SUMMARY
Transcription factors and DNA regulatory binding motifs are fundamental components of the gene regulatory network. Here, by using genome-wide binding profiling, we show extensive occupancy of transcription factors of myogenesis (MyoD and Myogenin) at extragenic enhancer regions coinciding with RNA synthesis (i.e. eRNA). In particular, multiple regions were transcribed to eRNA within regulatory region of MYOD1, including previously characterized Distal Regulatory Regions (DRR) and Core Enhancer (CE). While CERNA enhanced RNA polymerase II (PolII) occupancy and transcription at MYOD1, DRRRNA acted to activate the downstream myogenic genes. The deployment of transcriptional machinery to appropriate loci is contingent on chromatin accessibility, a rate-limiting step preceding PolII assembly. By nuclease sensitivity assay, it appear that eRNAs regulate genomic access of the transcriptional complex to defined regulatory regions. In conclusion, our data suggest that eRNAs contribute to establishing a cell-type-specific transcriptional circuitry by directing chromatin-remodeling events.
INTRODUCTION
Recent studies have documented a pervasive transcription (up to 80%) of the human genome (ENCODE project, http://encodeproject.org/ENCODE). This fraction goes well beyond the ~1% corresponding to protein-coding exons, implying that the transcript, rather than the gene (defined as protein-coding sequence), more accurately represents the fundamental unit of the genome (Djebali et al., 2012, Stamatoyannopoulos, 2012). The widespread transcription generates a vast collection of non-protein-coding transcripts including ribosomal, small nuclear/nucleolar, antisense, micro- and long intergenic non-coding RNAs. Recent discoveries demonstrate that RNAs transcribed from regulatory elements constitute another class of non-coding transcripts and are proposed to take part in the gene regulatory networks (Orom and Shiekhattar, 2011).
Acting as primary determinant of cell-type specificity, DNA enhancer elements increase transcriptional output of protein-coding counterparts over large genomic distances in an orientation-independent manner (Bulger and Groudine, 2011, Maston et al., 2006). Additionally, enhancers are characterized by a defined chromatin signature consisting of high H3K4me1-to-H3K4me3 ratio, p300 acetyltransferase binding, acetylated histones and increased sensitivity to nucleases (Bernstein et al., 2012, Biddie et al., 2011, Creyghton et al., 2010, Cui et al., 2009, Djebali et al., 2012, Ernst et al., 2011, Ghisletti et al., 2010, Heintzman et al., 2007, Melgar et al., 2011, Rada-Iglesias et al., 2011). PolII occupies enhancers of stimulated macrophages, neurons, keratinocytes and breast cancer cells resulting in localized transcription of eRNAs (De Santa et al., 2010, Hah et al., 2013, Kim et al., 2010, Lam et al., 2013, Li et al., 2013, Melo et al., 2013, Orom et al., 2010, Ostuni et al., 2013). Currently, eRNAs are described as a rare population of ~0.5–5kb transcripts, some of which undergo polyadenylation (based on polyA+ sequencing) (De Santa et al., 2010, Djebali et al., 2012, Hah et al., 2013, Kim et al., 2010, Melgar et al., 2011, Orom et al., 2010). The significance and molecular mechanisms by which eRNAs exert their function are currently under investigation.
Specification and differentiation of skeletal muscle are carried out by the activity of DNA-binding basic helix-loop-helix (bHLH) myogenic regulatory factors (MRFs) Myf5, MyoD, Myogenin (MyoG), and MRF4 (Rudnicki et al., 2008, Tapscott, 2005). MyoG, a downstream target of MyoD gene regulatory network, is essential for muscle differentiation (Hasty et al., 1993, Nabeshima et al., 1993). MRFs directly bind to their DNA consensus elements (E-box, CANNTG), trigger mRNA transcription and thereby orchestrate the processes of muscle cell formation (Rudnicki et al., 2008, Tapscott, 2005). Recent genome-wide studies show that MRFs predominantly occupy the extragenic regions (>10kb from protein-coding genes), marked by acetylated histones and co-occupied by PolII (Blum et al., 2012, Cao et al., 2010, Soleimani et al., 2012).
To characterize the extragenic areas occupied by MRFs, we utilized a genome wide analysis of MyoD/MyoG binding, PolII occupancies, histone modifications and corresponding transcriptome. Here, we report that MyoD and MyoG occupy thousands of extragenic locations with active enhancer signature (i.e. high H3K4me1-to-H3K4me3 ratio, acetylated histones and PolII-occupied) and generated RNA. Regulatory sequences contained within ~24kb upstream region of MYOD1 control its spatiotemporal expression during embryogenesis (Chen et al., 2001, Goldhamer et al., 1992). Within this region, the concerted activity of two DNA enhancer elements, CE and DRR, specifies the level of MyoD expression in the myogenic lineage (Asakura et al., 1995, Chen et al., 2001, Faerman et al., 1995, Goldhamer et al., 1992, Kucharczuk et al., 1999, Tapscott et al., 1992). We found transcripts corresponding to CE and DRR enhancers (i.e. CERNA and DRRRNA, respectively) in C2C12 and skeletal muscle satellite cells. Remarkably, CERNA and DRRRNA depletion resulted in reduced chromatin accessibility and PolII residency at MYOD1 and MYOG loci, respectively. Thus, our data suggest a role for eRNAs in establishing cell-type-specific chromatin-restructuring and consequently transcriptional circuitry.
RESULTS
Genome-Wide Binding of MyoD and MyoG
We mapped genome-wide occupancies of MyoD and MyoG in mouse C2C12 skeletal muscle cells, which closely resemble primary myoblasts and have been extensively used as a model of myogenesis (Blais et al., 2005, Cao et al., 2010). Similar to primary skeletal muscle cells, MyoD protein was detected in proliferative myoblasts (MB, ~50–70% confluency) and terminally differentiated myotubes (MT, 24–96hrs in DM), whereas MyoG protein was observed at the onset of differentiation (Figure S1A) (Rudnicki et al., 2008, Tapscott, 2005). We verified the specificity of commercially available antibodies (Figure S1B) and performed Chromatin ImmunoPrecipitation followed by high-throughput Sequencing (ChIP-Seq) accordingly (i.e. MyoD from MB/MT and MyoG from MT). The sequence reads were pooled from at least two independent runs. We utilized Model-based Analysis for ChIP-Seq (MACS) algorithm to call for MyoD/MyoG peaks (Zhang et al., 2008). Using specified parameters (p≤10−6 and FDR≤1%), selected MyoD+ peaks were 18,142 and 39,700 in MB and MT, respectively, MyoG+ regions were 35,273 in MT (Table S1) and overlapped (~77%) with MyoD+ binding sites. Because of this significant co-occupancy, henceforth we will refer to MyoD+/MyoG+ peaks as simply MyoG+. We observed ~5% of MyoG+ peaks at promoters (~1kb from Transcriptional Start Sites, TSS), ~4% in exons, ~42% in introns and ~49% in the extragenic regions (i.e. excluding ~1kb of UCSC annotated protein-coding genes) (Figure 1A). These distributions are in general agreement with recent reports, and differences may arise based on peak discovery methodology, genomic compartmentalization (i.e. boundaries considered) and other experimental variability (Cao et al., 2010, Soleimani et al., 2012). When distribution was normalized to DNA length (in kb) of distinct genomic partition (# of peaks/kb), higher peak density was apparent at the promoter regions with majority of MyoG+ genes marked by H3K4me3 (transcriptionally active chromatin modification) and bound by PolII (Figure S1C–F).
Figure 1. Transcription at MyoD+/MyoG+ Extragenic Regions.
A. Genome-wide distribution of MyoG occupancy. B. Positive correlation of MyoD+/MyoG+ tags with H3K4me1+ peaks. Binding sites were binned for presentation. C. H3K4me1, PolII and H3K4me3 occupancy profiles at MyoG+ (2,170) enhancer sites. D. Number of MyoG+ peaks in the extragenic regions and their occurrence with H3K4me1, H3K27ac, PolII and RNA transcripts. E. RNA read profile at MyoG+ enhancers. F. eRNA-assigned upregulated genes (eRNA+) have higher mRNA levels than those without eRNA assigned (eRNA−). Data are represented as mean +/− SEM. G. PolII occupancy profile at eRNA+ and eRNA- upregulated genes in 48hrs MT.
RNA Synthesis at the Extragenic MyoD/MyoG Binding Sites
Considering that a significant fraction (~49%) of total MyoG binding occurred in the extragenic compartment (Figure 1A), we asked whether these peaks corresponded to known extragenic chromatin landmarks associated with enhancers. In this context, we surveyed for the enhancer signature (i.e. H3K4me1) within ~1–2kb of MyoG+ sites. We obtained and analyzed ChIP-Seq data for H3K4me1 (Table S1) and observed the majority (~80%) of the extragenic MyoG+ peaks within H3K4me1+ domains (Figure 1B–D). In addition to H3K4me1, active enhancers are characterized by acetyltransferase occupancy (p300/CBP) and acetylated histones. Using published data (Blum et al., 2012), we found ~90% of the MyoG+/H3K4me1+ extragenic regions were also marked by acetylated H3K27 (Figure 1D, H3K27ac). Next, we observed that a sizable fraction (~36%) of MyoG+/H3K4me1+/H3K27ac+ sites were occupied by PolII (Figure 1D) (Mousavi et al., 2012). MyoG+/H3K4me1+/H3K27ac+/PolII+ locations were associated with low or no H3K4me3 mark (Figure 1C), thus excluding un-annotated promoters in our analysis. To ascertain whether these regions were transcriptionally active, we performed paired-end RNA-Seq (PE-Seq) from ribosome-depleted RNA fraction from MT (76bp/end, see Table S1 for number of reads). Approximately 72% of the MyoG+/H3K4me1+/H3K27ac+/PolII+ extragenic regions were RNA+ (≥5 reads within −/+2kb), while under the same criterion less than 20% of negative control regions (i.e. MyoG+/H3K4me1-/H3K27ac-) exhibited RNA reads (Table S2). RNA originating from MyoG+/H3K4me1+/H3K27ac+/PolII+ regions occurred in sense and/or antisense orientation extending ~1–2kb from MyoG+ sites (Figure 1E). Unless noted elsewhere, we will refer to this RNA population as “eRNA”. While we detected a fraction of eRNA+ as polyA+ (Figure 1E), this may have been a consequence of indirect purification. Therefore, another method was devised to further probe for polyadenylation of specific eRNAs (see below). Overall, these data are consistent with recent publications demonstrating the presence of eRNAs at the extragenic MyoD+ sites (Blum et al., 2012, Trapnell et al., 2010).
Subsequently, we verified the presence of eRNAs and assessed whether their expression depended on MyoD or MyoG. To this end, we chose random MyoG+/H3K4me1+/H3K27ac+/PolII+/eRNA+ regions on different chromosomes and performed conventional RT-qPCR and ChIP-qPCR following MyoD RNAi (MyoDi) or MyoG RNAi (MyoGi). MyoD and MyoG occupancies were reduced following RNAi at protein-coding promoters as well as eRNA+ locations (Figure S1G–H, J–K). Nonetheless, eRNA levels were preferentially reduced following MyoDi only (Figure S1I,L), signifying the distinctions between MyoD and MyoG in regulating the expression of these eRNAs.
Next, we assigned eRNA+ regions to the nearest downstream genes within 100kb. This way, 1,353 eRNA+ regions were assigned to 809 protein-coding genes (i.e. eRNA+ genes) (Table S3). While majority of these genes (595) were transcribed, a smaller fraction (214) were transcriptionally silent (transcription was assessed with a cut-off of 1 read per kilobase per million-RPKM) (Table S3). During C2C12 myogenic differentiation (MB to 48hrs MT), upregulated genes assigned to eRNAs (194) exhibited an average transcript level of ~91 RPKM (median=23.2), whereas those upregulated but not associated with eRNAs (1,561) had an average transcript level (RPKM) of ~35 (median=8.7) in MT (Figure 1F). Concordantly, eRNA-assigned genes had significantly higher PolII occupancy at their TSS and gene bodies than those upregulated genes without assigned eRNAs (Figure 1G).
eRNA Transcribed from the Distal Regulatory Region (DRR) near MYOD1
Two major gene ontology terms identified by the eRNA-assigned genes were “muscle organ development” and “regulation of transcription from PolII promoter” (Table S3). In the latter category, list of genes with multiple eRNA-assigned regions included master transcription factors MEF2A, FOXO1, MYOD1 and MYOG (Table S3). Given the central role of MyoD in establishing muscle cell lineage and extensive literature on its transcriptional control (Rudnicki et al., 2008, Tapscott, 2005), we initially focused on characterizing eRNAs in a genomic region spanning ~5–6kb upstream of MYOD1 encompassing an evolutionarily conserved enhancer element, DRR (Asakura et al., 1995, Chen and Goldhamer, 2004, Kablar et al., 1999) (Figure 2A). Our ChIP-Seq data demonstrated de novo recruitment of MyoD/MyoG at DRR (Figure 2A, highlighted near light-blue rectangle) in differentiating C2C12 MT coinciding with the appearance of H3K4me1 (absence of H3K4me3) and PolII occupancy (Figure 2A). PolyA+ RNA-Seq demonstrated eRNA synthesis at DRR in MT (Figure 2A). Moreover, PE-Seq from differentiated C2C12 cells and ribosome-depleted RNA-Seq (Single run, 50 bases) from purified muscle satellite cells [by Fluorescence-Activated Cell Sorting (FACS)], revealed the synthesis of eRNA at DRR (i.e. DRRRNA) with four distinct eRNA peaks (Figure 2A, highlighted in blue). Analyses of cell fractionation, myogenic differentiation, qPCR (with oligo dT) and endonuclease treatment suggested that DRRRNA was mainly nuclear (other eRNAs were shown to be nuclear as well), upregulated during differentiation, and potentially subjected to polyadenylation and single-stranded (Figure 2B–C, Figure S2). Similar to other nuclear eRNAs examined (Figure S1G–I), DRRRNA levels were significantly reduced following MyoDi and to a lesser extent by MyoGi (Figure 2D), suggesting that its transcriptional regulation is under the concerted control of these transcription factors.
Figure 2. eRNA Synthesis at DRR of MYOD1 (DRRRNA).
A. Occupancies of MyoD, MyoG, H3K4me1, H3K4me3, PolII and presence of RNA (polyA+ and ribosome-depleted) in regions within ~5–6kb of MYOD1 in C2C12 MB, MT and activated satellite cells. For better visualization, RNA scales are set to logarithmic. Region highlighted in light blue depicts DRRRNA-coding region. B. Nuclear (Nuc) distribution of DRRRNA, assessed by fractionation from cytoplasmic (Cyto) compartment. Data are represented as mean +/− SEM. C. Upregulation of DRRRNA during myogenic differentiation in C2C12 and primary cells (MB, 50–70% proliferating myoblasts, MT, 48hrs differentiated myotubes). Data are represented as mean +/− SEM. D. Significant reduction of MyoD and DRRRNA transcript levels following MyoDi (RT-qPCR), and reduction of MyoG and DRRRNA transcript levels after MyoGi (RT-qPCR). Biological replicates (n) and p-value are shown, data are represented as mean +/− SEM.
eRNA from the Core Enhancer (CE) Promotes MyoD Expression
A ~24kb genomic fragment upstream of MYOD1 controls spatiotemporal expression of MyoD during embryogenesis (Chen et al., 2001, Goldhamer et al., 1992). To expand our analysis, we inspected this region as well as its surrounding area (~50kb in total) and observed multiple (~10, including DRR) overlapping MyoD/MyoG peaks with enclosing H3K4me1, little or no H3K4me3 modification (except at MYOD1) and corresponding PolII occupancy (Figure 3A). Moreover, PE-Seq revealed a prevalent presence of eRNAs (in forward and reverse orientation) throughout ~50kb region in C2C12 MT as well as in FACS-sorted muscle progenitors (Figure 3A, data not shown). Genomic locations where eRNAs were originating within the MYOD1 regulatory region are highly conserved in mammals (Figure 3A, Placental Mammal Conservation by PhastCons), and some of these conserved eRNA+ regions corresponded to DNaseI sensitivity in mouse skeletal muscle (Figure 3A, Skeletal Muscle DNaseI HS from ENCODE/University of Washington), thus further supporting a dynamic chromatin structure and active transcription within these locations.
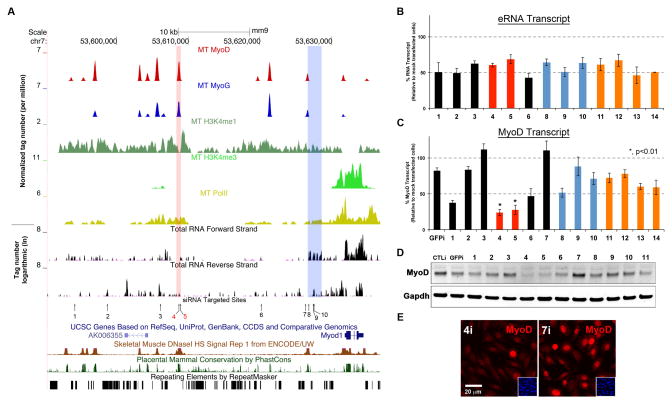
Figure 3. CERNA is Required for MyoD Expression.
A. MyoD, MyoG, H3K4me1, H3K4me3, PolII occupancy profiles as well as PE-Seq reads (ribosome-depleted) in forward and reverse strand within ~50kb regions of MYOD1. CE and DRR are highlighted in red and blue bars, respectively. The sites of siRNAs are labeled 1–10. Sites 11–14 are located elsewhere on other chromosomes (see Figure S3). As well, sites of DNaseI sensitivity (ENCODE/UW), mammalian conservation (PhastCons) and repetitive elements (RepeatMasker) are shown. B. Targeted eRNA levels (measured by RT-qPCR) after RNAi. Data are represented as mean +/− SEM. C–D. Relative MyoD transcript levels (measured by RT-qPCR) and protein levels (shown by western blotting) following eRNAi, respectively. In C, data are represented as mean +/− SEM. E. MyoD immunofluorescence detection (red) after eRNAi against regions 4 and 7 in Panel A. Insets are DAPI-labeled nuclei in the same field of view.
In an attempt to gain insight into their function, we considered depletion of eRNAs and assessment of ensuing impact on the MyoD transcript levels. To this end, we devised a screening approach where ten different small interfering RNA duplexes (siRNA) were designed to target various regions upstream of MYOD1 with proximity to MyoD+/MyoG+/H3K4me1+ peaks (Figure 3A, #1–10). As controls, siRNAs were designed to target four eRNA+ regions on other chromosomes (Figure S3) and another against green fluorescent protein (GFPi). Transfected C2C12 cells were placed in differentiation media (DM) for 4–6hrs and harvested thereafter for analysis. While all siRNAs tested resulted in uniform depletion of targeted eRNAs (~50%, Figure 3A–B), MyoD transcript and protein levels were severely diminished (>70%) only with those targeting CE (CEi) (Figure 3A, highlighted in red, Figure 3B–E), siRNAs against DRR (DRRi, Figure 3A, #8,9,10, highlighted in blue) did not significantly impact MyoD transcript and protein levels at early differentiation time-points (Figure 3B–D). These results suggest that eRNA from CE (i.e. CERNA) is critical for MYOD1 expression.
DRRRNA Activates the Myogenic Gene Regulatory Network
While DRR, in conjunction with MyoD promoter, has been associated with enhancing reporter gene expression (Asakura et al., 1995, Chen et al., 2001, Chen et al., 2002), it is also found to be dispensable for endogenous MyoD expression in embryonic and neonatal stages (Chen et al., 2002) (Figure 3). Given the suggestion that DRR may be essential for the early myogenic differentiation program (Chen et al., 2002), we wondered whether DRRRNA plays a broader role within the myogenic gene regulatory network (i.e. activating MyoD targets). To this end, we examined the expression of MyoD target genes (i.e. MyoD, MyoG and Myh) by immunofluorescence in late differentiating cells (24–48hrs in DM). While GFPi had little consequence on the induction of myogenic genes as compared to mock-transfected control cells, DRRi cells failed to activate MyoG and Myh (Figure 4A). In fact, during early differentiation time-point (4–6hrs), MyoG transcript levels were significantly and consistently reduced by three non-overlapping siRNAs against DRRRNA (Figure 4B, Figure S4A). Moreover, DRRi hindered the myogenic differentiation program (without apparently affecting other pathways), as confirmed by inadequate induction of myogenic genes (RNA-Seq from polyA+ fraction) as compared to GFPi cells (Figure 4B, left).
Figure 4. DRRRNA Promotes Myogenic Differentiation.
A. Immunofluorescence detection (red) of MyoD, MyoG, Myh in control, GFPi and DRRi cells. Note the reduction of MyoG and Myh in DRRi cells. B. Heat maps depicting the relative RNA levels (PolyA+ RNA-Seq) of myogenic genes in DRRi as compared to GFPi, and along side, relative transcript levels of myogenic genes from overexpressing cells (DRR1.2ox) as compared those expressing GFP (GFPox) are shown. Red arrows highlight MyoD and MyoG. C. Profiles of MyoD and MyoG occupancies as well as RNA (PE-Seq ribosome-depleted), the lengths of DRR fragments for overexpression are shown below. D. Increase in MyoG mRNA levels following overexpression of DRR1.2 and DRR2.0 fragments as compared to DRR0.5, a fragment −12kb upstream of MyoD (−12kb) and GFP. Biological replicates (n) and p-value are shown, data are represented as mean +/− SEM.
To further investigate the role of DRRRNA within the myogenic gene regulatory network, we also performed complementary experiments in which several lengths of DRR (Figure 4C, DRR fragments are shown), were constructed for over-expression. C2C12 MB cells were transduced (using retroviral preparations) with either empty vector, GFP, randomly chosen fragment ~12kb upstream of MyoD (−12kb) and various lengths of DRR and probed for their effect on MyoD and MyoG expression. Ectopic expression of GFP, −12kb and DRR0.5 did not significantly modify MyoD or MyoG transcript levels (Figure 4D, Figure S4B–C). Remarkably, over-expression of 1.2kb and 2.0kb fragments of DRR (DRR1.2 and DRR2.0), in trans, activated MyoG expression and the rest of the myogenic gene regulatory network without influencing MyoD transcript levels (Figure 4C–D, Figure S4C). Hence, these observations suggest that the effects exerted by DRRRNA require the first half of DRRRNA region (Figure 4C) and may occur irrespective of its genomic locality (i.e. in trans), and that perhaps its genuine position confers efficiency within the myogenic gene regulatory network.
eRNAs Regulate PolII Occupancy at Specific Promoter Regions
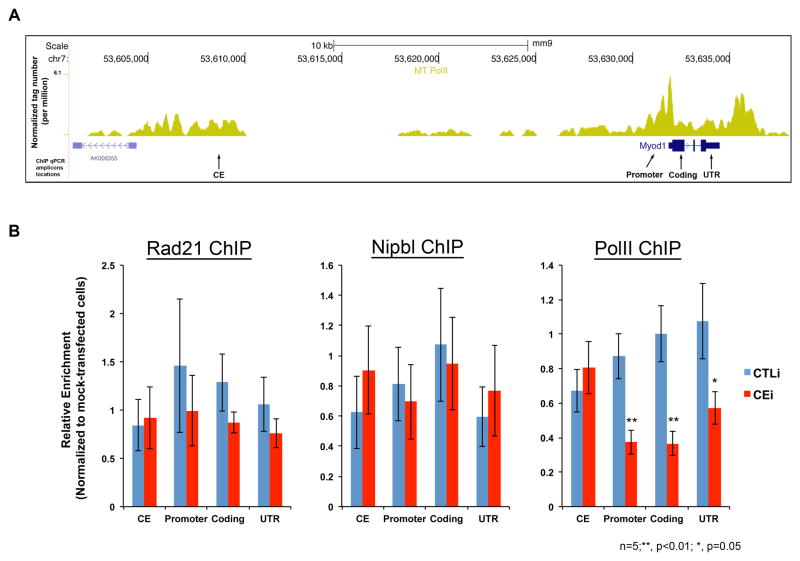
In an attempt to decode the molecular mechanisms involved in CERNA-mediated enhancement of MyoD expression, we set out to assess the chromatin signature at CE and MYOD1 proximal regions in C2C12 cells (Figure 5A). To this end, cells were treated with siRNA 4 (CEi) and 7 (CTLi) or mock-transfected (Figure 3A) and assessed for various occupancies by ChIP-qPCR (Figure 5A, ChIP-qPCR amplicons). Given the depletion of MyoD following CEi (Figure 3C), we initially considered the involvement of a silencing mechanism (i.e. chromatin regions acquiring H3K27me3 modification) in this process. In this setting, we envisioned the CERNA to act as anti-repressive element at MYOD1. Accordingly, we evaluated H3K27me3 occupancy (by ChIP-qPCR) at CE and MYOD1 and found no significant gain of this modification following CEi (Figure S5). Recently, long-range chromatin interactions have been demonstrated between enhancers and promoters (Li et al., 2012, Sanyal et al., 2012). Nevertheless, as evident from the literature and argued by Taberlay et al., the available genome-wide assays for chromatin conformation analyses are unreliable for interpreting connectivity at distances nearing ~20kb (i.e. length between CE and MYOD1) (Dekker et al., 2002, Taberlay et al., 2011). However, the chromatin occupancies of the cohesin complex, which facilitates enhancer-promoter looping, may infer such interactions (Kagey et al., 2010, Li et al., 2013, Phillips-Cremins et al., 2013, Taberlay et al., 2011). A member of cohesin complex, Rad21, is recruited to CE and MYOD1 upon its transcriptional activation (Taberlay et al., 2011). Therefore, we also assessed the occupancy of Rad21 and a cohesin-loading factor (Nipbl) and again found no significant alteration upon CEi at CE or MYOD1 (Figure 5B). Lastly, given the PolII-centered long-range chromatin interaction (Li et al., 2012), we asked whether CERNA is functionally associated with transcriptional machinery loading and assembly. In examining this link, we found that while CEi did not disrupt PolII occupancy at CE, it severely reduced PolII occupancy at MYOD1 proximal regions (Figure 5B, PolII ChIP). Altogether, these results suggest that while CERNA does not influence the loading of the cohesin complex, it specifically influences PolII residency at MYOD1.
Figure 5. CERNA Influences PolII Occupancy at MYOD1.
A. PolII occupancy (ChIP-Seq) profile at regulatory regions of MYOD1. Arrows are locations of ChIP-qPCR amplicons as CE, promoter, coding and 3′ untranslated region (UTR). B. Charts show relative ChIP-qPCR enrichment in CTLi (control) and CEi (CERNA siRNA) cells normalized to mock-transfected control samples. Biological replicates (n) and p-value are shown, data are represented as mean +/− SEM.
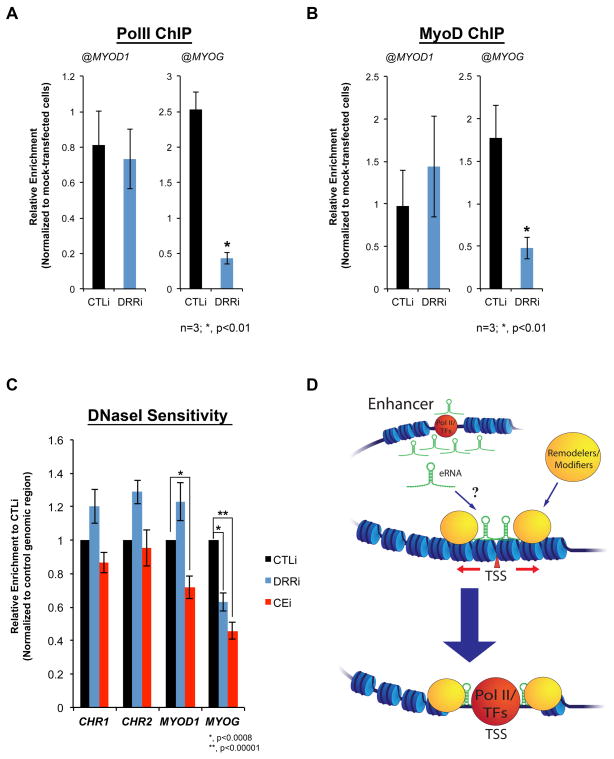
The above findings prompted us to evaluate whether RNA-assisted PolII recruitment could also explain the effect of DRRRNA on MyoG expression (Figure 4A,B). Indeed, ChIP experiments revealed that DRRi resulted in the reduction of PolII at MYOG, but not at MYOD1 (Figure 6A). Sequence-specific DNA binding factors may, in part, mediate the chromatin recruitment of PolII complex (Kadonaga, 2004, Stargell and Struhl, 1996, Weake and Workman, 2010). In this context, reduced PolII occupancy at the MYOG in DRRi cells may be a consequence of suboptimal MyoD recruitment (Rudnicki et al., 2008, Tapscott, 2005). Therefore, we examined and found that despite similar recruitment kinetics at MYOD1 and MYOG promoters during differentiation (Figure 2A, Figure S6), MyoD occupancy was surprisingly only altered at MYOG in DRRi cells (Figure 6B), suggesting distinct engagement mechanisms. Overall, these observations suggest that eRNAs are involved in the assembly of transcriptional system onto regulatory regions.
Figure 6. eRNAs Regulate Chromatin Accessibility.
A–B. Charts represent relative enrichment (normalized to mock-transfected) of PolII and MyoD occupancies, respectively, at MYOD1 and MYOG in CTLi (control) and DRRi (DRRRNA siRNA) cells. Biological replicates (n) and p-value are shown, data are represented as mean +/− SEM. C. Chart shows relative enrichment of amplicons from CTLi, DRRi and CEi cells [values were normalized to a control genomic region with no accessibility (mm9, chr15:15,304,393-15,304,469). Values represent three biological replicates and 3–5 technical replicates per biological sample (p-value are shown, data are represented as mean +/− SEM). D. A drawing depicting a potential role of eRNAs (either direct or accompanied by chromatin remodelers/modifiers) in directing chromatin accessibility at protein-coding promoters.
eRNAs Promote Chromatin Accessibility at Specified Loci
Given the reduction of PolII at MYOD1 and MYOG upon CEi and DRRi respectively, and selective impairment of MyoD recruitment at the MYOG promoter in DRRi cells, we wondered whether eRNAs impact molecular events preceding the assembly of active PolII complex (i.e. the pre-initiation complex). In particular, we sought to assess chromatin accessibility as an indication for remodeling events, and the prerequisite to PolII assembly (Biddie et al., 2011, Boeger et al., 2005, Clapier and Cairns, 2009, Kornberg and Lorch, 1992, Lelli et al., 2012, Paranjape et al., 1994). Prior to terminal differentiation of myogenic cells, MYOG locus is inaccessible to various nucleases and requires the activity of remodelers for its transcriptional activation (de la Serna et al., 2005, Gerber et al., 1997). Therefore, we also assessed MYOG sensitivity (as well as other loci) to DNaseI treatment following DRRi. Whereas control genomic regions (CHR1:162143164-162143229 at CACYBP, and CHR2:22923544-22923659 at ACBD5, mm9) as well as MYOD1 appeared equally sensitive to DNaseI in DRRRNA-depleted and control cells, DRRi resulted in a significant reduction in DNaseI accessibility at MYOG (Figure 6C, blue columns), suggesting differences in chromatin remodeling at this locus. Given the above observations, we wondered whether CERNA is involved in the chromatin-remodeling events at MYOD1. In parallel experiments, CEi cells were shown to be less accessible to DNaseI treatment at MYOD1 as well as MYOG, as an indirect consequence of MyoD/DRRRNA reduction (Figure 6C, red columns). Therefore, these data suggest that eRNAs from MYOD1 regulatory region promote chromatin remodeling and PolII assembly at defined loci within the myogenic gene regulatory network, consequently establishing cell identity.
DISCUSSION
A fundamental question in biology is how DNA enhancer elements exert their preeminent regulatory function. Studies suggest that enhancers behave as modular platforms for assembly of long-range interactions in which transcription factors, chromatin regulators, cohesin/Mediator complex and PolII elevate gene transcription (Dean, 2006, Deng et al., 2012, Kagey et al., 2010, Li et al., 2012, Phillips-Cremins et al., 2013). In addition to their participation in increasing mRNA transcription, enhancers are also sites of active transcription (this study) (Blum et al., 2012, De Santa et al., 2010, Kim et al., 2010, Lai et al., 2013, Orom et al., 2010). Whether eRNAs are simply byproducts of adjoining transcriptional processes or active components of the gene regulatory network is currently under investigation. Our studies as well as recent reports are in favor of the latter where regulatory RNAs, including eRNAs, positively impact the expression of target protein-coding genes (Lai et al., 2013, Lam et al., 2013, Li et al., 2013, Ling et al., 2004, Orom et al., 2010).
In this study, we report pervasive binding of MyoD and MyoG throughout the genome, supporting and extending previous findings to demonstrate their co-occupancies at sites with distinct chromatin signatures (Cao et al., 2010, Soleimani et al., 2012). In conjunction with a recent report, we provide evidence for the presence of PolII and eRNAs at thousands of MyoD+/MyoG+ extragenic sites, thereby demonstrating active transcription at these loci (Blum et al., 2012). Our association study suggests higher expression of genes near eRNA+ sites (Figure 1F). If we assume their direct interaction, then our assertions are in agreement with a recent ENCODE project demonstrating higher expression of genes that physically interact with enhancer elements (Sanyal et al., 2012). The higher occupancy of PolII at eRNA-assigned genes could also be the consequence, rather than the cause, of corresponding eRNA expression. In fact, we demonstrate that CERNA is essential for elevated PolII occupancy at MYOD1 (Figure 5B). These observations suggest that eRNAs facilitate molecular events culminating in higher PolII occupancy and engagement at protein-coding loci.
The MYOD1 regulatory region (~50 kb) is recently labeled as a “super-enhancer”, representing a relatively large genomic region with high occupancy of master regulators and Mediator complex (Whyte et al., 2013) (Figure 3A). In our study, an eRNA from this super-enhancer (i.e. CERNA) regulated the transcription of MYOD1 while another (i.e. DRRRNA) activated MYOG expression. Therefore, one can envision that the myogenic and perhaps other super-enhancers serve as templates for numerous eRNAs that, in concert with master regulators, establish a cell-type-specific gene regulatory network.
While DRR enhancer directs gene expression in early-differentiated myogenic cells (Asakura et al., 1995, Kablar et al., 1997, Tapscott et al., 1992), CE propels mRNA transcription in somitic precursors during embryogenesis (Faerman et al., 1995, Goldhamer et al., 1995, Goldhamer et al., 1992). Our findings are consistent in that we detected DRRRNA, associated with enhancer function, in early differentiating C2C12 cells, whereas CERNA is also transcribed in proliferating myoblasts. While our data suggest that DRRRNA may be processed and polyadenylated, direct confirmation of these properties awaits cloning of the DRRRNA. DRRRNA promotes the expression of gene downstream of MyoD gene regulatory network (i.e. MyoG) on another chromosome, suggesting that DRRRNA functions in trans. However, given the autoregulatory nature of the myogenic gene regulatory network, we cannot exclude the possibility that DRRRNA may also indirectly influence MyoD transcription at later stages of cell differentiation. While there are demonstrated examples of in trans regulatory roles for non-coding RNAs (Rinn et al., 2007, Sanchez-Elsner et al., 2006), it is reasonable to speculate that their specificity is implemented, at least in part, by RNA-RNA or RNA-DNA pairing.
Both DRRRNA and CERNA increase PolII occupancy at different loci. Given the molecular steps preceding PolII recruitment (Cosma, 2002, Narlikar et al., 2002), we examined events prior to PolII assembly and discovered reduction in chromatin accessibility at promoters upon eRNA depletion. These observations suggest that eRNAs function at the level of chromatin restructuring and/or decondensation conceivably by: 1) Targeting chromatin to facilitate nucleosome rearrangement or 2) Recruiting remodelers/modifiers (Figure 6D). While several chromatin remodelers (e.g. SWI/SNF complex) and modifiers (e.g. histone acetyltransferases) have been implicated in the myogenic gene activation (McKinsey et al., 2001, Puri and Mercola, 2012), future studies should address whether eRNAs prime the chromatin and orchestrate the recruitment of various complexes. In this context, eRNA-mediated recruitment of chromatin remodelers may occur in a complementarity-dependent fashion (i.e. RNA-RNA or RNA-DNA) to stabilize DNA conformation conducive for transcription.
In conclusion, our findings provide insights into hierarchy within the myogenic gene regulatory network. Together with previous findings, our data suggest that transcription at CE occurs upstream of MyoD gene regulatory network (Taberlay et al., 2011). CERNA promotes chromatin restructuring at MYOD1, thereby resulting in PolII recruitment and expression (Figure 6D). Upon MyoD gene activation and during differentiation, DRRRNA facilitates the necessary chromatin modifications at MYOG for its expression, culminating in a coherent feed-forward loop for the myogenic program. Thus, eRNAs direct gene activation and dosage by modulating chromatin remodeling at distinct regulatory genomic regions.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
All cells were cultured at 37°C with 5% CO2. All cell media were supplemented with 500 μg/ml Penicillin-streptomycin-glutamine (Gibco). C2C12 cells (ATCC) were grown in 1x Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% qualified fetal bovine serum (FBS) (26140, Gibco). For differentiation, FBS was replaced with 2% horse serum and 1x insulin-transferrin-selenium (Gibco). For overexpression of enhancer fragments, cells were transduced and puromycin-selected.
Antibodies
Antibodies used were anti-Myh (MF20, DSHB), anti-MyoD (C-20, Santa Cruz), anti-Myogenin (F5D, Santa Cruz), anti-RNA polymerase II (8WG16, Covance), anti-Rad21 (ab992, Abcam) and anti-Nipbl (A301-779A, Bethyl). Alexa Fluor® 594 goat anti-mouse (H+L) or anti-rabbit (Molecular Probes) was used as secondary antibodies for immunofluorescence. Dynabeads® magnetic beads (100-04D, Invitrogen) were used for ChIP experiments. For other ChIP antibodies, refer to Table S1.
Plasmid Construction
DRR was subcloned (BamHI/XhoI) from MD6.0-lacZ (Asakura et al., 1995). Insert parental construct was restricted (KpnI-blunted/XhoI) and was inserted into pHAN retroviral vector (BamHI-blunted/SalI). Primers were designed (Table S4) to subclone DRR2.0 fragment into pHAN retroviral vector. DRR0.5 (BamHI/XmnI, 567bp) was inserted into pHAN (BamHI/EcoRI-blunted).
Satellite Cell Preparation and Fluorescence Activated Cell Sorting (FACS)
Satellite cell isolation was performed according to published protocols (Joe et al., 2010), with minor modifications described in the Supplemental Information.
RNA Digestion, Fractionation and RT-qPCR
Purified RNA extracts were used for digestion with RNase-free DNase I (NEB, M0303), RNase H (NEB, M0297) and RNase If (NEB, M0243) for ~1h and used thereafter for reverse transcription/quantitative PCR (RT/qPCR) as previously described (Mousavi et al., 2012). Briefly, RT was performed using 100 ng of total RNA according to manufacturer’s protocol (Applied Biosystems, High Capacity cDNA Reverse Transcription kit), followed by qPCR with Power SYBR Green master mix. RNA was isolated from nuclear and cytoplasmic fractions with the PARIS kit according to the manufacturer’s instructions (Life Technologies).
ChIP-Seq and RNA-Seq
ChIP-Seq and mRNA-seq (polA+ fraction) procedures are described elsewhere (Mousavi et al., 2012). For ribosomal depletion, 1–2 μg of total RNA was used for Ribo-Zero™ rRNA Removal kit (Epicenter) was used according to manufacturer’s protocol. Ribosomal-depleted library was processed for sequencing according to manufacturer’s protocol (Illumina). The quality and quantity of total RNA and final libraries were assessed using RNA 6000 Nano chips and Agilent Technologies 2100 Bioanalyzer. Data for PolII, H3K4me3 ChIP-Seq and PolyA+ RNA-Seq were previously deposited to Gene Expression Omnibus (GEO) database with an accession number GSE25549.
DNaseI Sensitivity Assay
Cells (~5 x 107) were trypsinized and washed twice in PBS with protease inhibitors. For nuclei isolation, cells were dounced in buffer A [15 mM Tris-HCl pH 8.0, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.5 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine (sigma) and protease inhibitors] and NP-40 (0.03%) was added for 3 minutes. Nuclei were washed and resuspended in buffer A. DNaseI treatment was performed in digestion buffer [15 mM Tris-HCl pH 8.0, 6mM CaCl2, 90 mM NaCl, 60 mM KCl, 0.5 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine (sigma)] for 3 mins at 37°C followed by addition of one volume stop buffer [50 mM Tris-HCl pH 8.0, 100 mM NaCl, 0.1% SDS, 100 mM EDTA pH 8.0, 1 mM spermidine, RNase A]. Proteinase K (50 μg/ml) was added and samples were digested at 55°C overnight followed by phenol/chloroform extraction. Extracted DNA was size-fractionated in 9% sucrose solution (1 M NaCl, 20 mM Tris-HCl pH8.0, 0.5 mM EDTA) at 25,000 rpm for 24hrs with no brake. After agarose gel, DNA fractions (50–500bp) were analyzed by qPCR.
Data Analysis
ChIP-Seq data generated from Illumina Genome Analyzers (GAII and Hi-Seq 2000) were mapped to the mouse genome (USCS browser, mm9 version) using ELAND algorithm integrated within Illumina parallel sequencing analyzer software. The total sequence reads were obtained by pooling the results obtained from several independent runs. To control for false positives, ChIP-Seq data generated from mock DNA immunoprecipitates (input DNA) were used against the sample data in calling enriched regions. Mapped tags for MyoD and MyoG were used in MACS package to call peaks with p-value set to 10−6 and FDR to 1%. H3K4me1, H3K4me3 and PolII enriched regions were detected using SICER algorithm, the window size, gap size and FDR were set to 200bp, 600bp and 5% respectively (Zang et al., 2009). This algorithm has been shown to be more powerful in detecting broad enriched regions such as those occupied by PolII or modified histones. All of the downstream analysis has been done using MATLAB. For RNA-Seq data, we used TOPHAT package (Trapnell et al., 2009), to map the reads and spliced reads to the genome. Transcript assembly and final transcript levels of all UCSC known genes were calculated in unit of Reads Per Kilo base pair per Million (RPKM) by Cufflink (Trapnell et al., 2010). For the purpose of differential expression analysis, genes with <1 RPKM in both MB and MT were filtered out.
Supplementary Material
HIGHLIGHTS.
Extensive MyoD and MyoG occupancy in the extragenic regions
RNA synthesis at MyoD+/MyoG+ extragenic enhancer sites
Enhancer RNAs (eRNA) enhance gene expression
eRNAs promote chromatin access to RNA polymerase II at defined genomic loci
Acknowledgments
We thank Dr. Hong-Wei Sun (Biodata Mining and Discovery Section, NIAMS) for initial MyoD binding analysis, Dr. Stephen J. Tapscott for sharing reagents, Drs. Rahul Roychoudhuri, Ofir Hakim and Weiqun Peng for stimulating discussions, and Dr. Miroslav Koulnis for assistance with illustrations. This work was supported in part by the Intramural Research Program of the National Institute of Arthritis, Musculoskeletal, and Skin Diseases of the National Institutes of Health.
Footnotes
AUTHOR CONTRIBUTIONS
KM and VS conceived the experiments, KM, SDO, LG, GG-C and AD performed the experiments and analyzed data, HZ designed and conducted the analyses. KM, HZ, GLH and VS wrote the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asakura A, Lyons GE, Tapscott SJ. The regulation of MyoD gene expression: conserved elements mediate expression in embryonic axial muscle. Dev Biol. 1995;171:386–398. doi: 10.1006/dbio.1995.1290. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Molecular cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R, Vethantham V, Bowman C, Rudnicki M, Dynlacht BD. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes & development. 2012;26:2763–2779. doi: 10.1101/gad.200113.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger H, Bushnell DA, Davis R, Griesenbeck J, Lorch Y, Strattan JS, Westover KD, Kornberg RD. Structural basis of eukaryotic gene transcription. FEBS letters. 2005;579:899–903. doi: 10.1016/j.febslet.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Goldhamer DJ. The core enhancer is essential for proper timing of MyoD activation in limb buds and branchial arches. Dev Biol. 2004;265:502–512. doi: 10.1016/j.ydbio.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Chen JC, Love CM, Goldhamer DJ. Two upstream enhancers collaborate to regulate the spatial patterning and timing of MyoD transcription during mouse development. Dev Dyn. 2001;221:274–288. doi: 10.1002/dvdy.1138. [DOI] [PubMed] [Google Scholar]
- Chen JC, Ramachandran R, Goldhamer DJ. Essential and redundant functions of the MyoD distal regulatory region revealed by targeted mutagenesis. Developmental biology. 2002;245:213–223. doi: 10.1006/dbio.2002.0638. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Cosma MP. Ordered recruitment: gene-specific mechanism of transcription activation. Molecular cell. 2002;10:227–236. doi: 10.1016/s1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. From the Cover: Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean A. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 2006;22:38–45. doi: 10.1016/j.tig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faerman A, Goldhamer DJ, Puzis R, Emerson CP, Jr, Shani M. The distal human myoD enhancer sequences direct unique muscle-specific patterns of lacZ expression during mouse development. Developmental biology. 1995;171:27–38. doi: 10.1006/dbio.1995.1257. [DOI] [PubMed] [Google Scholar]
- Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes & Development. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Goldhamer DJ, Brunk BP, Faerman A, King A, Shani M, Emerson CP., Jr Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development. 1995;121:637–649. doi: 10.1242/dev.121.3.637. [DOI] [PubMed] [Google Scholar]
- Goldhamer DJ, Faerman A, Shani M, Emerson CP. Regulatory elements that control the lineage-specific expression of myoD. Science. 1992;256:538–542. doi: 10.1126/science.1315077. [DOI] [PubMed] [Google Scholar]
- Hah N, Murakami S, Nagari A, Danko C, Kraus WL. Enhancer Transcripts Mark Active Estrogen Receptor Binding Sites. Genome Res. 2013 doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nature cell biology. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development. 1997;124:4729–4738. doi: 10.1242/dev.124.23.4729. [DOI] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Tapscott SJ, Goldhamer DJ, Rudnicki MA. Myogenic determination occurs independently in somites and limb buds. Dev Biol. 1999;206:219–231. doi: 10.1006/dbio.1998.9126. [DOI] [PubMed] [Google Scholar]
- Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010 doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Chromatin structure and transcription. Annu Rev Cell Biol. 1992;8:563–587. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- Kucharczuk KL, Love CM, Dougherty NM, Goldhamer DJ. Fine-scale transgenic mapping of the MyoD core enhancer: MyoD is regulated by distinct but overlapping mechanisms in myotomal and non-myotomal muscle lineages. Development. 1999;126:1957–1965. doi: 10.1242/dev.126.9.1957. [DOI] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013 doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelli KM, Slattery M, Mann RS. Disentangling the many layers of eukaryotic transcriptional regulation. Annu Rev Genet. 2012;46:43–68. doi: 10.1146/annurev-genet-110711-155437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013 doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Ainol L, Zhang L, Yu X, Pi W, Tuan D. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. The Journal of biological chemistry. 2004;279:51704–51713. doi: 10.1074/jbc.M404039200. [DOI] [PubMed] [Google Scholar]
- Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- Melgar MF, Collins FS, Sethupathy P. Discovery of active enhancers through bidirectional expression of short transcripts. Genome biology. 2011;12:R113. doi: 10.1186/gb-2011-12-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Molecular cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Wang AH, Sartorelli V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Molecular cell. 2012;45:255–262. doi: 10.1016/j.molcel.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Shiekhattar R. Noncoding RNAs and enhancers: complications of a long-distance relationship. Trends Genet. 2011;27:433–439. doi: 10.1016/j.tig.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Paranjape SM, Kamakaka RT, Kadonaga JT. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell. 2013 doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Mercola M. BAF60 A, B, and Cs of muscle determination and renewal. Genes & development. 2012;26:2673–2683. doi: 10.1101/gad.207415.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki MA, Le Grand F, McKinnell I, Kuang S. The molecular regulation of muscle stem cell function. Cold Spring Harb Symp Quant Biol. 2008;73:323–331. doi: 10.1101/sqb.2008.73.064. [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani VD, Yin H, Jahani-Asl A, Ming H, Kockx CE, van Ijcken WF, Grosveld F, Rudnicki MA. Snail Regulates MyoD Binding-Site Occupancy to Direct Enhancer Switching and Differentiation-Specific Transcription in Myogenesis. Molecular cell. 2012;47:457–468. doi: 10.1016/j.molcel.2012.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA. What does our genome encode? Genome Res. 2012;22:1602–1611. doi: 10.1101/gr.146506.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stargell LA, Struhl K. Mechanisms of transcriptional activation in vivo: two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- Taberlay PC, Kelly TK, Liu CC, You JS, De Carvalho DD, Miranda TB, Zhou XJ, Liang G, Jones PA. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell. 2011;147:1283–1294. doi: 10.1016/j.cell.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ, Lassar AB, Weintraub H. A novel myoblast enhancer element mediates MyoD transcription. Mol Cell Biol. 1992;12:4994–5003. doi: 10.1128/mcb.12.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.