Abstract
Rett syndrome (RTT) is a severe neurological disorder that is caused by mutations in the MECP2 gene. Many missense mutations causing RTT are clustered in the DNA-binding domain of MeCP2, suggesting that association with chromatin is critical for its function. We identified a second mutational cluster in a previously uncharacterized region of MeCP2. We found that RTT mutations in this region abolished the interaction between MeCP2 and the NCoR/SMRT co-repressor complexes. Mice bearing a common missense RTT mutation in this domain exhibited severe RTT-like phenotypes. Our data are compatible with the hypothesis that brain dysfunction in RTT is caused by a loss of the MeCP2 ‘bridge’ between the NCoR/SMRT co-repressors and chromatin.
RTT is one of a small group of clinically discrete disorders in the autism spectrum that is caused by single gene mutations1. It therefore provides an opportunity to understand a brain disorder at the molecular level, linking the specific genetic lesion to its downstream pathology. The role of MeCP2 is also of fundamental biological interest, as the protein appears to link DNA methylation with chromatin structure by mediating epigenetic events that alter genome function2. MeCP2 was initially identified by virtue of its specific binding to DNA containing the methylated dinucleotide CG, and many of the mutations underlying RTT affect this interaction3,4. Early work implicated MeCP2 in the recruitment of the SIN3A histone deacetylase (HDAC) complex to chromatin, suggesting that it promotes a deacetylated chromatin structure that inhibits transcription2. Analysis of transcription in mice lacking the Mecp2 gene, however, suggests that this model is incomplete, as gene expression patterns are perturbed in complex ways that have so far defied simple explanation5.
One way of elucidating protein function is to seek specific binding partners. Using a variety of approaches, at least 12 candidate partner proteins for MeCP2 have been identified6, but functional annotation is still at an early stage. Specifically, in no case have mutations causing RTT been shown to interfere with any of these interactions. We found that contact between MeCP2 and the NCoR/SMRT co-repressor complexes occurs at a discrete site in the MeCP2 protein. Notably, we observed that missense mutations causing RTT abolished this interaction. Mice in which one of these mutations, Mecp2R306C, replaced the endogenous wild-type gene showed pronounced RTT-like phenotypes. These findings suggest that MeCP2 can bridge between DNA and the NCoR/SMRT co-repressors and that loss of this bridging function gives rise to RTT.
RESULTS
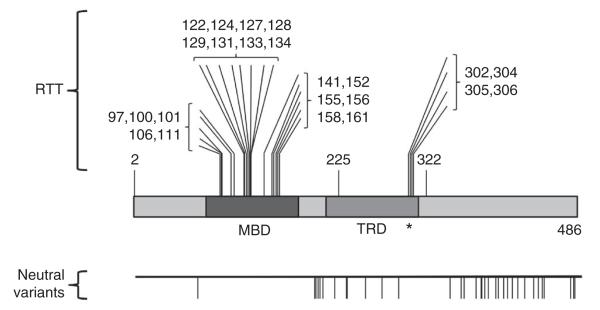
It is commonly considered that RTT is a result of mutations distributed throughout the MeCP2 protein (RettBASE, http://mecp2.chw.edu.au). We evaluated this notion by collating MeCP2 mutations for which published parental analysis confirmed a de novo origin. We focused on missense mutations, as they have the potential to precisely localize important functional motifs, unlike nonsense and frameshift mutations, which truncate the protein. Verified missense mutations causing classical RTT predominantly fall into two discrete clusters: those localizing to the well-characterized methyl-CpG binding domain (MBD), which often disrupt the association of MeCP2 with methylated DNA4,7, and a previously unknown mutation hotspot at the C-terminal extremity of the transcriptional repression domain (TRD)8, which includes amino acids 302–306 (Fig. 1). We also analyzed the distribution of amino acid substitutions in the general population by collating DNA sequence variants in the NHLBI GO ESP Exome Variant Server (http://evs.gs.washington.edu/EVS). These polymorphic variants in a population of 6,503 individuals were distributed broadly across the MeCP2 sequence (Fig. 1), but were absent from the two regions that are mutated in RTT. The reciprocal pattern of polymorphisms versus disease mutations in MeCP2 supports the view that amino acid substitutions in the MBD and C-terminal region of the TRD are deleterious.
Figure 1.
Missense RTT mutations in the MECP2 gene occur predominantly in two discrete clusters. Missense mutations present in RTT patients, but absent in their parents, map to the MBD and the C terminus of the TRD. The asterisk indicates the domain that we analyzed. The map below shows neutral variants that are present in a population sampled by exome sequencing.
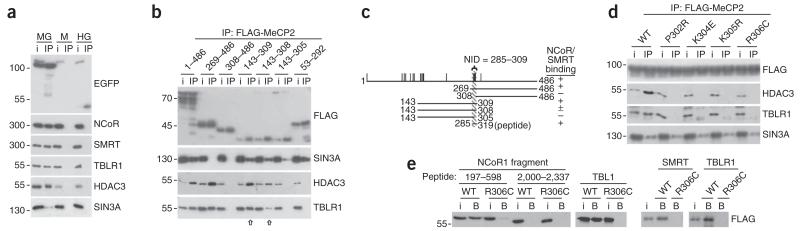
We hypothesized that the 302–306 cluster of RTT mutations represents a recruitment surface for a critical mediator of MeCP2 function. To seek potential partners, we purified MeCP2 from the brains of Mecp2-EGFP knock-in mice (Supplementary Fig. 1) and identified associated factors by mass spectrometry. Five of the top seven proteins identified were subunits of the known NCoR/SMRT co-repressor complexes9 (Supplementary Fig. 2). This finding was validated on western blots by probing MeCP2-EGFP immunoprecipitates with antibodies to NCoR1, SMRT, TBLR1 and HDAC3 (Fig. 2a). Antibodies to untagged MeCP2 also immunoprecipitated NCoR components from mouse brain (see below). The analysis confirmed a previously reported interaction with the SIN3A co-repressor complex2 (Fig. 2a).
Figure 2.
MeCP2 interacts with the NCoR/SMRT corepressor complex via a domain that coincides with a previously unknown cluster of RTT mutations. (a) Immunoprecipitation (IP) of EGFP, NCoR1, SMRT, TBLR1, HDAC3 and SIN3A from extracts of Mecp2-EGFP (MG) knock-in mouse brain using an antibody to EGFP. Immunoprecipitations from an extract from wild-type (M) brain (lacking fused EGFP) and H2B-EGFP (HG) brain are controls. i, inputs. (b) Mapping of the NID by expression of FLAG-tagged fragments of MeCP2 in HeLa cells. Immunoprecipitations with antibody to FLAG were probed on western blots. Arrows compare immunoprecipitation efficiency of the 143–309 and 143–308 fragments. (c) Map summarizing the results shown in b. Vertical lines denote missense mutations and the striped bar corresponds to the minimal NID. (d) Missense mutations that cause RTT abolish the interaction between MeCP2 and NCoR/SMRT. Full-length FLAG-tagged MeCP2 containing the point mutations shown was expressed in HeLa cells, purified with antibody to FLAG and probed on western blots. (e) A biotin-tagged peptide comprising residues 285–319 of MeCP2 bound (B) to an in vitro–translated FLAG-tagged N-terminal domain of NCoR1 (197–598) and SMRT (187–586) and to full-length TBL1 and TBLR1, but failed to bind the C-terminal domain of NCoR1 (2,000–2,337). Interactions were all greatly reduced or abolished by the R306C mutation. WT, wild type. All blots in this figure are representative of at least two biological replicate experiments involving recovery based on protein affinity from independent mouse brains, HeLa cell transfections or in vitro translation reactions. Uncropped blots are presented in Supplementary Figure 8.
NCoR and SMRT were previously found to interact with MeCP2, but the binding site was not defined10,11. By immunopurifying exogenously expressed FLAG-tagged MeCP2 deletion fragments from HeLa cells, we found that only amino acids 269–309 of MeCP2 were necessary for binding to components of NCoR/SMRT (Fig. 2b,c). As the 269–309 domain contains the 302–306 cluster of missense RTT mutations, we tested each mutant for NCoR/SMRT subunit binding and found that the MeCP2P302R, MeCP2K304E, MeCP2K305R and MeCP2R306C mutations each abolished this association (Fig. 2d). Binding to SIN3A was unaffected by these mutations and did not depend on this region (Fig. 2b,d). To determine the region of NCoR/SMRT that interacts with MeCP2, we coexpressed overlapping fragments of the subunits of this complex as FLAG-tagged polypeptides with EGFP-MeCP2 in HeLa cells. Following immunopurification using antibodies to GFP, both TBL1 and an N-terminal region of NCoR1 were found to interact with MeCP2 (Supplementary Fig. 3). A peptide comprising residues 285–319 of MeCP2 bound directly to in vitro–translated N-terminal regions of NCoR1 and SMRT and their shared homodimeric subunits TBL1 and TBLR1 (ref. 9), further supporting multiple MeCP2 binding sites on NCoR/SMRT complexes. An MeCP2R306C mutation abolished the interaction of this peptide with NCoR/SMRT components (Fig. 2e). Taken together, these results define an NCoR/SMRT interaction domain (NID) of MeCP2.
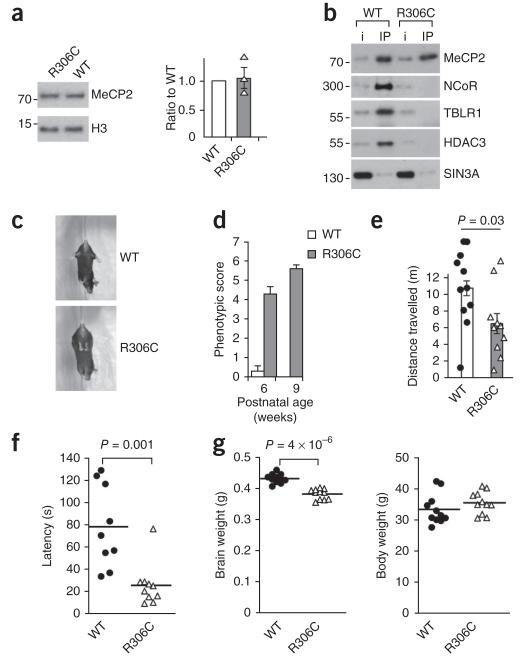
To assess the biological relevance of the NID, we generated a mouse bearing the most frequent mutation in this domain, MeCP2R306C, which accounts for approximately 5% of all classical RTT cases. The expression level of MeCP2R306C was indistinguishable from that in wild-type brain extracts (Fig. 3a). Immunoprecipitation of MeCP2 in extracts from littermate wild-type and mutant brains revealed that MeCP2R306C did not interact with NCoR/SMRT components (Fig. 3b). By postnatal week 6, these mice developed a severe phenotype resembling that of Mecp2-null mice12. We used an established scoring method that allows assessment of phenotypic features in unison, rather than singly13. Impairments regarding general condition, mobility, hindlimb clasp and tremor (Fig. 3c,d) were apparent, leading to a high aggregate score in independent cohorts aged 6 and 9 weeks. More specifically, we also observed significant defects in performance with respect to distance traveled in an open field (P = 0.03; Fig. 3e) and latency to fall from an accelerating rotarod (P = 0.001; Fig. 3f). We conclude that, as in Mecp2-null mice, mobility and motor coordination were both substantially compromised by the MeCP2R306C mutation. RTT patients often present with a reduced head circumference, and reduced brain weight has been observed in Mecp2-null mice14. This feature was recapitulated in Mecp2R306C mice, which displayed an 11% reduction in brain weight, but no change in body weight, when compared with age-matched control mice (Fig. 3g). Notably, Mecp2R306C knock-in mice also showed an early death phenotype, with 12 of 27 males (44%) failing to survive beyond 18.5 weeks. This combination of phenotypic features has been reported in Mecp2-null mice.
Figure 3.
Mice with an Mecp2R306C knock-in allele lose the MeCP2-NCoR/SMRT interaction and exhibit a severe neurological phenotype. (a) Representative western blot of whole brain wild-type MeCP2 and MeCP2R306C from 5-week-old mice. Histone H3 is a control for equal loading. The histogram shows the ratio of MeCP2R306C to wild-type protein (1.05) averaged over three biological replicates based on separate protein preparations from independent pairs of mouse brains (s.e.m. = 0.17). The ratios from individual pairs of brains are also plotted. (b) Extracts from brain nuclei of 5-week-old wild-type and Mecp2R306C mice were immunoprecipitated with antibody to MeCP2 and the resulting western blots probed with antibodies to the NCoR/SMRT components NCoR1, TBLR1 and HDAC3. The weak interaction with the SIN3A co-repressor was not affected by this mutation. Blots are representative of two biological replicate experiments involving separate immunopurifications from independent mouse brain samples. Uncropped blots are presented in Supplementary Figure 8. (c) Mice expressing knocked-in Mecp2R306C showed a hindlimb clasping phenotype at 6 weeks of age. (d) Phenotypic scoring (see Online Methods) was performed blind to genotype on littermates from two independent cohorts of mice at postnatal ages of 6 (n = 11) and 9 (n = 12) weeks. Phenotypic scores are mean ± s.e.m. (e) Distance traveled in an open field during a 30-min interval was measured and the data were analyzed for wild-type (n = 9) and Mecp2R306C (n = 10) 15–18-week-old littermates. Plotted data points correspond to individual mice. (f) Latency to fall from an accelerating rotarod was measured and the data analyzed for wild-type (n = 9) and Mecp2R306C (n = 10) 15–18-week-old littermates. Plotted data points correspond to individual mice. (g) Brain weight (left) and whole body weight (right) were determined for wild-type (n = 11) and Mecp2R306C (n = 11) 15–18-week-old mice. Plotted data points correspond to individual mice. The P values shown in e–g were obtained by two-sided t test.
The genetic data suggest that the inability to recruit NCoR/SMRT co-repressors is highly deleterious. Compatible with this notion, we found that a published allele of the mouse Mecp2 gene, which causes a relatively mild phenotype15, shows intermediate binding to NCoR/SMRT. The Mecp21–308 allele terminates at the C-terminal edge of the NID and immunoprecipitated reduced amounts of NCoR/SMRT in both transfected HeLa cells (Fig. 2b and Supplementary Fig. 4) and extracts from Mecp21–308 mouse brain (Supplementary Fig. 4). Although missing the C-terminal third of the protein, Mecp21–308 is not a reported RTT mutation and ~90% of male mice with this allele survive beyond 1 year. We propose that the absence of a severe phenotype in Mecp21–308 mice is a result of the retention of binding to NCoR/SMRT co-repressors, albeit at a reduced level.
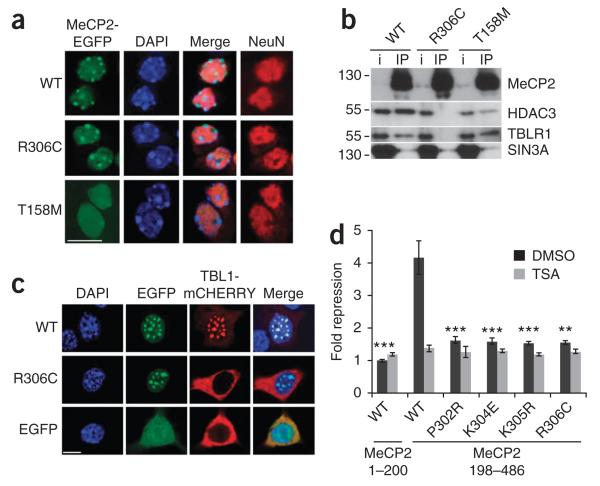
We visualized the chromatin binding of MeCP2 in neurons derived from embryonic stem (ES) cells expressing EGFP-tagged MeCP2 (Supplementary Fig. 1). In addition to wild-type Mecp2, we replaced the endogenous gene with two common RTT mutations16: one in the NID (MeCP2R306C) and one in the MBD (MeCP2T158M). Wild-type, Mecp2R306C and Mecp2T158M knock-in ES cells yielded neurons with high efficiency, as assessed by NeuN staining (Fig. 4a). The MeCP2R306C mutant and wild-type proteins correctly localized to highly methylated heterochromatic foci6, whereas MeCP2T158M was distributed diffusely as expected of a DNA binding mutant (Fig. 4a). Conversely, both MeCP2T158M and wild-type MeCP2 interacted with NCoR/SMRT, whereas MeCP2R306C failed to bind. The MeCP2-SIN3A interaction was unaffected by the MeCP2R306C mutation (Fig. 4b). We conclude that the MeCP2T158M and MeCP2R306C mutations inactivate either the MBD or the NID of MeCP2.
Figure 4.
Mutations in either the MBD or NID interfere with localization and transcriptional repression by MeCP2. (a) Mouse ES cells carrying knocked-in wild-type Mecp2, Mecp2T158M or Mecp2R306C expressed as a fusion with EGFP differentiated to >85% neurons as measured by NeuN staining. DAPI-stained spots correspond to heterochromatin rich in 5-methylcytosine. Scale bar represents 10 μm. (b) Immunoprecipitation with antibody to EGFP from neuronal nuclear extracts of the three genotypes. MeCP2-bound endogenous proteins were assayed on western blots. i, input. Uncropped blots are presented in Supplementary Figure 8. (c) Wild-type MeCP2-EGFP recruited the shared NCoR/SMRT subunit TBL1-mCherry to densely methylated heterochromatic foci in mouse fibroblasts (top row), but the MeCP2R306C NID mutation prevented recruitment (middle row). Data is from duplicate experiments showing efficient TBL1 recruitment in 55% of cells expressing wild-type MeCP2-EGFP (n = 219) compared with 0% recruitment by both MeCP2R306C-EGFP (n = 106) and EGFP alone (n = 100). Scale bar represents 10 μm. (d) N-terminal and C-terminal halves of MeCP2 were fused to GAL4 DNA-binding domains and co-transfected with a luciferase reporter gene containing GAL4-binding sites. The effect of RTT missense mutations on luciferase expression was monitored (dark gray bars). TSA, trichostatin A. Error bars represent s.e.m. on the basis of 4–9 replicate experiments. **P < 0.01 and ***P < 0.001, compared with repression by the wild-type C-terminal half of MeCP2 (two-sided t test).
To test whether MeCP2 can recruit NCoR/SMRT components to DNA, we used a cell-imaging strategy. TBL1 lacks a canonical nuclear localization signal, and a TBL1-mCherry fusion protein expressed in mouse fibroblasts accumulated in the cytoplasm. In the presence of exogenous MeCP2-EGFP, TBL1-mCherry relocated to densely methylated nuclear foci. In contrast, MeCP2R306C-EGFP targeted nuclear foci, but did not colocalize with TBL1 (Fig. 4c). We conclude that MeCP2 can recruit NCoR/SMRT to methylated DNA in vivo. Colocalization of NCoR/SMRT with MeCP2 across the genome could not be confirmed. Detection of the dispersed MeCP2 profile by chromatin immunoprecipitation (ChIP) depends on its high abundance17, but we found that HDAC3 was ~300-fold less abundant than MeCP2 in brain (Supplementary Fig. 5a). In addition, formaldehyde cross-linking abolished the interaction of MeCP2 with NCoR/SMRT (Supplementary Fig. 5b), further complicating conventional ChIP analysis.
As NCoR/SMRT complexes are co-repressors, we tested the effect of NID mutations on transcriptional silencing. A C-terminal fragment of MeCP2 repressed transcription of a reporter gene (Supplementary Fig. 6), but missense RTT mutations that prevent binding to NCoR/SMRT greatly reduced this activity (Fig. 4d). Trichostatin A, an HDAC inhibitor, relieved repression by MeCP2, demonstrating that silencing requires a catalytic activity known to be associated with NCoR/SMRT complexes (Fig. 4d).
DISCUSSION
We report, to the best of our knowledge, the first example of a protein-protein interaction that is disrupted by mutations causing RTT. Our findings explain the presence of a discrete group of RTT mutations in the C-terminal half of MeCP2 that disrupt the NID, a surface that interacts with the NCoR/SMRT co-repressor complexes. Together with the cluster of MBD mutations, which often disrupt DNA binding, these amino acid substitutions account for most of the missense mutations that cause this disorder. The paucity of missense mutations elsewhere in the protein, coupled with the relative abundance of neutral polymorphic amino acid substitutions in other domains, emphasizes the importance of these interactions in preventing this clinical condition. It is notable that weak binding to SIN3A was not disrupted by NID mutations, questioning the relevance of this co-repressor interaction for RTT.
For the majority of human genetic diseases, mutations involving deamination of cytosine in a CG context are the most frequent18. One of the mutations in the NID, MeCP2R306C, is of this type, and accounts for >200 RTT cases, or ~5% of the total19. Mice in which the wild-type allele of Mecp2 was replaced with Mecp2R306C lost the interaction between MeCP2 and NCoR/SMRT in the brain. Accordingly, the mice exhibited a RTT-like phenotype. Based on initial phenotypic analysis, the severity of the R306C phenotype resembled that of Mecp2-null mice, as behavioral defects were fully penetrant at 6 weeks of age and approximately half of the mice failed to survive beyond 20 weeks. It is possible that future direct comparison on a homogeneous genetic background will reveal further differences that may be informative, although the large number of clinical cases already attests to the consequences of this single amino acid change19.
Correlation of specific RTT mutations with clinical severity has been hindered by the heterogeneity of this disorder, as, even among patients with the same mutation, symptom severity varies greatly. By combining data from many patients, however, a subtle genotype-phenotype correlation is discernable for the most common RTT mutations16. According to this ranking, MeCP2R306C is more severe on average than MeCP2R133C, but somewhat less severe than MeCP2T158M, MeCP2R168X and MeCP2R255X. It is noteworthy that a mouse model carrying MeCP2T158A (ref. 20) shows destabilization of the mutated MeCP2 protein, whereas no such destabilization was observed for the MeCP2R306C mutation (Fig. 3a). Thus, it is possible that weak residual functions of the intact MeCP2R306C protein slightly mitigate the severity of this mutation in humans.
On the basis of the genetic and biochemical data, a simple, but testable, working model is that loss of the DNA-MeCP2-NCoR/SMRT bridge is a common feature of most or all cases of RTT (Supplementary Fig. 7). The majority of nonsense and frameshift RTT mutations fit with this proposal, as they eliminate the NID and/or the MBD. Potentially incompatible with the model, however, are RTT cases involving C-terminal truncations that would potentially leave both domains intact. A requirement of the bridge model is that these truncations either destabilize MeCP2 protein, leading to its degradation, or cause abnormal protein folding that interferes with NID and/or MBD function. Other models are also compatible with the data. For example, the activity of NCoR/SMRT co-repressor complexes recruited to chromatin by other proteins may be regulated via NID-mediated binding of MeCP2. Future work is required to assess these possible roles.
MeCP2 has been implicated in several biological processes, including activation5 and repression8 of transcription, control of alternative splicing21, regulation of global chromatin structure22,23 and control of protein synthesis24. Our data suggest that co-repressor recruitment to DNA is a core MeCP2 function that is disturbed in RTT. Could the loss of this bridge compromise brain function by preventing transcriptional repression, as suggested by earlier experiments2,8? Gene expression analyses in Mecp2-null brains have revealed numerous potentially deleterious changes, but these are not confined to the increases in transcription that might be expected following the loss of a repressor. Numerous examples of decreased gene expression have also been observed6. Alternatively, elevated transcription of repetitive DNA in Mecp2-null brains suggests that MeCP2 can globally inhibit transcriptional noise17. Specifically, the transcription and transposition of L1 elements is elevated in MeCP2-deficient mice25. A further possibility is that dynamic changes in MeCP2 phosphorylation triggered by neuronal activity modulate MeCP2-mediated silencing of specific genes in the brain26. Indeed, a recent study27 found that the NCoR/SMRT interaction described here is blocked by activity-dependent phosphorylation of MeCP2 at a nearby amino acid residue (T308). Switching of the MeCP2–co-repressor interaction in this way may provide regulatory flexibility whose loss through mutation could contribute to aberrant brain function. These and other potential roles will need to be explored in future attempts to understand the molecular basis of RTT.
ONLINE METHODS
Mutation analysis
Missense mutations in the RettBASE data set28 were extracted. Mutations found in healthy individuals or in male patients were excluded. Mutations were then considered verified if at least one report confirmed the mutation to be de novo by analysis of DNA from both parents.
Protein expression
Human MeCP2 fragments and mouse NCoR/SMRT complex component fragments were cloned into p3xFLAG-CMV-10 (Sigma). Plasmid pEGFP-C1-MeCP2 was the same as used previously29. HeLa cells in 15-cm dishes were transfected using JetPei (Polyplus Transfection) and harvested after 24 h.
Nuclear extracts
Mouse brain nuclei were isolated as described previously30. Nuclei from HeLa cells or cultured neurons were obtained by Dounce homogenization in NE10 buffer (20 mM HEPES (pH 7.5), 10 mM KCl, 1 mM MgCl2, 0.1% Triton X-100 (vol/vol), protease inhibitors (Roche), 15 mM β-mercaptoethanol) followed centrifugation for 5 min at 500 g. Nuclei were washed in NE10 buffer and then incubated at 25 °C for 5 min with 250 units benzonase (Sigma) per 107 nuclei. Nuclei were resuspended in NE150 buffer (NE10 supplemented with 150 mM NaCl), except in the case of EGFP-H2B extraction, where NE300 buffer was used. After mixing for 20 min, lysates were cleared by centrifugation at 16,000 g for 20 min and supernatants were used for subsequent protein purifications. Where applicable, cells were subjected to formaldehyde cross-linking as described previously29 before the preparation of nuclear extracts.
Protein purification
Proteins were captured by 30 min mixing with either the GFP-Trap_A (Chromotek)31 or with antibody to FLAG M2 affinity gel (Sigma), and, after four washes with NE150 (NE300 for purification of GFP-H2B for mass spectrometry), proteins were eluted with SDS-PAGE sample buffer or 0.5 mg ml−1 3× FLAG peptide (Sigma). The use of 150 mM rather than 300 mM NaCl was necessary to preserve the interaction between MeCP2 and NCoR/SMRT.
Protein identification
Proteins were identified by mass spectrometry using an LTQ-Orbitrap mass spectrometer (Thermofisher Scientific). Proteins were only considered if they were identified by two or more peptides in both independent MeCP2-EGFP purifications. Proteins were discarded if they were identified by one or more peptides in either of the two purifications from wild-type brains lacking fused GFP or in the single purification from H2B-GFP brains32.
Antibodies
For immunoblotting, we used antibodies to MeCP2 (Sigma, M6818 and M7443), NCoR1 (Bethyl, A301-146A), SMRT (Bethyl, A301-147A), TBLR1 (Bethyl, A300-408A), HDAC3 (Santa Cruz, sc-11417), HDAC3 (Sigma, 3E11), Sin3A (Bethyl, A300-726A) and FLAG (Sigma, F3165) (all at a 1:1,000 dilution). We visualized proteins by chemiluminescence and quantitative infrared imaging (LI-COR Odyssey, LI-COR Biosciences). For immunofluorescence, we used antibody to NeuN (Millipore, MAB377) at 1:100 dilution. For immunoprecipitation, we employed 5 μg of antibody to MeCP2 (Sigma, M6818) or 4 μl of homemade anti-MeCP2 crude serum (674)8.
Peptide binding assay
Peptides corresponding to amino acids 285–319 of wild-type and mutant MeCP2 were synthesized with an N-terminal biotin tag (Thermo). FLAG-tagged TBL1 and fragments of NCoR were produced by in vitro translation using the TNT Quick system (Promega). Complexes were captured with streptavidin sepharose (GE Healthcare) and after four washes with NE150, bound proteins were eluted with SDS-PAGE sample buffer.
Knock-in of Mecp2 alleles
Targeting vectors to create EGFP-tagged alleles for wild-type Mecp2, Mecp2T158M and Mecp2R306C were constructed12 using a 7.2-kb plasmid subclone of mouse 129/Ola genomic DNA, including Mecp2 exon 3 and part of exon 4 (Supplementary Fig. 1). The coding sequence of EGFP was fused in-frame to the end of the coding sequence of Mecp2 in exon 4 followed by a loxP-flanked NeoStop cassette33 as a selectable marker, retaining the first 1.8 kb of the Mecp2 3′ UTR. Point mutations T158M and R306C were introduced into the wild-type Mecp2-EGFP targeting vector. Linearized constructs were electroporated into 129/Ola E14 TG2a mouse ES cells and correctly targeted clones identified by PCR screening and Southern blotting. The loxP-flanked selection cassette was removed by electroporation of targeted clones with pCAGGS-Cre plasmid34, with deleted clones identified initially by their G418 sensitivity, followed by Southern blotting. Mice expressing MeCP2 with a C-terminal EGFP tag (Mecp2-EGFP) were generated from ES cells by standard procedures. Mice carrying the knocked-in Mecp2R306C allele without any tag were generated by homologous recombination in ES cells essentially as described previously35.
Phenotypic analysis
All mice were group housed on a 7-h:7-h light-dark cycle and were given food and water ad libitum. The mice tested were males and were of first generation backcrossed to C57Bl/6. For habituation, mice were brought into the room 30 min before all tests. The progression of the experiments was phenotypic scoring and then open field followed by rotarod with at least 2 d rest between testing. Phenotypic scoring was performed blind to genotype on littermate males with or without the Mecp2R306C allele according to published procedures13. Open field activity was measured in a 42- × 42- × 42-cm plexiglass box. The box was illuminated at 250 lx and wrapped around all four sides with opaque bench paper. Each mouse was placed into the box for 30 min, and their movement was recorded with a low-light camera. Total distance traveled was analyzed using tracking software (Ethovision XT, Noldus). For motor coordination testing, each mouse was placed on an accelerating rotarod (ENV-577M, Med Associates) set to 4.0–40 r.p.m. over a period of 5 min. A fall was called either when a mouse fell from the rod or rotated twice without recovery, with a 300 s maximum latency. Each mouse was given three trials at 1-h intervals. All experiments with mice were approved by the Animal Care and Use Committee of Harvard Medical School.
Neuronal cultures
We produced neurons from ES cells using a modified version of published protocols36,37. ES cells were cultured in Petri dishes in the absence of leukemia inhibitory factor for 8 d. The medium was changed every 2 d and 5 μM retinoic acid was added after 4 d. The resulting embryoid bodies were treated with trypsin and cells were then resuspended in DMEM/F-12 medium with N2 supplement (Invitrogen) before being passed through a 40-μm cell strainer (Falcon) and plated in dishes coated with poly-l-ornithine hydrobromide (Sigma) and laminin (Roche). After 24 h, the medium was replaced with a 50:50 mixture of N2 medium and Neurobasal medium with B27 supplement (Invitrogen). After every 3 d, half of the medium was removed and replaced with Neurobasal/B27 medium. Cells were harvested 8 d after plating. We performed two independent neuronal differentiation and observed equivalent results on both occasions.
Repression assays
NIH-3T3 cells in 24-well format were transfected using JetPei with the following amounts of plasmid: 10 ng GAL4 DBD-MeCP2 (ref. 2), 1 μg pEGFP-C1, 100 ng pRL-TK and 1 μg TK-Firefly (containing 5 GAL4 UAS sites; Supplementary Fig. 6). The use of limiting amounts of MeCP2 was critical to reveal the failure of repression by RTT mutants. Specifically, we found that commonly used concentrations of reporter constructs (1 μg per transfection) gave repression for all mutant forms, suggesting that the expressed protein was in large excess. Titration revealed that ~100-fold lower concentrations still gave effective repression with wild-type, but not mutant, forms of MeCP2. We propose that overexpression of R306C masked its defective repression in previous assays38. Where indicated 50 ng ml−1 TSA (Sigma) was applied. After 48 h, cells were harvested and reporter gene expression was quantified using the Dual-Luciferase reporter assay system (Promega). Transfection efficiencies were normalized using Renilla luciferase levels. Fold repression of the Firefly luciferase reporter was calculated relative to a sample without MeCP2.
Statistical methods
No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those generally employed in the field. Data distribution was assumed to be normal but this was not formally tested. We determined statistical significance using the t test procedure.
Supplementary Material
ACKNOWLEDGMENTS
We thank Harrison Gabel for advice and materials, and Martha Koerner, Thomas Clouaire and Sabine Lagger for comments on the manuscript. The work was supported by a grant to A.B. and M.E.G. from the Rett Syndrome Research Trust and by grants from the Wellcome Trust (to A.B.) and the NIH R01NS048276 (to M.E.G.). D.H.E. was supported by NIH grant K08MH90306. The Mouse Gene Manipulation Facility of the Boston Children’s Hospital Intellectual and Developmental Disabilities Research Center (IDDRC) was supported by grant NIHP30-HD 18655. R.E. and J.N. were funded by Wellcome Trust 4 year PhD studentships and J.R. holds a Wellcome Trust Senior Fellowship.
Footnotes
Note: Supplementary information is available in the online version of the paper.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG–binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 2.Nan X, et al. Transcriptional repression by the methyl-CpG–binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 3.Lewis JD, et al. Purification, sequence and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 4.Kudo S, et al. Heterogeneity in residual function of MeCP2 carrying missense mutations in the methyl CpG binding domain. J. Med. Genet. 2003;40:487–493. doi: 10.1136/jmg.40.7.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol. 2011;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- 7.Yusufzai TM, Wolffe AP. Functional consequences of Rett syndrome mutations on human MeCP2. Nucleic Acids Res. 2000;28:4172–4179. doi: 10.1093/nar/28.21.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nan X, Campoy J, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 9.Watson PJ, Fairall L, Schwabe JW. Nuclear hormone receptor co-repressors: structure and function. Mol. Cell Endocrinol. 2012;348:440–449. doi: 10.1016/j.mce.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kokura K, et al. The Ski protein family is required for MeCP2-mediated transcriptional repression. J. Biol. Chem. 2001;276:34115–34121. doi: 10.1074/jbc.M105747200. [DOI] [PubMed] [Google Scholar]
- 11.Stancheva I, Collins AL, Van den Veyver IB, Zoghbi H, Meehan RR. A mutant form of MeCP2 protein associated with human Rett syndrome cannot be displaced from methylated DNA by notch in Xenopus embryos. Mol. Cell. 2003;12:425–435. doi: 10.1016/s1097-2765(03)00276-4. [DOI] [PubMed] [Google Scholar]
- 12.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 13.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 15.Shahbazian M, et al. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 16.Schanen C, et al. Phenotypic manifestations of MECP2 mutations in classical and atypical Rett syndrome. Am. J. Med. Genet. A. 2004;126A:129–140. doi: 10.1002/ajmg.a.20571. [DOI] [PubMed] [Google Scholar]
- 17.Skene PJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper DN, Krawczak M. The mutational specturm of single base-pair substitutions causing human genetic disease: patterns and predictions. Hum. Genet. 1990;85:55–74. doi: 10.1007/BF00276326. [DOI] [PubMed] [Google Scholar]
- 19.Fyfe S, Cream A, de Klerk N, Christodoulou J, Leonard H. InterRett and RettBASE: International Rett Syndrome Association databases for Rett syndrome. J. Child Neurol. 2003;18:709–713. doi: 10.1177/08830738030180100301. [DOI] [PubMed] [Google Scholar]
- 20.Goffin D, et al. Rett syndrome mutation MeCP2 T158A disrupts DNA binding, protein stability and ERP responses. Nat. Neurosci. 2012;15:274–283. doi: 10.1038/nn.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young JI, et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc. Natl. Acad. Sci. USA. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 23.Agarwal N, et al. MeCP2 Rett mutations affect large scale chromatin organization. Hum. Mol. Genet. 2011;20:4187–4195. doi: 10.1093/hmg/ddr346. [DOI] [PubMed] [Google Scholar]
- 24.Ricciardi S, et al. Reduced AKT/mTOR signaling and protein synthesis dysregulation in a Rett syndrome animal model. Hum. Mol. Genet. 2011;20:1182–1196. doi: 10.1093/hmg/ddq563. [DOI] [PubMed] [Google Scholar]
- 25.Muotri AR, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Z, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebert DH, et al. Activity-dependent phosphorylation of MeCP2 T308 regulates interaction with NCoR. Nature. 2013 doi: 10.1038/nature12348. doi:10.1038/nature12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christodoulou J, Grimm A, Maher T, Bennetts B. RettBASE: The IRSA MECP2 variation database: a new mutation database in evolution. Hum. Mutat. 2003;21:466–472. doi: 10.1002/humu.10194. [DOI] [PubMed] [Google Scholar]
- 29.Schmiedeberg L, Skene P, Deaton A, Bird A. A temporal threshold for formaldehyde crosslinking and fixation. PLoS ONE. 2009;4:e4636. doi: 10.1371/journal.pone.0004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klose RJ, Bird AP. MeCP2 behaves as an elongated monomer that does not stably associate with the Sin3a chromatin remodelling complex. J. Biol. Chem. 2004;279:46490–46496. doi: 10.1074/jbc.M408284200. [DOI] [PubMed] [Google Scholar]
- 31.Rothbauer U, et al. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y, Matevossian A, Huang HS, Straubhaar J, Akbarian S. Isolation of neuronal chromatin from brain tissue. BMC Neurosci. 2008;9:42. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dragatsis I, Zeitlin S. A method for the generation of conditional gene repair mutations in mice. Nucleic Acids Res. 2001;29:E10. doi: 10.1093/nar/29.3.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Araki K, Araki M, Miyazaki J, Vassalli P. Site-specific recombination of a transgene in fertilized eggs by transient expression of Cre recombinase. Proc. Natl. Acad. Sci. USA. 1995;92:160–164. doi: 10.1073/pnas.92.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen S, et al. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72:72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bain G, Kitchens D, Yao M, Huettner JE, Gottlieb DI. Embryonic stem cells express neuronal properties in vitro. Dev. Biol. 1995;168:342–357. doi: 10.1006/dbio.1995.1085. [DOI] [PubMed] [Google Scholar]
- 37.Li M, Pevny L, Lovell-Badge R, Smith A. Generation of purified neural precursors from embryonic stem cells by lineage selection. Curr. Biol. 1998;8:971–974. doi: 10.1016/s0960-9822(98)70399-9. [DOI] [PubMed] [Google Scholar]
- 38.Drewell RA, Goddard CJ, Thomas JO, Surani MA. Methylation-dependent silencing at the H19 imprinting control region by MeCP2. Nucleic Acids Res. 2002;30:1139–1144. doi: 10.1093/nar/30.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.