Abstract
The Trypanosoma brucei cysteine protease cathepsin B (TbCatB), which is involved in host protein degradation, is a promising target to develop new treatments against sleeping sickness, a fatal disease caused by this protozoan parasite. The structure of the mature, active form of TbCatB has so far not provided sufficient information for the design of a safe and specific drug against T. brucei. By combining two recent innovations, in vivo crystallization and serial femtosecond crystallography, we obtained the room-temperature 2.1 angstrom resolution structure of the fully glycosylated precursor complex of TbCatB. The structure reveals the mechanism of native TbCatB inhibition and demonstrates that new biomolecular information can be obtained by the “diffraction-before-destruction” approach of x-ray free-electron lasers from hundreds of thousands of individual microcrystals.
Over 60 million people are affected by human African trypanosomiasis (HAT), also known as sleeping sickness, which causes ~30,000 deaths per year (1). The protozoan parasite Trypanosoma brucei, transmitted by tsetse flies, infects the blood and the lymphatic system before invading the brain. Severe clinical manifestations occur within weeks or months. Current treatments of HAT rely on antiparasitic drugs developed during the last century, without knowledge of the biochemical pathways. These treatments are limited in their efficacy and safety, and drug resistance is increasing (2–4). Thus, new compounds that selectively inhibit vital pathways of the parasite without adverse affects to the host are urgently required. A promising strategy is to target lysosomal papainlike cysteine proteases that are involved in host-protein degradation, such as cathepsin B (5). The knockdown of this essential enzyme in T. brucei resulted in clearance of parasites from the blood of infected mice and cured the infection (6), which qualify cathepsin B as a suitable drug target. Cysteine proteases are synthesized as inactive precursors with N-terminal propeptides that act as potent and selective intrinsic inhibitors until the proteases enter the lysosome (7), where the propeptide is released and forms the mature active enzyme. Such native propeptide-inhibited structures have been used to develop species-specific protease inhibitors against proteases of other Trypanosoma species, e.g., cruzipain of T. cruzi (causing human Chagas disease in America) and congopain of T. congolense (causing nagana in cattle) (8, 9). This approach could not be explored for T. brucei cathepsin B (TbCatB) because of the lack of structural information on the mode of propeptide inhibition and the large extent of structural conservation at the active site between mammalian and trypanosome cathepsin B (10–12). Previously solved mature T. brucei and human CatB structures show differences at the S2 and in part of the S1′ subsite of the substrate-binding cleft (Fig. 1C) and have been suggested as possible targets for the development of species-specific CatB inhibitors (10). Together with the natively inhibited human procathepsin B structure (13), our work fills the gap to understand the structural basis for species-specific inhibition.
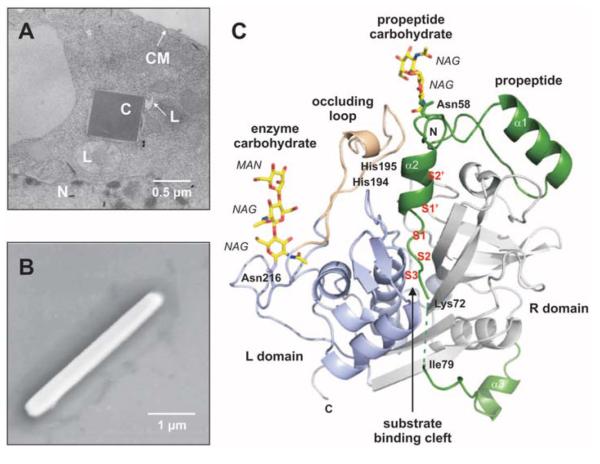
Fig. 1.
In vivo grown crystals and three-dimensional structure of the TbCatB-propeptide complex. (A) Transmission electron microscopy (EM) of an infected Sf9 insect cell showing a crystal of overexpressed TbCatB inside the rough endoplasmic reticulum that is cut perpendicular to its long axis. N, nucleus; L, lysosome; C, crystal; CM, cell membrane. (B) Scanning EM of a single TbCatB crystal after isolation. (C) Cartoon plot of the TbCatB-propeptide complex exhibiting the typical papain-like fold of cathepsin B–like proteases (supplementary text S1). Gray, R domain; blue, L domain; beige, occluding loop. The native propeptide (green) blocks the active site. The subsites of the substrate-binding cleft N-terminal (nonprime: S2, S3) and C-terminal (prime: S1′, S2′) to the active site (S1) have been identified by comparison with the human CatB structure (13) and labeled (red) according to Schechter and Berger (27). Two N-linked carbohydrate structures (yellow) consist of N-acetylglucosamine (NAG) and mannose (MAN) residues (yellow, carbon atoms; blue, nitrogen atoms; red, oxygen atoms).
The growth of large well-ordered protein crystals is one of the major bottlenecks in structure determination by x-ray crystallography—with important biological targets, such as integral membrane proteins and posttranslationally modified proteins, proving particularly challenging to crystallize (14). Sizable crystals are required to obtain measurable high-resolution diffraction data within an exposure that is limited by the accumulation of radiation damage (15). Although microfocus beamlines enable the collection of diffraction data from micron-sized protein crystals (16), the tolerable dose limit of less than 30 MGy for cryogenically cooled protein crystals remains, which limits the achievable signal. The tolerable dose for room temperature measurements is about 1 MGy (15). We have previously shown that micron-sized crystals of glycosylated TbCatB spontaneously form in insect cells during protein over-expression (11). Such crystals are extremely well suited for the new method of serial femtosecond crystallography (SFX) (17). X-ray free-electron laser (FEL) pulses of less than 100-fs duration allow the dose to individual crystals to exceed the ~1 MGy limit by over a thousand times because of the “diffraction-before-destruction” principle (17, 18). Diffraction data are recorded for each pulse as crystals are continually replenished by a microcrystal suspension in aqueous buffer flowing across the FEL beam in a vacuum in a fine liquid jet.
The Coherent X-ray Imaging (CXI) beamline (19) at the Linac Coherent Light Source (LCLS) enables high-resolution data collection using the SFX approach (20). We used this instrument to obtain diffraction data from in vivo grown crystals of TbCatB produced in the baculovirus-infected Spodoptera frugiperda (baculovirus-Sf9) insect cell system (11) (Fig. 1, A and B). Crystals with average dimensions of about 0.9 by 0.9 by 11 μm3 (fig. S1) were sent in a 4-μm-diameter column of buffer fluid at room temperature, at a flow rate of 10 μl/minute, by using a liquid microjet (21). X-ray pulses from the FEL were focused onto this column to a spot 4 μm in diameter, before the breakup of the jet into drops (fig. S2). Single-pulse diffraction patterns of randomly oriented crystals that, by chance, were present in the interaction region, were recorded at a 120-Hz repetition rate by a Cornell-SLAC pixel array detector (CSPAD) (19, 20) at 9.4-keV photon energy (1.3 Å wavelength). An average pulse energy of 0.6 mJ at the sample (4 × 1011 photons per pulse) with a duration of less than 40 fs gave an x-ray intensity above 1017 W/cm2 and a maximum dose of about 31 MGy per crystal. This dose exceeds that tolerable at room temperature with conventional data collection approaches because of the radically different time scales and dose rates. The electron and photon beam parameters are summarized in table S1. Almost 4 million individual “snap-shot” diffraction patterns were collected. Of these, 293,195 snap-shots contained crystal diffraction (fig. S3), from which 178,875 (61%) diffraction patterns were indexed and combined into a three-dimensional data set of structure factors by “Monte Carlo” integration of partial reflections from each randomly oriented microcrystal (22, 23). The resulting complete set of structure factors contains 25,969 reflections in a resolution range from 20 to 2.1 Å. The high quality of the merged data set is indicated by an Rsplit of 10.2% (which is a quality measure for SFX instead of Rmerge) (23). Data statistics are summarized in table S2, table S3, and fig. S4. The structure was solved by molecular replacement using the coordinates of the previously determined in vitro crystallized mature TbCatB structure (Protein Data Bank ID, 3MOR) (11) as a search model.
The refined SFX TbCatB structure (R factor = 18.1%, Rfree = 21.4%) shares the papainlike fold that is characteristic of cathepsin B–like proteases (Fig. 1C and supplementary text S1) (24), with a root mean square deviation of 0.4 Å for equivalent Cα atoms of the mature TbCatB structure determined at 100 K and refined to 2.55 Å resolution (11). The molecular replacement solution reveals electron density that is not part of the search model, which we identified as the coordinated, cleaved main part of the propeptide (residues 26 to 72) (Fig. 2A), and as two-carbohydrate structures (Fig. 2, B and C). Proteolytic cleavage of the expressed precursor occurs within the propeptide between Ser78 and Ile79, as revealed by mass spectrometry, which leaves 15 propeptide residues bound to the N terminus of mature TbCatB (supplementary text S2). The preceding residues Lys73 to Ser78 are disordered in the crystal structure, owing to a rise in flexibility, which shows up as a gap in the electron density in this region of the propeptide. The cleavage may be part of the initial maturation step within the activation process of TbCatB. The final model of glycosylated TbCatB in complex with its processed, but still-bound, propeptide contains 62 propeptide residues and 247 mature enzyme residues, as well as 98 solvent and 5 carbohydrate molecules. No electron density is observed for 11 flexible amino acid side chains or the eight atoms of the carbohydrate structures.
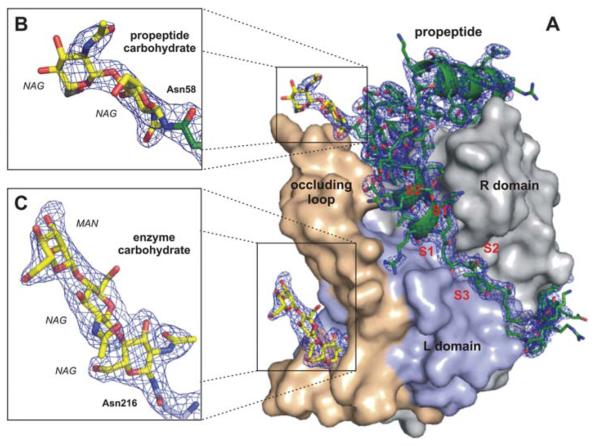
Fig. 2.
Quality of the calculated electron density. (A) Surface representation of the TbCatB-propeptide complex solved by molecular replacement using the mature TbCatB structure (11) as a search model. The solution revealed additional electron density (2Fobs – Fcalc, 1σ, blue) of the propeptide (green) that is bound to the V-shaped substrate-binding cleft and of two carbohydrate structures (yellow) N-linked to the propeptide (B) and to the mature enzyme (C). The propeptide, as well as both carbohydrates, are well-defined within the electron density map (blue), which confirms that the phases are not biased by the search model. Color codes correspond to Fig. 1C.
The SFX TbCatB structure shows that the inhibitory mechanism observed for mammalian papainlike protease-precursors remains largely conserved in T. brucei, including the overall conformation of the propeptide (supplementary text S3 and fig. S5). The active site of TbCatB is blocked by the propeptide, which tightly binds in a direction the reverse of the substrate’s (fig. S6) (25). A detailed comparison of the propeptide-enzyme contact area with that observed for human procathepsin B (Protein Data Bank ID, 3PBH) (13) indicates an interface enlarged by ~310 Å2 within the TbCatB-propeptide complex (supplementary text S4). Tight binding of the T. brucei propeptide to the enzyme interface through three conserved epitopes is maintained by 21 intermolecular polar and ionic interactions (fig. S7). These are eight fewer interactions than for human procathepsin B.
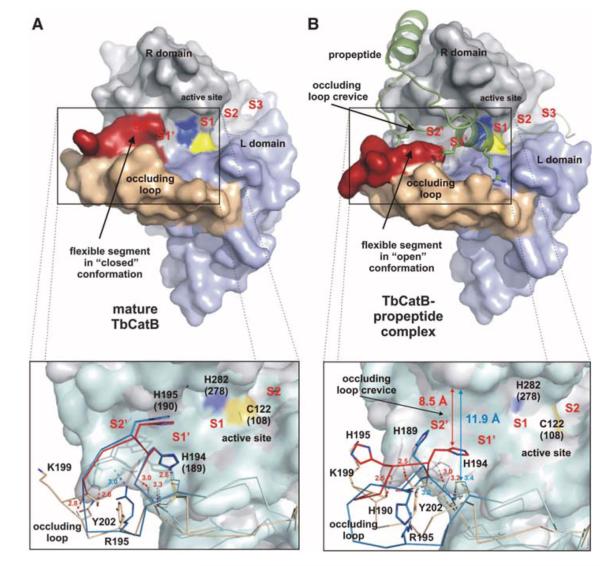
The most significant difference between the structures of mature TbCatB and the natively inhibited propeptide complex occurs in the “occluding loop” region (residues 193 to 207) (fig. S8). This highly flexible loop is a structural element characteristic of cathepsin B–like enzymes that confers exopeptidase activity (removal of dipeptide units from the C terminus of the substrate), which supplements the endopeptidase (nonterminal substrate cleavage) activity common to all papainlike proteases (26). In mature CatB, the occluding loop is in the “closed” conformation and buries an essential part of the prime subsite (S1′ and S2′ positions) at the substrate cleft (Fig. 3A) (27) and competes for binding with large substrates with an affinity that depends sensitively on pH (28). As a consequence of propeptide binding, the occluding loop is reoriented into an “open” conformation, exposing the entire S1′ and S2′ subsite of the substrate-binding cleft in TbCatB (Fig. 3B). This mirrors the open and closed confirmations observed in human CatB; however, the trypanosomal occluding loop is more rigid. The displaced loop segment comprises only 4 residues rather than the 10 observed in human CatB (13). This results in a narrower exposed S2′ subsite ~8.5 Å wide compared with ~11.9 Å for human CatB (supplementary text S5). In particular, the side chain of His194 is only slightly shifted compared with the closed loop conformation and still extends into the open cleft. Thus, His194 not only establishes steric constraints for the substrates but also provides a prominent polar anchor in the otherwise largely hydrophobic S2′ and S1′ subsites that are highly conserved between trypanosome and human CatB (fig. S9). In human CatB, the larger exposed S2′ subsite in the open loop conformation is less restricted by the corresponding His189 residue. This suggests that smaller hydrophobic substituents could target the prime site (S1′ and S2′ positions) in TbCatB, which is also supported by the propeptide structures: The bulky Phe residue that sticks into the S2′ subsite of human CatB is replaced by the smaller Met of the T. brucei propeptide.
Fig. 3.
Occluding loop conformations of mature and propeptide-inhibited TbCatB. (A) Surface representation of mature TbCatB (11) showing the occluding loop (rigid part, beige; flexible part, red) in the closed conformation. A loop segment blocks the S2′ site and part of the S1′ site of the substrate-binding cleft. (B) Propeptide binding (green) shifts the flexible occluding loop segment (red) into an open conformation, which exposes the entire prime subsite. The insets compare the Tb (gray) and human (cyan) surface representations of mature and propeptide bound CatB. Almost superimposable Cα chains of the human (blue) and Tb (beige/red) occluding loops indicate a conservation of the closed loop conformation in the mature enzymes (A), whereas the open conformations show significant differences (B). Four H bonds maintained during conformational transition restricted the flexible loop segment to four residues (red) in TbCatB, which opens a crevice of ~8.5 Å. In contrast, only one H bond is maintained in the open loop conformation of human procathepsin B (13). Thus, the mobile segment of the human occluding loop (blue) comprises 10 residues and exposes an enlarged occluding loop crevice (supplementary text S5). Residues are labeled according to TbCatB, referring to the corresponding human CatB numbering in parentheses, if applicable.
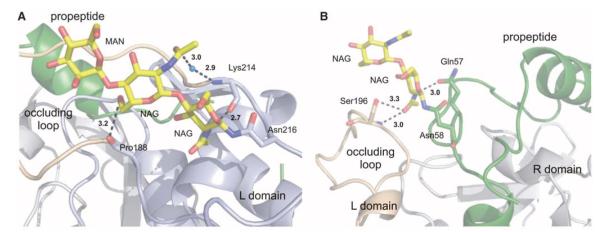
The occluding loop conformation is further stabilized by two carbohydrate structures identified in the TbCatB complex, as shown in Fig. 4. The enzyme carbohydrate chain interacts with both strands of the occluding loop at the loop termini (Fig. 4A), which supports the increased loop rigidity in TbCatB mentioned above. The propeptide carbohydrate connects the tip of the open occluding loop and stabilizes the open conformation (Fig. 4B). Although N-linked oligosaccharide substitution has been detected in human procathepsin B, the predicted glycosylation sites differ from our observations in TbCatB (28, 29). Therefore, it is unlikely that the occluding loop is stabilized in a similar way in the human case (supplementary text S6). Differential glycosylation between the human and T. brucei precursors along with the differences in the occluding loop conformation could be exploited for synthetic parasite-specific inhibition.
Fig. 4.
Glycosylation of the TbCatB-propeptide complex. (A) Enzyme carbohydrate structure comprising two NAG and one MAN residue (yellow) N-linked to Asn216 C-terminal of the occluding loop (beige). The carbohydrate structure connects both occluding loop strands by two direct and one water-bridged H bond (black dashed lines). (B) Propeptide glycosylation site comprising two NAG units (yellow) at Asn58 within the kinked region of the propeptide (green). The propeptide carbohydrate structure forms an H bond to Gln57 of the propeptide and two H bonds to Ser196 at the tip of the occluding loop (beige), which stabilize its open conformation. Color codes correspond to Fig. 1C.
As illustrated by the room-temperature glycosylated TbCatB-propeptide structure determined here, the combination of in vivo grown microcrystals with the diffraction-before-destruction technique of x-ray FELs provides a compelling path to obtain macromolecular structures from challenging samples. This methodology could vastly speed up structure determination by removing the need for large well-diffracting crystals and providing a suitable amount of crystals of posttranslationally modified proteins, in their biologically functional form.
Supplementary Material
Acknowledgments
Experiments were carried out in February 2011 at the LCLS, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. This work was supported by the following agencies: the German Federal Ministry for Education and Research (grants 01KX0806 and 01KX0807), the Hamburg Ministry of Science and Research and Joachim Herz Stiftung as part of the Hamburg Initiative for Excellence in Research and the Hamburg School for Structure and Dynamics in Infection (SDI), the Deutsche Forschungsgemeinschaft (DFG) Cluster of Excellence “Inflammation at interfaces” (EXC 306), the DFG, the Landesgraduiertenförderung Baden-Württemberg, the Max Planck Society, the Swedish Research Council, the Swedish Strategic Research Foundation, the Swedish Foundation for International Cooperation in Research and Higher Education, the U.S. Department of Energy Office of Basic Energy Sciences through PULSE Institute at SLAC, the U.S. Department of Energy through Lawrence Livermore National Laboratory under the contract DE-AC52-07NA27344 and supported by the University of California Office of the President Lab Fee Program (award no. 118036), the NSF (award MCB-1021557 and MCB-1120997), and the NIH (award 1R01GM095583).
Footnotes
Author contributions: H.N.C., J.C.H.S., S.B., P.F., I.S., M.D., L.R., and C.B. conceived the experiment, which was designed with A.B., R.A.K., J.C.H.S., D.P.D., U.W., R.B.D., M.J.B., R.L.S., and H.F.; R.K., S.M., and T.B. performed the in vivo crystallization experiments under the supervision of M.D.; FEL samples were prepared and characterized by F.S., K.N., L.R, and D.R. under the supervision of C.B. and H.N.C.; SFX experiments were carried out by K.N., L.R., H.N.C., D.P.D., F.S., M.L., T.A.W., A.A., M.J.B., U.W., A.B., L.G., S. Bajt, R.A.K., R.B.D., R.L.S., L.L., D.A., L.C.J., C.C., R.N., G.K., C.K., P.F., D.W., I.G., R.F., T.C., N.A.Z., N.T., M.S.H., M.F., J.S., S.B., M.M., M.M.S., and G.J.W. Beamline setup was done by S.B., G.J.W., and M.M. The development and operation of the sample delivery system was performed by R.B.D., D.P.D., U.W., R.L.S, L.L., J.S., and J.C.H.S.; K.N., T.A.W., R.A.K., A.A., A.V.M., L.L., S.K., T.R.M.B., I.S., and H.N.C. analyzed the data. K.N., L.R., and C.B. performed molecular replacement, refined the structure, and calculated the electron density maps. The manuscript was prepared by L.R., C.B., K.N., M.D, I.S., A.B., and H.N.C. with discussions and improvements from all authors. The structure factors and coordinates have been deposited with the Protein Data Bank (accession code 4HWY). The Arizona Board of Regents, acting for and on behalf of Arizona State University and in conjunction with R.B.D., U.W., D.P.D., and J.C.H.S., has filed U.S. and international patent applications on the nozzle technology applied herein. One of the patents was granted on 25 September 2012, U.S. 8,272,576 “Gas dynamic virtual nozzle for generation of microscopic droplet streams.”
Supplementary Materials www.sciencemag.org/cgi/content/full/science.1229663/DC1
References and Notes
- 1.Frearson JA, et al. Nature. 2010;464:728. doi: 10.1038/nature08893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett MP, Boykin DW, Brun R, Tidwell RR. Br. J. Pharmacol. 2007;152:1155. doi: 10.1038/sj.bjp.0707354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alsford S, et al. Nature. 2012;482:232. doi: 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fairlamb AH. Trends Parasitol. 2003;19:488. doi: 10.1016/j.pt.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Bryant C, et al. Bioorg. Med. Chem. Lett. 2009;19:6218. doi: 10.1016/j.bmcl.2009.08.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdulla MH, et al. PLoS Negl. Trop. Dis. 2008;2:e298. doi: 10.1371/journal.pntd.0000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lecaille F, Kaleta J, Brömme D. Chem. Rev. 2002;102:4459. doi: 10.1021/cr0101656. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto Y, Kurata M, Watabe S, Murakami R, Takahashi SY. Curr. Protein Pept. Sci. 2002;3:231. doi: 10.2174/1389203024605331. [DOI] [PubMed] [Google Scholar]
- 9.Dubin G. Cell. Mol. Life Sci. 2005;62:653. doi: 10.1007/s00018-004-4445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerr ID, Wu P, Marion-Tsukamaki R, Mackey ZB, Brinen LS. PLoS Negl. Trop. Dis. 2010;4:e701. doi: 10.1371/journal.pntd.0000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koopmann R, et al. Nat. Methods. 2012;9:259. doi: 10.1038/nmeth.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomoo K. Curr. Top. Med. Chem. 2010;10:696. doi: 10.2174/156802610791113441. [DOI] [PubMed] [Google Scholar]
- 13.Podobnik M, Kuhelj R, Turk V, Turk D. J. Mol. Biol. 1997;271:774. doi: 10.1006/jmbi.1997.1218. [DOI] [PubMed] [Google Scholar]
- 14.Bill RM, et al. Nat. Biotechnol. 2011;29:335. doi: 10.1038/nbt.1833. [DOI] [PubMed] [Google Scholar]
- 15.Southworth-Davies RJ, Medina MA, Carmichael I, Garman EF. Structure. 2007;15:1531. doi: 10.1016/j.str.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Riekel C. J. Synchrotron Radiat. 2004;11:4. doi: 10.1107/s090904950302541x. [DOI] [PubMed] [Google Scholar]
- 17.Chapman HN, et al. Nature. 2011;470:73. doi: 10.1038/nature09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barty A, et al. Nat. Photonics. 2012;6:35. doi: 10.1038/nphoton.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boutet S, Williams GJ. New J. Phys. 2010;12:035024. [Google Scholar]
- 20.Boutet S, et al. Science. 2012;337:362. [Google Scholar]
- 21.Weierstall U, Spence JC, Doak RB. Rev. Sci. Instrum. 2012;83:035108. doi: 10.1063/1.3693040. [DOI] [PubMed] [Google Scholar]
- 22.Kirian RA, et al. Opt. Express. 2010;18:5713. doi: 10.1364/OE.18.005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White TA, et al. J. Appl. Cryst. 2012;45:335. [Google Scholar]
- 24.McGrath ME. Annu. Rev. Biophys. Biomol. Struct. 1999;28:181. doi: 10.1146/annurev.biophys.28.1.181. [DOI] [PubMed] [Google Scholar]
- 25.Chagas JR, Ferrer-Di Martino M, Gauthier F, Lalmanach G. FEBS Lett. 1996;392:233. doi: 10.1016/0014-5793(96)00822-8. [DOI] [PubMed] [Google Scholar]
- 26.Illy C, et al. J. Biol. Chem. 1997;272:1197. doi: 10.1074/jbc.272.2.1197. [DOI] [PubMed] [Google Scholar]
- 27.Schechter I, Berger A. Biochem. Biophys. Res. Commun. 1967;27:157. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 28.Quraishi O, et al. Biochemistry. 1999;38:5017. doi: 10.1021/bi981950o. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Plouffe C, Ménard R, Storer AC. FEBS Lett. 1996;393:24. doi: 10.1016/0014-5793(96)00847-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.