of these polymorphisms are related to a low-dose phenotype – i.e patients with variant alleles require a less-than-average stable dose of warfarin to achieve a therapeutic INR, though rare VKORC1 polymorphisms have been associated with a high-dose phenotype. This association was intially noted from several clinical studies but subsequently has been confirmed in a series of genome-wide association studies (GWAS).[13, 22, 23]
Polymorphisms in CYP2C9 (rs1799853, rs1057910) were the first to be associated with warfarin metabolism and account for up to 18% of the variability in stable warfarin dosing.[1] One of the initial studies to demonstrate this association was a small case-control study from a British anticoagulation clinic, in which 36 patients who required a significantly lower warfarin dose than control patients in the same clinic were 6 times more likely to have a variant CYP2C9 allele, and four times more likely to have a major bleeding complication during the induction phase of therapy.[7] This paved the way for larger studies that confirmed the association between CYP2C9 genotype and warfarin metabolism.[9, 10, 14, 24]
CYP2C9 is highly polymorphic, resulting in multiple functional variants which can significantly affect drug metabolism. Patients with low enzyme activity may be at increased risk for adverse drug events due to poor drug clearance. Polymorphisms in CYP2C9, therefore, can lead to delayed clearance of warfarin and thus require a lower dose to reach the goal INR. CYP2C9*1 is the most prevalent or major allele, while CYP2C9*2 and CYP2C9*3 represent the most frequent variant alleles. Patients can be heterozygous and carry any combination of the alleles, and the variant alleles have an additive effect. However, a prospective study demonstrated that while CYP2C9 variant alleles affect the stable warfarin dose, they did not as contribute as much as VKORC1 to the variability in the time required to initially reach a therapeutic INR.[14]
The CYP2C9*2 variant (430C>T; rs1799853) results in a missense mutation with significantly decreased enzyme activity secondary to an alteration in the interaction between the CYP enzyme molecule and P450 reductase.[21] Patients with this genotype have a maximum clearance rate of warfarin that is approximately 50% lower than patients with CYP2C9*1. This variant allele tends to be ethnicity-dependent, with allele frequencies of 10–20% in the Caucasian population, ~1–5% in the African-American and Asian populations, and 3–10% in the Hispanic population.[25]
The CYP2C9*3 variant (1075A>C; rs1057910) results in a missense mutation causing conformational changes in the substrate binding site at residue 359 is located with resultant reduced enzyme activity.[21] Patients heterozygous for this allele have approximately 50% of the activity of a CYP2C9*1*1 patient, and homozygotes are rare. This variant occurs less commonly than the CYP2C9*2 variant, with an allele frequency of ~2.5–8% in Caucasians, ~1–3% in African-Americans, ~1.6% in Asians, and ~3–10% inHispanic populations.[25]
However, only a relatively small percentage of warfarin dose variability was accounted for by CYP2C9 polymorphisms alone, and soon thereafter VKORC1 was identified as a key determinant of warfarin sensitivity. A cohort of 147 patients, originally evaluated for information regarding CYP2C9 polymorphisms and warfarin dose, was genotyped for VKORC1 polymorphisms.[6] A similar association to that of CYP2C9 was identified: patients with one or more variant alleles for VKORC1 required a lower stable dose of warfarin to maintain their INR in therapeutic range.
Multiple polymorphisms have been noted in VKORC1. A retrospective study of 186 European-Americans on long-term warfarin demonstrated that the presence of any of these polymorphisms on at least one allele places the patient in haplotype group A, which is associated with a low-dose phenotype.[11] The major allele was subsequently referred to as haplotype group B or G, depending on the source of the nomenclature. Thus, patients may be designated A/A (homozygous for a variant allele), A/B (heterozygous for a variant allele), or B/B (homozygous for the major allele). The effect of haplotype A is additive, in that A/A patients require a low dose while A/B patients require an intermediate dose as compared to B/B patients.[11]
Haplotype A includes seven single nucleotide polymorphisms (SNPs) in linkage disequilibrium (1173 C>T, rs9934438; 497 T>G, rs2884737; 1542 G>C, rs8050894; 3730 G>A, rs7294, 2255C>T, rs2359612; −4451C>A, rs17880887; −1639 G>A, rs9923231) but is tagged with −1639 G>A (rs9923231), in the promoter region of VKORC1. [11, 26, 27] This SNP appears to alter a transcription factor binding site and reduces the amount of transcribed VKORC1 mRNA, leading to fewer functional copies of the mature VKORC1 protein. VKORC1 mRNA levels in human liver tissue measured by Rieder et al were three times higher in B/B patients as compared to A/A patients, further correlating the polymorphism with the clinical phenotype.[11]
−1639 G>A is present in ~34–47% of the Caucasian population, with similar frequencies in the Hispanic population, and has been identified as the majority allele in the Asian population, with a frequency of approximately ~67–90%, depending on the heterogeneity of the Asian population being studied.[25] It is less frequent in the African-American population, where approximately ~10–30% of patients express the variant allele.[25]
Recent studies have provided evidence that another cytochrome P450 enzyme, CYP4F2, may play a role in warfarin metabolism. It has been hypothesized that a SNP in CYP4F2 (1297G>A, rs2108622) may result in decreased Vitamin K1 (VK1, a form of vitamin K) oxidation activity, leading to higher hepatic levels of VK1 and a higher required warfarin dose. The clinical utility of this association has not been well-delineated at this time.[28] However, the association between CYP2C9 and VKORC1 genotype and stable warfarin dose in the adult population has been clearly defined.
In 2007 the FDA approved a labeling change for warfarin which describes the reported effects of these genetic variants on dose requirements, stating, "lower initiation doses should be considered for patients with certain genetic variations in CYP2C9 and VKORC1 enzymes”.[2] In addition, the FDA approved clinical testing for these genetic variants to inform warfarin dosing. Subsequently, in 2009, the International Warfarin Pharmacogenomics Consortium (IWPC) released a study examining the utility of a dosing algorithm for adults which included genotype along with other clinical factors to determine the most appropriate starting dose of warfarin. The algorithm was created using data from over 4000 patients and was validated with a cohort of over 1000 patients. This algorithm was found to be significantly more accurate in predicting the stable therapeutic dose of warfarin than either fixed-dose regimens or strictly clinical algorithms.[17] Despite this, the overall clinical utility and cost benefit ratio of using genotype to direct warfarin dosing has not been clearly determined. There are several major issues, including cost benefit analysis of reducing under- or overdosing based on routine genotyping, number needed to screen and turnaround time of widespread genotyping, before pharmacogenetics-based warfarin dosing algorithms may be widely implemented in clinical practice.
WARFARIN DISPOSITION IN THE PEDIATRIC POPULATION
Current warfarin dosing algorithms in the pediatric population are extrapolated from adult algorithms, and do not take genotype into account. The American College of Chest Physicians Evidence-Based Clinical Practice Guidelines are based on an age-specific, weight-based algorithm which starts at 0.2 mg/kg/day and is then adjusted based on a nomogram using subsequent INRs.[29] While these algorithms are relatively effective in clinical practice, they are not ideal in light of the inherent differences between adult and pediatric hemostatic systems, which affect the pathophysiology of thrombosis and the response to anticoagulation.[29–31]
The levels of several coagulation factors in children differ from adults significantly in infancy, with differences persisting through adolescence.[32, 33] While anti-thrombin and heparin cofactor II, the two primary physiologic inactivators of thrombin, reach adult levels around 6 months of age – prior to the time when most pediatric patients are started on warfarin – protein C and S do not reach adult levels until adolescence.[32, 33] In addition, children up to the age of 16 have a decreased capacity to generate thrombin as compared to adults.[33]
These physiologic differences may make it difficult to accurately extrapolate a dosing algorithm for children based on adult pharmacogenetic-based dosing algorithms that include genotype, as was demonstrated in one study in which the IWPC algorithm tended to overestimate the actual required dose in pediatric patients.[34] Current practice uses the patient’s age, weight, and serial INRs to dictate initial warfarin dose and dose adjustment to therapeutic levels.[29] As most pediatric physicians who have prescribed and managed warfarin can attest, this method can be time-consuming, inaccurate, and frustrating, leading to both sub- and supra-therapeutic periods with risks for therapeutic failure or adverse effects. Whether or not genotype would provide a valuable addition to current warfarin dosing algorithms in children is currently unknown.
WARFARIN PHARMACOGENETICS IN PEDIATRIC PATIENTS
Between 1995 and 2008, there were fewer than 10 multinational randomized trials initiated addressing the issues of anticoagulation in the pediatric population, and the majority of these have failed to recruit an adequate number of patients to address the primary study question.[30] The incidence of thromboembolic events is significantly less in children compared to adults. Hence, conducting clinical trials in children evaluating the outcome of long-term anticoagulation is difficult both in design and execution due to small numbers of patients. However, warfarin pharmacogenetic discoveries in adults have led to increased efforts to evaluate similar associations in children.
A study comparing warfarin pharmacokinetics in Japanese children and adults found that among patients with most prevalent CYP2C9 allele, pre-pubertal children had a decreased clearance of warfarin as compared to adults of the same genotype (367 ± 226 mL/min vs 667 ± 282 mL/min). In contrast, there was no significant difference in the clearance of warfarin between pre-pubertal children and adults heterozygous for CYP2C9*3 (212 ± 50 mL/min vs 213 ± 62 mL/min, respectively).[35] While the study size was small, this may indicate that the variant alleles for CYP2C9 prevent the full development and maturation of enzyme activity as observed in patients homozygous for the major allele, though the entire scope of the ontogeny of CYP2C9 in the context of a variant allele is unknown. [35].
Recent studies have evaluated the relationship of polymorphisms in CYP2C9 and VKORC1 to warfarin response in children. The first was a prospective cohort of 59 German patients aged 3 months or older who were either taking warfarin or phenprocoumon, another vitamin K antagonist. Nowak-Gottl et al. identifed a significant association between VKORC1 A/A haplotype and lower required doses of warfarin as compared to B/B patients, while CYP2C9 variants had no significant effect. Despite this significant association, age accounted for more variability in stable warfarin dose (28.3%) than genotype (3.7% for VKORC1 and 0.4% for CYP2C9), leading the authors to suggest that the effect of age “overwhelms” the effect of genotype in children.[36] Prior studies had also noted age as an important factor in the dosing of warfarin or acenocoumarin in children.[20, 37] It should be noted however that the study by Nowak-Gottl et al. was limited by a small sample size and that age was highly correlated with weight and height, both of which were excluded from the final regression model. Nevertheless, this study suggested that the most influential contributors to warfarin dosing in children may be related to age or developmental changes associated with age.
A similar study was conducted in pediatric patients in Japan examining the relationship between genotype and stable warfarin dose. The study enrolled 48 patients (ages 0.42 – 19.25 years) and only 1 patient was found to have a CYP2C9 variant, so only the effects of VKORC1 variants were examined. A multiple regression analysis model to predict INR values which included age, weight, VKORC1 genotype and daily warfarin dose demonstrated that patients with a homozygous variant genotype (A/A) required a 28% lower warfarin dose than the patients with heterozygous or homozygous for the major allele (B/B or A/B). The homozygous variant patients also had a higher mean INR than the other patients, despite their lower doses.[38]
In a French study, Moreau et al. examined a cohort of 83 pediatric cardiology patients on warfarin (ages 3 months-18 years, median age 12 years) and 35 patients on fluindione, another vitamin K antagonist. The relative contribution of non-genetic factors and VKORC1/CYP2C9/CYP4F2 genotype to stable warfarin dose and time spent with sub- or supra-therapeutic levels was evaluated by multivariate analysis. Height, target INR, VKORC1 and CYP2C9 genotypes were the main determinants of warfarin dose requirement, accounting for 48.1%, 4.4%, 18.2% and 2.0%, respectively, and explaining 69.7% of the variability in warfarin dose. This suggested non-genetic factors, in this case height, may have more influence on stable warfarin dose than genetic factors and in contrast to what had been noted from the majority of pharmacogenetic studies in adults. In addition, there was no association between any of the covariates and time spent with sub- or supra-therapeutic INR, though patients within their cohort spent 80% of the study time within therapeutic INR range, which may have limited their ability to detect factors that contributed to this clinical endpoint. There was no association between CYP4F2 genotype and warfarin dose requirement. This information was also used to create a model for warfarin dose prediction, which was accurate to within 7 mg/week in 86.7% of patients. The same group is currently evaluating this model in a validation cohort of pediatric patients receiving warfarin.[39]
In a multi-center cross-sectional study of 120 pediatric patients ages 1–18 years old in Canada and the United Kingdom, Biss et al. similarly found that 72.4% of the interindividual variability in warfarin dosing could be attributed to height (29.8% contribution), VKORC1 genotype (26.6%), CYP2C9 genotype (12.8%), and indication for warfarin therapy (3.2%), including both cardiac patients and patients with thromboembolism. This was the most ethnically diverse study population, with 75.8% Caucasian, 8.3% Indian, 5% black, 5% Asian, and 5.8% other. This study also noted a difference in the dose as predicted by the IWPC algorithm versus the patient’s actual maintenance dose, with the IWPC consistently overestimating the dose. Similar to the findings of Moreau et al, CYP4F2 had no effect on stable warfarin dose.[34]
Recently, a small prospective cohort study (n=37) examining the role of genotype in warfarin dosing among pediatric patients with heart disease found that VKORC1 genotype makes a significant contribution to stable warfarin dose, with CYP2C9 genotype accounting for only 5% of the variability. Age, height, and goal INR were also found to contribute to stable warfarin dose. While this study is small and limited to a select sub-population of pediatric patients, it serves as further evidence to support the genotype-phenotype correlation in children.[40]
As previously noted, the Chest guidelines utilize a weight-based dosing algorithm to estimate starting doses of warfarin (0.2 mg/kg/day).[29] In a recent study, Hamberg et al. compared dosing algorithms for warfarin in children, noting that the fixed dose weight-based dosing regimen predicted ideal maintenance dose 33% of time while pharmcogenetics-based algorithms derived from studies conducted by Nowak-Gottl et al., Moreau et al. and Biss et al. predicted ideal maintenance dose 35%, 33% and 41% of time, respectively.[41] However, the fixed dose regimen, while intended to be used to estimate starting doses, overestimated maintenance dose a majority of the time (59%). Although these studies point to significant associations between VKORC1 and/or CYP2C9 genotype and warfarin dose in the pediatric population, they are also all limited by size, lack of ethnic diversity, and lack of information on potential confounders in regards to warfarin disposition, such as diet and vitamin K intake. These limitations make it difficult to conclusively determine whether or not genotype is likely to provide clinically meaningful information in estimating therapeutic warfarin dose, and more importantly, if knowledge and use of genotype could improve the clinical care and outcomes in this population.
CONCLUSIONS
Overall, there is a paucity of published information regarding pharmacogenetics and warfarin response in children. Given the evidence supporting the correlation of commonly occurring CYP2C9 and VKORC1 polymorphisms and warfarin sensitivity in the adult population, the increasing number of pediatric patients on warfarin, and inherent differences in adult and pediatric hemostatic physiology, this is an area in which further investigation may or may not provide information with clinical benefit.
All pediatric studies to date have been limited in scope by sample size as compared to the majority of adult studies conducted. Prospective multicenter trials offer the most potential to determine the true utility of warfarin pharmacogenomics in the pediatric population, allowing for a more accurate representation of the distribution of genotypes among children of multiple ethnicities, and may help to determine the relative contribution of both nongenetic and genetic factors to warfarin dosing. However, the question remains as to the clinical utility of this information, which while physiologically interesting, may ultimately offer limited benefit to clinical practice. Therefore, it may be worthwhile to perform meta-analyses of existing studies via an international pediatric warfarin consortium, similar to what has been established for adults, along with cost-benefit analyses, prior to embarking on costly multicenter trials.
If the relationship of CYP2C9 and VKORC1 polymorphisms to warfarin response in children can be more clearly defined and shown to have clinical relevance, the opportunity will exist for future prospective and randomized trials examining the utility of a warfarin dosing algorithm which includes patient genotype. These future trials may allow for a safer and more accurate method of dosing warfarin in the pediatric population, thus improving clinical care for this already at-risk patient population.
Contributor Information
Susan I. Vear, Vanderbilt University/Monroe Carell Jr Children's Hospital
C. Michael Stein, Vanderbilt University Medical Center
Richard H. Ho, Vanderbilt University/Monroe Carell Jr Children's Hospital.
REFERENCES
- 1.Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 Genotypes and Warfarin Dosing. Clin Pharmacol Ther. 2011;90:625–629. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.U.S. Food and Drug Administration. Warfarin FDA label. [Accessed February 4, 2012];Drugs@FDA home page. http://www.accessdata.fda.gov/drugsatfda_docs/label/2011/009218s107lbl.pdf. Published October 4, 2011.
- 3.Bauman ME, Black K, Kuhle S, et al. KIDCLOT©: The importance of validated educational intervention for optimal long term warfarin management in children. Thromb Res. 2009;123:707–709. doi: 10.1016/j.thromres.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Ageno W, Gallus AS, Wittkowsky A, et al. Oral Anticoagulant Therapy: Antithrombotic Therapy and Prevention of Thrombosis, 9th ed: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest. 2012;141:e44S–e88S. doi: 10.1378/chest.11-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan E, McLachlan A, Pegg M, et al. Disposition of warfarin enantiomers and metabolites in patients during multiple dosing with rac-warfarin. British Journal of Clinical Pharmacology. 1994;37:563. doi: 10.1111/j.1365-2125.1994.tb04305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Andrea G. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105:645–649. doi: 10.1182/blood-2004-06-2111. [DOI] [PubMed] [Google Scholar]
- 7.Aithal GP, Day CP, Kesteven PJL, et al. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. The Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 8.Loebstein R. Interindividual variability in sensitivity to warfarin-Nature or nurture? Clin Pharmacol Ther. 2001;70:159–164. doi: 10.1067/mcp.2001.117444. [DOI] [PubMed] [Google Scholar]
- 9.Taube J, Halsall D, Baglin T. Influence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatment. Blood. 2000;96:1816–1819. [PubMed] [Google Scholar]
- 10.Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–1698. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 11.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1Haplotypes on Transcriptional Regulation and Warfarin Dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 12.Aquilante C, Langaee T, Lopez L, et al. Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clin Pharmacol Ther. 2006;79:291–302. doi: 10.1016/j.clpt.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Cooper GM, Johnson JA, Langaee TY, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz UI, Ritchie MD, Bradford Y, et al. Genetic Determinants of Response to Warfarin during Initial Anticoagulation [Internet] N Engl J Med. 2008;358:999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gage B, Eby C, Johnson J, et al. Use of Pharmacogenetic and Clinical Factors to Predict the Therapeutic Dose of Warfarin. Clin Pharmacol Ther. 2008;84:326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wadelius M, Chen LY, Lindh JD, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113:784–792. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Consortium TIWP. Estimation of the Warfarin Dose with Clinical and Pharmacogenetic Data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong IY, Tirona RG, Schwarz UI, et al. Prospective evaluation of a pharmacogenetics-guided warfarin loading and maintenance dose regimen for initiation of therapy. Blood. 2011;118:3163–3171. doi: 10.1182/blood-2011-03-345173. [DOI] [PubMed] [Google Scholar]
- 19.Moyer TP, O'Kane DJ, Baudhuin LM, et al. Warfarin sensitivity genotyping: a review of the literature and summary of patient experience. Mayo Clin Proc. 2009;84:1079–1094. doi: 10.4065/mcp.2009.0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonduel MM. Oral anticoagulation therapy in children. Thromb Res. 2006;118:85–94. doi: 10.1016/j.thromres.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 21.Zhou S-F, Zhou Z-W, Huang M. Polymorphisms of human cytochrome P450 2C9 and the functional relevance. Toxicology. 2010;278:165–188. doi: 10.1016/j.tox.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Teichert M, Eijgelsheim M, Rivadeneira F, et al. A genome-wide association study of acenocoumarol maintenance dosage. Human Molecular Genetics. 2009;18:3758–3768. doi: 10.1093/hmg/ddp309. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi F, McGinnis R, Bourgeois S, et al. A Genome-Wide Association Study Confirms VKORC1, CYP2C9, and CYP4F2 as Principal Genetic Determinants of Warfarin Dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CR, Goldstein JA, Pieper JA. Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002;12:251–263. doi: 10.1097/00008571-200204000-00010. [DOI] [PubMed] [Google Scholar]
- 25.Scott SA, Khasawneh R, Peter I, et al. Combined CYP2C9, VKORC1and CYP4F2frequencies among racial and ethnic groups [Internet] Pharmacogenomics. 2010;11:781–791. doi: 10.2217/pgs.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L, Ge W, Yu F, et al. Impact of VKORC1 gene polymorphism on interindividual and interethnic warfarin dosage requirement--a systematic review and meta analysis. Thromb Res. 2010;125:e159–e166. doi: 10.1016/j.thromres.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 27.Limdi NA, Wadelius M, Cavallari L, et al. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood. 2010;115:3827–3834. doi: 10.1182/blood-2009-12-255992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang R, Wang C, Zhao H, et al. Influence of CYP4F2 genotype on warfarin dose requirement–a systematic review and meta-analysis. Thromb Res. 2011 doi: 10.1016/j.thromres.2011.11.043. [DOI] [PubMed] [Google Scholar]
- 29.Monagle P, Chan AKC, Goldenberg NA, et al. Antithrombotic Therapy in Neonates and Children: Antithrombotic Therapy and Prevention of Thrombosis, 9th ed: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest. 2012;141:e737S–e801S. doi: 10.1378/chest.11-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monagle P, Chalmers E, Chan A, et al. Antithrombotic Therapy in Neonates and Children: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines (8th Edition) Chest. 2008;133:887S–968S. doi: 10.1378/chest.08-0762. [DOI] [PubMed] [Google Scholar]
- 31.ClinicalTrials.gov. Clarification of optimal anticoagulation through genetics (COAG) [Accessed November 18, 2012];COAG study information page. http://www.clinicaltrials.gov/ct2/show/NCT00839657?term=coag&rank=1. Published May 17, 2012.
- 32.Andrew M, Paes B, Milner R, et al. Development of the human coagulation system in the full-term infant. Blood. 1987;70:165–172. [PubMed] [Google Scholar]
- 33.Andrew M, Vegh P, Johnston M, et al. Maturation of the hemostatic system during childhood. Blood. 1992;80:1998–2005. [PubMed] [Google Scholar]
- 34.Biss TT, Avery PJ, Brandao LR, et al. VKORC1 and CYP2C9 genotype and patient characteristics explain a large proportion of the variability in warfarin dose requirement among children. Blood. 2012;119:868–873. doi: 10.1182/blood-2011-08-372722. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi H. Developmental changes in pharmacokinetics and pharmacodynamics of warfarin enantiomers in Japanese children. Clin Pharmacol Ther. 2000;68:541–555. doi: 10.1067/mcp.2000.110977. [DOI] [PubMed] [Google Scholar]
- 36.Nowak-Gottl U, Dietrich K, Schaffranek D, et al. In pediatric patients, age has more impact on dosing of vitamin K antagonists than VKORC1 or CYP2C9 genotypes. Blood. 2010;116:6101–6105. doi: 10.1182/blood-2010-05-283861. [DOI] [PubMed] [Google Scholar]
- 37.Streif W, Andrew M, Marzinotto V, et al. Analysis of warfarin therapy in pediatric patients: a prospective cohort study of 319 patients. Blood. 1999;94:3007–3014. [PubMed] [Google Scholar]
- 38.Kato Y, Ichida F, Saito K, et al. Effect of the VKORC1 genotype on warfarin dose requirements in Japanese pediatric patients. Drug Metab Pharmacokinet. 2011;26:295–299. doi: 10.2133/dmpk.DMPK-10-NT-082. [DOI] [PubMed] [Google Scholar]
- 39.Moreau C, Bajolle F, Siguret V, et al. Vitamin K antagonists in children with heart disease: height and VKORC1 genotype are the main determinants of the warfarin dose requirement. Blood. 2012;119:861–867. doi: 10.1182/blood-2011-07-365502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen N, Anley P, Yu MY, et al. Genetic and Clinical Determinants Influencing Warfarin Dosing in Children With Heart Disease. Pediatr Cardiol. 2012 doi: 10.1007/s00246-012-0592-1. [DOI] [PubMed] [Google Scholar]
- 41.Hamberg A-K, Friberg LE, Hanséus K, et al. Warfarin dose prediction in children using pharmacometric bridging—comparison with published pharmacogenetic dosing algorithms. Eur J Clin Pharmacol. 2013 doi: 10.1007/s00228-012-1466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


