Introduction
In 30 years, the HIV-1/AIDS pandemic has evolved into an increasingly complex disease composed of multiple epidemics, each influenced by a complex array of biological, behavioural and cultural factors [1–4]. The concentrated subtype B epidemics in Western world settings have been largely restricted to MSM and IDU populations [3]. The generalized heterosexual (HET) epidemics in Africa and Asia have expanded and diversified to include nine major HIV-1 subtypes (A–D, F–H, Jand K) and mosaic circulating recombinant forms (e.g. CRF01_AE and CRF02_AG) [1,5,6]. Migration and globalization has contributed to the spread of non-B subtypes contributing to 20–60% of new infections in Europe, Asia and America [1,2,7].
Highly active antiretroviral therapy (HAART) remains the key to the management of epidemics, reducing mortality, opportunistic diseases and HIV-1 transmission [8–17]. Global health initiatives to scale up antiretroviral therapy (ART) in Africa and Asia have led to population-level reductions in HIV-1 transmission despite weak healthcare systems [8,10,18,19]. The HIV Prevention Trials Network (HPTN) 052, CAPRISA 004, Preexposure Prophylaxis Initiative (iPREX) and Botswana Preprophylaxis Trial (TDF2) randomized clinical trials have shown the potential benefit of early ART initiation, microbicides and prexposure prophylaxis (PrEP) in averting HIV-1 transmission [10,13–16,20,21]. This has led to revisions in treatment guidelines recommending early initiation of ART to all HIV-infected persons and PrEP for high-risk HIV-seronegative populations [15,16]. The failure of the Female Preprophylaxis Trial (FEM-PrEP) underscores the importance of ART adherence and retention [13,14,21,22]. The resurgence of MSM epidemics in the post-HAARTera emphasizes the importance of linking ART with other prevention control modalities [23,24].
The development, implementation and maintenance of effective prevention and treatment interventions will require a thorough understanding of the driving forces of individual epidemics [25,26]. Phylogenetics is a molecular epidemiological strategy that characterizes epidemics on the basis of the genetic interrelatedness of HIV-1 viral sequences, capturing the underlying structure of transmission networks or ‘clusters’ within a given population [27–32]. Molecular phylogenetic analysis can be combined with epidemiological, demographical and behavioural data to describe the spatial, temporal and biological dynamics of individual epidemics [7,27–34]. Phylogenetics provides an important dimension in HIV-1 surveillance by delineating:
The introduction and dissemination of HIV-1 viral subtypes in different regional settings;
The range of transmission patterns fuelling HET, MSM and IDU epidemics;
The interrelationships of biological, demographical and social determinants on viral ‘cluster’ networks;
The role of disease stage in transmission dynamics; and
Underlying trends in regional epidemics, important in the selection of control interventions to limit HIV transmission [26,35,36].
Biological basis of phylogenetic ‘clustering’
High genetic diversity within and across HIV-1 viruses and subtypes can be ascribed to rapid rates of viral replication and turnover with an error-prone reverse transcriptase having high recombination potential [37–41]. Viral evolution within and across HIV-1 subtypes occurs under host, immunological and antiretroviral drug selection pressures, contributing to up to 40, 20 and 10% variations in HIV-1 envelope (env), gag and pol domains, respectively [37,41,42]. Open access websites, such as the Los Alamos (http://www.hiv-lanl.gov), Stanford (http://hivdb.stan-ford.edu) and REGAdb (http://jose.med.kuleuven.be/subtypetool/html/) databases, have sequence datasets and computer tools for classification of viral subtypes, immune epitopes and antiretroviral drug resistance pathways [5,7,43].
Antithetically, despite the amazing potential of HIV to diversify, surprisingly few viral variants and subtypes have contributed to individual infections and regional epidemics [27,44,45]. Severe bottlenecks in HIV transmission lead to a single transmitted/founder (T/F) virus seeding and dominating the virus population (quasispe-cies) [46–54]. Single-genome amplification and next-generation sequencing technologies have shown that 80% of MSM and HET infections may be attributed to a single virus [48,49,55–59]. An understanding of the genotypic and phenotypic signatures of these ancestral strains will be important in understanding critical events contributing to their preferential establishment of infection and evolution in the earliest stages of infection [50,52,56,60–62].
The critical determinant of HIV-1 transmission is viral burden in blood, genital fluid and semen [46,63,64]. Acute/primary stage infection has been postulated to disproportionately contribute to onward transmission related in part to high viraemia (>4.5 log copies/ml) and homogeneity of T/F variants [11,46,57,65,66]. ART limits transmission of chronic stage infection by reducing viral load at an individual and population level [11]. Geospatial bottlenecks add a further constraint, limiting the range of viral subtypes in different risk populations and demographic groups [67–71].
Taken together, high genetic diversity coupled with stringent transmission bottlenecks contribute a single ancestral species seeding infections. Phylogenetic ‘clustering’ elucidates transmission dynamics of individual epidemics and the geospatial expansion of viral subtypes in different regional settings.
Geospatial expansion of HIV-1 subtypes
Molecular phylogeny approaches were first used to classify HIV-1 subtypes and circulating recombinant forms (CRFs) [1,2,5,6,72]. In Western world settings, subtype B epidemics have predominated; in Africa, non-B subtypes have expanded and diversified. Phylogenetic analysis can follow the introduction and spread of specific subtypes and CRFs into local MSM, IDU and HET epidemics [2,71,73–80].
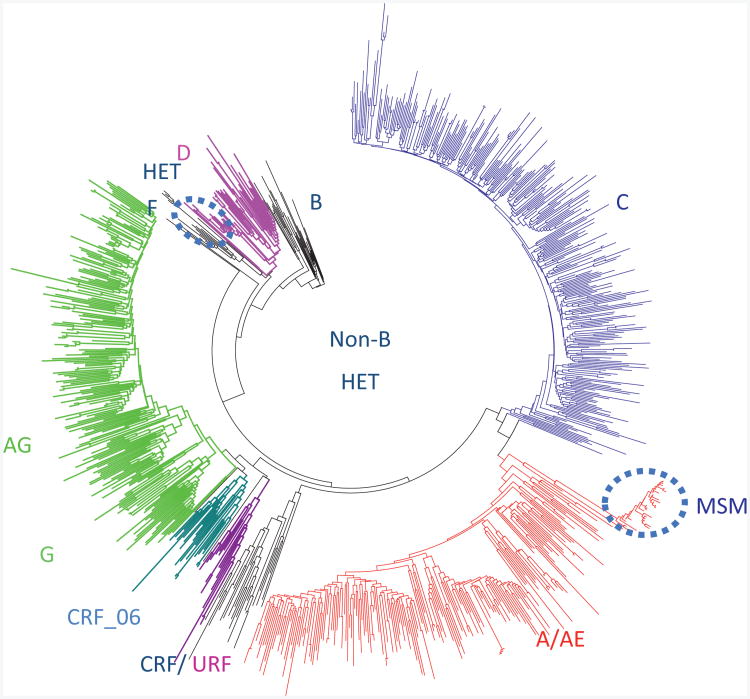
The subtype diversity of the global pandemic is reflected in the distribution of non-B subtypes in Quebec originating from francophone countries in West and Central Africa (Fig. 1) [1,2,6]. Subtype C infections account for half of infections worldwide, whereas subtypes A (12%), CRF01_AE (5%), CRF02_AG (8%) and G (5%) collectively contribute to 30% of the global pandemic [1,2,6]. The remainder of the non-B pandemic is composed of subtype D (Uganda), subtype F (Angola, Romania and South America) and CRFs [1,6]. Subtypes B MSM epidemics dominate in Europe and America, accounting for 9% of global infections. The intermixing of viral subtypes leads to the expansion of novel recombinant mosaics in different regions.
Fig. 1. Phylogenetic analysis of the Quebec non-B subtype epidemic (n=858) originating from West and Central Africa.
Clustering (27%) was limited to conjugal families (n=2–4infections/cluster). Domestic spread of a subtype D (n=13) heterosexual cluster and a CRF_AB (n=30) MSM transmission clusters have been circled.
The fastest growing epidemics worldwide are the IDU epidemics in Eastern Europe where subtypes A1 and CRF03_AB are most prevalent [81]. In heavily populated regions, including India, China and Southeast Asia, epidemics have rapidly shifted from predominant IDU to HET epidemics with selective expansion of subtype C, CRF07_BC, CRF08_BC and CRF01_AE subtypes [82,83].
Non-B subtype epidemics comprise 20–60% of new infections in Europe, Asia and America [71,73–78,84]. Crossover of non-B subtype epidemics into domestic MSM, IDU and/or HETepidemics has been observed in the United Kingdom, France, Germany, Spain, Israel, Greece, the Balkans and Asia [74,84–90]. Domestic spread of non-B subtype infections was rarer in Holland, Switzerland and Quebec [91,92]. As illustrated in Fig. 2, secondary spread of non-B subtypes into Quebec has been limited to one founderCRF_AB(n=33)MSM clusterand one subtype D (n=9) HET cluster. This highlights the importance of providing healthcare to migrant populations.
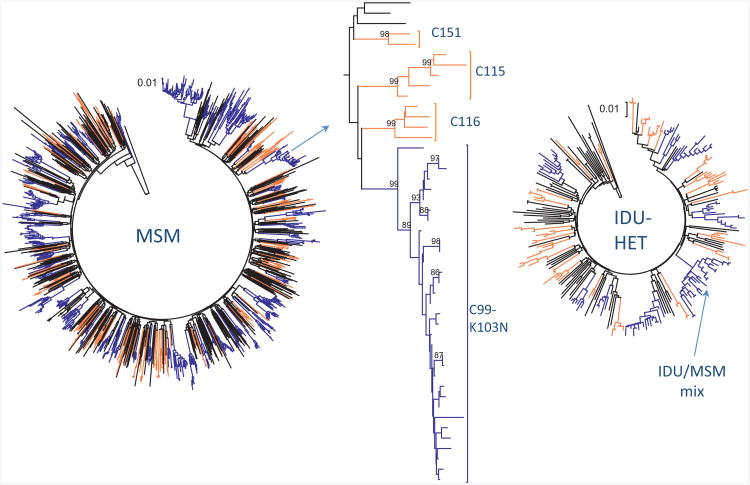
Fig. 2. Phylogenetic clustering of MSM and IDU epidemics in Montreal.
(a) Phylogenetic tree of MSM infections (n=1359) with large clusters [6–30primary HIV infection (PHI)/cluster, n=553] and small clusters (2–4 PHI, n=272) denoted in blue and orange, respectively. (b) Section of MSM tree (arrowed) showing three unique transmissions, three small clusters and one large cluster harbouring the K103N resistance mutations. (c) Phylogenetic tree of IDU infections (n=244) with small (n=90) and large (n=77) clusters denoted in blue and orange, respectively. One mixed cluster (n=17) is arrowed.
Transmission dynamics of MSM epidemics
Phylogenetic analysis has provided a framework for an in-depth evaluation of transmission dynamics of MSM epidemics. Drug-resistance programmes, introduced in the 2000s, have provided large pol (RT/protease) sequence datasets for analysis of transmission trends of regional epidemics [27–29,31,90,93–96]. Molecular phylogeny approaches using neighbour-joining, maximum-likelihood and/or Bayesian (BEAST) methodologies have retraced transmission ‘clusters’ or networks that could not be otherwise identified [28,32].
Systematic surveillance has enhanced our understanding of MSM transmission dynamics by highlighting the role of primary/recent stage infection in onward spread of regional epidemics (Table 1) [27,29,90,93,94,97–101]. The temporal spread of clustered transmissions has been evaluated on the basis of chronological and stage of infection time scales.
Table 1. Comparison of the phylodynamic patterns of MSM, IDU and non-B heterosexual epidemics.
| Frequency of infections in cluster groups (% cases) | Cluster interval | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Risk group | Location | Inclusion criteria | Unique | Small 2–10 | Large +10 | Median (months) | <6 months (% cases) |
| MSM | Montreal | Acute/PHIa | 30 | 42 | 28 | 15 | 27 |
| MSM | Amsterdam | Recent < 18 months | 43 | 57 | 0 | 17 | 25 |
| MSM | UK/London | PHI/Chronic | 60 | 28 | 12 | 22/14 | 16/25 |
| MSM | France | PHI (Cohort) | 83 | 17 | 0 | 15 | 30 |
| MSM | Denmark | PHI/Chronic | 51 | 18 | 31 | ||
| MSM | Belgium | Recent <1 year | 53 | 28 | 19 | ||
| MSM | Switzerland | Acute/PHI/Recent | 64 | 0 | 36 | ||
| Mixed | North Carolina | PHI/Chronic | 66 | 29 | 4 | ||
| IDU/HET | Switzerland | Acute/PHI/Recent | 41 | 0 | 63 | ||
| IDU/HET | Montreal | Acute/PHI/Recent | 30 | 45 | 25 | ||
| HET | Denmark | PHI/Chronic | 85 | 9 | 6 | ||
| Non-B | Montreal | PHI/Chronic | 74 | 24 | 2 | ||
| Non-B | UK | PHI/Chronic | 75 | 20 | 5 | ||
| Non-B | Switzerland | Acute/PHI/Recent | 78 | 22 | 0 | ||
| Non-B | Belgium | Recent (<1 year) | 87 | 12 | 0 | ||
Data were extracted from studies performed in Quebec [27,29], Netherlands [94,182], United Kingdom [31,90,93], France [105], Denmark [101], Belgium [111], Switzerland [86,103] and North Carolina [106] to elaborate the relative role of nonclustered (unique), small clustered (2–10) and large clustered (10+) networks in different settings. The median cluster intervals and the percentage of linked infections occurring within 6-month intervals substantiate the relative role of early infection. HET, heterosexual; PHI, primary HIV infection.
The Montreal study included drug-naive and treated chronic populations to establish cluster size but excluded these samples from analysis of cluster intervals.
Comparative analysis of MSM transmission dynamics has been confounded by the use of different selection criteria and methodology. Molecular phylogeny studies have been performed on different groups: acute/primary HIV infection (PHI), PHI (<6 months), recent infection (<12–18 months) and combined PHI and chronic cohorts. Statistical criteria have varied combining high bootstrap values (>95–98%) and low genetic distance (<0.015–0.045) for the designation of ‘clusters’ [28,29,93,101]. Correction for drug resistance is necessary when using chronic treatment experienced populations. Robust criteria delineate ‘transmission clusters’ with internal controls for repeat sampling and include known source-index partnerships [28,29,93,101]. Clustering can be cross-validated by analysis of sequence congruency in natural polymorphisms and reproduction of pol gene clustering in gag, env and integrase domains [28,29,93,101].
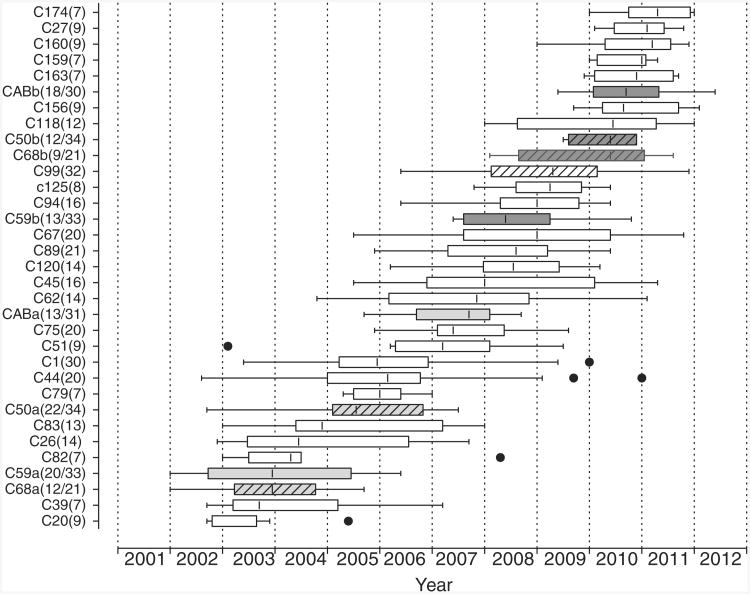
The expansion of the MSM epidemic in Montreal evaluated clustering of primary infections (PHI < 6 months) over the last decade. Three patterns of onward spread of MSM epidemics were observed: unique transmissions, small clusters (2–4 PHI) and large clusters (5–60 PHI) (Fig. 2a, b) [27,29]. Large clusters were the driving force sustaining the MSM epidemic, increasing from 25% of primary infections in 2005 to 51% of infections by the end of 2011. The high rates of PHI-related clustering may be related to suppressed viraemia at a population level wherein 85% of diagnosed MSM are receiving HAART. Episodic expansion of large clusters (7–60, median 14 PHI) has arisen through the stepwise formation of new clusters unrelated to prior clusters, as well as secondary outbreaks of older clusters (Fig. 3) [27,29]. Whereas small clusters expanded over 4.75-month intervals (1–11.5 interquartiles), large clusters expanded over 11-month intervals (3.5–25.5 interquar-tiles).
Fig. 3. Temporal expansion of 29 large MSM clusters (7+ primary infections) over the last decade.
Box and whiskers plots represent the episodic transmission intervals of each cluster, with Tukey whiskers showing 1.5 time interquartile intervals and severe outliers (●). Four transmission clusters showed biphasic expansion with initial (light grey) and secondary outbreaks (dark grey). Clusters harbouring resistance to nonnucleoside reverse transcriptase inhibitors have been dashed. The sizes of each transmission cluster are denoted in parenthesis. Clusters 68, 50 and 99 harbouring transmitted resistance to NNRTIs are dashed.
Phylodynamic features of other regional MSM epidemics were consistent with that observed in Montreal (Table 1) [27,29,31,93,94,102–104]. Cluster membership (17– 70%) and size of clusters showed regional variation based, in part, on the depth of population sampling and incidence rates (Table 1) [27,31,93,101,102,105–110]. Transmission clustering was highest in concentrated urban settings and lowest in diffuse nationwide epidemics.
Time-resolved phylogenies on date-stamped sequences in the United Kingdom, Netherlands, France and Montreal cohorts indicated that 25–30% of clustering arose over 6-month time spans with median cluster intervals occurring over 14–17 month time spans (Table 1) [27,29,31,93,94,102]. Phylogenetic clustering in other studies was also related to acute/PHI/recent infection status and high CD4 cell count [29,101,103,111].
Temporal dynamics of MSM transmissions
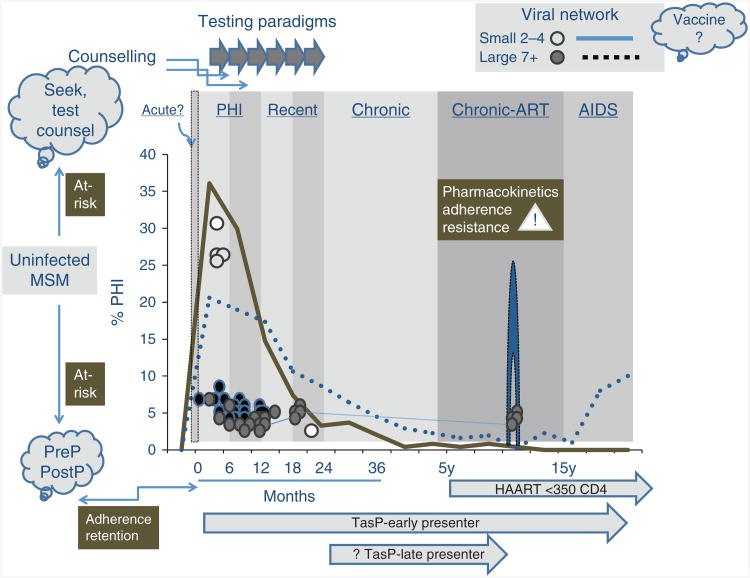
Phylogenetic inferences project a role of primary infection (<6 months) and recent stage infection (<14–18 months) in sustaining MSM epidemics (Table 1, Fig. 4). This time span goes well beyond empirical and mathematical models that emphasize a disproportionate contribution role of the 3-week infectious period of ‘acute’ infection in onward transmission [46,112,113]. Newly infected persons in primary and recent stage infection may harbour homogeneous pools of ancestral founder viruses that may have a higher transmissible ‘dose’ of virus for an extended period of time. These early disease stages are distinct from chronically infected source partners that harbour a vast array of quasispecies, including variants having impaired replicative competence and/or fitness [54,114].
Fig. 4. Phylogenetic insights on future possibilities in HIV prevention.
Temporal expansion of small (2–4 PHI) and large (5–60) clusters are denoted by solid and dashed lines, respectively. Testing must be frequent to capture individuals in a timely fashion for ‘Treatment for as Prevention’ (TasP) paradigms. Antiretroviral drug (ART) pharmacokinetics and patient adherence to ART is necessary to prevent viral bound and chronic stage transmissions.
Accurate dating remains a serious caveat in time-resolved phylogenies and modelling of transmission dynamics. Acute infection is rarely detected in real time, as newly infected persons are often symptom-free and unaware of their status [115,116]. Those unaware of their status frequently engage in higher risk behaviour associated with a 3.5-fold increased risk for HIV transmission [117–120].
Control interventions to limit HIV transmission will be predicated on early diagnosis [46,113,121–126]. Serological incidence and p24 assays remain imprecise measures of recency of infection and are limited to those persons recruited into PHI cohorts [68,127–131]. Viral sequence-based assays may assist in estimating recency of infection by monitoring time-dependent evolution from a ‘clonal’ founder event [128,129,132,133]. The frequency of ambiguous calls in bulk sequencing can serve as a surrogate marker that distinguishes recent infection (<0.44% ambiguity in first year) from chronic infection (predictive value 98.7%) [129,133]. Next generation sequencing and high-resolution melting assays may be applied in dating archived specimens [128,134,135].
There remain caveats in Bayesian analysis, time-resolved phylogenies and sequence diversity assays. Issues pertinent to phylogenetic metrics include sequence quality; interval between seroconversion and first genotyping, especially for chronic populations; and effects of ART on quasispecies dynamics. ART suppresses viral replication and quasispecies evolution; treatment failure leads to limited expansion of drug-resistant variants; treatment cessation leads to rebound of previously transmitted ancestral variants [114,136].
Universal genotyping at presentation will become increasingly more important, particularly with the introduction of test–treat paradigms. Genotyping is relatively inexpensive (∼$125, homebrew assays), the frequency of transmitted drug resistance is high (10–20%) and subtype diversity is common.
Transmission patterns of IDU and heterosexual epidemics
Despite the overall decline in IDU epidemics associated with harm reduction and needle-exchange programmes and antiviral therapy ART, IDU epidemics continue to emerge in North America, Eastern Europe and Asia [85,110,137–141]. Transmission clustering has been implicated in the spread of IDU epidemics, including the spread of drug-resistant subepidemics (Table 1) [110,142–144]. IDU epidemics may bridge the crossover of HET and MSM epidemics, as well as contribute to the introduction and spread of non-B subtype infections and HIV/hepatitis C coinfections [110,137,145–148].
Phylogenetic clustering has been frequently observed in IDU epidemics [103,110,146,149,150] (Fig. 2c). Single-genome analysis has shown that 40–60% of IDU transmissions are associated with a single T/F variant, with remaining infections harbouring three to 16 closely related monophyletic strains [58]. Higher multiplicity of infection among IDU populations may be associated with an absence of the mucosal barriers, prison settings and poor general or mental health [58,149,151].
There is a paucity of data on the phylodynamics of HET non-B subtype epidemics, particularly in endemic Third World settings (Table 1) [57,152,153]. Single T/F viruses were observed in 68% of subtype C primary infections from Botswana, similar to that observed for MSM cohorts [153]. The overall membership size of transmission clusters (two to four infections/cluster) among HET cohorts has been far lower than that reported for MSM populations (Table 1) [42,154]. The role of early stage infection may also be less pronounced in HET partnerships (Table 1) [155,156]. Domestic spread of subtype A and C HET infections (cluster size >10) in the United Kingdom occurred over cluster intervals of 27 months, with only 2% of transmissions occurring over 6-month periods [90].
Individuals with extended high viraemia have been observed to disproportionately contribute to expansion of the subtype C epidemic in Botswana [9,154]. A substantial fraction (34%) of newly infected persons maintain extended high viraemia (>100 000 copies/ml) for median periods of 318 (282–459) days [57]. This suggests that treatment as prevention (TasP) initiatives may be selectively targeted to those persons showing extended viraemia [9,154].
Phylogenetics were important in randomized control prevention trials, showing that 10–30% of HIV-1 transmissions among serodiscordant partnerships were unlinked and likely involved third partners [157–160]. Analysis of virus transmitted in serodiscordent partnerships in the Rakai cohort showed that there was a preferential transmission of the ancestral founder strain from the source partner's quasispecies [114].
Transmitted drug resistance
Genotypic drug resistance testing programmes have been instrumental in identifying mutations conferring resistance to nucleoside and nonnucleoside reverse transcriptase (NRTIs and NNRTIs), protease inhibitors and integrase inhibitors (INIs) [5]. Transmitted drug resistance (TDR) remains a serious public health concern, representing 10–20% of new infections among treatment-naive populations in different regional settings [161–164].
TDR is particularly noteworthy for NNRTIs wherein single point mutations (K103N, G190A, Y181C and E138K) confer high level resistance with a little impact on viral replicative fitness [5,162,163,165]. There is innate resistance to NNRTIs in subtype O and HIV-2, as well as a signature V106M pathway for subtype C [5,166,167]. The use of nevirapine to prevent vertical transmission may lead to NNRTI resistance in infants [168,169]. Another concern is the facilitated development of K65R in subtype C, in association with stavudine (d4T), didanosine (ddI) and/or nevirapine (NVP)-based regimens [170,171]. TDR may emerge more rapidly in Third World settings due to drugs having poor pharmacodynamic properties (d4T and ddI) [5].
High rates of TDR among drug-naive MSM and IDU populations have been related to clustering [5,162,163,172,173]. TDR to NNRTIs (17%) in Montreal can be ascribed to six MSM clusters harbouring K103N, V108I or G190A (n=9–60) (Figs. 1a and 2) [162]. Prospective monitoring of the expansion of drug-resistant subepidemics may help guide local treatment policies. Other frequently transmitted species include the L90M protease mutation and thymidine analogue mutations (e.g. M41L, D67N, T215 revertants and K219Q) [109,162,163,174–177].
Phylogenetic insights on future possibilities in HIV prevention
The HPTN 052 and PreP randomized control trials have advanced the vision that TasP paradigms may dramatically reduce HIV incidence and prevalence at a population level [10,13–16,20,21]. The development of prevention interventions will require metrics that characterize the drivers of individual epidemics [86,91]. Realistic models on how control interventions will impact on local networks will require a comprehensive understanding of transmission patterns of MSM, IDU and HETepidemics in different regional settings [27–29,31,90,93–96].
The cornerstone to HIV prevention is timely diagnosis of HIV in high-risk populations [15,16]. Acute infection is rarely detected in real-time, and half of newly diagnosed individuals are ‘late presenters’ (>1 year postseroconversion) [178–180]. Phylogenetic inferences project a median duration of 6 and 11–15 months of small and large clusters, respectively (Table 1, Fig. 4) [27,29,90,93,94,97–100]. This period substantiated a plausible benefit of early ART initiation to avert HIV clustering [46]. Seek–test–counsel paradigms are needed to address issues of poor testing habits and late presentation (Fig. 4).
The efficacy of ART requires adherence and retention, as viral rebound will occur on treatment cessation that may contribute to episodic bursts in transmission [136]. The observation of secondary outbreaks in the Montreal and Swiss epidemics has been linked to time frames when treatment interruption trials (e.g. SMART) were in place (Fig. 4) [136]. Individuals receiving PreP and early ART must be counselled on adherence and treatment continuity.
There have been few studies characterizing endemic non-B HETepidemics. In contrast to that observed for MSM and IDU epidemics, studies on HET populations show infrequent clustering and low cluster size. This would suggest that TasP would not have as dramatic an impact on reducing transmission. The Botswana study suggests a potential benefit of TasP for extended high viraemics [57].
It is important to recognize that phylogenetics define viral networks that are not necessarily synonymous to sex networks and individual-level risk. There is mixing of high and low-risk populations having high and low awareness of status [112,181]. Data from the Montreal SPOT rapid testing site show that although seropositivity was related to casual partnerships, cluster membership and size of cluster was related to poor testing habits. It has been estimated that 7–20% of MSM infections may be acquired abroad, contributing in part to unique transmissions [24,182].
Testing, treatment and other prevention interventions require major public health commitments. The integration of phylogenetic, epidemiological clinical and demographic data will be important in delineating the role of linkage to care, behaviour, socioeconomic factors and migration on transmission dynamics [96]. Universal genotyping of all newly infected persons could provide critical information needed to reconstruct underlying structures of local epidemics necessary for the design and evaluation of control interventions. Baseline resistance profiles could assess transmitted resistance and guide HIV-1 management. Molecular phylogeny strategies could help elucidate the role of disease stage and virus-specific determinants in transmission dynamics [128,129,132,133]. This analysis is particularly important for the design of cost-effective treatment and prevention combinations in resource-poor settings.
Whereas early stage infection may dominate in regional settings having universal access to healthcare and ART coverage, significant contributions of chronic stage infections may be related to socioeconomic factors, including lower awareness of status and poor linkage to care and treatment [115,116,183–186]. Phylodemo-graphics can be of importance in surveillance of the rise MSM epidemics among young adults (13–29 years) and racial/ethic minorities [109,184,187]. Phylogenetic inferences of local epidemics may assist in the design of targeted educational campaigns and prevention policies for distinct HIV-infected populations.
Future research may broaden our knowledge of underlying mechanisms leading to the preferential selection and expansion of transmitted ancestral strains. Phylodynamic inferences will be important in the design, implementation and assessment of candidate public health, therapeutic and behavioural interventions and the ultimate pursuit of HIV vaccines.
Acknowledgments
We thank the patients, clinicians and research staff from all centres participating in the Montreal PHI cohort study and SPOT study groups and the Quebec genotyping programme, including principal investigators Jean-Pierre Routy, Joanne Otis, Robert Rousseau, Cecile Tremblay and Hugues Charest, and participating clinicians/investigators J. Allan, F. Asselin, M. Blais, M. Boissonnault, J.-G. Baril, J. Bruneau, L. Charest, M.A. Charron, M. Couillard, J. Cox, P. Côté, A. de Pokomandy, A. Dascal, H. Dion, S. Dufresne, G. Emond, G. Fadel, J. Falutz, C. Fortin, N. Gilmore, G. Godin, M.S. Joyal, P. Junod, M. Klein, R.G. Lalonde, G. Lambert, F. Laplante, S. Lavoie, N. Lapointe, R. Leblanc, D. Legault, B. Lessard, M. Legault, D. Longpré, P.J. Maziade, M.E. Morin, M. Munoz, D. Murphy, V.K. Nguyen, R. O'Brien, J. Otis, M.Y. Parent, D. Poirier, Potter, D. Rouleau, R. Rousseau, É. Sasseville, A. Talbot, D. Tessier, R. Thomas, C. Tremblay, B. Trottier, C. Tsoukas and S. Vézina.
B.B. wrote this review. M.A.W. and M.R. are coinvestigators in the Montreal phylogenetic study and reviewed the contents of this study.
Studies on HIV-1 transmission dynamics in Montreal have been supported by grants from the Canadian Institutes for Health Research (CIHR), Fonds de la Recherche en Santé Québec (FRSQ) and the National Institute of Health (USA) (Grant R01 A1078752).
Footnotes
Conflicts of interest: The authors have no conflicts of interest to declare.
References
- 1.Tebit DM, Arts EJ. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis. 2011;11:45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- 2.Lihana RW, Ssemwanga D, Abimiku A, Ndembi N. Update on HIV-1 diversity in Africa: a decade in review. AIDS Rev. 2012;14:83–100. [PubMed] [Google Scholar]
- 3.Beyrer C, Baral SD, van Griensven F, Goodreau SM, Chariya-lertsak S, Wirtz AL, et al. Global epidemiology of HIV infection in men who have sex with men. Lancet. 2012;380:367–377. doi: 10.1016/S0140-6736(12)60821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beyrer C, Sullivan PS, Sanchez J, Dowdy D, Altman D, Trap-ence G, et al. A call to action for comprehensive HIV services for men who have sex with men. Lancet. 2012;380:424–438. doi: 10.1016/S0140-6736(12)61022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wainberg MA, Zaharatos GJ, Brenner BG. Development of antiretroviral drug resistance. N Engl J Med. 2011;365:637–646. doi: 10.1056/NEJMra1004180. [DOI] [PubMed] [Google Scholar]
- 6.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS. 2011;25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castro-Nallar E, Perez-Losada M, Burton GF, Crandall KA. The evolution of HIV: inferences using phylogenetics. Mol Phylogenet Evol. 2012;62:777–792. doi: 10.1016/j.ympev.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnell D, Baeten JM, Kiarie J, Thomas KK, Stevens W, Cohen CR, et al. Heterosexual HIV-1 transmission after initiation of antiretroviral therapy: a prospective cohort analysis. Lancet. 2010;375:2092–2098. doi: 10.1016/S0140-6736(10)60705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novitsky V, Essex M. Using HIV viral load to guide treatment-for-prevention interventions. Curr Opin HIV AIDS. 2012;7:117–124. doi: 10.1097/COH.0b013e32834fe8ff. [DOI] [PubMed] [Google Scholar]
- 10.Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365:493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds SJ, Makumbi F, Nakigozi G, Kagaayi J, Gray RH, Wawer M, et al. HIV-1 transmission among HIV-1 discordant couples before and after the introduction of antiretroviral therapy. AIDS. 2011;25:473–477. doi: 10.1097/QAD.0b013e3283437c2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das M, Chu PL, Santos GM, Scheer S, Vittinghoff E, McFarland W, et al. Decreases in community viral load are accompanied by reductions in new HIV infections in San Francisco. PLoS One. 2010;5:e11068. doi: 10.1371/journal.pone.0011068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant RM, Lama JR, Anderson PL, McMahan V, Liu AY, Vargas L, et al. Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N Engl J Med. 2010;363:2587–2599. doi: 10.1056/NEJMoa1011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdool Karim Q, Abdool Karim SS, Frohlich JA, Grobler AC, Baxter C, Mansoor LE, et al. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science. 2010;329:1168–1174. doi: 10.1126/science.1193748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson MA, Aberg JA, Hoy JF, Telenti A, BensonC, Cahn P, et al. Antiretroviral treatment of adult HIV infection: 2012 recommendations of the International Antiviral Society-USA Panel. JAMA. 2012;308:387–402. doi: 10.1001/jama.2012.7961. [DOI] [PubMed] [Google Scholar]
- 16.Labarga P. New DHHS guidelines recommend antiretroviral therapy to all HIV-infected persons. AIDS Rev. 2012;14:154. [PubMed] [Google Scholar]
- 17.Baeten JM, Donnell D, Ndase P, Mugo NR, Campbell JD, Wangisi J, et al. Antiretroviral prophylaxis for HIV prevention in heterosexual men and women. N Engl J Med. 2012 doi: 10.1056/NEJMoa1108524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montaner JS, Lima VD, Barrios R, Yip B, Wood E, Kerr T, et al. Association of highly active antiretroviral therapy coverage, population viral load, and yearly new HIV diagnoses in British Columbia, Canada: a population-based study. Lancet. 2010;376:532–539. doi: 10.1016/S0140-6736(10)60936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bendavid E, Grant P, Talbot A, Owens DK, Zolopa A. Cost-effectiveness of antiretroviral regimens in the World Health Organization's treatment guidelines: a South African analysis. AIDS. 2011;25:211–220. doi: 10.1097/QAD.0b013e328340fdf8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okwundu CI, Uthman OA, Okoromah CA. Antiretroviral preexposure prophylaxis (PrEP) for preventing HIV in high-risk individuals. Cochrane Database Syst Rev. 2012;7:CD007189. doi: 10.1002/14651858.CD007189.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thigpen MC, Kebaabetswe PM, Paxton LA, Smith DK, Rose CE, Segolodi TM, et al. Antiretroviral preexposure prophylaxis for heterosexual HIV transmission in Botswana. N Engl J Med. 2012;367:423–434. doi: 10.1056/NEJMoa1110711. [DOI] [PubMed] [Google Scholar]
- 22.Van Damme L, Corneli A, Ahmed K, Agot K, Lombaard J, Kapiga S, et al. Preexposure prophylaxis for HIV infection among African women. N Engl J Med. 2012;367:411–422. doi: 10.1056/NEJMoa1202614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeGruttola V, Smith DM, Little SJ, Miller V. Developing and evaluating comprehensive HIV infection control strategies: issues and challenges. Clin Infect Dis. 2010;50(Suppl 3):S102–S107. doi: 10.1086/651480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Sighem A, Vidondo B, Glass TR, Bucher HC, Vernazza P, Gebhardt M, et al. Resurgence of HIV infection among men who have sex with men in Switzerland: mathematical modelling study. PLoS One. 2012;7:e44819. doi: 10.1371/journal.pone.0044819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin F, Jansson J, Law M, Prestage GP, Zablotska I, Imrie JC, et al. Per-contact probability of HIV transmission in homosexual men in Sydney in the era of HAART. AIDS. 2010;24:907–913. doi: 10.1097/QAD.0b013e3283372d90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith K, Powers KA, Kashuba AD, Cohen MS. HIV-1 treatment as prevention: the good, the bad, and the challenges. Curr Opin HIV AIDS. 2011;6:315–325. doi: 10.1097/COH.0b013e32834788e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brenner BG, Roger M, Stephens D, Moisi D, Hardy I, Weinberg J, et al. Transmission clustering drives the onward spread of the HIV epidemic among men who have sex with men in Quebec. J Infect Dis. 2011;204:1115–1119. doi: 10.1093/infdis/jir468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hue S, Clewley JP, Cane PA, Pillay D. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS. 2004;18:719–728. doi: 10.1097/00002030-200403260-00002. [DOI] [PubMed] [Google Scholar]
- 29.Brenner BG, Roger M, Routy JP, Moisi D, Ntemgwa M, Matte C, et al. High rates of forward transmission events after acute/early HIV-1 infection. J Infect Dis. 2007;195:951–959. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 30.Pillay D, Fisher M. Primary HIV Infection, phylogenetics, and antiretroviral prevention. J Infect Dis. 2007;195:924–926. doi: 10.1086/512090. [DOI] [PubMed] [Google Scholar]
- 31.Leigh Brown AJ, Lycett SJ, Weinert L, Hughes GJ, Fearnhill E, Dunn DT. Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis. 2011;204:1463–1469. doi: 10.1093/infdis/jir550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam TT, Hon CC, Tang JW. Use of phylogenetics in the molecular epidemiology and evolutionary studies of viral infections. Crit Rev Clin Lab Sci. 2010;47:5–49. doi: 10.3109/10408361003633318. [DOI] [PubMed] [Google Scholar]
- 33.Grenfell BT, Pybus OG, Gog JR, Wood JL, Daly JM, Mumford JA, et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 34.Pybus OG, Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nat Rev Genet. 2009;10:540–550. doi: 10.1038/nrg2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delva W, Wilson DP, Abu-Raddad L, Gorgens M, Wilson D, Hallett TB, et al. HIV treatment as prevention: principles of good HIV epidemiology modelling for public health decision-making in all modes of prevention and evaluation. PLoS Med. 2012;9:e1001239. doi: 10.1371/journal.pmed.1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delva W, Eaton JW, Meng F, Fraser C, White RG, Vickerman P, et al. HIV treatment as prevention: optimising the impact of expanded HIV treatment programmes. PLoS Med. 2012;9:e1001258. doi: 10.1371/journal.pmed.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Preston BD. Error-prone retrotransposition: rime of the ancient mutators. Proc Natl Acad Sci U S A. 1996;93:7427–7431. doi: 10.1073/pnas.93.15.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang M, Foley B, Schultz AK, Macke JP, Bulla I, Stanke M, et al. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology. 2010;7:25. doi: 10.1186/1742-4690-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, et al. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 40.Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol Mol Biol Rev. 2009;73:451–480. doi: 10.1128/MMBR.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preston BD, Dougherty JP. Mechanisms of retroviral mutation. Trends Microbiol. 1996;4:16–21. doi: 10.1016/0966-842x(96)81500-9. [DOI] [PubMed] [Google Scholar]
- 42.Novitsky V, Wang R, Lagakos S, Essex M. HIV-1 subtype C phylodynamics in the global epidemic. Viruses. 2010;2:33–54. doi: 10.3390/v2010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmes EC, Grenfell BT. Discovering the phylodynamics of RNA viruses. PLoS Comput Biol. 2009;5:e1000505. doi: 10.1371/journal.pcbi.1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke DK, Duarte EA, Moya A, Elena SF, Domingo E, Holland J. Genetic bottlenecks and population passages cause profound fitness differences in RNA viruses. J Virol. 1993;67:222–228. doi: 10.1128/jvi.67.1.222-228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arien KK, Troyer RM, Gali Y, Colebunders RL, Arts EJ, Vanham G. Replicative fitness of historical and recent HIV-1 isolates suggests HIV-1 attenuation over time. AIDS. 2005;19:1555–1564. doi: 10.1097/01.aids.0000185989.16477.91. [DOI] [PubMed] [Google Scholar]
- 46.CohenMS, Shaw GM, McMichael AJ, Haynes BF. Acute HIV-1 Infection. N Engl J Med. 2011;364:1943–1954. doi: 10.1056/NEJMra1011874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee HY, Giorgi EE, Keele BF, Gaschen B, Athreya GS, Salazar-Gonzalez JF, et al. Modeling sequence evolution in acute HIV-1 infection. J Theor Biol. 2009;261:341–360. doi: 10.1016/j.jtbi.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferrari G, Korber B, Goonetilleke N, Liu MK, Turnbull EL, Salazar-Gonzalez JF, et al. Relationship between functional profile of HIV-1 specific CD8 T cells and epitope variability with the selection of escape mutants in acute HIV-1 infection. PLoS Pathog. 2011;7:e1001273. doi: 10.1371/journal.ppat.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wood N, Bhattacharya T, Keele BF, Giorgi E, Liu M, Gaschen B, et al. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 2009;5:e1000414. doi: 10.1371/journal.ppat.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kearney M, Maldarelli F, Shao W, Margolick JB, Daar ES, Mellors JW, et al. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J Virol. 2009;83:2715–2727. doi: 10.1128/JVI.01960-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kong X, West JT, Zhang H, Shea DM, M'Soka TJ, Wood C. The human immunodeficiency virus type 1 envelope confers higher rates of replicative fitness to perinatally transmitted viruses than to nontransmitted viruses. J Virol. 2008;82:11609–11618. doi: 10.1128/JVI.00952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, Karita E, et al. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 2009;5:e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, et al. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J Virol. 2012;86:2715–2728. doi: 10.1128/JVI.06157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Novitsky V, Ndung'u T, Wang R, Bussmann H, Chonco F, Makhema J, et al. Extended high viremics: a substantial fraction of individuals maintain high plasma viral RNA levels after acute HIV-1 subtype C infection. AIDS. 2011;25:1515–1522. doi: 10.1097/QAD.0b013e3283471eb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bar KJ, Li H, Chamberland A, Tremblay C, Routy JP, Grayson T, et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol. 2010;84:6241–6247. doi: 10.1128/JVI.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abrahams MR, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, Ping LH, et al. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a nonpoisson distribution of transmitted variants. J Virol. 2009;83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gnanakaran S, Bhattacharya T, Daniels M, Keele BF, Hraber PT, Lapedes AS, et al. Recurrent signature patterns in HIV-1 B clade envelope glycoproteins associated with either early or chronic infections. PLoS Pathog. 2011;7:e1002209. doi: 10.1371/journal.ppat.1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Freel SA, Picking RA, Ferrari G, Ding H, Ochsenbauer C, Kappes JC, et al. Initial HIV-1 antigen-specific CD8+ T cells in acute HIV-1 infection inhibit transmitted/founder virus replication. J Virol. 2012;86:6835–6846. doi: 10.1128/JVI.00437-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, Salazar-Gonzalez JF, et al. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4beta7. PLoS Pathog. 2012;8:e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pilcher CD, Tien HC, Eron JJ, Jr, Vernazza PL, Leu SY, Stewart PW, et al. Brief but efficient: acute HIV infection and the sexual transmission of HIV. J Infect Dis. 2004;189:1785–1792. doi: 10.1086/386333. [DOI] [PubMed] [Google Scholar]
- 64.Wawer MJ, Gray RH, Sewankambo NK, Serwadda D, Li X, Laeyendecker O, et al. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J Infect Dis. 2005;191:1403–1409. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- 65.Hecht FM, Hartogensis W, Bragg L, Bacchetti P, Atchison R, Grant R, et al. HIV RNA level in early infection is predicted by viral load in the transmission source. AIDS. 2010;24:941–945. doi: 10.1097/QAD.0b013e328337b12e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hollingsworth TD, Laeyendecker O, Shirreff G, Donnelly CA, Serwadda D, Wawer MJ, et al. HIV-1 transmitting couples have similar viral load set-points in Rakai, Uganda. PLoS Pathog. 2010;6:e1000876. doi: 10.1371/journal.ppat.1000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brenner BG, Routy JP, Petrella M, Moisi D, Oliveira M, Detorio M, et al. Persistence and fitness of multidrug-resistant human immunodeficiency virus type 1 acquired in primary infection. J Virol. 2002;76:1753–1761. doi: 10.1128/JVI.76.4.1753-1761.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamlyn E, Jones V, Porter K, Fidler S. Antiretroviral treatment of primary HIV infection to reduce onward transmission. Curr Opin HIV AIDS. 2010;5:283–290. doi: 10.1097/COH.0b013e32833a6b11. [DOI] [PubMed] [Google Scholar]
- 69.Skar H, Hedskog C, Albert J. HIV-1 evolution in relation to molecular epidemiology and antiretroviral resistance. Ann N Y Acad Sci. 2011;1230:108–118. doi: 10.1111/j.1749-6632.2011.06128.x. [DOI] [PubMed] [Google Scholar]
- 70.Brenner BG, Turner D, Wainberg MA. HIV-1 drug resistance: can we overcome? Expert Opin Biol Ther. 2002;2:751–761. doi: 10.1517/14712598.2.7.751. [DOI] [PubMed] [Google Scholar]
- 71.Gray RR, Tatem AJ, Lamers S, Hou W, Laeyendecker O, Serwadda D, et al. Spatial phylodynamics of HIV-1 epidemic emergence in east Africa. AIDS. 2009;23:F9–F17. doi: 10.1097/QAD.0b013e32832faf61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kantor R. Impact of HIV-1 pol diversity on drug resistance and its clinical implications. Curr Opin Infect Dis. 2006;19:594–606. doi: 10.1097/QCO.0b013e3280109122. [DOI] [PubMed] [Google Scholar]
- 73.Paraskevis D, Nikolopoulos G, Tsiara C, Paraskeva D, Antoniadou A, Lazanas M, et al. HIV-1 outbreak among injecting drug users in Greece, 2011: a preliminary report. Euro Surveill. 2011;16:1–4. doi: 10.2807/ese.16.36.19962-en. [DOI] [PubMed] [Google Scholar]
- 74.Gonzalez-Alba JM, Holguin A, Garcia R, Garcia-Bujalance S, Alonso R, Suarez A, et al. Molecular surveillance of HIV-1 in Madrid, Spain: a phylogeographic analysis. J Virol. 2011;85:10755–10763. doi: 10.1128/JVI.00454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paraskevis D, Magiorkinis E, Magiorkinis G, Sypsa V, Paparizos V, Lazanas M, et al. Increasing prevalence of HIV-1 subtype A in Greece: estimating epidemic history and origin. J Infect Dis. 2007;196:1167–1176. doi: 10.1086/521677. [DOI] [PubMed] [Google Scholar]
- 76.Paraschiv S, Otelea D, Batan I, Baicus C, Magiorkinis G, Paraskevis D. Molecular typing of the recently expanding subtype B HIV-1 epidemic in Romania: evidence for local spread among MSMs in Bucharest area. Infect Genet Evol. 2012;12:1052–1057. doi: 10.1016/j.meegid.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salemi M, Lamers SL, Huysentruyt LC, Galligan D, Gray RR, Morris A, et al. Distinct patterns of HIV-1 evolution within metastatic tissues in patients with non-Hodgkins lymphoma. PLoS One. 2009;4:e8153. doi: 10.1371/journal.pone.0008153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pang W, Zhang C, Duo L, Zhou YH, Yao ZH, Liu FL, et al. Extensive and complex HIV-1 recombination between B', C and CRF01_AE among IDUs in south-east Asia. AIDS. 2012;26:1121–1129. doi: 10.1097/QAD.0b013e3283522c97. [DOI] [PubMed] [Google Scholar]
- 79.Kuiken C, Korber B, Shafer RW. HIV sequence databases. AIDS Rev. 2003;5:52–61. [PMC free article] [PubMed] [Google Scholar]
- 80.Maljkovic Berry I, Athreya G, Kothari M, Daniels M, Bruno WJ, Korber B, et al. The evolutionary rate dynamically tracks changes in HIV-1 epidemics: application of a simple method for optimizing the evolutionary rate in phylogenetic trees with longitudinal data. Epidemics. 2009;1:230–239. doi: 10.1016/j.epidem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stanojevic M, Alexiev I, Beshkov D, Gokengin D, Mezei M, Minarovits J, et al. HIV1 molecular epidemiology in the Balkans: a melting pot for high genetic diversity. AIDS Rev. 2012;14:28–36. [PubMed] [Google Scholar]
- 82.He X, Xing H, Ruan Y, Hong K, Cheng C, Hu Y, et al. A comprehensive mapping of HIV-1 genotypes in various risk groups and regions across china based on a nationwide molecular epidemiologic survey. PLoS One. 2012;7:e47289. doi: 10.1371/journal.pone.0047289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Neogi U, Bontell I, Shet A, De Costa A, Gupta S, Diwan V, et al. Molecular epidemiology of HIV-1 subtypes in India: origin and evolutionary history of the predominant subtype C. PLoS One. 2012;7:e39819. doi: 10.1371/journal.pone.0039819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lawyer G, Schulter E, Kaiser R, Reuter S, Oette M, Lengauer T. Endogenous or exogenous spreading of HIV-1 in Nordrhein-Westfalen, Germany, investigated by phylodynamic analysis of the RESINA Study cohort. Med Microbiol Immunol. 2012;201:259–269. doi: 10.1007/s00430-011-0228-8. [DOI] [PubMed] [Google Scholar]
- 85.Yerly S, Vora S, Rizzardi P, Chave JP, Vernazza PL, Flepp M, et al. Acute HIV infection: impact on the spread of HIV and transmission of drug resistance. AIDS. 2001;15:2287–2292. doi: 10.1097/00002030-200111230-00010. [DOI] [PubMed] [Google Scholar]
- 86.von Wyl V, Kouyos RD, Yerly S, Boni J, Shah C, Burgisser P, et al. The role of migration and domestic transmission in the spread of HIV-1 non-B subtypes in Switzerland. J Infect Dis. 2011;204:1095–1103. doi: 10.1093/infdis/jir491. [DOI] [PubMed] [Google Scholar]
- 87.Chemtob D, Grossman Z. Epidemiology of adult and adolescent HIV infection in Israel: a country of immigration. Int J STD AIDS. 2004;15:691–696. doi: 10.1177/095646240401501011. [DOI] [PubMed] [Google Scholar]
- 88.Yebra G, de Mulder M, Martin L, Rodriguez C, Labarga P, Viciana I, et al. Most HIV type 1 non-B infections in the Spanish cohort of antiretroviral treatment-naive HIV-infected patients (CoRIS) are due to recombinant viruses. J Clin Microbiol. 2012;50:407–413. doi: 10.1128/JCM.05798-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leoz M, Chaix ML, Delaugerre C, Rivoisy C, Meyer L, Rou-zioux C, et al. Circulation of multiple patterns of unique recombinant forms B/CRF02_AG in France: precursor signs of the emergence of an upcoming CRF B/02. AIDS. 2011;25:1371–1377. doi: 10.1097/QAD.0b013e328347c060. [DOI] [PubMed] [Google Scholar]
- 90.Hughes GJ, Fearnhill E, Dunn D, Lycett SJ, Rambaut A, Leigh Brown AJ. Molecular phylodynamics of the heterosexual HIV epidemic in the United Kingdom. PLoS Pathog. 2009;5:e1000590. doi: 10.1371/journal.ppat.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xiridou M, van Veen M, Prins M, Coutinho R. How patterns of migration can influence the heterosexual transmission of HIV in The Netherlands. Sex Transm Infect. 2011;87:289–291. doi: 10.1136/sti.2010.048512. [DOI] [PubMed] [Google Scholar]
- 92.Xiridou M, van Veen M, Coutinho R, Prins M. Can migrants from high-endemic countries cause new HIV outbreaks among heterosexuals in low-endemic countries? AIDS. 2010;24:2081–2088. doi: 10.1097/QAD.0b013e32833a6071. [DOI] [PubMed] [Google Scholar]
- 93.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5:e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bezemer D, van Sighem A, Lukashov VV, van der Hoek L, Back N, Schuurman R, et al. Transmission networks of HIV-1 among men having sex with men in the Netherlands. AIDS. 2010;24:271–282. doi: 10.1097/QAD.0b013e328333ddee. [DOI] [PubMed] [Google Scholar]
- 95.Yerly S, Junier T, Gayet-Ageron A, Amari EB, von Wyl V, Gunthard HF, et al. The impact of transmission clusters on primary drug resistance in newly diagnosed HIV-1 infection. AIDS. 2009;23:1415–1423. doi: 10.1097/QAD.0b013e32832d40ad. [DOI] [PubMed] [Google Scholar]
- 96.Levy I, MorZ, Anis E, Maayan S, Leshem E, Pollack S, et al. Men who have sex with men, risk behavior, and HIV infection: integrative analysis of clinical, epidemiological, and laboratory databases. Clin Infect Dis. 2011;52:1363–1370. doi: 10.1093/cid/cir244. [DOI] [PubMed] [Google Scholar]
- 97.Pao D, Fisher M, Hue S, Dean G, Murphy G, Cane PA, et al. Transmission of HIV-1 during primary infection: relationship to sexual risk and sexually transmitted infections. AIDS. 2005;19:85–90. doi: 10.1097/00002030-200501030-00010. [DOI] [PubMed] [Google Scholar]
- 98.Hue S, Clewley JP, Cane PA, Pillay D. Investigation of HIV-1 transmission events by phylogenetic methods: requirement for scientific rigour. AIDS. 2005;19:449–450. doi: 10.1097/01.aids.0000161778.15568.a1. [DOI] [PubMed] [Google Scholar]
- 99.German D, Sifakis F, Maulsby C, Towe VL, Flynn CP, Latkin CA, et al. Persistently high prevalence and unrecognized HIV infection among men who have sex with men in Baltimore: the BESURE study. J Acquir Immune Defic Syndr. 2011;57:77–87. doi: 10.1097/QAI.0b013e318211b41e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Paraskevis D, Pybus O, Magiorkinis G, Hatzakis A, Wensing AM, van de Vijver DA, et al. Tracing the HIV-1 subtype B mobility in Europe: a phylogeographic approach. Retrovirology. 2009;6:49. doi: 10.1186/1742-4690-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Audelin AM, Cowan SA, Obel N, Nielsen C, Jorgensen LB, Gerstoft J. Phylogenetics of the Danish HIV epidemic: the role of Very Late Presenters in sustaining the epidemic. J Acquir Immune Defic Syndr. 2013;62:102–108. doi: 10.1097/QAI.0b013e318276becc. [DOI] [PubMed] [Google Scholar]
- 102.Bezemer D, de Wolf F, Boerlijst MC, van Sighem A, Hollingsworth TD, Prins M, et al. A resurgent HIV-1 epidemic among men who have sex with men in the era of potent antiretroviral therapy. AIDS. 2008;22:1071–1077. doi: 10.1097/QAD.0b013e3282fd167c. [DOI] [PubMed] [Google Scholar]
- 103.Kouyos RD, von Wyl V, Yerly S, Boni J, Taffe P, Shah C, et al. Molecular epidemiology reveals long-term changes in HIV type 1 subtype B transmission in Switzerland. J Infect Dis. 2010;201:1488–1497. doi: 10.1086/651951. [DOI] [PubMed] [Google Scholar]
- 104.Yerly S, von Wyl V, Ledergerber B, Boni J, Schupbach J, Burgisser P, et al. Transmission of HIV-1 drug resistance in Switzerland: a 10-year molecular epidemiology survey. AIDS. 2007;21:2223–2229. doi: 10.1097/QAD.0b013e3282f0b685. [DOI] [PubMed] [Google Scholar]
- 105.Frange P, Meyer L, Deveau C, Tran L, Goujard C, Ghosn J, et al. Recent HIV-1 infection contributes to the viral diffusion over the French territory with a recent increasing frequency. PLoS One. 2012;7:e31695. doi: 10.1371/journal.pone.0031695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dennis AM, Hue S, Hurt CB, Napravnik S, Sebastian J, Pillay D, et al. Phylogenetic insights into regional HIV transmission. AIDS. 2012;26:1813–1822. doi: 10.1097/QAD.0b013e3283573244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aldous JL, Pond SK, Poon A, Jain S, Qin H, Kahn JS, et al. Characterizing HIV transmission networks across the United States. Clin Infect Dis. 2012;55:1135–1143. doi: 10.1093/cid/cis612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ambrosioni J, Junier T, Delhumeau C, Calmy A, Hirschel B, Zdobnov E, et al. Impact of HAART on the molecular epidemiology of newly diagnosed HIV infections in Geneva, Switzerland. AIDS. 2012;26:2079–2086. doi: 10.1097/QAD.0b013e32835805b6. [DOI] [PubMed] [Google Scholar]
- 109.Dennis AM, Hue S, Hurt CB, Napravnik S, Sebastian J, Pillay D, et al. Phylogenetic insights into HIV transmission in North Carolina. AIDS. 2012;26:1813–1822. doi: 10.1097/QAD.0b013e3283573244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ragonnet-Cronin M, Ofner-Agostini M, Merks H, Pilon R, Rekart M, Archibald CP, et al. Longitudinal phylogenetic surveillance identifies distinct patterns of cluster dynamics. J Acquir Immune Defic Syndr. 2010;55:102–108. doi: 10.1097/QAI.0b013e3181e8c7b0. [DOI] [PubMed] [Google Scholar]
- 111.Chalmet K, Staelens D, Blot S, Dinakis S, Pelgrom J, Plum J, et al. Epidemiological study of phylogenetic transmission clusters in a local HIV-1 epidemic reveals distinct differences between subtype B and non-B infections. BMC Infect Dis. 2010;10:262. doi: 10.1186/1471-2334-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Volz EM, Koopman JS, Ward MJ, Brown AL, Frost SD. Simple epidemiological dynamics explain phylogenetic clustering of HIV from patients with recent infection. PLoS Comput Biol. 2012;8:e1002552. doi: 10.1371/journal.pcbi.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pilcher CD, Joaki G, Hoffman IF, Martinson FE, Mapanje C, Stewart PW, et al. Amplified transmission of HIV-1: comparison of HIV-1 concentrations in semen and blood during acute and chronic infection. AIDS. 2007;21:1723–1730. doi: 10.1097/QAD.0b013e3281532c82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Redd AD, Collinson-Streng AN, Chatziandreou N, Mullis CE, Laeyendecker O, Martens C, et al. Previously transmitted HIV-1 strains are preferentially selected during subsequent sexual transmissions. J Infect Dis. 2012;206:1433–1442. doi: 10.1093/infdis/jis503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sifakis F, Flynn CP, Metsch L, LaLota M, Murrill C, Koblin BA, et al. HIV prevalence, unrecognized infection, and HIV testing among men who have sex with men: five U.S. cities, June 2004-April 2005. MMWR Morb Mortal Wkly Rep. 2005;54:597–601. [PubMed] [Google Scholar]
- 116.Hernandez HL, Prejean J, Doshani M, Linley L, Ziebell R, Branson BM, et al. Previous HIV testing among adults and adolescents newly diagnosed with HIV infection: National HIV Surveillance System, 18 Jurisdictions, United States, 2006–2009. MMWR Morb Mortal Wkly Rep. 2012;61:441–445. [PubMed] [Google Scholar]
- 117.Marks G, Crepaz N, Janssen RS. Estimating sexual transmission of HIV from persons aware and unaware that they are infected with the virus in the USA. AIDS. 2006;20:1447–1450. doi: 10.1097/01.aids.0000233579.79714.8d. [DOI] [PubMed] [Google Scholar]
- 118.Lauby JL, Millett GA, LaPollo AB, Bond L, Murrill CS, Marks G. Sexual risk behaviors of HIV-positive, HIV-negative, and serostatus-unknown Black men who have sex with men and women. Arch Sex Behav. 2008;37:708–719. doi: 10.1007/s10508-008-9365-6. [DOI] [PubMed] [Google Scholar]
- 119.Dawson JM, Fitzpatrick RM, Reeves G, Boulton M, McLean J, Hart GJ, et al. Awareness of sexual partners' HIV status as an influence upon high-risk sexual behaviour among gay men. AIDS. 1994;8:837–841. [PubMed] [Google Scholar]
- 120.Lavoie E, Alary M, Remis RS, Otis J, Vincelette J, Turmel B, et al. Determinants of HIV seroconversion among men who have sex with men living in a low HIV incidence population in the era of highly active antiretroviral therapies. Sex Transm Dis. 2008;35:25–29. doi: 10.1097/OLQ.0b013e31814fb113. [DOI] [PubMed] [Google Scholar]
- 121.Cohen MS, Fidler S. HIV prevention 2010: where are we now and where are we going? Curr Opin HIV AIDS. 2010;5:265–268. doi: 10.1097/COH.0b013e32833acafa. [DOI] [PubMed] [Google Scholar]
- 122.Powers KA, Ghani AC, Miller WC, Hoffman IF, Pettifor AE, Kamanga G, et al. The role of acute and early HIV infection in the spread of HIV and implications for transmission prevention strategies in Lilongwe, Malawi: a modelling study. Lancet. 2011;378:256–268. doi: 10.1016/S0140-6736(11)60842-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fisher M, Pao D, Brown AE, Sudarshi D, Gill ON, Cane P, et al. Determinants of HIV-1 transmission in men who have sex with men: a combined clinical, epidemiological and phylo-genetic approach. AIDS. 2010;24:1739–1747. doi: 10.1097/QAD.0b013e32833ac9e6. [DOI] [PubMed] [Google Scholar]
- 124.Jacquez JA, Koopman JS, Simon CP, Longini IM., Jr Role of the primary infection in epidemics of HIV infection in gay cohorts. J Acquir Immune Defic Syndr. 1994;7:1169–1184. [PubMed] [Google Scholar]
- 125.Kim JH, Riolo RL, Koopman JS. HIV transmission by stage of infection and pattern of sexual partnerships. Epidemiology. 2010;21:676–684. doi: 10.1097/EDE.0b013e3181e6639f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Miller WC, Rosenberg NE, Rutstein SE, Powers KA. Role of acute and early HIV infection in the sexual transmission of HIV. Curr Opin HIV AIDS. 2010;5:277–282. doi: 10.1097/COH.0b013e32833a0d3a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pilcher CD, Christopoulos KA, Golden M. Public health rationale for rapid nucleic acid or p24 antigen tests for HIV. J Infect Dis. 2010;201(Suppl 1):S7–S15. doi: 10.1086/650393. [DOI] [PubMed] [Google Scholar]
- 128.Poon AF, McGovern RA, Mo T, Knapp DJ, BrennerB, Routy JP, et al. Dates of HIV infection can be estimated for seropreva-lent patients by coalescent analysis of serial next-generation sequencing data. AIDS. 2011;25:2019–2026. doi: 10.1097/QAD.0b013e32834b643c. [DOI] [PubMed] [Google Scholar]
- 129.Kouyos RD, von Wyl V, Yerly S, Boni J, Rieder P, Joos B, et al. Ambiguous nucleotide calls from population-based sequencing of HIV-1 are a marker for viral diversity and the age of infection. Clin Infect Dis. 2011;52:532–539. doi: 10.1093/cid/ciq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rosenberg NE, Kamanga G, Phiri S, Nsona D, Pettifor A, Rutstein SE, et al. Detection of acute HIV infection: a field evaluation of the determine(R) HIV-1/2 Ag/Ab combo test. J Infect Dis. 2012;205:528–534. doi: 10.1093/infdis/jir789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Branson BM, Stekler JD. Detection of acute HIV infection: we can't close the window. J Infect Dis. 2012;205:521–524. doi: 10.1093/infdis/jir793. [DOI] [PubMed] [Google Scholar]
- 132.Giorgi EE, Funkhouser B, Athreya G, Perelson AS, Korber BT, Bhattacharya T. Estimating time since infection in early homogeneous HIV-1 samples using a poisson model. BMC Bioinformatics. 2010;11:532. doi: 10.1186/1471-2105-11-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Maldarelli F, Shao W, Dewar R, Rehman T, Kearney M, Rehm C, et al. New bioinformatic algorithm to identify recent HIV-1 infection. Antiviral Ther. 2010;15:A97. [Google Scholar]
- 134.Yang J, Xia X, He X, Yang S, Ruan Y, Zhao Q, et al. A new pattern-based method for identifying recent HIV-1 infections from the viral env sequence. Sci China Life Sci. 2012;55:328–335. doi: 10.1007/s11427-012-4312-0. [DOI] [PubMed] [Google Scholar]
- 135.Cousins MM, Ou SS, Wawer MJ, Munshaw S, Swan D, Magaret CA, et al. Comparison of a high-resolution melting assay to next-generation sequencing for analysis of HIV diversity. J Clin Microbiol. 2012;50:3054–3059. doi: 10.1128/JCM.01460-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rieder P, Joos B, von Wyl V, Kuster H, Grube C, Leemann C, et al. HIV-1 transmission after cessation of early antiretroviral therapy among men having sex with men. AIDS. 2010;24:1177–1183. doi: 10.1097/QAD.0b013e328338e4de. [DOI] [PubMed] [Google Scholar]
- 137.Volz E, Frost SD, Rothenberg R, Meyers LA. Epidemiological bridging by injection drug use drives an early HIV epidemic. Epidemics. 2010;2:155–164. doi: 10.1016/j.epidem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 138.Tagliamonte M, Naderi HR, Tornesello ML, Farid R, Buonaguro FM, Buonaguro L. HIV type 1 subtype A epidemic in injecting drug user (IDU) communities in Iran. AIDS Res Hum Retroviruses. 2007;23:1569–1574. doi: 10.1089/aid.2007.0169. [DOI] [PubMed] [Google Scholar]
- 139.Balode D, Skar H, Mild M, Kolupajeva T, Ferdats A, Rozentale B, et al. Phylogenetic analysis of the Latvian HIV-1 epidemic. AIDS Res Hum Retroviruses. 2012;28:928–932. doi: 10.1089/aid.2011.0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Perez-Losada M, Jobes DV, Sinangil F, Crandall KA, Arenas M, Posada D, et al. Phylodynamics of HIV-1 from a phase III AIDS vaccine trial in Bangkok, Thailand. PLoS One. 2011;6:e16902. doi: 10.1371/journal.pone.0016902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen JH, Wong KH, Chan KC, To SW, Chen Z, Yam WC. Phylodynamics of HIV-1 subtype B among the men-having-sex-with-men (MSM) population in Hong Kong. PLoS One. 2011;6:e25286. doi: 10.1371/journal.pone.0025286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kao CF, Chang SY, Hsia KT, Chang FY, Yang CH, Liu HR, et al. Surveillance of HIV type 1 recent infection and molecular epidemiology among different risk behaviors between 2007 and 2009 after the HIV type1 CRF07_BC outbreak in Taiwan. AIDS Res Hum Retroviruses. 2011;27:745–749. doi: 10.1089/aid.2010.0244. [DOI] [PubMed] [Google Scholar]
- 143.Lemstra M, Rogers M, Thompson A, Moraros J, Buckingham R. Risk indicators associated with injection drug use in the Aboriginal population. AIDS Care. 2012 doi: 10.1080/09540121.2011.650678. [DOI] [PubMed] [Google Scholar]
- 144.Martin LJ, Houston S, Yasui Y, Wild TC, Saunders LD. All-cause and HIV-related mortality rates among HIV-infected patients after initiating highly active antiretroviral therapy: the impact of Aboriginal ethnicity and injection drug use. Can J Public Health. 2011;102:90–96. doi: 10.1007/BF03404154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen JH, Wong KH, Li P, Chan KC, Lee MP, Lam HY, et al. Molecular epidemiological study of HIV-1 CRF01_AE transmission in Hong Kong. J Acquir Immune Defic Syndr. 2009;51:530–535. doi: 10.1097/QAI.0b013e3181aac516. [DOI] [PubMed] [Google Scholar]
- 146.Pilon R, Leonard L, Kim J, Vallee D, De Rubeis E, Jolly AM, et al. Transmission patterns of HIV and hepatitis C virus among networks of people who inject drugs. PLoS One. 2011;6:e22245. doi: 10.1371/journal.pone.0022245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ciccozzi M, Lai A, Ebranati E, Gabanelli E, Galli M, Mugosa B, et al. Phylogeographic reconstruction of HIV type 1B in Montenegro and the Balkan Region. AIDS Res Hum Retroviruses. 2012 doi: 10.1089/aid.2011.0138. [DOI] [PubMed] [Google Scholar]
- 148.Bernini F, Ebranati E, De Maddalena C, Shkjezi R, Milazzo L, Lo Presti A, et al. Within-host dynamics of the hepatitis C virus quasispecies population in HIV-1/HCV coinfected patients. PLoS One. 2011;6:e16551. doi: 10.1371/journal.pone.0016551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Masharsky AE, Dukhovlinova EN, Verevochkin SV, Toussova OV, Skochilov RV, Anderson JA, et al. A substantial transmission bottleneck among newly and recently HIV-1-infected injection drug users in St Petersburg, Russia. J Infect Dis. 2010;201:1697–1702. doi: 10.1086/652702. [DOI] [PubMed] [Google Scholar]
- 150.Yerly S, Rickenbach M, Popescu M, Taffe P, Craig C, Perrin L. Drug resistance mutations in HIV-1-infected subjects during protease inhibitor-containing highly active antiretroviral therapy with nelfinavir or indinavir. Antivir Ther. 2001;6:185–189. [PubMed] [Google Scholar]
- 151.Hilton BA, Thompson R, Moore-Dempsey L, Janzen RG. Harm reduction theories and strategies for control of human immunodeficiency virus: a review of the literature. J Adv Nurs. 2001;33:357–370. doi: 10.1046/j.1365-2648.2001.01672.x. [DOI] [PubMed] [Google Scholar]
- 152.Duri K, Gumbo F, Kristiansen K, Mapingure M, Munjoma M, Chirenje M, et al. Phylogenetic analysis of human immunodeficiency virus type1subtypeCenv gp120 sequences among four drug-naive families following subsequent heterosexual and vertical transmissions. AIDS Res Hum Retroviruses. 2012;28:885–893. doi: 10.1089/AID.2011.0217. [DOI] [PubMed] [Google Scholar]
- 153.Novitsky V, Wang R, Margolin L, Baca J, Rossenkhan R, Moyo S, et al. Transmission of single and multiple viral variants in primary HIV-1 subtype C infection. PLoS One. 2011;6:e16714. doi: 10.1371/journal.pone.0016714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Novitsky V, Wang R, Bussmann H, Lockman S, Baum M, Shapiro R, et al. HIV-1 subtype C-infected individuals maintaining high viral load as potential targets for the ‘test-and-treat’ approach to reduce HIV transmission. PLoS One. 2010;5:e10148. doi: 10.1371/journal.pone.0010148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Koopman JS, Jacquez JA, Welch GW, Simon CP, Foxman B, Pollock SM, et al. The role of early HIV infection in the spread of HIV through populations. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;14:249–258. doi: 10.1097/00042560-199703010-00009. [DOI] [PubMed] [Google Scholar]
- 156.Hollingsworth TD, Anderson RM, Fraser C. HIV-1 transmission, by stage of infection. J Infect Dis. 2008;198:687–693. doi: 10.1086/590501. [DOI] [PubMed] [Google Scholar]
- 157.Trask SA, Derdeyn CA, Fideli U, Chen Y, Meleth S, Kasolo F, et al. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J Virol. 2002;76:397–405. doi: 10.1128/JVI.76.1.397-405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gray RH, Wawer MJ. Probability of heterosexual HIV-1 transmission per coital act in sub-Saharan Africa. J Infect Dis. 2012;205:351–352. doi: 10.1093/infdis/jir751. [DOI] [PubMed] [Google Scholar]
- 159.Eshleman SH, Hudelson SE, Redd AD, Wang L, Debes R, Chen YQ, et al. Analysis of genetic linkage of HIV from couples enrolled in the HIV Prevention Trials Network 052 trial. J Infect Dis. 2011;204:1918–1926. doi: 10.1093/infdis/jir651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Campbell MS, Mullins JI, Hughes JP, Celum C, Wong KG, Raugi DN, et al. Viral linkage in HIV-1 seroconverters and their partners in an HIV-1 prevention clinical trial. PLoS One. 2011;6:e16986. doi: 10.1371/journal.pone.0016986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jain V, Liegler T, Vittinghoff E, Hartogensis W, Bacchetti P, Poole L, et al. Transmitted drug resistance in persons with acute/early HIV-1 in San Francisco, 2002–2009. PLoS One. 2010;5:e15510. doi: 10.1371/journal.pone.0015510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Brenner BG, Roger M, Moisi DD, Oliveira M, Hardy I, Turgel R, et al. Transmission networks of drug resistance acquired in primary/early stage HIV infection. AIDS. 2008;22:2509–2515. doi: 10.1097/QAD.0b013e3283121c90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hue S, Gifford RJ, Dunn D, Fernhill E, Pillay D. Demonstration of sustained drug-resistant human immunodeficiency virus type 1 lineages circulating among treatment-naive individuals. J Virol. 2009;83:2645–2654. doi: 10.1128/JVI.01556-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vercauteren J, Wensing AM, van de Vijver DA, Albert J, Balotta C, Hamouda O, et al. Transmission of drug-resistant HIV-1 is stabilizing in Europe. J Infect Dis. 2009;200:1503–1508. doi: 10.1086/644505. [DOI] [PubMed] [Google Scholar]
- 165.Wittkop L, Gunthard HF, de Wolf F, Dunn D, Cozzi-Lepri A, de Luca A, et al. Effect of transmitted drug resistance on virolo-gical and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. Lancet Infect Dis. 2011;11:363–371. doi: 10.1016/S1473-3099(11)70032-9. [DOI] [PubMed] [Google Scholar]
- 166.Brenner B, Turner D, Oliveira M, Moisi D, Detorio M, Carobene M, et al. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to nonnucleoside reverse transcriptase inhibitors. Aids. 2003;17:F1–F5. doi: 10.1097/00002030-200301030-00001. [DOI] [PubMed] [Google Scholar]
- 167.Grossman Z, Istomin V, Averbuch D, Lorber M, Risenberg K, Levi I, et al. Genetic variation at NNRTI resistance-associated positions in patients infected with HIV-1 subtype C. AIDS. 2004;18:909–915. doi: 10.1097/00002030-200404090-00008. [DOI] [PubMed] [Google Scholar]
- 168.Charpentier C, Gody JC, Tisserand P, Matta M, Pere H, Fournier J, et al. Surveillance of antiretroviral drug resistance mutations in untreated young children living in the Central African Republic. Antivir Ther. 2011;16:1347–1350. doi: 10.3851/IMP1896. [DOI] [PubMed] [Google Scholar]
- 169.Tshabalala M, Manasa J, Zijenah LS, Rusakaniko S, Kadzirange G, Mucheche M, et al. Surveillance of transmitted antiretro-viral drug resistance among HIV-1 infected women attending antenatal clinics in Chitungwiza, Zimbabwe. PLoS One. 2011;6:e21241. doi: 10.1371/journal.pone.0021241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Brenner BG, Coutsinos D. The K65R mutation in HIV-1 reverse transcriptase: genetic barriers, resistance profile and clinical implications. HIV Ther. 2009;3:583–594. doi: 10.2217/hiv.09.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Brenner BG, Oliveira M, Doualla-Bell F, Moisi DD, Ntemgwa M, Frankel F, et al. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS. 2006;20:F9–F13. doi: 10.1097/01.aids.0000232228.88511.0b. [DOI] [PubMed] [Google Scholar]
- 172.Castor D, Low A, Evering T, Karmon S, Davis B, Figueroa A, et al. Transmitted drug resistance and phylogenetic relationships among acute and early HIV-1 infected individuals in New York City. J Acquir Immune Defic Syndr. 2012 doi: 10.1097/QAI.0b013e31825a289b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Castro E, Khonkarly M, Ciuffreda D, Burgisser P, Cavassini M, Yerly S, et al. HIV-1 drug resistance transmission networks in southwest Switzerland. AIDS Res Hum Retroviruses. 2010;26:1233–1238. doi: 10.1089/aid.2010.0083. [DOI] [PubMed] [Google Scholar]
- 174.Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One. 2009;4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.van de Vijver DA, Wensing AM, Angarano G, Asjo B, Balotta C, Boeri E, et al. The calculated genetic barrier for antire-troviral drug resistance substitutions is largely similar for different HIV-1 subtypes. J Acquir Immune Defic Syndr. 2006;41:352–360. doi: 10.1097/01.qai.0000209899.05126.e4. [DOI] [PubMed] [Google Scholar]
- 176.Grgic I, Lepej SZ, Lunar MM, Poljak M, Vince A, Vrakela IB, et al. The prevalence of transmitted drug resistance in newly diagnosed HIV-infected individuals in Croatia: the role of transmission clusters of men who have sex with men carrying the T215S surveillance drug resistance mutation. AIDS Res Hum Retroviruses. 2012 doi: 10.1089/aid.2012.0191. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Buskin SE, Ellis GM, Pepper GG, Frenkel LM, Pergam SA, Gottlieb GS, et al. Transmission cluster of multiclass highly drug-resistant HIV-1 among 9 men who have sex with men in Seattle/King County, WA, 2005-2007. J Acquir Immune Defic Syndr. 2008;49:205–211. doi: 10.1097/QAI.0b013e318185727e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Stolte IG, de Wit JB, Kolader ME, Fennema HS, Coutinho RA, Dukers NH. Low HIV-testing rates among younger high-risk homosexual men in Amsterdam. Sex Transm Infect. 2007;83:387–391. doi: 10.1136/sti.2005.019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Brenner B, Moodie E, Otis J, Ohnona F, Wainberg MA, Rousseau R, et al. Phylogenetic analysis of the SPOT cohort reveals risk determinants implicated in the spread of the Montreal Male-Sex-Male (MSM) epidemic. Can J Infect Dis Med Microbiol. 2012;23:79. [Google Scholar]
- 180.Le Vu S, Velter A, Meyer L, Peytavin G, Guinard J, Pillonel J, et al. Biomarker-based HIV incidence in a community sample of men who have sex with men in Paris, France. PLoS One. 2012;7:e39872. doi: 10.1371/journal.pone.0039872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Alam SJ, Romero-Severson E, Kim JH, Emond G, Koopman JS. Dynamic sex roles among men who have sex with men and transmissions from primary HIV infection. Epidemiology. 2010;21:669–675. doi: 10.1097/EDE.0b013e3181e9e901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bezemer D, de Wolf F, Boerlijst MC, van Sighem A, Hollings-worth TD, Fraser C. 27 years of the HIV epidemic amongst men having sex with men in the Netherlands: an in depth mathematical model-based analysis. Epidemics. 2010;2:66–79. doi: 10.1016/j.epidem.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 183.Chen M, Rhodes PH, Hall IH, Kilmarx PH, Branson BM, Valleroy LA. Prevalence of undiagnosed HIV infection among persons aged >/=13 years: National HIV Surveillance System, United States, 2005–2008. MMWR Surveill Summ. 2012;61:57–64. [PubMed] [Google Scholar]
- 184.Oster AM, Wiegand RE, Sionean C, Miles IJ, Thomas PE, Melendez-Morales L, et al. Understanding disparities in HIV infection between black and white MSM in the United States. AIDS. 2011;25:1103–1112. doi: 10.1097/QAD.0b013e3283471efa. [DOI] [PubMed] [Google Scholar]
- 185.Ayala G, Bingham T, Kim J, Wheeler DP, Millett GA. Modeling the impact of social discrimination and financial hardship on the sexual risk of HIV among Latino and Black men who have sex with men. Am J Public Health. 2012;102(Suppl 2):S242–S249. doi: 10.2105/AJPH.2011.300641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Aldous JL, Pond SK, Poon A, Jain S, Qin H, Kahn JS, et al. Characterizing HIV transmission networks across the United States. Clin Infect Dis. 2012 doi: 10.1093/cid/cis612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Oster AM, Pieniazek D, Zhang X, Switzer WM, Ziebell RA, Mena LA, et al. Demographic but not geographic insularity in HIV transmission among young black MSM. AIDS. 2011;25:2157–2165. doi: 10.1097/QAD.0b013e32834bfde9. [DOI] [PubMed] [Google Scholar]