Abstract
Interstrand cross-links (ICLs) are very severe lesions as they are absolute blocks of replication and transcription. This property of interstrand cross-linking agents has been exploited clinically for the treatment of cancers and other diseases. ICLs are repaired in human cells by specialized DNA repair pathways including components of the nucleotide excision repair pathway, double-strand break repair pathway and the Fanconi anemia pathway. In this report, we identify the role of RECQL5, a member of the RecQ family of helicases, in the repair of ICLs. Using laser-directed confocal microscopy, we demonstrate that RECQL5 is recruited to ICLs formed by trioxalen (a psoralen-derived compound) and ultraviolet irradiation A. Using single-cell gel electrophoresis and proliferation assays, we identify the role of RECQL5 in the repair of ICL lesions. The domain of RECQL5 that recruits to the site of ICL was mapped to the KIX region between amino acids 500 and 650. Inhibition of transcription and of topoisomerases did not affect recruitment, which was inhibited by DNA-intercalating agents, suggesting that the DNA structure itself may be responsible for the recruitment of RECQL5 to the sites of ICLs.
Introduction
Interstrand cross-linking (ICL) agents are used clinically as chemotherapeutic tools (1). One class of these agents, psoralens, are photoactive planar tricyclic compounds that intercalate into DNA and upon exposure to ultraviolet (UVA) light (365 nm) can form covalent bonds primarily with 5, 6 double bonds of thymines (2). ICLs between the two strands of DNA are one of the most toxic DNA lesions because they act as definitive blocks to transcription and replication (3,4).
Although primarily used in the treatment of psoriasis, a chronic skin disease marked by uncontrolled epidermal cell proliferation, psoralens are also used against cutaneous T-cell lymphomas (1,5). Treatment with psoralen plus UVA light (PUVA) can lead to both squamous and basal cell carcinoma, as well as malignant melanoma (6–9). This is thought to be a side effect of the ICL DNA repair process through the generation of point mutations, deletions and translocations that modulate genetic stability (1). A clear understanding of the DNA repair process is, therefore, pivotal to understand the process of tumorigenesis after ICL formation.
Mammalian ICL repair is a complex process (10). Briefly, the first cross-link ‘unhooking’ step in mammalian cells involves dual incisions on either side of the lesion, followed by processing, gap filling and the removal of the ‘unhooked’ DNA. The precise mechanism, and the requisite enzymes involved, has yet to be clarified. Recent studies of the RecQ family of helicases demonstrate that these enzymes facilitate repair of many types of DNA damage in eukaryotic cells. Although there is only one RecQ helicase in Escherichia coli, human cells express five RecQ helicases: Werner syndrome protein (WRN), Bloom syndrome protein (BLM), RECQL1, RECQL4 and RECQL5 (11), all of which interact with and can help ‘resolve’ complex DNA structures that arise due to damaged DNA (12). Humans with RecQ helicase disorders suffer from premature aging and cancer. RecQ helicase-deficient cells are also hypersensitive to DNA damaging agents, and WRN- and BLM-deficient cells are hypersensitive to ICLs (13–15), further suggesting that RecQ helicases play important roles in repairing DNA damage (16). WRN interacts with other proteins that facilitate repair of DNA ICLs, including BRCA1, Pso4, RAD51, RAD54, RAD54B and ATR (17–19), and is thought to unwind duplex DNA near the ICL (19). BLM interacts with proteins in the Fanconi anemia pathway and plays a role during the recombination phase of ICL repair (20). Much less is known about the role of RECQL1, RECQL4 and RECQL5 in ICL repair. RECQL5 is of particular interest as the depletion of this protein causes enhanced cell death in cultured cells (21), and RECQL5 knockout mice suffer from elevated cancer formation (22).
In this study, we used a recently developed method to selectively generate PUVA-induced ICLs in subnuclear regions of U2OS cells (23). DNA damage-containing cells were then used to investigate the role of RecQ helicases in the repair of PUVA-induced ICLs. The results show that RECQL5, WRN and BLM are recruited to and promote repair of ICLs in U2OS cells. Domain mapping experiments showed that the ‘KIX’ (amino acids 501–650) region of RECQL5 is required for recruitment. The recruitment does not require the KIX domain to interact with RNA polymerase II because a single-point mutant E584D within the KIX domain, which was previously demonstrated to lack domain interaction with RNA polymerase II (24), is still recruited to the site of ICL. Additionally, the RNA polymerase inhibitors α-amanitin and 5,6-dichloro-β-d-ribofuranosylbenzimidazole (DRB) do not inhibit recruitment of RECQL5 to ICLs. However, DNA-intercalating agents, such as actinomycin-D (Act-D) and ethidium bromide (EtBr), do inhibit RECQL5 recruitment to ICLs. These data suggest that RECQL5 binds to and facilitates repair of ICLs in the absence of active transcription, but that the structure or flexibility of DNA may influence recruitment and/or retention of RECQL5 at sites of DNA damage.
Materials and methods
Cell lines
U2OS and HEK293T cells were obtained from American Type Culture Collection (ATCC, Manassas, VA) and were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin (Pen–Strep) at 37°C in a humidified atmosphere with 5% CO2. Human glioblastoma cell lines M059K (containing normal levels DNA-PKcs) and M059J (lacking expression of DNA-PKcs) were also obtained from American Type Culture Collection and were grown in Dulbecco’s modified Eagle’s medium and F-12 medium with 15% fetal bovine serum. CS1AN vector and Cockayne syndrome group B-expressing cells were grown as described previously (25). Xeroderma pigmentosum complementation group XPC patient fibroblasts (GM16370), SV40-transformed normal fibroblasts (GM637), WRN patient fibroblasts (AG11395), BLM patient fibroblasts (GM08505), ATR (Seckel) patient fibroblasts (GM18366) and ATM patient fibroblasts (GM5849) were obtained from Coriell Cell Repositories (Camden, NJ). The cells were cultured in modified Eagle’s media supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin along with 1× non-essential and essential amino acids, vitamins and 2mM glutamine.
Chemicals and reagents
Trioxalen, α-amanitin, actinomycin-D, DRB, etoposide and camptothecin were purchased from Sigma–Aldrich (St Louis, MO) and used at the concentrations indicated. EtBr was purchased from Bio-Rad (Hercules, CA).
Generation of lentivirus
Stable RECQL5 lentiviral knockdown cells were generated as described previously (21). pLKO.1 vector harboring short hairpin RNA (shRNA) construct targeting human RECQL5 or RECQL4 was obtained from Sigma–Aldrich. shRNA targeting the coding region of RECQL5 gene, 5′-CCGGCCCTAAAGGTACGAGTAA GTT CTCGAGAA CTTACTC GTACCTTTAGGGTTTTTG- 3′, or shRNA targeting the coding region of RECQL4 gene, 5′-CCGGCCTCGATTCCATTATCATTTA CTCGAGTAAATGATAATGGA ATCGAGGTTTTTG-3′, was used. shRNA construct expressing scrambled sequence was purchased from Addgene (Plasmid 1864, deposited by Sabatini lab (26)). pCMV ΔR8.2 (Addgene plasmid 8455) and pCMV spike glycoprotein of the vesicular stomatitis virus (Addgene plasmid 8454) both deposited by Weinberg lab (27) were used to generate second-generation spike glycoprotein of the vesicular stomatitis virus pseudotyped lentiviruses by transient cotransfection of HEK293T cells with a three-plasmid combination.
Laser irradiation and confocal microscopy
Cells were seeded in 35 mm glass bottom dishes from MatTek (Ashland, MA) or ibidi (Verona, WA) 24h before transfection. Transfections were performed using Lipofectamine LTX (Invitrogen, Grand Island, NY) according to the manufacturer’s protocol. Prior to targeting, cells were exposed to 6 µM trioxalen (Sigma–Aldrich) for 20 min. Where mentioned, the appropriate chemical treatment was also added. We used a Nikon Eclipse 2000E microscope with a Yokogawa CSU 10 Spinning Disk head for confocal microscopy (Improvision/PerkinElemer, Waltham, MA) with an attached Photonic MicroPoint ablation system (Photonic Instruments, St Charles, IL) adjusted to a 365 nm wavelength. The power of the laser was attenuated through Improvison’s Volocity software 6.0.2 (Improvision/PerkinElmer) in terms of percent intensity to 1.7% to generate ICL’s, and the areas were struck twice in succession to drive in the ICLs. A 3 × 20 pixel region internal to the nuclei of the cells was targeted via a Plan Fluor 60×/1.25d n.a. oil objective. Images were captured at various time points and analyzed using Volocity. The microscope slide chamber is encased in an environmental chamber (Solent Scientific Ltd) to maintain the normal physiology of the cells.
Determination of ICL-induced incision by alkaline comet assay
The comet assay was performed under alkaline conditions to detect the presence of ICLs as reported previously with the slight modification (25). Cells treated with trioxalen and irradiated with UVA (365nm) were washed with phosphate-buffered saline (PBS) and scraped immediately 0, 6 or 16 h after the UVA treatment and suspended in PBS. Approximately, 1.5×104 cells were mixed with 75 μl of 1.5% low-melting point agarose and spread on a microscope slide precoated with 1% agarose. Low-melting-point agarose (90 μl) was applied on top of the sample layer as the last step of slide preparation. Slides were placed in cold lysis buffer (10mM Tris–HCl pH 7.4, 2.5M NaCl, 100mM ethylenediaminetetraacetic acid, 1% Triton X-100 and 10% dimethyl sulfoxide) for at least 1 h at 4°C, followed by rinsing three times in PBS for 5min. After lysis, slides were incubated in dimethyl sulfoxide-free lysis buffer containing 1mg/ml of proteinase K for 2h at 37°C. Slides were washed in PBS three times before incubation in cold unwinding solution (300mM NaOH, pH 13 and 1mM ethylenediaminetetraacetic acid) in the dark at 4°C for 45min. Electrophoresis was carried out under the same condition at 25V for 30min. Slides were then rinsed in neutralizing solution (0.4M Tris–HCl, pH 7.5) three times for 5min and fixed in 100% ethanol before staining with EtBr. Images of ~100 cells per sample were obtained by using a fluorescence microscope (Axiovert 200M, Carl Zeiss) and Axiovision 4.2 software. Individual comet images were evaluated by using Komet 5 image software (Kinetic Imaging). The removal of ICL was analyzed by comparing the tail intensity of the UVA-irradiated trioxalen-treated cells with that of the UVA-irradiated control cells.
Western blot analysis
Cells were washed twice with PBS, harvested and subsequently lysed in RIPA buffer (50mM Tris–HCl pH 8.0, 150mM NaCl, 0.1% sodium dodecyl sulfate and 0.5% sodium deoxycholate) supplemented with 1× protease and phosphatase inhibitors (Roche). Cell debris was then removed by centrifugation. Protein concentration of the cell lysates was determined using a protein assay kit manufactured by Bio-Rad (Bradford method). A 30 μg total protein per sample was applied to precast 4–12% sodium dodecyl sulfate–polyacrylamide gels (Invitrogen), transferred to polyvinylidene difluoride membrane (Invitrogen) and probed with the antibodies for primary.
Antibodies
The following antibodies were used in western blots and immunofluorescence studies: RECQL5 (Abcam, 1:1000), RECQL5 (Custom rabbit, 1:1000) (21), WRN (Custom mouse, 1:1000) (28), BLM (Abcam, rabbit, 1:1000), RECQL4 (Custom rabbit, 1:1000) (29), ATM (Abcam, mouse, 1:1000), ATR (Bethyl Laboratories, rabbit, 1:1000), XPC (GeneTex, mouse, 1:1000), FANCD2 (GeneTex, N1, rabbit, 1:1000), DNAPK (Santa Cruz Biotechnology, mouse, 1:1000), γ-H2AX (Abcam, mouse, 1:1000), green fluorescent protein (GFP) (Abcam, mouse 1:1000), β-actin (Sigma, mouse, 1:10 000) and FLAG (Sigma, rabbit, 1:180).
Construction of the GFP-tagged RECQL5 fragments
GFP-RECQL5 plasmid was constructed as follows: RECQL5 gene was PCR amplified from pPG10 plasmid (30), using primers RQ5-Met-KpnI-F, 5ʹ-GAGATTGGTACCTAT GAGCAGCCACCATACAACCT-3ʹ and RQ5-nostop-XbaI-R, 5ʹ-GAGATTTCTAGA TCTCTGGGGGCCACA CAG-3ʹ. PCR products were digested and ligated between KpnI and XbaI sites of the pcDNA3.1-CT-GFP plasmid (Invitrogen) generating RQ5/pcDNA3.1-CT-GFP construct. The RECQL5 gene fragments corresponding to the amino acid residues 1–240, 1–500, 501–650 and 853–991 were amplified by PCR. The nucleotide sequence corresponding to the SV40 nuclear localization signal (SV40 NLS) was added onto the RECQL5 fragments in PCR primers for these truncated fragments, except the 853–991 which contains RECQL5’s original NLS sequence. Each of the PCR products was inserted into the pcDNA3.1-CT-GFP-TOPO plasmid according to the manufacturers’ protocol (Invitrogen) (31). The flag-tagged constructs used for the recruitment analysis of the wild-type and E584D KIX proteins were kind gifts from Dr W.Wang (National Institute on Aging) (24).
Cell viability assays
For the cytotoxicity assays, 5000 cells were plated in triplicate onto 96-well microtiter plates in complete growth medium. After 24h of incubation, the cells were treated with indicated concentration of trioxalen for 20 min and then treated with UVA (365 nm). The medium was removed, and then, 100 μl of fresh medium was added, followed by 10 μl of WST-1 (Roche). After appropriate time of incubation in WST-1, the absorbance at 450nm was determined. For cell growth assays, 2000 cells were plated in triplicate into 96 well microtiter plates. Cell numbers were measured each day by replacing growth medium with medium containing WST-1 as described above.
Results
WRN, BLM and RECQL5 are recruited to ICLs
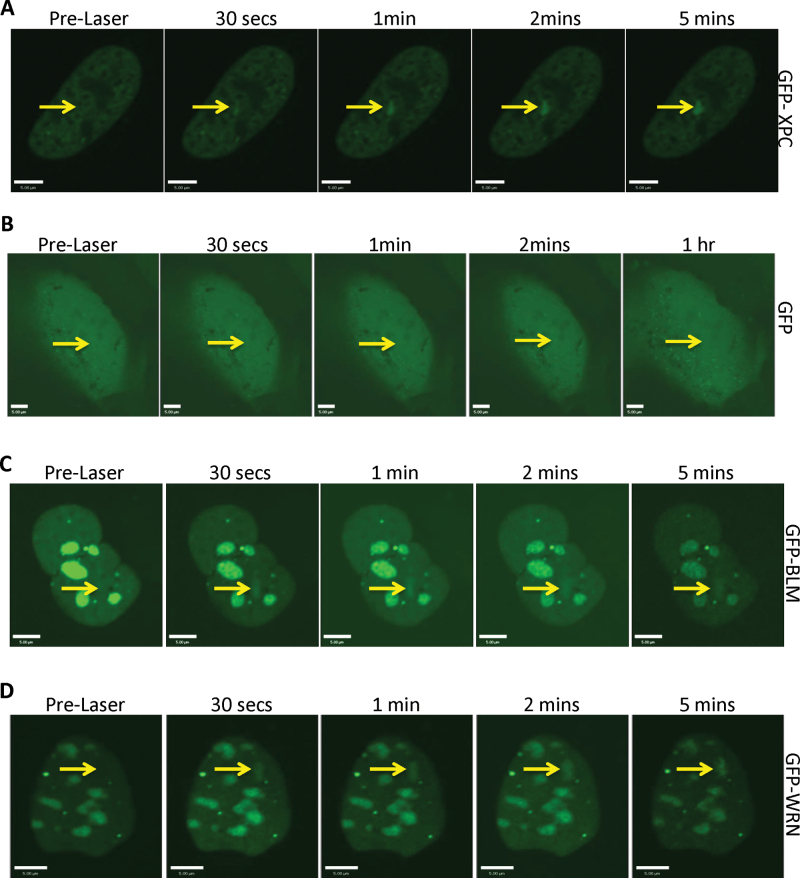
To generate ICLs, U2OS cells were exposed to 4-, 5-, 8-trimethylpsoralen (trioxalen) for 20 min, followed by 365nm laser irradiation restricted to a defined region of the cell nuclei (32). The recruitment of GFP-tagged XPC was followed as a measure of ICL formation. GFP-XPC accumulated at sites exposed to the laser–trioxalen combination (Figure 1A); the recruitment is indicated by arrows in Figure 1A and in subsequent figures. Control experiments using a GFP plasmid indicated no recruitment of GFP protein to the sites of the ICLs generated by PUVA (Figure 1B). Next, we investigated the recruitment pattern of the different RecQ helicases to the PUVA-generated ICLs within 5 min of the damage. As expected, we observed recruitment of the GFP-tagged BLM and WRN to the sites of damage (Figure 1C and D). This is consistent with previous reports that have used other approaches to determine that WRN and BLM participate in ICL processing (17–20). Interestingly, we also observed the localization of GFP-tagged RECQL5 (GFP-RECQL5) to the PUVA-generated ICLs (Figure 1E). Under similar conditions, GFP-tagged RECQL1 and RECQL4 failed to recruit to the cross-links (Figure 1F and G). Because the localization of RECQL1 and RECQL4 to DSBs has been reported, the observed recruitment of RECQL5 to the targeted regions was not due to breaks in the duplex DNA (33). To confirm that the recruitment of RECQL5 was due to the presence of ICLs, we irradiated the cells expressing GFP-RECQL5 with laser in the absence of trioxalen. In this study, we did not observe the recruitment of RECQL5 (Figure 1H).
Fig. 1.
WRN, BLM and RECQL5 are recruited to ICLs. (A) U2OS cells were transfected with 2 µg of GFP-XPC; 24h posttransfection, the cells were incubated with 6 µM trioxalen for 20 min and then laser photoactivated in the nuclear region in individual cells in a defined time sequence. (B) U2OS cells were transfected with 2 µg of GFP expressing plasmid and treated as in panel (A). U2OS cells were transfected with 2 µg of (C) GFP-BLM, (D) GFP-WRN, (E) GFP-RECQL5, (F) GFP-RECQL1 or (G) GFP-RECQL4, and the cells were processed as in panel (A). (H) U2OS cells were transfected with 2 µg of GFP-RECQL5 and treated as in panel (A). The targeted regions are indicated by arrows. Bar, 5 µM.
We next investigated if there was recruitment of endogenous RECQL5 to the site of the ICLs. After laser targeting, cells were fixed and probed with either of two different RECQL5 antibodies as described in Materials and methods. Indeed, we observed recruitment of the endogenous RECQL5 to the ICLs; the recruitment was confirmed by costaining for γ-H2AX (Supplementary Figure 1A and B, available at Carcinogenesis Online). Furthermore, by depleting RECQL5 in cells using shRNAs (Figure 2C), we found a reduction in the number of cells where RECQL5 was targeted to the ICLs, as detected by XPC recruitment, 52 versus 22% RECQL5 response in shScramble versus RECQL5 KD cells (Supplementary Figure 1C and D, available at Carcinogenesis Online). These results suggest that RECQL5, like WRN and BLM, may play a role in the repair of ICLs.
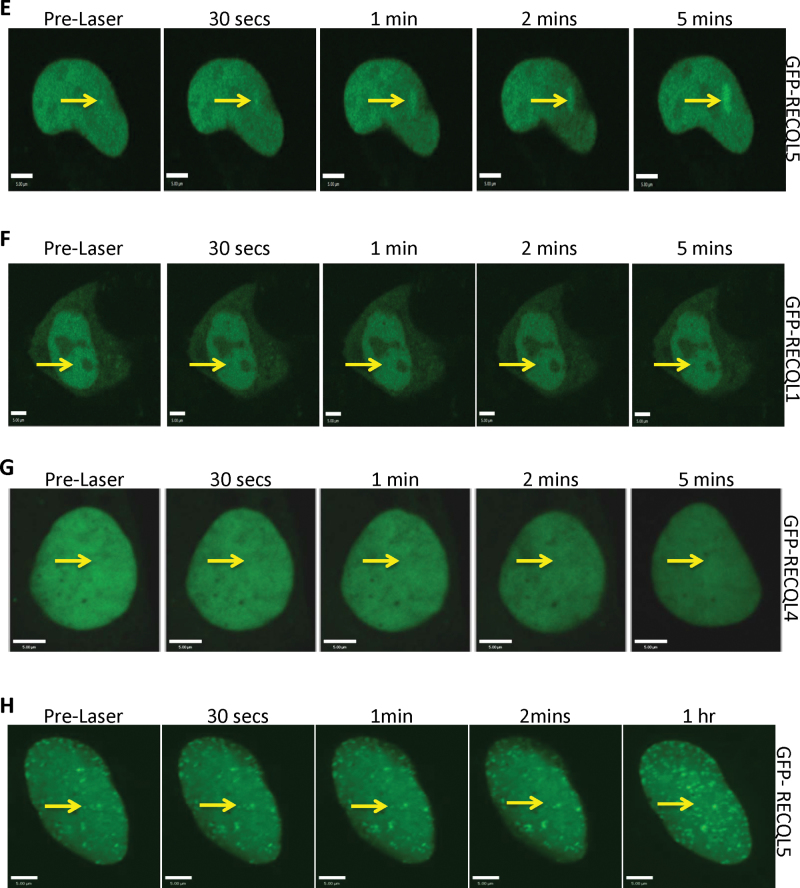
Fig. 2.
RECQL5 facilitates repair of ICLs. (A) Representative images from an alkaline comet tail recovery assay of shScrambled and shRECQL5 U2OS cells with and without trioxalen plus UVA treatment and fixed immediately (0h), 6 or 16 h later. Untreated represents the nuclear DNA from cells unexposed to trioxalen. (B) Quantitation of the comet tail. Mean tail moment was measured as described in Materials and methods and was normalized relative to 0 h measurement of each sample. More than 100 nuclear DNAs were analyzed in each experiment. Two biologically independent experiments were performed and error bars represent ± SD. ***P < 0.001, analyzed with Student’s t-test. (C) Western blot showing reduced expression of RECQL5 in cellular extracts of U2OS shRECQL5-treated cells compared with control shScrambled-treated cells. Equal loading was confirmed by probing with a β-actin-specific antibody. (D) Western blot showing reduced expression of RECQL4 in cellular extracts of U2OS shRECQL4-treated cells compared with control shScrambled-treated cells. Equal loading was confirmed by probing with a β-actin-specific antibody. (E) Cells were plated onto 96-well dishes and exposed to 0.06 µM trioxalen plus UVA. Survival, measured as WST-1 reduction, was calculated relative to the survival of cells exposed to trioxalen alone at the days indicated. Error bars represent ±SD (n = 3).
Kinetics of RECQL5 binding to ICLs
Kinetic experiments showed that GFP-RECQL5 accumulated rapidly at the site of laser irradiation (in <5min) and appeared to remain bound at ICLs for up to 8 h (Supplementary Figure 2A, available at Carcinogenesis Online). Upon investigating the recruitment kinetics, we found that the recruitment of RECQL5 (Supplementary Figure 2B, available at Carcinogenesis Online) was nearly as rapid as WRN to the site of the ICLs (Supplementary Figure 2C, available at Carcinogenesis Online); the apparent difference is the time taken for the GFP intensity of RECQL5 to plateau compared with WRN. We believe this is due to the variable expression level of GFP-RECQL5, in comparison with GFP-WRN, which required high image capture frame rates and this causes photo bleaching. This also explains why we see a consistent increase in the GFP signal in the 8 h experiment (Supplementary Figure 2A, available at Carcinogenesis Online) (much slower frame rate) but see the signal decreasing at around 2 min in the 5 min graphs (Supplementary Figure 2B and C, available at Carcinogenesis Online). This was also apparent when we measured the recruitment kinetics of BLM using a slower frame rate (Supplementary Figure 2D, available at Carcinogenesis Online). Our results indicate a fast response of RECQL5 to the site of ICL suggesting an early role at the repair site, whereas the retention study suggested that it may also be involved in the later steps of the DNA repair after the unhooking of the ICL (17,23).
RECQL5 participates in repair of PUVA-induced DNA damage
To address whether RECQL5 plays a role in the repair of ICLs, we depleted RECQL5 and performed alkaline comet assays to determine the DNA repair kinetics. Under the alkaline condition, damaged DNA from cells without any treatment, especially the DNA with single-strand breaks, migrates faster in the alkaline gel than undamaged DNA, nuclear matrix (Figure 2A top panels; untreated). DNA from cells treated with cross-linking agents fails to migrate into the alkaline gel (Figure 2A, 0 h treatment), but incision of the cross-linked genomic DNA during repair restores the comet tail (Figure 2A, 16 h treatment). Therefore, the result of the assay is evaluated by the tail moment, a measure of the relative DNA electrophoretic mobility. The tail moment at different time points after PUVA treatment was measured to determine the ICL repair kinetics (34). The results showed that the mean tail moment in the control cells gradually increased between 0 (when the cross-links were generated) and 16 h, indicating repair of the ICLs in these cells. In comparison, the rate of increase in the mean tail moment was significantly slower in the shRECQL5 cells (Figure 2A and B, at 6 h where P < 0.001), suggesting that RECQL5 is required to efficiently repair the ICL lesions. After 16 h, the difference in mean tail moment between shRECQL5 and shScrambled cells became smaller, indicating that the repair of ICL lesions was ongoing in both cells. The extent of RECQL5 knockdown was confirmed by western blot (Figure 2C) and showed almost complete depletion. Although we recognize that PUVA creates monoadducts and ICL, the comet assay specifically measures unhooking or removal of ICLs and not repair of monoadducts, and thus, we can state that RECQL5 contributes to repair of PUVA ICLs.
Next, we measured the effect of the RECQL5 depletion on cellular proliferation in response to PUVA-generated ICLs. For comparison, we also depleted RECQL4 using the same strategy as used to deplete RECQL5 (Figure 2D) as RECQL4 was not recruited to the ICL (Figure 1G). We compared the effect of RECQL4 and RECQL5 depletion on cell proliferation after ICL generation. Previously, we reported a slower proliferation rate of cells with RECQL5 depletion (21). Therefore, the data were corrected for the slower growth in this analysis. The result demonstrated that depletion of RECQL5 reduced the proliferation of cells over a period of 3 days after PUVA treatment (Figure 2E). On the other hand, depletion of RECQL4 did not affect the growth of cells after PUVA treatment and they grew similar to PUVA-treated control population (Figure 2E). This was also the case when we measured proliferation in the presence of increasing damage (using increasing dosage of trioxalen, Supplementary Figure 3, available at Carcinogenesis Online). These results, together with the recruitment data, suggest a more significant role of RECQL5 than that of RECQL4 in ICL processing.
The KIX domain of RECQL5 recruits to the sites of ICLs
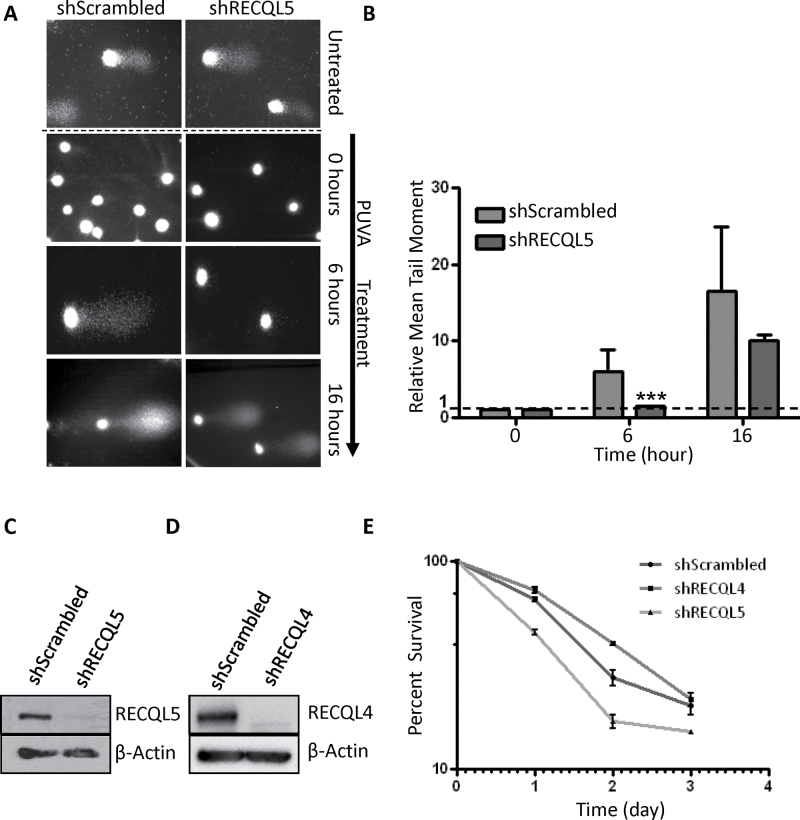
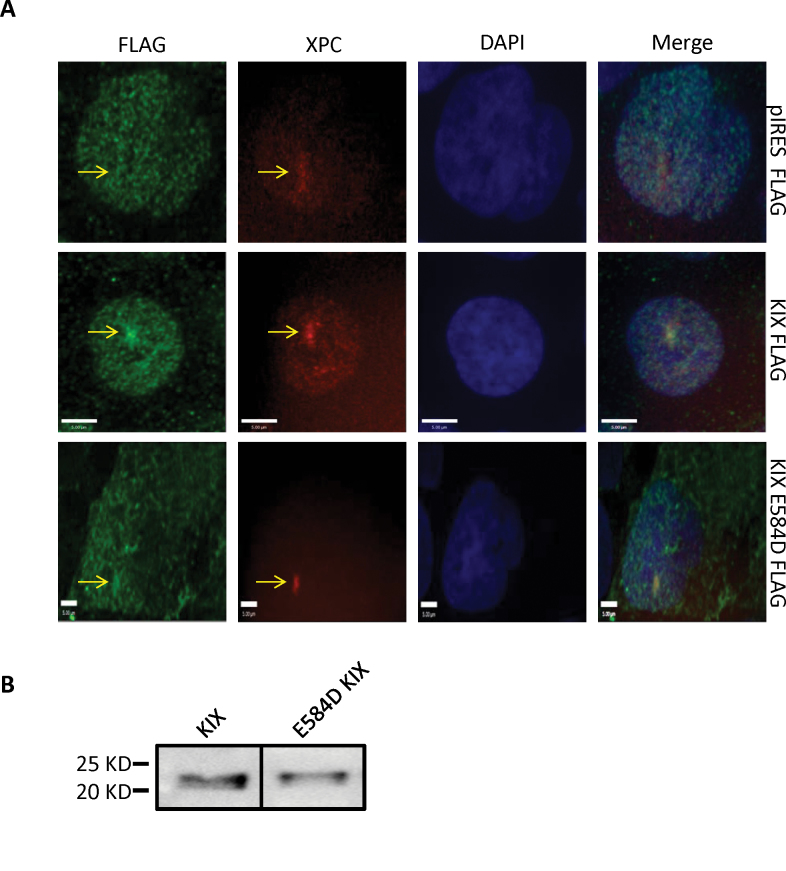
In order to identify the domain responsible for the recruitment of RECQL5 to the sites of ICLs, we generated deletion fragments of RECQL5 with a GFP tag at the C terminus (Figure 3A). The expression of the GFP-RECQL5 fragments was confirmed by western blot using cells expressing each of these deletion fragments (Figure 3B). Analysis of the recruitment of the deletion mutants indicated that the full-length RECQL5 (1–991 aa) and the fragment corresponding to the region 501–650 aa (containing the KIX domain) were recruited to the ICLs (Figure 3C) (24). The other fragments (1–240, 1–500 and 853–991) were not recruited to the ICL sites (Figure 3C).
Fig. 3.
Interaction of RECQL5 deletion mutants with ICLs. (A) Schematic illustration of the wild-type and deletion mutants of RECQL5. The helicase, RecQ C-terminal (RQC), KIX and SRI domains are shown along with the nuclear localization signal (NLS) in endogenous RECQL5. The ribbon diagrams below show which GFP-fusion constructs recruit to ICLs. (B) Western blot showing the cellular expression of the various deletion mutants. (C) U2OS cells were transfected with 2 µg of indicated plasmid; 24h posttransfection, the cells were incubated with 6 µM trioxalen for 20 min and then laser photoactivated in the nuclear region in individual cells in a defined time sequence. The targeted regions are indicated by arrows. Bar, 5 µm.
Inhibition of transcription does not perturb recruitment of RECQL5 to ICLs
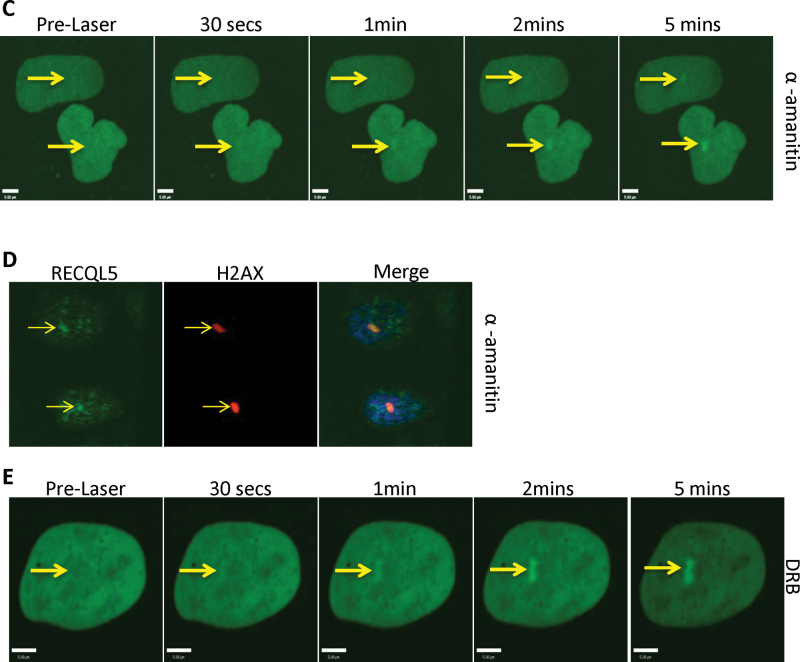
RECQL5 fragment 501–650 shares amino acid sequence homology with the KIX domain, often found in transcription factors such as CREB-binding protein (24). Interestingly, unlike the full-length RECQL5, which interacts with both the promoter-bound RNA polymerase II (RNAP IIa) and the elongating form (RNAP IIo), the KIX domain interacts only with RNAP Iia (35–37). This suggested that the recruitment of RECQL5 to the ICL sites might be independent of active transcription. To address this hypothesis, we carried out recruitment experiments using a point mutant of the KIX domain (E584D), which was shown previously to disrupt its binding to RNA polymerase (24). We observed localization of both the KIX domain and the E584D mutant at the sites of ICLs, suggesting that the recruitment to ICL was not dependent on the interaction with RNA polymerase II (Figure 4A). The KIX domain and its mutant form were expressed stably (Figure 4B). Furthermore, we tested the recruitment of RECQL5 to ICLs using two different agents known to arrest transcription: α-amanitin and DRB (38,39). Even with the high concentrations of α-amanitin (50 μg/ml, Figure 4C, to 200 µg/ml, data not shown), GFP-RECQL5 recruitment to the ICLs was still observed in live cells. This was also the case when we generated ICLs in the presence of α-amanitin and then fixed the cells and stained for endogenous RECQL5 (Figure 4D). Similarly, RECQL5 recruitment to the ICLs was also observed after DRB treatment (Figure 4E). As both α-amanitin and DRB are potent inhibitors of active transcription which require the presence of RNAP IIo, we inferred that active transcription was dispensable for recruitment of RECQL5 to sites of ICLs.
Fig. 4.
Inhibition of transcription does not perturb recruitment of RECQL5 to ICLs. (A) U2OS cells were transfected with the indicated plasmids. After 24h, the cells were treated with 6 µM trioxalen for 20 min, then laser photoactivated in the nucleus in individual cells, fixed after 5 min and immunostained for FLAG (green) to detect the KIX-Flag protein and endogenous XPC (red) to indicate a damage signal. The targeted regions are indicated by the arrows. (B) Western blot showing the cellular expression of the KIX domain and the E584D KIX constructs. (C) U2OS cells were transfected with 2 µg GFP-RECQL5. Twenty-four hours posttransfection, the cells were incubated with α-amanitin at 50 µg/ml for 2 h and with 6 µM trioxalen for 20 min, then laser photoactivated in the nuclear region in individual cells in a defined time sequence. (D) U2OS cells were incubated with α-amanitin at 50 µg/ml for 2 h and with 6 µM trioxalen for 20 min, then laser photoactivated in the nucleus in individual cells, fixed after 5 min and immunostained for endogenous RECQL5 (green) and for the γ-H2AX (red) for damage signal. (E) U2OS cells were transfected with 2 µg GFP-RECQL5. Twenty-four hours posttransfection, the cells were incubated with DRB at 80 µg/ml for 2 h and with 6 µM trioxalen for 20 min, then laser photoactivated in the nuclear region in individual cells in a defined time sequence. The targeted regions are indicated by arrows. Bar, 5 µm.
Topoisomerase poisons do not inhibit recruitment of RECQL5 to ICLs
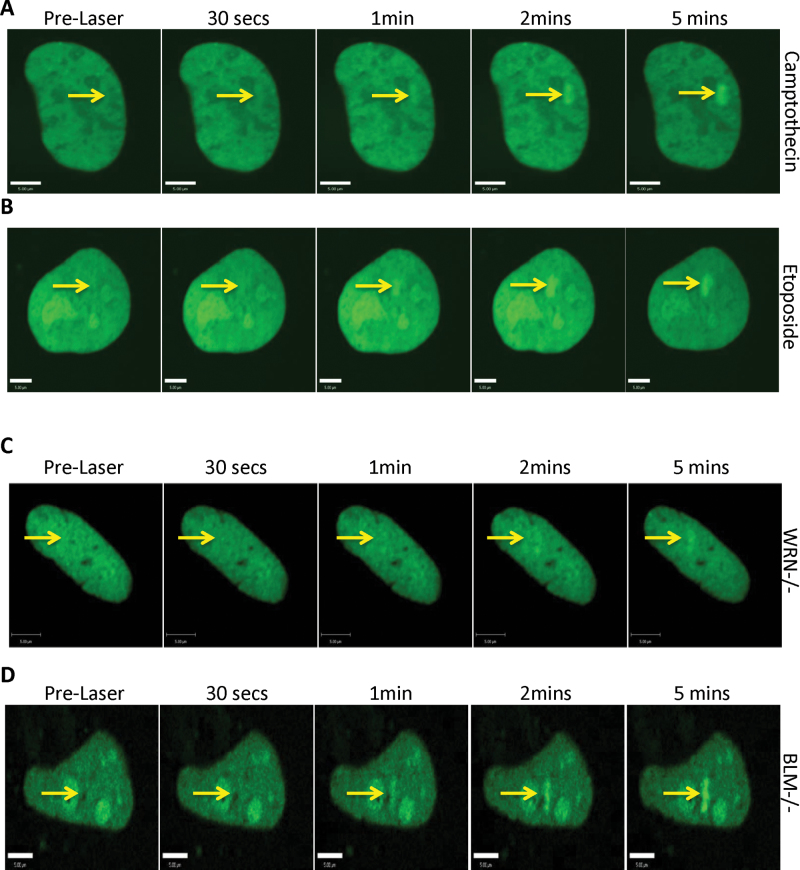
RecQ helicases cooperate physically and functionally with type I and type II topoisomerases (21,40–42). The KIX domain of RECQL5 was reported to be required for cellular resistance to camptothecin, a topoisomerase I inhibitor (24). As the KIX domain was recruited to the sites of ICLs (Figure 3), we explored if treatment with topoisomerase inhibitors, camptothecin or etoposide (topoisomerase II inhibitor) would disrupt the recruitment of RECQL5 to the ICL sites. Our results indicated that neither inhibition of topoisomerase I nor inhibition of topoisomerase II prevented RECQL5 from recruiting to the sites of the ICLs (Figure 5A and B).
Fig. 5.
Topoisomerase poisons and other RecQ helicases do not influence recruitment of RECQL5 to the site of ICLs. U2OS cells were transfected with 2 µg GFP-RECQL5. Twenty-four hours posttransfection, the cells were incubated with either 500 nM camptothecin (A) or 6 µM etoposide (B) for 2 h and with 6 µM trioxalen for 20 min, then laser photoactivated in the nuclear region in individual cells in a defined time sequence. (C) WRN patient fibroblasts (AG11395) and (D) BLM patient fibroblasts (GM08505) were incubated with 6 µM trioxalen for 20 min and then laser photoactivated in the nuclear region in individual cells in a defined time sequence. The targeted regions are indicated by arrows. Bar, 5 µm.
Effect of other related DNA repair proteins on RECQL5’s recruitment to ICLs
In order to further explore the requirements for RECQL5’s recruitment to ICLs, we expressed GFP-RECQL5 in different RecQ helicase and DNA repair deficient backgrounds. First, we investigated if recruitment of RECQL5 required the expression of the related RecQ family members, WRN or BLM. This was done by expressing GFP-RECQL5 in fibroblasts derived from either Werner syndrome patients (Figure 5C) or Bloom syndrome patients (Figure 5D) and generating ICLs as before. We observed recruitment of RECQL5 in both mutant backgrounds, suggesting that recruitment of RECQL5 was independent of these two proteins. Further experiments revealed that the recruitment of RECQL5 was independent of XPC, Cockayne syndrome group B protein, Fanconi anemia group D2 protein, ATM, ATR, DNAPKcs and γ-H2AX (Supplementary Figure 4A–G, available at Carcinogenesis Online) (43–52). Thus, GFP-RECQL5 is recruited early and independently of several other DNA damage and response proteins.
Effect of DNA-intercalating agents on binding of RECQL5 to ICLs
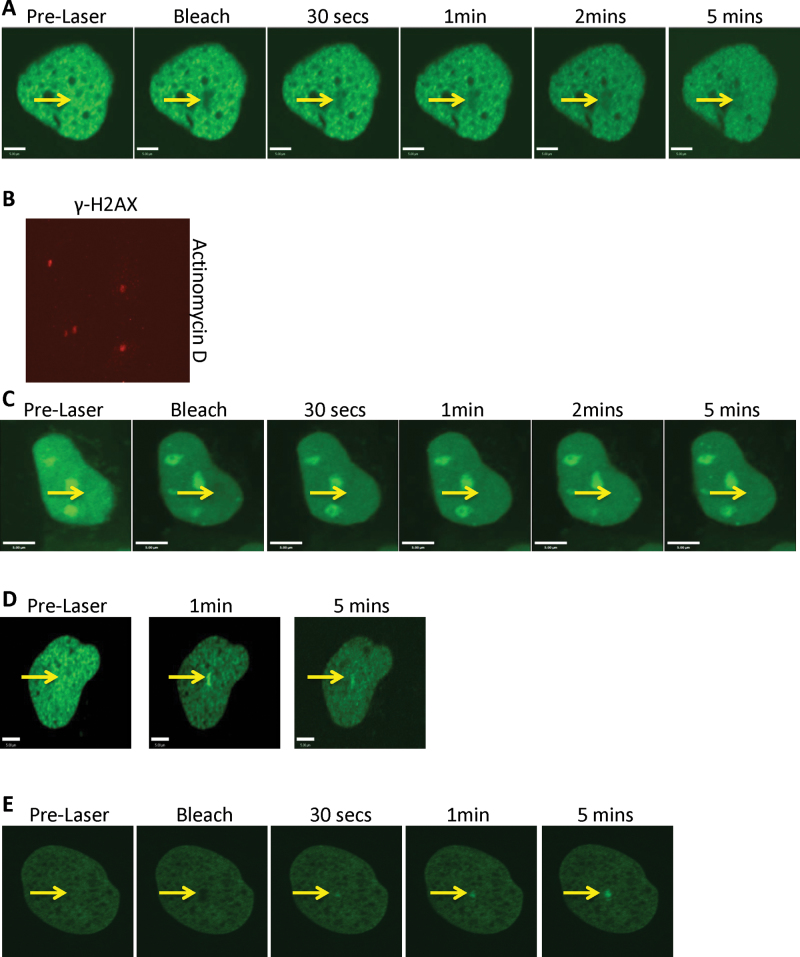
Although testing RECQL5’s recruitment in the presence of transcription inhibitors, we noticed a failure to recruit in the presence of actinomycin-D (Act-D), an intercalating agent that also inhibits transcription (Figure 6A) (53). To confirm the introduction of ICLs in the targeted cells, we fixed the cells after the laser irradiation and stained for γ-H2AX. The positive staining of γ-H2AX indicated that ICLs were formed in the presence of Act-D (Figure 6B). In order to test if the failure of RECQL5’s recruitment to the ICLs was due to the intercalation of DNA by Act-D, we performed a similar experiment using another intercalating agent, EtBr (54). As we observed with Act-D treatment, there was no recruitment of RECQL5 in the presence of EtBr to the PUVA-induced ICLs (Figure 6C). In stark contrast, we observed localization of XPC after Act-D (Figure 6D) and EtBr (Figure 6E) treatment. Taken together, we infer that the DNA-intercalating agents prevented recruitment of RECQL5 to the ICL sites.
Fig. 6.
Intercalating agents affect the recruitment of RECQL5 to the sites of ICLs. U2OS cells were transfected with either 2 µg GFP–RECQL5 (A) or 2 µg GFP-XPC (D). Twenty-four hours posttransfection, the cells were incubated with actinomycin-D at 0.05 µg/ml for 2 h and with 6 µM trioxalen for 20 min, then laser photoactivated in the nuclear region in individual cells in a defined time sequence. (B) U2OS cells were incubated with actinomycin-D at 0.05 µg/ml for 2 h and with 6 µM trioxalen for 20 min, then laser photoactivated in the nucleus in individual cells, fixed after 5 min and immunostained for γ-H2AX (red) to indicate a damage signal. The targeted regions are indicated by arrows. U2OS cells were transfected with either 2 µg GFP-RECQL5 (C) or 2 µg GFP-XPC (E). Twenty-four hours posttransfection, the cells were incubated with EtBr at 5 µg/ml for 2 h and with 6 µM trioxalen for 20 min, then laser photoactivated in the nuclear region in individual cells in a defined time sequence. The targeted regions are indicated by arrows. Bar, 5 µm.
Discussion
Recql5 − /− mice show elevated incidences of cancer (nearly 52% of all animals) and therefore the protein is thought to play a critical role in the maintenance of genomic stability (22). Direct evidence of its role in double-strand break repair comes from laser microscopy studies where RECQL5 was shown to interact with the MRE11 complex (55). Furthermore, we have recently demonstrated its role in single-strand break repair (56). In this study, we report the participation of RECQL5 in ICL repair. It is not surprising to observe DNA repair proteins localize to variety of lesions and play important roles in the resolution of DNA damage. Indeed, among the RecQ family of helicases, WRN and BLM have been reported to function in multiple repair pathways, including DNA double-strand break repair, ICL repair and base excision repair (11,16,57). However, we also report that other RecQ family members, RECQL1 and RECQL4, do not recruit to the sites of ICLs by our methods. This could illustrate a degree of specificity among the RecQ helicases. Further studies could shed light on this and bring about the ‘division of labor’ concept that probably explains why mammals have five RecQ helicase members, whereas lower eukaryotes only have one (57).
Recruitment of RECQL5 to the sites of PUVA-induced ICLs was relatively early. Also, as evidenced by the comet assays, depletion of RECQL5 significantly affects the critical unhooking step in the repair of ICLs. From our studies using various mutant cell lines, it appears that RECQL5 acts independently of other proteins known to recruit early to ICLs, including members of NER, the Fanconi anemia pathway, DNA damage signaling proteins and DNA damage markers (Supplementary Figure 4, available at Carcinogenesis Online). Additionally, the recruitment of RECQL5 was independent of the WRN and BLM status in cells, suggesting an independent recruitment requirement from its other family members (Figure 5C and D). Together, these data suggest a possible role for RECQL5 in the early steps of ICL repair.
Interestingly, the ‘KIX’ domain of RECQL5 was recruited to the sites of ICLs. The KIX domain has been shown to interact with RNAP II and provide resistance to camptothecin treatment. Mutation of the hydrogen-bound network of RECQL5 KIX (E584D) abolished the interaction of the KIX domain with RNAP II (24). We used this mutant in our recruitment study and found that it localized to the ICL similarly to the intact KIX domain, suggesting that RNAP II interaction was not required for RECQL5 localization to ICLs. Also, neither inhibition of RNAP II mediated transcription nor treatment with camptothecin inhibited the recruitment of RECQL5. Li et al. reported that the previously observed interaction between the KIX domain of RECQL5 and active RNAP II was observed because of the high concentration of Triton used in the immunoprecipitation (35). The recruitment of the full length and the KIX domain fragment of RECQL5 to the ICLs implies binding to or around the lesion generated. Consistent with our interpretation, we did not observe accumulation of RNAP II to the sites of damage (data not shown).
It should be noted that we observed extranucleolar localization of GFP-RECQL5 in all the cells treated with Act-D (Figure 6A). A similar localization pattern was also observed when the cells were treated with camptothecin (Figure 5A). Therefore, we inferred that the subcellular localization did not influence the recruitment of RECQL5 to the sites of ICLs. This is further supported by the results that RECQL5’s recruitment to the ICL sites is inhibited in the presence of either Act-D or EtBr even though the subcellular localization of RECQL5 in the presence of Act-D is different from that in the presence of EtBr (Figure 6A and C).
The structure of DNA is important in the recognition of damage, for example, the minor groove enlargement is thought to be a signal for XPC recruitment in NER (58). Further, structural studies showed that although carcinogen–base stacking interactions contributed to the local DNA stability, they could prevent the successful insertion of an XPC β-hairpin into the duplex and the normal recruitment of other downstream NER factors (59).
Similarly, the structural integrity of the DNA might correlate with RECQL5 recruitment to sites of ICLs and further retention. In other words, the drastic structural changes in DNA caused by the introduction of the lesions may directly influence RECQL5’s ability to recognize the damaged DNA site or influence the DNA-binding proteins, which recruit RECQL5 to the damage sites, and thereby affect indirectly the recruitment of RECQL5. This argues for the direct recruitment of RECQL5 to the damaged DNA or binding to a DNA-binding protein that is influenced by the DNA structure. This argument is strengthened by the results that RECQL5 did not recruit in the presence of intercalating agents which alters the local structure of the DNA (Figure 6). Biochemical studies suggest that RECQL5 may bind DNA in a structure-specific manner not in a sequence-specific manner (30,60). Furthermore, the binding of other RecQ helicases to DNA is thought to be through the sugar-phosphate backbone and the RQC region as shown in the WRN crystal structure in complex with DNA (61). However, it should be noted that RECQL5 lacks the winged helix domain in the RQC region but retains the Zn2+-binding motif (30). Interestingly, we note the recruitment of XPC to ICL’s even in the presence of Act-D or EtBr (Figure 6D and E). This would suggest that RECQL5 is more sensitive to intercalators compared with XPC.
In conclusion, our results expand the importance of the RecQ family of helicases in the ICL repair pathway and add to their repertoire as the ‘guardians of the genome’ (57). The multiple roles played by this family of proteins in DNA repair processes are the reason why they are considered potent tumor suppressors (57). As mentioned before, this is clearly evident from knockout mouse studies in the case of RECQL5 (22). At this time, we do not know specifically how RECQL5 participates in ICL repair, but we have clearly identified a role for RECQL5 here and report a novel division of labor among the human RecQ helicases. Further studies will be required to delineate the exact roles played by each of the RecQ helicases in ICL repair and understand where they cooperate to bring about specific DNA repair functions.
Supplementary material
Supplementary Figures 1–4 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health Intramural Program of the National Institute on Aging (Z01-AG000726-17).
Supplementary Material
Acknowledgements
We thank Dr D.K.Singh and Dr E.Fang for critically reading this article. M.R., A.M. and V.A.B. conceived and designed the study; M.R., A.M., T.T. and V.P. performed the experiments and analyzed the data; M.R., M.M.S., D.C. and V.B. interpreted the data; M.R. wrote the article with comments from D.L.C., M.S. and V.A.B. We would also like to thank Dr W.Wang for sharing the RECQL5 KIX domain mutant plasmids.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- Act-D
actinomycin-D
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3
- BLM
Bloom syndrome protein
- DRB
5,6-dichloro-β-d-ribofuranosylbenzimidazole
- EtBr
ethidium bromide
- GFP
green fluorescent protein
- ICL
interstrand cross-links
- PBS
phosphate-buffered saline
- PUVA
psoralen plus UVA
- shRNA
short hairpin RNA
- UV
ultraviolet
- WRN
Werner syndrome protein
- XPC
xeroderma pigmentosum complementation.
References
- 1. Deans A.J., et al. (2011). DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer, 11, 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cimino G.D., et al. (1985). Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu. Rev. Biochem., 54, 1151–1193 [DOI] [PubMed] [Google Scholar]
- 3. Akkari Y.M., et al. (2000). DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell. Biol., 20, 8283–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Song P.S., et al. (1979). Photochemistry and photobiology of psoralens. Photochem. Photobiol., 29, 1177–1197 [DOI] [PubMed] [Google Scholar]
- 5. Parrish J.A., et al. (1974). Photochemotherapy of psoriasis with oral methoxsalen and longwave ultraviolet light. N. Engl. J. Med., 291, 1207–1211 [DOI] [PubMed] [Google Scholar]
- 6. Gasparro F.P. (2000). The role of PUVA in the treatment of psoriasis. Photobiology issues related to skin cancer incidence. Am. J. Clin. Dermatol., 1, 337–348 [DOI] [PubMed] [Google Scholar]
- 7. Stern R.S., et al. (1997). Noncutaneous malignant tumors in the PUVA follow-up study: 1975-1996. J. Invest. Dermatol., 108, 897–900 [DOI] [PubMed] [Google Scholar]
- 8. Stern R.S., et al. (1997). Malignant melanoma in patients treated for psoriasis with methoxsalen (psoralen) and ultraviolet A radiation (PUVA). The PUVA Follow-Up Study. N. Engl. J. Med., 336, 1041–1045 [DOI] [PubMed] [Google Scholar]
- 9. Stern R.S., et al. (1998). Oral psoralen and ultraviolet-A light (PUVA) treatment of psoriasis and persistent risk of nonmelanoma skin cancer. PUVA Follow-up Study. J. Natl Cancer Inst., 90, 1278–1284 [DOI] [PubMed] [Google Scholar]
- 10. Muniandy P.A., et al. (2010). DNA interstrand crosslink repair in mammalian cells: step by step. Crit. Rev. Biochem. Mol. Biol., 45, 23–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bohr V.A. (2008). Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci., 33, 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bachrati C.Z., et al. (2008). RecQ helicases: guardian angels of the DNA replication fork. Chromosoma, 117, 219–233 [DOI] [PubMed] [Google Scholar]
- 13. Poot M., et al. (2001). Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J., 15, 1224–1226 [DOI] [PubMed] [Google Scholar]
- 14. Poot M., et al. (2002). Werner syndrome diploid fibroblasts are sensitive to 4-nitroquinoline-N-oxide and 8-methoxypsoralen: implications for the disease phenotype. FASEB J., 16, 757–758 [DOI] [PubMed] [Google Scholar]
- 15. Hook G.J., et al. (1984). Sensitivity of Bloom syndrome fibroblasts to mitomycin C. Mutat. Res., 131, 223–230 [DOI] [PubMed] [Google Scholar]
- 16. Brosh R.M., Jr, et al. (2007). Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res., 35, 7527–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng W.H., et al. (2006). Collaboration of Werner syndrome protein and BRCA1 in cellular responses to DNA interstrand cross-links. Nucleic Acids Res., 34, 2751–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Otterlei M., et al. (2006). Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J. Cell Sci., 119,(Pt 24)5137–5146 [DOI] [PubMed] [Google Scholar]
- 19. Zhang N., et al. (2005). The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J. Biol. Chem., 280, 40559–40567 [DOI] [PubMed] [Google Scholar]
- 20. Meetei A.R., et al. (2003). A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol., 23, 3417–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramamoorthy M., et al. (2012). RECQL5 cooperates with Topoisomerase II alpha in DNA decatenation and cell cycle progression. Nucleic Acids Res., 40, 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu Y., et al. (2007). RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev., 21, 3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muniandy P.A., et al. (2009). Repair of laser-localized DNA interstrand cross-links in G1 phase mammalian cells. J. Biol. Chem., 284, 27908–27917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Islam M.N., et al. (2010). RecQL5 promotes genome stabilization through two parallel mechanisms–interacting with RNA polymerase II and acting as a helicase. Mol. Cell. Biol., 30, 2460–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Selzer R.R., et al. (2002). Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells. Nucleic Acids Res., 30, 782–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sarbassov D.D., et al. (2005). Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science., 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 27. Stewart S.A., et al. (2003). Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA, 9, 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Opresko P.L., et al. (2002). Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem., 277, 41110–41119 [DOI] [PubMed] [Google Scholar]
- 29. Rossi M.L., et al. (2010). Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair (Amst)., 9, 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garcia P.L., et al. (2004). Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J., 23, 2882–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Popuri V, et al. (2012). Recruitment and retention dynamics of RECQL5 at DNA double strand break sites. DNA Repair (Amst.), 11, 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thazhathveetil A.K., et al. (2007). Psoralen conjugates for visualization of genomic interstrand cross-links localized by laser photoactivation. Bioconjug. Chem., 18, 431–437 [DOI] [PubMed] [Google Scholar]
- 33. Singh D.K., et al. (2010). The involvement of human RECQL4 in DNA double-strand break repair. Aging Cell, 9, 358–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rothfuss A., et al. (2004). Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol. Cell. Biol., 24, 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li M., et al. (2011). The SET2-RPB1 interaction domain of human RECQ5 is important for transcription-associated genome stability. Mol. Cell. Biol., 31, 2090–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aygün O., et al. (2008). A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl Acad. Sci. USA., 105, 8580–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kanagaraj R., et al. (2010). RECQ5 helicase associates with the C-terminal repeat domain of RNA polymerase II during productive elongation phase of transcription. Nucleic Acids Res., 38, 8131–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Duensing A., et al. (2007). RNA polymerase II transcription is required for human papillomavirus type 16 E7- and hydroxyurea-induced centriole overduplication. Oncogene, 26, 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tamm I. (1983). Recovery of HeLa cell population growth after treatment with 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB). J. Cell. Physiol., 116, 26–34 [DOI] [PubMed] [Google Scholar]
- 40. Laine J.P., et al. (2003). Werner protein stimulates topoisomerase I DNA relaxation activity. Cancer Res., 63, 7136–7146 [PubMed] [Google Scholar]
- 41. Russell B., et al. (2011). Chromosome breakage is regulated by the interaction of the BLM helicase and topoisomerase IIalpha. Cancer Res., 71, 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu L., et al. (2000). The Bloom’s syndrome gene product interacts with topoisomerase III. J. Biol. Chem., 275, 9636–9644 [DOI] [PubMed] [Google Scholar]
- 43. Sugasawa K., et al. (1998). Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell., 2, 223–232 [DOI] [PubMed] [Google Scholar]
- 44. van H.A., et al. (1993). Deficient repair of the transcribed strand of active genes in Cockayne’s syndrome cells. Nucleic Acids Res., 21, 5890–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Venema J., et al. (1990). The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc. Natl Acad. Sci. USA, 87, 4707–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang W. (2007). Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet., 8, 735–748 [DOI] [PubMed] [Google Scholar]
- 47. Shiloh Y. (2003). ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer, 3, 155–168 [DOI] [PubMed] [Google Scholar]
- 48. Abraham R.T. (2001). Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev., 15, 2177–2196 [DOI] [PubMed] [Google Scholar]
- 49. Yang J., et al. (2003). ATM, ATR and DNA-PK: initiators of the cellular genotoxic stress responses. Carcinogenesis, 24, 1571–1580 [DOI] [PubMed] [Google Scholar]
- 50. Yang J., et al. (2004). ATM and ATR: sensing DNA damage. World J. Gastroenterol., 10, 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paull T.T., et al. (2000). A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol., 10, 886–895 [DOI] [PubMed] [Google Scholar]
- 52. Rogakou E.P., et al. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem., 273, 5858–5868 [DOI] [PubMed] [Google Scholar]
- 53. Jao C.Y., et al. (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl Acad. Sci. USA., 105, 15779–15784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu L.F., et al. (1975). On the degree of unwinding of the DNA helix by ethidium. II. Studies by electron microscopy. Biochim. Biophys. Acta, 395, 401–412 [PubMed] [Google Scholar]
- 55. Zheng L., et al. (2009). MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res., 37, 2645–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tadokoro T., et al. (2012). Human RECQL5 participates in the removal of endogenous DNA damage. Mol. Biol. Cell, 23, 4273–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hickson I.D. (2003). RecQ helicases: caretakers of the genome. Nat. Rev. Cancer, 3, 169–178 [DOI] [PubMed] [Google Scholar]
- 58. Cai Y., et al. (2010). Distant neighbor base sequence context effects in human nucleotide excision repair of a benzo[a]pyrene-derived DNA lesion. J. Mol. Biol., 399, 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reeves D.A., et al. (2011). Resistance of bulky DNA lesions to nucleotide excision repair can result from extensive aromatic lesion-base stacking interactions. Nucleic Acids Res., 39, 8752–8764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kanagaraj R., et al. (2006). Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res., 34, 5217–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kitano K., et al. (2010). Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure, 18, 177–187 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.