Abstract
Under various physiological and patho-physiological conditions, spectrin breakdown reactions generate several spectrin breakdown products (SBDPs) - in particular SBDP of 150 kDa (SBDP150) and of 120 kDa (SBDP120). Recently, numerous studies have shown that reactions leading to SBDPs are physiologically relevant, well regulated, and complex. Yet molecular studies on the mechanism of the SBDP formation are comparatively scarce. We have designed basic systems to allow us to follow the breakdown of αII-spectrin model proteins by caspase-3 in detail with gel electrophoresis, fluorescence and mass spectrometry methods. Amongst the predicted and reported sites, our results show that caspase-3 cleaves after residues D1185 and D1478, but not after residues D888, D1340 and D1475. We also found that the cleavage at these two sites are independent of each other. It may be possible to inhibit one site without affecting the other site. Cleavage after residue D1185 in intact αII-spectrin leads to SBDP150, and cleavage after D1478 site leads to SBDP120. Our results also show that the cleavage after the D1185 residue is unusually efficient, with a kcat/KM value of 40,000 M−1 sec−1, and the cleavage after the D1478 site is more similar to most of the other reported caspase-3 substrates, with a kcat/KM value of 3,000 M−1 sec−1. We believe that this study lays out a methodology and foundation to study caspase-3 catalyzed spectrin breakdown to provide quantitative information. Molecular understanding may lead to better understanding of brain injuries and more precise and specific biomarker development.
Keywords: nonerythroid (brain) spectrin, caspase-3, spectrin breakdown products, brain injury biomarkers
1. Introduction
Caspase-3 is the most abundant caspase in cells (around 200 nM), with its activity often dominant over the activities of other members in the caspase family (Denault and Salvesen, 2001). While many proteins are cleaved during apoptosis, a prominent target of caspase-3 action is αII- and βII-spectrin, also known as fodrin (Moon and McMahon, 1990). During apoptosis, spectrin is the substrate for caspase-3 in lymphocytes, hematopoietic cells and neurons (Janicke et al., 1998; Machnicka et al., 2012; Wang et al., 1998). The spectrin breakdown products, SBDPs, characterized and named by their electrophoretic masses, are found to be SBDP150 and SBDP120 (Cryns et al., 1996; Li et al., 2010a; Martin et al., 1995). In the last few years, numerous studies report detecting SBDPs in several brain related injuries, such as traumatic brain injury (d'Avella et al., 2002; Beer et al., 2000; Hall et al., 2005; Hyman and Yuan, 2012; Kupina et al., 2003; Newcomb et al., 1997; Pike et al., 2003), mechanical stretch injury (Pike et al., 2000), ischemia (Nath et al., 1998; Zhang et al., 2002), and degenerative brain diseases, such as Alzheimer's Disease, as well as other age-related degenerative diseases (Cotman et al., 2005; Hyman and Yuan, 2012; Marx, 2001). Furthermore, SBDP120, or its antibody activity, has been shown in the salivary gland, and plays a crucial role as an auto-antigen in the development of primary Sjögren's Syndrome (Haneji et al., 1997). SBDP120 is also found in the sera of patients with normal tension glaucoma (Grus et al., 2008). There is also increased evidence for non-apoptotic roles of caspase-3 in the brain, including the regulation of synaptic plasticity (Machnicka et al., 2012) and of neuronal morphology via local remodeling of the spectrin cytoskeleton (Westphal et al., 2010). A recent “fragment generation” hypothesis suggests that Ca++ activated calpain/caspase fragmentation of spectrin, as well as of other proteins, plays a role in sleep since the fragmentation process decreases during sleep while fragment-destroying pathways are up-regulated (Varshavsky, 2012). Furthermore, it has been shown that cleavages of specific neuronal proteins play a role in long-term memory (Li et al., 2010b; Lynch and Baudry, 1984; Shimizu et al., 2007). Thus, caspase-3 catalyzed spectrin breakdown reactions appear to be physiologically important, well regulated, complicated and sometimes confusing. SBDPs have been proposed to be potential biomarkers for neurodegenerative diseases (Yan and Jeromin, 2012). These SBDPs are often separated by gel electrophoresis and detected by monoclonal antibody immunoblotting (Pike et al., 2003). Due to the low mass resolution of gel electrophoresis method, many SBDPs with similar masses may appear as a single breakdown product. It is generally considered that SBDP150 is the C-terminal fragment of αII-spectrin after caspase-3 cleavage, and a subsequent cleavage of SBDP150 gives SBDP120 (Wang et al., 1998). The potential use of SBDPs as biomarkers in monitoring brain injuries or diseases or in elucidating their potential involvement in long-term memory requires a molecular level understanding of spectrin breakdown reactions and of SBDPs. Yet molecular and quantitative studies of these reactions are comparatively rare. Without quantitative molecular information, it is difficult to study the reaction mechanisms as well as factors affecting spectrin breakdown reactions, as in regulation or inhibition, for example. It will also be difficult to develop accurate and selective biomarkers/antibodies that do not exhibit cross-reactivity to other SBDP.
There are multiple predicted and reported caspase-3 cleavage sites in the middle of αII-spectrin (after residues D888, D1185, D1340, D1475 and D1478 (Wang et al., 1998; Dix et al., 2008; noted there D1475 as D1483 due to a different amino acid numbering system; Versputen et al., 2009; Song et al., 2010; Ayyash et al., 2012). In this study, we followed the αII-spectrin breakdown reactions catalyzed by caspase-3 that yield SBDP150 and SBDP120. We have developed simple reproducible systems of model αII-spectrin proteins representing the mid-section of αII-spectrin containing the residues that have been reported to be cleavage sites. We used five αII-spectrin fragments of specific lengths and recombinant caspase-3 to follow the breakdown reactions with fluorescence, gel electrophoresis and mass spectrometry methods. The gel electrophoresis methods allow us to compare our breakdown reactions with those published. Using mass spectrometry analysis, we unequivocally identified two cleavage sites (after D1185 and after D1478, but not after D1475). With model proteins including only one of the two sites and with a protein including both sites, we showed that the cleavages at these two sites are independent of each other. Using fluorescence methods, we obtained the first kinetic information on these cleavage reactions. The kinetic data showed that these two sites exhibited very different caspase-3 catalytic rates. The cleavage after D1185, leading to SBDP150 in intact αII-spectrin (or 45 kDa fragment in our model protein D10-D13), was unusually rapid but not necessarily essential to the cleavage after D1478, leading to SBDP120, and a SBDP of 37 kDa (SBDP37) from intact αII-spectrin (or 10 kDa and 37 kDa from D10-D13). We applied the method to a mutated (D1185E) model protein, and found that the mutant protein was cleaved by caspase-3, but with a much reduced kcat/KM value. We believe that this study lays out a methodology and foundation for quantitatively studying caspase-3 catalyzed spectrin breakdown. A better molecular understanding may lead to better understanding of brain injuries and more precise and specific biomarker development.
2. Results
2.1. Protein Characterization
2.1.1. αII-spectrin
The model proteins representing specific segments of αII-spectrin were designed not only to include the sites cleaved by caspase-3, but also to resemble the folding of spectrin structural domains (triple helical bundles), as indicated in the Experimental section. These model protein samples, D8-D11, D10-D11, D10-D13, D12-D13 and D13 (Table 1), were more than 90% pure, as indicated by SDS gel electrophoresis data. As noted in our previous publication (Lusitani et al., 1994), spectrin fragments with proper boundaries consisting of full triple helical bundle structural domain(s) are stable and represent well the full-length spectrin of particular regions for functional studies (Mehboob et al., 2010).
TABLE 1.
The mass predicted from sequence (massseq), obtained from gel electrophoresis (massele) and obtained from high-resolution mass spectrometry methods (massms) of αII-spectrin model proteins (see text for protein identity) used in the study (left aligned, in bold). The first set of fragments obtained from caspase-3 cleavage (center aligned) and the fragments from a subsequent cleavage of a fragment, if any, (right aligned) are also listed.
| Protein | Sequence | Massseq (Da) | Massele (kDa) | Massms (Da) |
|---|---|---|---|---|
| D8-D11 | L780-F1344 | 62,358.4 | 66 | 62,357.6 |
| L780-D1185 | 44,171.3 | 46 | ||
| S1186-F1344 | 18,205.0 | 19 | ||
| D10-D11 | L1087-F1344 | 29,830.0 | 36 | 29,829.9 |
| L1087-D1185 | 11,642.9 | 17 | ||
| S1186-F1344 | 18,205.0 | 19 | ||
| D10-D13 | L1087-F1556 | 54,265.5 | 62 | 54,265.6 |
| L1087-D1185 | 11,642.9 | 17 | 11,642.2 | |
| S1186-F1556 | 42,640.5 | 45 | ||
| S1186-D1478 | 33,690.3 | 37 | 33,689.8 | |
| S1479-F1556 | 8,968.2 | 10 | 8,967.7 | |
| D10-D13(D1185E) | L1087-F1556 | 54,279.5 | 62 | 54,279.8 |
| L1087-D1478 | 45,329.3 | 53 | 45,330.1 | |
| L1087-E1185 | 11,657.0 | 17 | 11,655.7 | |
| S1186-D1478 | 33,690.3 | 37 | ||
| S1479-F1556 | 8,968.2 | 10 | 8,967.8 | |
| D12-D13 | L1335-F1556 | 25,766.8 | 28 | 25,766.1 |
| L1335-D1478 | 16,816.6 | 17 | ||
| S1479-F1556 | 8,968.2 | 10 | 8,967.8 | |
| D13 | L1441-F1556 | 13,550.1 | 15 | 13,549.9 |
| L1441-D1478 | 4,600 | 5 | 4,599.1 | |
| S1479-F1556 | 8,968.2 | 10 | 8,967.8 |
Sequence alignment of residues L1087 to F1238 (designated as Domain 10) aligned with residues L40 to Y154 in Domain 1 (Mehboob et al., 2010), with 22% sequence identity and 41% similarity (Fig 1A). In this alignment, the region V1167-H1200 did not align with the sequence in the template, and is an insertion. However, when the insertion region is removed, the alignment scores improved to 29% identity and 52% similarity (Fig 1B). Secondary structural prediction showed Helix A (following the nomenclatures in previous publication, Mehboob et al., 2010; each structural domain in spectrin consists of Helices A, B and C to form a triple helical bundle) in Domain 10 as residues K1092-L1114, Helix B as residues V1126-E1160, followed by an unusually long, 40-residue loop (residues G1161-H1200) (Fig 1C) and then Helix C as residues T1201-G1230 (Fig 1B). It should be noted that the two conserved tryptophan residues found in Helices A and C in most triple helical bundles were aligned with those in Domain 1. More importantly, these predictions showed that the residue D1185, one of the reported cleavage site, was in the middle of this long loop (Fig 1C) between Helix B and Helix C. The COILS program predicted regions similar to those mentioned above as coiled helices except for the region consisting of L1162-F1205. This region exhibited a very low probability (<3%) of forming coiled helix, whereas 20 residues prior to this region, for example, exhibited very high probability (>90%) of forming coiled helix. The PONDR analysis of the region from G1161 to P1191 predicted this region to be disordered. Thus, these structural analyses showed that Domain 10 in αII-spectrin was folded into a triple helical bundle similar to other structural domains in spectrin, but with a very long loop between Helices B and C, and D1185 was in the middle of this long loop (V1167-H1200 ). Previous studies of a fragment consisting of residues 1172-1213 show unstructured prior binding to calmodulin, and residues 1191-1210 folds into helical conformation upon binding to calmodulin (Simonovic et al., 2006).
Figure 1.
Secondary structure predictions. (A) Sequence alignment of D10 (L1087-F1238, See Table 1) with domain 1 (L40-Y154), with D10 exhibiting 22% sequence identity and 41% similarity. The D1185 residue (bold and double underline) is located in the middle of the long, 34-residue segment (V1167-H1200, dotted line) that does not align with D1. Two conserved tryptophan residues found in Helices A and C are shown in grey. (B) D10Δ, excluding residues from V1167-H1200, sequence aligned with domain 1 exhibiting 29% sequence identity and 52% similarity. The high sequence homology with domain 1 allows us to predict that Helix A in D10 as residues K1092-L1114 (underlined), Helix B as residues V1126 - E1160, and Helix C as residues T1201-G1230. Also shown is the sequence alignment of D13 (L1441-F1556) with D1. The alignment shows 28% identity and 49% similarity between D13 and D1. We assigned L1445-F1467 in D13 as Helix A (underlined), V1480-A1514 as Helix B, and K1519-G1548 as Helix C. The D1478 residue (bold, and double underline) is at the end of the segment connecting Helix A and Helix B. (C) A 40-residue segment (G1161 -H1200) located between Helix B and Helix C in D10.
For Domain 13, the sequence alignment was relatively straightforward, with L1441-F1556 aligned with L40-Y154 to give 28% identity and 49% similarity, and we assigned L1445-F1467 as Helix A, V1480-A1514 as Helix B, and K1519-G1548 as Helix C in Domain 13 (Fig 1B). Again the conserved tryptophan residues were also found in Domain 13. The other reported D1475 and D1478 cleavage sites were found at the end of the 12-residue (LNTEDKGD*SLD*S) loop between Helix A and Helix B. In summary, simple structural analyses suggested that Domain 10 folds into a triple helical coiled coil structure as in other spectrin domains, but with a very long unstructured loop between Helix B and Helix C, with the D1185 residue in the middle of this long loop. D13 was also predicted to fold into a triple helical coiled coil structure, but with D1475 and D1478 residues at the end of a shorter loop between Helix A and Helix B. Obviously, the actual structures of these two domains await experimental data.
The CD spectra showed that the helical contents of αII-spectrin model proteins were between 60 - 70% (spectra not shown), in good agreement with previously published values for similar (triple helical bundles) spectrin model proteins (Mehboob et al., 2010), suggesting that the proteins used were well folded into a triple helical domain. It should be mentioned that an SH3 domain (residues 965-1025) is nested in Domain 9 (Musacchio et al., 1992; Rotter et al., 2004).
2.1.2. Caspase-3
The CD spectra for the recombinant caspase-3 prepared for this study showed two minima with similar signal amplitudes at 208 and 215 nm (spectra not shown), in good agreement with published spectra (Pop et al., 2001). Using the substrate Z-DEVD-AFC, the kcat value of our caspase-3 was 10.6/sec, with KM 16 μM, and kcat/KM 662,500 M−1sec−1, values in good agreement with published values (kcat of 11.63/sec, KM of 21.1 μM, and kcat/KM value of 551,342 M−1sec−1) where a similar substrate, Ac-DEVD-AFC, was used (Timmer et al., 2009).
2.2. Predicted Cleavage sites in αII-spectrin by Caspase-3
Three of prediction algorithms (See Experimental section) showed D1185 and D1478 sites with high scores (Table 2). An observed D1475 site (Dix et al., 2008) was predicted to be the third highest by one method (CAT3, Table 23). One prediction (SitePrediction, Table 23) scored the D888 site higher than the D1475 site while the D1340 site was scored as high as the D1478 site, with the D888 site just slightly lower (Cascleave, Table 23). However, when we used the mutated sequence for prediction by these algorithms, the score for E1185 site was much lower than those for the D888, D1340, or D1475 sites (Table 23). Yet we did not observe any cleavage at these sites in D1185E mutant. It appears that our current understanding of caspase-3 cleavage site preferences is still limited and could not be reliably used to predict cleavage sites, at least for spectrin, without experimentation.
TABLE 2.
Caspase/Caspase-3 cleavage site predictions on αII-spectrin (L780-V1560) using algorithms from Cascleave (Song et al., 2010), CAT3 (Caspase Analysis Tool 3) (Ayyash et al., 2012), and SitePrediction (SP) (Versputen et al., 2009) servers.
| Predicted Structural Features | P1 Res # | P4 - P4′ | WT - D1185 | D1185E | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cascleaveb | CAT3c | SPc | Cascleaveb | CAT3c | SPc | |||||
| Domain | Helix | Loop | Scored | Scoree | Scoref | Scored | Scoree | Scoref | ||
| 8 | A | 795 | DVED*EETW | 0.85 | 9 | 3 | 0.75 | 9 | 3 | |
| 8-9 | CA | 888 | DLED*SLQA | 1.04 | 31 | 137 | 0.83 | 31 | 137 | |
| 9 | AB | 917 | GSTD*YGKD | 0.37 | 3 | 5 | 0.57 | 3 | 5 | |
| 9 | AB | 923 | KDED*SAEA | 0.28 | 1 | 3 | 0.03 | 1 | 3 | |
| 9 | B | 936 | LMSD*LSAY | 0.37 | - | 1 | 0.01 | - | 1 | |
| SH3 | 977 | ALYD*YQEK | 0.00 | - | 0 | 0.22 | - | 0 | ||
| 9 | C | 1050 | EQID*NQTR | 0.67 | 2 | 5 | 0.63 | 2 | 5 | |
| 10 | BC | 1185 | DETD*SKTA | 1.10 | 98 | 3012 | 0.06 | 0 | 6 | |
| 11 | B | 1281 | FERD*LAAL | 0.22 | - | 2 | 0.34 | - | 2 | |
| 11 | C | 1309 | SAED*LQEK | 0.07 | - | 1 | 0.10 | - | 1 | |
| 11-12 | CA | 1340 | DSHD*LQRF | 1.05 | 31 | 8 | 1.12 | 31 | 8 | |
| 12 | A | 1350 | DFRD*LMSW | 0.81 | 21 | 2 | 0.87 | 21 | 2 | |
| 12 | AB | 1365 | VSSD*ELAK | 0.17 | 3 | 1 | 0.21 | 3 | 1 | |
| 12 | B | 1389 | TEID*ARAG | 0.94 | 11 | 21 | 1.04 | 11 | 21 | |
| 12 | C | 1424 | DILD*QERA | 0.70 | 21 | 2 | 0.72 | 21 | 2 | |
| 13 | AB | 1475 | DKGD*SLDS | 0.70 | 46 | 22 | 0.65 | 46 | 22 | |
| 13 | AB | 1478 | DSLD*SVEA | 1.05 | 76 | 940 | 1.03 | 76 | 940 | |
| 13 | B | 1481 | DSVE*ALIK | 0.07 | - | 2 | 0.03 | - | 2 | |
| 13 | B | 1491 | EDFD*KAIN | 0.22 | - | 1 | 0.42 | - | 1 | |
| 13 | C | 1531 | EVLD*RWRR | 0.02 | 3 | 0 | 0.23 | 3 | 0 | |
astructural predictions based on the amino acid sequence alignment with domain 1 (see results for details)
for all caspases
specific to caspase-3
Cascleave - cleavage probability score
CAT3 - score
SitePrediction - average score.
2.3. Cleavage of αII-spectrin Proteins by Caspase-3
2.3.1. Gel Electrophoresis and Mass Spectrometry Studies
For the sample D10-D11 in the presence of caspase-3, the band at 36 kDa clearly decreased as [casp-3] increased above 0.5 nM (Fig 2A). No significant changes were detected in samples with lower [casp-3] = 0, 0.005, 0.01, and 0.05 nM. Two bands at 17 and 19 kDa were clearly detected with [casp-3] = 3 nM. The band intensity plot as a function of [casp-3] (log scale for presentation convenience) (Fig 2B) showed the gradual disappearance of D10-D11 upon increasing [casp-3]. The [E]1/2 value determined from this plot was about 3 nM. The average value of several runs was 2.9 ± 0.6 nM (n = 3), and the corresponding average value of kcat/KM was found to be 67,700 ± 13,200 M−1sec−1 (Table 3).
Figure 2.
Caspase-3 proteolysis of αII-spectrin model protein D10-D11 and D8-D11. (A) A typical SDS-PAGE for the substrate D10-D11 (1 μM) in the presence of various [casp-3] (0, 0.005, 0.01, 0.05, 0.5, 10, 50, 100, and 500 nM). Each sample of D10-D11 and caspase-3 was incubated at 37 °C for 1 hr precisely before gel electrophoresis runs (see text). Polyacrylamide gel (4-20% in Tris-HEPES-SDS, from Pierce) was used. See Table 1 for the masses of the proteins and fragments obtained from sequence, gel electrophoresis and mass spectrometry methods. Molecular markers are shown in the left most lane. At low concentrations of caspase-3, the 36 kDa (D10-D11) band is clearly seen. For samples with higher concentrations of caspase-3, bands at 17 and 19 kDa are detected. For the sample consisting of 500 nM caspase-3, the bands of caspase-3 at 17 kDa (corresponding to a mass of 16,614.8 Da) and 13 kDa (12,960.7 Da) are also visible. Similar caspase-3 bands at 500 nM were observed in samples without D10-D11(data not shown). The insets are gels with [casp-3] = 0.05, 0.5, 1, 3, 5, 7, 8, 9, and 10 nM. (B) The intensities of the 36 kDa band in (A), converted to fractions of that without caspase-3 (%) (solid circle), at different [casp-3] show the disappearance of D10-D11 at [casp-3] > 0.05 nM. The value of [casp-3] to give 50% of D10-D11 (dash line), [E]1/2, was read from the plot directly with the plotting software (Origin) as 3.3 nM. The kcat/KM value was then determined with the half-life equation kcat/KM = ln2/t [E]1/2 (see text) with t = 1 hr or 3600 sec, as 58,346 M−1 sec−1. The open circle symbols correspond to the data for D10-D11'w used in fluorescence studies. The [E]1/2 value for D10-D11'w, from the plot, is 4.0 nM to give a kcat/KM value of 48,135 M−1 sec−1. (C) A typical SDS-PAGE for the substrate D8-D11 (1 μM). Details are similar to (A). (D) The intensities of the 66 kDa band in (C) at different [casp-3] were converted to fractions of that without caspase-3 (%) (solid diamond). The [E]1/2 value from the plot is 9.0 nM with a kcat/KM value of 21,393 M− sec−.
TABLE 3.
Equilibrium ([E]1/2 = [Caspase-3] to give 50% cleavage of an αII-spectrin model protein) and kinetic (kcat/KM) results of caspase-3 cleavage at a specific site in αII-spectrin model proteins, as detected by gel electrophoresis and tryptophan fluorescence methods. See text for details in experimental set-up. The number of runs to give the listed average values and standard deviations was 3.
| Cleavage Site | Proteins | [E]1/2 (nM) | kcat/KM (M−1sec−1) | |
|---|---|---|---|---|
| Electrophoresis | Electrophoresis | Fluorescence | ||
| D1185 | D10-D11 | 2.9 ± 0.6 | 67,700 ± 13,200 | |
| D8-D11 | 7.7 ± 1.4 | 25,400 ± 4,300 | ||
| D10-D13a | 4.3 ± 1.1 | 47,200 ± 15,000 | ||
| D10-D11'w | 4.2 ± 0.5 | 46,800 ± 3,900 | 35,000 | |
| E1185 | D10-D13(D1185E) | 172.7 ± 27.0 | 1,100 ± 180 | |
| D1478 | D13 | 57.8 ± 3.1 | 3,300 ± 200 | |
| D12-D13 | 62.2 ± 10.6 | 3,100 ± 500 | ||
| D10-D13b | 58.2 ± 10.1 | 3,400 ± 650 | ||
| D10-D13(D1185E)c | 73.0 ± 22.8 | 2,800 ± 900 | ||
| D13'w | 2,900 | |||
D10-D13 protein also consisted of the D1478 site.
D10-D13 protein also consisted of the D1185 site.
D10-D13(D1185E) protein also consisted of the E1185 site.
For the larger model protein, D8-D11 (66 kDa), two bands at 19 and 46 kDa were also clearly detectable at [casp-3] = 3 nM (Fig 2C). From the 66 kDa-band intensity plot (the disappearance of D8-D11) as a function of [casp-3] (Fig 2D), the [E]1/2 value was found to be about 10 nM. The average value was 7.7 ± 1.4 nM (n = 2), which corresponded to kcat/KM of 25,400 ± 4,300 M−1 sec−1 (Table 3).
For the model protein D13 (15 kDa), only one product band was detected at the 10-kDa position, with a smaller fragment presumably migrating off the gel (Fig 3A). As [casp-3] increased, the intensity of the 10 kDa band was seen to increase, while that of the parent band at 15 kDa decreased (Fig 3A). From the 15 kDa-band intensity plots (a typical plot is shown in Fig 3B), the average value of [E]1/2 value for the disappearance of D13 was found to be 57.8 ± 3.1 nM (n = 3), corresponding to a kcat/KM value of 3,300 ± 200 M−1sec−1. Mass spectrometry analysis of the cleaved products showed two fragments of 8,967.8 and 4,599.1 Da (Table 1) indicating that D13 protein was cleaved after residue D1478 (Wang et al., 1998; Dix et al., 2008) and not after D1475 (Dix et al., 2008) since the C-terminal fragment of the D1475 site cleavage, S1476-F1556 with an expected mass from the sequence as 9,283.5 Da, was not found in our mass spectrometry results.
Figure 3.
Caspase-3 proteolysis of αII-spectrin model proteins D13 and D12-D13. (A) A typical SDS-PAGE for the substrate D13 (1 μM) in the presence of various [casp-3] with conditions similar to those in Fig 2(A). See Table 1 for the masses of proteins and fragments obtained from sequence, gel electrophoresis and mass spectrometry methods. Molecular markers are shown in the left most lane. At low concentrations of caspase-3, the 15 kDa (D13) band is clearly seen. For higher concentrations of caspase-3, a band at 10 kDa is detected. For the sample consisting of 500 nM caspase-3, the bands of caspase-3 at 17 kDa (corresponding to a mass of 16,614.8 Da) and 13 kDa (12,960.7 Da) are also visible. (B) The intensities of the 15 kDa band in (A), converted to fractions of that without caspase-3 (%) (right-half shaded circle), at different [casp-3] show the disappearance of D13 at [casp-3] > 100 nM. The [casp-3] value to give 50% fraction (dash line) of D13 ([E]1/2) was read from the plot directly with the plotting software (Origin) to give a value of 57.2 nM. The kcat/KM value was then determined to be 3,366 M−1 sec−1. (C) A typical SDS-PAGE for the substrate D12-D13. Details are similar to (A). (D) The intensities of the 28 kDa band in (C) at different [casp-3], converted to fractions of that without caspase-3 (%), as a function of [casp-3] (left-half shaded hexagon). The [E]1/2 value from the plot is 55.6 nM with a kcat/KM value of 3,463 M−1 sec−1.
Upon cleavage of the larger D12-D13 protein (28 kDa), two fragments (10 and 17 kDa) were generated (Fig 3C, [casp-3] = 50 nM, for example). From the band intensity plots (e.g., Fig 3D), the average [E]1/2 value for the disappearance of D12-D13 was found to be 62.2 ± 10.6 nM (n = 2), corresponding to a kcat/KM value of 3,100 ± 500 M−1sec−1. These values were very similar to those found for the D13 protein ([E]1/2 of 57.8 nM and kcat/KM of 3,300 M−1sec−1, Table 32).
Finally, the D10-D13 protein (62 kDa) (Fig 4A) generated various fragments upon caspase-3 cleavage. For [casp-3] = 0.5 nM, fragments of 17 and 45 kDa appeared (Fig 4A). When [casp-3] >10 nM, the 45 kDa band began to disappear while two other fragments at 10 and 37 kD appeared. The mass spectrometry analysis of the final three cleaved products (10, 17 and 37 kDa; Table 1) indicated that D10-D13 protein was cleaved after residue D1185 and after D1478, as reported (Wang et al., 1998), but not after D1475, as reported (Dix et al., 2008). A typical plot of the relative intensity of the 62 kDa-band (Fig 4B) showed an [E]1/2 of about 3 nM. The average [E]1/2 value was 4.3 ± 1.2 nM (n = 3) (Table 3). With this value, the kcat/KM was found to be 47,200 ± 15,000 M−1sec−1. The [E]1/2 value for the appearance of 37 kDa was about 58.2 ± 10.1 nM (n = 3) and the kcat/KM value was 3,400 ± 650 M−1sec−1. For comparison, the relative intensity plot of D10-D11 (Fig 2B) was also plotted in Fig 4B to show that caspase-3 cleaved D10-D11 similarly to the D1185 site cleavage in D10-D13, with comparable [E]1/2 and kcat/KM values (Table 3).
Figure 4.
Caspase-3 proteolysis of αII-spectrin model proteins D10-D13 and D10-D13(D1185E). (A) A typical SDS-PAGE for the substrate D10-D13 (1 μM) in the presence of various [casp-3], with conditions similar to Fig 2(A). See Table 1 for the masses of the proteins and fragments obtained from sequence, gel electrophoresis and mass spectrometry methods. Molecular markers are shown in the left most lane. At low [casp-3] values, the 62 kDa (D10-D13) band is clearly seen in each lane. For higher [casp-3] values, bands at 45, 37, 17, and 10 kDa are detected. Again, for the sample consisting of 500 nM caspase-3 or higher, the bands of caspase-3 at 17 kDa and 13 kDa are also visible. The inset gels are with [casp-3] = 0, 10, 50, 100, 500, 700, 1000, 1200 and 1500 nM. (B) The intensities of the 62 kDa band in (A), converted to fractions of that without caspase-3 (%), at different [casp-3] (solid stars) show the disappearance of D10-D13 at [casp] > 10 nM. The [E]1/2 value was read from the plot directly with the plotting software to give a value of 3.1 nM with a kcat/KM value of 62,110 M−1sec−1. Also plotted are the appearance a 45 kDa fragment (solid triangle). When the [casp-3] >10 nM, the concentration of the 45 kDa fragment decreases. A fragment at 37 kDa band (upside down left-half shaded triangle) becomes visible at [casp-3] ~ 10 nM, and its concentration continues to increase as [casp-3] increases. For comparison, the relative intensity plot of D10-D11 (solid circle) from Fig 2(A) was also plotted. (C) A typical SDS-PAGE for the substrate D10-D13(D1185E), with conditions similar to (A). (D) The intensities of the 62 kDa band in (C) at different [casp-3], converted to fractions of that without caspase-3 (%) as a function of [casp-3] (open star). The [E]1/2 value from the plot is 57.4 nM with kcat/KM value of 3,354 M−1sec−1. Also plotted are the appearance of a 53 kDa fragment as a function of [casp-3] (open triangle). When the [caspase-3] >100 nM, the concentration of the 53 kDa fragment decreases. The 37 kDa band (upside down open triangle) becomes visible at [casp-3] ~ 100 nM, and its concentration continues to increase as [casp-3] increases.
The kcat/KM values for D10-D11 and D8-D11 agreed well with the values of the D1185 site cleavage for D10-D13, and the values for D13 and D12-D13 agreed well with the values of the cleavage, after the D1478 site for D10-D13. This finding indicated that the efficiency of caspase-3 cleavage of these two sites were very different, and the two cleavages were independent of each other.
For the D10-D13(D1185E) mutant, its 62 kDa band intensity decreased at much higher values of [casp-3] and the first set of fragments detected were 10 and 53 kDa (Fig 4C and D). The [E]1/2 value was 73.0 ± 22.8 nM (n = 2) (Fig 4D), and the kcat/KM value was 2,800 ± 900 M−1sec−1. These values were very similar to those found in the D13 protein ([E]1/2 of 57.8 nM and kcat/KM of 3,300 M− sec−1), and in wild-type D10-D13 ([E]1/2 of 58.2 nM and kcat/KM of 3,400 M−1sec−1) (Table 3). The 53 kDa fragment was further cleaved into two fragments (17 and 37 kDa) at [casp-3] > 50 nM. The average [E]1/2 value for this cleavage (from the appearance of 37 kDa plot) was 172.7 ± 27.0 nM (n = 3) with a corresponding kcat/KM of 1,100 ± 180 M−1sec−1 (Table 3). The efficiency for the E1185 site cleavage was lower than that for the D1478 site. The mass spectrometry analysis of the cleaved products indicated that D10-D13(D1185E) was cleaved after E1185 and D1478 to give 10, 17, and 37 kDa fragments (Table 1). Again cleavage after D1475 (Dix et al., 2008) was not observed in this mutant, suggesting that cleavage at this site, if occurring under the conditions we used, would be even less efficient than at the E1185 site.
2.3.2. Fluorescence Studies
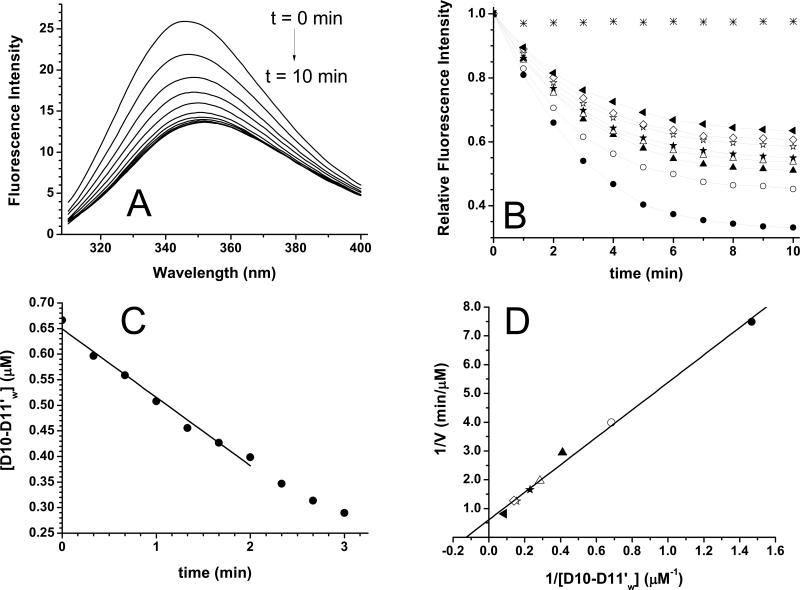
To measure the rates of caspase-3 cleavage of the various recombinant αII-spectrin model proteins, we modified the model proteins such that their tryptophan fluorescence signal was sensitive to caspase cleavage (see Experimental section). The fluorescence intensities at 347 nm of D10-D11'w-1 and D10-D11'w-2 samples (see the Experimental section for the identities of these proteins) showed no change with the addition of caspase-3, at either t = 0 or t = 10 min at 37 °C (data not shown). However, the intensity of the D10-D11'w sample (at ~ 0.7 μM, with [casp-3] = 100 nM) decreased as a function of time (Fig 5A). Similarly, the intensity of the D13'w sample (at ~ 0.9 μM, with [casp-3] = 700 nM) also decreased as a function of time (Fig 6A). No intensity decreases in both samples occurred in the absence of caspase-3 (Figs 5B and 6B). Thus D10-D11'w and D13'w appeared to be good model proteins to be used to follow the kinetics of caspase-3 cleavage at D1185 and D1478 sites, respectively. We also checked the caspase-3 cleavage of D10-D11'w by gel electrophoresis and found the average value for [E]1/2 to be 4.2 ± 0.5 nM with a kcat/KM of 46,800 ± 3,900 M−sec− (Fig 2B; Table 3), very similar to the values for D10-D11. Thus, tryptophan-residue replacement in this protein did not appear to affect the caspase-3 cleavage.
Figure 5.
Cleavage efficiency at D1185 site. (A) Intrinsic tryptophan fluorescent spectra of a sample D10-D11'w (see the Methods and Table 1) at 0.7 μM with [casp-3] = 100 nM at 37 °C show that the intensity decreases as a function of time (at 1 min interval), with slight λmax shift, from 347 to 351 nm. The excitation wavelength was 295 nm. (B) Relative intensities at 347 nm of the spectra in (A), of [D10-D11'w] = 0.7 μM (solid circles), as a function of time, with the intensity at t = 0 as 1. Also shown are those for [D10-D11'w] = 2.4 (open circles), 3.5, 4.4, 6.5, 7.1, and 11.7 μM (solid triangle). The data for D10-D11'w without caspase-3 (*) show no decrease in intensity as a function of time. (C) A linear decrease of [D10-D11'w], converted from the intensities at 347 nm of the sample in (A) at a time interval of 20 sec, was found for the first 2 min. The linear fit (R2 = 0.98) gives a slope, or the initial rate V, of 0.13352 μM/min. (D) The Lineweaver-Burk (1/V vs 1/[D10-D11'w]) plot of the data from (B). The Vmax value from the y-intercept was 1.4 μM/min, the KM value from the x-intercept was 6.3 μM, and the kcat from Vmax/[casp-3] was 14/min, or 0.23/sec. The kcat/KM value was then 37,000 M−1sec−1.
Figure 6.
Cleavage efficiency at D1478 site. (A) Intrinsic tryptophan fluorescent spectra of D13'w (see the Methods; Table 1) at 0.9 μM with [casp-3] = 700 nM at 37 °C show decreasing intensity as a function of time (at 1 min interval), with slight λmax shift, from 347 to 349 nm. The excitation wavelength was 295 nm. (B) Relative intensities at 347 nm of the spectra in (A), with the intensity at t = 0 as 1 (solid circles, [D13'w] = 0.9 μM). Also shown are those for [D13'w] = 1.8 (open circles), 2.6, 3.5, 4.7, 5.5, 6.5, 7.4, and 8.2 μM (solid triangle). The data for D13'w without caspase-3 (*) show no decrease in intensity as a function of time. (C) A linear decrease of [D13'w], converted from the intensities at 347 nm of the sample in (A) at a time interval of 20 sec, was found for the first 2 min. The linear fit (R2 = 0.99) gives a slope, or the initial rate V, of 0.08759 μM/min. (D) The Lineweaver-Burk (1/V vs 1/[D13'w]) plot of the data from (B). The Vmax from the y-intercept was 0.5 μM/min, the KM from the x-intercept was 3.8 μM and the kcat from Vmax/[casp-3] was 0.7/min, or 0.01/sec. The kcat/KM value was then 2,900 M−1sec−1.
A plot of the relative fluorescence intensity at 347 nm as a function of time for samples of D10-D11'w at a series of concentrations (from ~ 0.7 to 11.7 μM), each with [casp-3] = 100 nM, showed corresponding decreases in intensities (Fig 5B). The initial cleavage rate (V) for D10-D11'w at 0.7 μM, determined from the slope of the linear fit of the data in Fig 5C, was 0.1335 μM/min. The V values at other concentrations of D10-D11'w were determined, and the Lineweaver-Burk plot (1/V vs 1/[D10-D11'w]) (Fig 5D) showed that the data exhibited linear dependence, indicating that the kinetics of caspase-3 cleavage of D10-D11'w followed Michaelis-Menten kinetics. The Vmax from the y-intercept was 1.6 μM/min, with a KM from the x-intercept of 7.8 μM, and a kcat from Vmax/[casp-3] of 16/min, or 0.27/sec. The kcat/KM value was then 35,000 M−sec−1 (Table 3). This value agreed well with the gel electrophoresis finding of 25,000 - 68,000 M−1sec−1 for cleavage at the D1185 site.
Similarly, Figure 6B shows a series of curves for D13'w samples with concentrations ranging from 0.9 to 8.2 μM, and [casp] = 700 nM, each with intensity decreasing as a function of time. The V value obtained from Fig 6C for [D13'w] = 0.9 μM was 0.08759 μM/min. The Lineweaver-Burk plot (Fig 6D) was again linear, demonstrating Michaelis-Menten kinetics for caspase cleavage of D13'w.The Vmax value was 0.7 μM/min, with the KM 7.3 μM, and the kcat of 1/min. The kcat/KM value was then 2,900 M−1sec−1 (Table 32). This value agreed well with the gel electrophoresis finding of 2,800 - 3,400 M−1sec−1 for the cleavage at the D1478 site, values much lower that those for the cleavage at the D1185 site.
3. Discussion
It has been reported that the rank-order of substrate cutting in lysate is similar to that in apoptotic cells, suggesting that cellular structures do not dramatically alter the substrate accessibility (Agard et al., 2012), providing justification and confidence in using model proteins to study SBDPs in cells. In this study, we showed that intact αII-spectrin was well represented by our αII-spectrin model proteins, in the breakdown reactions catalyzed by caspase-3. Our results from αII-spectrin model proteins of five different lengths that contained various reported and predicted caspase-3 cleavage sites (Ayyash et al., 2012; Dix et al., 2008; Song et al., 2010; Versputen et al., 2009; Wang et al., 1998), individually or together, clearly showed two specific cleavage sites in αII-spectrin, one cleaved after the D1185 residue (D1185 site), and another after the D1478 residue (D1478 site), but not after D888 (predicted), D1340 (predicted), and D1475 (reported) residues. We found the kcat/KM value for the D1185 site to be 40,000 M−1sec−1 and that for the D1478 site to be 3,000 M−1sec−1. Recent proteomic studies (Agard et al., 2012) find that only 2% of substrate proteins studied exhibit efficiency between 40,000 - 70,000 M−1sec−1, and most (76%) substrate proteins are less than 5,000 M−1sec−1. Thus, for αII-spectrin, the D1185 site is an unusually efficient site, whereas the D1478 site is more similar to most protein substrates, with an average efficiency/rate.
It has been suggested that the caspase-3 cleavage of αII-spectrin proceeds in a two-step mechanism, first at the D1185 site to cause conformational changes to expose the D1478 site for subsequent cleavage (Wang et al., 1998; Williams et al., 2003). This suggestion is not compatible with an ex vivo experiment, in which the D1478 cleavage occurs during apoptosis independently of the D1185 site, since proteins with the D1185 site deleted were still cleaved after the D1478 residue (Meary et al., 2007). Our results quantitatively and unequivocally showed that the cleavage of these two sites were independent of each other; with cleavage after the residue D1478 occurring even when the D1185 region was not yet cleaved, as in the D1185E mutant. We observed a kcat/KM value of ~ 1,000 M−1 sec−1 for the D1185E mutant. Thus, genetic abnormalities, such as point mutations or deletions, at the D1185 cleavage site would change the SBDP profiles, shifting the first cleavage site to D1478, with only SBDP120, and no SBDP150 detected.
Our findings indicate that, for intact αII-spectrin in the presence of caspase-3, both spectrin breakdown products, SBDP150 and SBDP120, will form. However, due to the difference in the catalytic rates, the amount of SBDP120 is much lower than that of SBDP150 since the accumulation of SBDP120 is much slower than that of SBDP150. Only a very sensitive method will be able to detect SBDP120 in the presence of SBDP150. Further cleavage of SBDP150 at the D1478 site produces SBDP120, as well as a SBDP37.
Our results indicate that the cleavage after residue 1185 in intact αII-spectrin gives a slightly smaller N-terminal segment (residues 1-1185; 134,563.7 Da) and a C-terminal segment (residues 1186-2452; 147,735.9 Da; the SBDP150). Cleavage after residue 1478 in intact αII-spectrin gives a larger N-terminal segment (residues 1-1478; 168,236.0 Da), harboring the D1185 site and a C-terminal segment (residues 1479-2452; 114,063.6 Da; the SBDP120). Since the rate of cleavage at the D1185 site is rapid, this N-terminal segment (residues 1-1478) will not continue to accumulate, but will subsequently be fragmented at the D1185 site, with high efficiency, into an N-terminal segment (residues 1-1185) and a C-terminal segment (residues 1186-1478; 33,672.3 Da; the SBDP37 ). This mechanism indicates that the final SBDPs should include the N-terminal segment (134,563.7 Da), the middle segment (33,672.3 Da; SBDP37) and the C-terminal segment (114,063.6 Da; SBDP120). Little is known about the “134,563.7 Da” segment and the SBDP37.
Recent studies show that the vast majority of proteolyzed proteins yielded persistent fragments that correspond to discrete protein domains, suggesting that the generation of active effector proteins may be a principal function of apoptotic proteolytic cascades (Dix et al., 2008). Additional functional and structural studies of SBDPs will allow us to further understand both the apoptotic and non-apoptotic events, e.g., traumatic brain injury, mechanical stretch injury, ischemia, and brain diseases, such as Alzheimer's Disease, as well as other age-related degenerative diseases, Sjögren's Syndrome, normal tension glaucoma as well as the regulation of synaptic plasticity and of neuronal morphology via local remodeling of the spectrin cytoskeleton.
It is possible that the cleavages at these two sites may represent different cellular events, with SBDP150 and SBDP120 involved in different functions. It may be possible to inhibit one site, such as the efficient site, without affecting the other site. It appears that our model system is well-suited for studying the different factors affecting the caspase-3 catalyzed spectrin breakdown, from simple mutation to more complicated regulation pathways. Individual factors may be studied quantitatively to provide a better understanding of the action of caspase-3 on αII-spectrin. With knowledge of specific fragments generated under specific conditions, more precise biomarker detection methods, such as those using fragment-specific antibodies (Zhang et al., 2009) to detect specific fragments, can be developed to replace the use of an antibody directed against αII-spectrin in Western blot analysis (e.g. Weber et al., 2013). Additional specific fragments, such as the N-terminal segment (134,563.7 Da) and/or the “middle” segment (33,672.3 Da; SBDP37) mentioned above, may also be tested and developed as biomarkers. We believe that this study lays out a methodology and foundation to study caspase-3 catalyzed spectrin breakdown to provide quantitative information. Molecular understanding may lead to better understanding of brain injuries and more precise and specific biomarker development.
4. Experimental Procedures
4.1. Chemicals
Ampicillin, dithiothreitol (DTT), EDTA, 3[(3-cholamido propyl) dimethyl ammonio]-propane sulfonic acid (CHAPS), sucrose, 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), and sodium dodecyl sulfate (SDS) were obtained from Thermo Fisher Scientific (Waltham, MA). Imidazole and piperazine-N,N’-bis(2-ethanesulfonic acid) were obtained from Sigma-Aldrich (St. Louis, MO). All other reagents were either similar to those used in our previous studies of spectrin (e.g., Mehboob et al., 2010), or are specified below.
4.2. DNA Plasmids Preparation
Two plasmids from ATCC (Manassas, VA): (1) pCMV-SPORT6 vector (product no 10436486) with the cDNA of human αII-spectrin (GenBank BC053521) and (2) pET-23b vector (product no 99625) with the cDNA of pro-caspase-3 (GenBank U26943) were obtained. Five αII-spectrin plasmids, using the spectrin cDNA from the pCMV-SPORT6 vector, were cloned into pDEST-15 vector (Invitrogen, Grand Island, NY), following the methods provided by the company's user manual, to give five αII-spectrin model protein fragments in the region of residues 780-1556 (Table 1) consisting of residues (1) L780-F1344 (domain 8 to domain 11, or D8-D11. It should be noted that domain 10 is being referred to as “repeat 11” in Wang et al., 1998), (2) L1087-F1344 (D10-D11), (3) L1087-F1556 (D10-D13), (4) H1335-F1556 (D12-D13) and (5) L1441-F1556 (D13). The boundaries of these fragments were designed to ensure stable protein folding (see below). A thrombin recognition site (LVPRGS) between GST and the spectrin fragment was introduced in the primer sequence. All primers were purchased from Integrated DNA Technologies (Coralville, IA). A pDEST-15 plasmid with D1185E mutation (D10-D13(D1185E)) was also prepared, using primer-based site directed mutagenesis methods (Mehboob et al., 2010). In addition, several plasmids for mutants of D10-D11 and D13 were prepared to replace tryptophan with phenylalanine residues. In D10-D11, residues 1106, 1192, 1215, 1248 and 1321 are tryptophan. The following mutants were prepared: (1) 1106F-1215F-1248F-1321F (only residue 1192 remained as W; designated as D10-D11'w-1), (2) 1215F-1248F-1321F (residues 1106 and 1192 remained as W; D10-D11'w), (3) 1106F-1248F-1321F (residues 1192 and 1215 remained as W; D10-D11'w-2). In D13, W1533 was mutated to F, leaving only residue 1460 as W (D13'w). All DNA constructs were verified by DNA sequencing analysis (services provided by the Research Resources Center (RRC) at the University of Illinois at Chicago).
4.3. Protein Design, Expression, Purification and Characterization
In the design of the model proteins, the boundaries of each triple helical structural domain (Mehboob et al., 2010; Lusitani et al., 1994) were predicted, first using the program in the FASTA package (EMBOSS Needle) for sequence analysis to align residues 780-1556 to αII-spectrin residues 40-147, since the atomic structure of this region (domain 1) is known (Mehboob et al., 2010). Furthermore, the alignment was also guided by the fact that the 16th and 88th residues in each typical spectrin triple helical domain are tryptophan residues (Mehboob et al., 2010). After sequence alignment, the secondary structural elements (helices and loops) of αII-spectrin domain 1 were assigned to the aligned regions in D8-D13. Each model protein consisted of predicted αII-spectrin domain(s) plus the previous 4 residues and GS from thrombin cleavage to give a 6-residue overhang at the N-terminus and the 6 residues following the predicted boundary of the last structural domain to give an overhang at the C-terminus (Table 1) to enhance proper protein folding.
All plasmids, stored in DH5α E. coli cells (Zymo Research Corporation; Irvine, CA), were extracted and transformed into E. coli BL21-CodonPlus (DE3)-RIL competent cells (Agilent; Santa Clara, CA) for protein expression.
The αII-spectrin model proteins, expressed as fusion proteins with an N-terminal GST tag followed by a thrombin cleavage site, were prepared with standard procedures developed in our laboratory (Mehboob et al., 2010; Li and Fung, 2009). Caspase-3, with a C-terminal His-tag, was prepared from a pro-caspase-3 expression vector, following published methods (Wolan et al., 2009). To optimize the yield of caspase-3 from pro-caspase-3, the protein expression induction time was extended from 20 min to 3 hr (Stennicke and Salvesen, 1999). The electrophoretic mass and the purity of proteins were determined by 16% SDS-PAGE (Li and Fung, 2009). Protein concentrations were determined using absorbance values at 280 nm, with extinction coefficients determined from the sequence of each protein. The mass of each protein, including D10-D11'w and D13'w, was analyzed by mass spectrometry, with a LTQ-FT Ultra mass spectrometer, at the RRC. Only those proteins with the mass values within 1.5 Da of the expected mass values, as calculated from the sequence, were used.
The folding of each protein (10 μM in 5 mM phosphate buffer containing 150 mM sodium chloride at pH 7.4, PBS) was analyzed by circular dichroism spectroscopy methods (JASCO 810; Easton, MD), scanning from 200 to 250 nm at 20 °C with a 0.1-cm path-length. Mean residue molar ellipticity values at 222 nm were used to calculate the helical content using a value of 36,000 deg cm2dmol−1 for a 100% helical content (Mehboob et al., 2010).
Recombinant caspase-3 activity was determined in a standard “caspase activity buffer” (20 mM piperazine-N,N’-bis-(2-ethanesulfonic acid)) with 100 mM NaCl, 10 mM DTT, 1 mM EDTA, 0.1% CHAPS, and 10% sucrose at pH 7.2) with fluorogenic substrate, Z-DEVD-AFC (EMD Biosciences; Billerica, MA), following a published method (Denault and Salvesen, 2008).
4.4. Structural and Cleavage Site Predictions
4.4.1. Structure Prediction
In addition to the prediction of domain boundaries and secondary structural elements as mentioned above, the sequence of Domain 10 (D10) was also analyzed for the probability of forming coiled coil structure using COILS (Lupas et al., 1991) and the sequence of a part of D10 (L1157-E1210) was analyzed for structural dis-orderness using PONDR (Romero et al., 2001).
4.4.2. Predicting Caspase-3 Cleavage Sites
Recently, several algorithms have become available to predict caspase-3 cleavage sites. These algorithms were developed based on knowledge from structural features of some of the caspase substrates, such as preferred primary structure (sequence logo DxxD) (Dix et al., 2008, Timmer et al., 2009; Pop and Salvesen, 2009; Mahrus et al., 2008; Weber et al., 2008; Luthi and Martin, 2007; Timmer and Salvesen, 2007; Fischer et al., 2003; Nicholson, 1999; Talanian et al., 1997; Schneider and Stephens, 1990), secondary structure (unstructured loop) (Garay-Malpartida et al., 2005), solvent accessible surface (Pop and Salvesen, 2009; Xu et al., 2001), and phosphorylation at the P3 position (Dix et al., 2012), etc.
4.5. Cleavage of αII-spectrin Fragments by Caspase-3
4.5.1. SDS-PAGE Detection
A previously published gel electrophoresis method (Stennicke and Salvesen, 1999; Timmer et al., 2009), including the published caspase-3 concentrations (Timmer et al., 2009), was followed to allow for the study of a wide range of caspase-3 cleavage efficiency in αII-spectrin model proteins. Briefly, a sample, 1 μM final concentration, of each of the six αII-spectrin proteins (D8-D11, D10-D11, D12-D13, D13, D10-D13, and D10-D13(D1185E)) was incubated with various amounts of caspase-3 (final concentration ranging from 0 to 500 nM) for exactly 1 hr (t = 1 hr) at 37 °C in the “caspase activity buffer.” The reaction was terminated by boiling the sample for 10 min in the presence of the loading dye. Samples were loaded to 4 - 20% SDS-PAGE Precise Protein Gels (Thermo Fisher Scientific), and the electrophoresis was done in a running buffer (0.1 M Tris Base, 0.1 M HEPES, and 0.1% (w/v) SDS at pH 8) with constant current (50 mA). The protein staining was carried out using AcquaStain from Bulldog Bio (Portsmouth, NH). The protein band intensities at different caspase-3 concentrations ([casp-3]) were determined with Alphaimager HP (Protein Simple; Santa Clara, CA). The intensity values for samples with [casp-3] = 0.005, 0.01 and 0.05 nM were essentially the same as those without caspase-3 ([casp-3] = 0). Thus, the band intensities of these four points was averaged and set to represent the band intensity without caspase-3 (100%). The values for samples consisting of [casp-3] > 0.05 nM were converted to fractions (%) accordingly. For the samples consisting of high [casp-3], such as 500 nM, (Fig 2A and C, Fig 3A and C, and Fig 4A and C), the 13 and 17 kDa bands of caspase-3 were also visible on the gel. For band intensity analysis, the caspase-3 bands at corresponding concentrations were subtracted from each lane accordingly. A plot of the fraction of the remaining intact protein, or of the cleaved product(s), as a function of [casp-3] was made, and the value of [casp-3] corresponding to 50% of a spectrin protein disappeared/remained, or cleaved product appeared, [E]1/2, was read from the plot directly, and the apparent value for kcat/KM was then determined with the half-life equation: kcat/KM = ln2/t[E]1/2, where t = 1 hr (39).
The masses of some cleaved products were also determined by high-resolution mass spectrometry methods at the RRC (see Table 1).
4.5.2. Fluorescence Detection
In fluorescence studies, a substrate protein (αII-spectrin model proteins) with tryptophan fluorescence signal sensitive to caspase cleavage is needed. Thus, we monitored the fluorescence intensities of tryptophan mutants D10-D11'w, D10-D11'w-1, D10-D11'w-2 and D13'w (see above, “DNA Plasmids Preparation” section) at 347 nm (with excitation at 295 nm) as a function of time in the “caspase activity buffer” in the presence of caspase-3 at 37 °C. The measurements were done with a Jasco Fluorometer (FP-6200; Easton, MD). Those model proteins sensitive to caspase-3 cleavage were selected for kinetics studies. A selected protein (D10-D11'w) was also studied by SDS-PAGE method to verify that the mutation does not affect the catalytic rate.
For kinetics studies, fluorescence spectra of the selected proteins at various concentrations in the “caspase activity buffer,” incubated with caspase-3, were obtained as a function of time. The initial cleavage rate (V) at each protein concentration was measured from the linear portion of the plot of fluorescence intensity versus time. From the Lineweaver-Burk plot (1/V vs. 1/[protein]), where [protein] was the concentration of αII-spectrin model protein, we obtained 1/Vmax value from the y-intercept of the linear fit of the data and −1/KM from the x-intercept. The kcat value was obtained since kcat = Vmax/[casp-3].
OriginPro 8.5 software was used for data processing both in gel electrophoresis and in fluorescence studies.
Highlights.
spectrin breakdown products are detected under various brain conditions
kinetics studies of breakdown reactions of spectrin model proteins by caspase-3
more efficient cleavage to generate the 150 kDa fragment than the 120 kDa fragment
The formations of 150 kDa and 120 kDa fragments are independent of eachother
development of more precise biomarkers to monitor spectrin degradation
Acknowledgments
This work was supported, in part, by National Institutes of Health grants (GM68621 to LF and T32DE018381 to L. DiPietro, program director with MAW as a trainee).
Abbreviations
- casp-3
the concentration of recombinant caspase-3
- caspase activity buffer
20 mM piperazine-N,N’-bis-(2-ethanesulfonic acid) with 100 mM NaCl, 10 mM DTT, 1 mM EDTA, 0.1% CHAPS, and 10% sucrose at pH 7.2
- D8-D11
a recombinant protein consisting of residues 780-1344 of αII-spectrin plus GS as the first two residues
- D10-D11
a recombinant protein consisting of residues 1087-1344 of αII-spectrin plus GS as the first two residues
- D10-D11'w-1
D10-D11 with all its tryptophan residues except W1192 replaced with phenylalanine residues
- D10-D11'w-2
D10-D11 with all its tryptophan residues except W1192 and W1215 replaced with phenylalanine residues
- D10-D11'w
D10-D11 with all its tryptophan residues except W1106 and W1192 replaced with phenylalanine residues
- D10-D13
a recombinant protein consisting of residues 1087-1556 of αII-spectrin plus GS as the first two residues
- D12-D13
a recombinant protein consisting of residues 1335-1556 of αII-spectrin plus GS as the first two residues
- D13
a recombinant protein consisting of residues 1441-1556 of αII-spectrin plus GS as the first two residues
- D13'w
D13 with W1533F mutation
- SBDP
spectrin breakdown product
- SBDP37
the SBDP with an electrophoretic mass of 37 kDa
- SBDP120
the SBDP with an electrophoretic mass of 120 kDa
- SBDP150
the SBDP with an electrophoretic mass of 150 kDa
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agard NJ, Mahrus S, Trinidad JC, Lynn A, Burlingame AL, Wells JA. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1913–1918. doi: 10.1073/pnas.1117158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Avella D, Servadei F, Scerrati M, Tomei G, Brambilla G, Angileri FF, Massaro F, Cristofori L, Tartara F, Pozzati E, Delfini R, Tomasello F. Traumatic intracerebellar hemorrhage: clinicoradiological analysis of 81 patients. Neurosurgery. 2002;50:16–25. doi: 10.1097/00006123-200201000-00004. [DOI] [PubMed] [Google Scholar]
- Ayyash M, Tamimi H, Ashhab Y. Developing a powerful In Silico tool for the discovery of novel caspase-3 substrates: a preliminary screening of the human proteome. BMC Bioinformatics. 2012;13:1–14. doi: 10.1186/1471-2105-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer R, Franz G, Srinivasan A, Hayes RL, Pike BR, Newcomb JK, Zhao X, Schmutzhard E, Poewe W, Kampfl A. Temporal profile and cell subtype distribution of activated caspase-3 following experimental traumatic brain injury. J. Neurochem. 2000;75:1264–1273. doi: 10.1046/j.1471-4159.2000.0751264.x. [DOI] [PubMed] [Google Scholar]
- Cotman CW, Poon WW, Rissman RA, Blurton-Jones M. The role of caspase cleavage of tau in Alzheimer disease neuropathology. J. Neuropathol. Exp. Neurol. 2005;64:104–112. doi: 10.1093/jnen/64.2.104. [DOI] [PubMed] [Google Scholar]
- Cryns VL, Bergeron L, Zhu H, Li H, Yuan J. Specific cleavage of alpha-fodrin during Fas- and tumor necrosis factor-induced apoptosis is mediated by an interleukin-1beta-converting enzyme/Ced-3 protease distinct from the poly(ADP-ribose) polymerase protease. J. Biol. Chem. 1996;271:31277–31282. doi: 10.1074/jbc.271.49.31277. [DOI] [PubMed] [Google Scholar]
- Denault JB, Salvesen GS. Caspases. Curr. Protocols in Protein Sci. 2001;21:8.1–8.16. doi: 10.1002/0471140864.ps2108s26. [DOI] [PubMed] [Google Scholar]
- Denault JB, Salvesen GS. Apoptotic caspase activation and activity. Methods in Mol. Biol. 2008;414:191–220. doi: 10.1007/978-1-59745-339-4_15. [DOI] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–691. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Wang C, Okerberg E, Patricelli MP, Craves BF. Functional interplay between caspase cleavage and phosphorylation sculpts the apoptotic proteome. Cell. 2012;150:426–440. doi: 10.1016/j.cell.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Janicke RU, Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garay-Malpartida HM, Occhiucci JM, Alves J, Belizario JE. CaSPredictor: a new computer-based tool for caspase substrate prediction. Bioinformatics. 2005;21:169–176. doi: 10.1093/bioinformatics/bti1034. [DOI] [PubMed] [Google Scholar]
- Grus FH, Joachim SC, Sandmann S, Thiel U, Bruns K, Lackner KJ, Pfeiffer N. Transthyretin and complex protein pattern in aqueous humor of patients with primary open-angle glaucoma. Mol. Vis. 2008;14:1437–1445. [PMC free article] [PubMed] [Google Scholar]
- Hall ED, Sullivan PG, Gibson TR, Pavel KM, Thompson BM, Scheff SW. Spatial and temporal characteristics of neurodegeneration after controlled cortical impact in mice: more than a focal brain injury. J. Neurotrauma. 2005;22:252–265. doi: 10.1089/neu.2005.22.252. [DOI] [PubMed] [Google Scholar]
- Haneji N, Nakamura T, Takio K, Yanagi K, Higashiyama H, Saito I, Noji S, Sugino H, Hayashi Y. Identification of α-fodrin as a candidate autoantigen in primary Sjögren's Syndrome. Science. 1997;276:603–607. doi: 10.1126/science.276.5312.604. [DOI] [PubMed] [Google Scholar]
- Hyman BT, Yuan J. Apoptotic and non-apoptotic roles of caspases in neuronal physiology and pathophysiology. Nature Reviews - Neuroscience. 2012;13:395–406. doi: 10.1038/nrn3228. [DOI] [PubMed] [Google Scholar]
- Janicke RU, Ng P, Sprengart ML, Porter AG. Caspase-3 is required for α-fodrin cleavage but dispensable for cleavage of other death substrates in apoptosis. J. Biol. Chem. 1998;273:15540–15545. doi: 10.1074/jbc.273.25.15540. [DOI] [PubMed] [Google Scholar]
- Kupina NC, Detloff MR, Bobrowski WF, Snyder BJ, Hall ED. Cytoskeletal protein degradation and neurodegeneration evolves differently in males and females following experimental head injury. Exp. Neurol. 2003;180:55–73. doi: 10.1016/s0014-4886(02)00048-1. [DOI] [PubMed] [Google Scholar]
- Li J, Li X-Y, Feng DF, Pan DC. Biomarkers associated with diffuse traumatic axonal injury: Exploring pathogenesis, early diagnosis and prognosis. J. Trauma. 2010a;69:1610–1618. doi: 10.1097/TA.0b013e3181f5a9ed. [DOI] [PubMed] [Google Scholar]
- Li Q, Fung LW-M. Structural and dynamic study of the tetramerization region of non-erythroid α-spectrin: a frayed helix revealed by site-directed spin labeling electron paramagnetic resonance. Biochemistry. 2009;48:206–215. doi: 10.1021/bi8013032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jo J, Jia J-M, Lo S-C, Whitcomb DJ, Jiao S, Cho K, Sheng M. Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell. 2010b;141:859–871. doi: 10.1016/j.cell.2010.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Lusitani DM, Qtaishat N, LaBrake CC, Yu RN, Davis J, Kelley MR, Fung LW-M. The first human α-spectrin structural domain begins with serine. J. Biol. Chem. 1994;269:25955–25958. [PubMed] [Google Scholar]
- Luthi AU, Martin SJ. The CASBAH: a searchable database of caspase substrates. Cell Death Differ. 2007;14:641–650. doi: 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- Lynch G, Baudry M. The biochemistry of memory: a new and specific hypothesis. Science. 1984;224:1057–1063. doi: 10.1126/science.6144182. [DOI] [PubMed] [Google Scholar]
- Machnicka B, Grochowalska R, Boguslawska DM, Sikorski AF, Lecomte MC. Spectrin-based skeleton as an actor in cell signaling. Cell. Mol. Life Sci. 2012;69:191–201. doi: 10.1007/s00018-011-0804-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SJ, O'Brien AG, Nishioka WK, McGahon AJ, Mahboubi A, Saido TC, Green DR. Proteolysis of fodrin (non-erythroid spectrin) during apoptosis. J. Biol. Chem. 1995;270:6425–6428. doi: 10.1074/jbc.270.12.6425. [DOI] [PubMed] [Google Scholar]
- Marx J. New leads on the ‘how’ of Alzheimer's. Science. 2001;21:2192–2194. doi: 10.1126/science.293.5538.2192. [DOI] [PubMed] [Google Scholar]
- Meary F, Metral S, Ferreira C, Eladari D, Colin Y, Lecomte M-C, Nicolas G. A mutant αII-spectrin designed to resist calpain and caspase cleavage questions the functional importance of this process in vivo. J. Biol. Chem. 2007;282:14226–14237. doi: 10.1074/jbc.M700028200. [DOI] [PubMed] [Google Scholar]
- Mehboob S, Song Y, Witek M, Long F, Santarsiero BD, Johnson ME, Fung LW-M. Crystal structure of the nonerythroid α-spectrin tetramerization site reveals differences between erythroid and nonerythroid spectrin tetramer formation. J. Biol. Chem. 2010;285:14572–14584. doi: 10.1074/jbc.M109.080028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon RT, McMahon AP. Generation of diversity in nonerythroid spectrins. J. Biol. Chem. 1990;265:4427–4433. [PubMed] [Google Scholar]
- Musacchio A, Noble M, Pauptit R, Wierenga R, Saraste M. Crystal structure of Src-homology 3 (SH3) domain. Nature. 1992;359:851–855. doi: 10.1038/359851a0. [DOI] [PubMed] [Google Scholar]
- Nath R, Probert A, Jr., McGinnis KM, Wang KKW. Evidence for activation of caspase-3-like protease in excitotoxin- and hypoxia/hypoglycemia-injured neurons. J. Neurochem. 1998;71:186–195. doi: 10.1046/j.1471-4159.1998.71010186.x. [DOI] [PubMed] [Google Scholar]
- Newcomb JK, Kamfl A, Posmantur RM, Zhao X, Pike BR, Liu SJ, Clifton GL, Hayes RL. Immunohistochemical study of calpain-mediated breakdown products to alpha-spectrin following controlled cortical impact injury in the rat. J. Neurotrauma. 1997;14:369–383. doi: 10.1089/neu.1997.14.369. [DOI] [PubMed] [Google Scholar]
- Nicholson DW. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- Pike BR, Zhao X, Newcomb JK, Glenn CC, Anderson DK, Hayes RL. Stretch injury causes calpain and caspase-3 activation and necrotic and apoptotic cell death in septo-hippocampal cell cultures. J. Neurotrauma. 2000;17:283–298. doi: 10.1089/neu.2000.17.283. [DOI] [PubMed] [Google Scholar]
- Pike BR, Flint J, Dave JR, Lu X-CM, Wang KKK, Tortella FC, Hayes RL. Accumulation of caplain and caspase-3 proteolytic fragments of brain-derived αII-spectrin in cerebral spinal fluid after middle cerebral artery occlusion in rats. J. Cerebral Blood Flow & Metabolism. 2003;24:98–106. doi: 10.1097/01.WCB.0000098520.11962.37. [DOI] [PubMed] [Google Scholar]
- Pop C, Chen Y-R, Smith B, Bose K, Bobay B, Tripathy A, Franzen S, Clark CA. Removal of the pro-domain does not affect the conformation of the procaspase-3 dimer. Biochemistry. 2001;40:14224–14235. doi: 10.1021/bi011037e. [DOI] [PubMed] [Google Scholar]
- Pop C, Salvesen GS. Human caspases: activation, specificity, and regulation. J. Biol. Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins: Struct., Funct., Bioinf. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Rotter B, Kroviarski Y, Nicolas G, Dhermy D, Lecomte M-C. αII-Spectrin is an in vitro target for caspase-2, and its cleavage is regulated by calmodulin binding. Biochem. J. 2004;378:161–168. doi: 10.1042/BJ20030955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acid Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Phan T, Mansuy IM, Storm DR. Proteolytic degradation of SCOP in the hippocampus contributes to activation of map kinase and memory. Cell. 2007;128:1219–1229. doi: 10.1016/j.cell.2006.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonovic M, Zhang Z, Cianci CD, Steitz TA, Morrow JS. Structure of the calmodulin αII-spectrin complex provides insight into the regulation of cell plasticity. J. Biol. Chem. 2006;281:34333–34340. doi: 10.1074/jbc.M604613200. [DOI] [PubMed] [Google Scholar]
- Song J, Tan H, Shen H, Mahmood K, Boyd SE, Webb GI, Akutsu T, Whisstock JC. Cascleave: towards more accurate prediction of caspase substrate cleavage sites. Bioinformatics. 2010;26:752–760. doi: 10.1093/bioinformatics/btq043. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Salvesen GS. Caspases: preparation and characterization. Methods. 1999;17:313–319. doi: 10.1006/meth.1999.0745. [DOI] [PubMed] [Google Scholar]
- Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, Ghayur T, Brady KD, Wong WW. Substrate specificities of caspase family proteases. J. Biol. Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- Timmer JC, Salvesen GS. Caspase substrates. Cell Death Differ. 2007;14:66–72. doi: 10.1038/sj.cdd.4402059. [DOI] [PubMed] [Google Scholar]
- Timmer JC, Zhu W, Pop C, Regan T, Snipas SJ, Eroshkin AM, Riedl SJ, Salvesen GS. Structural and kinetic determinants of protease substrates. Nat. Struct. Mol. Biol. 2009;16:1101–1109. doi: 10.1038/nsmb.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. Augmented generation of protein fragments during wakefulness as the molecular casue of sleep: a hypothesis. Protein Sci. 2012;21:1634–1661. doi: 10.1002/pro.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verspurten J, Gevaert K, Declercq W, Vandenabeele P. SitePredicting the cleavage of proteinase substrates. Trends Biochem Sci. 2009;7:319–323. doi: 10.1016/j.tibs.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Wang KKW, Posmantur R, Nath R, McGinnis K, Whitton M, Talanian RV, Glantz SB, Morrow JS. Simultaneous degradation of αII- and βII-spectrin by caspase-3 (CPP32) in apoptotic cells. J. Biol. Chem. 1998;273:22490–22497. doi: 10.1074/jbc.273.35.22490. [DOI] [PubMed] [Google Scholar]
- Weber I, Fang B, Agniswamy J. Caspases: structure-guided design of drugs to control cell death. Mini-Reviews in Medicinal Chemistry. 2008;8:1154–1162. doi: 10.2174/138955708785909899. [DOI] [PubMed] [Google Scholar]
- Weber H, Muller L, Schult C, Sparmann G, Schuff-Werner P. Calpain mediates caspase-dependent apoptosis initiated by hydrogen peroxide in pancreatic acinar AR42J cells. Free Radic Res. Epub ahead of print. PMID. 2013:23495712. doi: 10.3109/10715762.2013.785633. [DOI] [PubMed] [Google Scholar]
- Westphal D, Sytnyk V, Schachner M, Leshchyns'ka I. Clustering of the neural cell adhesion molecule (NCAM) at the neuronal cell surface induces caspase-8 and -3-dependent changes of the spectrin meshwork required for NCAM-mediated neurite outgrowth. J. Biol. Chem. 2010;285:42046–42057. doi: 10.1074/jbc.M110.177147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ST, Smith AN, Cianci CD, Morrow JS, Brown TL. Identification of the primary caspase 3 cleavage site in alpha II-spectrin during apoptosis. Apoptosis. 2003;8:353–361. doi: 10.1023/a:1024168901003. [DOI] [PubMed] [Google Scholar]
- Wolan DW, Zorn JA, Gray DC, Wells JA. Small-molecule activators of a proenzyme. Science. 2009;326:853–858. doi: 10.1126/science.1177585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Cirilli M, Huang Y, Rich RL, Myszka DG, Wu H. Covalent inhibition revealed by the crystal structure of the caspase-8/p35 complex. Nature. 2001;410:494–497. doi: 10.1038/35068604. [DOI] [PubMed] [Google Scholar]
- Yan X, Jeromin A. Spectrin breakdown products (SBDPs) as potential biomarkers for neurodegenerative diseases. Curr. Tran. Geriatr. Gerontol. Rep. 2012;1:85–93. doi: 10.1007/s13670-012-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Siman R, Xu YA, Mills AM, Frederick JR, Neumar RW. Comparison of calpain and caspase activities in the adult rat brain after transient forebrain ischemia. Neurobiol. Dis. 2002;10:289–305. doi: 10.1006/nbdi.2002.0526. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Larner SF, Liu MC, Zheng W, Hayes RL, Wang KKW. Multiple alpha II-spectrin breakdown products distinguish calpain and caspase dominated necrotic and apoptotic cell death pathways. Apoptosis. 2009;14:1289–1298. doi: 10.1007/s10495-009-0405-z. [DOI] [PubMed] [Google Scholar]