Abstract
Despite recent advances, understanding of molecular genetic alterations underlying thyroid carcinogenesis remains unclear. One key question is how dynamic temporal changes in global genomic expression affect carcinogenesis as the disease progresses. To address this question, we used a mouse model that spontaneously develops follicular thyroid cancer similar to human cancer (Thrb PV/PV mice). Using complementary DNA microarrays, we compared global gene expression profiles of thyroid tumors of Thrb PV/PV mice with the age- and gender-matched thyroids of wild-type mice at 3 weeks and at 2, 4, 6 and 14 months. These time points covered the pathological progression from early hyperplasia to capsular invasion, vascular invasion and eventual metastasis. Microarray data indicated that 462 genes were upregulated (Up-cluster genes) and 110 genes were downregulated (Down-cluster genes). Three major expression patterns (trending up, cyclical and spiking up and then down) and two (trending down and cyclical) were apparent in the Up-cluster and Down-cluster genes, respectively. Functional clustering of tumor-related genes followed by Ingenuity Pathways Analysis identified the transforming growth factor β (TGF β)-mediated network as key signaling pathways. Further functional analyses showed sustained activation of TGFβ receptor–pSMAD2/3 signaling, leading to decreased expression of E-cadherin and increased expression of fibronectin, vimentin, collagens and laminins. These TGFβ-induced changes facilitated epithelial-to-mesenchymal transition, which promotes cancer invasion and migration. Thus, complex temporal changes in gene expression patterns drive thyroid cancer progression, and persistent activation of TGFβ–TGFRβII–pSMAD2/3 signaling leads to EMT, thus promoting metastasis. This study provides new understanding of progression and metastatic spread of human thyroid cancer.
Introduction
Thyroid cancers constitute the most frequent endocrine neoplasia. The incidence rate of thyroid cancer has been rising sharply since the mid-1990s, and it is the fastest increasing cancer in both men and women (1). The majority of thyroid cancers are differentiated thyroid cancers with papillary and follicular types (2,3). Papillary thyroid carcinomas commonly metastasize to lymph nodes and are often multifocal, whereas follicular carcinomas show blood-borne metastases. The prognosis for the vast majority of individuals diagnosed with differentiated thyroid cancers is excellent, with current treatment that includes surgery, radioactive iodine ablation and postoperative suppression of thyroid-stimulating hormone. Unfortunately, in some patients, recurrence of the tumor with metastasis becomes the major cause for thyroid cancer-related death (3). Recent progress in the identification of genetic changes associated with thyroid cancer has advanced our understanding of its molecular basis. Less clear, however, is how signaling pathways undergoing temporal changes as thyroid cancer progresses ultimately lead to metastasis. Animal models that can be interrogated during thyroid carcinogenesis would be a powerful approach to elucidate molecular changes that propel thyroid cancer progression.
To this end, we took advantage of an animal model that spontaneously develops metastatic follicular thyroid cancer (FTC). This mouse harbors a knockin dominant negative mutation, known as PV, in the Thrb gene locus (4). The PV mutation was identified in a patient suffering from resistance to thyroid hormone (5). As Thrb PV/PV mice age, their thyroids undergo pathological changes from hyperplasia to capsular and vascular invasion, anaplasia and eventual metastasis to the lung (6). The pathological progression, route and frequency of metastasis in Thrb PV/PV mice are similar to that in human FTC. Extensive molecular analyses of altered signaling pathways during thyroid carcinogenesis further confirmed that the Thrb PV/PV mouse is a preclinical mouse model of FTC. As found in human FTC, Thrb PV/PV mice exhibit aberrant signaling pathways that include constitutive activation of phosphatidyl inositol 3-kinase/Akt (7,8) and integrin–Src–mitogen-activated protein kinase signaling (9) and aberrant accumulation of the oncogenic pituitary tumor-transforming gene protein (PTTG) (10,11) and β-catenin (12). Thus, the Thrb PV/PV mouse model faithfully recapitulates the molecular aberrations found in human thyroid cancer and is suitable for elucidation of temporal genomic changes during thyroid cancer progression. Using microarrays, we identified dynamic global genomic changes in expression patterns during pathological progression from hyperplasia to capsular and vascular invasion and eventual metastasis. We further elucidated that TGFβ-mediated signaling was one of the powerful networks that drove the progression of thyroid cancer and epithelial-to-mesenchymal transition (EMT) to promote metastasis.
Materials and methods
Mouse strains
The animal studies were carried out according to the protocol approved by the National Cancer Institute Animal Care and Use Committee. Mice harboring the ThrbPV gene (Thrb PV/PV mouse) were generated and genotyped as described earlier (4). Littermates with a similar genetic background were used as a control in experiments.
Microarray analysis
The mouse arrays contained 20 000 complementary DNAs. Hybridization, scanning and image analysis were performed as described previously (www.nhgri.nih.gov/DIR/microarray) (13–15). Briefly, fluorescent-labeled complementary DNA was synthesized from ∼20 μg of pooled RNA of five mice by oligo(dT)-primed polymerization in the presence of aminoallyl-deoxyuridine triphosphate (Amersham Pharmacia Biotech, Piscataway, NJ) and coupled with either Cy-3 or Cy-5. Image analyses were performed using DeArray software (Signal Analytics, Vienna, VA) (15). The two fluorescent images (red and green channels) obtained from one array constitute the raw data used to calculate differential gene expression ratio values. The ratios of the red intensity to the green intensity (R/G) for all targets were determined, and the data were stored in a FileMaker Pro database (FileMaker, Santa Clara, CA).
RNA extraction and real-time PCR
Thyroid lobes from wild-type (WT) and Thrb PV/PV mice were homogenized in the TRIzol (Invitrogen, CA) solution. Total RNA was extracted as indicated in the manufacturer’s protocol. The total amount of RNA was measured and real-time reverse transcription (RT)–PCR was carried out as described previously (16). The primer sequences used will be provided upon request.
Western blot analysis
Thyroid tissues preparation from WT and Thrb PV/PV mice and western analysis were carried out as described previously (12). The antibodies were total anti-SMAD2 (1:1000 dilution, #5678, Cell Signaling, MA), anti-GAPDH (1:1000 dilution, #2118, Cell Signaling, MA), anti-TIMP3 (1:1000 dilution, #5673, Cell Signaling, MA), anti-TGFBRII (1:1000 dilution, #3713, Cell Signaling, MA), anti-TGFBI (1:300 dilution, 10188-1-AP, Proteintech, IL), anti-vimentin (1:1000 dilution, #5741, Cell Signaling, MA), anti-E-cadherin (1:1000 dilution, #4065, Cell Signaling, MA), anti-fibronectin (1:250 dilution, ac-6952, Santa Cruz, CA), anti-LAMC1 (1:1000, sc-5584, Santa Cruz, CA), anti-pSMAD2/3 (1:1000 dilution, sc-11769, Santa Cruz, CA) and anti-collagen type IV (1:200 dilution, AB-769, Millipore, MA).
Immunohistochemical staining
Immunohistochemical staining was performed as described previously (17). Antibodies were TGFBRII (1:100 dilution, #3713, Cell Signaling, MA), pSMAD2/3 (1:1000 dilution, sc-11769, Santa Cruz, CA) and Ki-67 (1:50 dilution, Thermo Scientific, NJ).
Determination of serum TGFβ1
The serum levels of TGFβ1 were determined using a TGF beta1 Mouse ELISA Kit (ab119557, Abcam, MA) according to the manufacturer’s instructions.
Confocal fluorescence microscopy
Primary cultured thyroid cells were obtained from WT and Thrb PV/PV mice as described previously (7). Cells (4 × 104 cells/well) were seeded in a chambered coverglass (Thermo Scientific, NY, #155411) and were cultured in Coon’s modified Ham’s F-12 medium supplemented with 5% bovine serum containing 10 μg/ml insulin, 0.4ng/ml cortisol, 5 μg/ml transferrin, 10ng/ml glycyl-l-histidyl-l-lysine acetate, 10ng/ml somatostatin and 1 milliunit/ml thyroid-stimulating hormone (Sigma) for 24 h. Then the cells were treated with or without TGFβ1 (10ng/ml) (Cell Signaling, MA, #8915) for 48 h. All subsequent steps were carried as described previously (17). Antibodies were anti-vimentin (1: 100 dilution, #5741, Cell Signaling, MA), anti-FN1 (1:400 dilution, ab6328, Abcam, MA), anti-TTF1 (1:100, sc-13040, Santa Cruz, CA) and anti-PAX8 (1:100 dilution, 10336-1-AP, Proteintech, IL) for 1 h.
Wound healing assay
Primary cultured thyroid cells obtained from WT and Thrb PV/PV mice were seeded in a 12-well plate in culture media as described above. After 24 h, cells were wounded with a p200 tip and washed with 1× PBS, and then media was replaced with culture media supplemented with 2% bovine serum with or without TGFβ1 (10ng/ml). Pictures were taken at time point 0, 24 and 48 h after TGFβ1 treatment. The distance between cells was measured using ImageJ software (NIH).
Statistical analysis
Data are expressed as mean ± standard error of the mean. Significant differences between groups were calculated using Student’s t-test via GraphPad Prism 5 (GraphPad Software, San Diego, CA). P < 0.05 was considered statistically significant.
Results
Complex temporal changes in gene expression patterns during thyroid carcinogenesis
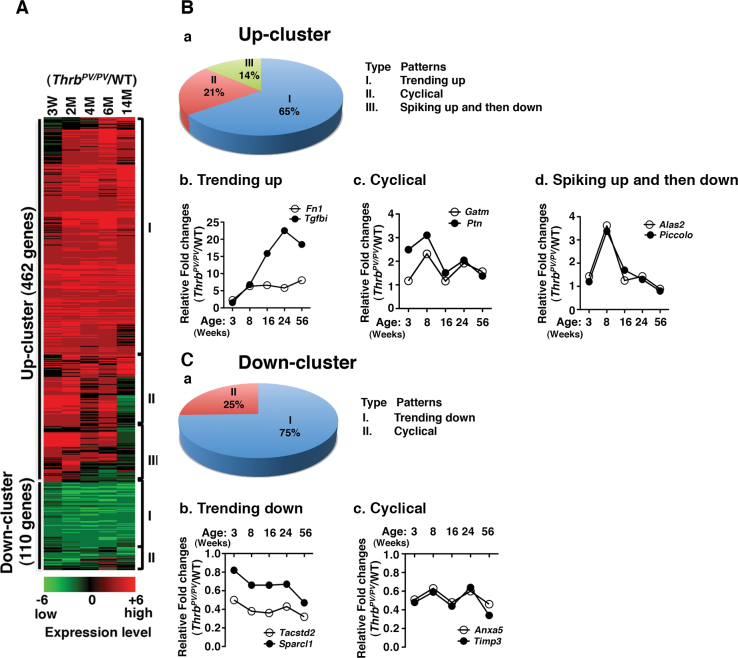
To ascertain the changes in global gene expression during thyroid cancer progression, we analyzed the microarray data obtained from comparison of thyroid tumors of male Thrb PV/PV mice with the age- and gender-matched thyroids of WT mice (n = 3 for each genotype) at the ages of 3 weeks and at 2, 4, 6 and 14 months. These time points covered the pathological progression from early hyperplasia beginning at 3 weeks to capsular and vascular invasion occurring at 5–6 months and to metastasis detectable at 8–9 months and onward (6). A total of 572 genes with expression ratios >2 were selected for further analysis (Figure 1A). Two major gene clusters, termed Up-cluster and Down-cluster, of altered gene expression profiles were apparent during thyroid carcinogenesis of Thrb PV/PV mice. Four hundred and sixty-two genes remained mostly upregulated in the Up-cluster and 110 genes remain mostly downregulated in the Down-cluster. The extent of changes was reflected in the intensities of the heat bar shown in Figure 1A. The complete list of 572 genes with fold of changes in a time-dependent manner is shown in Supplementary Table A, available at Carcinogenesis Online.
Fig. 1.
Identification of complex temporal changes in global gene expression patterns by microarray analysis. (A) Total RNA was extracted from thyroids of age- and gender-matched WT and Thrb PV/PV mice at 3, 8, 16, 24 and 56 weeks of age, and gene expression ratios of Thrb PV/PV to WT were calculated and grouped as either Up-cluster or Down-cluster genes. Each upregulated or downregulated gene is marked in either red or green, respectively and shown as a heat map in log2 scale. (B-a and C-a) Both the Up-cluster and the Down-cluster genes were further classified according to their gene expression patterns. The Up-cluster genes have three subgroups; the Down-cluster genes have two. (B-b, c and d) Two representatives from subgroups of the Up-cluster and two examples from the Down-cluster (C-b and c) genes were selected, and their expression patterns are shown. Data are expressed as ratios of fold changes in the expression of genes in Thrb PV/PV to those of WT mice.
Further analyses of the gene expression profiles in the two clusters revealed major patterns of changes during thyroid carcinogenesis from 3 weeks of age up to 14 months. Three major patterns were identified from the Up-cluster (Figure 1B-a) and two from the Down-cluster (Figure 1C-a). As shown in Figure 1B-a, for the Up-cluster the most frequent pattern of gene expression was ‘trending up’ (65%; Type I). Two genes representative of this pattern of expression, fibronectin (Fn1) and TGFβ induced (Tgfbi), are shown in Figure 1B-b. Less frequent were the ‘cyclical’ pattern (21%; Type II) and the ‘spiking up and then down’ pattern (14%; Type III). Two representative genes with the ‘cyclical’ pattern are glycine amidinotransferase (Gatm) and pleiotrophin (Ptn) (Figure 1B-c). The aminolevulinic acid synthase 2 (Alas2) and the presynaptic cytomatrix protein (Piccolo) genes exhibited the pattern of spiking up at 8 weeks, and then decreasing (Figure 1B-d). For the Down-cluster genes, 75% displayed the ‘trending up’ pattern (Figure 1C-a). As shown in Figure 1C-b, the tumor-associated calcium signal transducer (Tacstd2) and the SPARC-like 1 (Sparcl1) genes are two representative examples of this pattern. Only 25% of the genes displayed the ‘cyclical’ pattern of expression (Figure 1C-a). Two representative genes with this pattern are annexin A5 (Anxa5) and TIMP metallopeptidase inhibitor (Timp3) (Figure 1C-c). These data indicate that complex alterations in the expression of genes in a temporal-dependent manner drive the progression of thyroid cancer from hyperplasia to tumor invasion and metastasis.
Identification of activated TGFβ signaling network in thyroid carcinogenesis of ThrbPV/PV mice
Functional clustering analysis of the 572 genes showed that genes with altered expression functioned as key regulators in cell death/survival, cell proliferation/growth and cell development (22% for each category) and in cellular movement and morphology (17% for each category). To elucidate oncogenic genes that undergo temporal alterations in expression, which contribute to thyroid carcinogenesis, we used Ingenuity Pathways Analysis (IPA) to search for genes known to be involved in cancer development. Among those genes identified by IPA, we found 23 in the Up-cluster and 15 in the Down-cluster, which play key roles (Table I). These genes exhibited different temporal patterns (Types I and II in the Up-cluster and Types I and II in the Down-cluster) and were involved in a wide range of critical cellular functions such as proliferation, apoptosis, invasion, metastasis and migration (Table I). Among these genes, serpin peptidase inhibitor, clade F, member 1, (Serpinf1) (18), Fn1 (19), vimentin (Vim) (19,20), pericentriolar material 1 (Pcm1) (21), the prolactin receptor (Prlr) (22) and Timp3 (23) are reported to be involved in human thyroid cancer.
Table I.
Genes associated with tumor development
| Gene | Gene name | Maximal fold changes | Type | Proliferation | Apoptosis | Invasion | Metastasis | Migration | Thyroid cancer |
|---|---|---|---|---|---|---|---|---|---|
| Up-cluster | |||||||||
| Lcn2 | Lipocalin 2 | 55.02 | I | * | * | * | * | * | |
| Tgfbi | Transforming growth factor, beta-induced, 68kDa | 22.5 | I | * | * | * | * | ||
| Serpinf1 | Serpin peptidase inhibitor, clade F, member 1 | 9.33 | I | * | * | * | * | ||
| Anxa1 | Annexin A1 | 9.21 | I | * | * | * | * | * | |
| Fn1 | Fibronectin 1 | 8.05 | I | * | * | * | * | * | * |
| Lbp | Lipopolysaccharide binding protein | 7.84 | I | * | |||||
| Ltf | Lactotransferrin | 5.95 | I | * | * | * | * | ||
| Camk1d | Calcium/calmodulin- dependent protein kinase ID | 5.07 | I | * | * | ||||
| Igf1 | Insulin-like growth factor 1 | 4.41 | I | * | * | * | * | * | |
| Smarcb1 | SWI/SNF-related matrix-associated actin-dependent regulator chromatin, subfamily b, member 1 | 4.26 | II | * | * | * | |||
| Vim | Vimentin | 4.16 | II | * | * | * | * | * | * |
| Col18a1 | Collagen, type XVIII, alpha 1 | 3.99 | I | * | * | * | * | * | |
| Grn | Granulin | 3.82 | II | * | * | * | * | ||
| Lamc1 | Laminin, gamma 1 | 3.7 | I | * | * | * | |||
| Col4a1 | Collagen, type IV, alpha 1 | 3.67 | I | * | * | * | * | ||
| Ehf | Ets homologous factor | 3.61 | II | * | * | ||||
| Tyms | Thymidylate synthetase | 3.6 | II | * | * | * | |||
| Nr2f2 | Nuclear receptor subfamily group F, member 2 | 3.56 | II | * | * | ||||
| Ucp2 | Uncoupling protein 2 | 3.53 | I | * | * | * | |||
| Bgn | Biglycan | 3.38 | I | * | * | * | |||
| Tfrc | Transferrin receptor | 3.28 | I | * | * | ||||
| Col3a1 | Collagen, type III, alpha 1 | 3.2 | II | * | |||||
| Ptn | Pleiotrophin | 3.1 | II | * | * | * | * | ||
| Down-cluster | |||||||||
| Agr2 | Anterior gradient 2 homolog | 0.5 | I | * | |||||
| Esr1 | Estrogen receptor 1 | 0.5 | II | * | * | * | * | * | |
| Smo | Smoothened frizzled family receptor | 0.49 | I | * | * | * | * | ||
| Pcm1 | Pericentriolar material 1 | 0.49 | I | * | |||||
| Gnas | GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus | 0.48 | I | * | * | ||||
| Flrt2 | Fibronectin leucine rich transmembrane protein 2 | 0.47 | I | * | |||||
| Anxa5 | Annexin A5 | 0.46 | II | * | * | * | |||
| Nfkbia | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | 0.45 | I | * | * | * | * | ||
| Prlr | Prolactin receptor | 0.38 | I | * | * | * | * | ||
| Lmo7 | LIM domain 7 | 0.38 | I | * | * | ||||
| Timp3 | TIMP metallopeptidase inhibitor 3 | 0.34 | II | * | * | * | * | * | |
| Tacstd2 | Tumor-associated calcium signal transducer 2 | 0.32 | I | * | |||||
| Timp4 | TIMP metallopeptidase inhibitor 4 | 0.31 | II | * | * | * | |||
| Fasn | Fatty acid synthase | 0.31 | I | * | * | ||||
| Txnip | Thioredoxin interacting protein | 0.29 | I | * | * | * | |||
| Maximal fold changes (bold values): the ratio of highest value versus lowest value at five selective time points in mice aged from 3 weeks to 14 months. | |||||||||
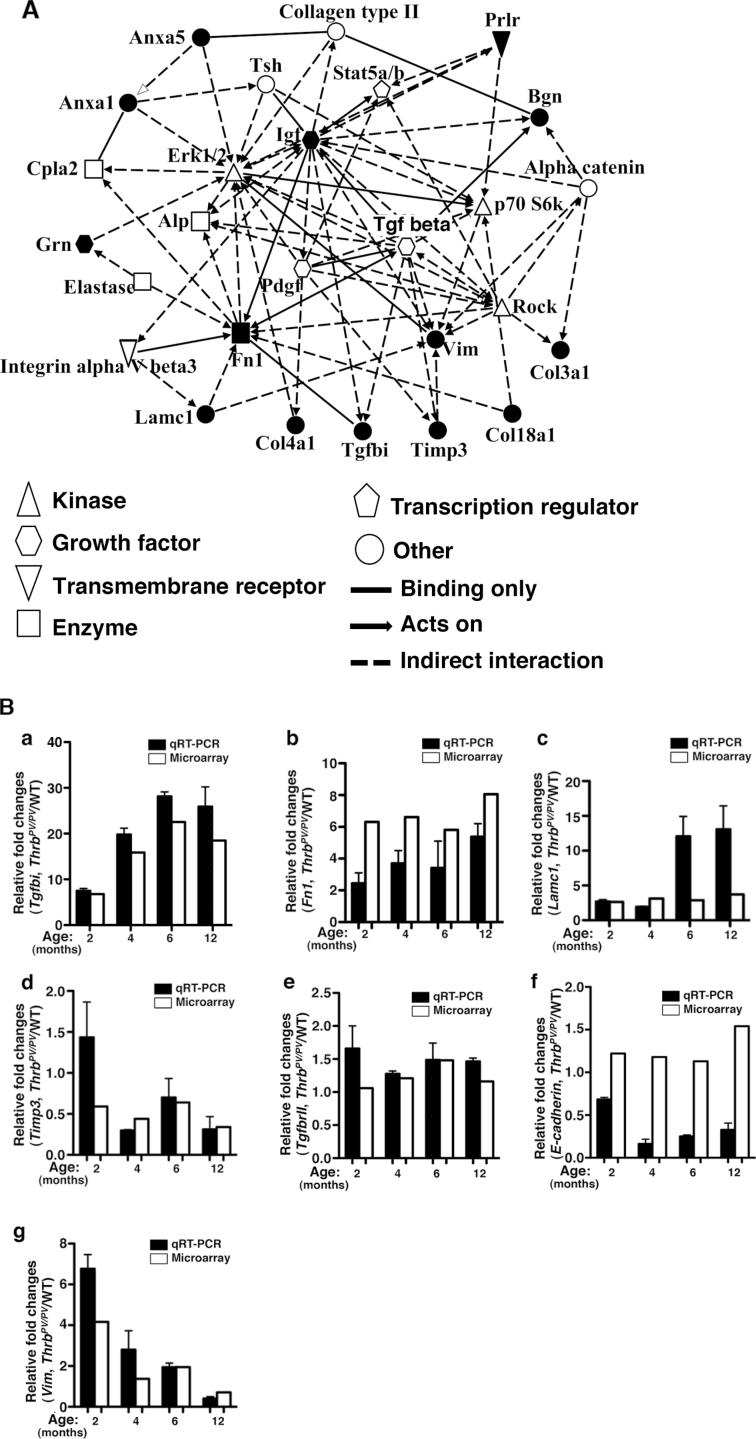
The data reported thus far suggest complex networks of genes undergo temporal changes as thyroid cancer progresses. We therefore used IPA to further ascertain how the genes in Table I could be interconnected to drive thyroid carcinogenesis. IPA identified four potential networks with top functions involved in cancer/cell movement/cardiovascular system (network 1), cancer/connective tissue disorder/inflammatory diseases (network 2), connective tissue disorder/embryonic development (network 3) and cell movement/tissue development/cellular growth (network 4). IPA-computed scores of 31, 25, 11 and 10 for networks 1, 2, 3 and 4, respectively, indicate a >95% confidence that these results were not by random chance.
Further analysis showed that among the 35 regulators predicted by IPA to be involved in network 1, 14 were identified in our analysis as shown in Table I (40% of total 38 genes listed in Table I). In Figure 2A, these 14 genes are marked by solid black (as opposed to open) symbols. However, a lower number of genes, 12, 6 and 6 genes listed in Table I were found to participate in the activities of networks 2, 3 and 4, respectively.
Fig. 2.
(A) Predicted signaling network of genes associated with tumor development in Thrb PV/PV mice. Genes related to tumor development listed in Table I were assessed by IPA. Solid black (filled) symbols indicate the genes identified from arrays and shown in Table I. The functional category of each gene is denoted by either open or filled symbols. The functional relationship among genes is shown with a solid line to indicate binding only or a broken line to indicate indirect interaction, and with arrowheads attached to indicate acts on. (B) Validation of the extent of gene expression from arrays by quantitative real-time PCR. Total RNA was extracted from thyroids of WT and Thrb PV/PV mice at the indicated age, as described in Materials and methods. Seven representative genes were selected for gene validation. Three of them were from the Up-cluster (a–c), one from the Down-cluster (d) and three (e–g) were genes related with TGFβ signaling: (a) Tgfbi, (b) Fn1, (c) Lamc1, (d) Timp3, (e) TgfbrII, (f) E-cadheirn and (g) Vim. Open bars indicate microarray data; filled bars indicate data from quantitative real-time PCR.
Therefore, we focused our in-depth analysis on the role of network 1 in thyroid carcinogenesis.
Central to network 1 is the signaling pathway mediated by the TGFβ (Figure 2A). TGFβ is a key player in cell proliferation, differentiation and apoptosis, and its aberrant signaling is associated with cancer development (24). Among the genes listed in Table I, 14 genes in the Up-cluster (Tgfb1, Anxa1, Fn1, Lbp, Ltf, Igf1, Vim, Lamc1, Col4a1, Nr2f2, Ucp2, Bgn, Tfrc and Col3a1) and seven genes in the Down-cluster (Agr2, Nfkbia, Lmo7, Timp3, Tacstd2, Timp4 and Fasn) are affected directly or indirectly by TGFβ signaling (25–44).
We therefore used qRT–PCR to validate the time-dependent gene expression profiles obtained by array analysis. Examples of genes used for validation are shown in Figure 2B. Consistent with array data, qRT–PCR shows that the expression of the TGFβ-induced, 68kDa (Tgfbi), exhibited the pattern of ‘trending up’ (Up-cluster, Type I) (Figure 2B-a). Similarly, the expression pattern of Fn1 and laminin γ 1 as determined by qRT–PCR was in line with that determined by arrays (Figure 2B-b and -c, respectively). Figure 2B-d shows the expression pattern of the Timp3 gene shown by qRT–PCR was consistent with that found by microarray data (Figure 1C-c) (Down-cluster; Type II). In addition, examination of other genes involved in TGFβ signaling shows that the expression patterns of TgfbrII (Figure 2B-e) and Vim (Figure 2B-g) were consistent with those of microarray data, but expression level of E-cadherin determined by qRT/PCR was lower than that determined by microarray analysis (Figure 2B-f).
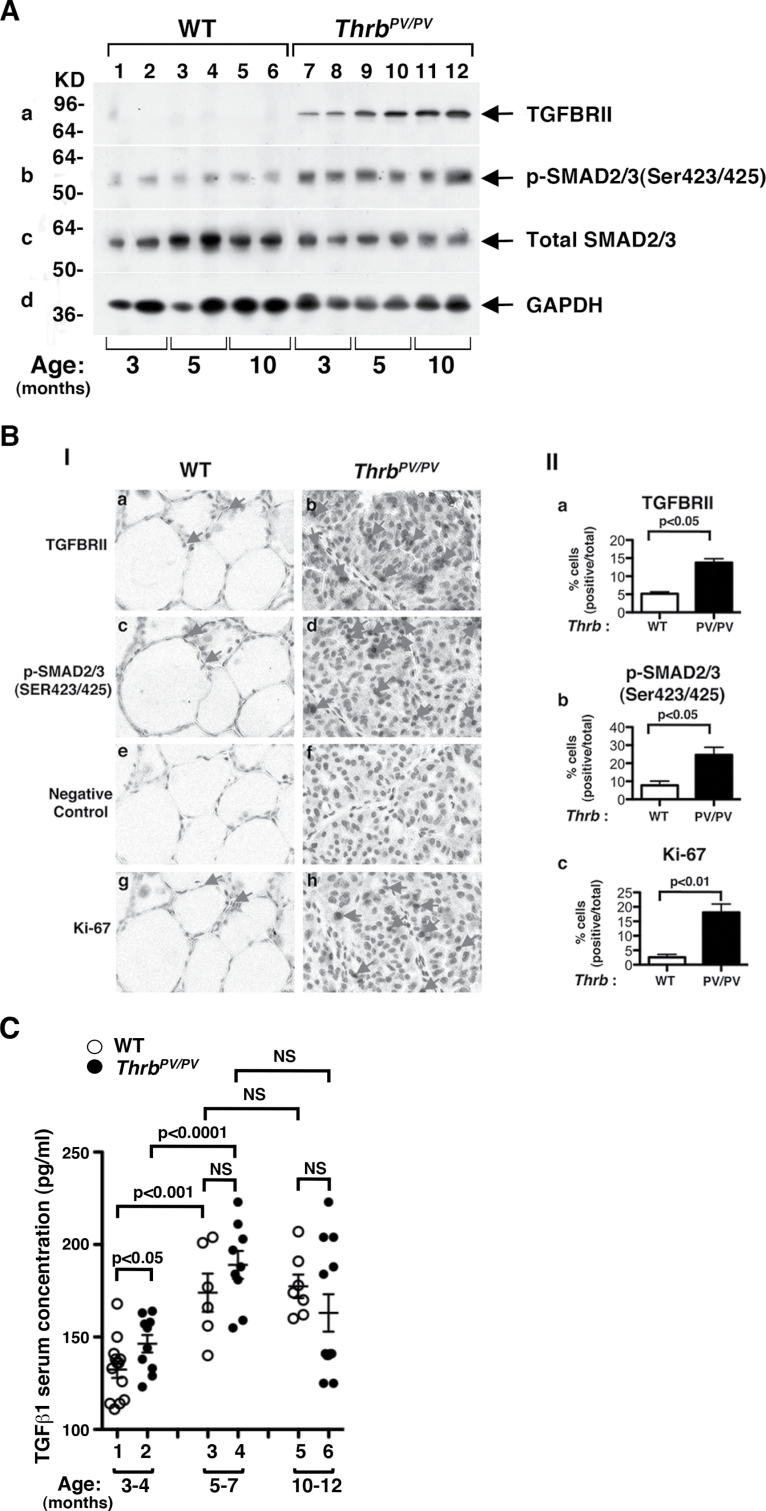
In tumors, TGFβ acts as either a tumor suppressor or a proto-oncogene, depending on cell context and tumor stage (45). Cancer cells often evade the growth inhibitory effects of TGFβ but retain the TGFβ-mediated cellular functions that promote tumor progression. Accordingly, we next sought to elucidate how TGFβ signaling could affect thyroid carcinogenesis of Thrb PV/PV mice. The TGFβ canonical signaling is via the TGFβ receptors (TGFβRs)–SMAD pathways. Upon binding to TGFβ, TGFβRs dimerize and phosphorylate intracellular SMAD proteins. Phosphorylated SMAD (pSMAD) proteins translocate into the nucleus to regulate TGFβ-dependent target genes. Our western blot analysis (Figure 3A) showed that TGFβRII protein abundance was markedly higher (lanes 7–12) in Thrb PV/PV mice than in WT mice (lanes 1–6) at all ages; and moreover, this abundance increased as mice aged (Figure 3A-a). The increased abundance of TGFβRII protein was accompanied by a concomitant age-dependent increase of pSMAD2/3 (Figure 3A-b). However, there was no apparent difference between WT and Thrb PV/PV mice in protein abundance of total SMAD2/3 (compare Figure 3A-c, lanes 1–7 to 8–12). Panel d shows the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) loading control.
Fig. 3.
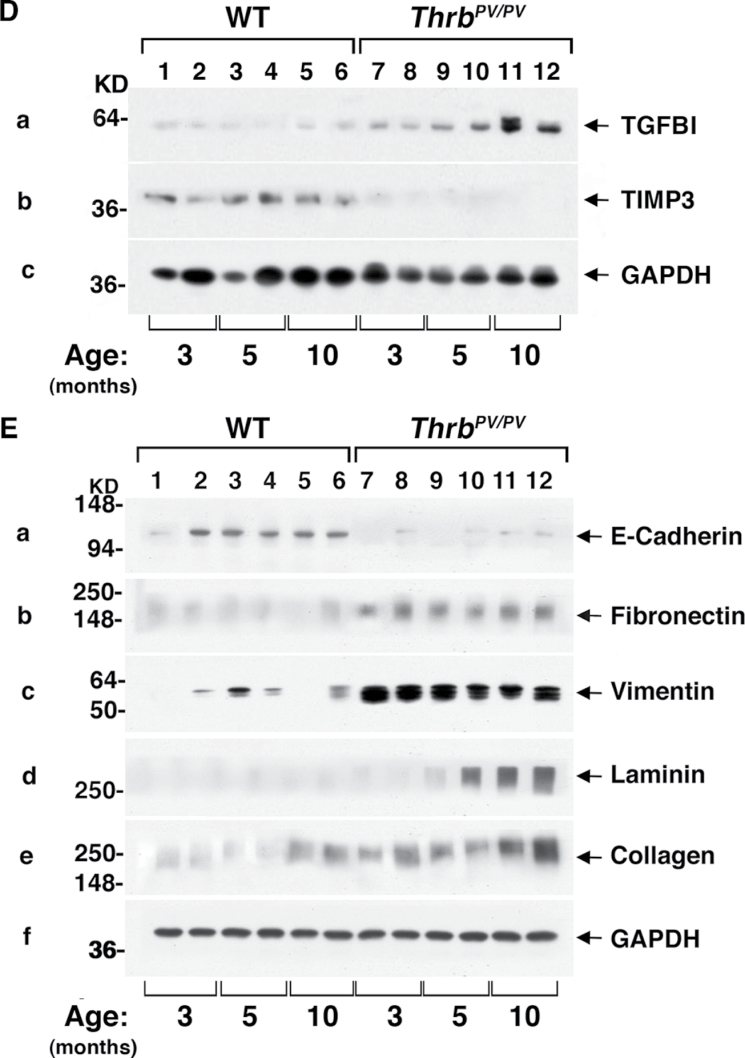
Activation of TGFβ-mediated signaling pathway in thyroids of Thrb PV/PV mice. (A) Total tissue extracts of thyroids from WT and Thrb PV/PV mice were prepared as described in Material and methods. Thirty micrograms of total protein was used to determine the protein abundance of TGFBRII (A-a), the phosphorylated form of SMAD2/3 (Ser423/425) (A-b) and total SMAD2/3 (A-c) by western blot analysis. GAPDH was used as a loading control (A-d). (B) Immunohistochemical analysis of TGFβRII and pSMAD2/3 in thyroid tumors of Thrb PV/PV mice. (B-I) Each paraffin section obtained from WT (a, c, e and g) and Thrb PV/PV mice (b, d, f and h) was subjected to immunohistochemical analysis using anti-TGFBRII (panels a and b), anti-pSMAD2/3(Ser423/425) (panels c and d), no primary antibody (negative control; panels e and f) and anti-Ki-67 antibodies (panels g and h). Arrows point to the positive staining for each antibody. (B-II) The positively stained cells were counted, and the data are shown as % of positively stained cells versus total cells (n = 3). The genotypes are marked. (C) Comparison of serum levels of TGFβ1 in Thrb PV/PV and WT mice. The serum concentrations of TGFβ1 were determined, as described in Materials and methods, using sera obtained from WT and Thrb PV/PV mice at the indicated age. Data are expressed as mean ± standard error of the mean (n = 6–13) with P values shown. NS, not significant. (D) Thyroid extracts (30 μg) of WT and Thrb PV/PV mice cells were analyzed by western blot analysis using anti-TGFBI (panel a), anti-TIMP3 (panel b) and anti-GAPDH as a loading control (panel c). (E) Analysis of protein abundance of key regulators in EMT in the thyroid tumors of Thrb PV/PV mice. Thyroid extracts (30 μg) of WT and Thrb PV/PV mice cells were analyzed by western blot analysis using anti-E-cadherin (panel a), anit-FN1 (panel b), anti-vimentin (panel c), anti-Laminin (panel d), anti-collagen (panel e) and anti-GAPDH as a loading control (panel f).
Immunohistochemical analysis also revealed increased expression of TGFβRII protein in the thyroid lesions. Figure 3B-I shows intense staining of TGFBRII protein in the hyperplastic follicular cells of Thrb PV/PV mice (panel b) but not in the normal follicular cells of WT mice (panel a). The staining shown in panel b was specific because very little background signaling was detected when no primary antibodies were used in either the normal follicular cells (panel e) or the tumor cells (panel f). A count of the cells stained positively with TGFBRII showed ~3-fold more hyperplastic cells in Thrb PV/PV mice than normal cells in WT mice (Figure 3B-II-a). Consistent with this observation, Figure 3B-I shows more tumor cells of Thrb PV/PV mice (panel d) than normal cells of WT mice (panel c) when intensely stained with anti-pSMAD2/3. As indicated in the counting of positively stained cells (Figure 3B-II-b), ~3-fold higher percentage of cells stained with pSMAD2/3 was detected in thyroids of Thrb PV/PV mice than in WT mice. These data indicate an activated TGFβ signaling during thyroid carcinogenesis of Thrb PV/PV mice. It is known that activation of TGFβ signaling leads to increased cell proliferation at various cancers (46–48). We therefore further stained the thyroids with nuclear proliferation marker, Ki-67. Panel h (Figure 3B-I) shows that tumor cells were intensively stained with anti-Ki-67 antibodies than in the normal follicular cells of WT mice (panel g). The Ki-67-positive cells were counted and the quantitative data are shown in Figure 3B-II-c, indicating ~7.5-fold increase in the Ki-67 stained cells in thyroids of Thrb PV/PV mice.
We further assessed whether serum TGFβ1 levels of Thrb PV/PV mice were elevated to stimulate TGFBRII–SMAD signaling. Indeed, Figure 3C shows that at 3–4 months of age, serum TGFβ1 levels in Thrb PV/PV mice were significantly higher than those in WT mice (compare data group 2 with data group 1). TGFβ1 serum levels of Thrb PV/PV and WT mice were further increased at age 5–7 months. Although the TGFβ1 serum level of Thrb PV/PV mice was not significantly different from that of WT mice at age 5–7 months (data groups 3 and 4), the TGFβ1 serum level of Thrb PV/PV mice tended to be higher (data group 3 versus 4). No additional increases in TGFβ1 serum levels were detected in mice aged 10–12 months. These data indicate that in addition to the elevated protein abundance of TGFBRII, increased serum TGFβ1 levels also contributed to the activation of TGFBRII–SMAD signaling. Other evidence of TGFβ–TGFBRII–SMAD signaling activation was the increased expression of its downstream target gene Tgfbi (Up-cluster, Table I) at the messenger RNA level (Figure 1B-b) and at the protein level shown in Figure 3D-a. Likewise, the expression of the Timp3 gene (Down-cluster; Table I, Type II), which is negatively regulated by the TGFβ–TGFBRII–SMAD pathway, was also reduced at the messenger RNA level (Figure 1C-c) and at the protein level (Figure 3D-b). Taken together, these data indicate that TGFβ–TGFBRII–SMAD signaling was activated to promote thyroid carcinogenesis of Thrb PV/PV mice.
Induction of EMT by activated TGFβ signaling to increase invasion and metastasis
In advanced cancers, it is known that TGFβ elicits EMT by transcriptional and posttranscriptional regulation of key regulators that affect cell adhesion, cell mobility and migration (49). As shown in Table I and Figure 2A, several genes regulated by TGFβ signaling were involved in cell adhesion and migration, such as Fn1 (50), Vim (51), Col18a1 (52), Lamc1 (53), Col4a1 (54), Col3a1 (55), Timp3 (23) and Timp4 (56). Such gene expression profiles raised the question of whether activated TGFβ signaling in the thyroid of Thrb PV/PV mice could lead to induction of EMT to increase cell invasion and metastasis. To test this possibility, we determined the protein abundance of E-cadherin because its repressed expression is the hallmark of EMT (57). Indeed, Figure 3E-a shows that the protein abundance of E-cadherin in thyroids of Thrb PV/PV mice (lanes 7–12) was very low at all ages compared with WT mice (lanes 1–6). Moreover, several mesenchymal proteins such as fibronectin (Figure 3E, panel b), vimentin (Figure 3E-c), laminin (Figure 3E-d) and collagen (Figure 3E-e) were elevated during cancer progression. These expressed protein data support the notion that activated TGFβ signaling led to EMT.
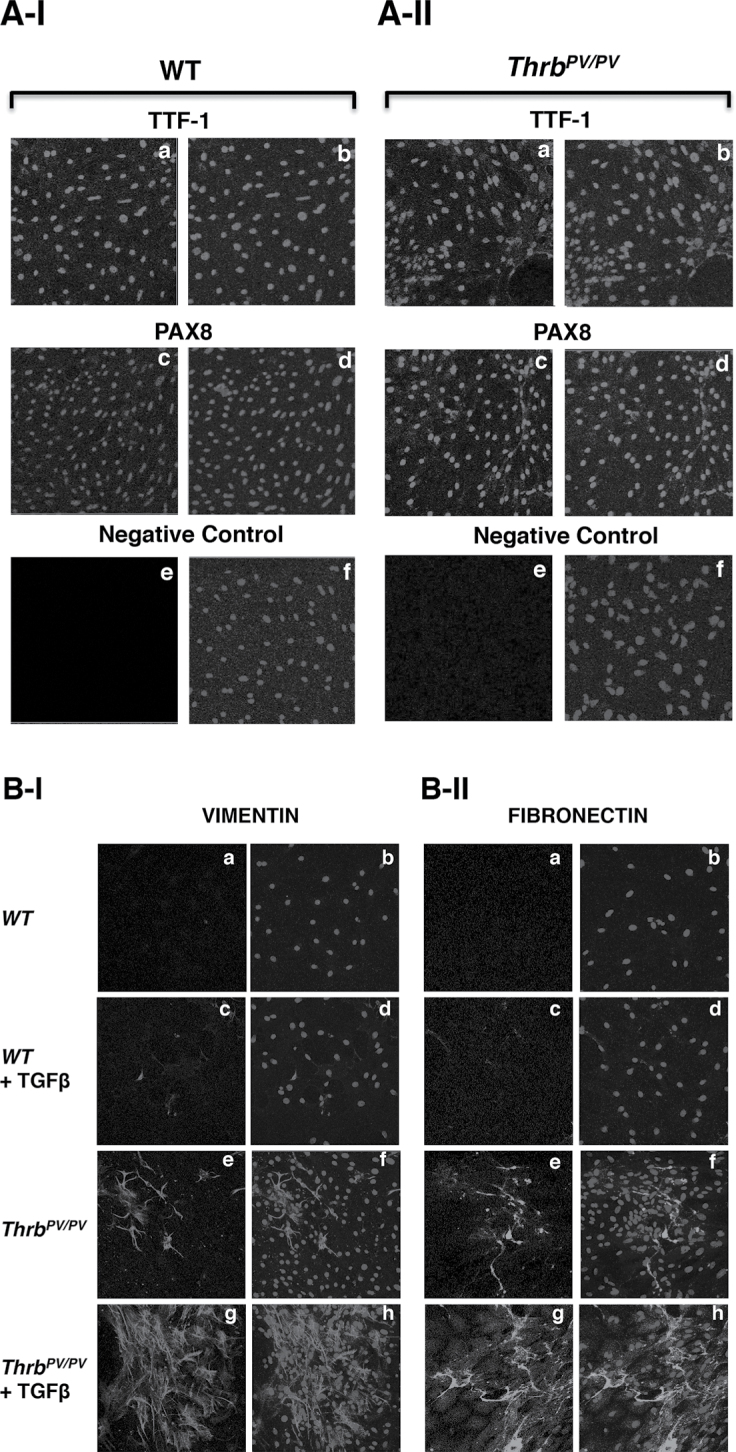
We carried out immunochemical analysis to examine directly whether TGFβ stimulation led to expression of its target genes critical for EMT, such as the Vim and Fn1 genes. We isolated primary tumor cells from thyroids of Thrb PV/PV mice and treated the cells with or without TGFβ1. Figure 4A-I and -II show that the normal thyrocytes and tumor cells isolated from thyroids of WT and Thrb PV/PV mice, respectively, were indeed from follicular origins as the cells expressed the follicular cell transcription factor, TTF1, shown in panel a and panel b (co-stained with nuclear Hoechst dye). Panels c and d (Figure 4A-I and -II for normal thyrocytes from WT mice and tumor cells from Thrb PV/PV mice, respectively) show that another follicular transcription factor PAX8 was also similarly expressed in the normal and tumor cells (Figure 4A-I and -II, respectively). The signals detected in panels a and b for TTF1 and panels c and d were specific because when no primary antibodies were used, no signals were detected as shown in the negative controls (panels e and f). At basal state (without treatment of TGFβ1), no vimentin-stained cells were observed in follicular cells from WT mice (Figure 4B-I, panels a and b). In the presence of TGFβ1, only a few normal follicular cells were stained with anti-vimentin antibodies (panels c and d). In contrast, in tumor cells isolated from thyroids of Thrb PV/PV mice, even without TGFβ1 treatment, more vimentin-stained cells (panel e) were clearly visible than those in normal follicular cells (compare panels a and c with panel e). Upon stimulation by TGFβ1, dramatically increased vimentin abundance was detected in thyroid tumor cells of Thrb PV/PV mice (panels g and h). We also similarly evaluated another mensenchymal target, fibronectin. We found visible staining by anti-fibronectin antibodies in the thyroid tumor cells in the absence of TGFβ1 (Figure 4B-II, panel e), but not in the normal follicular cells of WT mice (Figure 4B-II, panel a). In the presence of TGFβ1, a marked increase in fibronectin protein abundance in thyroid tumor cells was observed in Thrb PV/PV mice (Figure 4B-II, panel g) compared with normal follicular cells of WT mice (Figure 4B-II, panel c). These results provided direct evidence to indicate that TGFβ-mediated signaling led to increased expression of genes critical for EMT to promote invasion and metastasis of thyroid tumor cells.
Fig. 4.
TGFβ1 induces EMT in thyroid tumors of Thrb PV/PV mice. (A) Primary cultured thyroid cells were plated in a chamber slide, and cells were cultured for 48 h before immunostaining. The purity of the isolated thyroid cells from WT mice (A-I) and tumor cells (A-II) was determined using antibodies for thyroid-specific markers: anti-TTF1 (A-I and A-II, a and b) and anti-PAX8 (A-I and A-II, c and d). More than 95% of cells showed positive staining of thyroid-specific markers. (B). Increased EMT activities by TGFβ1 treatment in Thrb PV/PV mice. Antibodies for vimentin (B-I) and FN1 (B-II) were used to examine the EMT progression in primary cultured cells obtained from WT and Thrb PV/PV mice. Primary cultured thyroid cells were treated with (B-I, and B-II, c, d, g and h) or without (B-I, and B-II, a, b, e and f) TGFβ1 for 48 h, as described in Materials and methods.
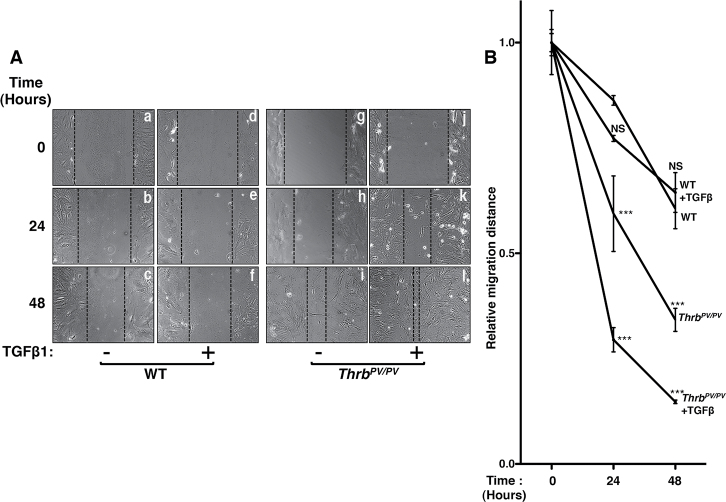
Using wound-healing assays, we further carried out functional analysis by evaluating migration activity of primary tumor cells from Thrb PV/PV mice treated with or without TGFβ1 (Figure 5). As shown in Figure 5A, normal follicular cells from WT mice exhibited very slow migration without (panels a, b and c) or with TGFβ1 (panels d, e and f). In contrast, thyroid tumor cells from Thrb PV/PV mice showed a higher migration activity than that of normal follicular cells (compare panels g, h and i with panels a, b c). A marked increase in cell migration activity of thyroid tumor cells was clearly evident upon stimulation by TGFβ1 (panels j, k and l). The migration distance of cells was measured. The quantitative data (Figure 5B) show that tumor cells exhibit a stronger migration activity than the normal follicular cells and that TGFβ1 further stimulates the migration activity of tumor cells by ~2- to 3-fold. Taken together, these results indicate that activated TGFβ signaling promotes thyroid carcinogenesis by increasing cell migration activity and EMT.
Fig. 5.
Increased migration activity of primary thyroid tumor cells induced by TGFβ1. (A) A wound healing assay was performed using primary cultured thyroid tumor cells isolated from thyroids of Thrb PV/PV mice (n = 3). Cells were treated (d, e, f, j, k and l) or not treated with TGFβ1 (a, b, c, g, h and i). An Olympus microscope at ×100 magnification was used to take images before and after treatment with TGFβ1 as indicated time. (B) The relative distance between cells was measured using Image J software, and data were analyzed by a two-way analysis of variance followed by Bonferroni post hoc tests. ***P < 0.001 when compared with WT; NS, not significant.
Discussion
The molecular genetic changes identified in patients frequently represent a snapshot of the disease at the time of diagnosis and analysis in an individual patient. More difficult to know are the detailed molecular genetic alterations leading to cancer at the time of diagnosis and what deleterious changes could be continuously occurring in patients if cancer is not diagnosed and treated. Our studies addressed these important questions by using Thrb PV/PV mice because it would be difficult to assess in patients. Thrb PV/PV mice spontaneously develop FTC, thereby allowing us to identify global alterations of gene expression patterns during cancer progression from onset of disease to the stage when death occurred. We found dynamic temporal changes in gene expression with major patterns of ‘trending up’, ‘cyclical’ and ‘spiking up and then down’ for the upregulated genes and ‘trending down’ and ‘cyclical’ patterns for the downregulated genes during cancer progression. These results indicate that dynamic temporal changes in gene expression are occurring via complex regulation. Therefore, analysis of the expression of the same genes in patients with the same type of cancer, but at different disease stages, could yield different results. These findings could explain the variable results from analyses of identical genes from different groups of patients at different disease stages (58).
Our analysis of tumor-related genes with altered expression (Table I) uncovered an activated TGFβ-mediated network. Persistently elevated TGFβRII and its downstream effector, pSMAD2/3, were detected in the thyroid of Thrb PV/PV mice aged 3 to 10–12 months, indicative of sustained signaling via the network to drive tumor progression. We found elevated TGFβRII protein during thyroid carcinogenesis of Thrb PV/PV mice. However, the precise mechanism, leading to the increased TGFβRII protein, was not clear. In cancers, TGFβ acts to promote cancer progression by inducing EMT (49). Indeed, we found genes with reported functions in EMT were closely associated with an activated TGFβ-mediated network (Figure 2A and Table I). Loss of expression of E-cadherin, a hallmark of EMT (57), was clearly evident during cancer progression (Figure 3E). Several mesenchymal protein-encoding genes such as laminins and collagens were persistently overexpressed during cancer progression (Table I) (59–64). Other genes that have been shown to regulate EMT, such as fibronectin, vimentin, collagens, lipocalin, insulin-like growth factor 1, annexin A, calcium/calmodulin-dependent insulin-like growth factor 1 and pleiotrophin, were highly expressed (Table I) (26,61,62,65–69). The altered expression patterns of these genes would promote the loss of cell–cell adhesion and facilitate cell motility and invasion. Indeed, we showed that TGFβ stimulated the migration of primary tumor cells isolated from Thrb PV/PV mice. Thus, sustained activation of TGFβ-mediated signaling via the network promotes EMT to increase metastasis of thyroid tumor cells in Thrb PV/PV mice.
EMT is a multiple step process involving reorganization of cellular matrix components and altering expression of key regulators by repression of epithelial markers and upregulation of mesenchymal proteins (70). At present, the mechanism by which EMT was initiated in thyroid tumor cells of Thrb PV/PV mice is not clearly understood. However, secreted TGFβ1 in the serum was higher as early at age 3 months and persistently higher at 5 months in Thrb PV/PV mice than in WT mice (Figure 3C). This finding together with a concomitant elevated expression of TGFβRII suggests that one of the early events leading to EMT could be driven by elevated TGFβ1 activity. This notion is consistent with the gene expression profile-based network analysis discussed above (also see Figure 2A and Table I). However, it is important to point out that we have shown previously that PV-activated Src–focal adhesion kinase signaling promotes thyroid carcinogenesis in Thrb PV/PV mice (9) and that inhibition of Src activity by SKI-606 prevents repression of E-cadherin and activated expression of vimentin and slug, thereby decreasing EMT to delay cancer progression and reducing metastasis (71). This outcome is consistent with findings by others that indicate, via regulation of E-cadherin, increased Src activity promotes EMT, whereas Src inhibition suppresses EMT in cancers (72–74). So, it is reasonable to suggest that PV-activated Src–focal adhesion kinase signaling could work independently and/or in collaboration with activated TGFβ signaling, driving EMT to increase cell invasion and metastasis in thyroid carcinogenesis of Thrb PV/PV mice.
It is well recognized that during thyroid carcinogenesis, there are ongoing genetic molecular changes from the initiating events in primary lesions to distant metastasis and from differentiated tumors to undifferentiated cancer. However, detailed systematic analysis of gene expression profiles undergoing temporal changes during carcinogenesis would not be possible in patients for ethical and practical reasons. Using Thrb PV/PV mice the present studies provided evidence to indicate that thyroid carcinogenesis is driven by complex progressive changes in the expression of many genes with defined patterns. Therefore, after the initiating events, temporal interconnected alterations in gene expression direct the tumor cells on the paths toward metastasis and toward loss of differentiation. Efforts have long been directed toward identifying prognostic factors to better define the disease state and to predict the possible outcome. The importance of signature-recognizable patterns of multiple genes could be considered to be far more reliable prognostic indicators than sole reliance on a single gene. Our study was the first of this kind of gene expression analysis from a mouse model with pathological progression similar to human FTC. Thus, the temporal gene expression patterns we uncovered could be considered for use as potential prognostic signatures. Moreover, on the basis of recognizable expression of multiple genes at a certain disease state, a better choice of effective treatment modality could be achieved by targeting the affected aberrant pathways.
Supplementary material
Supplementary Table A can be found at http://carcin.oxfordjournals.org/
Funding
Intramural Research Program at the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
Supplementary Material
Acknowledgement
We thank Dr Fumihiko Furuya for the contributions in early phase of this present work.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- EMT
epithelial-to-mesenchymal transition
- FTC
follicular thyroid cancer
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- IPA
Ingenuity Pathways Analysis
- RT–PCR
reverse transcription–PCR
- TGF
transforming growth factor
- TGFβRs
TGFβ receptors
- WT
wild type.
References
- 1. Siegel R., et al. (2012). Cancer treatment and survivorship statistics, 2012. CA. Cancer J. Clin., 62, 220–241 [DOI] [PubMed] [Google Scholar]
- 2. Handkiewicz-Junak D., et al. (2010). Molecular prognostic markers in papillary and follicular thyroid cancer: Current status and future directions. Mol. Cell. Endocrinol., 322, 8–28 [DOI] [PubMed] [Google Scholar]
- 3. Xing M. (2008). Recent advances in molecular biology of thyroid cancer and their clinical implications. Otolaryngol. Clin. North Am., 41, 1135–46, ix [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaneshige M., et al. (2000). Mice with a targeted mutation in the thyroid hormone beta receptor gene exhibit impaired growth and resistance to thyroid hormone. Proc. Natl Acad. Sci. USA, 97, 13209–13214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parrilla R., et al. (1991). Characterization of seven novel mutations of the c-erbA beta gene in unrelated kindreds with generalized thyroid hormone resistance. Evidence for two “hot spot” regions of the ligand binding domain. J. Clin. Invest., 88, 2123–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suzuki H., et al. (2002). Mice with a mutation in the thyroid hormone receptor beta gene spontaneously develop thyroid carcinoma: a mouse model of thyroid carcinogenesis. Thyroid, 12, 963–969 [DOI] [PubMed] [Google Scholar]
- 7. Furuya F., et al. (2006). Activation of phosphatidylinositol 3-kinase signaling by a mutant thyroid hormone beta receptor. Proc. Natl Acad. Sci. USA, 103, 1780–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ringel M.D., et al. (2001). Overexpression and overactivation of Akt in thyroid carcinoma. Cancer Res., 61, 6105–6111 [PubMed] [Google Scholar]
- 9. Lu C., et al. (2010). Growth activation alone is not sufficient to cause metastatic thyroid cancer in a mouse model of follicular thyroid carcinoma. Endocrinology, 151, 1929–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ying H., et al. (2006). Aberrant accumulation of PTTG1 induced by a mutated thyroid hormone beta receptor inhibits mitotic progression. J. Clin. Invest., 116, 2972–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim C.S., et al. (2007). The pituitary tumor-transforming gene promotes angiogenesis in a mouse model of follicular thyroid cancer. Carcinogenesis, 28, 932–939 [DOI] [PubMed] [Google Scholar]
- 12. Guigon C.J., et al. (2008). Regulation of beta-catenin by a novel nongenomic action of thyroid hormone beta receptor. Mol. Cell. Biol., 28, 4598–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Y., et al. (1997). Ratio-based decisions and the quantitative analysis of cDNA microarray images. J. Biomed. Opt., 2, 364–374 [DOI] [PubMed] [Google Scholar]
- 14. Khan J., et al. (2001). Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat. Med., 7, 673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nimmakayalu M., et al. (2000). Simple method for preparation of fluor/hapten-labeled dUTP. Biotechniques, 28, 518–522 [DOI] [PubMed] [Google Scholar]
- 16. Guigon C.J., et al. (2010). Tumor suppressor action of liganded thyroid hormone receptor beta by direct repression of beta-catenin gene expression. Endocrinology, 151, 5528–5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim D.W., et al. (2012). Thyroid hormone receptor β suppresses SV40-mediated tumorigenesis via novel nongenomic actions. Am. J. Cancer Res., 2, 606–619 [PMC free article] [PubMed] [Google Scholar]
- 18. Kebebew E., et al. (2005). Diagnostic and prognostic value of angiogenesis-modulating genes in malignant thyroid neoplasms. Surgery, 138, 1102–9; discussion 1109. [DOI] [PubMed] [Google Scholar]
- 19. Vasko V., et al. (2007). Gene expression and functional evidence of epithelial-to-mesenchymal transition in papillary thyroid carcinoma invasion. Proc. Natl Acad. Sci. USA, 104, 2803–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamamoto Y., et al. (1992). An immunohistochemical study of epithelial membrane antigen, cytokeratin, and vimentin in papillary thyroid carcinoma. Recognition of lethal and favorable prognostic types. Cancer, 70, 2326–2333 [DOI] [PubMed] [Google Scholar]
- 21. Corvi R., et al. (2000). RET/PCM-1: a novel fusion gene in papillary thyroid carcinoma. Oncogene, 19, 4236–4242 [DOI] [PubMed] [Google Scholar]
- 22. Costa P., et al. (2006). Expression of prolactin receptor and prolactin in normal and malignant thyroid: a tissue microarray study. Endocr. Pathol., 17, 377–386 [DOI] [PubMed] [Google Scholar]
- 23. Anania M.C., et al. (2011). TIMP3 regulates migration, invasion and in vivo tumorigenicity of thyroid tumor cells. Oncogene, 30, 3011–3023 [DOI] [PubMed] [Google Scholar]
- 24. Kubiczkova L., et al. (2012). TGF-β - an excellent servant but a bad master. J. Transl. Med., 10, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Skonier J., et al. (1992). cDNA cloning and sequence analysis of beta ig-h3, a novel gene induced in a human adenocarcinoma cell line after treatment with transforming growth factor-beta. DNA Cell Biol., 11, 511–522 [DOI] [PubMed] [Google Scholar]
- 26. de Graauw M., et al. (2010). Annexin A1 regulates TGF-beta signaling and promotes metastasis formation of basal-like breast cancer cells. Proc. Natl Acad. Sci. USA, 107, 6340–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Isono M., et al. (2002). Smad pathway is activated in the diabetic mouse kidney and Smad3 mediates TGF-beta-induced fibronectin in mesangial cells. Biochem. Biophys. Res. Commun., 296, 1356–1365 [DOI] [PubMed] [Google Scholar]
- 28. Hallatschek W., et al. (2004). Inhibition of hepatic transcriptional induction of lipopolysaccharide-binding protein by transforming-growth-factor beta 1. Eur. J. Immunol., 34, 1441–1450 [DOI] [PubMed] [Google Scholar]
- 29. Okazaki R., et al. (1995). Transforming growth factor-beta and forskolin increase all classes of insulin-like growth factor-I transcripts in normal human osteoblast-like cells. Biochem. Biophys. Res. Commun., 207, 963–970 [DOI] [PubMed] [Google Scholar]
- 30. Rogel M.R., et al. (2011). Vimentin is sufficient and required for wound repair and remodeling in alveolar epithelial cells. FASEB J., 25, 3873–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiang Y., et al. (2005). Transforming growth factor-beta1 regulation of laminin gamma1 and fibronectin expression and survival of mouse mesangial cells. Mol. Cell. Biochem., 278, 165–175 [DOI] [PubMed] [Google Scholar]
- 32. Grande J., et al. (1993). Transforming growth factor-beta 1 induces collagen IV gene expression in NIH-3T3 cells. Lab. Invest., 69, 387–395 [PubMed] [Google Scholar]
- 33. Calonge M.J., et al. (2004). Opposite Smad and chicken ovalbumin upstream promoter transcription factor inputs in the regulation of the collagen VII gene promoter by transforming growth factor-beta. J. Biol. Chem., 279, 23759–23765 [DOI] [PubMed] [Google Scholar]
- 34. Sayeed A., et al. (2010). Negative regulation of UCP2 by TGFβ signaling characterizes low and intermediate-grade primary breast cancer. Cell Death Dis., 1, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heegaard A.M., et al. (2004). Transforming growth factor beta stimulation of biglycan gene expression is potentially mediated by sp1 binding factors. J. Cell. Biochem., 93, 463–475 [DOI] [PubMed] [Google Scholar]
- 36. Smeland E.B., et al. (1987). Transforming growth factor type beta (TGF beta) inhibits G1 to S transition, but not activation of human B lymphocytes. Exp. Cell Res., 171, 213–222 [DOI] [PubMed] [Google Scholar]
- 37. Kenyon N.J., et al. (2003). TGF-beta1 causes airway fibrosis and increased collagen I and III mRNA in mice. Thorax, 58, 772–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Norris A., et al. (2012). AGR2 is a SMAD4-suppressible gene that modulates MUC1 levels and promotes the initiation and progression of pancreatic intraepithelial neoplasia. Oncogene. http//dx..org/10.1038/onc.2012.394 (3 September 2012, date last accessed). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ghafoori P., et al. (2009). Increased IkappaB alpha expression is essential for the tolerogenic property of TGF-beta-exposed APCs. FASEB J., 23, 2226–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakamura H., et al. (2005). Transforming growth factor-beta1 induces LMO7 while enhancing the invasiveness of rat ascites hepatoma cells. Cancer Lett., 220, 95–99 [DOI] [PubMed] [Google Scholar]
- 41. Wang B., et al. (2010). TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene, 29, 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eisenwort G., et al. (2011). Identification of TROP2 (TACSTD2), an EpCAM-like molecule, as a specific marker for TGF-β1-dependent human epidermal Langerhans cells. J. Invest. Dermatol., 131, 2049–2057 [DOI] [PubMed] [Google Scholar]
- 43. Huang W., et al. (2011). Enhanced expression of tissue inhibitor of metalloproteinases-4 gene in human osteoarthritic synovial membranes and its differential regulation by cytokines in chondrocytes. Open Rheumatol. J., 5, 81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lu Z., et al. (2001). Transcriptional regulation of the lung fatty acid synthase gene by glucocorticoid, thyroid hormone and transforming growth factor-beta 1. Biochim. Biophys. Acta, 1532, 213–222 [DOI] [PubMed] [Google Scholar]
- 45. Sharma V., et al. (2011). Enhancement of TGF-β signaling responses by the E3 ubiquitin ligase Arkadia provides tumor suppression in colorectal cancer. Cancer Res., 71, 6438–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yan Z., et al. (2001). Oncogenic Ki-ras confers a more aggressive colon cancer phenotype through modification of transforming growth factor-beta receptor III. J. Biol. Chem., 276, 1555–1563 [DOI] [PubMed] [Google Scholar]
- 47. Park B.J., et al. (2000). Mitogenic conversion of transforming growth factor-beta1 effect by oncogenic Ha-Ras-induced activation of the mitogen-activated protein kinase signaling pathway in human prostate cancer. Cancer Res., 60, 3031–3038 [PubMed] [Google Scholar]
- 48. Jonson T., et al. (2001). Altered expression of TGFB receptors and mitogenic effects of TGFB in pancreatic carcinomas. Int. J. Oncol., 19, 71–81 [PubMed] [Google Scholar]
- 49. Heldin C.H., et al. (2012). Regulation of EMT by TGFβ in cancer. FEBS Lett., 586, 1959–1970 [DOI] [PubMed] [Google Scholar]
- 50. Obara M., et al. (2010). The third type III module of human fibronectin mediates cell adhesion and migration. J. Biochem., 147, 327–335 [DOI] [PubMed] [Google Scholar]
- 51. McInroy L., et al. (2007). Down-regulation of vimentin expression inhibits carcinoma cell migration and adhesion. Biochem. Biophys. Res. Commun., 360, 109–114 [DOI] [PubMed] [Google Scholar]
- 52. Marneros A.G., et al. (2005). Physiological role of collagen XVIII and endostatin. FASEB J., 19, 716–728 [DOI] [PubMed] [Google Scholar]
- 53. Gu Y.C., et al. (2003). Laminin isoform-specific promotion of adhesion and migration of human bone marrow progenitor cells. Blood, 101, 877–885 [DOI] [PubMed] [Google Scholar]
- 54. Olivero D.K., et al. (1993). Type IV collagen, laminin, and fibronectin promote the adhesion and migration of rabbit lens epithelial cells in vitro . Invest. Ophthalmol. Vis. Sci., 34, 2825–2834 [PubMed] [Google Scholar]
- 55. Lisman T., et al. (2006). A single high-affinity binding site for von Wille brand factor in collagen III, identified using synthetic triple-helical peptides. Blood, 108, 3753–3756 [DOI] [PubMed] [Google Scholar]
- 56. Tummalapalli C.M., et al. (2001). Tissue inhibitor of metalloproteinase-4 instigates apoptosis in transformed cardiac fibroblasts. J. Cell. Biochem., 80, 512–521 [DOI] [PubMed] [Google Scholar]
- 57. Teng Y., et al. (2007). Transcriptional regulation of epithelial-mesenchymal transition. J. Clin. Invest., 117, 304–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Scheumman G.F., et al. (1995). Clinical significance of E-cadherin as a prognostic marker in thyroid carcinomas. J. Clin. Endocrinol. Metab., 80, 2168–2172 [DOI] [PubMed] [Google Scholar]
- 59. Sherman-Baust C.A., et al. (2003). Remodeling of the extracellular matrix through overexpression of collagen VI contributes to cisplatin resistance in ovarian cancer cells. Cancer Cell, 3, 377–386 [DOI] [PubMed] [Google Scholar]
- 60. Tang L., et al. (2006). Aberrant expression of collagen triple helix repeat containing 1 in human solid cancers. Clin. Cancer Res., 12, 3716–3722 [DOI] [PubMed] [Google Scholar]
- 61. Shintani Y., et al. (2008). Collagen I promotes epithelial-to-mesenchymal transition in lung cancer cells via transforming growth factor-beta signaling. Am. J. Respir. Cell Mol. Biol., 38, 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Espinosa Neira R., et al. (2012). Native type IV collagen induces an epithelial to mesenchymal transition-like process in mammary epithelial cells MCF10A. Int. J. Biochem. Cell Biol., 44, 2194–2203 [DOI] [PubMed] [Google Scholar]
- 63. De Arcangelis A., et al. (2001). Overexpression of laminin alpha1 chain in colonic cancer cells induces an increase in tumor growth. Int. J. Cancer, 94, 44–53 [DOI] [PubMed] [Google Scholar]
- 64. Fujita M., et al. (2005). Overexpression of beta1-chain-containing laminins in capillary basement membranes of human breast cancer and its metastases. Breast Cancer Res., 7, R411–R421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yang J., et al. (2009). Lipocalin 2 promotes breast cancer progression. Proc. Natl Acad. Sci. USA, 106, 3913–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Walsh L.A., et al. (2011). IGF-1 increases invasive potential of MCF 7 breast cancer cells and induces activation of latent TGF-β1 resulting in epithelial to mesenchymal transition. Cell Commun. Signal., 9, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bergamaschi A., et al. (2008). CAMK1D amplification implicated in epithelial-mesenchymal transition in basal-like breast cancer. Mol. Oncol., 2, 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Perez-Pinera P., et al. (2006). Pleiotrophin disrupts calcium-dependent homophilic cell-cell adhesion and initiates an epithelial-mesenchymal transition. Proc. Natl Acad. Sci. USA, 103, 17795–17800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zeisberg M., et al. (2009). Biomarkers for epithelial-mesenchymal transitions. J. Clin. Invest., 119, 1429–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grünert S., et al. (2003). Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat. Rev. Mol. Cell Biol., 4, 657–665 [DOI] [PubMed] [Google Scholar]
- 71. Kim W.G., et al. (2012). SKI-606, an Src inhibitor, reduces tumor growth, invasion, and distant metastasis in a mouse model of thyroid cancer. Clin. Cancer Res., 18, 1281–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nagathihalli N.S., et al. (2012). Src-mediated regulation of E-cadherin and EMT in pancreatic cancer. Front. Biosci., 17, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 73. Cicchini C., et al. (2008). TGFbeta-induced EMT requires focal adhesion kinase (FAK) signaling. Exp. Cell Res., 314, 143–152 [DOI] [PubMed] [Google Scholar]
- 74. Avizienyte E., et al. (2004). Src SH3/2 domain-mediated peripheral accumulation of Src and phospho-myosin is linked to deregulation of E-cadherin and the epithelial-mesenchymal transition. Mol. Biol. Cell, 15, 2794–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.