Abstract
Ribosomes are RNA-protein complexes responsible for protein synthesis. A dominant structural motif in the rRNAs is an RNA helix capped with a four-nucleotide loop, called a tetraloop. The sequence of the tetraloop is invariant at some positions in the rRNAs but is highly variable at other positions. The biological reason for the conservation of the tetraloop sequence at specific positions in the rRNAs is not clear. In the 16S rRNA the GAAA tetraloop in helix 8 and the UACG tetraloop in helix 14 are highly conserved and located near the binding site for EF-Tu and EF-G. To investigate whether the structural stability of the tetraloop or the precise sequence of the tetraloop is important for function, we separately changed the GAAA tetraloop in helix 8 to a UACG tetraloop, and the UACG tetraloop in helix 14 to a GAAA tetraloop. The effects of the tetraloop replacements on protein synthesis were analyzed in vivo and in vitro. Replacement of the tetraloops in helices 8 and 14 did not significantly affect the growth rate of E. coli (Δ7rrn) strain. However, the mutant ribosomes showed a slightly reduced rate of protein synthesis in vitro. In addition, we observed a two-fold increase in the error rate of translation with the mutant ribosomes, which is consistent with an earlier report. Our results suggest that the tetraloops in helices 8 and 14 are highly conserved mainly for their structural stability and the precise sequence of these tetraloops are not critical for protein synthesis.
Ribosomes are megadalton-sized complexes made up of both RNA and protein. In bacteria the 30S ribosomal subunit is composed of a 16S rRNA and about 21 proteins, and the 50S ribosomal subunit is composed of 23S rRNA, 5S rRNA and about 30 proteins. The three-dimensional structure of the 30S and 50S subunits, to a large extent, is established by the folded structure of the rRNAs (1). A common structural motif in the rRNAs is the tetraloop (2). A tetraloop is a small loop composed of four residues connecting the two anti-parallel chains of an RNA helix (Figure 1). Tetraloops have exceptionally high thermodynamic stability and function as nucleation sites for folding large RNA molecules (3, 4). Based on their consensus sequence there are four major types of tetraloops: GNRA, UNCG, CUYG, and GANC (where N is any of the four nucleotides, R is a purine and Y is a pyrimidine) (2, 3, 5, 6).
Figure 1.
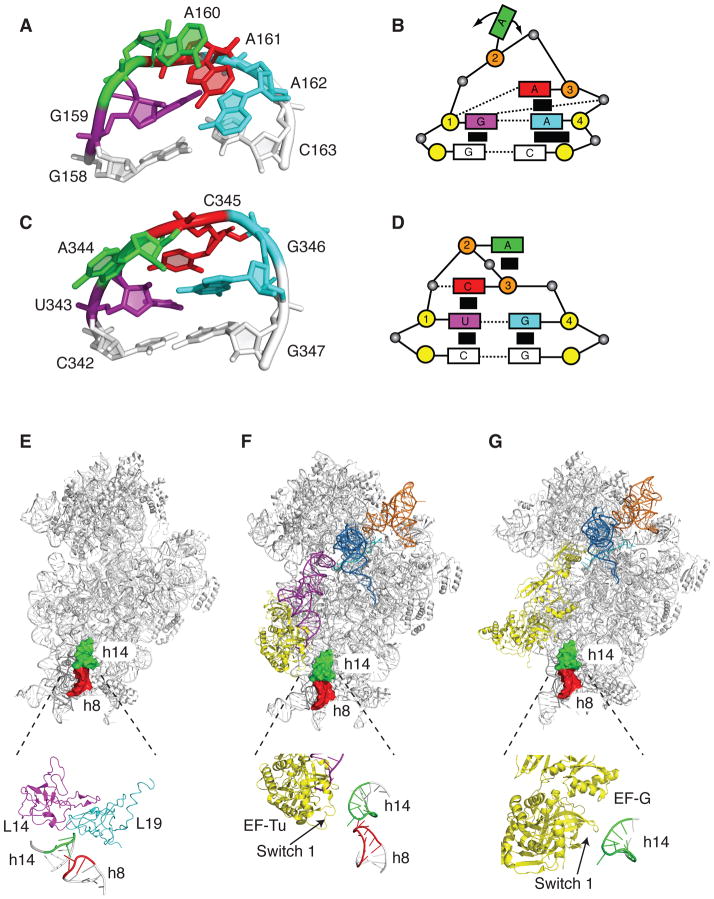
Structural features of the two studied tetraloops and their location in the ribosome. (A) Tertiary structure of the GAAA tetraloop in helix 8 (Protein Data Bank entry 2AVY) (15). The bases that form the tetraloop are G159 (purple), A160 (green), A161 (red), and A162 (cyan). The G-C base pair of the RNA stem is shown in grey. (B) Schematic representation of the GAAA tetraloop. Shown are the bases (rectangles, which are color coded as above), phosphates (black spheres) and sugars (yellow circles for 3′-endo sugar conformation and orange circles for 2′-endo sugar conformation). The hydrogen bonds are indicated by the dotted lines and the stacking interactions are indicated by the black rectangles. Base G1 is in 5′ stack, and A3 and A4 are in 3′ stack. (C) Tertiary structure of the UACG tetraloop in helix 14 (Protein Data Bank entry 2AVY) (15). The bases that form the tetraloop are U343 (purple), A344 (green), C345 (red), and G346 (cyan). The C-G base pair of the RNA stem is shown in grey. (D) Schematic representation of the UACG tetraloop. The coloring scheme and labels are as described above for the GAAA tetraloop. Bases U1 and C3 are in 5′ stack and base G4 is in 3′ stack. (E) Structure of the 30S subunit showing the location of helices 8 and 14 of the 16S rRNA (Protein Data Bank entry 2AVY) (15). Shown are the 30S subunit (grey), the GAAA tetraloop in h8 (red), and the UACG tetraloop in h14 (green). The interaction of helix 14 with the 50S subunit ribosomal proteins L14 (magenta) and L19 (cyan) to form the bridge B8 is shown below. (F) Interaction of EF-Tu with helices 8 and 14 of the 16S rRNA (Protein Data Bank entry 3FIH) (20). Shown are the 30S subunit (grey), the GAAA tetraloop in h8 (red), the UACG tetraloop in h14 (green), EF-Tu (yellow), A-tRNA (purple), P-tRNA (sky blue), E-tRNA (orange). A close-up of the interaction between the switch 1 loop in EF-Tu and the junction of helices 8 and 14 is shown below. (G) Interaction of EF-G with helices 8 and 14 of the 16S rRNA in the post-translocation state (Protein Data Bank entry 2WRI) (46). Shown are the 30S subunit (grey), the GAAA tetraloop in h8 (red), the UACG tetraloop in h14 (green), EF-G (yellow), P-tRNA (sky blue), E-tRNA (orange). A close-up of the interaction between the switch 1 loop in EF-G and helix 14 is shown below (Protein Data Bank entry 2OM7) (22).
Interestingly, nearly 90% of all the tetraloops in the 16S rRNA are GNRA or UNCG (2). The overall fold adopted by the GNRA and UNCG tetraloops is similar (7, 8) (Figure 1A to 1D). In both GNRA and UNCG, the first and fourth bases form hydrogen bonds, the non-conserved second base is flipped out of the loop, and the phosphate backbone between the second and the third nucleotides is extended for closing the loop. Nevertheless, the GNRA and UNCG tetraloops differ significantly in their local structure and dynamics (3). In the GNRA tetraloop the base G is in a 5′ stack and the last two bases are in a 3′ stack. In contrast, in the UNCG tetraloop the first base U and the third base C are in a 5′ stack and the fourth base G is in a 3′ stack. The second and third nucleotides in the GNRA loop have highly flexible sugar conformation alternating equally between the 2′-endo and 3′-endo conformers, whereas in the UNCG loop the second and third nucleotides remain essentially in the 2′-endo sugar conformation. These differences make the UNCG loop thermodynamically more stable but less dynamic than the GNRA loop. By being more dynamic, the GNRA loop often can form long-range tertiary interactions, unlike the UNCG loop (3, 9–11). Thus, the greater flexibility of the GNRA loop permits it to play a more specialized role in the folding and function of large RNAs.
Despite these differences in the structure and dynamics of the GNRA and UNCG tetraloops, a previous study showed that a GNRA loop found in the signal recognition particle RNA could be functionally interchanged with an UNCG loop suggesting that the tetraloops sometimes act as a unit (12). In contrast, studies with group I self-splicing introns showed that a GNRA loop cannot be functionally interchanged with an UNCG loop (9, 13). Here, the GNRA loop is important for function because it forms a tertiary interaction that is required for stabilizing the folded state of the self-splicing intron. Therefore, in some cases, the precise sequence of the tetraloop is more important for function than a thermodynamically stable structure. In the ribosome, there are several highly conserved tetraloops at particular positions in the rRNAs (2, 14). The reason for the conservation of a GNRA verses an UNCG tetraloop at a particular position in the rRNAs is not clear. They may be conserved purely for their structural stability, the ability to form tertiary interactions, or for some functional role during protein synthesis such as binding translation factors.
To determine whether the thermodynamic stability and the ability to form sequence specific tertiary interactions account for the conservation of a tetraloop sequence at a particular position in the rRNAs, we decided to analyze two highly conserved tetraloops in 16S rRNA. We selected the GAAA tetraloop in helix 8 and the UACG tetraloop in helix 14 of 16S rRNA, which are located in the shoulder domain of the 30S subunit (Figure 1E). Interestingly, the GAAA tetraloop in helix 8 has the precise geometry for making an A-minor tertiary interaction with helix 14 except it appears to be out of contact range in the crystal structures (15). Noller has made the intriguing proposal that some of the GNRA loops that are out of contact range of their receptor helix may form tertiary interactions during defined steps of translation to stabilize distinct conformational states of the ribosome (16). The UACG tetraloop in helix 14 contacts the large ribosomal subunit proteins L14 and L19 to form the intersubunit bridge B8 (Figure 1E) (15, 17–19). Cryo-EM structures showed that the switch I domain of elongation factor Tu (EF-Tu) contacts the junction of helices 8 and 14 (Figure 1F) (20, 21). In addition, switch I of elongation factor G (EF-G) interacts with helix 14 (22, 23) (Figure 1G).
Consistent with the structural data, mutations in helices 8 and 14 have been shown to increase the error rate of translation indicating a functional role for these tetraloops during tRNA selection (24). However, mutations can change the structure and thermodynamic stability of the tetraloop leading to more pronounced functional defects. We instead, chose to replace as a unit one type of tetraloop with another type of tetraloop so as to maintain the overall structure and stability of the hairpin but with a different sequence. We analyzed the effect of these tetraloop replacements using biochemical assays. Our results show that the helix 8 and helix 14 tetraloop replacements do not significantly affect the function of the ribosome suggesting that the precise sequence of these two tetraloops are not essential for translation.
Experimental Procedures
Site-Directed Mutagenesis of 16S rRNA
Site-directed mutagenesis was performed with a QuickChange PCR mutagenesis kit (Stratagene). Plasmid pLK35-16S-MS2 was used as the template to replace 158g(GAAA)c163 with c(UACG)g in helix 8, and 343UACG346 with GAAA in helix 14 of 16S rRNA (25, 26). All clones were verified by automated DNA sequencing of the entire 16S rRNA operon.
Plasmid-Replacement Strategy
Plasmid replacement was performed as described previously (27, 28). Briefly, E. coli strain SQZ10 (Δ7rrn) containing plasmid pHK-rrnC+sacB (kanamycin resistant) was transformed with pLK35-16S-MS2 containing the desired mutations. The transformants were grown overnight in LB medium (10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl per litre of medium) with 100 μg/mL ampicillin at 37 °C with shaking. The cultures were diluted and plated on 2YT agar plates with 8% sucrose and 100 μg/mL ampicillin. The colonies on the plates were screened for sensitivity to kanamycin by replica plating. Plasmid replacement was confirmed by isolating plasmids and automated DNA sequencing. Cell stocks were prepared in 15% glycerol and stored at −80 °C.
Growth Curve
Growth of plasmid-replaced E. coli strain SQZ10 (Δ7rrn) expressing the wild-type or mutant 16S rRNA occurred in 200 μL of LB medium in the presence of 100 μg/mL ampicillin at 37 °C with continuous shaking in a plate reader (Genios, Tecan) (26). Each culture was inoculated with the same number of cells from overnight starter cultures. The absorbance at 600 nm was automatically measured at 10 min interval by the plate reader. The data were fit to an exponential growth curve equation (Y=Aebx), and the doubling time was calculated as ln(2)/b using Prism (GraphPad Prism, San Diego, CA).
Ribosome Profile
Ribosome profiles of E. coli strain SQZ10 (Δ7rrn) expressing the wild-type or mutant 16S rRNA were determined essentially as described previously (26).
In Vitro Translation of Reporter Protein
The activity of the purified ribosome was analyzed by in vitro translation of the reporter protein Renilla luciferase as described previously (25, 26). Briefly, activated ribosomes were added to the S-100 in vitro translation mix and transferred to a 96-well plate. The 96-well plate was incubated at 37 °C in a plate reader (Genios, Tecan), and the synthesis of the luciferase enzyme was monitored in real time by measuring the luminescence every 2 min. Duplicates of the samples were used for each experiment, and the assays were repeated at least two times.
Peptidyl Transferase Assay by Quench-Flow Methods
Peptidyl transferase reactions were performed in HiFi buffer [50 mM Tris-HCl (pH 7.5), 70 mM NH4Cl, 30 mM KCl, 3.5 mM MgCl2, 8 mM putrescine, 0.5 mM spermine and 2 mM DTT] as described previously (29). Initiation complexes were prepared in HiFi buffer (with 7 mM Mg2+) by incubation of the activated 70S ribosome (2.5 μM) with mRNA (5 μM) at 37 °C for 10 min. f[35S]Met-tRNAfMet charging mixture was prepared in HiFi buffer (3.75 μM tRNAfMet, 3 mM ATP, 0.25 μM [35S]-L-methionine, 3.75 μM L-methionine 0.4 mM N10-formyltetrahydrofolic acid, 10 μg of formyl transferase, and 10 μg of MetRS incubated at 37 °C for 20 min) was added directly to the ribosome-mRNA complex, and the incubation was continued at 37 °C for 10 min. Unbound f[35S]Met-tRNAfMet was removed by ultrafiltration using Microcon Centrifugal Filter Devices (Amicon; 100000 MWCO) and by washing six times with 400 μL of HiFi buffer (7 mM Mg2+). The initiation complexes were recovered after washing, and the concentration of Mg2+ was adjusted to 3.5 mM via addition of HiFi buffer lacking Mg2+. To prepare the EF-Tu·GTP·Phe-tRNAPhe ternary complex, the EF-Tu·GTP complex was formed via incubation of 1 mM GTP, 3 mM phosphoenolpyruvate, 0.25 μg/μL pyruvate kinase, and 1.5 μM EF-Tu at 37 °C for 20 min in HiFi buffer. Phe-tRNAPhe (0.5 μM) was then added, and the incubation was continued for an additional 20 min. To determine the rate of the peptidyl transferase reaction, 15 μL of the 70S initiation complex was rapidly mixed with 15 μL of the ternary complex and quenched with 1 M KOH in a quench-flow instrument (μQFM-400, BioLogic). The dipeptide was resolved by electrophoresis on cellulose TLC plates and quantified using a phosphorimager (BioRad).
Estimation of missense error and nonsense error by in vivo reporter assay
To estimate the frequency of missense errors, the activity of Renilla luciferase (RL) mutant (Glu144Asp) relative to that of wild-type RL (control) was determined using for E. coli strain SQZ10 (Δ7rrn) expressing the wild type or mutant 16S rRNA. The mutation of the catalytic residue Glu144 (codon GAA) to Asp (codon GAT) decreases the reporter activity to 5% (30). Any miscoding at codon GAT will be reflected as increased reporter activity. A similar approach was used to estimate the frequency of UGA read-through (nonsense errors). To estimate the nonsense error, the activity of Renilla luciferase mutant (Leu156Stop) relative to that of wild type RL (control) was determined for E. coli strain SQZ10 (Δ7rrn) expressing the wild type or mutant 16S rRNA. The stop codon is in the middle of the catalytic tried of Renilla luciferase D120, E144 and H285 (30) and it abolishes luciferase activity completely. Read-through of the stop codon will be reflected as increased reporter activity.
In vitro Fidelity Experiment
The in vitro fidelity experiments were performed as described previously (29).
Translocation Experiment
Pre-translocation complexes were made and translocation was monitored by the toeprinting assay as described previously (31). Rapid kinetic experiments were performed essentially as described (26, 32).
Results
Replacing the tetraloops in helices 8 and 14 does not affect growth rate
To study the significance for the conservation of the GAAA tetraloop in helix 8 and the UACG tetraloop in helix 14, we separately replaced these tetraloop sequences by site-directed mutagenesis in plasmid pLK35-MS2-16S (33). The GAAA tetraloop in helix 8 was replaced with the UACG tetraloop to create the h8 mutant (h8). Replacement of the GNRA tetraloop with the UNCG tetraloop is expected to disfavor the formation of the A-minor tertiary interaction with helix 14. The UACG tetraloop in helix 14 was replaced with a GAAA tetraloop to create the h14 mutant (h14). Replacement of the UNCG tetraloop with the GNRA tetraloop is expected to disrupt any sequence specific interactions between helix 14 and EF-Tu and EF-G. In addition, the bridge B8 may also get disrupted resulting in weakened association of the 30S subunit to the 50S subunit.
We used the E. coli strain SQZ10 (Δ7rrn) to analyze the effect of the tetraloop replacements on cell growth. The E. coli strain SQZ10 (Δ7rrn) lacks the seven rRNA operons in its genome and survives by having the rRNA genes expressed from a plasmid (27, 28). The h8 and h14 mutant plasmids were transformed into the E. coli strain SQZ10 (Δ7rrn) and selected using the appropriate antibiotic. Our plasmid replacement studies showed that the E. coli strain SQZ10 could grow normally with either the h8 or the h14 mutant plasmids exclusively expressing the rRNA genes (data not shown). Growth rate studies in liquid cultures at 37 °C and 42 °C further confirmed that the growth rates of the h8 and h14 mutants are similar to wild type (doubling time at 37 °C are WT = 60 ± 1 min, h8 = 62 ± 1 min, and h14 = 62 ± 3 min). Thus, the replacement of h8 or h14 with a different type of tetraloop does not appreciably affect the growth rate.
Replacing the h14 tetraloop causes a modest defect in subunit association
We analyzed the ribosome profile of h8 and h14 mutant cells to identify defects in 30S subunit assembly and in their association with the 50S subunit to form 70S ribosome. Ribosome profile showed that the h8 mutant has no defects in 30S assembly and in subunit association (Figure 2). In contrast, the h14 mutant showed a small shoulder to the left of the 70S peak suggesting that the mutant 30S subunit does not bind tightly to the 50S subunit and dissociate during the centrifugation (Figure 2C). Thus, the h14 mutant likely has subtle defects in subunit association. It is well known that increasing the Mg2+ ion concentration from 10 mM to 20 mM can improve subunit association. Therefore, we repeated the ribosome analysis in the presence of 20 mM Mg2+ ion. As expected, the h14 mutant 30S subunit showed normal subunit association at the higher Mg2+ ion concentration (Figure 2E).
Figure 2.
Ribosome profile of cells expressing mutant 16S rRNAs. (A) Wild type. (B) h8 mutant. (C) h14 mutant. (D) Wild type (20 mM MgCl2). (E) h14 mutant (20 mM MgCl2). The wild type and mutant 16S rRNAs were expressed in E. coli strain SQZ10 (Δ7rnr). Experiments A to C were performed in buffer containing 10 mM MgCl2. Experiments E and F were performed in buffer containing 20 mM MgCl2. Labels: 30S (small ribosomal subunit), 50S (large ribosomal subunit) and 70S (ribosome). The dotted line indicates the position of the 70S ribosome in the sucrose gradient.
Inhibition of in vitro translation by the tetraloop replacements
We analyzed the activity of the mutant ribosomes in protein synthesis using an in vitro translation assay. The time course of Renilla luciferase synthesis by the wild type and mutant ribosomes was monitored by bioluminescence (25) (Figure 3). The experiment was done at several ribosome concentrations. Our results showed that the wild type ribosomes efficiently synthesize luciferase at 0. 1 μM final concentration (Figure 3A). In contrast, 0.2 μM of h8 mutant ribosome and 0.4 μM h14 mutant ribosomes are required for achieving a similar rate of luciferase synthesis (Figure 3B and 3C). Thus, the in vitro translation assay indicates that the mutant ribosomes are slightly defective in in vitro protein synthesis.
Figure 3.
In vitro protein synthesis by mutant ribosomes. Representative time courses showing the synthesis of luciferase enzyme at the indicated concentrations of the wild type and mutant ribosomes. (A) Wild type ribosome. (B) h8 mutant ribosome. (C) h14 mutant ribosome. The in vitro protein synthesis was carried out at 6 mM MgCl2 concentration. The maximum signal that we can observe with this instrument is ≈65,000 relative luminescence units (RLU) because of detector saturation.
Tetraloop replacements do not affect the peptidyl transferase reaction
Structural data showed that the switch 1 loop of EF-Tu interacts with the junction of helix 8 and helix 14 (20, 21). Therefore, to determine whether the replacement of the h8 and h14 tetraloops affected the rate of peptide bond formation we carried out pre-steady state kinetic experiments. Ribosome with f-[35S]-Met-tRNAfMet in the P site and a phenylalanine codon in the A site was rapidly mixed with a nearly saturating concentration of EF-Tu•GTP•Phe-tRNAPhe ternary complex with a quench-flow instrument (29). The product of the reaction, f-[35S]-Met-Phe dipeptide, was analyzed by electrophoretic TLC (eTLC) (Figure 4). The time course of fMet-Phe dipeptide formation was fitted to a single-exponential equation to determine the rate of peptide bond formation (WT = 6.1 ± 0.3 s−1, h8 = 8.5 ± 1.2 s−1, and h14 = 9.9 ± 2.0 s−1). The rate of peptide bond formation was similar for wild type, h8 and h14 mutant ribosomes indicating that tRNA selection is not significantly affected by the mutations.
Figure 4.

Kinetics of peptide bond formation. Representative electrophoretic TLC plates showing the time course of dipeptide formation by wild-type and mutant ribosomes. (A) Wild-type ribosome. (B) h8 mutant ribosome. (C) h14 mutant ribosome. (D) Graph showing the time course of dipeptide formation by wild-type ribosome (●), h8 mutant ribosome (◆), and h14 mutant ribosome (■). Data were fit to a single-exponential equation to determine the apparent rate of peptide bond formation.
Interchanging the tetraloops decreases the fidelity of translation
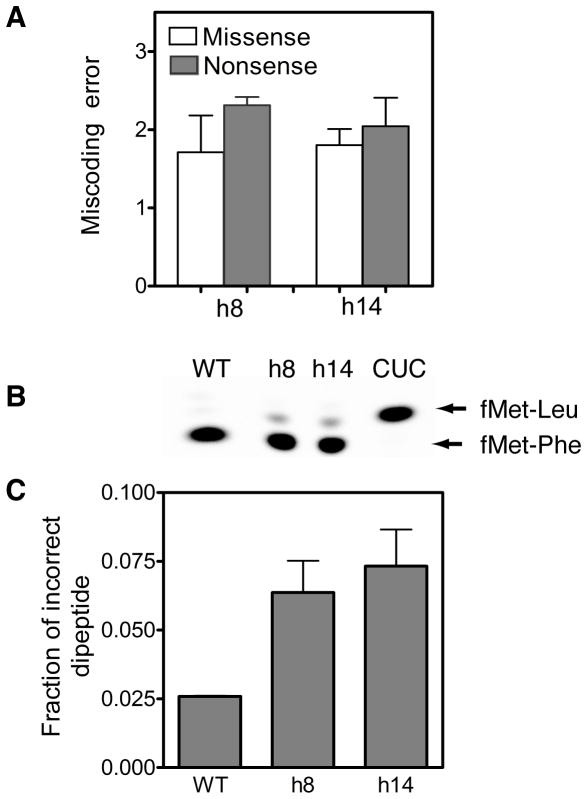
A previous study showed that mutations in h8 and h14 increase the error frequency of protein synthesis by 2- to 3-fold (24). Therefore, we determined the in vivo missense error rates of cells expressing h8 or h14 replacement. The Renilla luciferase gene was used as the reporter for determining the fidelity of translation. The catalytic triad Asp120, Glu144 and His285 in Renilla luciferase are essential for the activity of the enzyme (30). Mutation of these catalytic residues decreases the activity of Renilla luciferase to less than 5% (30). We measured the missense error rates using a Renilla luciferase gene with a point mutation that changes the essential glutamic acid at position 144 to an aspartic acid (codon GAA to GAU). An increase in the misincorporation of glutamic acid at position 144 will increase the amount of active luciferase produced by the cell. Luciferase activity can be detected easily and with high sensitivity by monitoring bioluminescence. We found that both h8 and h14 mutant cells had a missense error rate that was about 2-fold higher than wild type cells (Figure 5A).
Figure 5.
Effects of 16S rRNA mutations on miscoding by the ribosome. (A) Bar graph showing the miscoding error determined using an in vivo reporter assay. Missense and nonsense errors were calculated from Renilla luciferase activity in E. coli strain SQZ10 (Δ7rnr) expressing the wild type or mutant 16S rRNAs. The error rate by cells expressing the wild type 16S rRNA was normalized to 1.0. Values on the X-axis reflect the fold increases in miscoding error by cells expressing the h8 and h14 mutant 16S rRNAs compared to cells expressing the wild type 16S rRNA. The data shown is the average ± SD from three independent experiments. (B) A representative electrophoretic TLC plate showing the products of the in vitro fidelity experiment. Binding of the cognate Phe-tRNAPhe to the ribosomal A site produces the [35S]fMet-Phe dipeptide, whereas binding of the near-cognate Leu-tRNALeu to the ribosomal A site produces [35S]fMet-Leu dipeptide. Label CUC shows a control reaction with the wild type ribosome having a CUC codon in the A site that was performed in parallel to serve as a marker for the [35S]fMet-Leu dipeptide. (C) Bar graph showing the fraction of incorrect [35S]fMet-Leu dipeptide formed by wild-type (WT), h8 and h14 ribosomes.
We next determined the rate of nonsense suppression by h8 and h14 mutants. For this, we used a Renilla luciferase gene with a point mutation that changes a leucine at position 156 to a stop codon (codon UGG to UGA). Leucine 156 is not essential for the activity of luciferase and substitution by any other amino acid is tolerated at this position (30). However, termination of protein synthesis at position 156 will lead to a truncated luciferase enzyme that is inactive. This permitted us to determine the rate of nonsense suppression during protein synthesis. We found that the nonsense suppression rate was increased by 2-fold with the h8 and h14 mutant cells compared to the wild type cells (Figure 5A).
To confirm the increased error rate of protein synthesis by the h8 and h14 mutant ribosomes we used a simplified in vitro miscoding assay (29). For the in vitro miscoding assay, we formed ribosomal complex containing f-[35S]-Met-tRNAfMet in the P site and a phenylalanine codon (UUU) in the A site. The ribosome complex was mixed with EF-Tu•GTP•aminoacyl-tRNA ternary complex formed from a mixture of E. coli total tRNA. The expected dipeptides are f-[35S]-Met-Phe for cognate and f-[35S]-Met-Leu for near-cognate binding of the ternary complex to the ribosome. We analyzed the dipeptides formed by the ribosome by eTLC (Figure 5B). Our results showed that h8 and h14 mutant ribosomes had an error frequency that was 2- to 3-fold higher than the wild type ribosome and are consistent with the error rates measured in vivo (Figure 5C).
Tetraloop replacements do not affect translocation of the mRNA-tRNA complex
We analyzed whether the tetraloop replacements in h8 and h14 affected EF-G-dependent mRNA-tRNA translocation by the ribosome. Initially, we used the toeprinting assay to monitor the translocation of the mRNA-tRNA complex by the ribosome (31, 34). Pre-translocation complexes were programmed with a fragment of the T4 gene 32 mRNA and contained tRNAfMet in the P site and tRNAPhe in the A site. The extension of a DNA primer hybridized to the 3′-end of the mRNA by reverse transcriptase resulted in a toeprint band that is characteristic for the pre-translocation complex (Figure 6A). The addition of EF-G and GTP to the pre-translocation complexes resulted in the appearance of a new toeprint band that corresponds to the post-translocation complex. Both h8 and h14 mutants gave a similar extent of translocation as the wild type ribosome.
Figure 6.
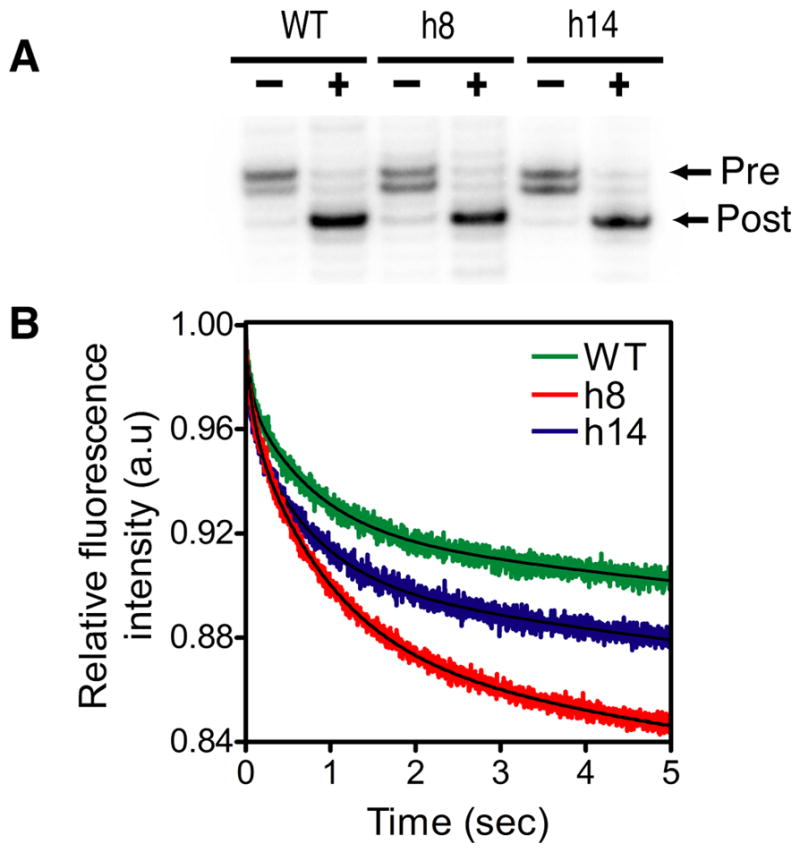
Translocation of mRNA-tRNA complex. (A) The extent of mRNA-tRNA translocation was determined using a toeprinting assay. Minus and plus indicate the absence and presence of EF-G, respectively. Toeprinting bands corresponding to the pre-translocation complex (Pre) and the post-translocation complex (Post) are indicated. (B) Pre-steady state kinetic analysis of EF-G-dependent translocation of wild type and mutant ribosomes. The decrease in fluorescence corresponds to EF-G-dependent translocation of the mRNA-tRNA complex. The data was fit to a double-exponential equation to determine the apparent rate of translocation.
The rate of translocation by the h8 and h14 mutant ribosomes was determined using a fluorescence-based, pre-steady state kinetic assay (32). Pre-translocation complex contained a short mRNA having a pyrene dye attached to the 3′ terminus, tRNAfMet in the P site, and fMet-Phe-tRNAPhe in the A site. The pre-translocation complex was rapidly mixed with a 10-fold excess of EF-G with a stopped-flow instrument and the decrease in fluorescence intensity of the pyrene dye because of translocation was recorded (Figure 6B). The change in fluorescence intensity showed two phases with the apparent rate constants kobs1 and kobs2 for the fast and slow phases, respectively. The reason for the two phases is not clear but has been reported earlier (26, 35–37). The rate of the rapid phase is consistent with the rate of translocation measured using fluorescently labeled tRNAs (38). Furthermore, translocation inhibitors such as viomycin and neomycin inhibit the rapid phase (32, 37). The slow phase may be caused by sample heterogeneity or a conformational change in the ribosome that occurs after mRNA-tRNA translocation. The data were analyzed by fitting the decrease in fluorescence intensity to a double exponential equation to calculate the kobs1 and kobs2 for the wild type, h8 and h14 ribosomes (WT kobs1 = 15.8 ± 0.3 s−1 and kobs2 = 1.3 ± 0.01 s−1) (h8 kobs1 = 11.3 ± 0.3 s−1 and kobs2 = 1.1 ± 0.1 s−1) and (h14 kobs1 = 18.7 ± 0.6 s−1 and kobs2 = 1.2 ± 0.1 s−1). The rate of translocation by the h8 and the h14 mutants showed less than a 2-fold difference from the wild type suggesting that the mutations do not substantially affect translocation.
Discussion
The ribosome is a precisely assembled molecular machine sculpted out of RNA and protein. The rRNAs not only provide the framework for the overall shape of the ribosome but also are intimately involved in forming the functional centers of the ribosome (39, 40). A dominant secondary structural element of the rRNAs is the hairpin (2, 14, 41, 42). About 70% of the 16S rRNA folds into stem-loop structures (2, 14). The number and sequence of the nucleotides that form the loop are variable; however, the most common loops in 16S rRNA are tetraloops with the consensus sequence GNRA or UNCG (2). These tetraloops form compact structures and have very high thermodynamic stability (3, 4, 7, 8). Despite their similarity in shape and stability, the rRNAs show a strong preference for the GNRA and the UNCG loops at specific positions in the rRNA (2). The basis for this conservation of the tetraloop sequence at particular positions in the rRNAs is not clear. Woese and co-workers have suggested that the stability of a particular tetraloop sequence itself may be the primary determinant for their conservation at some positions in the rRNAs (2).
To test whether the primary sequence or the structural stability of the tetraloops is important for ribosome function, we examined two tetraloops in the 16S rRNA that are >99% conserved. We picked the GNRA-type tetraloop in h8 and the UNCG-type tetraloop in h14 because structural studies have suggested that they might be important for translation. Cryo-EM structures showed that the junction between h8 and h14 tetraloops interacts with the switch I region in the GTPase domain of EF-Tu (20, 21) (Figure 1F). The interaction of the switch I of EF-Tu with the junction of helices h8/h14 of the 16S rRNA was proposed to open the “hydrophobic gate” in EF-Tu and reorient the catalytic His84 to induce GTP hydrolysis (20, 21). Similarly, the switch I regions of EF-G-GMPPNP (Figure 1G), and eEF2 trapped in the transition state interacts with h14 of the 16S rRNA (22, 23). The switch I region of EF4 (LepA) also interacts with h14 (43). However, the interaction of EF-Tu and EF-G with helices 8 and 14 were not observed in recent crystal structures (44–46). It is possible that these crystal structures represent a state that is different from the cryo-EM structures.
Another function attributed to the helix 14 is the formation of an intersubunit bridge. The UACG tetraloop in helix 14 contacts ribosomal proteins L14 and L19 in the 50S subunit to form the intersubunit bridge B8 (15, 17–19) (Figure 1E). Interestingly, the 30S subunit can undergo a ratchet-like rotation relative to the 50S subunit spontaneously (47, 48). Furthermore, the binding of some translational factors such as EF-G, RF3 and RRF induce the ratchet-like rotation of the ribosome to varying degrees (49–52). Recent structural data showed that the interactions that h14 makes with L14 and L19 changes when the 30S subunit undergoes a ratchet-like rotation relative to the 50S subunit. For example, in the classical state, bridge B8 is formed by nucleotide C345 in the h14 tetraloop interacting with Ser116 and Ala118 in L14 and Arg39 and Arg41 in L19 (50). In the 70S•RF3 complex, bridge B8 rearranges and nucleotide C345 in h14 tetraloop interacts only with Ser35 and Lys36 in L19 (50). In addition, nucleotide G347 in h14 make new contacts with Leu118 and Ala119 in L14 (50). The phosphate non-bridging oxygens atoms of C345 and G347 mainly mediate these new bridge interactions in the rotated state (50). Nevertheless, the integrity of bridge B8 may depend on the local structure and thermodynamic stability of the h14 tetraloop.
Genetic and biochemical studies also indicate a functional role for helices 8 and 14. Previously, A161G mutation in h8 tetraloop was isolated in a dominant negative screen, and this mutation reduced translation activity in vivo by ≈25% (53). Another genetic screen isolated several mutations in h8 and h14 tetraloops that increase misreading of sense and stop codons (24). These mutations in h8 and h14 also reduced translation activity in vivo by 2- to 4-fold. Interestingly, they showed that shortening helix 14 by 2 base pairs decreased translation by 5-fold, whereas lengthening helix 14 by 2 base pairs abolished translation in vivo (24). Lengthening helix 14 is probably not tolerated because it disrupts the bridge B8 and prevents subunit association. Furthermore, 2 or 3 base pair deletions in helix 8 or 2 base pair deletion in helix 14 increase the missense error rate by 10-fold compared to the wild type ribosome (24). The increased missense error rate is caused by an increase in the rate of GTP hydrolysis by the near-cognate EF-Tu ternary complex binding to the h8 and h14 mutant ribosomes compared to the wild type ribosome (24). Thus, the biochemical data suggest that h8/h14 junction negatively regulates GTP hydrolysis on EF-Tu (24). Mutations in helices 8 and 14 may weaken the bridge B8 and lower the energetic cost for the conformational changes that induce GTP hydrolysis on EF-Tu. This will lead to an increase in the error rate because even near-cognate EF-Tu ternary complex can trigger GTP hydrolysis and be accepted by the ribosome (24).
We find that the complete replacement of the conserved tetraloops in h8 and h14 with a tetraloop having a different sequence is better tolerated by the cells compared to the single base mutations, deletions and insertions discussed above. We did not observe any defects in the growth rates of E. coli strain (Δ7 rrn) expressing the h8 and h14 mutant plasmids. This is consistent with the idea that the GNRA and UNCG tetraloops have a similar fold and comparable thermodynamic stability and can be functionally interchanged if the precise sequence is not critical for function (3, 12). In contrast, mutations, deletions and insertions show increased defects in translation because they may be more disruptive to the local or global structure of the ribosome. However, we note that the growth rate of E. coli strain (Δ7 rrn) is slower than wild-type E. coli strains and minor defects in protein synthesis may not significantly affect the growth rate of E. coli strain (Δ7 rrn).
We observed a slight defect in subunit association with the h14 mutant at 10 mM Mg2+ ion concentration, which is consistent with h14 forming the intersubunit bridge B8. However, the subunit association defect was overcome at 20 mM Mg2+ ion concentration. It is possible that Mg2+, polyamines and the high concentrations of 30S and 50S subunits present in vivo may stabilize subunit association even with a GNRA loop in helix 14. This is consistent with our observation that increasing the concentration of the h8 and h14 mutant 30S subunits could increase the rate of in vitro translation. Interestingly, the rate of in vitro translation is slower with the h8 and h14 mutant ribosomes. Since these experiments were performed at 6 mM MgCl2 concentration, it is possible that during the initiation step of translation the rate of association of the 50S subunit to the mutant 30S subunits is slower than to the wild type 30S subunit explaining the slow rate of in vitro translation. More studies are necessary to understand whether the h8 and h14 tetraloop replacements inhibit translation initiation.
Our studies showed that the h8 and h14 mutants have rates of peptide bond formation and translocation that are similar to the wild type ribosome. Thus, the precise sequence of h8 and h14 is not essential for these steps in the elongation cycle of protein synthesis. Nevertheless, the h8 and h14 mutants showed about a 2-fold increased error rate of translation. The increased rate of miscoding, however, is much lower than the 5- to 10-fold increase in miscoding reported earlier with single base substitutions and the deletion mutants (24). Therefore, in helices 8 and 14, changing the conserved sequence of the tetraloop as a unit with a different tetraloop sequence is better tolerated by the ribosome. This may be a useful strategy to determine whether the conservation of a tetraloop sequence at a particular position in the rRNAs is essential for protein synthesis. Finally, our results suggest that the conservation of a tetraloop sequence at a particular position in the rRNAs may come from evolutionary pressures to maintain the stability of the tetraloop structure itself instead of for a defined function during protein synthesis.
Acknowledgments
We thank Kurt Fredrick, Ulrich Muller, and Gourisankar Ghosh for comments on the manuscript.
Abbreviations
- rRNA
ribosomal RNA
- mRNA
messenger RNA
- tRNA
transfer RNA
- EF
elongation factor
Footnotes
This work was supported by National Institutes of Health Grant GM065265 to S.J.
References
- 1.Ramakrishnan V, Moore PB. Atomic structures at last: the ribosome in 2000. Curr Opin Struct Biol. 2001;11:144–154. doi: 10.1016/s0959-440x(00)00184-6. [DOI] [PubMed] [Google Scholar]
- 2.Woese CR, Winker S, Gutell RR. Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops”. Proc Natl Acad Sci U S A. 1990;87:8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varani G. Exceptionally stable nucleic acid hairpins. Annu Rev Biophys Biomol Struct. 1995;24:379–404. doi: 10.1146/annurev.bb.24.060195.002115. [DOI] [PubMed] [Google Scholar]
- 4.Uhlenbeck OC. Tetraloops and RNA folding. Nature. 1990;346:613–614. doi: 10.1038/346613a0. [DOI] [PubMed] [Google Scholar]
- 5.Butcher SE, Pyle AM. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc Chem Res. 2011;44:1302–1311. doi: 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- 6.Keating KS, Toor N, Pyle AM. The GANC tetraloop: a novel motif in the group IIC intron structure. J Mol Biol. 2008;383:475–481. doi: 10.1016/j.jmb.2008.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheong C, Varani G, Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5′ GGAC(UUCG)GUCC. Nature. 1990;346:680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- 8.Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 9.Jaeger L, Michel F, Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J Mol Biol. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 10.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. Embo J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pley HW, Flaherty KM, McKay DB. Model for an RNA tertiary interaction from the structure of an intermolecular complex between a GAAA tetraloop and an RNA helix. Nature. 1994;372:111–113. doi: 10.1038/372111a0. [DOI] [PubMed] [Google Scholar]
- 12.Selinger D, Liao X, Wise JA. Functional interchangeability of the structurally similar tetranucleotide loops GAAA and UUCG in fission yeast signal recognition particle RNA. Proc Natl Acad Sci U S A. 1993;90:5409–5413. doi: 10.1073/pnas.90.12.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy FL, Cech TR. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J Mol Biol. 1994;236:49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- 14.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Müller KM, Pande N, Shang Z, Yu N, Gutell RR. The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 16.Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 17.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 18.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 19.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 20.Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, Frank J. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci U S A. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuette JC, Murphy FVt, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, Ramakrishnan V, Spahn CM. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. Embo J. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, Dabrowski M, Mielke T, Fucini P, Yokoyama S, Spahn CM. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25:751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 23.Sengupta J, Nilsson J, Gursky R, Kjeldgaard M, Nissen P, Frank J. Visualization of the eEF2-80S ribosome transition-state complex by cryo-electron microscopy. J Mol Biol. 2008;382:179–187. doi: 10.1016/j.jmb.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClory SP, Leisring JM, Qin D, Fredrick K. Missense suppressor mutations in 16S rRNA reveal the importance of helices h8 and h14 in aminoacyl-tRNA selection. RNA. 2010;16:1925–1934. doi: 10.1261/rna.2228510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Ortega L, Stephen J, Joseph S. Precise alignment of peptidyl tRNA by the decoding center is essential for EF-G-dependent translocation. Mol Cell. 2008;32:292–299. doi: 10.1016/j.molcel.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Shi X, Chiu K, Ghosh S, Joseph S. Bases in 16S rRNA important for subunit association, tRNA binding, and translocation. Biochemistry. 2009;48:6772–6782. doi: 10.1021/bi900472a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asai T, Condon C, Voulgaris J, Zaporojets D, Shen B, Al-Omar M, Squires C, Squires CL. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J Bacteriol. 1999;181:3803–3809. doi: 10.1128/jb.181.12.3803-3809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asai T, Zaporojets D, Squires C, Squires CL. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc Natl Acad Sci U S A. 1999;96:1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hetrick B, Khade PK, Lee K, Stephen J, Thomas A, Joseph S. Polyamines accelerate codon recognition by transfer RNAs on the ribosome. Biochemistry. 2010;49:7179–7189. doi: 10.1021/bi1009776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woo J, Howell MH, von Arnim AG. Structure-function studies on the active site of the coelenterazine-dependent luciferase from Renilla. Protein Sci. 2008;17:725–735. doi: 10.1110/ps.073355508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joseph S, Noller HF. EF-G-catalyzed translocation of anticodon stem-loop analogs of transfer RNA in the ribosome. EMBO J. 1998;17:3478–3483. doi: 10.1093/emboj/17.12.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studer SM, Feinberg JS, Joseph S. Rapid Kinetic Analysis of EF-G-dependent mRNA Translocation in the Ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 33.Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 35.Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. Proc Natl Acad Sci U S A. 2008;105:9192–9197. doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol. 2011;18:457–462. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khade PK, Joseph S. Messenger RNA interactions in the decoding center control the rate of translocation. Nat Struct Mol Biol. 2011;18:1300–1302. doi: 10.1038/nsmb.2140. [DOI] [PubMed] [Google Scholar]
- 38.Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 39.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 41.Noller HF, Kop J, Wheaton V, Brosius J, Gutell RR, Kopylov AM, Dohme F, Herr W, Stahl DA, Gupta R, Waese CR. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981;9:6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noller HF, Woese CR. Secondary structure of 16S ribosomal RNA. Science. 1981;212:403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- 43.Connell SR, Topf M, Qin Y, Wilson DN, Mielke T, Fucini P, Nierhaus KH, Spahn CM. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1469. [DOI] [PubMed] [Google Scholar]
- 44.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVt, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munro JB, Altman RB, Tung CS, Cate JH, Sanbonmatsu KY, Blanchard SC. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc Natl Acad Sci U S A. 2010;107:709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 50.Zhou J, Lancaster L, Trakhanov S, Noller HF. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. Rna. 2012;18:230–240. doi: 10.1261/rna.031187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin H, Kelley AC, Ramakrishnan V. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc Natl Acad Sci U S A. 2011;108:15798–15803. doi: 10.1073/pnas.1112185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yassin A, Fredrick K, Mankin AS. Deleterious mutations in small subunit ribosomal RNA identify functional sites and potential targets for antibiotics. Proc Natl Acad Sci U S A. 2005;102:16620–16625. doi: 10.1073/pnas.0508444102. [DOI] [PMC free article] [PubMed] [Google Scholar]