Abstract
The burgeoning of phosphoinositide-binding domains and proteins in cellular signaling and trafficking has drawn laboratories from a wide variety of fields into the study of lipid interactions with peripheral membrane proteins. Many different approaches have been developed to assess phosphoinositide binding, some of which are more problematic than others, and some of which can be quantitated more readily than others. With a focus on the methods used in our laboratory, we describe here the considerations that need to be taken into account when establishing – and quantitating – the specific binding of a protein or domain to phosphoinositides in membranes. We also discuss briefly a few examples in which no clear consensus has yet been reached as to the specificity of a given domain or protein because of discrepancies between different commonly-used approaches.
Keywords: phosphoinositide, PH domain, surface plasmon resonance, centrifugation, lipid binding, vesicle, membrane, inositol
1. Introduction
The number of proteins and small domains known to bind membrane phosphoinositides has increased dramatically over the past decade [1, 2]. Of the 13 most populous classes of signaling interaction domain found in the human proteome [3], members of more than half have been reported to drive reversible membrane association by such interactions [1, 2, 4, 5]. Domains that have been implicated in head group-specific recognition of phosphoinositides include pleckstrin homology (PH) domains [6, 7]; phagocyte oxidase (phox) homology (PX) domains [8]; FYVE domains (for Fab1, YOTB, Vac1 and EEA1) [9]; epsin or AP180 N-terminal homology (ENTH/ANTH) domains [10, 11]; and plant homeodomain (PHD) zinc fingers [12]. In addition, phosphoinositide binding has been reported for PDZ (for Postsynaptic density protein, Disc large, Zona occludens) domains [13], FERM (for band Four-point-one, Ezrin, Radixin, Moesin) domains [14], Tubby [15], and MARCKS [16] proteins.
It has not always been straightforward to reach a consensus as to whether a given domain is capable of specific and high affinity phosphoinositide binding. It is likely that the literature harbors several examples of reported phosphoinositide interactions that are not functionally important. Moreover, there are several cases in which the precise headgroup specificity of a particular domain is not agreed upon. With the burgeoning use of purportedly specific phosphoinositide-binding domains as cellular probes for analyzing distribution of the respective lipids [5, 17], defining these specificities is especially critical. Active laboratories in this field differ significantly in their preferred methods for assessing the affinity and specificity of the phosphoinositide binding domains listed in the previous paragraph. The lack of ‘standardization’ of methods has advantages and disadvantages. An important benefit is that well-established phosphoinositide-binding domains have been studied in many different ways, so that affinities and specificities have been compared and reassessed under a plethora of conditions. A negative, however, is that proposals of new specificities have often been based on application of a single commonly-used method that has known drawbacks. In any case, it should be insisted upon that any report of phosphoinositide-binding specificity should utilize two or more of the commonly used approaches, with at least one being applied to determine an apparent dissociation constant for the binding event. In the following we discuss some of the most commonly used methods, describing the approaches used in the Lemmon laboratory. We also cite examples in which there is a consensus among different laboratories as to the specificity of a particular phosphoinositide-binding domain or protein, as well as cases in which there is disagreement between groups.
2. Description of Methods
2.1 Lipid state and context: an important consideration
The first issue to consider when comparing methods for analyzing phosphoinositide specificity and affinity is the state and context of the phospholipid that is presented to the putative binding domain. Some commonly applied methods assess protein binding to pure phosphoinositide that has been immobilized in one way or another. For example, commonly used ‘fat blots’ [18, 19] or dot-blots [20] employ pure dipalmitoyl phosphoinositide that has been dried on to a nitrocellulose support. Other immobilization procedures can be used [21]. For example, biotinylated phosphoinositides can be immobilized on solid streptavidin beads or plate supports, and protein binding to these substrates can be assessed [22]. An alternative approach, used by Rameh et al. [23] in one of the first assessments of PH domain specificity, is to employ labeled soluble short-chain (dioctanoyl or dibutanoyl) phosphoinositides and to assess their binding to immobilized protein. In each of these cases it should be appreciated that the phosphoinositide is presented to its potential binding partner in a clearly non-physiological context. First, the phosphoinositide is not present in a lipid bilayer as it would be in any cellular situation. Second, the effective local concentration of the phosphoinositide when used in the majority of these approaches will be substantially greater than is ever likely to be reached in vivo, where phosphoinositides are effectively ‘diluted’ by much more abundant components of cellular membranes including phosphatidylcholine and phosphatidylserine. Although it seems reasonable to expect that approaches utilizing pure phosphoinositide in this way will be prone to artifacts, these methods do lend themselves to a relatively high throughput, and so can nonetheless be of great value for first-pass assessments of phosphoinositide binding and potential specificity. It should be stressed, however, that their value is limited to this first pass.
The real biological question (in most cases) is whether or not a given domain or protein can be recruited specifically to an intracellular membrane in which a particular phosphoinositide accounts for perhaps 1% (at the very most) of all phospholipid molecules. The other >99% of ‘background’ phospholipid molecules (using values for the inner leaflet of the erythrocyte plasma membrane) are phosphatidylcholine (~35%), phosphatidylserine (~25%), phosphatidylethanolamine (~40%). A good argument can therefore be made for using physiologically ‘representative’ lipid mixtures in membrane bilayer form for assessing phosphoinositide binding by different domains and proteins. Several laboratories have taken this approach. We have often used 1–3% (mole/mole) of the phosphoinositide of interest in a background of pure phosphatidylcholine, in order to reduce background binding to negatively charged phosphatidylserine and for ease and reproducibility in vesicle preparations. Although the validity of any particular lipid mixture must always be questioned, a strong argument can be made that any vesicle-based assay is much more representative of the in vivo situation than any approach based on pure phosphoinositides. It is therefore essential to insist that any study that suggests specific phosphoinositide recognition and is based on studies with pure phosphoinositides be repeated with membrane mimetics. Both approaches have their place, but the key is to use multiple techniques.
2.2 Fat blots/dot-blots/lipid Westerns for analyzing binding to pure immobilized phosphoinositides
Perhaps the most commonly used (and arguably abused) method of assessing phosphoinositide binding specificity is to spot phosphoinositides onto nitrocellulose membranes, and to determine the extent to which a protein of interest will interact specifically with the phosphoinositide-bearing spots [18, 20, 24]. A similar approach was employed for immunological detection of glycosphingolipids over a decade previously [25]. Based on the studies of Dowler et al. [18], Echelon Inc. began in the late 1990s to market PIP Strips™, in which an array of phosphoinositides are pre-spotted (100 pmoles per spot) on nitrocellulose. We have assessed several different approaches for lipid dot-blots in our laboratory, focusing primarily on protein detection issues and minimizing the length of time for which the lipid-bearing nitrocellulose filters are incubated in aqueous buffer. We produce our own lipid-bearing nitrocellulose filters, and probe them with 32P-labeled GST fusion proteins containing the domain of interest. Although this approach has the disadvantage of utilizing radioactivity, we have found the direct visualization method (with 32P-labeled protein) to be more reproducible than alternatives involving indirect detection of the fusion protein with, for example, anti-GST antibodies.
As with all related approaches, this approach is very sensitive, but cannot be quantitated reliably. It should therefore only be used as a first-pass to determine whether binding to phosphoinositides (or other lipids) can be detected at all. As described in our genome-wide analyses of S. cerevisiae PH and PX domains [26, 27], we have often detected phosphoinositide binding using this method that is too weak to be measured by any of the more quantitative approaches outlined later in this article. Other laboratories have reported similar findings [28]. A ‘positive’ result in this assay, while suggestive, must therefore be viewed with great skepticism. Such an outcome may simply reflect very low affinity binding that has no physiological relevance in the context of real membrane. Certainly, given this fact, data from dot-blots/lipid overlay studies alone cannot be relied upon to evaluate phosphoinositide binding – although there has been a worrying trend in the literature not to go further than this crude assay.
Where phosphoinositide binding seen in 32P-based dot-blots can also be detected with other methods, we find that the dot-blots faithfully report binding specificity. As discussed in Section 2.2.3, apparent specificity can be distorted to some extent (e.g. [20]) as a result of the different water solubility of the phosphoinositides – so that they differ in the stability of their association with the nitrocellulose filter. If this effect is accounted for, specificity is accurately reported according to our experience.
2.2.1. Lipid Western/dot-blot assay procedure
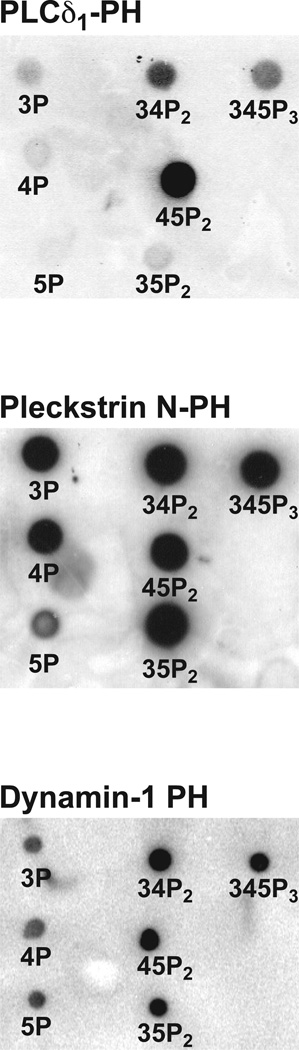
To generate the filters to be probed with 32P-labeled GST fusion protein, we spot onto nitrocellulose sheets (NitroBind: MSI) 2 nmoles of each lipid from a 2 mM lipid solution in chloroform or (for phosphoinositides) chloroform:methanol (1:1) containing 0.1% HCl. We obtain all phosphoinositides from Cell Signals (Lexington, KY), Echelon Inc. (Salt Lake City, UT), or Matreya (Pleasant Gap, PA). Phosphatidylcholine and other lipids are from Sigma or Avanti Polar Lipids (Birmingham, AL). As always when treating lipids in organic solvents, all manipulations are performed in glass vials, using glass positive displacement pipettes (Drummond), and lipid solutions are stored at –80˚C under nitrogen. After spotting lipids onto nitrocellulose in the pattern shown in Figure 1, the filter is dried and blocked for 1–4 h at 4˚C in detergent-free tris-buffered saline (TBS) containing 3% bovine serum albumin (BSA). After blocking, the filter is simply probed at room temperature for 30 min with a 32P-labeled GST fusion protein of the domain of interest (in TBS/BSA) at a concentration of 10 µg/ml. Filters are then washed 5 times with TBS (5 min each), dried by blotting with Whatman paper, and bound radioactivity is visualized using a Phosphorimager (Molecular Dynamics, Sunnyvale, CA). Example dot-blots for PH domains are shown in Figure 1 ([20] and unpublished data).
Figure 1.
Representative dot-blot results for the PH domains from PLCδ1, pleckstrin (N-terminal) and human dynamin-1. Methods are described in the text. The phosphoinositide spotted at each position is labeled. Whereas the PLCδ1 PH domain is specific for PtdIns(4,5)P2 (labeled 45P2), the pleckstrin and dynamin PH domains are quite promiscuous, and appear to bind similarly to all phosphoinositides. Data are from [20] or are unpublished. Note for the dynamin-1 PH domain that centrifugation experiments show that PtdIns3P and PtdIns4P bind ~10-fold more weakly than PtdIns(3,4)P2 and PtdIns(4,5)P2 [51].
2.2.2. Preparation of 32P-labeled probe
To produce 32P-labeled GST fusion proteins, we subclone the coding region for the relevant domain into pGEX-2TK (Amersham) or pGSTag [29], which encode a protein kinase A (PKA) site between GST and the domain of interest. The bacterially-expressed GST fusion protein is then phosphorylated while bound to glutathione-agarose beads as described [20, 30]. Approximately 10 µg of GST-fusion protein, bound to glutathione-agarose beads in a slurry of ~60 µl is washed three times with DK buffer [50 mM potassium phosphate, pH 7.15, containing 10 mM MgCl2, 5 mM NaF and 4.5 mM dithiothreitol (DTT)]. Ten units of PKA (catalytic subunit, Sigma P-2645), which has been dissolved in 6 mg/ml DTT 5 min earlier, and 0.75 mCi of [γ-32P]ATP (6000 Ci/mmol, 150 mCi/ml: unpurified, from Perkin Elmer NEG035C) are then added, and the 75 µl reaction is nutated at room temperature for 30 min. After labeling, the beads are washed 5 times with phosphate-buffered saline (containing 1 mM DTT and 1 mM PMSF), and labeled GST fusion protein is eluted with 15 mM reduced glutathione in 50 mM Tris-HCl, pH 8.0, containing 5 mM NaF, 1 mM EDTA, 0.5 mg/ml BSA, 1 mM DTT, 1 mM PMSF. It is very important in this procedure to ensure that glutathione addition has not acidified the elution buffer (this will depend on the glutathione source), so pH of the final elution buffer must be checked before use. It is also important not to spin the glutathione-agarose beads faster that ~1000 rpm (setting #1 on an Eppendorf microfuge), or elution efficiency is greatly reduced.
Once the GST fusion protein is labeled and eluted, it should be filtered through a 0.22 µm filter and used immediately to probe the nitrocellulose filter as described in Section 2.2.1.
2.2.3. Problems to consider
We have discussed above several of the limitations of this approach – which are significant. First, the state of the phosphoinositides in the spots on the nitrocellulose filter is far from representative of that found in vivo. Second, experience has shown that in many cases phosphoinositide binding that appears robust in dot-blots cannot be detected using any of the other commonly used approaches to study lipid binding [26–28]. Conversely, the examples of dynamin’s PH domain [31–33] and the PH domains from S. cerevisiae proteins Ynl047cp and Yil105cp [27, 34] illustrate that phosphoinositide binding that is very weak (and only detectable with dot-blots) can nonetheless be physiologically relevant. This can certainly not be assumed, however. In our genome-wide analysis of yeast PH domains [27], there are 17 examples (~50% of all S. cerevisiae PH domains) for which we see phosphoinositide binding by dot-blots, but with no supporting evidence from other techniques or from studies of subcellular localization. It remains quite possible that the apparent phosphoinositide binding seen in these cases is simply an artifact of the dot-blot approach.
We alluded in Section 2.2 to the issue of selective dissociation of phosphoinositides from nitrocellulose filters, and the effect that this has upon apparent binding specificity when using dot-blots. It is well known that PtdIns(3,4,5)P3 is significantly more water-soluble than any other natural phosphoinositide. Similarly, the water solubilities of PtdIns(4,5)P2, PtdIns(3,4)P2, and PtdIns(3,5)P2 (which are very similar) are greater than those of PtdIns3P, PtdIns4P or PtdIns5P. The result of this can be seen directly if equal quantities of each phosphoinositide are spotted onto nitrocellulose and the remaining bound lipid is visualized with molybdenum blue after overnight incubation in TBS. After such treatment, significantly more of the PtdInsP isomers than the PtdInsP2 isomers remains on the filter, and PtdIns(3,4,5)P3 is seen to have been washed away quite substantially. The dot-blot signal strength reflects both the binding affinity and the amount of lipid present in the dot at the end of the experiment. Since monophosphorylated phosphoinositides are the most stably associated with the nitrocellulose, they will tend to give the strongest signals even for proteins that do not distinguish between phosphoinositides. We discussed this issue previously for the N-terminal PH domain from phospholipase C-γ1 (PLCγ1) [20]. The PLCγ1 PH domain showed a clear preference in dot-blots for PtdIns3P over PtdIns(3,4)P2, which in turn gave a stronger signal than PtdIns(3,4,5)P3. However, in quantitative centrifugation-based vesicle binding experiments (described below), the PLCγ1 PH domain bound equally well to all of these phosphoinositides. We suggest that the apparent PtdIns3P specificity in this case was artifactual, simply reflecting the fact that there was more of it on the filters used for the dot-blot experiments. As discussed below there are several related examples in the literature, and apparent specificity for monophosphorylated phosphoinositides is quite frequently argued in the literature based on dot-blot studies.
2.3 Studies of binding to phosphoinositides in membranes – vesicle studies
Whereas dot-blots and related approaches have the advantage of convenience, they have the disadvantages that the physical state of the lipid is not clear, and that they cannot be quantitated reliably. In this Section, we outline methods that we have used for studying the binding of domains and proteins to phosphoinositides embedded in bilayer membranes. These include isothermal titration calorimetry (ITC), vesicle sedimentation approaches, and surface plasmon resonance (SPR).
2.3.1 Direct binding studies in solution – titration calorimetry
Ideally, one would like to monitor the binding of domains to phosphoinositides in membranes directly, without having to perturb the equilibrium in any way. Two methods that have been employed for this are fluorescence resonance energy transfer (FRET) and isothermal titration calorimetry (ITC) [35, 36]. Corbin et al. [37] have followed vesicle association of the Grp1 PH domain by monitoring FRET between tryptophans in the PH domain and dansylated lipid incorporated into the vesicles. A similar FRET approach has also been employed for assessing the binding of Texas Red labeled MARCKS peptide to vesicles containing fluorescent PtdIns(4,5)P2 [38]. These approaches are very powerful, but require the appropriate labeled lipids and/or proteins.
We previously employed isothermal titration calorimetry (ITC) to assess binding of the PLCδ1 PH domain to artificial membranes containing PtdIns(4,5)P2 [39]. Limit-sonicated dimyristoylphosphatidylcholine (DMPC) vesicles containing 5% (mole/mole) dipalmitoyl PtdIns(4,5)P2 were placed in the calorimeter cell, and aliquots of purified PLCδ1 PH domain (at 120 µM) were injected during the titration. A 1:1 stoichiometry was suggested if 57% of the PtdIns(4,5)P2 in each vesicle was assumed to be accessible on the outside of the small vesicles (and 43% inaccessible on the inside aspect). These experiments gave a mean KD for PtdIns(4,5)P2 binding by the PLCδ1 PH domain of 1.66 ± 0.8 µM. Measurements using different techniques by several other laboratories, including those of McLaughlin [38], Rebecchi [40] and Hirose [41] all fall within a range of 1 µM – 2.8 µM. This is perhaps the most well characterized PH domain/phosphoinositide interaction.
Although ITC is often considered the ‘gold standard’ for affinity determinations, it has the disadvantage that it requires a large amount of material. For PLCδ1-PH/PtdIns(4,5)P2 titrations, each single experiment consumes 50 µg of PtdIns(4,5)P2 and nearly 1 milligram of PH domain. The method is therefore not accessible for expensive lipids or for proteins that cannot readily be produced in milligram quantities. In our laboratory, we have therefore sought other ways to measure protein binding to phosphoinositide-containing vesicles, always using the PLCδ1-PH/PtdIns(4,5)P2 interaction as a test case so that results with alternative methods can be validated directly by reference to our earlier ITC measurements.
2.3.2. Vesicle sedimentation
A convenient approach for measuring binding of proteins to vesicles that requires no special equipment involves quantitatively pelleting the vesicles from a lipid/protein mixture and determining the extent to which the protein partitions into the vesicle pellet. The key to this experiment is to increase the density of the vesicles so that they can be pelleted completely by ultracentrifugation. One useful approach for achieving this is to use large unilamellar vesicles loaded with sucrose solution [42]. We have employed an alternative method introduced by Tortorella and London [43] in which the primary lipid in the vesicles is a brominated form of phosphatidylcholine, which has a significantly higher density than the normal phospholipids and therefore pellets readily. We generate limit-sonicated small unilamellar vesicles (SUVs) containing the phosphoinositide (or other lipid of interest) at 3% (mole/mole) in a background of di(9,10-dibromostearoyl)phosphatidylcholine, which is available from Avanti Polar Lipids, Inc. (Birmingham, AL). A series of different concentrations of vesicles are incubated with a fixed protein concentration. Vesicles are then pelleted, and the partitioning of protein into the vesicle pellet is assessed using standard protein assays. One important shortcoming of this approach is that if the protein of interest has any tendency to precipitate, or if the presence of lipid promotes protein precipitation, the extent of binding can be dramatically overestimated (precipitated protein will appear as ‘bound’ protein). For this reason, we only use this method for highly purified protein that we know is well behaved, and does not precipitate. A control with brominated PC alone is, of course, also critical. Where there is some question about the tendency of a protein to precipitate, an approach in which vesicles are ‘floated’ in a sucrose gradient may be more appropriate (e.g. [44]), if a little more cumbersome to quantitate. Precipitated protein will pellet in a sucrose gradient, while vesicle-bound protein will migrate (float) with the less-dense vesicles and can thus be readily distinguished (provided that the extent of protein binding does not significantly increase vesicle density).
2.3.2.1. Vesicle preparation
We mix di(9,10-dibromostearoyl)phosphatidylcholine and the phosphoinositide of interest at a 97:3 molar ratio in 1:1 CHCl3:CH3OH containing 0.1% HCl in a glass vial. The phospholipid mixture is then dried under a stream of dry N2 gas, and exposed to high vacuum for 1–2h to complete drying. The dried lipid mixture is then rehydrated in 25 mM HEPES, pH 7.2, containing 100 mM NaCl (HBS) to give a final total lipid concentration of 25 mM (i.e. 0.75 mM phosphoinositide). Following vigorous vortexing, the hydrated lipid mixture is subjected to between 10 and 20 rounds of freezing (in liquid N2) and thawing (in a 45˚C sonicating water bath). After the first 1–2 rounds, the pH of the hydrated mixture is checked (by spotting 1 µl onto pH strips), and corrected to pH 7.2 with 1M NaOH if necessary. Intensive bath sonication and freeze/thawing is then continued until the vesicle suspension is optically clear.
2.3.2.2. Vesicle-binding assay
Vesicles are pelleted in a 20-place TLA-100 rotor spun in an Optima TLX benchtop ultracentrifuge (Beckman). Using thick-walled polycarbonate tubes for this rotor (Beckman), assay volumes of just 50 µl can be used, thus minimizing lipid and protein usage. The purified protein used for the experiment must be free of DTT, glycerol, glutathione, or any other components that will interfere with protein assays. We therefore dialyze it exhaustively against HBS prior to use.
Centrifugation assay samples (50 µl) are set up containing 10 µM (or 5 µM, or some other concentration, depending on the particular case) purified, dialyzed, protein plus limit-sonicated SUVs at various concentrations ranging from 0 to 4 mM total lipid (equivalent to 0–120 µM total phosphoinositide). The samples are then spun at 85,000 rpm for 1h at 25˚C. After pelleting the vesicles, 35 µl of the supernatant is transferred to a fresh tube (this is ‘sup’). The remaining supernatant is carefully removed from the pellet and discarded, the pellet is resuspended in 50 µl HBS by brief bath sonication, and 35 µl of this suspension is then transferred to a fresh tube (this is ‘pell’). We use the bicinchonic acid (BCA) protein assay (Pierce, Rockford IL) to determine protein concentration in the ‘sup’ and ‘pell’ samples according to the manufacturers’ instructions. To avoid artifacts arising from light scattering by the vesicles, SDS is added (to a final concentration of 0.5%) to both ‘sup’ and ‘pell’ samples after incubation with the BCA reagent, but before measuring absorbances. A protein standard curve is generated in parallel for each protein in order to determine what percentage of total added protein has pelleted with the vesicles. Molar partition coefficients (K), as defined by Peitzsch and McLaughlin [45] can then be estimated by fitting the data to the following equation:
where [lipid] is the concentration of available lipid (>[protein]bound), approximated by one half of the total lipid concentration (assuming that only 50% is available on the outer leaflet). Determination of K makes no assumption of stoichiometry and has units M−1. However, in cases such as that of the PLCδ1 PH domain where the stoichiometry is known to be 1:1, the apparent dissociation constant (KD) can be estimated as (mole ratio)/K. Thus a KD of 1.7 µM for binding of the PLCδ1 PH domain to PC vesicles containing 3% (mole/mole) PtdIns(4,5)P2 would correspond to a molar partition coefficient in this approach of approximately 17,000 M−1.
We have used this method extensively for studying binding of PH and FYVE domains to vesicles of different lipid compositions [20, 46]. In these cases, the KD estimates agree very well with results obtained for binding to equivalent vesicles from ITC (for PLCδ1) and surface plasmon resonance studies (see Section 2.3.3.5). We also made extensive use of the approach for assessing the ability of the PH domain from human dynamin to bind phosphoinositides, and the importance of its oligomerization, as illustrated in Figure 2.
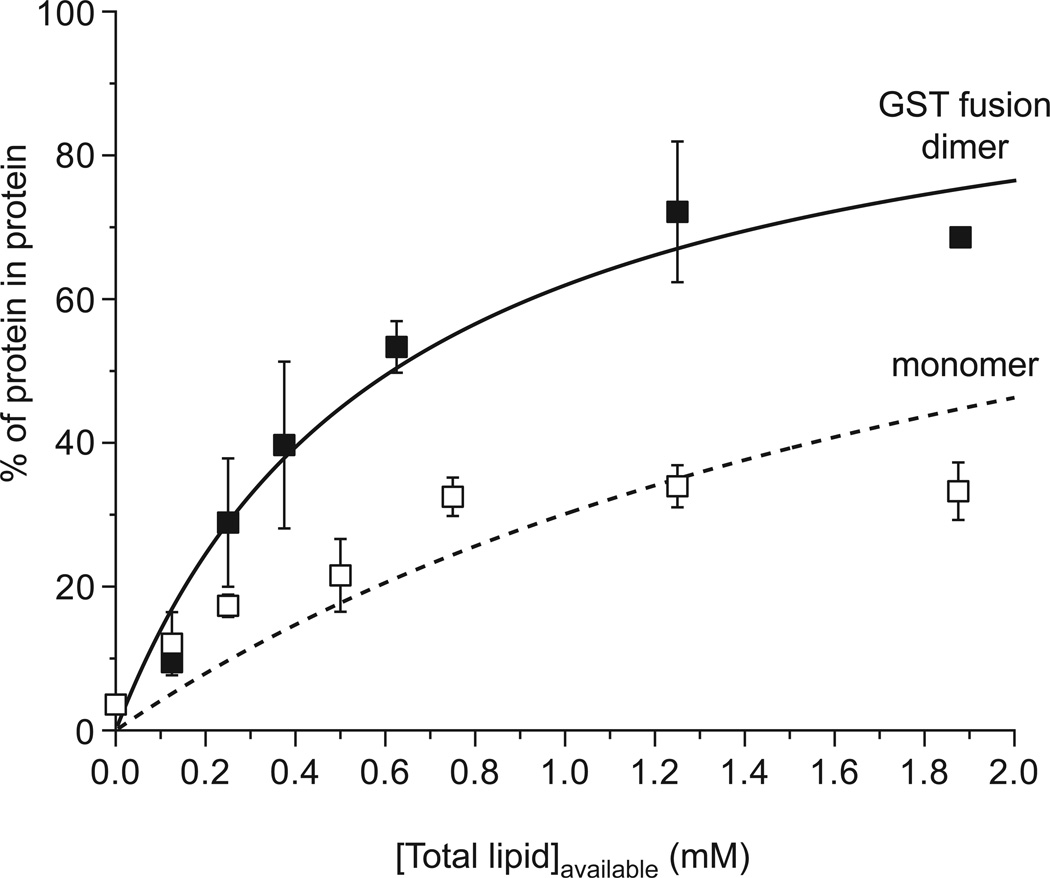
Figure 2.
Representative data for analysis of phosphoinositide binding by vesicle centrifugation. Using the approach described in the text, binding to vesicles containing 3% (mole/mole) PtdIns(4,5)P2 is compared for monomeric and dimeric forms of the dynamin-1 PH domain. Dimerization is induced by fusion to GST. Fits to the equation described in Section 2.3.2.2. are plotted. According to this analysis, the monomeric PH domain binds vesicles with a K value of approximately 479 M−1 (KD ~ 65 µM if 1:1 stoichiometry is assumed), while the largely dimeric GST fusion gives a K value of 1626 M−1, corresponding to an estimated KD in the 18 µM range. Data are taken from [51].
2.3.2.3. Oligomerization and avidity. Considerations when using GST
The case of the dynamin PH domain nicely illustrates how important it is to consider oligomerization and avidity effects when studying the binding of proteins to membranes. There was some confusion in the literature as to whether the isolated dynamin PH domain binds phosphoinositides. Our own dot-blot approaches suggested that dynamin’s PH domain binds all phosphoinositides (see Figure 1), but our attempts to quantitate its binding to vesicles containing PtdIns(4,5)P2 had failed [39], suggesting that the interaction may be very weak. NMR-based KD measurements for binding of the PH domain to isolated PtdIns(4,5)P2 headgroup were in the millimolar range [47, 48], consistent with this observation. By contrast, separate studies by Salim et al. [48] suggested a strong interaction between the dynamin PH domain and PtdIns(4,5)P2-containing vesicles. The primary difference between these studies and ours was that we investigated vesicle binding by an isolated, monomeric form of the dynamin PH domain, whereas Salim et al. studied binding of vesicles to immobilized PH domain. Since multiple PH domains can bind to a single vesicle, the latter experimental set-up (with the PH domain ‘pre-oligomerized’ by virtue of its immobilization) enhances the avidity of the interaction. By artificially dimerizing the dynamin PH domain with introduced disulfide bonds or by fusion to GST (which forms tight dimers [49, 50]), we were able to demonstrate with the centrifugation assay described above that crosslinking two PH domains is sufficient to bring the apparent KD for PtdIns(4,5)P2 binding into the 20 µM range [51]. This is illustrated by the binding curves presented in Figure 2. Since dynamin forms tetramers [52], this avidity effect will be even more exaggerated in the intact molecule. It is possible, but has not been shown, that other weak phosphoinositide-binding domains utilize similar avidity effects for their in vivo function. It should be stressed, though, that the use of GST or other means of promoting oligomerization can dramatically increase the apparent membrane-binding affinity of a given domain or protein regardless of whether this is of physiological importance. In considering strengths of binding, it is therefore critical that careful attention is paid to the oligomeric state of the protein (and lipid) components used. In this context, immobilization should be considered as a means of artificially oligomerizing a protein or lipid.
2.3.3. Surface plasmon resonance (SPR)
Our favored approach for assessing phosphoinositide binding of protein domains is surface plasmon resonance (SPR) using a Biacore instrument. This approach has the disadvantage that one needs access to an expensive instrument, but has many advantages including automation, convenience, and the consumption of relatively small amounts of material. In deciding how we would employ this approach in our laboratory, we began by comparing different SPR experimental protocols to study binding of the PLCδ1 PH domain to vesicles containing 3% (mole/mole) PtdIns(4,5)P2 and of the Hrs1 FYVE domain to vesicles containing 3% (mole/mole) PtdIns3P. This allowed us to compare results obtained using ITC, centrifugation assays, and SPR directly.
Practically, the SPR approach involves immobilizing lipid vesicles on a commercial biosensor chip surface as described below, and using the phenomenon of surface plasmon resonance to detect binding of proteins to this surface when flowed past it. Achieving this amounts simply to injecting the relevant samples into the Biacore instrument. The instrument effectively monitors the refractive index (and thus mass concentration of protein or lipid) at the sensor chip surface by measuring the surface plasmon resonance angle (output as resonance or response units) as different samples are flowed across it in a microfluidic chamber. For physical details of the SPR experiment, readers are directed elsewhere [53]. An important advantage of the SPR approach is that the resonance angle (and thus refractive index changes at the experimental surface) can be monitored in real time. A refractive index increase is detected every time a sample is injected, simply because the refractive index of a protein solution is higher than that of the mobile phase buffer. In order to distinguish refractive index changes that arise from protein binding to the sensorchip surface from those that merely reflect bulk protein concentration, a ‘blank’ surface is monitored at the same time as the experimental surface for each sample, and this blank or reference signal is subtracted in real time. In the Biacore 3000, three experimental surfaces can be monitored simultaneously along side a fourth blank or control surface.
2.3.3.1. Kinetics versus saturation analysis by Biacore
The output from the SPR apparatus, in resonance units (or RUs), is directly proportional to the mass of protein bound to the experimental binding surface (once corrected for bulk refractive index effects as described above). For a series of injections of protein at different concentrations, an array of sensorgrams such as those shown in Figure 3A are obtained. There are two prevailing approaches for extracting affinity information from these data. In many cases, Biacore software is used to fit (globally) the time-dependent association and dissociation curves for several different experiments to equations describing a simple 1:1 interaction, so that association and dissociation rate constants (kon and koff respectively) can be estimated. In many cases, however, the shapes of the experimental curves make it clear that the simple kinetic model applied is not valid. Alternatively (or in addition), the steady state values reached by the association curve for each protein concentration can be taken as representing the equilibrium binding at that particular (free) protein concentration. In this case, a simple plot of steady-state RU value against protein concentration yields a saturation binding curve (see Figure 3B) that can be fit to simple binding equations as described below. It has been shown for studies of EGF binding to the extracellular domain of its receptor that these two approaches give essentially identical results (where the kinetic assumptions etc are presumably correct) [54, 55]. However, when PH or FYVE domain binding to phosphoinositides is studied, there appears to be more of a discrepancy. Monitoring steady state RU signals, we achieve excellent agreement in KD values (0.6–3 µM) for PtdIns(4,5)P2 binding by the PLCδ1 PH domain between our SPR studies [56, 57], ITC [39], and centrifugation approaches [20]. Similarly, estimates for PtdIns3P binding by the Hrs1 FYVE domain are within 2-fold when comparing results from SPR (KD = 4.8 µM [57]) and centrifugation assays (KD = 2.5 µM [46]). By contrast, using koff/kon ratios, Cho and colleagues report a 100-fold higher apparent affinity for the Hrs1/PtdIns3P interaction [58] than we have measured, the origin (or validity) of which is not clear. Indeed, SPR-based KD estimates for phosphoinositide binding by PH and other domains that rely upon determination of kinetic parameters tend, in general, to overestimate affinities by 10–100 (or more) fold compared with our equilibrium-based determinations. Although its origin is not clear, we suggest that this discrepancy reflects problems with the kinetic model used, and we have chosen to focus only on steady state or equilibrium determination of KD values. The differences in the literature make it critical, when comparing lipid-binding affinities, to consider whether any apparent differences arise from the techniques and assumptions employed, or reflect actual differences in molecular properties.
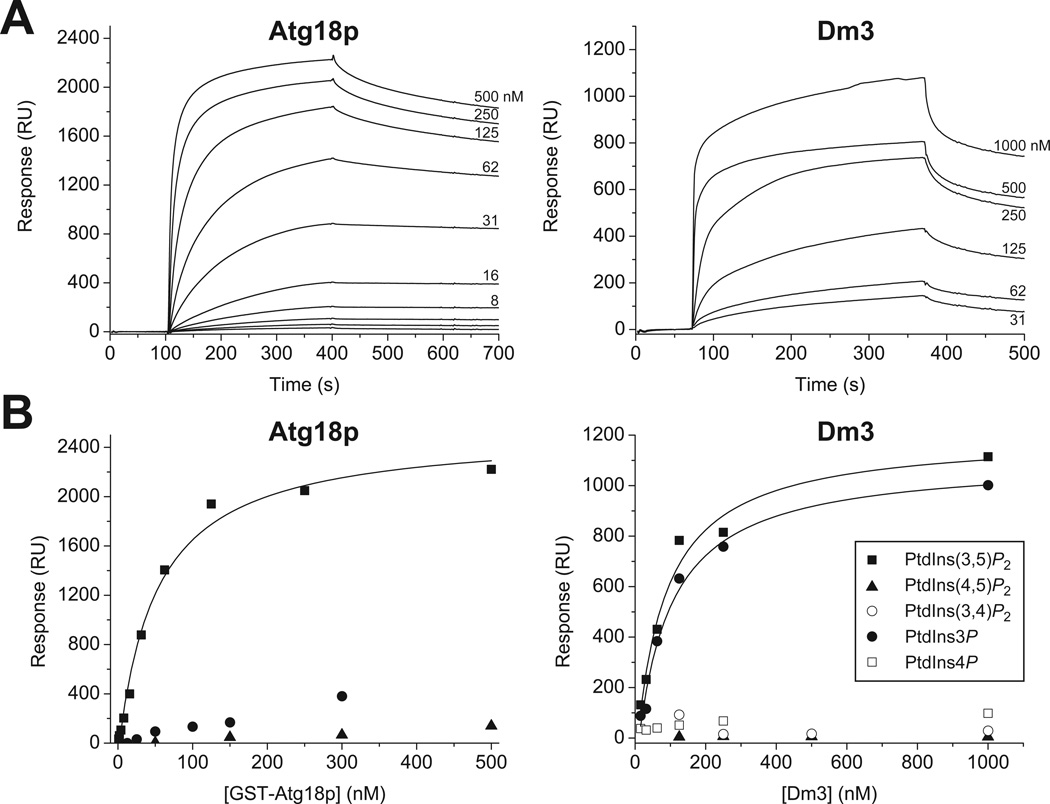
Figure 3.
SPR analysis of phosphoinositide binding by the β-propeller proteins [57] Atg18p (from S. cerevisiae) and Dm3 (from D. melanogaster), using approaches described in the text. Experiments were performed in HBS-N containing 0.5 mM MgCl2. Atg18p experiments employed a bacterially-expressed GST fusion protein, which leads to enhanced binding affinities due to dimerization (see [57]). Dm3 experiments utilized monomeric protein produced in Sf9 cells infected with recombinant baculovirus. (A), individual sensorgrams are shown for a representative set of experiments. Each is labeled with the protein concentration used. Steady state response levels for each sensorgram were assessed and plotted against the relevant protein concentration in (B) to give binding curves that can be fit for KD determination as described in the text. The data shown in (B) illustrate the clear tendency of Atg18p to bind specifically to PtdIns(3,5)P2, but not PtdIns(4,5)P2 or PtdIns3P (contrary to the suggestions of [70]). Dm3 binds much more significantly to PtdIns3P, but does not bind detectably to PtdIns(4,5)P2, PtdIns(3,4)P2, or PtdIns4P.
2.3.3.2. Preparing sensorchip surfaces
Our SPR experiments all employ a Biacore 3000 or a Biacore X instrument, in which the same sensorchips are used. Lipid vesicles are immobilized on Pioneer L1 sensorchips (Biacore, Inc) essentially as described by Erb et al. [26, 59]. Dioleoylphosphatidylcholine (DOPC) from Sigma is codissolved with the phosphoinositide of interest at a molar ratio of 97:3 (DOPC:PtdInsPn), in 1:1 chloroform: methanol containing 0.1% HCl. The mixture is then dried under a stream of nitrogen, followed by exposure to high vacuum for 30 minutes. The dried lipids are then hydrated by addition of ‘HBS-N’ buffer (25 mM HEPES, pH 7.4, 150 mM NaCl) containing no surfactant to generate a suspension containing 10 mM total phospholipid, vortexed thoroughly, and are bath sonicated. Once all of the lipid is in suspension, the pH is checked using pH strips, and corrected if necessary using 1 µl aliquots of 1 M NaOH. The resuspended lipid mixture is then subjected to 8 rounds of freeze (liquid N2) – thaw (in a bath sonicator set at 40˚C). Vesicles are then prepared by extrusion (10 passages) through a 0.1 µm polycarbonate filter, using an Avanti MiniExtruder according to the manufacturers’ instructions. A vesicle suspension containing 1.5 mM total lipid is then prepared for generation of the sensorchip surface.
To prepare the surface (two can be used per chip in the Biacore X, four in the Biacore 3000), all surfaces are washed at 25˚C at a flow rate of 10 µl/min (HBS-N as mobile phase) with 20 µl of 1% (w/v) β-octylglucoside, and then another 20 µl of 0.5% SDS. Another wash with 10 µl of 1% (w/v) β-octylglucoside is performed, and residual detergent is then rinsed away with a 10 µl injection of 30% ethanol. Once the surface is ‘primed’ in this way, 55 µl of the extruded 1.5 mM (total) lipid suspension is injected over a contact time of 5.5 minutes, during which time a increase in signal of 5,000–10,000 RU is seen as the vesicles associate with the (proprietary) hydrophobic tails linked to the carboxymethylated dextran layer in the L1 sensor chip. A final wash with 20 µl of 0.1 M NaOH is then performed to remove lipid associated through other than hydrophobic interactions. Atomic force microscopy studies performed by Erb et al. [59] indicate that vesicles bound to the L1 sensor chip in this way fuse with one another to form a continuous lipid layer on the chip surface. On the other hand, Höning et al. [60] have recently reported that the L1 surface appears to be decorated with individual vesicles in negative-stain electron microscopy images, although this could reflect differences in sample preparation. In either case, it appears reasonable to argue that the immobilized vesicles maintain or adopt a membrane-like structure that is relevant for studies of protein/membrane interactions.
2.3.3.3. SPR analysis of phosphoinositide binding
In a simple experiment, we immobilize vesicles containing only DOPC (the background/reference) to flow cell 1 of the sensorchip, and vesicles containing, for example, 3% (mole/mole) PtdIns(3,5)P2 (in DOPC) to flow cell 2. The mobile phase is passed sequentially across flow cell 1 followed by 2 in the Biacore instrument. It is important to set up the experiment in this way (with the reference cell first in the flow path) so that phosphoinositide-containing vesicles (or phosphoinositide itself) cannot migrate to the reference cell. We have found that such migration can create significant problems over a day of experiments if this rule is not followed. Once the lipid surfaces are established, the protein of interest is injected at a series of different concentrations into the Biacore instrument at 25˚C with HBS-N as the mobile phase. A substantial response is seen for both flow cells, reflecting both the bulk refractive index increase due to the protein sample and binding to the lipid surfaces. The SPR response that reflects binding due to the phosphoinositide present on flow cell 2 is obtained directly by subtracting the response measured for flow cell 2 from that measured simultaneously for the DOPC-only flow cell 1 reference. This background correction subtracts both bulk refractive index effects and any non-specific binding to the DOPC surface alone (which needs to be assessed using other methods). Example corrected sensorgrams for several different phosphoinositides and a variety of protein concentrations are shown for PtdIns(3,5)P2-binding proteins in Figure 3A). The lipid surface is washed with an injection of 10–20 µl of 100 mM NaOH between samples, in order to remove residually bound protein.
For each protein sample, Figure 3A shows that the SPR response approaches a steady state value within about 250 seconds after sample injection. This steady state value is recorded and plotted against the protein concentration used in the sample, to give the saturation binding curves shown in Figure 3B. These data are then analyzed as described in Section 2.3.3.5.
2.3.3.4. Experimental considerations
Since SPR instruments pass the sample sequentially though the 1–4 flow cells, and phosphoinositides can migrate from one prepared L1 chip surface to another, it is very important to have the reference cell as flow cell 1 – to prevent migration of phosphoinositide to that surface (and resulting ‘contamination’ of the reference). The fact that phosphoinositide can migrate in this way (dissociating from one surface and binding to another) argues that one should ideally restrict studies to two flow cells (and thus one phosphoinositide) at a time. We therefore usually use only two flow cells whether doing experiments on the Biacore X or Biacore 3000 instruments.
The fact that phosphoinositide can dissociate from the L1 chip surfaces also implies that the surfaces have a limited lifetime. Indeed, with the PLCδ1 PH domain and Hrs FYVE domain as test cases, we have found that a PtdIns3P- or PtdIns(4,5)P2-containing surface loses most of its binding activity after approximately 24 hours of constant flow. Over a period of approximately 8 hours, however, the saturation binding signal does not drop more than 10–20%. We therefore limit the use of prepared surfaces to around 8 hours before washing the surface with detergent and regenerating a fresh experimental surface. Saturation binding signals from newly regenerated surfaces are found to be within ~10% of one another. We have not yet detected a limit to the number of times that a Biacore L1 chip can be washed and regenerated following the procedure described in Section 2.3.3.2.
One issue that requires attention is the contact time allowed before steady state is assumed to have been reached. For very high affinity interactions, significant binding is detected even at very low protein concentrations, and under these conditions mass transport can limit the rate at which steady state is reached. In these cases (including some of the lower concentration samples in Figure 3A) a longer contact time should be employed so as not to distort the low concentration region of the binding curve. Needless to say, at the other end of the spectrum (high protein concentration) clear evidence for saturation should be obtained before any attempt is made to fit the data to a binding equation.
2.3.3.5. Data analysis
We fit binding curves such as those shown in Figure 3B to a simple binding equation that provides an estimate of apparent KD. We assume that each protein molecule binds independently to a single class of binding site present on the sensorchip surface (which may be one lipid molecule, or may be made up of several – or even many). We also assume at steady state that there is no net transport of protein from the injected sample to the sensorchip surface (association and dissociation rates are equal), so that the protein concentration in the sample injected during the ~250 second period equals the free protein concentration.
For a simple interaction, the apparent KD is equal to the concentration of protein required to reach half-maximal binding, so that the data can be fit to the following equation:
Where B is the binding signal obtained at a particular concentration of free protein ([P]free) following the assumption outlined above, and Bmax is the binding signal obtained at saturation, which must be measured directly.
For the PLCδ1 PH domain, our studies with this SPR approach have given KD estimates of 0.7 ± 0.3 µM for PtdIns(4,5)P2 binding [27], very similar to our ITC-derived value [39] of 1.66 ± 0.8 µM. Similarly, for PtdIns3P binding by the Hrs1 FYVE domain, this approach gives a KD value of 3.4 ± 0.6 µM, compared with a value of 2.5 µM estimated from the vesicle centrifugation approach outlined above. Similar agreement has been seen in studies of many phosphoinositide-binding and other domains and proteins in our laboratory. We therefore feel quite confident that the SPR approach described in Section 2.3.3. provides KD values that can be compared directly with those obtained using other equilibrium methods. Moreover, specificity as detected in our SPR studies has always agreed with centrifugation and ITC analysis where compared directly.
3. Examples in which consensus has yet to be reached: PtdIns4P and PtdIns(3,5)P2 binding proteins
As hinted earlier in this article, there are a few cases in which a proposed phosphoinositide-binding specificity is not agreed upon. One example is a class of PH domains reported to recognize PtdIns4P with high specificity. Another is a set of proteins suggested in the literature to bind specifically to PtdIns(3,5)P2. In most cases in which there is disagreement, the initial suggestion of the proposed specificity came from studies using dot-blots or other approaches utilizing pure phosphoinositides in a clearly non physiologically-representative context. Although further work is required to reach a consensus on the phosphoinositide-binding properties of these examples, it is possible that dot-blots and similar approaches may have been misleading with regard to specificity.
3.1 PtdIns4P-specific PH domains
The PH domain from FAPP1 was reported from dot-blot/lipid overlay studies to recognize PtdIns4P with high specificity [28]. This PH domain and several with closely similar sequences were found to be localized specifically to the Golgi when studied as green fluorescent protein/PH domain fusion proteins [61]. The related PH domains include those from oxysterol binding protein (OSBP), its S. cerevisiae homologs Osh1p and Osh2p, and the Goodpasture antigen binding protein (GPBP). Several studies have shown that the Golgi localization of these PH domains is dependent on the production of PtdIns4P in the Golgi [61–64], and it was argued that the FAPP1 PH domain is a highly specific PtdIns4P-specific domain that can be used to monitor the distribution of this phosphoinositide in living cells [63, 64].
That PtdIns4P is not exclusively responsible for Golgi targeting of these PH domains was suggested by the finding that Arf1 is also critical for their localization at this site [61, 62]. Perhaps more importantly, studies in our own laboratory ([27] and unpublished data) and those of Levine and Munro [61, 65] argue that the FAPP1, OSBP, GPBP, Osh1p and Osh2p PH domains are not PtdIns4P-specific at all in vitro. Vesicle centrifugation experiments using raffinose-loaded DOPC vesicles containing 2.5% (mole/mole) phosphoinositide showed that the FAPP1, OSBP and GPBP PH domains all bind PtdIns(4,5)P2 and PtdIns4P with affinities within 2-fold of one another [61]. Our own unpublished experiments using Biacore agree for FAPP1 and OSBP, and we have published that the Osh1p and Osh2p PH domains also bind almost identically to PtdIns(4,5)P2 and PtdIns4P. In fact, our studies suggest that these PH domains bind to all phosphoinositides with very similar affinities (KD values of 1–10 µM)
These data argue that the apparent PtdIns4P specificity observed in vivo does not arise from the phosphoinositide-binding properties of these PH domains per se. Rather, the specific subcellular localization of these domains is defined by the coincidence of phosphoinositide and Arf1 in the Golgi membranes. The apparent specificity for PtdIns4P arises simply because PtdIns4P is the predominant phosphoinositide in the Golgi. The origin of the reported PtdIns4P specificity for the FAPP1 PH domain in lipid overlay studies remains unclear. It is also difficult to explain the Biacore data presented by Dowler et al. [28] that suggest a similar specificity – although these data are difficult to evaluate as presented.
3.2 PtdIns(3,5)P2-specific binding proteins
Similar discrepancies have arisen in the study of potential targets for PtdIns(3,5)P2, a phosphoinositide involved in intracellular trafficking and vacuolar homeostasis [66, 67]. In a similar vein to that described for the FAPP1 PH domain and PtdIns4P, we have been unable, with the vesicle-based techniques described above, to detect PtdIns(3,5)P2 specificity for two interesting classes of protein: mammalian Vps24p [22] (a component of the ESCRT III complex), and Ent3p/Ent5p from S. cerevisiae [11, 68]. In both of these cases, the primary evidence in support of PtdIns(3,5)P2 specificity came from studies in which the phosphoinositide was studied in isolation. Phage display was used to identify mVps24p as a protein that binds to immobilized biotinylated PtdIns(3,5)P2 [22]. Lipid overlay/dot-blot approaches provided the initial suggestion that the ENTH domains of Ent3p and Ent5p bind PtdIns(3,5)P2 [11, 68]. Both sets of studies also employed vesicle sedimentation approaches using 5% (mole/mole) phosphoinositide in a reasonable lipid background. These studies appeared to support PtdIns(3,5)P2 specificity for Ent3p/Ent5p, but showed more promiscuity for mVps24p, which also bound detectably to PtdIns(3,4)P2 and PtdIns3P in vesicles [22]. It should be noted, though, that nothing was done to increase vesicle density in these studies, so only a fraction of the lipid (that with highest density – from protein binding and/or lipid precipitation) will be found in the pellet. Moreover, these studies did not measure binding curves or provide any other quantitative evaluation of lipid binding. Our own quantitative studies of these proteins using SPR and the vesicle centrifugation assay described above indicate substantially less specificity for PtdIns(3,5)P2 than suggested in the published reports. In our studies, mVps24p appears to bind similarly (with KD > 50 µM) to both PtdIns(4,5)P2 and PtdIns(3,5)P2 – although both of these bound significantly better than the phosphatidylinositol monophosphates [69]. In our hands, the Ent3p and Ent5p ENTH domains also do not appear to distinguish the more abundant PtdIns(4,5)P2 from PtdIns(3,5)P2. In this regard they appear more like other PtdIns(4,5)P2-binding ENTH domains [10], arguing against the function of these proteins as potential PtdIns(3,5)P2-effectors.
By contrast with these examples, a recent study by Dove et al. [57], including the SPR approach described here, identified the S. cerevisiae protein Atg18p (also called Svp1p) as a highly specific PtdIns(3,5)P2-binding protein. The clear specificity of Atg18p for PtdIns(3,5)P2 over PtdIns(4,5)P2 and PtdIns3P is illustrated in the SPR data shown in Figure 3B. A related Drosophila protein, known as Dm3, shows a similar specificity for PtdIns(3,5)P2 over PtdIns(4,5)P2 in this assay, but does not appear to distinguish PtdIns(3,5)P2 from PtdIns3P, which has significant functional implications. Atg18p/Svp1p was first identified as a possible PtdIns(3,5)P2 effector on the basis that its deletion in S. cerevisiae gives rise to a phenotype similar to that seen when yeast do not express Fab1p, the PtdIns3P 5-kinase that produces PtdIns(3,5)P2 [57]. The PtdIns(3,5)P2 specificity seen for Atg18p/Svp1p in our vesicle studies was also recapitulated in studies using pure phosphoinositides, although the dot-blot/lipid overlay approach appears to have greatly overestimated the extent to which it binds PtdIns3P – possibly as a result of the lipid solubility issue mentioned in Section 2.2.3. Strømhaug et al. [70] have made a similar observation with Atg18p and its two close relatives in S. cerevisiae, Atg21p and Hsv2p/Ygr223cp. Employing a dot-blot/lipid overlay approach with maltose binding protein fusions, Strømhaug et al. saw a significant signal for PtdIns(3,5)P2 and PtdIns5P binding, but stronger signals for binding to PtdIns4P and PtdIns3P [70]. Since the location of these proteins was altered in yeast cells with impaired PtdIns3P production, the authors proposed that Atg18p, Atg21p and Hsv2p are specific PtdIns3P-binding proteins. However, they reported no analysis with lipid vesicles containing physiologically reasonable levels of phosphoinositide, and it should be noted that PtdIns3P is also the precursor for PtdIns(3,5)P2 production. As mentioned previously, the reduced water solubility of PtdIns3P/PtdIns4P/PtdIns5P causes the apparent binding to these phosphoinositides to be over-represented in dot-blot/lipid overlay studies compared with binding to PtdInsP2 isomers including PtdIns(3,5)P2. We suggest that this provides the explanation for the discrepancy between the findings of Strømhaug et al. [70] and those presented in Figure 3B [57]. According to the methodological considerations presented in this article, we are confident that yeast Atg18p, Atg21p and Hsv2p/Ygr223cp are specific for PtdIns(3,5)P2 under physiological conditions. The Drosophila Dm3 homolog does bind similarly to PtdIns3P, as has also been described for a human homolog known as WIPI49 or WIPI-1 [71], so the phosphoinositide-binding characteristics of this family requires further detailed analysis.
4. Concluding Remarks
The key point to stress here is that any method employed to study phosphoinositide binding specificity and/or affinity has its weaknesses and strengths. Therefore, to establish any hypothesized binding characteristics it is very important to apply several techniques, preferably more than two. The different approaches should agree with one another, and at least one should be quantitative, so that a KD value can be discussed. This last criterion is frequently overlooked. The methods that we have outlined in this article each have their place. The dot-blot approach has the advantage of relatively rapid throughput, but has several weaknesses including the entirely qualitative nature of its results and its tendency to over-represent binding to PtdIns4P, PtdIns3P and PtdIns5P. The vesicle centrifugation approach is ideal for studying interactions with low affinities in the 5–40 µM range, but cannot be used if the protein or domain is prone to precipitation, and requires quite a large amount of material – although substantially less than isothermal titration calorimetry. Another difficulty here is that the brominated distearoyl PC used in our centrifugation approach has a very high phase transition temperature, so is performed with lipids in the gel phase. Finally, the surface plasmon resonance approach is our current method of choice. It can be quantitated readily, requires relatively small amounts of protein and lipid, and is highly reproducible. One disadvantage is that one needs access to the SPR instrument, another is that the precise nature of the immobilized lipid vesicles is still not completely clear. Given these considerations, we generally employ any two of these approaches – most often dot-blots and SPR.
Even if a clear consensus can be reached concerning phosphoinositide binding in vitro, it should be stressed that there is no guarantee that this will be relevant in vivo. Relevance in vivo can be assessed using methods not described in this article, but outlined elsewhere (e.g. [5]). Work from several laboratories in S. cerevisae has led to the accumulation of yeast strains that are temperature sensitive or defective in one or other (or more) of the kinases and phosphatases responsible for generating and degrading phosphoinositides [72]. Using such strains with depressed or elevated levels of a particular phosphoinositide(s) in combination with green fluorescent protein fusion proteins of the domains in which one is interested can be a particularly powerful way of assessing the in vivo relevance of in vitro-determined phosphoinositide binding specificities, as exemplified by our recent study of yeast PH domains [27].
Acknowledgements
Work in this area in the Lemmon laboratory is supported by NIGMS grant RO1-GM056846 (to M.A.L.). K.N. is supported by NRSA F32-GM073496 from NIGMS.
References
- 1.Lemmon MA. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 2.DiNitto JP, Cronin TC, Lambright DG. Sci. STKE 2003. 2003:re16. doi: 10.1126/stke.2132003re16. [DOI] [PubMed] [Google Scholar]
- 3.Yu JW, Lemmon MA. Curr. Opin. Chem. Biol. 2003;7:103–109. doi: 10.1016/s1367-5931(02)00008-x. [DOI] [PubMed] [Google Scholar]
- 4.Cho W, Stahelin RV. Annu. Rev. Biophys. Biomol. Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 5.Downes CP, Gray A, Lucocq JM. Trends Cell Biol. 2005;15:259–268. doi: 10.1016/j.tcb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Lemmon MA, Ferguson KM. Biochem. J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- 7.Lemmon MA, Ferguson KM. Biochem. Soc. Trans. 2001;29:377–384. doi: 10.1042/bst0290377. [DOI] [PubMed] [Google Scholar]
- 8.Ellson CD, Andrews S, Stephens LR, Hawkins PT. J. Cell Sci. 2002;115:1099–1105. doi: 10.1242/jcs.115.6.1099. [DOI] [PubMed] [Google Scholar]
- 9.Gillooly DJ, Simonsen A, Stenmark H. Biochem. J. 2001;355:249–258. doi: 10.1042/0264-6021:3550249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itoh T, Takenawa T. Curr. Top. Microbiol. Immunol. 2004;282:31–47. doi: 10.1007/978-3-642-18805-3_2. [DOI] [PubMed] [Google Scholar]
- 11.Friant S, Pecheur EI, Eugster A, Michel F, Lefkir Y, Nourrisson D, Letourneur F. Dev. Cell. 2003;5:499–511. doi: 10.1016/s1534-5807(03)00238-7. [DOI] [PubMed] [Google Scholar]
- 12.Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 13.Zimmermann P, Meerschaert K, Reekmans G, Leenaerts I, Small JV, Vandekerckhove J, David G, Gettemans J. Mol. Cell. 2002;9:1215–1225. doi: 10.1016/s1097-2765(02)00549-x. [DOI] [PubMed] [Google Scholar]
- 14.Barret C, Roy C, Montcourrier P, Mangeat P, Niggli V. J. Cell Biol. 2000;151:1067–1080. doi: 10.1083/jcb.151.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 16.McLaughlin S, Hangyas-Mihalyne G, Zaitseva I, Golebiewska U. Biochem. Soc. Symp. 2005;72:189–198. doi: 10.1042/bss0720189. [DOI] [PubMed] [Google Scholar]
- 17.Balla T, Varnai P. Sci. STKE 2002. 2002:PL3. doi: 10.1126/stke.2002.125.pl3. [DOI] [PubMed] [Google Scholar]
- 18.Dowler S, Currie RA, Downes CP, Alessi DR. Biochem. J. 1999;342(Pt 1):7–12. [PMC free article] [PubMed] [Google Scholar]
- 19.Dowler S, Kular G, Alessi DR. Sci. STKE 2002. 2002:PL6. doi: 10.1126/stke.2002.129.pl6. [DOI] [PubMed] [Google Scholar]
- 20.Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, Lemmon MA. J. Biol. Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, Hortsman H, Seet L, Wong SH, Hong W. Nat. Cell Biol. 2001;3:658–666. doi: 10.1038/35083051. [DOI] [PubMed] [Google Scholar]
- 22.Whitley P, Reaves BJ, Hashimoto M, Riley AM, Potter BV, Holman GD. J. Biol. Chem. 2003;278:38786–38795. doi: 10.1074/jbc.M306864200. [DOI] [PubMed] [Google Scholar]
- 23.Rameh LE, Arvidsson A, Carraway KL, Couvillon AD, Rathbun G, Crompton A, VanRenterghem B, Czech MP, Ravichandran KS, Burakoff SJ, Wang DS, Chen CS, Cantley LC., 3rd J. Biol. Chem. 1997;272:22059–22066. doi: 10.1074/jbc.272.35.22059. [DOI] [PubMed] [Google Scholar]
- 24.Stevenson JM, Perera IY, Boss WF. J. Biol. Chem. 1998;273:22761–22767. doi: 10.1074/jbc.273.35.22761. [DOI] [PubMed] [Google Scholar]
- 25.Towbin H, Schoenberger C, Ball R, Braun DG, Rosenfelder G. J. Immunol. Meth. 1984;72:471–479. doi: 10.1016/0022-1759(84)90015-2. [DOI] [PubMed] [Google Scholar]
- 26.Yu JW, Lemmon MA. J. Biol. Chem. 2001;276:44179–44184. doi: 10.1074/jbc.M108811200. [DOI] [PubMed] [Google Scholar]
- 27.Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Mol. Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 28.Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Biochem. J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ron D, Dressler H. Biotechniques. 1992;13:866–869. [PubMed] [Google Scholar]
- 30.Margolis B, Young RA. In: DNA Cloning. Glover DM, Hames BD, editors. Vol. 2. Oxford, UK: IRL Press; 1995. pp. 1–14. [Google Scholar]
- 31.Lee A, Frank DW, Marks MS, Lemmon MA. Curr. Biol. 1999;9:261–264. doi: 10.1016/s0960-9822(99)80115-8. [DOI] [PubMed] [Google Scholar]
- 32.Achiriloaie M, Barylko B, Albanesi JP. Mol. Cell. Biol. 1999;19:1410–1415. doi: 10.1128/mcb.19.2.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vallis Y, Wigge P, Marks B, Evans PR, McMahon HT. Curr. Biol. 1999;9:257–260. doi: 10.1016/s0960-9822(99)80114-6. [DOI] [PubMed] [Google Scholar]
- 34.Audhya A, Loewith R, Parsons AB, Gao L, Tabuchi M, Zhou H, Boone C, Hall MN, Emr SD. EMBO J. 2004;23:3747–3757. doi: 10.1038/sj.emboj.7600384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ladbury JE. Biotechniques. 2004;37:885–887. doi: 10.2144/04376TE01. [DOI] [PubMed] [Google Scholar]
- 36.Wiseman T, Williston S, Brandts JF, Lin LN. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 37.Corbin JA, Dirkx RA, Falke JJ. Biochemistry. 2004;43:16161–16173. doi: 10.1021/bi049017a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gambhir A, Hangyas-Mihalyne G, Zaitseva I, Cafiso DS, Wang J, Murray D, Pentyala SN, Smith SO, McLaughlin S. Biophys. J. 2004;86:2188–2207. doi: 10.1016/S0006-3495(04)74278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemmon MA, Ferguson KM, O’Brien R, Sigler PB, Schlessinger J. Proc. Natl. Acad. Sci. U. S. A. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia P, Gupta R, Shah S, Morris AJ, Rudge SA, Scarlata S, Petrova V, McLaughlin S, Rebecchi MJ. Biochemistry. 1995;34:16228–16234. doi: 10.1021/bi00049a039. [DOI] [PubMed] [Google Scholar]
- 41.Hirose K, Kadowaki S, Tanabe M, Takeshima H, Iino M. Science. 1999;284:1527–1530. doi: 10.1126/science.284.5419.1527. [DOI] [PubMed] [Google Scholar]
- 42.Rebecchi M, Peterson A, McLaughlin S. Biochemistry. 1992;31:12742–12747. doi: 10.1021/bi00166a005. [DOI] [PubMed] [Google Scholar]
- 43.Tortorella D, London E. Anal. Biochem. 1994;217:176–180. doi: 10.1006/abio.1994.1106. [DOI] [PubMed] [Google Scholar]
- 44.Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 45.Peitzsch RM, McLaughlin S. Biochemistry. 1993;32:10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- 46.Sankaran VG, Klein DE, Sachdeva MM, Lemmon MA. Biochemistry. 2001;40:8581–8587. doi: 10.1021/bi010425d. [DOI] [PubMed] [Google Scholar]
- 47.Zheng J, Cahill SM, Lemmon MA, Fushman D, Schlessinger J, Cowburn D. J. Mol. Biol. 1996;255:14–21. doi: 10.1006/jmbi.1996.0002. [DOI] [PubMed] [Google Scholar]
- 48.Salim K, Bottomley MJ, Querfurth E, Zvelebil MJ, Gout I, Scaife R, Margolis RL, Gigg R, Smith CI, Driscoll PC, Waterfield MD, Panayotou G. EMBO J. 1996;15:6241–6250. [PMC free article] [PubMed] [Google Scholar]
- 49.Tudyka T, Skerra A. Protein Sci. 1997;6:2180–2187. doi: 10.1002/pro.5560061012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maru Y, Afar DE, Witte ON, Shibuya M. J. Biol. Chem. 1996;271:15353–15357. doi: 10.1074/jbc.271.26.15353. [DOI] [PubMed] [Google Scholar]
- 51.Klein DE, Lee A, Frank DW, Marks MS, Lemmon MA. J. Biol. Chem. 1998;273:27725–27733. doi: 10.1074/jbc.273.42.27725. [DOI] [PubMed] [Google Scholar]
- 52.Binns DD, Barylko B, Grichine N, Atkinson MA, Helms MK, Jameson DM, Eccleston JF, Albanesi JP. J. Prot. Chem. 1999;18:277–290. doi: 10.1023/a:1021083211267. [DOI] [PubMed] [Google Scholar]
- 53.Jönsson U, Fägerstam L, Ivarsson B, Johnsson B, Karlsson R, Lundh K, Löfäs S, Persson B, Roos H, Rönnberg I, Sjölander S, Stenberg E, Stahlberg C, Malqvist M. Biotechniques. 1991;11:620–627. [PubMed] [Google Scholar]
- 54.Elleman TC, Domagala T, McKern NM, Nerrie M, Lonnqvist B, Adams TE, Lewis J, Lovrecz GO, Hoyne PA, Richards KM, Howlett GJ, Rothacker J, Jorissen RN, Lou M, Garrett TP, Burgess AW, Nice EC, Ward CW. Biochemistry. 2001;40:8930–8939. doi: 10.1021/bi010037b. [DOI] [PubMed] [Google Scholar]
- 55.Zhou M, Felder S, Rubinstein M, Hurwitz DR, Ullrich A, Lax I, Schlessinger J. Biochemistry. 1993;32:8193–8198. doi: 10.1021/bi00083a020. [DOI] [PubMed] [Google Scholar]
- 56.Flesch FM, Yu JW, Lemmon MA, Burger KN. Biochem. J. 2005;389:435–441. doi: 10.1042/BJ20041721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dove SK, Piper RC, McEwen RK, Yu JW, King MC, Hughes DC, Thuring J, Holmes AB, Cooke FT, Michell RH, Parker PJ, Lemmon MA. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W. J. Biol. Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- 59.Erb EM, Chen X, Allen S, Roberts CJ, Tendler SJ, Davies MC, Forsen S. Anal. Biochem. 2000;280:29–35. doi: 10.1006/abio.1999.4469. [DOI] [PubMed] [Google Scholar]
- 60.Höning S, Ricotta D, Krauss M, Spate K, Spolaore B, Motley A, Robinson M, Robinson C, Haucke V, Owen DJ. Mol. Cell. 2005;18:519–531. doi: 10.1016/j.molcel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 61.Levine TP, S M. Curr. Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- 62.Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. Nat. Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- 63.Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T. Mol. Biol. Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stefan CJ, Audhya A, Emr SD. Mol. Biol. Cell. 2002;13:542–557. doi: 10.1091/mbc.01-10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levine TP, Munro S. Curr. Biol. 1998;8:729–739. doi: 10.1016/s0960-9822(98)70296-9. [DOI] [PubMed] [Google Scholar]
- 66.Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 67.Dove SK, McEwen RK, Cooke FT, Parker PJ, Michell RH. Biochem. Soc. Trans. 1999;27:674–677. doi: 10.1042/bst0270674. [DOI] [PubMed] [Google Scholar]
- 68.Eugster A, Pecheur EI, Michel F, Winsor B, Letourneur F, Friant S. Mol. Biol. Cell. 2004;15:3031–3041. doi: 10.1091/mbc.E03-11-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baumeister MA. Ph.D. Thesis. Philadelphia, PA: University of Pennsylvania; 2005. [Google Scholar]
- 70.Strømhaug PE, Reggiori F, Guan J, Wang CW, Klionsky DJ. Mol. Biol. Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jeffries TR, Dove SK, Michell RH, Parker PJ. Mol. Biol. Cell. 2004;15:2652–2663. doi: 10.1091/mbc.E03-10-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Odorizzi G, Babst M, Emr SD. Trends Biochem. Sci. 2000;25:229–235. doi: 10.1016/s0968-0004(00)01543-7. [DOI] [PubMed] [Google Scholar]