Abstract
Peptide TZ1C2 can populate two distinct orientations: a staggered (out-of-register) fibril and an aligned (in-register) coiled-coil trimer. The coordination of two cadmium ions induces a registry shift that results in a reversible transition between these structural forms. This process recapitulates the self-assembly mechanism of native protein fibrils in which a ligand binding event gates a reversible conformational transition between alternate forms of a folded peptide structure.
Sequence-specific biomolecules, i.e., proteins and nucleic acids, confer significant advantages as substrates for the construction of structurally complex supramolecular materials.1–3 Foremost, the sequence-structure correlations that have been elucidated from physical analysis of native biomolecular structures provide a context for the rational design of novel materials.4–6 However, self-assembly of most designed peptide-based materials occurs commensurately with protein folding. In contrast, native protein assemblies undergo reversible self-association due to subtle shifts in conformation that are propagated hierarchically. We describe herein a mechanism to control supramolecular assembly of a synthetic peptide-based material through registry selection. This process recapitulates the native mechanism of fibril assembly in which a ligand binding event gates a reversible conformational transition between alternate forms of a folded peptide structure.
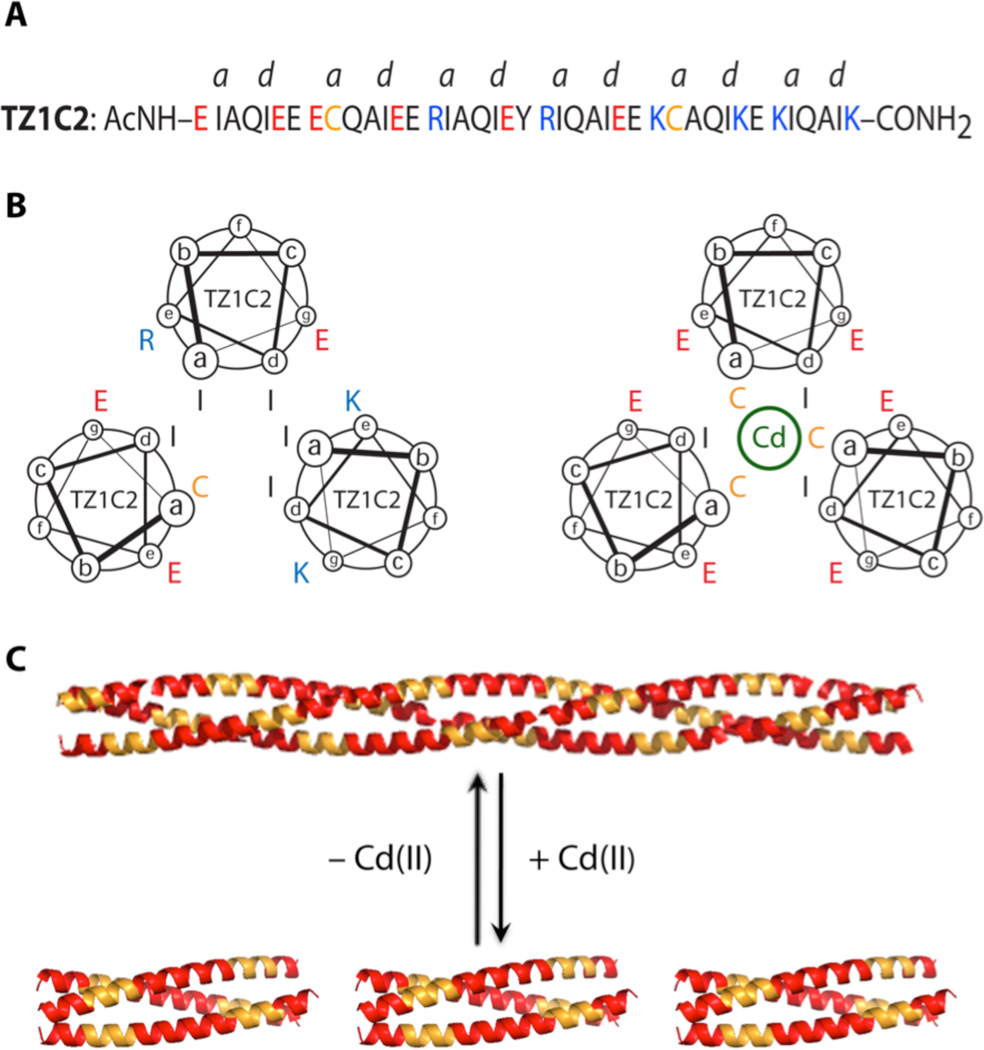
Peptide TZ1C2, a 41-residue sequence comprising six heptad repeats (Figure 1), was derived from modification of a previously reported trimeric coiled-coil TZ1.7 Sequence variants of peptide TZ1 have been shown to self-assemble into high aspect-ratio helical fibrils, in which lateral registration between adjacent helical protomers is enforced through Coulombic interactions between charged residues at the e- and g-positions of the heptad repeats. Peptide TZ1C2 differs from TZ1 in that two cysteine residues were introduced in place of isoleucines at core a-positions within the second and fifth heptad sequences. The sulfhydryl groups of the cysteine residues have the latent capacity to serve as ligands for the coordination of metal ions.8 However, in order to create effective binding sites, the cysteine residues should be oriented proximally across the helical interface, which requires an in-register alignment of helices within a trimeric coiled-coil structure.
Figure 1.
(A) Amino acid sequence of peptide TZ1C2. (B) Helical wheel representation of core layer packing within the second heptad of TZ1C2 in the staggered (left) and aligned (right) orientations. (C) Schematic representation of the registry shift that results from Cd(II) coordination to the cysteine residues of TZ1C2 (yellow: Cys-containing heptads and red: non-Cys-containing heptads).
Peptide TZ1C2 can populate two distinct orientations within this structural context. The preferred orientation of helices depends on the presence of a metal ion, such as Cd(II), that can form a stable complex with the thiolate ligands of TZ1C2. Metal ion coordination may provide sufficient thermodynamic driving force to overcome electrostatic repulsion between residues at the e/g-positions and drive re-alignment of the structure (Figure 1). In the absence of metal coordination, electrostatic interactions and steric complementation9 at core positions should favor the staggered alignment.7 Thus, the balance of intermolecular forces can be manipulated through changes in environmental conditions to shift the helices reversibly from a staggered (out-of-register)7,10–11 to aligned (in-register) orientation with a concomitant transition from a fibril to a discrete helical bundle.
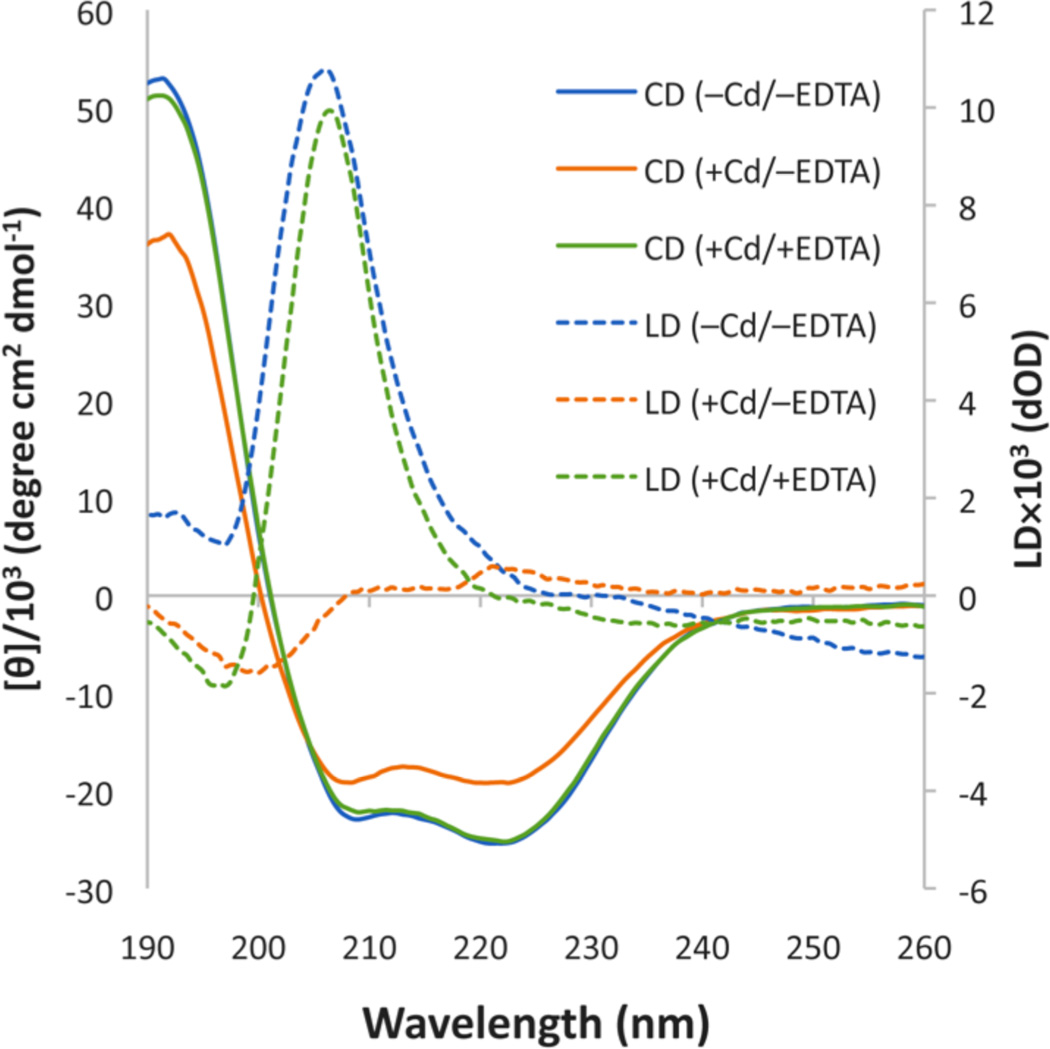
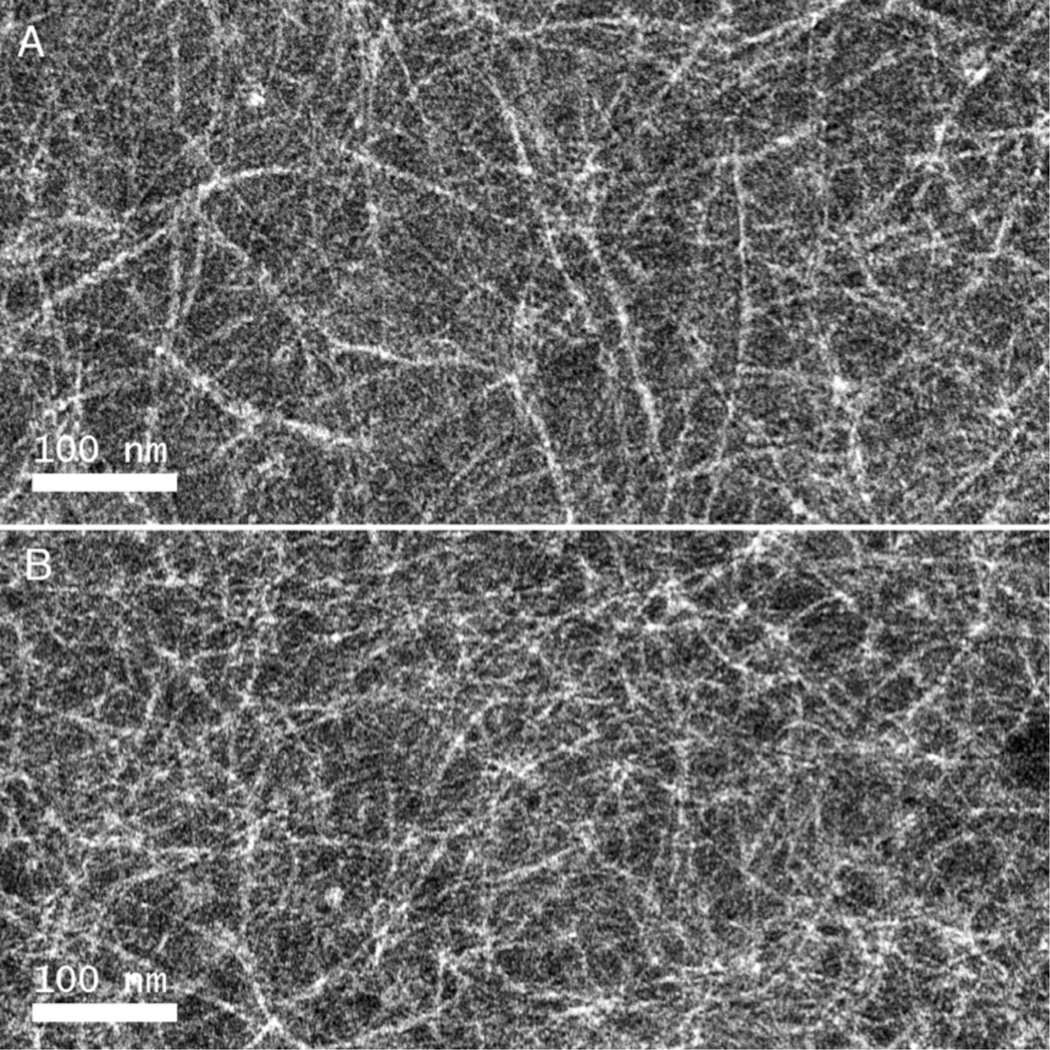
The circular dichroism (CD) spectrum of TZ1C2 (100 µM) in TAPS buffer was consistent with the presence of an α-helical conformation throughout the pH range from 6 to 9 (Figure 2). Flow linear dichroism (LD) of an aqueous solution of TZ1C2 (100 µM) in TAPS buffer (10 mM, pH 8.5) displays a strong positive signal at 206 nm under a Couette flow of 3,000 rpm (Figure 2). The observed spectroscopic response indicated the presence of extended helical assemblies of TZ1C2 that are stable to flow alignment. The flow LD data suggested a conformational arrangement of TZ1C2 in which the amide bonds, and consequently, the α-helices, are oriented parallel to the flow direction. Similar behavior has been observed in the flow LD spectra of synthetic coiled-coil fibers and tropomyosin under flow alignment.12 TEM analysis of assemblies derived from TZ1C2 confirmed the flow linear dichroism analysis. High aspect-ratio fibrils are observed in negatively stained specimens of TZ1C2 at concentrations ≥ 100 µM (Figure 3). The combined data support the hypothesis that TZ1C2 self-assembles into α-helical fibrils in which the peptides adopt a parallel, out-of-register orientation as observed previously for other TZ1 peptide derivatives.7
Figure 2.
Circular dichroism and flow linear dichroism spectra of peptide TZ1C2 (100 µM) in TAPS buffer (10 mM, pH 8.5) and NaCl (100 mM).
Figure 3.
Negative stain TEM images of TZ1C2 fibrils (500 µM in TAPS buffer (10 mM, pH 8.5)) and NaCl (100 mM) before Cd(II) addition (A) and after Cd(II) addition followed treatment with excess EDTA (B).
The scheme for dis-assembly of the TZ1C2 fibrils requires the presence of a metal ion to induce a registry shift that orients the cysteine residues within the same layer of the triple helical coiled-coil (Figure 1). Previous investigations have demonstrated that the chalcophilic cadmium(II) ion can bind to structurally related coiled-coil trimers in which cysteines have been substituted into core a- or d-positions within the heptad repeats.13–15 Cysteine residues were placed at the a-positions of the second and fifth heptad repeats of TZ1C2. Prior research had indicated a preference for Cd(II) complex formation at the trigonal binding sites at the a-postions of coiled-coil trimers.16 In addition, cadmium has the advantage that complexation can be monitored using 113Cd NMR spectroscopy17 and 111mCd perturbed angular correlation (PAC) spectroscopy,18 which are sensitive tools for analysis of ligation environment and coordination geometry of metal ion-binding sites in proteins.19
Aqueous solutions of peptide TZ1C2 (100 µM) were treated with two equivalents of cadmium(II) nitrate (67 µM) in TAPS buffer (10 mM, pH 8.5, 100 mM NaCl). Each peptide contains two cysteine residues that contribute one-third of a cadmium(II) binding site within the trimeric coiled-coil structure. CD titration experiments suggested that this concentration of cadmium should be sufficient to saturate the two trivalent binding sites created within the trimeric coiled-coil assembly (see Supporting Information). CD spectropolarimetry of Cd(II)-containing solutions of peptide TZ1C2 indicated that the α-helical conformation was retained in the complex, although the net helicity was reduced with respect to the corresponding value in the absence of cadmium ion (Figure 2).
The flow linear dichroism spectrum of TZ1C2 was significantly reduced in the presence of cadmium(II). The strong positive signal at 206 nm disappeared and was replaced with a weak negative signal at 200 nm (Figure 2). The spectroscopic behavior was consistent with disassembly of TZ1C2 from extended fibrillar structures into smaller species that could not be aligned under Couette flow. Similarly, TEM analysis did not detect the formation of fibrils in aqueous solutions of TZ1C2 in the presence of cadmium(II) (see Supporting Information). The dis-assembly was readily reversed in the presence of excess EDTA (200 µM), which can tightly bind to Cd(II) ion.19 The addition of EDTA restored the original flow LD signal (Figure 2) and induced the formation of fibrils that were observed in the TEM analysis (Figure 3).
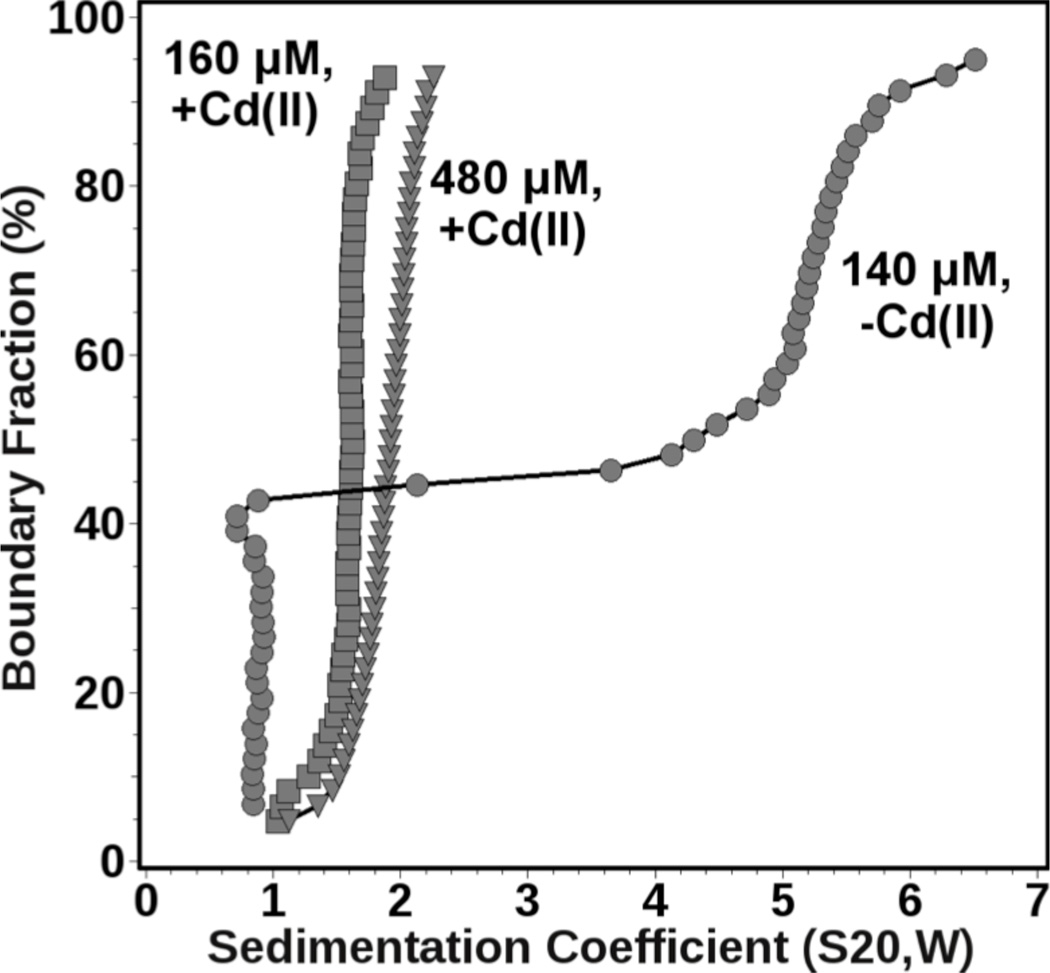
The oligomeric state of TZ1C2 in solution was investigated in the presence and absence of Cd(II) ion by sedimentation velocity analytical ultracentrifugation.21 The effect of Cd(II) ion was further examined as a function of TZ1C2 concentration. When TZ1C2 was measured under low loading concentration (160 µM) in the presence of Cd(II), a slightly heterogeneous sedimentation distribution with a weight-average s-value of as 1.68S was observed. Further analysis by genetic algorithm-Monte Carlo22 methods indicated the presence of a small amount of monomeric TZ1C2 species, as well as a major trimeric species (42% of absorption), and a small amount of a larger oligomer consistent with a hexameric species. Under higher loading concentration (480 µM), the weight average s-value shifted slightly to 1.89S, and the relative amount of trimer increased to 62% of absorption at the expense of the other species. This observation is consistent with a reversible mass action effect. In the absence of Cd(II), the sedimentation distributions changed drastically, and 38% of the absorbance corresponded to a monomeric species at low loading concentration (140 µM), while the remaining absorbance sedimented as a heterogeneous mixture with s-values ranging between 3–7S, corresponding to highly anisotropic aggregates ranging between 0.4–1 million Da in size (Figure 4). The latter species are consistent with the fibrils observed in the TEM images of TZ1C2. From these data it can be concluded that Cd(II) facilitates the formation of reversibly self-associating trimer, while, in the absence of Cd(II), TZ1C2 is partially monomeric and partially forms much larger, irreversible aggregates. In addition, non-denaturing nano-electrospray ionization mass spectrometry confirmed the identity of the TZ1C2 trimer as the di-cadmium adduct (Supporting FigureS7).
Figure 4.
Diffusion-corrected integral sedimentation coefficient distributions obtained from a van Holde-Weischet23 analysis. Shown are results for TZ1C2 in the presence of excess Cd(II) at 160 µM (squares) and 480 µM (triangles) and in the absence of Cd(II) at 140 µM (circles) loading concentration.
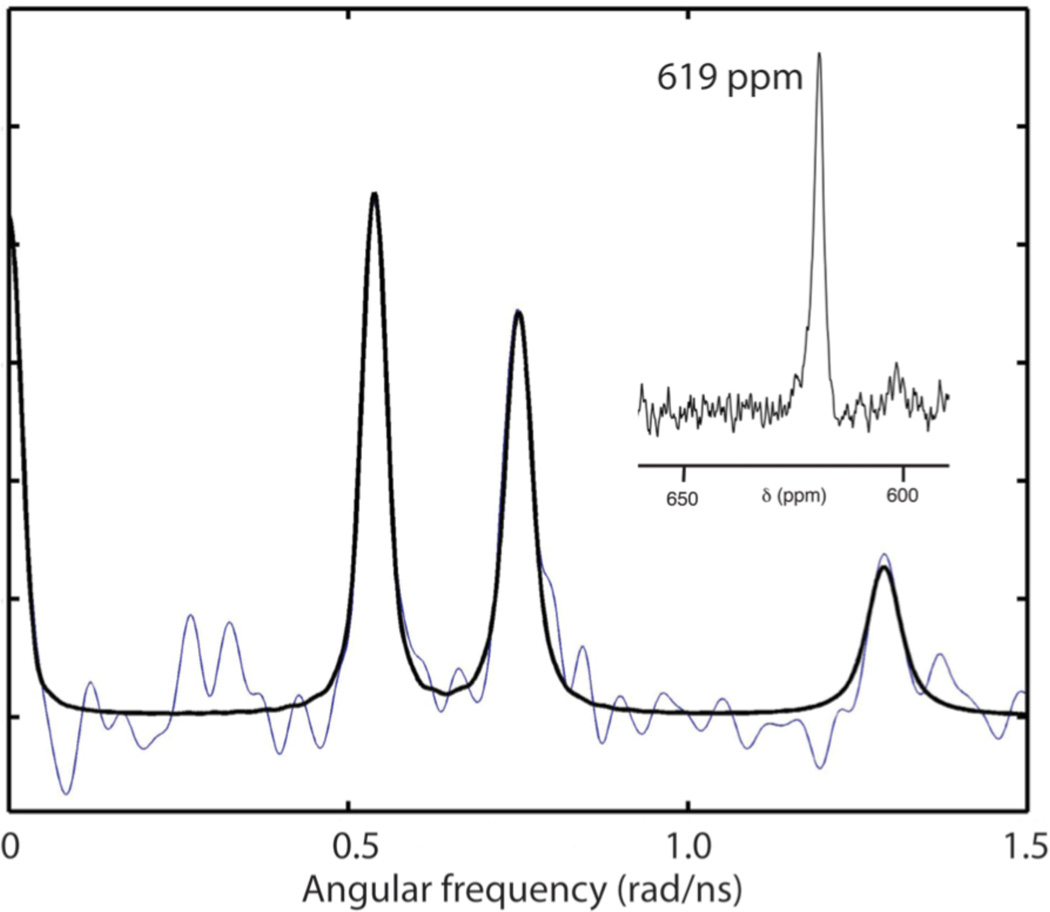
The structural role of the Cd(II) ion in the TZ1C2 assemblies was interrogated using a combination of 113Cd NMR spectroscopy and 111mCd PAC spectroscopy (Figure 5) performed on isotopically enriched specimens. A single resonance was observed at 619 ppm in the 113Cd NMR spectrum of an aqueous solution of peptide TZ1C2 that had been treated with 2 equivalents of 113CdCl2. The chemical shift was consistent with nearly identical CdS3X coordination environments for the two structurally similar metal ion binding sites in TZ1C2. The 113Cd NMR spectroscopic data were similar to that reported for Cd-(S-cysteinyl)3 complexes of structurally analogous coiled-coil trimers within the TRI series.13,19 These spectroscopic results were interpreted in terms of a dynamic equilibrium on the NMR time-scale for a Cd(II) complex that involved three cysteinyl thiolate groups and a reversibly bound water molecule (X).
Figure 5.
Fourier transformed 111mCd PAC spectroscopic data (blue: experimental data; black: fit) for the Cd(II) complex of TZ1C2. Inset: 113Cd NMR spectrum of the Cd(II) complex of TZ1C2 trimer.
However, the 111mCd PAC spectroscopic data appear to refute a similar scenario for the cadmium(II) complex of TZ1C2. The 111mCd PAC spectrum of fully complexed TZ1C2 gives a single very well defined nuclear quadrupole interaction (NQI), indicating that all Cd(II) ions are found in practically the same coordination geometry, i.e., the two Cd(II) binding sites are highly similar (Figure 5). The NQI, and thus the local structure, is different from those observed for Cd(II) complexes of the TRI peptide family. Both the frequency (ω0 about 0.399 rad/ns) and the asymmetry parameter (η about 0.61) are relatively high for the TZ1C2 complex. The high asymmetry parameter indicates the absence of axial symmetry, i.e., idealized trigonal planar CdS3, and tetrahedral CdS3X structures can be ruled out. The combined spectroscopic data suggest the presence of a single type of Cd(II) binding site, most likely of the CdS3X type, in which the X-ligand is not in dynamic exchange. The deviation from an axially symmetric coordination environment may arise from structural distortions due to electrostatic repulsion between similarly charged e/g-residues at the Cd(II) coordination sites within the inregister coiled-coil trimer (Figure 1).
These data demonstrate that metal ion coordination can reversibly control chain registry and, consequently, the assembly state of a designed nanomaterial. A similar phenomenon has been observed adventitiously for β-sheet assemblies,24–25 in which pH-dependent changes in strand registry can trigger reversible transitions between fibril and nanotube structures.24 In addition, experimental evidence suggests that a registry shift between helices in an anti-parallel coiled-coil dimer within the cytoskeletal motor protein dynein may underlie the mechanism of locomotion.26 Thus, control of chain alignment represents an attractive strategy for the design of dynamically reconfigurable nano-scale materials.
Supplementary Material
ACKNOWLEDGMENT
This research was supported from NSF and DOE grants (V.P.C) and the Danish Council for Independent Research | Natural Sciences (L.H.). The development of the UltraScan software is supported by the National Institutes of Health through grant RR022200 (to B.D.). Supercomputer time allocations were provided through NSF grant TGMCB070039 (to B.D.).
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental methods of peptide synthesis and characterization. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Hauser CA, Zhang S. Chem. Soc. Rev. 2010;39:2780–2790. doi: 10.1039/b921448h. [DOI] [PubMed] [Google Scholar]
- 2.Woolfson DN, Mahmoud ZN. Chem. Soc. Rev. 2010;39:3464–3479. doi: 10.1039/c0cs00032a. [DOI] [PubMed] [Google Scholar]
- 3.Tørring T, Voigt NV, Nangreave J, Yan H, Gothelf KV. Chem. Soc. Rev. 2011;40:5636–5646. doi: 10.1039/c1cs15057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King NP, Sheffler W, Sawaya MR, Vollmar BS, Sumida JP, André I, Gonen T, Yeates TO, Baker D. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lanci CJ, MacDermaid CM, Kang SG, Acharya R, North B, Yang X, Qiu XJ, DeGrado WF, Saven JG. Proc. Natl. Acad. Sci. U.S.A. 2012;109:7304–7309. doi: 10.1073/pnas.1112595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodin J, Ambroggio X, Tang C, Parent K, Baker T, Tezcan FA. Nat. Chem. 2012;4:375–382. doi: 10.1038/nchem.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Zimenkov Y, Dublin SN, Ni R, Tu RS, Breedveld V, Apkarian RP, Conticello VP. J. Am. Chem. Soc. 2006;128:6770–6771. doi: 10.1021/ja0605974. [DOI] [PubMed] [Google Scholar]; (b) Dublin SN, Conticello VP. J. Am. Chem. Soc. 2008;130:49–51. doi: 10.1021/ja0775016. [DOI] [PubMed] [Google Scholar]
- 8.Peacock AFA, Iranzo O, Pecoraro VL. Dalton Trans. 2009:2271–2280. doi: 10.1039/b818306f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnarr NA, Kennan AJ. J. Am. Chem. Soc. 2002;124:9779–9783. doi: 10.1021/ja0174940. [DOI] [PubMed] [Google Scholar]
- 10.Potekhin SA, Melnik TN, Popov V, Lanina NF, Vazina AA, Rigler P, Verdini AS, Corradin G, Kajava AV. Chem. Biol. 2001;8:1025–1032. doi: 10.1016/s1074-5521(01)00073-4. [DOI] [PubMed] [Google Scholar]
- 11.Papapostolou D, Smith AM, Atkins ED, Oliver SJ, Ryadnov MG, Serpell LC, Woolfson DN. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10853–10858. doi: 10.1073/pnas.0700801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulheller BM, Rodger A, Hicks MR, Dafforn TR, Serpell LC, Marshall KE, Bromley EH, King PJ, Channon KJ, Woolfson DN, Hirst JD. J. Am. Chem. Soc. 2009;131:13305–13314. doi: 10.1021/ja902662e. [DOI] [PubMed] [Google Scholar]
- 13.Matzapetakis M, Farrer BT, Weng TC, Hemmingsen L, Penner-Hahn JE, Pecoraro VL. J. Am. Chem. Soc. 2002;124:8042–8054. doi: 10.1021/ja017520u. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Suzuki K, Kanaori K, Tajima K, Kashiwada A, Hiroaki H, Kohda D, Tanaka T. Protein Sci. 2000;9:1327–1333. doi: 10.1110/ps.9.7.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kharenko OA, Ogawa MY. J. Inorg. Biochem. 2004;98:1971–1974. doi: 10.1016/j.jinorgbio.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 16.Matzapetakis M, Pecoraro VL. J. Am. Chem. Soc. 2005;127:18229–18233. doi: 10.1021/ja055433m. [DOI] [PubMed] [Google Scholar]
- 17.Hemmingsen L, Olsen L, Antony J, Sauer SP. J. Biol. Inorg. Chem. 2004;9:591–599. doi: 10.1007/s00775-004-0553-0. [DOI] [PubMed] [Google Scholar]
- 18.Hemmingsen L, Sas KN, Danielsen E. Chem. Rev. 2004;104:4027–4062. doi: 10.1021/cr030030v. [DOI] [PubMed] [Google Scholar]
- 19.Iranzo O, Jakusch T, Lee KH, Hemmingsen L, Pecoraro VL. Chemistry. 2009;15:3761–3772. doi: 10.1002/chem.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen O. Environ. Health Perspect. 1984;54:249–266. doi: 10.1289/ehp.8454249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demeler B. Curr. Protoc. Prot. Sci. 2010;Chapter 7(Unit 7.13) doi: 10.1002/0471140864.ps0713s60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.(a) Brookes E, Demeler B. GECCO '07 Proceedings of the 9th annual conference on Genetic and evolutionary computation. ACM New York; NY, USA. pp. 361–368. [Google Scholar]; (b) Demeler B, Brookes E. Colloid Polym. Sci. 2008;286:129–137. [Google Scholar]
- 23.Demeler B, vanHolde KE. Anal. Biochem. 2004;335:279–288. doi: 10.1016/j.ab.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 24.(a) Liang Y, Pingali SV, Jogalekar AS, Snyder JP, Thiyagarajan P, Lynn DG. Biochemistry. 2008;47:10018–10026. doi: 10.1021/bi801081c. [DOI] [PubMed] [Google Scholar]; (b) Mehta AK, Lu K, Childers WS, Liang Y, Dublin S, Dong J, Snyder JP, Skanthakumar S, Thiyagerajan P, Lynn DG. J. Am. Chem. Soc. 2008;130:9829–9835. doi: 10.1021/ja801511n. [DOI] [PubMed] [Google Scholar]
- 25.Petkova AT, Buntkowsky G, Dyda F, Leapman RD, Yau WM, Tycko R. J. Mol. Biol. 2004;335:247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 26.Kon T, Imamula K, Roberts AJ, Ohkura R, Knight PJ, Gibbons IR, Burgess SA, Sutoh K. Nat. Struct. Mol. Biol. 2009;16:325–333. doi: 10.1038/nsmb.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.