SUMMARY
Fatty acid and triglyceride synthesis is induced in response to feeding and insulin. This lipogenic induction involves coordinate transcriptional activation of lipogenic enzymes, including fatty acid synthase and glycerol-3-phosphate acyltransferase. We recently reported the importance of USF-1 phosphorylation and subsequent acetylation in insulin-induced lipogenic gene activation. Here, we show that Brg1/Brm-associated factor (BAF) 60c is a specific chromatin remodeling component for lipogenic gene transcription in liver. In response to insulin, BAF60c is phosphorylated at S247 by atypical PKCζ/λ, which causes translocation of BAF60c to the nucleus and allows a direct interaction of BAF60c with USF-1 that is phosphorylated by DNA-PK and acetylated by P/CAF. Thus, BAF60c is recruited to form the lipoBAF complex to remodel chromatin structure and to activate lipogenic genes. Consequently, BAF60c promotes lipogenesis in vivo and increases triglyceride levels, demonstrating its role in metabolic adaption to activate the lipogenic program in response to feeding and insulin.
INTRODUCTION
Understanding the regulation of lipogenesis is critical since dysregulated lipogenesis and triglyceride levels are often linked to pathological conditions including obesity, diabetes, and cardiovascular disease. Fatty acid and triglyceride synthesis is under tight nutritional and hormonal control (Wong and Sul, 2010). After a meal, insulin secretion increases causing lipogenic tissues such as liver and adipose to convert excess glucose to fatty acids (de novo lipogenesis), using NADPH as reducing equivalents. Fatty acids are then esterified into triglycerides. In liver, triglycerides are packaged into VLDL for secretion, whereas triglycerides in adipose tissue are stored and later released to the circulation as fatty acids, for use by other tissues during periods of energy demand. Enzymes involved in fatty acid and fat synthesis, such as fatty acid synthase (FAS), are coordinately regulated at the transcriptional level in response to changing conditions – transcription is low during fasting, but drastically increases in response to feeding/insulin. The effect of insulin on metabolic regulation is primarily mediated through activation of the PI3K pathway and its downstream kinases, including PKB/Akt, as well as through activation of protein phosphatase (PP1) (Brady and Saltiel, 2001; Taniguchi et al., 2006; Bakan and Laplante, 2012). However, the effect of insulin on chromatin remodeling that may be required for activation of the lipogenic program is not understood.
By catalyzing seven reactions in fatty acid synthesis, FAS plays a central role in de novo lipogenesis. Due to its regulation primarily at the transcriptional level, FAS provides an excellent model to dissect the transcriptional activation of lipogenesis in response to feeding/insulin (Paulauskis and Sul, 1988, 1989). We previously have shown that binding of the upstream stimulatory factor (USF)-1/2 heterodimer to the −65 E-box is required for FAS promoter activation in response to feeding/insulin (Moustaid et al., 1994; Moustaid et al., 1993). The critical role of USF in lipogenic gene transcription has been demonstrated in vivo in USF knockout mice that have significantly impaired lipogenic gene induction (Casado et al, 1999). Quantitative trait mapping studies have identified that USF-1 may be a candidate gene for familial combined hyperlipidemia (Pajukanta et al., 2004). In addition to USF, the role of sterol regulatory element-binding protein-1c (SREBP-1c) in lipogenesis is now well documented (Wong et al., 2010, Horton et al., 2002). In this regard, we have shown that SREBP-1c, when induced by insulin, binds to the SRE present nearby the E-box of the FAS promoter, but recruitment of SREBP-1c to the FAS promoter is dependent on USF binding to the −65 E-box and the direct interaction of USF with SREBP-1c (Latasa et al., 2003; Latasa et al., 2000; Griffin et al., 2007). We recently have shown that in response to feeding/insulin, USF-1 is phosphorylated at S262 by DNA-PK that is activated by PP1 and, upon phosphorylation, USF-1 recruits and is acetylated at K237 by P/CAF (Wong et al., 2009). Together, phosphorylation-dependent acetylation of USF-1 functions as a sensor for nutritional/insulin status to activate FAS transcription. Many other lipogenic genes contain closely spaced E-boxes and SREs in their proximal promoter regions, and therefore may also be subject to transcriptional regulation by USF-1. Along with FAS, by examining mitochondrial glycerol-3-phosphate acyltransferase (mGPAT) which catalyzes the initial esterification step for triglyceride synthesis, we have demonstrated that coordinate induction of lipogenic gene transcription in response to feeding/insulin occurs via this common mechanism (Griffin et al., 2007).
Dynamic chromatin structure plays an essential role in the control of gene transcription. Although not much is known, chromatin remodeling is predicted to be an important aspect governing metabolic gene regulation during different nutritional and hormonal conditions. The SWI/SNF complex carries out chromatin remodeling, and is evolutionarily conserved from yeast to mammals (Hargreaves and Crabtree, 2011, Becker and Horz, 2002). Within the mammalian SWI/SNF complex, the BAF (Brg1/Brm-associated factor) complex is composed of a core ATPase, Brg1/BAF190 or Brm, and other BAF subunits such as BAF155, BAF170 and BAF250 that act as modulators, as well as BAF57 and BAF60 that may provide the interaction of the BAF complex with transcriptional machinery (Hargreaves and Crabtree, 2011; Ryme et al., 2009). These BAF subunits are thought to be present in the nucleus to control transcription by forming BAF or the BAF-related chromatin remodeling complex. The mode of regulation of specific components of the BAF complex, and the signaling pathways that lead to recruitment of specific BAF complexes for chromatin remodeling, have not been well delineated.
BAF60 associates with Brg1/BAF190, potentially functioning as a bridge between DNA-binding transcription factors and other BAF subunits (Hsiao et al., 2003; Ito et al., 2001; Li et al., 2008). There are three isoforms of BAF60; BAF60a, b and c. These are expressed differentially in various tissues, suggesting a tissue-specific role for BAF60 isoforms, perhaps by association with distinct regulators. For example, BAF60a may be involved in tumor suppression and fatty acid metabolism by interacting with p53 and PGC1α, respectively (Li et al., 2008; Oh et al., 2008). On the other hand, BAF60c may play a role in heart development (Lickert et al., 2004; Takeuchi et al., 2007). BAF60c has also been reported to be important for muscle gene expression by interacting with MyoD (Forcales et al., 2011; Simone et al., 2004). However, how or whether BAF60 isoforms respond to physiological stimuli to associate with specific transcription factors is not well understood.
Here, we show that BAF60c, along with other BAF subunits that include Brg1/BAF190, BAF155 and BAF250, forms the lipoBAF complex for chromatin remodeling that is required for activation of the lipogenic program. USF-1, which is phosphorylated by DNA-PK and then acetylated by P/CAF, recruits BAF60c. BAF60c is phosporylated at S247 by aPKC in response to feeding/insulin. Phosphorylated BAF60c translocates from the cytosol to the nucleus and directly interacts with phosphorylated/acetylated USF, thus allowing recruitment of lipoBAF and remodeling of chromatin to activate lipogenic genes.
RESULTS
Identification of BAF60c as a USF interacting protein
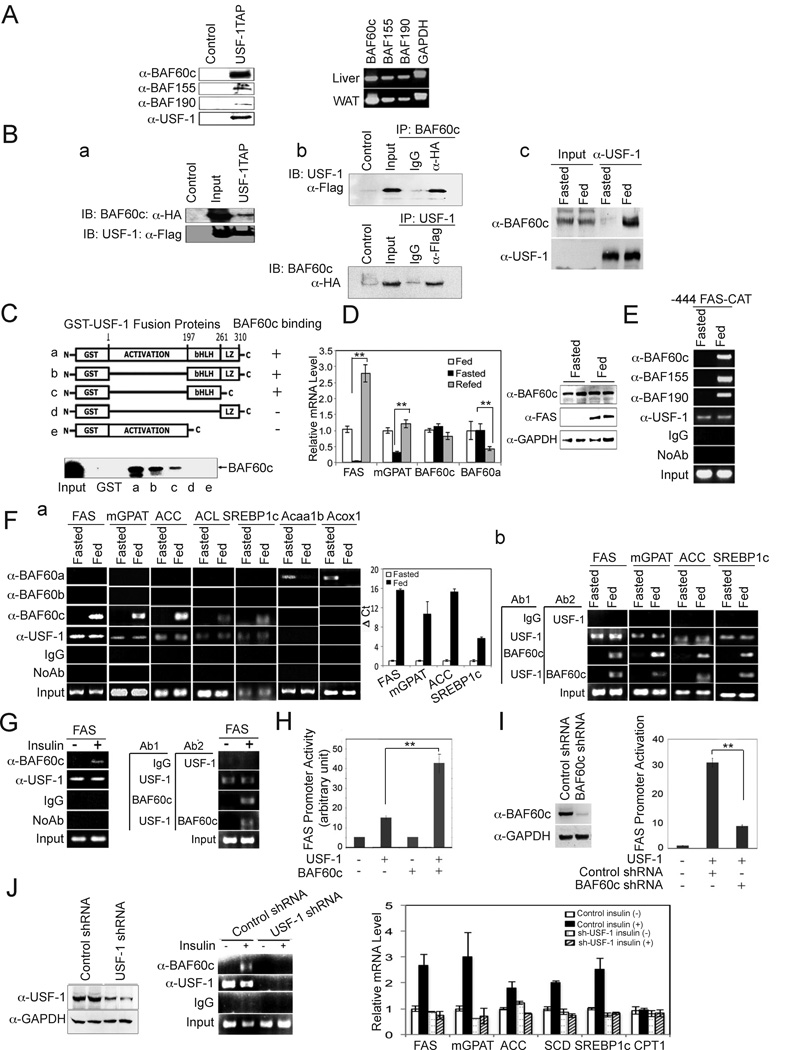
USF plays a pivotal role as a molecular switch by recruiting distinct transcription factors and coregulators in a fasting/feeding-dependent manner (Wong et al., 2009). To identify additional USF interacting factors, we performed tandem affinity purification (TAP) using USF-1 followed by mass spectrometry (MS) analysis. We identified three BAF subunits, BAF60c, BAF155 and Brg1/BAF190. Immunoblotting of the TAP eluates confirmed the presence of these BAF subunits that co-purified with TAP-tagged USF-1, but not with the control TAP-tag (Fig. 1A, left). We detected these BAF subunits in the lipogenic tissues, liver and adipose tissue (Fig. 1A, right), as well as in other tissues examined (Suppl. Fig. 1).
Figure 1. Identification of BAF subunits as USF-1 interacting proteins.
(A) Immunoblotting of TAP purified USF-1-associated proteins with indicated antibodies (left). RT-PCR for BAF subunits (right). (B) Immunoblotting of TAP eluates from 293F cells overexpressing BAF60c-HA and USF-1-Flag-TAP (a). Co-IP of cells overexpressing BAF60c-HA and USF-1-Flag (b). Immunoblotting of liver lysates from fasted and fed mice after IP with USF-1 antibody (c). (C) GST-USF-1 protein was incubated with in vitro translated 35S-labeled BAF60c before GST pull-down. (D) RT-qPCR. Means ± SEM. *p<0.05 (left). Immunoblotting of liver lysates from fasted and fed mice (right). (E) ChIP assay of livers of −444 FAS-CAT transgenic mice. (F) ChIP assay (a, left) and quantification for enrichment of BAF60c by qPCR (a, right). Re-ChIP using USF-1 antibody followed by BAF60c antibody (b). (G) ChIP on FAS in HepG2 cells (left) and re-ChIP (right). (H) FAS promoter activity in 293 cells upon BAF60c overexpression (I) FAS promoter activity in BAF60c knockdown cells treated with insulin. Means ± SEM. **p<0.01 (J) Immunoblotting of lysates from HepG2 cells infected with shUSF-1 lentivirus (left). ChIP assay (middle). Expression of lipogenic genes (right).
Since BAF60 is known to function as an anchor point between transcription factors and the BAF complex, we first tested the interaction of BAF60c with USF. In cells overexpressing HA-tagged BAF60c and Flag-tagged USF-1, BAF60c co-purified with USF-1 by TAP (Fig. 1Ba). We also detected an interaction between USF-1 and BAF60c in cells by co-immunoprecipitation (Co-IP) (Fig. 1Bb). The interaction between endogenous USF-1 and BAF60c was detected by Co-IP in liver extracts from fed mice, but not from fasted mice (Fig.1Bc). Next, by GST pull-down, we established the domain of USF-1 required for the interaction with BAF60c in vitro. BAF60c directly interacts with USF-1. Deletion of the bHLH domain of USF-1 prevented the interaction with BAF60c, and is therefore required (Fig. 1C). The bHLH domain of USF-1 contains K237 that we previously have shown is acetylated by P/CAF in response to feeding/insulin, suggesting a potential role for USF-1 K237 acetylation in the USF/BAF60 interaction. As predicted, hepatic mRNA levels of FAS and mGPAT were decreased upon fasting, and markedly increased in response to feeding (Fig. 1D, left). However, BAF60c mRNA levels did not change during fasting/feeding. We observed similar results at the protein level. Unlike FAS protein which was barely detectable in fasting but markedly increased in the fed condition, BAF60c protein levels did not change significantly with nutritional status (Fig. 1D, right). Parenthetically, expression of another BAF60 isoform, BAF60a, which has been reported to activate fatty acid oxidation, was decreased after feeding (Fig. 1D, left). Overall, we conclude that BAF60c expression remained the same in fasted and fed conditions.
BAF60c binds and activates the FAS promoter
We next investigated whether the BAF complex is recruited to the FAS promoter upon interaction of BAF60c with USF in response to feeding/insulin. Chromatin immunoprecipitation (ChIP) using livers from transgenic mice expressing a CAT reporter gene driven by the −444 FAS promoter (Latasa et al., 2003; Latasa et al., 2000; Moon et al., 2000) detected binding of USF to the FAS promoter in both the fasted and fed states (Wang and Sul, 1995). More importantly, the promoter region of the FAS-CAT transgene was occupied by all the identified USF-1 interacting BAF subunits, including BAF60c, BAF155 and Brg1/BAF190, (Fig. 1E) in the fed state only. We found that these BAF subunits are part of the BAF, but not the PBAF (Polybromo-associated BAF), complex (Wang et al., 1996; Thompson, 2009), because we detected BAF250 normally found in BAF complex, but not BAF180 or BAF200 which are hallmarks of the PBAF complex (Ryme et al., 2009) (Supp. Fig. 2, left). Co-IP showed strong interaction of BAF250, but a minimal interaction of BAF180 and BAF200, with USF-1 and BAF60c in livers of fed mice (Supp. Fig. 2, right). ChIP also detected recruitment of BAF60c to the proximal promoter region of the endogenous FAS gene only in the fed state (Fig. 1Fa). However, BAF60a and BAF60b were not detected. We also detected recruitment of BAF60c, but not other BAF60c isoforms, in the proximal promoter regions of other lipogenic genes including mGPAT, ACC, ACL, and SREBP-1c, which all contain an E-box. Thus, BAF60c is the specific isoform recruited to lipogenic promoters upon feeding. In contrast to the lipogenic genes, recruitment of BAF60a was detected on the FA oxidative genes, Acaa1b and Acoxl, only in the fasted state. Furthermore, re-ChIP experiments using USF-1 antibody followed by BAF60c antibody showed recruitment of USF-1 and BAF60c to the same lipogenic promoter regions (Fig. 1Fb). We also detected BAF60c bound to the FAS promoter in the presence of insulin in HepG2 cells, whereas USF-1 was bound both in the presence and absence of insulin (Fig. 1G left). The re-ChIP also showed recruitment of BAF60c and USF-1 at the same FAS promoter region in HepG2 cells treated with insulin (Fig. 1G right). Overall, these results demonstrate that BAF60c recruitment through USF-1 in response to feeding/insulin is specific to lipogenic genes but not oxidative genes.
We next tested the functional significance of BAF60c recruitment by USF to lipogenic gene promoters. Transfecting USF-1 along with the −444 FAS promoter-luciferase construct increased luciferase activity by approximately 3-fold. While cotransfection of BAF60c alone did not have a significant effect, cotransfection of both USF-1 and BAF60c resulted in a further 3-fold increase in FAS promoter activity (Fig. 1H). The degree of FAS promoter activation by BAF60c was dose-dependent (data not shown). Conversely, FAS promoter activity was decreased by more than 70% upon shRNA-mediated knockdown of BAF60c that decreased BAF60c protein levels by 80% (Fig. 1I). Apart from FAS and mGPAT genes that we previously studied (Wong et al., 2009), the role of USF in insulin-induced activation has not been documented for other lipogenic genes. We therefore performed USF-1 knockdown by using lentiviral infection of USF-1 shRNA in HepG2 cells, which decreased USF-1 protein level by more than 70% (Fig. 1J left). The recruitment of BAF60c on the FAS promoter by ChIP was markedly decreased in these cells even when treated with insulin (Fig. 1J middle). Expression of various lipogenic genes was increased upon insulin treatment of HepG2 cells, which was substantially blunted by USF-1 knockdown. Expression of CPT1, one of the oxidative genes, showed no change (Fig.1J right). Overall, these results show that BAF60c was recruited by USF-1 and the recruitment of BAF60c in response to feeding/insulin is linked to lipogenic gene promoter activation.
BAF60c phosphorylation at S247 brings its nuclear translocation in response to insulin/feeding
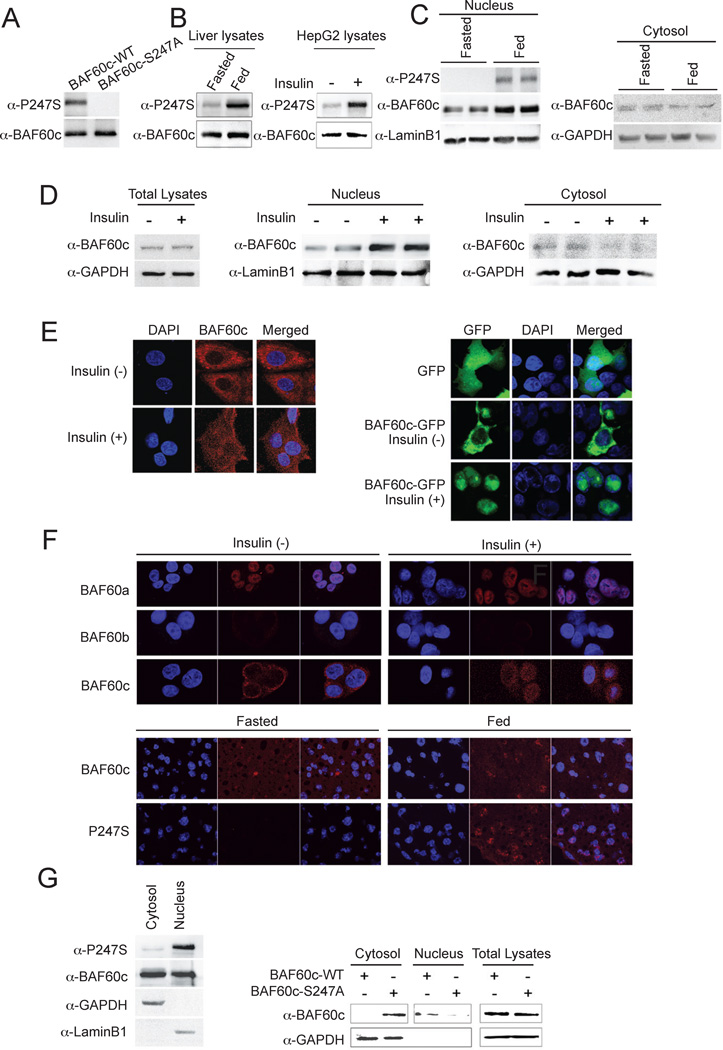
By MS analysis, we detected S247 phosphorylation of BAF60c. In fact, the NetPhos program indicated S247 as the most probable serine residue within BAF60c to be phosphorylated. Although S247 of BAF60c and its nearby amino acids are highly conserved among mammals (Suppl. Fig. 3, left), this specific serine residue is not conserved in lower organisms (Suppl. Fig. 3, right). Interestingly, S247 is also not found in other BAF60 isoforms, suggesting that S247 phosphorylation is unique to the BAF60c function. We generated an antibody against BAF60c phosphorylated at S247 peptide. The antibody specifically recognized the phosphorylated BAF60c since BAF60c S247A mutant did not show signal (Fig. 2A). Using this antibody, we could detect S247 phosphorylation of BAF60c in livers of fed, but not fasted, mice (Fig. 2B, left). A similar increase in S247 phosphorylation of BAF60c was also detected in HepG2 cells upon insulin treatment (Fig. 2B, right). These results demonstrate that BAF60c is phosphorylated at S247 in response to feeding/insulin.
Figure 2. Insulin/feeding induces S247 phosphorylation and nuclear localization of BAF60c.
(A) Immunoblotting of cell lyasates from cells transfected with wild type BAF60c (WT) or S247A BAF60c mutant (S247A) using S247 phosphorylation specific BAF60c antibody. (B) lysates of livers and HepG2 cells. (C) Immunoblotting for nuclear and cytosolic fractions of livers. (D) Immunoblotting for total cell lysates and nuclear and cytosolic fractions of HepG2 cells. (E) Confocal fluorescence microscopy for BAF60c localization in HepG2 cells (left) and GFP-BAF60c transfected 293 cells (right). (F) Immunofluorescence of endogenous BAF60a, b and c in HepG2 cells (top) and endogenous BAF60c and phosphorylated BAF60c (P247S) in livers (bottom). (G) Immunoblotting for phosphorylation at S247 in nuclear and cytosolic fractions of HepG2 cells upon insulin treatment (left) and BAF60c in total lysates as well as cytosolic and nuclear fractions of 293 cells transfected with wild type BAF60c or BAF60c S247A mutant after insulin treatment (right).
In general, BAF subunits have been regarded as nuclear proteins. We, however, detected BAF60c in both nuclear and cytosolic fractions after cell fractionation of livers (Fig. 2C). Moreover, we detected greater BAF60c levels in the nuclear fraction of livers from fed mice compared to fasted mice (Fig. 2C, left). An increase in BAF60c levels was also detected in the nuclear fraction of HepG2 cells after insulin treatment (Fig. 2D, middle). Conversely, a decrease in BAF60c levels was detected in the cytosolic fractions by feeding/insulin treatment (Fig. 2D right). A similar insulin-mediated BAF60c translocation to the nucleus was detected by immunostaining of HepG2 cells (Fig. 2E, left). In addition, we detected a diffuse signal of BAF60c-GFP throughout the cells upon transfection into 293FT cells in the absence of insulin, and insulin treatment induced localization of BAF60c-GFP in the nucleus (Fig. 2E, right). Furthermore, comparison of immunostaining of all three endogenous BAF60 isoforms in HepG2 cells clearly showed that, unlike BAF60c, BAF60a was mainly found in the nucleus at all times and BAF60b was barely detectable (Fig. 2F, top). BAF60c was also detected by immunostaining throughout cells in livers of fasted mice, but was higher in the nucleus of fed mice (Fig. 2F, bottom). We also detected BAF60c phosphorylated at S247 in the nuclear, but not in the cytosolic fraction in HepG2 cells treated with insulin (Fig. 2G, left). S247 phosphorylated BAF60c was also clearly detected in livers of fed, but not fasted, mice (Fig. 2C and F bottom). Furthermore, unlike wild type BAF60c, the nonphosphorylatable S247A mutant BAF60c remained in the cytosolic compartment even when treated with insulin (Fig. 2G), demonstrating the importance of S247 phosphorylation for its nuclear localization.
BAF60c phosphorylation allows its interaction with USF for FAS promoter activation
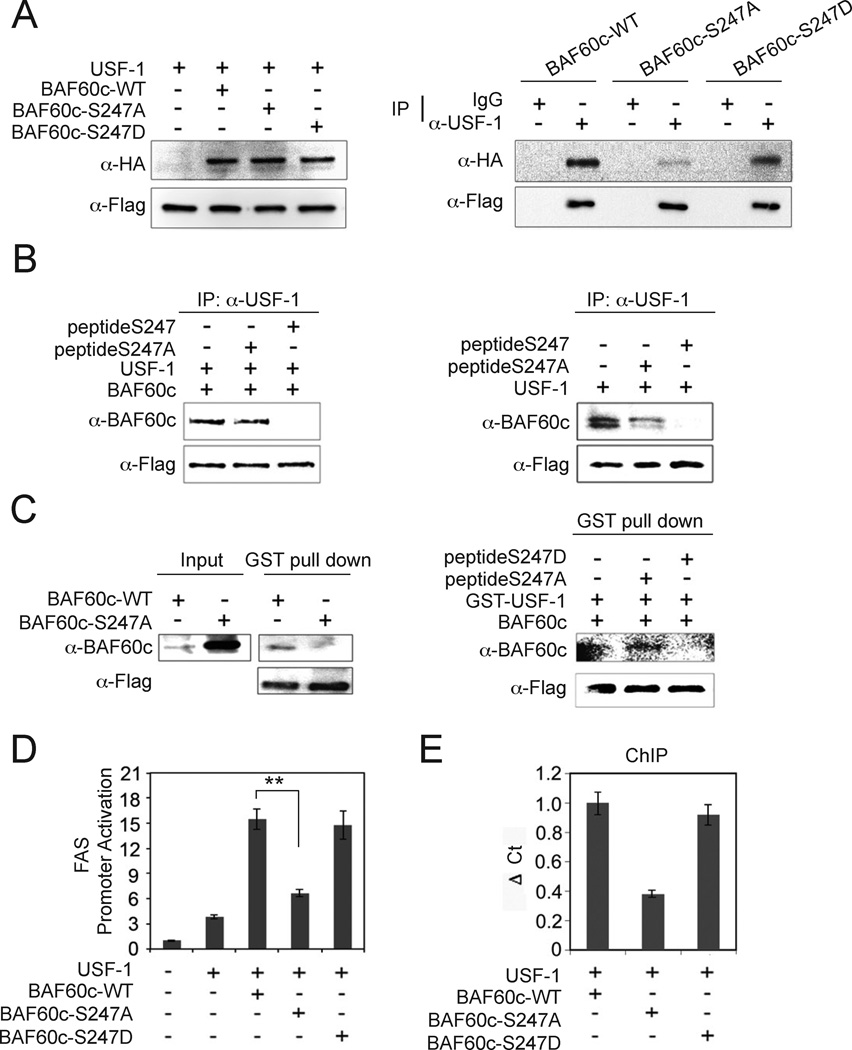
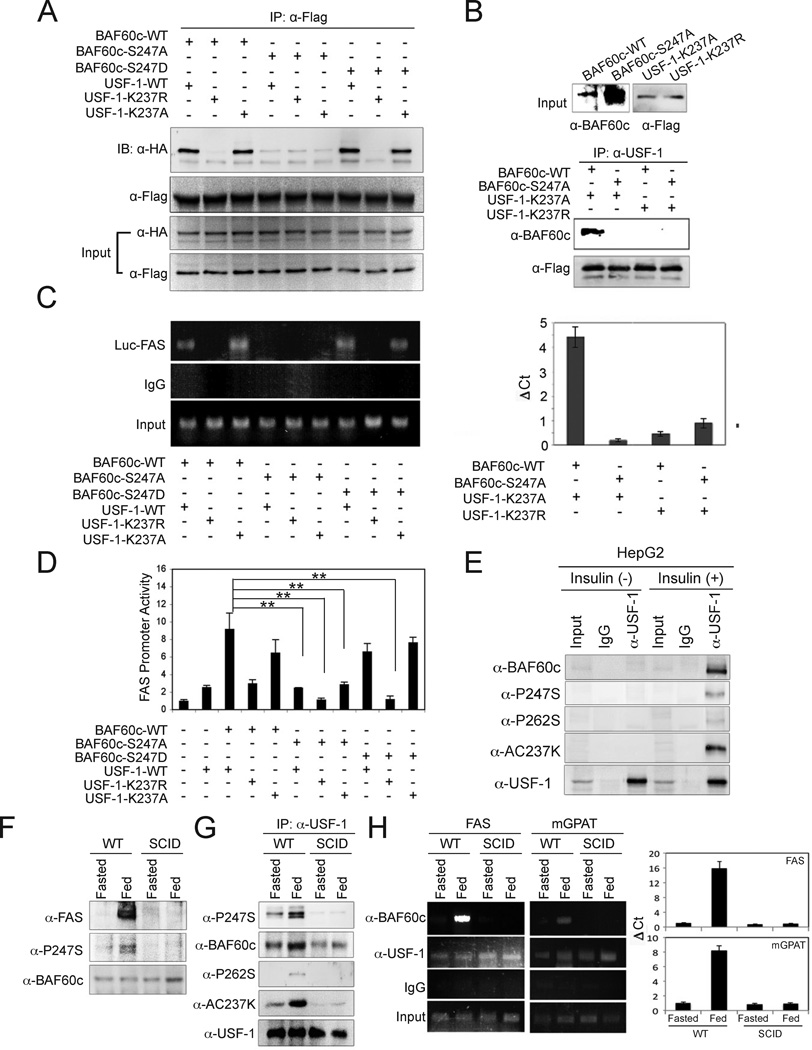
Next, we tested whether phosphorylation of BAF60c affects its interaction with USF-1. We cotransfected Flag-tagged USF-1 with HA-tagged wild type BAF60c, S247A, or phosphorylation-mimicking S247D mutant BAF60c (Fig. 3A, left) and the cells were treated with insulin. We verified similar levels of wild type, S247A and S247D BAF60c protein levels (Fig. 3A, left). We found that both the wild type and S247D mutant BAF60c were able to interact with USF-1, but the S247A mutant had a markedly decreased interaction (Fig. 3A, right), indicating that S247 phosphorylation of BAF60c is critical for its interaction with USF. In support of this, short peptides containing wild type S247, but not S247A, competed out the interaction between USF-1 and BAF60c (Fig. 3B, left). When cells were transfected with only USF-1, the interaction between USF-1 and endogenous BAF60c was also disrupted in the presence of S247 but not the S247A peptide (Fig. 3B, right). Overall, these data suggest that BAF60c phosphorylated at S247 interacts with USF-1 with a higher affinity, and the region containing S247 is the most likely site of interaction with USF-1. Next, we incubated bacterially expressed GST-USF-1 with in vitro translated wild type BAF60c or S247A BAF60c mutant. Compare to the wild type in vitro translated BAF60c that was efficiently pulled down by USF-1, the S247A mutant was poorly pulled down, even when we used a much higher amount (Fig. 3C, left). Furthermore, S247D short peptide, but not S247A peptide could compete out GST-USF-1 pull-down of BAF60c (Fig. 3C, right). These results further demonstrate the critical role of S247 phosphorylation of BAF60c in its interaction with USF.
Figure 3. S247 phosphorylation of BAF60c enhances the interaction with USF-1.
(A) Immunoblotting of lysates from 293 cells transfected with Flag-tagged USF-1 and HA-tagged BAF60c wild type (WT), S247A and S247D mutant BAF60c (left) and after IP with USF-1 antibody (right). (B) Lysates of cells transfected with Flag-tagged USF-1 and BAF60c were incubated with either wild type 247 peptide (peptide S247) or peptide containing S247A mutation (peptide S247A) before IP and immunoblotting (left). Lysates from cells transfected with only USF-1 were used (right). (C) GST-USF-1 fusion protein was used for pull-down of BAF60c using glutathione beads (left). GST-USF-1 was incubated with in vitro translated BAF60c in the presence of S247D or S247A peptides. (D) Luciferase activity. Means ± SEM. **P<0.01. (E) ChIP for BAF60c binding by qPCR. Means ± SEM.
We tested the functional significance of BAF60c S247 phosphorylation using FAS-promoter reporter assay. Wild type or the S247D BAF60c mutant both increased USF-dependent FAS promoter activation by 4-fold (Fig. 3D). In contrast, the S247A mutant BAF60c did not. Furthermore, using ChIP we detected decreased recruitment of the S247A mutant BAF60c to the FAS promoter in comparison to wild type or the S247D BAF60c mutant (Fig. 3E). Overall, these results indicate that S247 phosphorylation of BAF60c governs its nuclear localization and interaction with USF-1 for the activation of FAS promoter activity.
aPKC phosphorylates BAF60c at S247 in response to insulin
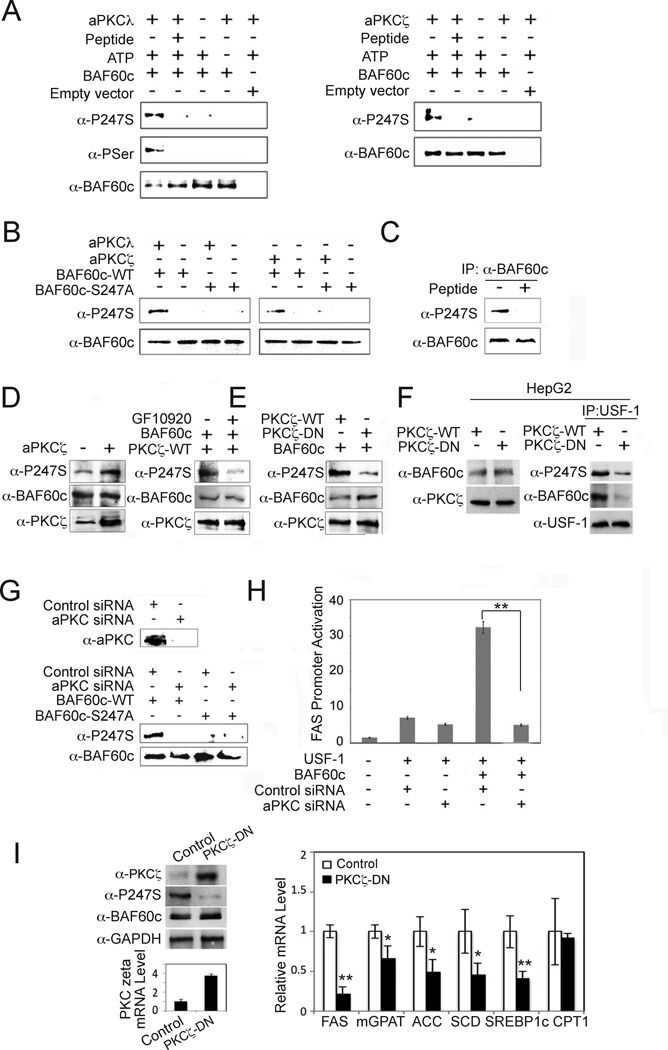
We next attempted to identify the specific kinase that catalyzes the phosphorylation of S247 in response to feeding/insulin. A review of phospho-protein databases predicted that a member of the protein kinase C (PKC) family may phosphorylate the S247 of BAF60c. Among PKC isoforms, atypical PKCζ and PKCλ are known to be activated by PI3K to mediate downstream insulin signaling (Taniguchi et al., 2006). Indeed, we detected in vitro phosphorylation of S247 of BAF60c by both PKCλ and PKCζ. Moreover, S247 phosphorylation was abolished upon addition of a myristoylated inhibitory peptide (Pep) (Farese and Sajan, 2010) of PKCλ and PKCζ (Fig. 4A). In addition, we easily detected phosphorylation of wild type BAF60c, but not the S247A mutant, by either PKCζ or PKCλ (Fig. 4B). Based on these results, we conclude that the S247 of BAF60c is a target of aPKC.
Figure 4. BAF60c is phosphorylated at S247 by aPKC.
(A) In vitro translated BAF60c-HA was in vitro phosphorylated with aPKCλ (left) and ζ (right) in the presence or absence of inhibitory peptide (100 mM) before immunoblotting. (B) In vitro translated wild type or S247A mutant BAF60c were in vitro phosphorylated before immunoblotting. (C) BAF60c-HA transfected cells were incubated with or without aPKC inhibitory peptide. (D) Lysates from 293 cells transfected with BAF60c, with or without PKCζ (left), PKCζ transfected cells treated with GF10920 at 5 mM or control (DMSO) for 30 min (right). (E) Immunoblotting of 293 cells cotransfected with BAF60c along with wild type or dominant negative PKCζ. (F) Lysates from HepG2 cells infected with adenovirus for wild type PKCζ or dominant negative PKCζ (PKCζ-DN) overexpression (left) and after IP with USF-1 antibody (right) before immunoblotting. (G) aPKC protein levels in cells transfected with aPKCζ and aPKCλ siRNA (aPKC siRNA) (top). Immunoblotting of lysates from 293 cells cotransfected with control or aPKC siRNA along with BAF60c or S247A mutant (bottom). (H) FAS promoter activity. Means± SEM. **p<0.01. (I) Immunoblotting of liver lysates from mice after administration of PKCζ-DN (left). Expression of lipogenic genes (right), Means ± SEM. *p<0.05, **p<0.01, n=3–4.
We then tested S247 phosphorylation of BAF60c in cells by aPKC in cultured cells. We clearly detected S247 phosphorylation of BAF60c in cells overexpression BAF60c, which was abolished in cells treated with cell permeable myristoylated inhibitory peptide (Fig. 4C). Additionally, overexpression of either PKCζ (Fig. 4D, left) or PKCλ (data not shown) along with BAF60c greatly increased BAF60c phosphorylation at S247, while, treating with the aPKC inhibitor GF109203X prevented S247 phosphorylation (Fig. 4D, right) (Uberall et al., 1999). Furthermore, in comparison to wild type PKCζ, co-transfection of dominant negative PKCζ into 293FT cells greatly diminished BAF60c phosphorylation (Fig. 4E), as well as the interaction between endogenous BAF60c and USF-1 (Fig. 4F) showing the functional role of phosphorylation for its interaction with USF-1. Moreover, BAF60c S247 phosphorylation was also decreased significantly upon siRNA-mediated knockdown of PKCζ and PKCλ by 80% (Fig. 4G) and mutant S247A BAF60c could not be phosphorylated either in control or aPKC siRNA transfected cells. Importantly, FAS promoter activation was markedly lower in aPKC siRNA transfected cells (Fig. 4H). By infecting adenoviral dominant negative PKCζ in mice, we also examined the function of aPKC-mediated phosphorylation of BAF60c in vivo. In these mice, expression level of dominant negative PKCζ was higher than endogenous PKCζ by 3.7-fold (Fig. 4I, left). Immunoblotting with an antibody that detects both the wild type and dominant negative PKCζ further confirmed the overexpression of dominant negative PKCζ. Although BAF60c levels remained the same, S247 phosphorylation of BAF60c was greatly diminished by overexpression of dominant negative PKCζ. In these mice, expression of lipogenic genes was substantially decreased, whereas CPT1 expression did not change (Fig. 4I, right). Overall, these results show that S247 of BAF60c is phosphorylated specifically by aPKC to mediate the effect of insulin on lipogenic gene activation.
Posttranslational modification of both BAF60c and USF are critical for their interaction and function
Since BAF60c interacts with USF-1 through the domain containing acetylated K237, we tested whether acetylation of USF-1 could affect its interaction with BAF60c. In Co-IP experiments, wild type USF and the K237A mutant that mimics hyper-acetylation, but not the K237R mutant clearly showed an interaction with the wild type and S247D mutant BAF60c (Fig. 5A). Similar results were obtained with hyperacetylation mimicking K237Q USF (data not shown), whereas S247A BAF60c could not interact with USF-1. In vitro interaction assays also showed that only hyperacetylated USF-1 was able to interact with wild type but not with the S247A BAF60c mutant, while nonacetylated USF failed to interact with either wild type or S247A BAF60c mutant (Fig. 5B).
Figure 5. Post-translational modification of both BAF60c and USF-1 mediates their interaction.
(A) IP of USF-1 with Flag-antibody of 293 cells co-transfected with Flag-tagged USF-1 (WT) or its acetylation mutants (K237R and K237Q) with HA-tagged BAF60 or its phosphorylation mutants (S247A and S247D) and immunoblotting with anti-HA antibody for BAF60c. (B) (top) Input of in vitro translated K237A and K237R mutant USF-1 and wild type and S247A mutant BAF60c. USF-1 and BAF60c were used for Co-IP followed by immunoblotting. (C) ChIP analysis for BAF60c bound to the FAS-Luc promoter in cells transfected with USF-1 or its acetylation mutants along with BAF60c or its mutant by PCR (left) or qPCR (right). (D) Luciferase activity in cells transfected with −444 FAS-Luc, USF-1 and BAF60c or its mutants. Means ± SEM. **p<0.01. (E) Immunoblotting of lysates from insulin-treated HepG2 cells after IP with anti-USF-1 antibody. (F) Immunoblotting of total liver lysates from 6 week-old SCID mice. (G) Immunoblotting of liver nuclear extracts after IP with USF-1 antibody. (H) ChIP for FAS and mGPAT promoters after IP with anti-BAF60c and anti-USF antibodies in livers from SCID mice (left) and quantification by qPCR (right). Means ± SEM.
Next, by ChIP, we detected wild type or S247D BAF60c mutant bound to the FAS promoter in cells transfected with wild type or K237A USF-1 mutant in insulin treated cells (Fig. 5C). S247A BAF60c mutant was not bound to the FAS promoter, regardless of the forms of USF-1 that were cotransfected. None of the BAF60c forms were detected in association with the FAS promoter region when K237R USF-1 mutant was cotransfected. Furthermore, the FAS promoter was activated to the highest degree only when wild type or the S247D BAF60c mutant were cotransfected with wild type or K237A USF-1 mutant (Fig. 5D), but not with the S247A BAF60c or K237R USF-1 mutant. In HepG2 cells, BAF60c phosphorylation at S247, and USF-1 phosphorylation at S262 and acetylation at K237, were detected only upon insulin treatment (Fig. 5E). These data clearly show that posttranslational modifications of both BAF60c (S247 phosphorylation) and USF (K237 acetylation) are required for their interaction as well as binding to the FAS promoter region for transcriptional activation.
In DNA-PK-deficient SCID mice, phosphorylation at S262 of USF is greatly reduced, which, in turn, results in a decrease in acetylation of K237 of USF. We thus examined whether recruitment of BAF60c to the FAS promoter by USF was altered in SCID mice. As we reported previously, unlike in wild type mice, FAS protein levels were not increased significantly by feeding in SCID mice. Total BAF60c levels remained the same in both wild type and SCID mice in fasted and fed states. However, while phosphorylation of BAF60c at S247 was significantly higher in wild type mice upon feeding, this feeding-induced BAF60c phosphorylation was not detected in SCID mice (Fig. 5F). As we have previously shown, total USF-1 protein levels remained the same, but K237 acetylation and S262 phosphorylation of USF-1 were substantially higher in wild type mice upon feeding. The total BAF60c or S247 phosphorylated BAF60c that coimmunoprecipitated with USF was substantially higher in wild type mice upon feeding also. In contrast, we could not detect 237K acetylation of USF in SCID mice in either the fasted or fed states. The amount of BAF60c that Co-IPed with USF remained the same in SCID mice, albeit lower than wild type mice. Furthermore, S247 phosphorylation of BAF60c was not detectable in either fasted or fed SCID mice (Fig. 5G). As expected, in ChIP, the total USF-1 bound to the FAS and mGPAT promoter regions remained the same in wild type and SCID mice in both fasted and fed conditions (Fig. 5H, left). And as we previously reported, the phosphorylated and acetylated USF was bound to the FAS promoter only in the fed state, but not in the fasted state. In contrast, BAF60c was bound to the FAS and mGPAT promoters only in wild type mice in the fed state, but not in the fasted state. More importantly, we could not detect BAF60c in either the fasted or fed SCID mice (Fig. 5H). These data show that USF phosphorylation by DNA-PK, which leads to subsequent USF acetylation, is required for the recruitment of BAF60c to lipogenic gene promoters.
BAF60c promotes chromatin remodeling of lipogenic genes in response to insulin
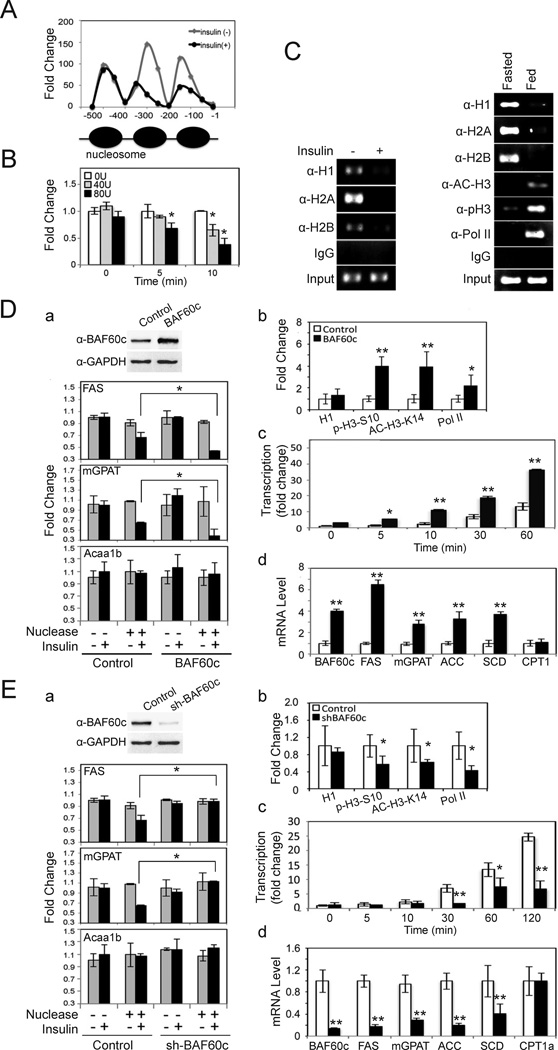
BAF60c has been proposed to serve as a bridge between transcription factors and the ATPase-dependent chromatin remodeling BAF complex. We therefore investigated whether chromatin accessibility is altered during lipogenic activation. Nuclear extracts from HepG2 cells were incubated with MNase that preferentially cuts DNA of nucleosome-free regions including regions between nucleosomes (Engel and Yamamoto, 2011). A periodic pattern of MNase accessibility was observed in control cells indicating nucleosome positioning at the proximal FAS promoter region (Fig. 6A). Treatment with insulin for 10 min caused a marked increase in accessibility around the first two nucleosomes from the transcription start site, indicating insulin-mediated remodeling of chromatin structure (Fig. 6A). Change in accessibility was detectable as early as 5 min after the addition of insulin (Fig. 6B). H3 phosphorylation and acetylation has been reported to play a role in recruitment of the Brg1 containing chromatin remodeling BAF complex (Drobic et al., 2010; Vicent et al., 2006; Vicent et al., 2009), so we examined occupancy of histones as well as Pol II in chromatin remodeling. By ChIP, we detected that H1, H2A and H2B were bound to the FAS promoter in control cells or in livers of fasted mice, but the signal was weak upon insulin treatment or feeding, respectively (Fig. 6C, left). Pol II and acetylated H3 at K14 and phosphorylated H3 at S10 were bound to the FAS promoter in feeding, but not in fasting (Fig. 6C, right). These results suggest that not only BAF complex but also histone modification occurs during chromatin remodeling of the FAS promoter with insulin/feeding.
Figure 6. BAF60c enhances chromatin modification of lipogenic genes in response to insulin.
(A) Nuclear extracts from HepG2 cells treated with insulin for 10 min were subjected to MNase assay using 80U/ml. qPCR using primers spanning 500 bp of the FAS promoter region. (B) MNase assay in HepG2 cells treated with insulin. Fold change over non-MNase treated cells. Means ± SEM. *p<0.05. (C) ChIP assay in HepG2 cells (left) and in mouse livers (right). (D) a; BAF60c protein levels in HepG2 cells infected with control or BAF60c adenovirus (top). Infected cells treated with insulin for 10 min for MNase assay (bottom). b; ChIP for the FAS promoter region in HepG2 cells after 10 min of insulin treatment using anti-H1, anti-p-H3-S10, anti-Ac-H3-K14, and anti-Pol II antibodies. qPCR for the FAS promoter. c; Nuclear run-on assay for FAS transcription in cells infected with BAF60c after insulin treatment. RT-qPCR for FAS run-on assay. d; RT-qPCR for lipogenic gene expression in cells overexpressing BAF60c. Means ± SEM. *p<0.05, **p<0.01. (E) a; BAF60c protein levels in shBAF60c adenovirus infected cells (top). MNase assays by qPCR (bottom). b; ChIP for FAS promoter in HepG2 cells after BAF60c knockdown. c; Run-on assay using RT-qPCR for FAS transcription. d; Lipogenic gene expression by RT-qPCR in BAF60c knockdown cells. Means ± SEM. *p<0.05, **p<0.01.
We then performed MNase assay in HepG2 cells after overexpression or knockdown of BAF60c. Overexpression of BAF60c by adenoviral infection increased BAF60c protein (Fig. 6Da, top) and mRNA levels (Fig. 6Dd). In the absence of insulin, there were no detectable differences in chromatin structure between control and BAF60c overexpressing cells as tested by digestion with MNase. Upon insulin treatment, control cells showed the accessibility of the proximal FAS promoter region increased by 30%, while in BAF60c overexpressing cells, accessibility increased by more than 50% (Fig. 6Da). Similar results were obtained with the mGPAT promoter showing its accessibility increasing by 65% in BAF60c overexpressing cells after insulin treatment versus only 40% in control cells. These increases in accessibility were lipogenic gene specific and were not seen in the Acaa1b promoter, either with or without BAF60c overexpression. In addition, ChIP analysis of the FAS promoter region showed a 4-fold increased binding of H3-S10 phosphorylation and H3-K14 acetylation in BAF60c overexpressing cells over control cells (Fig. 6Db). Pol II binding was increased by 2-fold also. In contrast, ChIP using a histone H1 antibody did not show a significant change. Nuclear run-on assays showed that FAS transcription in HepG2 cells started to increase significantly at 10 min after insulin addition, further increasing by 8- and 15-fold at 30 and 60 min, respectively. Importantly, cells overexpressing BAF60c showed an even greater increase in FAS transcription. A significant increase was easily detectable 5 min after insulin treatment and transcription was 19-, and 37-fold higher at 30, and 60 min, respectively, showing enhancement of insulin-mediated FAS transcription by BAF60c overexpression (Fig. 6Dc). Similarly, mRNA levels of various other lipogenic enzymes were significantly higher upon BAF60c overexpression, while mRNA levels of CPT1 did not change (Fig. 6Dd). In contrast, shRNA adenovirus mediated BAF60c knockdown cells that had 90% lower BAF60c showed a blunted insulin effect on FAS and mGPAT promoter accessibility (Fig. 6E). ChIP analysis showed that BAF60c knockdown decreased histone H3-S10 phosphorylation and H3-K14 acetylation as well as Pol II binding to the FAS promoter region, without any changes in binding of histone H1 (Fig. 6Eb). Consequently, BAF60c knockdown cells showed a significant blunting in the insulin mediated increase in FAS transcription (Fig. 6Ec). mRNA levels of lipogenic enzymes were all notably lower in BAF60c knockdown cells, while CPT1 mRNA levels were not altered (Fig. 6Ed). Overall, we conclude that BAF60c is specific for chromatin remodeling and transcription of lipogenic genes in response to insulin.
BAF60c induces lipogenesis in vivo
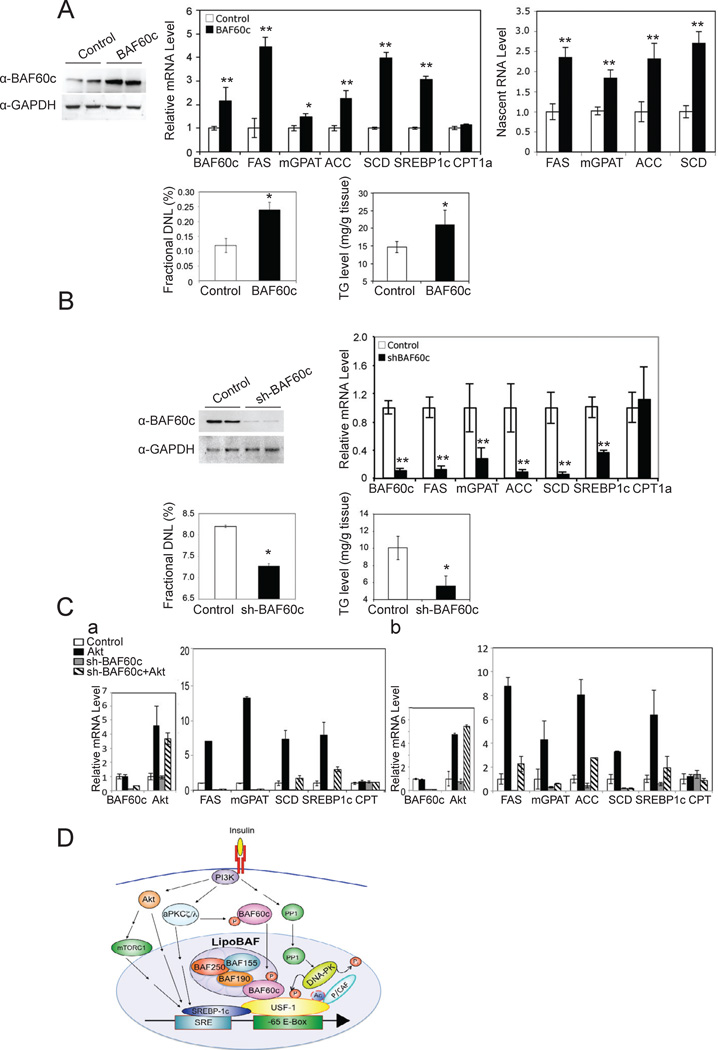
We next examined the biological function of BAF60c in vivo by adenoviral overexpression. Administration of BAF60c adenovirus by tail-vein injection increased liver BAF60c protein levels by approximately 3-fold (Fig. 7A, top left). In these mice, even in the fasted state, hepatic expression levels of lipogenic genes including FAS, ACC, SCD1, mGPAT and SREBP-1c, but not CPT1, were significantly higher than those in control mice (Fig. 7A, top middle). We also detected higher FAS protein levels upon BAF60c overexpression (data not shown). Nascent nuclear RNA levels of the lipogenic genes also showed a similar increase upon BAF60c overexpression (Fig. 7A, top right). Next, we measured de novo lipogenesis by employing a heavy water labeling technique. Although newly synthesized palmitate remained low in the fasted state in control mice as expected, BAF60c overexpression increased de novo fatty acid synthesis by approximately 2-fold (Fig. 7A, bottom left). Hepatic triglyceride levels also were significantly higher in livers of BAF60c overexpressing mice (Fig. 7A, bottom right). We performed microarray analysis comparing global liver gene expression patterns between wild type mice and mice overexpressing BAF60c. Indeed, we found that BAF60c overexpression activates the lipogenic program by increasing expression of a variety of lipogenic enzymes, including FAS, ACC, ACL, SCD1, mGPAT, as well as malic enzyme, G6PDH and gluconolactone dehydrogenase for NADPH synthesis (Suppl. Table S1). In addition, SREBP-1 which itself is induced upon feeding/insulin treatment and is involved in the activation of lipogenic genes (Horton et al., 1998, Horton et al., 2002) was also identified to be activated upon BAF60c overexpression. Thus, forced overexpression of BAF60c enhanced lipogenesis in vivo even in the fasted condition when no significant de novo lipogenesis is expected to normally occur.
Figure 7. BAF60c promotes lipogenesis in vivo.
(A) BAF60c protein levels in livers of mice 14 days after tail vein administration of BAF60c adenovirus (top left). mRNA levels (top middle) and nascent RNA levels (top right). Percent of newly synthesized fatty acids in liver (bottom left) and hepatic triglyceride levels (bottom right). Means ± SEM. *p<0.05, **p<0.01, n=3–5. (B) BAF60c protein levels in livers from mice 14 days after tail vein administration of shBAF60c adenovirus (top left). Expression of lipogenic genes (top right), de novo lipogenesis (bottom left) and triglyceride levels (bottom right) in the liver. Means ± SEM. *p<0.05, **p<0.01, n=3–5. (C) Expression levels of BAF60c and Akt 10 days after tail vein injection of adenoviral shBAF60c and adenoviral Akt in livers of mice (a, left). Expression levels of lipogenic genes in livers (b, right), n=4. Expression levels of BAF60c and Akt in HepG2 cells infected with adenoviral shBAF60c and adenoviral Akt (b, left). Expression levels of lipogenic genes 72 hrs post-infection (a, right). Means ± SEM. (D) Insulin signaling pathway for posttranslational modifications of BAF60c and USF-1 in chromatin remodeling and activation of lipogenic genes.
In contrast, adenovirus-mediated knockdown of BAF60c which caused an over 80% decrease in BAF60c protein levels resulted in a significantly lower hepatic expression of lipogenic genes even in the fed state, in comparison to control scrambled shRNA adenovirus (Fig. 7B, top right). Control CPT1a expression levels did not change. We also detected a significant decrease in de novo lipogenesis in BAF60c knockdown mice in the fed state (Fig. 7B, bottom left). The changes in de novo lipogenesis were reflected in total hepatic triglyceride levels that were decreased by approximately 50% in BAF60c knockdown mice (Fig. 7B, bottom right). Since we have previously shown that USF recruits SREBP-1c to nearby SREs by direct interaction and since PI3K-Akt is known to induce lipogenesis through SREBP-1c, we tested involvement of the PI3K-Akt pathway in BAF60c function in lipogenesis by adenoviral delivery for overexpression of Akt upon BAF60c knockdown. As above, BAF60c expression was decreased more than 80%, whereas Akt expression was increased by 4–5-fold in livers of mice upon adenovial infection of BAF60c shRNA or Akt, respectively (Fig. 7Ca, left). As expected, expression of lipogenic genes was decreased considerably upon BAF60 knockdown, whereas overexpression of Akt alone greatly increased expression levels. However, the increase from Akt overexpression was substantially diminished in BAF60c knockdown mice (Fig. 7Ca, right). Infection of BAF60 shRNA in HepG2 cells decreased BAF60 expression by 95% and Akt adenovirus increased Akt expression by 5-fold (Fig.7Cb, left). Similar to the in vivo results, overexpression of Akt alone markedly increased lipogenic gene expression in HepG2 cells, which was diminished in BAF60c knockdown cells (Fig.7Cb, right). These results suggest that PI3K-Akt pathway can induce lipogenic gene expression, but Akt cannot fully induce lipogenic genes in the absence of BAF60c. We predict that Akt increases SREBP-1c activation and/or expression to interact with USF. Overall, our in vivo results on BAF60c clearly show the critical role BAF60c plays in the feeding/insulin-induced activation of lipogenic genes.
DISCUSSION
A central issue in understanding metabolic regulation is the definition of coordinated molecular strategies that underlie the transition from fasting to feeding along specific signal transduction pathways. Transcriptional activation of lipogenic enzymes to induce lipogenesis is one such process. In this regard, chromatin remodeling by SWI/SNF related complexes, such as mammalian BAF, is indispensable for regulated transcription. However, regulation of the BAF complex in response to cellular signaling is not well understood. Here, we determine that BAF60c recruits other BAF subunits including BAF155 and BAF190 to form the lipoBAF complex to activate lipogenic gene transcription. We further demonstrate that this regulation is based on the phosphorylation-dependent nuclear translocation of BAF60c, and its interaction with USF-1. And, finally, we demonstrate that BAF60c phopshorylation by aPKC and USF-1 phosphorylation by DNA-PK are required for their interaction, converging two branches of insulin signaling for transcriptional activation of lipogenesis (Fig. 7C).
BAF60c and its associated subunits define the lipoBAF complex that is specific for chromatin remodeling for lipogenic gene transcription
BAF60 is thought to form a recruitment bridge between DNA binding factors and BAF subunits in regulating gene transcription. Three distinct isoforms of BAF60, BAF60a, BAF60b and BAF60c, have been identified and are able to form distinct complexes based on tissue and promoter context to regulate sophisticated and diverse cellular functions. Our present study shows that USF interacts with BAF60c, but not BAF60a or BAF60b, to recruit other BAF subunits, including BAF155, BAF190 and BAF250, to lipogenic gene promoters to increase lipogenesis in response to feeding/insulin treatment. Thus, the distinct binding pattern of BAF60c and other BAF subunits on the lipogenic gene promoters in response to feeding, but not in the fasted state, correlates with activation of lipogenic genes and alteration of chromatin structure. Since BAF60c is the specific isoform for lipogenic gene activation, we define the USF-1 interacting BAF subunit, BAF60c, for recruitment of other BAF subunits and thus formation of the lipogenesis specific BAF complex (lipoBAF). Moreover, similar binding patterns of the lipoBAF on lipogenic gene promoters indicate a common key mechanism to induce lipogenic gene transcription in response to fasting/feeding. Coordinate changes in nucleosome structure as detected by nuclease protection assay as well as recruitment of polymerase II to various lipogenic genes upon overexpression and knockdown of BAF60c further support a common mechanism. In this regard, the global gene expression pattern analysis confirmed activation of the lipogenic program by BAF60c in vivo. Furthermore, adenovirus-mediated overexpression and knockdown of BAF60c in mouse livers provided evidence for the BAF60c function in lipogenesis in vivo. The effects of BAF60c on transcription of various lipogenic enzymes were reflected in the changes in de novo lipogenesis, and consequently total hepatic triglyceride content. Overall, our studies demonstrate the importance of BAF60c in the recruitment of lipoBAF to promote lipogenesis.
Phosphorylation-dependent translocation of BAF60c in response to insulin functions as a novel regulation for BAF
Since BAF complexes govern chromatin remodeling events for transcription, BAF subunits are known to be found in the nucleus. Here, we demonstrate that BAF60c that can be found in the cytosol and is translocated to the nucleus dynamically in response to a physiological signal – feeding/insulin. This BAF60c translocation is regulated by aPKC mediated S247 phosphorylation. In this regard, cross talk between phosphorylation and translocation is well recognized for transcription factors. However, similar regulation has not previously been proposed or documented for subunits of chromatin remodeling complexes. We show here that translocation of BAF60c to the nucleus in response to feeding/insulin is dependent on S247 phosphorylation.
BAF60 has been proposed to bridge between DNA binding transcription factors and chromatin remodeling BAF complexes. However, regulatory mechanisms that permit BAF60 to interact with specific DNA binding transcription factors that will then allow recruitment of BAF complexes, have not previously been addressed. In this regard, while phosphorylation of BAF60a or BAF60b has not been reported previously, BAF60c was shown to be phosphorylated at T229 by p38 MAPK (Simone et al., 2004). However, this phosphorylation affects the interaction of BAF60c with the BAF component, Brg1. The S247 phosphorylation of BAF60c that allows interaction with USF-1 provides a mode by which the BAF complex can be recruited to lipogenic gene promoters in a feeding/insulin dependent manner. Interestingly, S247 of BAF60c, as well as nearby residues are conserved among mammalian species, but S247 is not found in other BAF60 isoforms. While lower organisms have only one BAF60 that does not contain S247, BAF60c in mammals appears to be the specific subunit involved in forming the lipoBAF complex for the regulation of lipogenesis. We propose that this regulation by BAF60c functions as a dynamic molecular switch in sensing the nutritional transition from fasting to feeding, providing an elegant way to fine tune mammalian lipogenic transcription.
BAF60c phosphorylation and USF acetylation converge the two branches of the insulin signaling pathway
It has been well established that PI3K primarily mediates insulin signaling for metabolic regulation. Insulin signaling is mediated by PI3K that activates PKB/Akt and aPKC (Taniguchi et al., 2006). The PI3K pathway also causes activation of PP1 to dephosphorylate target proteins such as DNA-PK that we have shown previously (Wong et al., 2009). Here, we provide evidence that BAF60c links aPKC to activation of lipogenic gene transcription. BAF60c is not regulated at the transcription level, since we did not detect any changes in BAF60c expression in vivo or in cultured cells upon feeding or insulin treatment. However, we detected an increase in BAF60c phosphorylation upon feeding/insulin. BAF60c is phosphorylated at S247 and our in vitro and in vivo studies clearly show that S247 phosphorylation is abolished by siRNA mediated knockdown of aPKC or by overexpression of aPKC-DN, pointing evidence to the notion that aPKC is the bona fide kinase for the S247 phosphorylation that occurs during the feeding/insulin condition. Although the link between aPKC and transcriptional activation of lipogenesis is not well understood, our study provides direct evidence that BAF60c bridges the gap. On the other hand, we previously reported that PP1 activates DNA-PK by dephosphorylation. Activated DNA-PK then phosphorylates USF-1 at S262, which then allows K237 acetylation by P/CAF. Although K237 acetylation is critical for lipogenic gene activation, its function was not clear. Here we show that K237 acetylation of USF-1 governs recruitment of BAF60c to the promoters of lipogenic genes. BAF60c can then recruit the other components of the BAF complex to lipogenic gene promoters for chromatin remodeling, an essential step for transcriptional activation. To our surprise, neither the phosphorylation of BAF60c, nor the USF phosphorylation at S247 and acetylation at K237 alone, is sufficient for their interaction. That interaction requires modification of both BAF60c and USF-1 through different insulin signaling pathways. The regulation of this interaction conceptually represents a "dance" like concept that resembles a biological circuit. A high output (interaction in this case) results only if both inputs (USF acetylation and BAF60c phosphorylation) to this biological circuit are high. This novel regulation provides a way to fine tune an interaction and it might also operate in other protein-protein interactions that govern various biological processes.
EXPERIMENTAL PROCEDURES
Purification of USF-1 interacting proteins, preparation of nuclear extracts, and cell fractionation
TAP and MS analysis were performed as described previously (Wong et al., 2009). Liver nuclear extracts were prepared by centrifugation through a sucrose cushion in the presence of NaF, and NE-PER ®Nuclear and Cytoplasmic extraction kit was used when cytosol was needed for experiments (Thermo).
Chromatin Immunoprecipitation
Livers from fasted or fed mice were fixed with disuccinimidyl glutarate (DSG) (Thermo) at 2 mM for 45 min at room temperature before formaldehyde cross-linking. ChIP was performed as described previously (Wong et al., 2009).
MNase assay
Cells were cross-linked with 1% formaldehyde, quenched by the addition of glycine, and scraped in NP-40 lysis buffer. Nuclei were collected, and treated with MNase. After the addition of proteinase K-containing stop buffer, mono-nucleosome DNA was purified with a gel extraction kit (Qiagen). As listed in Supplemental methods, qPCR primers were designed to span approximately 500 bp regions to generate PCR products of approximately 70 bp with 20 bp overlaps.
In vitro phosphorylation
In vitro phosphorylation was performed using recombinant/purified enzymes.
Immunoprecipitation, GST pull-down, luciferase reporter assays
Immunoprecipitation of nuclear extracts was performed under standard procedures. GST pull-down was performed as described previously (Griffin et al., 2007). Luciferase assays were performed in 293FT cells using Dual-Luc reagent (Promega). NIH image software was used to quantify immunoblotting data.
Confocal fluorescence microscopy
Briefly, HepG2 cells with or without insulin treatment were fixed with 4% paraformaldehyde in PBS for 10 min and permeabilized with 0.2% of Triton-X-100 for 10 min. After washing, cells were blocked with 1% BSA in PBS for 1 hr and incubated with anti-BAF60c antibody, followed by staining with Alexa Fluor 594 anti-rabbit IgG secondary antibody, and counterstained with DAPI for 5 min. Images were obtained using Zeiss LSM710 confocal microscopy.
RT-PCR analysis
RNA was isolated and reverse transcribed for PCR or qPCR.
De novo lipogenesis (DNL)
Fatty acids were formed during a 24 hr 2H2O body water labeling.
Measurement of Triglyceride levels
Total neutral lipids were extracted by the Folch method and lipids were solubilized in 1% Triton X-100. Triglyceride levels were measured using Infinity Reagent (Thermo).
Statistical analysis
The data are expressed as the means ± standard errors and Student's t test was used (*P < 0.05 and **P < 0.01).
Supplementary Material
BAF60c is required for feeding/insulin-mediated lipogenic gene activation.
BAF60c interacts with USF-1 to be recruited to lipogenic genes, such as FAS.
BAF60c is phosphorylated at S247 by aPKC to interact with P-S262, Act-K237 USF-1.
BAF60c forms lipoBAF complex on lipogenic promoters to remodel chromatin structure.
ACKNOWLEDGEMENTS
The work was supported in part by DK81098 (H.S.S.) from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental experimental procedures and supplemental results are included.
REFERENCES
- Bakan I, Laplante M. Connecting mTORC1 signaling to SREBP-1 activation. Curr Opin Lipidol. 2012;23:226–234. doi: 10.1097/MOL.0b013e328352dd03. [DOI] [PubMed] [Google Scholar]
- Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- Brady MJ, Saltiel AR. The role of protein phosphatase-1 in insulin action. Recent Prog Horm Res. 2001;56:157–173. doi: 10.1210/rp.56.1.157. [DOI] [PubMed] [Google Scholar]
- Casado M, VS Vallet VS, A Kahn A, Vaulont S. Essential role in vivo of upstream stimulatory factors for a normal dietary response of the fatty acid synthase gene in the liver. J Biol Chem. 1999;274:2009–2013. doi: 10.1074/jbc.274.4.2009. [DOI] [PubMed] [Google Scholar]
- Drobic B, Perez-Cadahia B, Yu J, Kung SK, Davie JR. Promoter chromatin remodeling of immediate-early genes is mediated through H3 phosphorylation at either serine 28 or 10 by the MSK1 multi-protein complex. Nucleic Acids Res. 2010;38:3196–3208. doi: 10.1093/nar/gkq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel KB, Yamamoto KR. The glucocorticoid receptor and the coregulator Brm selectively modulate each other's occupancy and activity in a gene-specific manner. Mol Cell Biol. 2011;31:3267–3276. doi: 10.1128/MCB.05351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese RV, Sajan MP. Metabolic functions of atypical protein kinase C: "good" and "bad" as defined by nutritional status. Am J Physiol Endocrinol Metab. 2010;298:E385–E394. doi: 10.1152/ajpendo.00608.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese RV, Sajan MP, Standaert ML. Atypical protein kinase C in insulin action and insulin resistance. Biochem Soc Trans. 2005;33:350–353. doi: 10.1042/BST0330350. [DOI] [PubMed] [Google Scholar]
- Forcales SV, Albini S, Giordani L, Malecova B, Cignolo L, Chernov A, Coutinho P, Saccone V, Consalvi S, Williams R, Wang K, Wu Z, Baranvskaya S, Miller A, Dilworth FJ, Puri PL. Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J. 2011;31:301–316. doi: 10.1038/emboj.2011.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin MJ, Wong RH, Pandya N, Sul HS. Direct interaction between USF and SREBP-1c mediates synergistic activation of the fatty-acid synthase promoter. J Biol Chem. 2007;282:5453–5467. doi: 10.1074/jbc.M610566200. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Research. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao PW, Fryer CJ, Trotter KW, Wang W, Archer TK. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol. 2003;23:6210–6220. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Yamauchi M, Nishina M, Yamamichi N, Mizutani T, Ui M, Murakami M, Iba H. Identification of SWI.SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J Biol Chem. 2001;276:2852–2857. doi: 10.1074/jbc.M009633200. [DOI] [PubMed] [Google Scholar]
- Latasa MJ, Moon YS, Kim KH, Sul HS. Nutritional regulation of the fatty acid synthase promoter in vivo: sterol regulatory element binding protein functions through an upstream region containing a sterol regulatory element. Proc Natl Acad Sci U S A. 2000;97:10619–10624. doi: 10.1073/pnas.180306597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latasa MJ, Griffin MJ, Moon YS, Kang C, Sul HS. Occupancy and function of the −150 sterol regulatory element and −65 E-box in nutritional regulation of the fatty acid synthase gene in living animals. Mol Cell Biol. 2003;23:5896–5907. doi: 10.1128/MCB.23.16.5896-5907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu C, Li N, Hao T, Han T, Hill DE, Vidal M, Lin JD. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008;8:105–117. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Moon YS, Latasa MJ, Kim KH, Wang D, Sul HS. Two 5'-regions are required for nutritional and insulin regulation of the fatty-acid synthase promoter in transgenic mice. J Biol Chem. 2000;275:10121–10127. doi: 10.1074/jbc.275.14.10121. [DOI] [PubMed] [Google Scholar]
- Moustaid N, Beyer RS, Sul HS. Identification of an insulin response element in the fatty acid synthase promoter. J Biol Chem. 1994;269:5629–5634. [PubMed] [Google Scholar]
- Moustaid N, Sakamoto K, Clarke S, Beyer RS, Sul HS. Regulation of fatty acid synthase gene transcription. Sequences that confer a positive insulin effect and differentiation-dependent expression in 3T3-L1 preadipocytes are present in the 332 bp promoter. Biochem J. 1993;292:767–772. doi: 10.1042/bj2920767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Sohn DH, Ko M, Chung H, Jeon SH, Seong RH. Baf60a ineracts with p53 to recruit the SWI/SNF complex. J Biol Chem. 2008;283:11924–11934. doi: 10.1074/jbc.M705401200. [DOI] [PubMed] [Google Scholar]
- Pajukanta P, Lilja HE, Sinsheimer JS, Cantor RM, Lusis AJ, Gentile M, Duan XJ, Soro-Paavonen A, Naukkarinen J, Saarela J, Laakso M, Ehnholm C, Taskinen MR, Peltonen L. Familial combined hyperlipidemia is associated with upstream transcription factor 1 (USF1) Nat Genet. 2004;36:371–376. doi: 10.1038/ng1320. [DOI] [PubMed] [Google Scholar]
- Paulauskis JD, Sul HS. Cloning and expression of mouse fatty acid synthase and other specific mRNAs. Developmental and hormonal regulation in 3T3-L1 cells. J Biol Chem. 1988;263:7049–7054. [PubMed] [Google Scholar]
- Paulauskis JD, Sul HS. Hormonal regulation of mouse fatty acid synthase gene transcription in liver. J Biol Chem. 1989;264:574–577. [PubMed] [Google Scholar]
- Ryme J, Asp P, Bohm S, Cavellan E, Farrants AK. Variations in the composition of mammalian SWI/SNF chromatin remodelling complexes. J Cell Biochem. 2009;108:565–576. doi: 10.1002/jcb.22288. [DOI] [PubMed] [Google Scholar]
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Lickert H, Bisgrove BW, Sun X, Yamamoto M, Chawengsaksophak K, Hamada H, Yost HJ, Rossant J, Bruneau BG. Baf60c is a nuclear Notch signaling component required for the establishment of left-right asymmetry. Proc Natl Acad Sci U S A. 2007;104:846–851. doi: 10.1073/pnas.0608118104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi CM, Kondo T, Sajan M, Luo J, Bronson R, Asano T, Farese R, Cantley LC, Kahn CR. Divergent regulation of hepatic glucose and lipid metabolism by phosphoinositide 3-kinase via Akt and PKClambda/zeta. Cell Metab. 2006;3:343–353. doi: 10.1016/j.cmet.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- Thompson M. Plybromo-1: The chromatin targeting subunit of the PBAF complex. Biochimie. 2009;91:309–319. doi: 10.1016/j.biochi.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SM, Murphy EJ, Neese RA, Antelo F, Thomas T, Agarwal A, Go C, Hellerstein MK. Measurement of TG synthesis and turnover in vivo by 2H2O incorporation into the glycerol moiety and application of MIDA. Am J Physiol Endocrinol Metab. 2003;285:E790–E803. doi: 10.1152/ajpendo.00402.2002. [DOI] [PubMed] [Google Scholar]
- Uberall F, Hellbert K, Kampfer S, Maly K, Villunger A, Spitaler M, Mwanjewe J, Baier-Bitterlich G, Baier G, Grunicke HH. Evidence that atypical protein kinase C-lambda and atypical protein kinase C-zeta participate in Ras-mediated reorganization of the F-actin cytoskeleton. J Cell Biol. 1999;144:413–425. doi: 10.1083/jcb.144.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell. 2006;24:367–381. doi: 10.1016/j.molcel.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Vicent GP, Zaurin R, Nacht AS, Li A, Font-Mateu J, Le Dily F, Vermeulen M, Mann M, Beato M. Two chromatin remodeling activities cooperate during activation of hormone responsive promoters. PLoS Genet. 2009;5:e1000567. doi: 10.1371/journal.pgen.1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Sul HS. Upstream stimulatory factors bind to insulin response sequence of the fatty acid synthase promoter. USF1 is regulated. J Biol Chem. 1995;270:28716–28722. doi: 10.1074/jbc.270.48.28716. [DOI] [PubMed] [Google Scholar]
- Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, Workman JL, Crabtree GR. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wong RH, Chang I, Hudak CS, Hyun S, Kwan HY, Sul HS. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell. 2009;136:1056–1072. doi: 10.1016/j.cell.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RH, Sul HS. DNA-PK: relaying the insulin signal to USF in lipogenesis. Cell Cycle. 2009;8:1977–1978. doi: 10.4161/cc.8.13.8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RH, Sul HS. Insulin signaling in fatty acid and fat synthesis: a transcriptional perspective. Curr. Opin. Pharmacol. 2010;10:684–691. doi: 10.1016/j.coph.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.