Abstract
Aims: The role of thioredoxin reductase (TrxR) in tumorigenesis has made it an attractive anticancer target. A systematic approach for development of novel compounds as TrxR inhibitors is currently lacking. Structurally diversified TrxR inhibitors share in common electrophilic propensities for the sulfhydryl groups, among which include the Michael reaction acceptors containing an α,β-unsaturated carbonyl moiety. We aimed to identify features among structurally diversified Michael acceptor-based compounds that would yield a strong TrxR inhibitory character. Results: Structurally dissimilar Michael acceptor-based natural compounds such as isobutylamides, zerumbone, and shogaols (SGs) were found to possess a poor TrxR inhibitory activity, indicating that a sole Michael acceptor moiety was insufficient to produce TrxR inhibition. The 1,7-diphenyl-hept-3-en-5-one pharmacophore in 3-phenyl-3-SG, a novel SG analog that possessed comparable TrxR inhibitory and antiproliferative potencies as 6-SG, was modified to yield 1,5-diphenyl-pent-1-en-3-one (DPPen) and 1,3-diphenyl-pro-1-en-3-one (DPPro, also known as chalcone) pharmacophores. These Michael acceptor-centric pharmacophores, when substituted with the hydroxyl and fluorine groups, gave rise to analogs displaying a TrxR inhibitory character positively correlated to their antiproliferative potencies. Lead analogs 2,2′-diOH-5,5′-diF-DPPen and 2-OH-5-F-DPPro yielded a half-maximal TrxR inhibitory concentration of 9.1 and 10.5 μM, respectively, after 1-h incubation with recombinant rat TrxR, with the C-terminal selenocysteine residue found to be targeted. Innovation: Identification of Michael acceptor-centric pharmacophores among diversified compounds demonstrates that a systematic approach to discover and develop Michael acceptor-based TrxR inhibitors is feasible. Conclusion: A strong TrxR inhibitory character correlated to the antiproliferative potency is attributed to structural features that include an α,β-unsaturated carbonyl moiety centered in a DPPen or DPPro pharmacophore bearing hydroxyl and fluorine substitutions. Antioxid. Redox Signal. 19, 1149–1165.
Introduction
Redox homeostasis is maintained by cellular antioxidant systems such as the thioredoxin (Trx) and glutathione (GSH) system. These thiol systems act as double-edged swords; they protect cells from oxidative stress, but in cancer, they contribute to tumor growth and progression. Perturbation of the redox state-sustaining systems in malignant cells can thereby form an important basis for chemotherapeutic strategies.
The Trx system consists of Trx, thioredoxin reductase (TrxR), and NADPH. TrxR catalyzes the NADPH-dependent reduction of a dithiol/disulfide active site (in human Trx: -Trp-Cys32-Gly-Pro-Cys35-Lys-) in oxidized Trx. Through the thiol–disulfide exchange, Trx reduces protein disulfides, which leads to protection of cellular proteins against an oxidative insult. As an electron donor to ribonucleotide reductase and peroxide scavengers peroxiredoxins, Trx also participates in DNA biosynthesis/repair and redox homeostasis, respectively. The redox protein is broadly involved in cell signaling by exerting redox control over the activity of several proteins, including activator protein-1, nuclear factor κB, and apoptosis signal-regulating kinase 1 (ASK1) (1, 19, 40). Homodimeric mammalian TrxR has each of the 55-kDa subunit containing an N-terminal–Cys59-Val-Asn-Val-Gly-Cys64- dithiol/disulfide active site and a C-terminal–Gly-Cys497-Sec498-Gly- active site. The C-terminal selenocysteine (Sec)-containing active site has a range of substrates, including Trx, 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) and protein disulfide isomerase (19, 33). In recent years, discovery and development of anticancer agents targeting the Trx system are gaining momentum, partly attributed to the evidence demonstrating that Trx and TrxR are overexpressed in aggressive tumors and such overexpression are related to cancer drug resistance and poor patient prognosis (5, 23, 24, 37, 43). Further, in mouse xenografts, TrxR is found to be essential for tumorigenesis (51), and in cancer cells, knockdown of TrxR retards self-sufficient growth and DNA replication (52). Indeed, mammalian TrxR is an attractive anticancer target of many electrophilic compounds due to its two unique structural properties (30, 45). First, being located at the flexible C-terminal tail, the Sec-containing active site is highly accessible for attack by inhibitors. Secondly, the low pKa of the free selenol group (-SeH) in Sec enables ionization to nucleophilic selenolate (Se−) at physiological pH. A number of chemotherapeutic agents used clinically or currently in clinical trials such as nitrosoureas (49), cyclophosphamide (47), gold complexes such as auranofin (8, 18), platinum complexes such as cisplatin (4), arsenic trioxide (29), and motexafin gadolinium (20) have been recognized to inhibit TrxR.
Innovation.
The structural diversity of Michael acceptor-based thioredoxin reductase (TrxR) inhibitors hinders carrying out of large-scale structure–activity relationship studies. Those possessing a weak TrxR inhibitory activity are also little known. In this present study, initial screening of a number of Michael acceptor-based natural compounds had not only demonstrated that a sole Michael acceptor moiety was insufficient to produce TrxR inhibition, but also led to the synthesis of compounds bearing 1,5-diphenyl-pent-1-en-3-one and 1,3-diphenyl-pro-1-en-3-one pharmacophores that displayed well-correlated TrxR inhibitory and antiproliferative activities. The findings have suggested the feasibility of a systematic approach to discover and develop Michael acceptor-based TrxR inhibitors.
Currently, there is no documentation of a systematic approach that allows a rational design and development of compounds as selective TrxR inhibitors. Structurally diversified small molecules acting on TrxR share in common electrophilic propensities for sulfhydryl groups, among which include metal complexes with reactive valencies and Michael reaction acceptors containing an α,β-unsaturated carbonyl moiety (7, 30, 45). Our laboratory has interest in discovering the Michael acceptor-based TrxR inhibitors. In this study, we report that a strong TrxR inhibitory character is attributed to structural features that include an α,β-unsaturated carbonyl moiety centered in a 1,5-diphenyl-pent-1-en-3-one (DPPen) or 1,3-diphenyl-pro-1-en-3-one (DPPro; also known as chalcone) pharmacophore bearing hydroxyl- and fluorine substitutions. Of note, the good correlation between the TrxR inhibitory and antiproliferative potencies among the DPPen and DPPro compounds suggests TrxR inhibition as the underlying mechanism of their antitumor effects. Taken together, the findings in this study have offered opportunities not only for possibly a more predictive identification and characterization of novel compounds as potential TrxR inhibitors, but also to be potentially applied for future design and development of TrxR inhibitors.
Results
Evaluation of naturally occurring α,β-unsaturated amides and unconjugated α,β-unsaturated ketones for in vitro TrxR inhibitory and antiproliferative activities
In an attempt to identify the structural features in the Michael acceptors that were required for TrxR inhibition, a number of structurally diversified natural products that bear a common α,β-unsaturated carbonyl moiety were selected for evaluation in the DTNB reduction assay. These compounds included α,β-unsaturated amides such as hexa-2,4-dienoic acid isobutylamide (Hexa-IBA) and dodeca-2(E),4(E)-dienoic acid isobutylamide (Dodeca-IBA) that are found in the species of Echinacea, as well as unconjugated α,β-unsaturated ketones such as zerumbone isolated from rhizomes of Zingiber zerumbet Smith and shogaols (SGs), active constituents isolated from rhizomes of Zingiber officinale Roscoe. As of yet, no studies have reported the possible effects of these compounds on the TrxR activity. The IBAs and zerumbone had failed to inhibit the TrxR activity even at a high concentration of 100 μM. Among the SGs, 4-SG and 6-SG containing shorter carbon-chain lengths displayed weak TrxR inhibition with a respective IC50 value of 64.5 and 88.0 μM at 1-h incubation, whereas 8-SG and 10-SG with longer chain lengths exhibited negligible TrxR inhibition (Table 1). We further synthesized a novel SG analog 3-phenyl-3-SG (3-Ph-3-SG; structure as shown in Table 1) and found that it possessed a comparable TrxR inhibitory activity as 6-SG (IC50 at 1-h incubation: 87.1 μM).
Table 1.
Mammalian TrxR Inhibitory and Antiproliferative Activities of Classes of Compounds Containing an α,β-Unsaturated Carbonyl Moiety
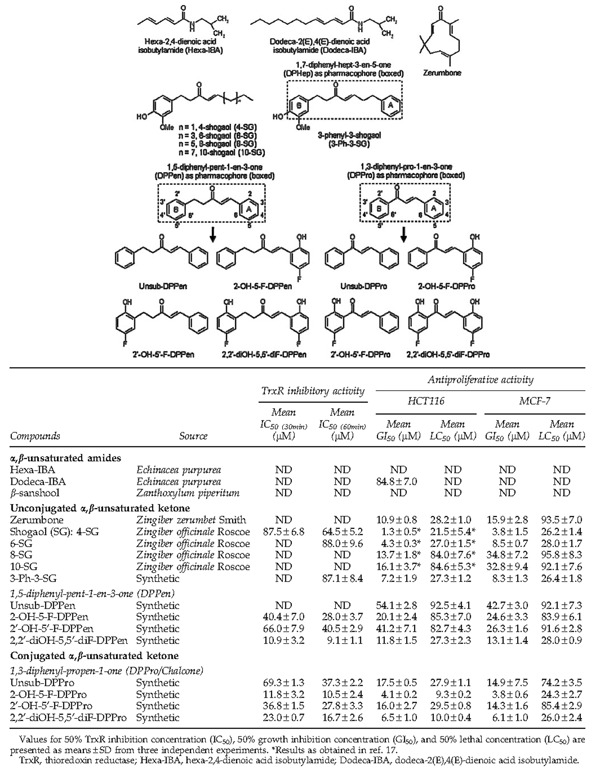 |
The effects of these compounds on the HCT 116 and MCF-7 cell viabilities were evaluated. The IBAs were found to possess negligible antiproliferative activities that were correlated to their poor TrxR inhibitory activities. On the other hand, in agreement with the studies reporting its antitumor effects (32, 35), zerumbone had displayed potent cytotoxic effects. For the series of SGs with increasing side-chain lengths, we had observed previously for HCT 116 cells (17) and in this present study for MCF-7 cells that the antiproliferative efficacies decreased as the chain length increased. This trend of diminishing antiproliferative activities was similar to the observed trend of decreasing TrxR inhibition. Similar to its TrxR inhibitory activity, the synthetic SG analog 3-Ph-3-SG possessed comparable antiproliferative potencies as 6-SG. Taken together, zerumbone and SGs had exhibited potent antiproliferative effects, but a lack of or weak TrxR inhibition, suggesting that TrxR targeting is unlikely the mechanism underlying their antitumor action.
Evaluation of analogs bearing DPPen or DPPro pharmacophore for in vitro TrxR inhibitory and antiproliferative activities
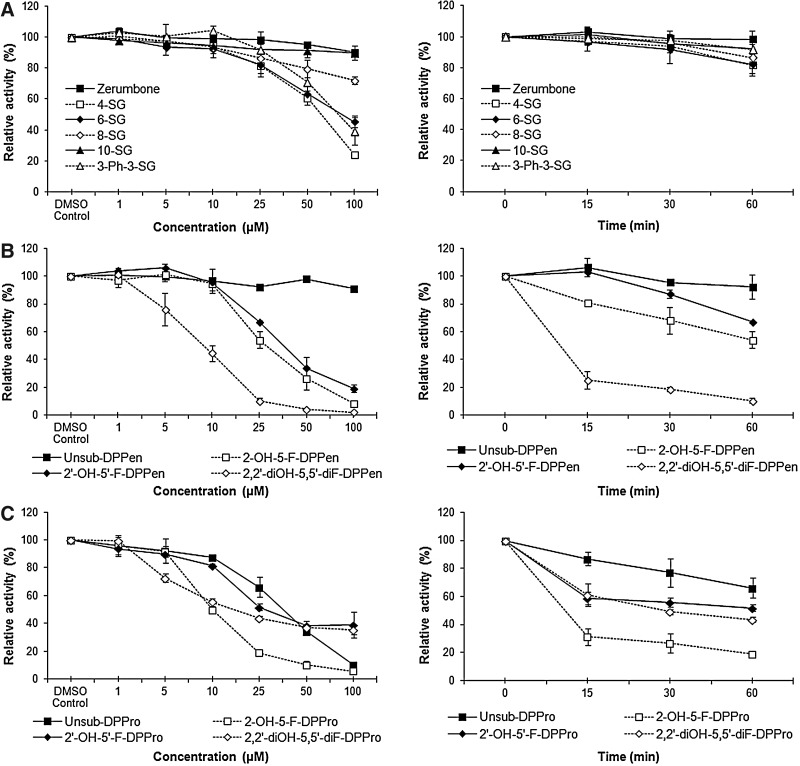
We wanted to synthesize the structural derivatives of either 6-SG or 3-Ph-3-SG that possessed comparable and weak TrxR inhibitory activities. 3-Ph-3-SG was selected as the chemical scaffold based on which derivatives were synthesized, because the two phenyl rings allowed feasible structural modifications via chemical synthesis. To investigate whether the distance between the two phenyl rings in 3-Ph-3-SG would affect TrxR inhibition, compounds containing shortened phenyl–phenyl distances, namely, those bearing a DPPen or DPPro pharmacophore, were synthesized. Analogs bearing the 2-hydroxy- and 5-fluorine substitutions on the phenyl ring were also synthesized (structures as shown in Table 1), and the TrxR inhibitory activities were evaluated by the in vitro DTNB reduction assay. For the series of DPPen-based compounds, the unsubstituted analog (unsub-DPPen) lacked the TrxR inhibitory activity. In contrast, analogs containing the hydroxyl and fluorine substituents on the phenyl rings displayed an elevated TrxR inhibitory activity with concentration- and time-dependent potencies stronger than the SGs (Fig. 1A, B and Table 1). It was found that the hydroxyl and fluorine substitutions at both Ring A and B of the DPPen pharmacophore produced the analog 2,2′-diOH-5,5′-diF-DPPen that displayed the strongest TrxR inhibitory effect (IC50 at 1-h incubation: 9.1 μM). Compounds containing a DPPro pharmacophore, also commonly known as chalcones, were found to exert a TrxR inhibition. Unlike the inactive unsub-DPPen, the unsubstituted DPPro analog (unsub-DPPro) possessed a greater TrxR inhibitory efficacy than the SGs, with hydroxyl and fluorine substitutions enhancing the inhibitory effects (Fig. 1A, C and Table 1). Among the DPPro-based analogs, 2-OH-5-F-DPPro produced the strongest dose- and time-dependent enzyme inhibition that was comparable to the inhibitory activity of the most active DPPen analog 2,2′-diOH-5,5′-diF-DPPen.
FIG. 1.
In vitro inhibitory activity of α,β-unsaturated carbonyl (Michael acceptor) moiety-bearing compounds on mammalian thioredoxin reductase (TrxR). Dose-dependent (left panels) and time-dependent (right panels) TrxR inhibitory activities of (A) Michael acceptor-based natural compounds, (B) analogs bearing a 1,5-diphenyl-pent-1-en-3-one (DPPen) pharmacophore, and (C) analogs bearing a 1,3-diphenyl-pro-1-en-3-one (DPPro) (also known as chalcone) pharmacophore. Enzyme activities were evaluated by the 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) assay, upon 1-h incubation of the compounds at indicated concentrations with 110 nM recombinant rat TrxR (for dose-dependence assay conditions) or incubation of 25 μM of the compounds with 110 nM recombinant rat TrxR for different times (for time-dependence assay conditions). All data points represent means±SD of at least two independent experiments.
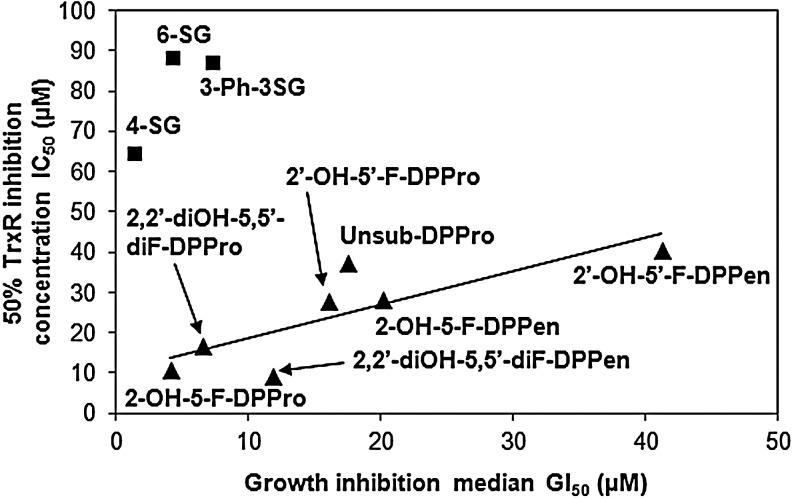
Evaluation of the effects of DPPen and DPPro compounds on the cell viability had revealed a positive correlation between the antiproliferative and TrxR inhibitory potencies (Fig. 2). The linear correlation coefficient r was calculated as 0.81. In contrast, these properties of SGs were poorly correlated. Expectedly, DPPen and DPPro analogs that displayed a greatest TrxR inhibition were also the most cytotoxic analogs. Of note, the DPPro-based compounds generally elicited greater antiproliferative activities than their DPPen counterparts.
FIG. 2.
Correlation of the TrxR inhibitory and antiproliferative potencies of Michael acceptor moiety-bearing compounds. The in vitro 50% TrxR inhibition concentration (IC50) values of 4-, 6-, and 3-Ph-3-SG, as well as the DPPen and DPPro analogs were plotted against their corresponding 50% growth inhibition concentration (GI50) values obtained in HCT 116 cells. The DPPen and DPPro analogs, but not the shogaols (SGs), showed a positive correlation with the linear correlation coefficient r=0.81.
Determination of effects of DPPen and DPPro analogs on TrxR, Trx, GSH reductase, and GSH peroxidase cellular activities
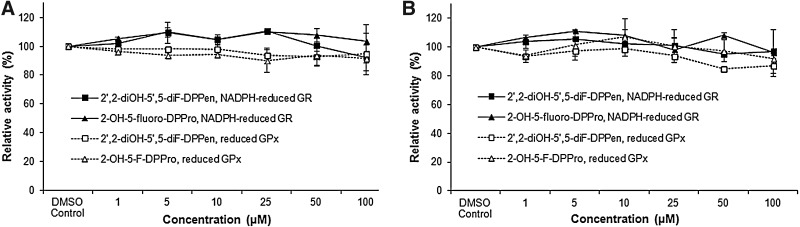
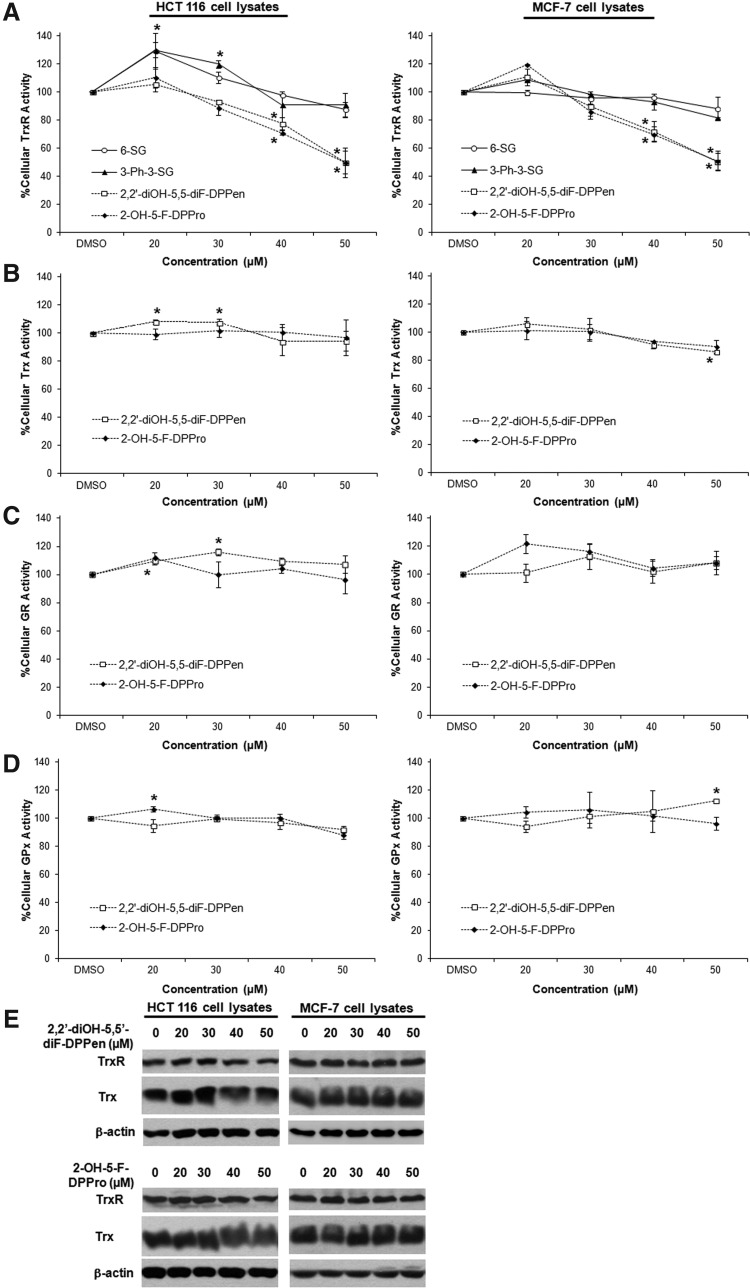
In vitro GSH reductase (GR) and GSH peroxidase (GPx) activity assays revealed that the lead DPPen/DPPro analogs 2,2′-diOH-5,5′-diF-DPPen and 2-OH-5-F-DPPro failed to inhibit both enzymes (Fig. 3A, B). The selectivity of their inhibitory effects against Trx- and GSH-related enzymes in HCT 116 and MCF-7 cells were next determined. As shown in Figure 4A, 6-SG and 3-Ph-3-SG were found to produce a marginal decline in the cellular TrxR activity. The cell-based results corroborated with the SGs' weak in vitro TrxR inhibitory activity. Conversely, 2,2′-diOH-5,5′-diF-DPPen and 2-OH-5-F-DPPro had caused a dose-dependent decrease in the TrxR activity, where lethal doses of 40 and 50 μM brought about a significant decline in the enzyme activity. The Western blot analyses showed that the protein levels of TrxR had remained relatively constant across the concentrations (Fig. 4E), which ruled out the possibility that reduction in the TrxR activity was attributed to a reduced enzyme expression. The Trx, GR, and GPx activities in the lysate samples were found to remain generally unsuppressed by the lead DPPen/DPPro analogs (Fig. 4B–D). The results obtained therefore indicate that the DPPen and DPPro analogs displayed selective TrxR inhibition.
FIG. 3.
In vitro effects of the DPPen and DPPro lead analogs on glutathione reductase (GR) and glutathione peroxidase (GPx) activities. The activity of yeast GR (20 nM) incubated with 200 μM NADPH and indicated compounds at various concentrations (1 to 100 μM) for (A) 30 min and (B) 1 h was determined by following the absorbance at 340 nm after adding glutathione disulfide and NADPH. The activity of bovine GPx (100 nM) incubated with 20 nM yeast GR, 200 μM NADPH, 1 mM glutathione, and indicated compounds at various concentrations for (A) 30 min and (B) 1 h was determined by after the absorbance at 340 nm after adding H2O2. All data points are means±SD of two independent experiments.
FIG. 4.
Effects of the DPPen and DPPro lead analogs on enzyme activities in HCT 116 and MCF-7 cells. Lysates of HCT 116 and MCF-7 cells treated with indicated concentrations of SG, DPPen, and DPPro analogs for ∼12 h were collected for enzyme activity and protein expression determination. TrxR (A), Trx (B), GR (C), and GPx (D) activities were expressed as a percentage of dimethyl sulphoxide control. All data points are means±SE of two to four independent experiments. *Statistically significant difference (p<0.05) in enzyme activity compared to control. (E) The cell lysates with equal protein were analyzed by Western blotting with indicated antibodies for TrxR and Trx expression. Western blot images are representative of at least two independent experiments.
Mechanistic characterization of cytotoxic activities of DPPen and DPPro compounds
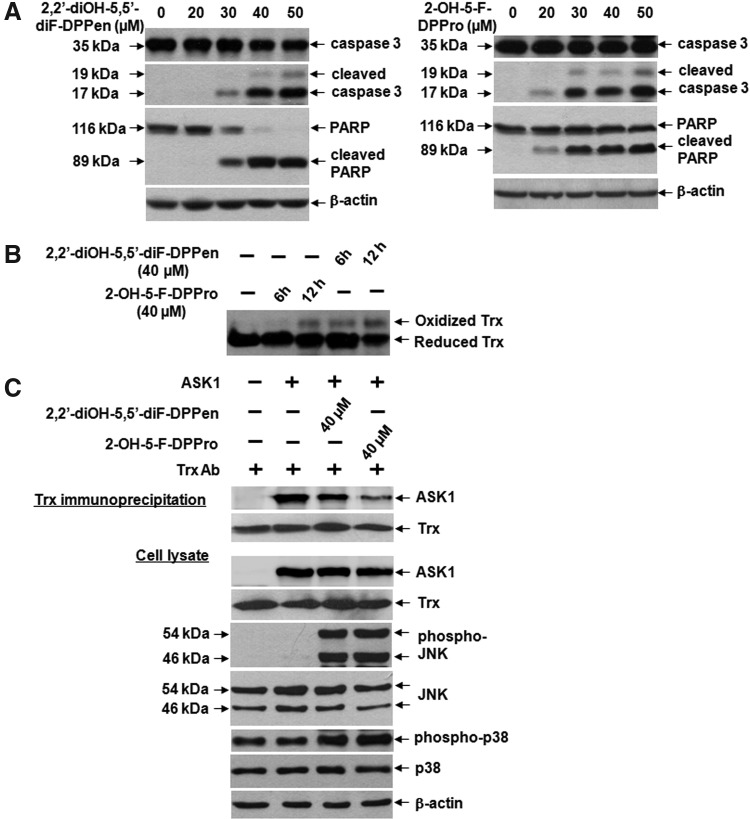
HCT 116 cells were selected for the examination of the cytotoxicities elicited by the DPPen and DPPro analogs. First, the Western blot analyses showed that the lead analogs 2,2′-diOH-5,5′-diF-DPPen and 2-OH-5-F-DPPro caused apoptosis dose-dependently, as indicated by cleaved forms of caspase 3 and poly (ADP-ribose) polymerase (PARP) (Fig. 5A). The compounds at a lethal concentration (40 μM) caused a time-dependent shift of reduced Trx to the oxidized form (Fig. 5B). One of the mechanisms by which Trx exerts antiapoptotic effects involves reduced Trx inhibiting ASK1 through a direct protein–protein interaction (40). ASK1 is a mitogen-activated protein kinase kinase kinase (MAPKKK) that lies upstream of the MAPK signaling pathway. Its activation by phosphorylation leads to activation (also by phosphorylation) of two downstream MAPK signaling cascades, the c-Jun N-terminal kinase (JNK) and p38 pathway, which eventually leads to apoptosis (22). The drug effects on the Trx-ASK1 interaction were examined by Trx immunoprecipitation reactions. As the endogenous ASK1 levels were nondetectable, the cells were transfected with pCMV-SPORT6-ASK1 to attain detectable ASK1 levels. As shown in Figure 5C, a decrease in the amount of coimmunoprecipitated ASK1 in the lysates of cells treated with 40 μM 2,2′-diOH-5,5′-diF-DPPen and 2-OH-5-F-DPPro was observed. This was accompanied by an increase in the phosphorylated forms of JNK and p38. Taken together, the results had suggested that cytotoxicity elicited by DPPen/DPPro-mediated TrxR inhibition was at least partly a result of dissociation of oxidized Trx from ASK1, leading to ASK1-dependent apoptosis induction.
FIG. 5.
Effects of the DPPen and DPPro lead analogs on the Trx-apoptosis signal-regulating kinase 1 (ASK1) interaction and apoptosis induction in HCT 116 cells. (A) Dose-dependent induction of apoptosis, indicated by presence of cleaved caspase 3 (17/19 kDa) and poly (ADP-ribose) polymerase (PARP) (89 kDa), after 12-h treatment with 2,2′-diOH-5,5′-diF-DPPen or 2-OH-5-F-DPPro. (B) Time-dependent changes in the redox state of Trx after treatment with 2,2′-diOH-5,5′-diF-DPPen or 2-OH-5-F-DPPro at a lethal concentration of 40 μM at indicated timepoints. (C) The collected lysates of ASK1-transfected cells treated or untreated for 8 h with 40 μM 2,2′-diOH-5,5′-diF-DPPen or 2-OH-5-F-DPPro were subjected with immunoprecipitation using an anti-human Trx antibody. Immunoprecipitates and aliquots of the cell lysates were analyzed by Western blotting with the indicated antibodies. The images shown are representative Western blot results of three independent experiments.
Characterization of TrxR inhibition by DPPen and DPPro compounds
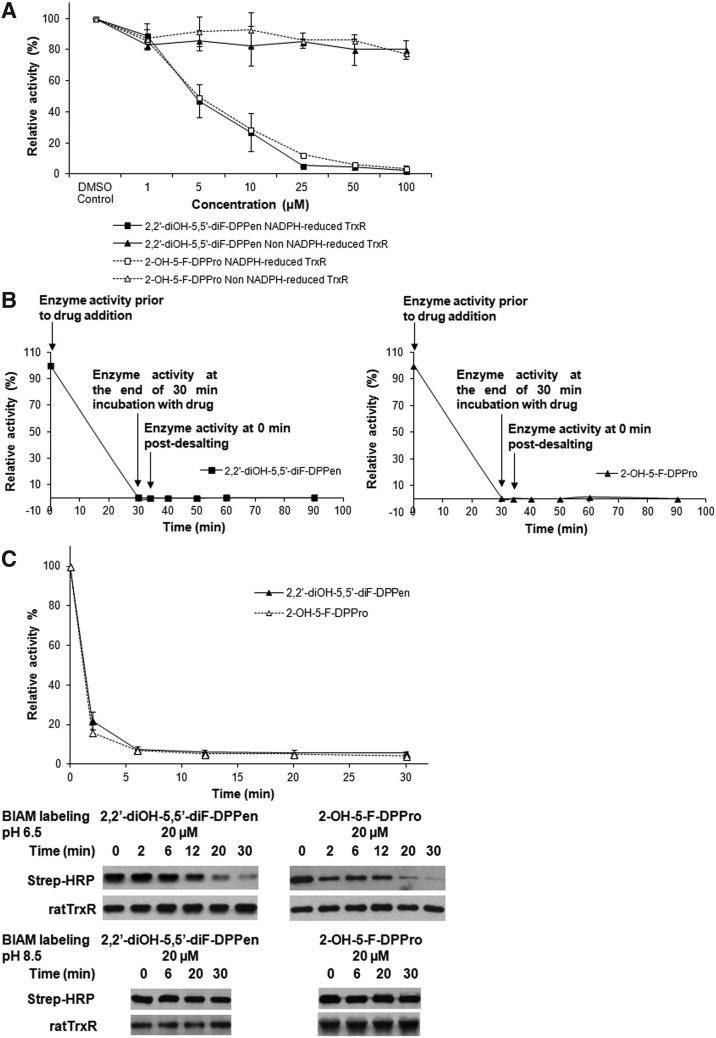
TrxR inhibition by the DPPen and DPPro compounds was NADPH dependent, with the activity of oxidized TrxR remaining relatively unaffected while the NADPH-reduced form was inhibited (Fig. 6A). To investigate whether the compounds inhibited the enzyme reversibly or irreversibly, recombinant rat TrxR that had activity completely inhibited by 20 μM of 2,2′-diOH-5,5′-diF-DPPen or 2-OH-5-F-DPPro was passed through an NAP-5-desalting column (GE Healthcare) to remove the excess drug. The enzyme activity failed to recover when aliquots of the eluted enzyme were assayed for activity (in the presence of NADPH) at various timepoints (Fig. 6B), indicating that TrxR was irreversibly inhibited by DPPen and DPPro compounds. We also found that the lead DPPen and DPPro analogs did not induce increased NADPH oxidase activity (data not shown), suggesting that DPPen and DPPro analogs are unlikely substrates of TrxR.
FIG. 6.
Irreversibility of TrxR inhibition and selenocysteine (Sec) residue targeting by the DPPen and DPPro lead analogs. (A) Oxidized recombinant rat TrxR (110 nM) in the presence or absence of 200 μM NADPH was incubated with different concentrations of 2,2′-diOH-5,5′-diF-DPPen or 2-OH-5-F-DPPro for 20 min, and the TrxR activity was determined by the DTNB assay. All data points are means of two independent experiments. (B) Recombinant rat TrxR (0.9 μM) was incubated with 20 μM of 2,2′-diOH-5,5′-diF-DPPen or 2-OH-5-F-DPPro for 20 min in a 50 mM Tris-HCl, 1 mN EDTA, pH 7.5 buffer containing 200 μM NADPH. The excess drug was removed by passing the protein through a NAP-5-desalting column, followed by determination of the activity of the eluted protein at indicated times using the DTNB assay. (C) Recombinant rat TrxR (0.9 μM) was incubated with 200 μM NADPH and 20 μM of 2,2′-diOH-5,5′-diF-DPPen or 2-OH-5-F-DPPro. At different timepoints, an aliquot of enzyme mixture was withdrawn for TrxR activity measurement by the DTNB assay and N-(Biotinoyl)-N′-(Iodoacetyl) Ethylenediamine (BIAM) labeling of Sec at pH 6.5 and 8.5. Top panel: time course of TrxR enzyme activity; middle panel: horseradish peroxidase (HRP)-conjugated streptavidin detection of BIAM labeling of free selenol at pH 6.5 at various incubation times; bottom panel: HRP-conjugated streptavidin detection of BIAM labeling of free selenol and sulfhydryls at pH 8.5 at various incubation times. Results shown are representative of three independent experiments.
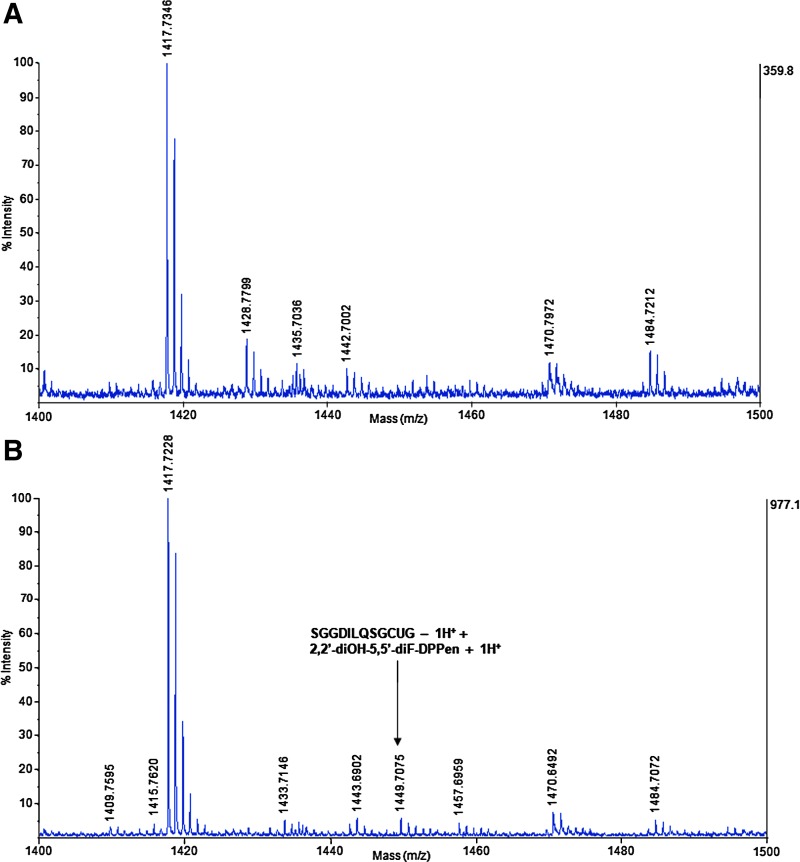
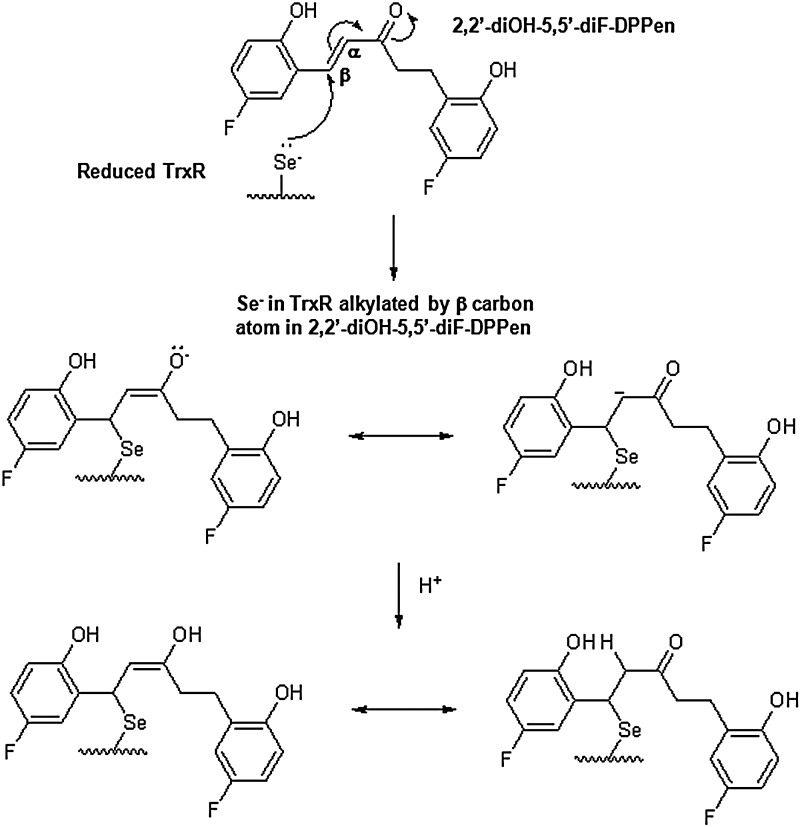
Whether the lead DPPen and DPPro compounds targeted the C-terminal Sec in mammalian TrxR was determined by the N-(Biotinoyl)-N′-(Iodoacetyl) Ethylenediamine (BIAM) labeling assay. In view that Sec and Cys possess a different pKa value of 5.2 and around 8, respectively, only Sec is available for BIAM alkylation at pH 6.5, whereas at pH 8.5, both Cys and Sec are available for BIAM labeling. During a 30-min incubation of TrxR protein with 2,2′-diOH-5,5′-diF-DPPen or 2-OH-5-F-DPPro, the enzyme showed a decline in the activity over time (top panel of Fig. 6C). The results were consistent with the observed progressive reduction in BIAM labeling at pH 6.5 (middle panel of Fig. 6C). In contrast, the same labeling experiment conducted at pH 8.5 showed no significant reduction in the BIAM labeling along time (bottom panel of Fig. 6C), suggesting that it was the C-terminal Sec, but not the Cys residues that was targeted by the compounds. In addition, the trypsin-digested peptide masses of TrxR protein treated with 2,2′-diOH-5,5′-diF-DPPen were analyzed by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. When the mass spectra obtained for untreated protein and drug-treated protein were compared, a mass peak of 1449.7075 had appeared in the mass spectrum of the latter sample (Fig. 7A, B). This peak corresponded to a calculated mass of 1448.7 of a peptide fragment containing the C-terminal Sec residue (SGGDILQSGCUG, mass peak of 1144.4–1 H) plus one molecule of 2,2′-diOH-5,5′-diF-DPPen, and plus 1 H (1143.4+304.3+1). The proposed mechanism of alkylation by 2,2′-diOH-5,5′-diF-DPPen at the Sec residue to form a Michael acceptor adduct is as illustrated in Figure 8. The results obtained from the BIAM labeling assay and MALDI mass spectrometry suggest that the penultimate Sec residue is targeted by the enone-containing compounds bearing the DPPen and DPPro pharmacophores.
FIG. 7.
Peptide mass analysis of TrxR-2,2′-diOH-5,5′-diF-DPPen protein adduct by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. NADPH-reduced recombinant rat TrxR (3 μM) was incubated at 37°C with or without 50 μM 2,2′-diOH-5,5′-diF-DPPen for 30 min, desalted to remove excess drug, denatured in urea, and trypsin-digested. Finally, MALDI mass spectrometry was performed to obtain the peptide mass spectrum of the (A) untreated and (B) 2,2′-diOH-5,5′-diF-DPPen-treated protein sample. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
FIG. 8.
Proposed mechanism of inhibition of mammalian TrxR by 2,2′-diOH-5,5′-diF-DPPen. The electrophilic β-carbon atom in 2,2′-diOH-5,5′-diF-DPPen undergoes a Michael addition reaction with the Sec residue in NADPH-reduced TrxR to form a Michael acceptor adduct.
Molecular simulation of the interaction between the C-terminal active site in TrxR with the SG, DPPen, and DPPro analogs
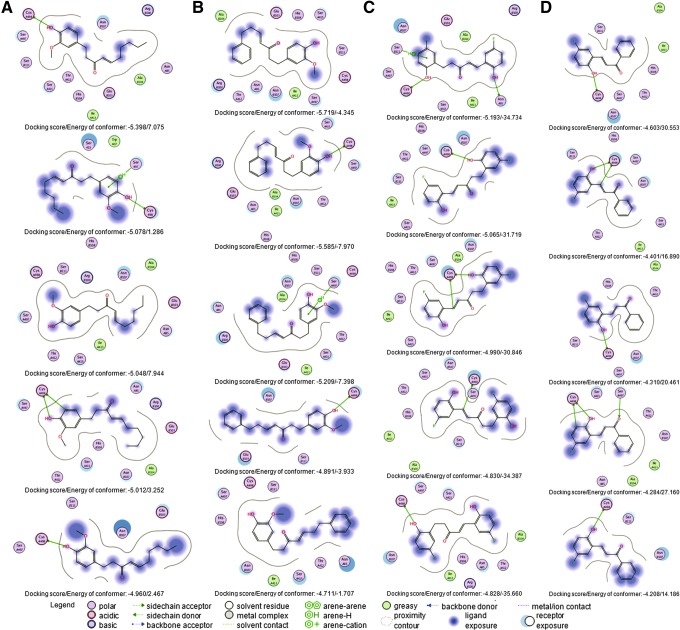
To study the plausible drug–protein interactions, molecular docking exercises were performed for 6-SG, 3-Ph-3-SG, 2,2′-diOH-5,5′-diF-DPPen and 2-OH-5-F-DPPro. In our docking studies, we hypothesized that the candidate inhibitors would interact with the charged Cys498 (i.e., U498C) and also block other residues within the TrxR active site that might be important for facilitating and/or stabilizing Trx binding. As observed in the first crystal structure of the human TrxR-Trx complex (15), besides the disulfide bond between C32 in Trx and U498C in TrxR, several amino acid interactions, such as those involving the Trx residues 58 to 74 and TrxR helical residues Glu103 to Glu122, could further stabilize the TrxR-Trx complex. Therefore, the smaller site at the protein surface that contained residues within the stretch of amino acids Glu103 to Glu122 was selected. Although the selected site differed from previous studies that had explored the docking of curcuminoids (42) and ethaselen (46) into the larger solvent-exposed 193-amino-acid site, it might be useful in the context of our studies to gain some insights into the interferences (if any) posed by potential TrxR inhibitors on the TrxR-Trx interaction. Analysis of the top five poses showed that the interactions of U498C in TrxR with 6-SG and 3-Ph-3-SG were often with the hydroxyl group in the vanilloid moiety. The remaining poses involving the SGs did not identify any other points of interaction with U498C. Notably, the α,β-unsaturated carbonyl group was found to be orientated away from U498C in all poses (Fig. 9A, B). Docking results obtained for 2,2′-diOH-5,5′-diF-DPPen revealed that the hydroxyl-group in Ring B and the β-carbon atom in the enone group could interact with U498C, whereas for 2-OH-5-F-DPPro, the hydroxy-group in Ring A and the β-carbon atom interacted with U498C (Fig. 9C, D). Additionally, a number of findings not observed for the SGs were noted for the DPPen and DPPro lead analogs. First, most of the poses showed that the enone group was proximal to and orientated toward U498C. Secondly, the DPPen and DPPro lead analogs established more points of interaction with residues within the docking site. Besides the interactions between the hydroxyl group and β-carbon atom with U498C, the top pose for 2,2′-diOH-5,5′-diF-DPPen revealed possible hydrogen bonding between the compound and TrxR's Ser111 (Fig. 9C), an amino acid residing within the stretch of TrxR helical residues (Glu103 to Glu122) identified to interact with the residues in Trx to stabilize the TrxR-Trx complex (15). Such interaction suggests that docking of the DPPen or DPPro analogs into TrxR's active site may result in (1) the Sec residue being targeted, and (2) a blocking effect to prevent formation of or to disrupt stabilization of the TrxR-Trx complex. Thirdly, the hydroxyl- and fluorine substituents on the lead DPPen and DPPro analogs showed a large solvent-accessible area, as indicated by the dark blue regions surrounding each atom on the compound in the ligand interaction map. Conversely, a large part of the respective nonpolar aliphatic and 3-phenyl (Ring A) side chain of 6-SG and 3-Ph-3-SG was much exposed to the solvent. Given that the active site was largely exposed to the solvent, the presence of the relatively small and hydrophilic substituents might favor the exposure of the compounds at the solvent surface, which in turn could help stabilize the drug–protein adduct. The docking results had suggested that 2,2′-diOH-5,5′-diF-DPPen and 2-OH-5-F-DPPro were docked into the TrxR C-terminal active site more favorably than the SGs, with an electrophilic β-carbon atom establishing the interaction with the Sec residue.
FIG. 9.
Molecular simulation of the interaction between C-terminal active site in TrxR with SGs, DPPen, and DPPro compounds. Molecular docking exercises were conducted for (A) 6-SG, (B) 3-Ph-3-SG, (C) 2,2′-diOH-5,5′-diF-DPPen, and (D) 2-OH-5-F-DPPro, with the ligand interaction maps of the top five poses obtained for each ligand presented in descending order.
Discussion
TrxRs belong to the flavoprotein family of the pyridine nucleotide–disulfide oxidoreductases that include GR. The types of TrxRs found in mammals include cytosolic TrxR1 (53), mitochondrial TrxR2 (28), and testis-specific Trx GR (44). All types of TrxR and GR have a common domain structure comprising a binding domain for flavin adenine dinucleotide and NADPH, as well as an interface domain. In addition, mammalian TrxR and GR possess a similar N-terminal -Cys-XXXX-Cys- active site and therefore share a similar reaction mechanism with regard to the reductive half-reaction (3, 41). The C-terminal Sec-containing active site, being absent in GR, is unique of mammalian TrxR. Upon reduction by accepting the reducing equivalents from the N-terminal dithiol active site, this C-terminal tail moves such that the selenothiol motif becomes solvent exposed and transfers electrons over to substrates (6, 10, 16, 41). Such unique properties have made TrxR susceptible to inhibition by electrophilic compounds that can react with the accessible and nucleophilic selenoate at the C-terminal active site. Another related enzyme GPx is also a selenoprotein, except that the Sec residue in the active site is not as solvent exposed as that in mammalian TrxR (27, 38). Efforts to discover and develop TrxR inhibitors are underway. For instance, high-throughput screening of the 1280-member LOPAC1280 collection of compounds has been conducted to identify the novel inhibitors and substrates of TrxR (36).
TrxR inhibitors are classified based on their mechanisms of enzyme inhibition and structural types (7, 30, 45). Among them are compounds containing α,β-unsaturated carbonyl groups, which allow them to undergo a Michael addition reaction between the β-carbon atom and the Se− of Sec in TrxR. Such electrophilic compounds, also called the Michael acceptors, are structurally diversified, which hinder the carrying out of large-scale structure–activity relationship studies. To add to the difficulty for the rational design of Michael acceptor-based TrxR inhibitors, Michael acceptors that lack the TrxR inhibitory effects are not clearly known. We hypothesized that despite the structural diversity, an α,β-unsaturated carbonyl moiety and other structural features formed a TrxR-inhibiting pharmacophore. Nature is a source of bioactive compounds that has led to discovery of Michael acceptor-based TrxR inhibitors such as palmarumycin (34, 48), curcumin (14), flavonoids (31), and trans-cinnamaldehyde (12). We therefore screened a number of commonly known Michael acceptor moiety-bearing natural compounds that as of yet have not been reported as TrxR inhibitors. These natural products, which serve as potential scaffolds for diversity-oriented synthesis, belong to distinct structural families: α,β-unsaturated amides and unconjugated α,β-unsaturated ketones. The α,β-unsaturated IBAs exhibited poor antiproliferative activities correlated to their lack of TrxR inhibitory activities. Conjugation of electrons on the amide nitrogen alleviated the electronegative effect of the carbonyl group, thus reducing the electrophilic character of the β carbon atom and in turn their ineffectiveness for TrxR inhibition. Unconjugated α,β-unsaturated ketones zerumbone and SGs displayed no or weak TrxR inhibitory activity in contrast to their potent antiproliferative effects. In these compounds, an electron-donating alkyl group linked to the Michael acceptor group had reduced electrophilicity of the β-carbon atoms, which explained the poor TrxR inhibition. The poor correlation between zerumbone's and SGs' inhibitory effects on cell proliferation and TrxR activity had indicated that TrxR targeting was unlikely the mechanism underlying their antitumor action. The results agreed with findings by published studies, which have reported the apoptotic activities of zerumbone and SGs in association with other signaling pathways (17, 21, 26, 32, 35). Molecular docking results obtained for the SGs also revealed plausible reasons for their poor TrxR inhibitory properties. First, the α,β-unsaturated carbonyl group was orientated away from the Sec residue, which reduced the probability of the interaction between the electrophilic β-carbon atom with Sec. Secondly, the high solvent exposure of the nonpolar aliphatic chain might discourage a stable drug interaction at the enzyme's active site. Collectively, the findings had suggested that having a Michael acceptor group could not unequivocally indicate the TrxR inhibitory activity; other structural features might be required for enzyme inhibition.
3-Ph-3-SG, a novel SG analog that possessed a comparable antiproliferative and TrxR inhibitory activity as 6-SG was selected as the compound on which structural derivatives were synthesized. Working along our hypothesis that other structural features were required for enzyme inhibition, we investigated whether a shortened distance between the two phenyl rings (Ring A and B) would affect the TrxR inhibitory activity. Compounds carrying the DPPen or DPPro pharmacophores were therefore synthesized. Based on the knowledge from published studies that Michael acceptor-based TrxR inhibitors such as quinones such as juglone (9), quinols such as PMX464 (11), 4-hydroxynonenal (13), palmarumycin CP1 (48), and flavonoids such as myricetin and quercetin (31) contain a hydroxyl group proximal to the α,β-unsaturated carbonyl group, and our recent findings that 2-hydroxy and 5-fluorine substitutions on the phenyl ring adjacent to the α,β-unsaturated carbonyl group led to an enhanced TrxR inhibitory activity (12), the DPPen and DPPro analogs carrying the hydroxyl and fluorine substitutions were synthesized. In the DPPen and DPPro series, both unsubstituted analogs registered weakest TrxR inhibition, whereas 2,2′-diOH-5,5′-diF-DPPen and 2-OH-5-F-DPPro were the respective analogs recording the strongest TrxR inhibitory effect. Clearly, 2-hydroxy and 5-fluorine substitutions at the phenyl ring adjacent to the α,β-unsaturated bond (Ring A) yielded a greater TrxR inhibitory activity than having similar substitutions at the phenyl ring closer to the carbonyl group (Ring B). This could be attributed to the stronger influence exerted by an electron-withdrawing fluorine substituent at Ring A on the β-carbon atom, making it more electrophilic for reaction with Se−. Molecular docking work had revealed high solvent exposure of the hydrophilic hydroxyl and fluorine substituents, which might contribute to the TrxR inhibition by favoring the stability of the ligand–protein adduct formed at the enzyme's active site. The hydroxyl and fluorine substitutions at Ring B, being distant from the β-carbon atom, would supposedly exert a diminished electron-withdrawing effect. Nonetheless, a favorable engagement of the substituents with solvent might have stabilized the compound's interaction with the active site, which possibly explained the greater inhibitory potencies of these analogs (2′-OH-5′-F-DPPen and 2′-OH-5′-F-DPPro) over their unsubstituted counterparts. On the combined bases of the favorable electronic and hydrophilic character, we would expect analogs bearing the 2-hydroxy and 5-fluorine substituents in both Ring A and B to display the strongest TrxR inhibitory activity. This was the case for the DPPen series of analogs, with 2,2′-diOH-5,5′-diF-DPPen found most potent. Unexpectedly, for the DPPro series of analogs, 2-OH-5-F-DPPro was found most potent instead, suggesting that the hydroxyl and fluorine substituents at Ring B might have impeding effects on TrxR inhibition. One proposed explanation was being adjacent to each other; 2′-hydroxy on Ring B and the carbonyl oxygen in 2,2′-diOH-5,5′-diF-DPPro were engaged in intramolecular hydrogen bonding, reducing electronegativity of the latter atom and in turn reduced electrophilicity on the β-carbon atom.
TrxR inhibitory efficacies of the DPPen and DPPro analogs were positively correlated to their antiproliferative potencies, and TrxR inhibition was found to be NADPH dependent and irreversible. The compounds' selectivity for TrxR was demonstrated in cell-based enzyme inhibition studies, where the lead DPPen and DPPro compounds caused a selective dose-dependent reduction in the TrxR activity with Trx, GR, and GPx activity minimally affected. Michael acceptor-based compounds tend to target multiple cellular pathways and are often multifunctional. Components of these signaling pathways usually contain soft nucleophiles such as thiols that are integral to protein functions. Selenothiol in Sec is also a soft nucleophile that the Michael acceptor moiety reacts with in preference over hard nucleophiles such as amino- and hydroxyl groups. Indeed, the BIAM labeling assay and MALDI mass spectrometry provided evidence that suggests that the C-terminal Sec residue in TrxR is the site of target by compounds that contain the DPPen or DPPro pharmacophores. Our study has therefore concluded that structural features that include an α,β-unsaturated carbonyl-centric core contained in a DPPen or DPPro pharmacophore, together with hydroxyl and fluorine substitutions, contribute toward a strong TrxR inhibitory character. TrxR inhibition caused Trx oxidation and dissociation of the Trx-ASK1 complex, resulting in ASK1-dependent activation of the JNK and p38 pathways that culminated in apoptosis induction. Nonetheless, the possibility that compound-induced TrxR inhibition mediates apoptosis induction independent of ASK1 signaling is not ruled out. As such, while the C-terminal active site of TrxR is known to broadly accommodate a structurally diversifying range of Michael acceptor-based compounds, our findings have suggested that a systematic approach for discovery of Michael acceptor-based TrxR inhibitors is feasible, and can be potentially applied for future design of novel TrxR inhibitors as anticancer agents.
Materials and Methods
Chemicals, materials, and cell culture
Dodeca-IBA and Hexa-IBA were from Chromadex. Synthesis of 4-, 6-, 8-, and 10-SG has been described previously (17). Dimethyl sulphoxide (DMSO) drug stocks of 50 mM were made and stored at −20°C. Recombinant rat TrxR was prepared as previously described (2). Yeast GR, bovine GPx, insulin, GSH disulfide (GSSG), and trypsin of proteomics grade were from Sigma. Recombinant human Trx and anti-human Trx antibody were obtained from IMCO Corporation. BIAM and horseradish peroxidase-conjugated streptavidin were from Molecular Probes. Antibodies against caspase 3, cleaved caspase 3, PARP, JNK, phospho-JNK (Thr183/Tyr185), p38, and phospho-p38 (Thr180/Tyr182) were from Cell Signaling Technology, while those specific for human TrxR, ASK1, and β-actin were from Santa Cruz Biotechnology. Polyclonal antibodies against rat TrxR1 were raised in rabbits and purified as described previously (39). Human-derived colon HCT 116 and MCF-7 breast carcinoma cells were maintained in a Rose Park Memorial Institute 1640 medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin and incubated at 37°C in an humidified atmosphere of 95% air and 5% CO2.
Extraction and purification of zerumbone
Zerumbone was extracted using a published method (25) with modifications. Slices of fresh rhizomes of Zingiber zerumbet Smith (1.1 kg) harvested in Chiang Mai, Thailand, were subjected to steam distillation. The distillate was kept for 1 or 2 days at room temperature to give a crystalline solid of an essential oil. The crystals were collected, and the remaining water–oil mixture was subjected to extraction by hexane. The pooled hexane layers were concentrated and used for recrystallization. Pure zerumbone was collected (yield: 0.19%).
Chemical synthesis
3-phenyl-3-shogaol
Vanillin was condensed with acetone in NaOH at room temperature for 12 h, and the product was hydrogenated to zingerone. The phenolic group in zingerone was protected by tetrahydropyran (THP). At −78°C, THP-protected zingerone was activated by lithium diisopropylamide in anhydrous THP, followed by an aldol condensation reaction with benzaldehyde. The formed gingerol was dehydrated by refluxing in benzene in the presence of toluene sulfonic acid to produce 3-Ph-3-SG. The compound was characterized by proton and carbon nuclear magnetic resonance (NMR), and its purity as determined by reverse-phase high-performance liquid chromatography (HPLC) was found to be 95%.
2-OH-5-F-DPPen and 2,2′-diOH-5,5′-diF-DPPen
To a mixture of benzylacetone or 2-hydroxy-5-fluoro-benzylacetone (2 mmol) and benzaldehyde or 2-hydroxy-5-fluoro-benzaldehyde (2 mmol) dissolved in ethanol (1.5 ml), two drops of piperidine were added. The mixture was stirred at room temperature for 2 weeks, with one drop of piperidine added every 3 days. The viscous reaction mixture was diluted with alcohol and made acidic with HCl. The pure product was isolated from silica-gel column chromatography with hexane/ethyl acetate as the mobile phase. Compounds were characterized by proton and carbon NMR, and their purities of at least 95% were determined by reverse-phase HPLC.
Unsub-DPPen and 2′-OH-5′-F-DPPen
A solution of benzylacetone (5 mmol) and benzaldehyde or 2-hydroxy-5-fluoro-benzaldehyde (5 mmol) was added to a solution of 85 mM KOH and stirred at 55°C for 4–6 h. The reaction mixture was then transferred to an ice bath with pH being adjusted to 2–3 by adding HCl. The mixture was extracted by ethyl acetate. The silica-gel column chromatography was performed to yield the desired DPPen compounds, whose structures were verified by proton and carbon NMR, and the purities of at least 95% were determined by reverse-phase HPLC.
DPPro compounds (also known as chalcones)
Benzaldehyde (2 mmol) and acetophenone (2 mmol) were condensed in 20 ml of 3% w/v NaOH in methanol at room temperature for 4 h. The reaction mixture was next submerged in ice bath, and HCl was added to adjust pH to 2–3. The mixture was stirred overnight, and the crude product was extracted by ethyl acetate. Further purification was done by recrystallization in ethanol. All DPPro analogs were characterized by proton and carbon NMR, and the purities as determined by reverse-phase HPLC were at least 95%.
Cell viability assay
The viabilities of HCT 116 and MCF-7 cells treated with various concentrations of indicated compounds were determined by reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide as described previously (12).
Determination of TrxR activity by in vitro DTNB reduction assay
TrxR activity was determined as previously described (11).
Preparation of cell lysates and Western blot analysis
The cell lysate was prepared, and the Western blot analysis was performed, as previously described (12).
Determination of TrxR and Trx activity in cell lysates
The TrxR and Trx activity in the cell lysates was determined by an endpoint insulin assay as previously described (12).
GR and GPx activity assays
The GR activity was determined in vitro as described previously (11). For determination of the cellular GR activity, 25 μg of cell lysate was mixed with a solution of GSSG and NADPH in 0.1 M sodium phosphate/2 mM EDTA, pH 7.5, to a final volume of 100 μl (final GSSG and NADPH concentration: 1 mM and 200 μM respectively). The enzyme activity was determined by measuring the decrease in absorbance at 340 nm during the initial 5 min and expressed as a percentage of the enzyme activity of that of the DMSO-treated sample.
For in vitro determination of GPx activity, to a mixture containing 100 nM GPx, 20 nM GR, 1 mM GSH, and 200 μM NADPH in 0.1 M sodium phosphate/2 mM EDTA, pH 7.5, H2O2 was added to a final concentration of 1.5 mM to initiate the reaction. The GPx activity was determined by measuring the decrease in absorbance at 340 nm during the initial 6 min and expressed as a percentage of enzyme activity of that of the DMSO-treated sample. The cellular GPx activity was determined similarly except to replace bovine GPx with 25 μg of cell lysates.
Preparation of lysates and determination of Trx redox state in cells
The Trx redox state in cells was determined as described previously (50) with slight modifications. Upon reaching 70%–80% confluency, HCT 116 seeded in 60-mm plates were treated with 40 μM 2,2′-diOH-5,5′-diF-DPPen, 2-OH-5-F-DPPro, or DMSO for 6 or 12 h. The lysates were collected in a lysis buffer comprised of 50 mM Tris–HCl (pH 8.3), 2 mM EDTA, 6 M guanidine HCl, 0.5% Triton-X, and 50 mM iodoacetamide. The lysates were incubated at room temperature in the dark for 30 min, followed by brief sonication and passing through a MicroSpin G-25 column (GE Healthcare). The desalted samples were subjected to native polyacrylamide gel electrophoresis (PAGE), followed by transfer of separated proteins onto nitrocellulose membranes for immunoblotting with an anti-human Trx antibody.
Immunoprecipitation
At 40% confluency, HCT 116 cells seeded in 60-mm plates were transfected with 1.5 μg of pCMV-SPORT6-ASK1 per plate. The transfected cells were allowed to grow to 70%–80% confluency, followed by treatment with 40 μM 2,2′-diOH-5,5′-diF-DPPen, 2-OH-5-F-DPPro, or DMSO for 8 h, and the lysates were collected. For each lysate sample, a volume containing equal amount of protein (1–1.2 mg protein) was mixed with 1.5 μg of Trx antibody coupled to 20 μl of protein G-sepharose (GE Healthcare) and tumbled at 4°C for 2 h. The sepharose beads were washed three times in a cold washing buffer containing 20 mM Tris (pH 7.5), 0.5 M NaCl, and 1 mM ethylene glycol-bis (β-amino ethyl ether)-N,N,N′N′-tetraacetic acid and once in buffer containing 50 mM Tris (pH 7.5). The immunoprecipitated proteins were denatured in a sodium dodecyl sulfate (SDS) sample buffer containing 5% β-mercaptoethanol and subjected to SDS-PAGE and Western blotting.
BIAM labeling assay
Biotin labeling through BIAM alkylation of the free selenol at pH 6.5, or the free selenol and sulfhydryls at pH 8.5, in recombinant rat TrxR was performed using the procedures as described previously (12).
MALDI mass spectrometric analysis of drug-treated recombinant rat TrxR protein
Recombinant rat TrxR (3 μM) in a 50 mM Tris-HCl, 1 mM EDTA, pH 7.5 buffer containing 200 μM NADPH was incubated at 37°C with 50 μM 2,2′-diOH-5,5′-diF-DPPen for 30 min. An aliquot of the protein was tested in the DTNB reduction assay to verify that enzyme activity was abolished. Excess drug and NADPH were removed using a Sephadex G-25-containing NAP-5-desalting column (GE Healthcare). An aliquot of the desalted enzyme was tested in the DTNB reduction assay to confirm the null activity. The enzyme was denatured in 8 M urea at 60°C for 1 h, diluted 10-fold in 50 mM Tris–HCl/1 mM CaCl2, pH 7.6, buffer, and concentrated using an Amicon Ultra 30K NMWL filter (Merck Millipore) to a final concentration range of 0.05–0.1 μg/μl. The protein was digested by trypsin (trypsin-to-protein ratio 1:10) at 37°C for 6 h, followed by desalting using C18 ZipTips (Merck Millipore) with the peptides eluted in 80% acetonitrile/0.1% trifluoroacetic acid (TFA). The desalted sample (0.5 μl) was spotted on a 384-well target plate and crystallized with 0.5 μl of α-cyano-4-hydroxycinnamic acid matrix solution in 50% acetonitrile/0.1% TFA. The sample was analyzed on the MALDI-Tof-Tof MS: 4800 MALDI TOF/TOF Analyzer (Applied Biosystems). The mass-over-charge ratio (m/z) data of the sample were acquired in the reflector mode using the reflectron method (accelerating voltage: 20000V; laser intensity: 3300–3600; acquisition mass range: 1100 to 1500). The peptide mass fingerprints were analyzed using the peptideCutter online tool (www.web.expasy.org).
Molecular modeling
The mammalian TrxR1 structure PDB ID 3QFA (resolution of 2.20 Å) carrying double mutations C497S and U498C was obtained from the Protein Data Bank. Docking simulations were performed using Molecular Operating Environment (MOE), version 2011.10. The 3QFA structure was processed using the recommended Structure Preparation application in MOE, which was used to diagnose and correct structural issues such as incorrect number of hydrogens and missing atoms. The S-atom in Cys498 was added a negative charge to mimic the nucleophilic selenoate ion for docking simulations. Visual inspection of the TrxR-Trx complex based on the crystal structure elucidated by Fritz-Wolf et al. (15) and the Site Finder results of MOE revealed two potential sites for molecular docking involving the charged residue. The first was a large site that had 193 residues accessible to solvent. The other potential site, being smaller, was located nearer at the protein surface and contained residues such as Asn104, Asn107, and Ser111. The latter site was chosen for docking with S− in Cys498 identified as an essential binding atom. During the automated docking process, each ligand was docked into the active site in different ways, creating different poses each with a docking score. The top five poses of each ligand were obtained and analyzed for their interactions with TrxR.
Abbreviations Used
- AP-1
activator protein-1
- ASK1
apoptosis signal-regulating kinase 1
- Dodeca-IBA
dodeca-2(E),4(E)-dienoic acid isobutylamide
- DPHep
1,7-diphenyl-hept-3-en-5-one
- DPPen
1,5-diphenyl-pent-1-en-3-one
- DPPro
1,3-diphenyl-pro-1-en-3-one
- DTNB
5,5′-dithiobis-(2-nitrobenzoic acid)
- GPx
glutathione peroxidase
- GR
glutathione reductase
- GSH
glutathione
- Hexa-IBA
hexa-2,4-dienoic acid isobutylamide
- JNK
c-Jun N-terminal kinase
- MALDI
matrix-assisted laser desorption/ionization
- MAPK
mitogen-activated protein kinase
- MOE
molecular operating environment
- Sec
selenocysteine
- SG
shogaol
- TFA
trifluoroacetic acid
- THP
tetrahydropyran
- Trx
thioredoxin
- TrxR
thioredoxin reductase
Acknowledgments
We thank Dr. Elias Arner and Dr. Qing Cheng (Division of Biochemistry, Karolinksa Institutet) for providing recombinant rat TrxR, and Dr. Thilo Hagen (Department of Biochemistry, Yong Loo Lin School of Medicine, National University of Singapore) for providing pCMV-SPORT6-ASK1. This work was supported by a National Medical Council Grant NMRC/NIG/0050/2009 and a National University of Singapore Academic Research Fund Tier 1 R-148-116-112.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Arnér ES. Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 2.Arnér ES. Sarioglu H. Lottspeich F. Holmgren A. Böck A. High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. J Mol Biol. 1999;292:1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 3.Arscott LD. Gromer S. Schirmer RH. Becker K. Williams CH., Jr The mechanism of thioredoxin reductase from human placenta is similar to the mechanisms of lipoamide dehydrogenase and glutathione reductase and is distinct from the mechanism of thioredoxin reductase from Escherichia coli. Proc Natl Acad Sci U S A. 1997;94:3621–3626. doi: 10.1073/pnas.94.8.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker K. Herold-Mende C. Park JJ. Lowe G. Schirmer RH. Human thioredoxin reductase is efficiently inhibited by (2,2′:6′,2′ ′-terpyridine)platinum(II) complexes. Possible implications for a novel antitumor strategy. J Med Chem. 2001;44:2784–2792. doi: 10.1021/jm001014i. [DOI] [PubMed] [Google Scholar]
- 5.Berggren M. Gallegos A. Gasdaska JR. Gasdaska PY. Warneke J. Powis G. Thioredoxin and thioredoxin reductase gene expression in human tumors and cell lines, and the effects of serum stimulation and hypoxia. Anticancer Res. 1996;16:3459–3466. [PubMed] [Google Scholar]
- 6.Biterova EI. Turanov AA. Gladyshev VN. Barycki JJ. Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism. Proc Natl Acad Sci U S A. 2005;102:15018–15023. doi: 10.1073/pnas.0504218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai W. Zhang L. Song Y. Wang B. Zhang B. Cui X. Hu G. Liu Y. Wu J. Fang J. Small molecule inhibitors of mammalian thioredoxin reductase. Free Radic Biol Med. 2012;52:257–265. doi: 10.1016/j.freeradbiomed.2011.10.447. [DOI] [PubMed] [Google Scholar]
- 8.Casini A. Messori L. Molecular mechanisms and proposed targets for selected anticancer gold compounds. Curr Top Med Chem. 2011;11:2647–2660. doi: 10.2174/156802611798040732. [DOI] [PubMed] [Google Scholar]
- 9.Cenas N. Nivinskas H. Anusevicius Z. Sarlauskas J. Lederer F. Arnér ES. Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian selenoprotein. J Biol Chem. 2004;279:2583–2592. doi: 10.1074/jbc.M310292200. [DOI] [PubMed] [Google Scholar]
- 10.Cheng Q. Sandalova T. Lindqvist Y. Arnér ES. Crystal structure and catalysis of the selenoprotein thioredoxin reductase 1. J Biol Chem. 2009;284:3998–4008. doi: 10.1074/jbc.M807068200. [DOI] [PubMed] [Google Scholar]
- 11.Chew EH. Lu J. Bradshaw TD. Holmgren A. Thioredoxin reductase inhibition by antitumor quinols: a quinol pharmacophore effect correlating to antiproliferative activity. FASEB J. 2008;22:2072–2083. doi: 10.1096/fj.07-101477. [DOI] [PubMed] [Google Scholar]
- 12.Chew EH. Nagle AA. Zhang Y. Scarmagnani S. Palaniappan P. Bradshaw TD. Holmgren A. Westwell AD. Cinnamaldehydes inhibit thioredoxin reductase and induce Nrf2: potential candidates for cancer therapy and chemoprevention. Free Radic Biol Med. 2010;48:98–111. doi: 10.1016/j.freeradbiomed.2009.10.028. [DOI] [PubMed] [Google Scholar]
- 13.Fang J. Holmgren A. Inhibition of thioredoxin and thioredoxin reductase by 4-hydroxy-2-nonenal in vitro and in vivo. J Am Chem Soc. 2006;128:1879–1885. doi: 10.1021/ja057358l. [DOI] [PubMed] [Google Scholar]
- 14.Fang J. Lu J. Holmgren A. Thioredoxin reductase is irreversibly modified by curcumin: a novel molecular mechanism for its anticancer activity. J Biol Chem. 2005;280:25284–25290. doi: 10.1074/jbc.M414645200. [DOI] [PubMed] [Google Scholar]
- 15.Fritz-Wolf K. Kehr S. Stumpf M. Rahlfs S. Becker K. Crystal structure of the human thioredoxin reductase-thioredoxin complex. Nat Commun. 2011;2:383. doi: 10.1038/ncomms1382. [DOI] [PubMed] [Google Scholar]
- 16.Fritz-Wolf K. Urig S. Becker K. The structure of human thioredoxin reductase 1 provides insights into C-terminal rearrangements during catalysis. J Mol Biol. 2007;370:116–127. doi: 10.1016/j.jmb.2007.04.044. [DOI] [PubMed] [Google Scholar]
- 17.Gan FF. Nagle AA. Ang X. Ho OH. Tan SH. Yang H. Chui WK. Chew EH. Shogaols at proapoptotic concentrations induce G(2)/M arrest and aberrant mitotic cell death associated with tubulin aggregation. Apoptosis. 2011;16:856–867. doi: 10.1007/s10495-011-0611-3. [DOI] [PubMed] [Google Scholar]
- 18.Gromer S. Arscott LD. Williams CH., Jr. Schirmer RH. Becker K. Human placenta thioredoxin reductase. Isolation of the selenoenzyme, steady state kinetics, and inhibition by therapeutic gold compounds. J Biol Chem. 1998;273:20096–20101. doi: 10.1074/jbc.273.32.20096. [DOI] [PubMed] [Google Scholar]
- 19.Gromer S. Urig S. Becker K. The thioredoxin system—from science to clinic. Med Res Rev. 2004;24:40–89. doi: 10.1002/med.10051. [DOI] [PubMed] [Google Scholar]
- 20.Hashemy SI. Ungerstedt JS. Zahedi Avval F. Holmgren A. Motexafin gadolinium, a tumor-selective drug targeting thioredoxin reductase and ribonucleotide reductase. J Biol Chem. 2006;281:10691–10697. doi: 10.1074/jbc.M511373200. [DOI] [PubMed] [Google Scholar]
- 21.Hu R. Zhou P. Peng YB. Xu X. Ma J. Liu Q. Zhang L. Wen XD. Qi LW. Gao N. Li P. 6-shogaol induces apoptosis in human hepatocellular carcinoma cells and exhibits anti-tumor activity in vivo through endoplasmic reticulum stress. PLoS One. 2012;7:e39664. doi: 10.1371/journal.pone.0039664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichijo H. Nishida E. Irie K. ten Dijke P. Saitoh M. Moriguchi T. Takagi M. Matsumoto K. Miyazono K. Gotoh Y. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 23.Kahlos K. Soini Y. Saily M. Koistinen P. Kakko S. Paakko P. Holmgren A. Kinnula VL. Up-regulation of thioredoxin and thioredoxin reductase in human malignant pleural mesothelioma. Int J Cancer. 2001;95:198–204. doi: 10.1002/1097-0215(20010520)95:3<198::aid-ijc1034>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 24.Kim SJ. Miyoshi Y. Taguchi T. Tamaki Y. Nakamura H. Yodoi J. Kato K. Noguchi S. High thioredoxin expression is associated with resistance to docetaxel in primary breast cancer. Clin Cancer Res. 2005;11:8425–8430. doi: 10.1158/1078-0432.CCR-05-0449. [DOI] [PubMed] [Google Scholar]
- 25.Kitayama T. Okamoto T. Hill RK. Kawai Y. Takahashi S. Yonemori S. Yamamoto Y. Ohe K. Uemura S. Sawada S. Chemistry of zerumbone. 1. Simplified isolation, conjugate addition reactions, and a unique ring contracting transannular reaction of its dibromide. J Org Chem. 1999;64:2667–2672. doi: 10.1021/jo981593n. [DOI] [PubMed] [Google Scholar]
- 26.Kundu JK. Na HK. Surh YJ. Ginger-derived phenolic substances with cancer preventive and therapeutic potential. Forum Nutr. 2009;61:182–192. doi: 10.1159/000212750. [DOI] [PubMed] [Google Scholar]
- 27.Ladenstein R. Epp O. Bartels K. Jones A. Huber R. Wendel A. Structure analysis and molecular model of the selenoenzyme glutathione peroxidase at 2.8 A resolution. J Mol Biol. 1979;134:199–218. doi: 10.1016/0022-2836(79)90032-9. [DOI] [PubMed] [Google Scholar]
- 28.Lee SR. Kim JR. Kwon KS. Yoon HW. Levine RL. Ginsburg A. Rhee SG. Molecular cloning and characterization of a mitochondrial selenocysteine-containing thioredoxin reductase from rat liver. J Biol Chem. 1999;274:4722–4734. doi: 10.1074/jbc.274.8.4722. [DOI] [PubMed] [Google Scholar]
- 29.Lu J. Chew EH. Holmgren A. Targeting thioredoxin reductase is a basis for cancer therapy by arsenic trioxide. Proc Natl Acad Sci U S A. 2007;104:12288–12293. doi: 10.1073/pnas.0701549104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J. Holmgren A. Thioredoxin system in cell death progression. Antioxid Redox Signal. 2012;17:1738–1747. doi: 10.1089/ars.2012.4650. [DOI] [PubMed] [Google Scholar]
- 31.Lu J. Papp LV. Fang J. Rodriguez-Nieto S. Zhivotovsky B. Holmgren A. Inhibition of mammalian thioredoxin reductase by some flavonoids: implications for myricetin and quercetin anticancer activity. Cancer Res. 2006;66:4410–4418. doi: 10.1158/0008-5472.CAN-05-3310. [DOI] [PubMed] [Google Scholar]
- 32.Murakami A. Takahashi D. Kinoshita T. Koshimizu K. Kim HW. Yoshihiro A. Nakamura Y. Jiwajinda S. Terao J. Ohigashi H. Zerumbone, a Southeast Asian ginger sesquiterpene, markedly suppresses free radical generation, proinflammatory protein production, and cancer cell proliferation accompanied by apoptosis: the alpha,beta-unsaturated carbonyl group is a prerequisite. Carcinogenesis. 2002;23:795–802. doi: 10.1093/carcin/23.5.795. [DOI] [PubMed] [Google Scholar]
- 33.Papp LV. Lu J. Holmgren A. Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid Redox Signal. 2007;9:775–806. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- 34.Powis G. Wipf P. Lynch SM. Birmingham A. Kirkpatrick DL. Molecular pharmacology and antitumor activity of palmarumycin-based inhibitors of thioredoxin reductase. Mol Cancer Ther. 2006;5:630–636. doi: 10.1158/1535-7163.MCT-05-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prasannan R. Kalesh KA. Shanmugam MK. Nachiyappan A. Ramachandran L. Nguyen AH. Kumar AP. Lakshmanan M. Ahn KS. Sethi G. Key cell signaling pathways modulated by zerumbone: role in the prevention and treatment of cancer. Biochem Pharmacol. 2012;84:1268–1276. doi: 10.1016/j.bcp.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 36.Prast-Nielsen S. Dexheimer TS. Schultz L. Stafford WC. Cheng Q. Xu J. Jadhav A. Arnér ES. Simeonov A. Inhibition of thioredoxin reductase 1 by porphyrins and other small molecules identified by a high-throughput screening assay. Free Radic Biol Med. 2011;50:1114–1123. doi: 10.1016/j.freeradbiomed.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raffel J. Bhattacharyya AK. Gallegos A. Cui H. Einspahr JG. Alberts DS. Powis G. Increased expression of thioredoxin-1 in human colorectal cancer is associated with decreased patient survival. J Lab Clin Med. 2003;142:46–51. doi: 10.1016/S0022-2143(03)00068-4. [DOI] [PubMed] [Google Scholar]
- 38.Ren B. Huang W. Akesson B. Ladenstein R. The crystal structure of seleno-glutathione peroxidase from human plasma at 2.9 A resolution. J Mol Biol. 1997;268:869–885. doi: 10.1006/jmbi.1997.1005. [DOI] [PubMed] [Google Scholar]
- 39.Rozell B. Hansson HA. Luthman M. Holmgren A. Immunohistochemical localization of thioredoxin and thioredoxin reductase in adult rats. Eur J Cell Biol. 1985;38:79–86. [PubMed] [Google Scholar]
- 40.Saitoh M. Nishitoh H. Fujii M. Takeda K. Tobiume K. Sawada Y. Kawabata M. Miyazono K. Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandalova T. Zhong L. Lindqvist Y. Holmgren A. Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc Natl Acad Sci U S A. 2001;98:9533–9538. doi: 10.1073/pnas.171178698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh DV. Misra K. Curcuminoids as inhibitors of thioredoxin reductase: a receptor based pharmacophore study with distance mapping of the active site. Bioinformation. 2009;4:187–192. doi: 10.6026/97320630004187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soini Y. Kahlos K. Napankangas U. Kaarteenaho-Wiik R. Saily M. Koistinen P. Paaakko P. Holmgren A. Kinnula VL. Widespread expression of thioredoxin and thioredoxin reductase in non-small cell lung carcinoma. Clin Cancer Res. 2001;7:1750–1757. [PubMed] [Google Scholar]
- 44.Sun QA. Kirnarsky L. Sherman S. Gladyshev VN. Selenoprotein oxidoreductase with specificity for thioredoxin and glutathione systems. Proc Natl Acad Sci U S A. 2001;98:3673–3678. doi: 10.1073/pnas.051454398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urig S. Becker K. On the potential of thioredoxin reductase inhibitors for cancer therapy. Semin Cancer Biol. 2006;16:452–465. doi: 10.1016/j.semcancer.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 46.Wang L. Yang Z. Fu J. Yin H. Xiong K. Tan Q. Jin H. Li J. Wang T. Tang W. Yin J. Cai G. Liu M. Kehr S. Becker K. Zeng H. Ethaselen: a potent mammalian thioredoxin reductase 1 inhibitor and novel organoselenium anticancer agent. Free Radic Biol Med. 2012;52:898–908. doi: 10.1016/j.freeradbiomed.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 47.Wang X. Zhang J. Xu T. Cyclophosphamide as a potent inhibitor of tumor thioredoxin reductase in vivo. Toxicol Appl Pharmacol. 2007;218:88–95. doi: 10.1016/j.taap.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 48.Wipf P. Hopkins TD. Jung JK. Rodriguez S. Birmingham A. Southwick EC. Lazo JS. Powis G. New inhibitors of the thioredoxin-thioredoxin reductase system based on a naphthoquinone spiroketal natural product lead. Bioorg Med Chem Lett. 2001;11:2637–2641. doi: 10.1016/s0960-894x(01)00525-x. [DOI] [PubMed] [Google Scholar]
- 49.Witte AB. Anestål K. Jerremalm E. Ehrsson H. Arnér ES. Inhibition of thioredoxin reductase but not of glutathione reductase by the major classes of alkylating and platinum-containing anticancer compounds. Free Radic Biol Med. 2005;39:696–703. doi: 10.1016/j.freeradbiomed.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 50.Yan C. Siegel D. Newsome J. Chilloux A. Moody CJ. Ross D. Antitumor indolequinones induced apoptosis in human pancreatic cancer cells via inhibition of thioredoxin reductase and activation of redox signaling. Mol Pharmacol. 2012;81:401–410. doi: 10.1124/mol.111.076091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoo MH. Xu XM. Carlson BA. Gladyshev VN. Hatfield DL. Thioredoxin reductase 1 deficiency reverses tumor phenotype and tumorigenicity of lung carcinoma cells. J Biol Chem. 2006;281:13005–13008. doi: 10.1074/jbc.C600012200. [DOI] [PubMed] [Google Scholar]
- 52.Yoo MH. Xu XM. Carlson BA. Patterson AD. Gladyshev VN. Hatfield DL. Targeting thioredoxin reductase 1 reduction in cancer cells inhibits self-sufficient growth and DNA replication. PLoS One. 2007;2:e1112. doi: 10.1371/journal.pone.0001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong L. Arnér ES. Ljung J. Aslund F. Holmgren A. Rat and calf thioredoxin reductase are homologous to glutathione reductase with a carboxyl-terminal elongation containing a conserved catalytically active penultimate selenocysteine residue. J Biol Chem. 1998;273:8581–8591. doi: 10.1074/jbc.273.15.8581. [DOI] [PubMed] [Google Scholar]