Abstract
Nanomaterials have been increasingly employed as drug(s)-incorporated vectors for drug delivery due to their potential of maximizing therapeutic efficacy while minimizing systemic side effects. However, there have been two main challenges for these vectors: (i) the existing synthetic approaches are cumbersome and incapable of achieving precise control of their structural properties, which will affect their biodistribution and therapeutic efficacies, and (ii) lack of an early checkpoint to quickly predict which drug(s)-incorporated vectors exhibit optimal therapeutic outcomes. In this work, we utilized a new rational developmental approach to rapidly screen nanoparticle (NP)-based cancer therapeutic agents containing a built-in companion diagnostic utility for optimal therapeutic efficacy. The approach leverages the advantages of a self-assembly synthetic method for preparation of two different sizes of drug-incorporated supramolecular nanoparticles (SNPs), and a positron emission tomography (PET) imaging-based biodistribution study to quickly evaluate the accumulation of SNPs at a tumor site in vivo and select the favorable SNPs for in vivo therapeutic study. Finally, the enhanced in vivo anti-tumor efficacy of the selected SNPs was validated by tumor reduction/inhibition studies. We foresee our rational developmental approach providing a general strategy in the search of optimal therapeutic agents among the diversity of NP-based therapeutic agents.
Keywords: Supramolecular assembly, Nanoparticles, Drug delivery, Positron emission tomography, Cancer therapeutics
1. Introduction
Nanoparticles (NPs) have been regarded as promising vectors for controlled delivery of anti-cancer drug(s), providing an emerging therapeutic strategy with enhanced anti-tumor efficacy and reduced systemic side effects [1–10]. However, even after decades of development, there are few successful examples that have reached clinical usage [2,11]. One of the major bottlenecks can be attributed to the expensive, time-consuming and labor-intensive pipeline adopted for the development of drug(s)-incorporated NPs, by which numerous optimization/evaluation cycles, including molecular design, multistep syntheses and in vitro/in vivo assays are repeated. Such a developmental pipeline does not guarantee generating drug(s)-incorporated NPs that exhibit improved therapeutic efficacy in clinic.
Herein, we utilize a new rational developmental approach to rapidly screen NP-based cancer therapeutic agents containing a built-in companion diagnostic utility for optimal therapeutic efficacy. The approach leverages the advantages of a self-assembly synthetic method for preparation of supramolecular nanoparticles (SNPs), and a positron emission tomography (PET) imaging-based biodistribution study to quickly evaluate the accumulation of SNPs at a tumor site in vivo. The self-assembly synthetic method enables a convenient, flexible and modular production of a small collection of drug-incorporated NPs with precisely controlled sizes ranging from 37 to 104 nm, below the threshold of NP size (200 nm) that allows the efficient extravasation of NPs into the leaky tumor vasculatures and accumulation of the NPs in the tumor tissues via enhanced permeability and retention (EPR) effect [12-17]. Subsequently, PET imaging-based biodistribution studies are employed to correlate the sizes of drug-incorporated NPs with their delivery performances. Thus, a specific size of drug-incorporated NPs that exhibit optimal accumulation and prolonged retention in tumor tissue can be readily identified, resulting in potentially the lead modality with improved therapeutic efficacy and reduced side effect that is subject to comprehensive pre-clinical studies.
Previously, we demonstrated a convenient, flexible, and modular self-assembly synthetic method for crafting SNPs as outstanding delivery vectors for highly efficient delivery of genes,[18,19] proteins [20] and inorganic nanoparticles [21]. Given the versatile utility of such a self-assembly synthetic method, we attempted to explore the use of SNP-based vectors for delivering an anti-cancer drug. In this case, camptothecin (CPT), a cytotoxic quinoline alkaloid that inhibits the topoisomerase I, was selected for the proof-of-concept demonstration. On the other hand, we have already demonstrated the incorporation of radioisotopes and a contrast agent into SNPs for PET [22] and magnetic resonance (MR) imaging, [23] respectively. PET imaging [24–26] is a sensitive, non-invasive technology that can be utilized to determine the biodistribution profiles of small molecules, polymers or NPs at a whole body level. The resulting biodistribution information [27] can be utilized to facilitate the implementation of pre-clinical studies. Moreover, once the drug-incorporated NPs with optimal therapeutic efficacy are identified, they will contain a built-in companion diagnostic utility that can be readily utilized if desired.
2. Materials and methods
2.1. General
Camptothecin (CPT) and other chemicals were purchased from Sigma–Aldrich (St. Louis, MO) and used as received without further purification unless otherwise noted. 1-Adamantanamine (Ad) hydrochloride and β-cyclodextrin (β-CD) were purchased from TCI America (San Francisco, CA). N-hydroxysuccinimide (SCM) and maleimido (MAL) hetero-functionalized poly(ethylene glycol) (SCM-PEG-MAL, MW = 5 kD) were obtained from NANOCS Inc (New York, NY). CD-grafted branched poly(ethylenimine) (CD-PEI), Ad-grafted poly(ethylene glycol) (Ad-PEG) and DOTA-grafted CD-PEI (CD-PEI-DOTA) were prepared via the method previously reported by our group [22,23]. Phosphate-Buffered Saline (PBS), Dulbecco’s Modified Eagle Medium (DMEM), Eagle’s Minimum Essential Medium (EMEM), and penicillin/streptomycin were obtained from Invitrogen (Carlsbad, CA). MCF7 breast cancer cell line and Lewis Lung Carcinoma (LLC) cell line were purchased from American Type Culture Collection (Manassas, VA). Fetal Bovine Serum (FBS) was obtained from Lonza Walkerrsville Inc (Walkerrsville, MD). 96 well BD Falcon culture plates were purchased from Fisher Scientific. CellTiter-Blue® Cell Viability Assay was purchased from Promega Corporation (Madison, WI). Human serum was purchased from Sigma–Aldrich.
The molecular weight of the PBLG were determined on a gel permeation chromatograph (GPC, also called size-exclusion chromatography (SEC)) equipped with an isocratic pump (Model 1100, Agilent Technology, Santa Clara, CA), a DAWN HELEOS 18-angle laser light scattering detector (Wyatt Technology, Santa Barbara, CA), and an Optilab rEX refractive index detector (Wyatt Technology, Santa Barbara, CA). The wavelength of the HELEOS detector was set at 658 nm. The size-exclusion columns used were serially connected on the GPC (Phenogel columns 100 Å, 500 Å, 103 Å and 104 Å, 5 μm, 300 × 7.8 mm, Phenomenex, Torrance, CA). DMF (HPLC grade) was used as the mobile phase for GPC. HPLC analysis was performed on a Beckman Gold system (Beckman Coulter, Fullerton, CA) equipped with a 126P solvent module, a System Gold 128 UV detector and an analytical C18 column (Luna C18, 250 × 4.6 mm, 5 μ, Phenomenex, Torrance, CA). NMR analyses were conducted on a Varian U500, VXR500 or UI500NB (500 MHz).
2.2. Synthesis of Poly(l-glutamic acid) (PGA)
Poly(γ-benzyl-l-glutamate) (PBLG50) was synthesized according to the procedure previously published. [28,29] The N-terminus of PBLG was capped by a carbobenzyloxy (Cbz) group. The Mn was 12,600 g/mol and the MW distributions (MWD = Mw/Mn) was 1.05 as determined by GPC (Fig. S1 in Supporting Information). The deprotection of PBLG50 was performed using standard HBr condition as described below: PBLG50 (500 mg, 2.28 mmol glutamate residues) was dissolved in TFA (15 mL) in an ice bath. HBr (33 wt% in HOAc, 4 mL) was added dropwise into stirred solution. The reaction mixture was stirred in the ice bath for an additional 2 h and then poured into cold ether (60 mL) in two 50-mL centrifuge tubes. The polymer precipitate was collected by centrifuge and washed with ether (30 mL × 3). The polymer was dried under vacuum to give the crude product. The polymer was dissolved in NaOH (2 M × 10 mL) and was stirred at room temperature (rt) overnight. The clear solution was acidified by 2 M HCl to pH 2. The product was purified by dialysis against DI water and dried by lyophilization to give a white powder. 1H NMR (D2O, 500 MHz): δ 4.86 (1H), 2.68 (2H), 2.34 (1H), 2.19 (1H).
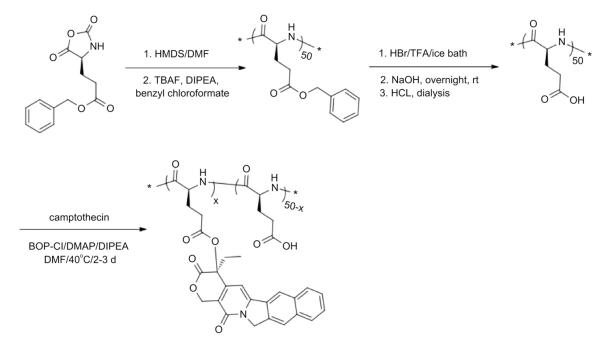
2.3. Synthesis of Poly(l-glutamic acid)-g-camptothecin (CPT-grafted PGA, CPT-PGA)
The synthesis of CPT-PGA (Scheme 2) was performed with a slightly modified protocol as reported before. [30] Bis(2-oxo-3-oxazolidinyl)phosphonic chloride (BOP-Cl, 175 mg, 0.68 mmol), DMAP (170 mg, 1.4 mmol), and diisopropyletheylamine (74 mg, 0.57 mmol) were added under nitrogen to a suspension of CPT (131 mg, 0.38 mmol) and dry PGA50 (310 mg, 2.4 mmol Glu) in anhydrous DMF (20 mL) cooled in an ice bath. The mixture was warmed to rt, stirred at 40 °C for 2 days, and cooled down in an ice bath. A 10% aqueous NaCl solution (40 mL) was added slowly to the reaction mixture under stirring. The resulting suspension was acidified to pH 2.5 by hydrochloric acid (0.5 M). The mixture was allowed to stir at rt for 1 h. The precipitate was filtered, washed with water (4 × 30 mL), dried under vacuum (<1 mm, 12 h), and ground to a powder. The precipitate was suspended in 2% MeOH-CH2Cl2 (10 mL), stirred for 3 h, and filtered. This process was repeated 4 times to effect complete removal of free CPT. The product was then dried under vacuum to give 220 mg of CPT-PGA (yield 56%). Loading of CPT was 20–22 wt% determined by 1H NMR (TFA-d, 500 MHz). The 1H NMR data of the obtained CPT-PGA was similar as reported by the literature. (Fig. S2 in Supporting Information).
Scheme 2.
Schematic representation of the synthesis of poly(l-glutamic acid)-g-camptothecin (CPT-PGA).
2.4. Synthesis of PGA⊂SNPs (drug-free vectors)
To a 200-μL solution of Ad-PEG (10 mg, 50 mg/mL), 10-μL DMSO solution of Ad-PAMAM (24 mg/mL) was slowly injected under vigorous stirring. Followed by addition of 120-μL CD-PEI (10.44 mg, 87 mg/mL) into the mixture and incubating at rt for 20 min. After the incubation, the mixture was slowly added into 700-μL CPT-PGA (10 mg/mL) solution and heated to 50 °C for an additional 20 min. PGA⊂SNPs with the size of 35 ± 5 nm were obtained after the solution cooled down.
2.5. Synthesis of CPT-PGA encapsulated SNPs (CPT-PGA⊂SNPs)
To a 200-μL solution of Ad-PEG (10 mg, 50 mg/mL), 10-μL DMSO solution of Ad-PAMAM with various concentrations (22 and 44 mg/mL) was slowly injected under vigorous stirring. Followed by addition of 120-μL CD-PEI (10.44 mg, 87 mg/mL) into the mixture and incubating at rt for 20 min. After the incubation, the mixture was slowly added into 700-μL CPT-PGA (10 mg/mL) solution and heated to 50 °C for an additional 20 min. Two different sizes of CPT-PGA⊂SNPs (37-nm and 104-nm CPT-PGA⊂SNPs) were obtained after the solution cooled down.
2.6. Synthesis of DOTA-grafted CPT-PGA⊂SNPs
To a 200-μL solution of Ad-PEG (10 mg, 50 mg/mL), 10-μL DMSO solution of Ad-PAMAM with two different concentrations (22, 44 mg/mL) was slowly injected under vigorous stirring followed by addition of 120-μL CD-PEI-DOTA (10 mg, 87 mg/mL) into the mixture. The mixture was incubated at rt for 20 min. After the incubation, the mixture was slowly added into 700-μL CPT-PGA (10 mg/mL) solution and heated to 50 °C for another 20 min. The resulting DOTA-grafted CPT-PGA⊂SNPs (37-nm and 104-nm, respectively) were obtained when the solution was cooled to rt.
2.7. 64Cu labeling of DOTA-grafted CPT-PGA⊂SNPs
All liquids were pretreated with Chelex-100 (Bio-Rad, Herchules, CA) to remove trace amount of metal contaminants. The 64Cu chloride (Washington university at St. Louis) was mixed with NH4OAc buffer (pH 5.5, I = 0.1 M); DOTA-grafted CPT-PGA⊂SNPs in 800-fold excess were added to the solution. The mixture was incubated for 1 h at 60 °C. The 64Cu-labeled DOTA-grafted CPT-PGA⊂SNPs product was purified by a molecular weight cut off filter (Centricon YM10, Billerica, MA) at 10,000 × g for 10 min. The labeling yield (>95%) was determined by measuring the radioactivity in the filter, the filtrate and the retaintate, respectively. The 64Cu-labeled DOTA-grafted CPT-PGA⊂SNPs were re-suspended in saline for in vivo injections.
2.8. Dynamic light scattering (DLS)
DLS experiments were performed with a Zetasizer Nano instrument (Malvern Instruments Ltd., United Kingdom) equipped with a 10-mW helium-neon laser (λ = 632.8 nm) and thermoelectric temperature controller. Measurements were taken at a 90° scattering angle.
2.9. Transmission electron microscope (TEM)
The morphology and sizes of CPT-PGA⊂SNPs were examined on a Philips CM 120 transmission electron microscope (TEM), operating at an acceleration voltage of 120 kV. The TEM samples were prepared by drop-coating 2-μL of CPT-PGA⊂SNPs solutions onto carbon-coated copper grids. Excess amounts of droplets were removed with filter paper after 45 s. Subsequently, the surface-deposited CPT-PGA⊂SNPs were negatively stained with 2% uranyl acetate for 45 s before TEM studies.
2.10. Zeta potential (ζ) measurements
Zeta potentials of CPT-PGA⊂SNPs were determined by photon correlation spectroscopy using a Zetasizer Nano instrument (Malvern Instruments, Malvern, Worcestershire, UK). The measurements were performed at 25 °C with a detection angle of 90°, and the raw data were subsequently correlated to Z average mean size using a cumulative analysis by the Zetasizer software package.
2.11. Drug encapsulation efficiency
Free CPT was removed from CPT-PGA⊂SNPs by centrifugation of CPT-PGA⊂SNPs solution at 1300 rpm for 30 min using centrifugal filter devices (3000 NMWL). After recovering the filtrate containing free CPT, CPT concentration was analyzed by ultraviolet absorption at a wavelength of 370 nm. The measurements were performed in triplicate. The amount of the CPT encapsulated in the SNPs was then calculated by the total loading amount of CPT subtracts the free CPT in the filtrate.
2.12. Time-dependent stability study
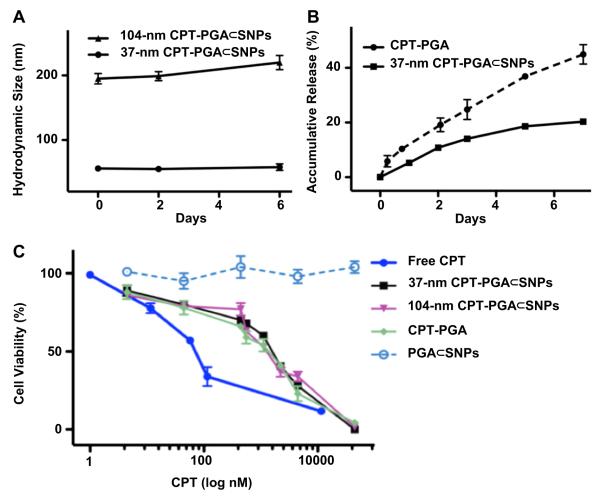
To ensure the in vivo stability of the resulting CPT-PGA⊂SNPs, it is critical to examine their size variation under a physiological ionic strength. Following the procedure described above, the 37-nm and 104-nm CPT-PGA⊂SNPs were prepared in PBS solutions (pH = 7.2, containing 1.5 mM KH2PO4, 155 mM NaCl and 2.7 mM Na2HPO4). After mixing the three molecular building blocks in their respective ratios, we employed real-time DLS measurements to monitor the hydrodynamic size variation of the 37-nm and 104-nm CPT-PGA⊂SNPs at different times. The sizes of CPT-PGA⊂SNPs were recorded up to 6 days.
2.13. Drug release profile
CPT-PGA (0.249 mg/mL) or 37-nm CPT-PGA⊂SNPs (0.977 mg/mL) was dispersed in 50% human serum (human serum:1 × PBS = 1:1, v/v) and equally distributed to 20 vials with 1 mL solution per vial, and then incubated at 37 °C. At selected time intervals, one selected vial of each group was taken out of the incubator. The solution was mixed with an equal volume of methanol (1 mL) and centrifuged at 15,000 rpm for 10 min. The supernatant (1 mL) was transferred to an eppendorf tube without disturbing the precipitates and brought to pH 2 with phosphoric acid (85%, 100 μL). The resulting solution was directly injected into an HPLC equipped with an analytical C18 column. A mixture of acetonitrile and water (containing 0.1% TFA) at a volume ratio of 1:3 was used as the mobile phase. The flow rate was set at 1 mL/min. The area of the HPLC peak of the released CPT (λabs = 370 nm) was intergraded for the quantification of CPT as compared to a standard curve of free CPT prepared separately. The accumulative release of CPT is shown in Fig. 2B.
Fig. 2.
(A) Time-dependent studies on the stability of 37-nm and 104-nm CPT-PGA⊂SNPs in 1 × PBS. (B) Release kinetics of CPT-PGA and 37-nm CPT-PGA⊂SNPs in 50% human serum (human serum:1 × PBS = 1:1, v/v). Released CPT was quantified by HPLC. (C) Dose-dependent cell viability studies of two different sizes of CPT-PGA⊂SNPs along with controls.
2.14. In vitro cell viability
Cell viability was measured by the MTT assay. After incubating MCF-7 breast cancer cell line with 100-μL 37-nm CPT-PGA⊂SNPs, 104-nm CPT-PGA⊂SNPs, free CPT, CPT-PGA and PGA⊂SNPs with respective concentrations in 96 well-plates for 48 h, the solutions were removed and washed with PBS three times. Fresh medium containing 20-μL CellTiter-Blue reagent was added to each well, followed by 3 h incubation at 37 °C. The cell viability results were quantified using fluorescent plate-reader.
2.15. Micro-PET imaging
C57BL/6 mice were purchased from DLAM Breeding Colony Services (Los Angeles, CA). All animal manipulations were performed with sterile technique and were approved by the University of California at Los Angeles Animal Research Committee (ARC protocol# 2006-135-12). C57BL/6 mice were injected subcutaneously in the right flank with LLC cells suspended in a 1:1 mixture of PBS buffer and matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Prior to use in tumor induction, LLC cells were cultured in DMEM medium. Tumors were grown for 9 days and then 100-μL 64Cu (150 μCi) labeled DOTA-grafted CPT-PGA⊂SNPs solution was injected via tail vein while the animal was anesthetized. Micro-PET imaging of the mice occurred 24 h post injection and was performed with a micro-PET FOCUS 220 PET scanner (Siemens, Malvern, PA). The mice were anesthetized by using 1.5–2% isoflurane in a heated (30 °C) induction chamber 15 min prior to imaging. The mice were then transferred to a heated isolation/imaging chamber for imaging. Static micro-PET scans (10 min) were then acquired. To determine the supramolecular nanoparticle concentration in various tissues, ellipsoid regions of interest were placed in the region that exhibited the highest 64Cu activity as determined by visual inspection using the Amide software. Relative intensity is expressed as the percentage of activity in the organ of interest to that of the sum of all activity in the organs listed.
2.16. In vivo study
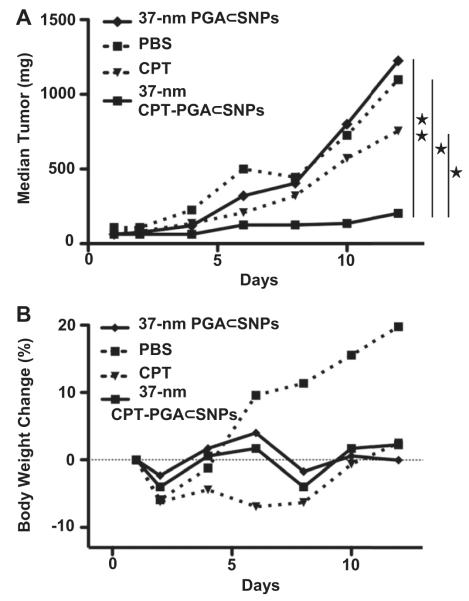
C57BL/6 mice (female) were purchased from Charles River, USA. Feed and water was available ad libitum. The animal study protocol was reviewed and approved by the Animal Care and Use Committee of University of Illinois at Urbana-Champaign. Female C57Bl/6 mice, 4–5 week old, were anesthetized, shaved, and prepared for implantation of the tumor cells. LLC cells were collected from culture, and 3 × 105 cells suspended in a 1:1 mixture of PBS buffer and matrigel were then injected subcutaneously into right flank of a mouse. After 6 days when tumors reached around 60–90 mm3 in size, mice were divided into 4 groups of five mice, minimizing weight and tumor size difference. Tumor-bearing mice were treated by intravenous injection of PBS, PGA⊂SNPs, CPT-PGA⊂SNPs (13.6 mg/kg CPT) or intraperitoneal injection of CPT (13.6 mg/kg). Three doses were administrated with 5-day interval, i.e. at day 1, day 6 and day 11, respectively (except for CPTgroup, which only received a single dose at day 1 due to emerging toxicity [31]). After injections, the animals were monitored closely, and measurements of the tumor size and body weight for each animal were performed at regular intervals using calipers without knowledge of which injection each animal had received. The tumor volume for each time point was calculated according to the formula, (length) × (width)2/2, where the long axis is the length, the short axis is the width. Tumor density is assumed as 1 mg/mm3. If body weight loss is beyond 20% of pre-dosing weight, the animals were euthanized. When the tumor load reached 1500 mm3 or the animal had become moribund, the mouse was sacrificed. The statistical analysis was undertaken using a Student’s t-test (two-tailed), and p-values < 0.05 were considered statistically significant, p < 0.01 were considered highly statistically significant. Median tumor growth curves prepared for each group depicted the median tumor size as a function of time (Fig. 4A).
Fig. 4.
(A) In vivo anti-tumor efficacy studies of 37-nm CPT-PGA⊂SNPs (13.6 mg CPT equivalent/kg) along with controls, i.e., free CPT (13.6 mg/kg), PBS and PGA⊂SNPs (equivalent to the amount of SNPs in the CPT-PGA⊂SNPs group). LLC tumor-bearing C57Bl/6 mice were treated with different groups via intravenous injection at day 1, 6 and 11 (except for CPT group, which was only treated once at day 1 via intraperitoneal injection). 37-nm CPT-PGA⊂SNPs showed delayed tumor growth from day 6 compared to PGA⊂SNPs group and also significantly outperformed CPT group from day 4 (*p < 0.05; **p < 0.01; Student’s t-test). (B) Body weight changes of the four groups over the course of treatments.
3. Results and discussion
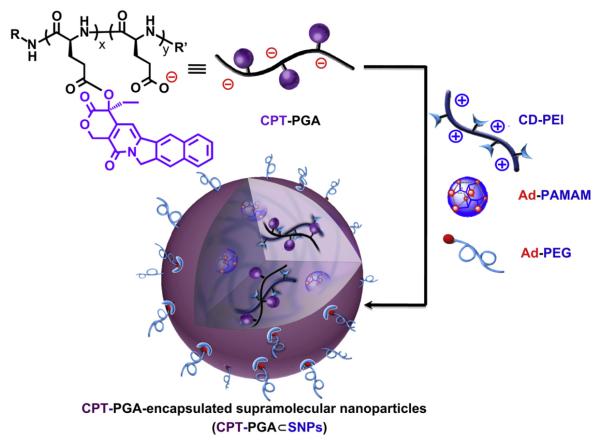
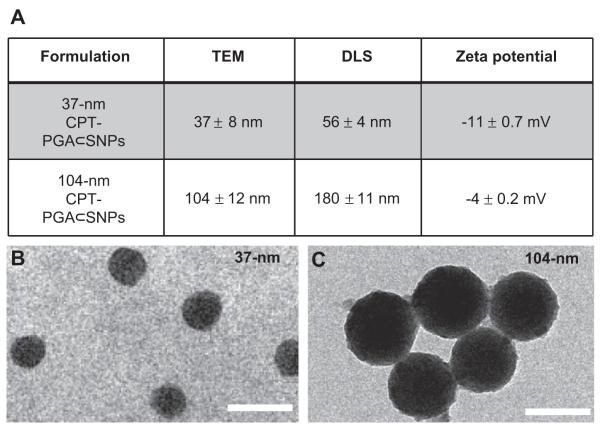
Similar to the self-assembly preparation of DNA encapsulated SNPs, [18,19] which used the coulombic interactions between the negatively charged DNA plasmid with the positively charged SNP vector, 5 KD anionic poly(l-glutamic acid) (PGA) [28,29] was employed as a carrier to covalently link with CPT molecules, enabling encapsulation into SNP vectors. Approximately five CPT molecules were conjugated to each PGA polymer chain (via ester bond formation) to give CPT-grafted PGA, denoted as CPT-PGA. [30] It is noteworthy that the connecting ester bonds can be degraded via esterase-mediated hydrolysis, which allows controlled release of CPT under physiological conditions. The encapsulation of CPT-PGA into SNP vectors to generate CPT-PGA encapsulated SNPs (CPT-PGA⊂SNPs) can be accomplished (Scheme 1) by simply mixing the drug conjugated polymer, CPT-PGA (Scheme 2), with the other two SNP building blocks (CD-PEI: CD-grafted branched polyethylenimine and Ad-PAMAM: Ad-grafted polyamidoamine dendrimer), as well as a solvation ligand (Ad-PEG: Ad-grafted poly(ethylene glycol)). By altering the mixing ratios between the two SNP building blocks (Ad-PAMAM/CD-PEI = 0.125 and 0.25), two different sizes (37 and 104 nm) of CPT-PGA⊂SNPs can be obtained with narrow size distributions after annealing at 50 °C for 20 min. Both transmission electron microscopy (TEM) and dynamic light scattering (DLS) measurements were performed to characterize the sizes and morphologies of the resulting CPT-PGA⊂SNPs and the results are summarized in Fig. 1A. The hydrodynamic sizes obtained from DLS for the two different formulations were slightly larger than what was observed by TEM. These size characterizations show that the resulting CPT-PGA⊂SNPs possessed homogeneous size distributions (Fig. 1B and C). Moreover, other studies, including zeta potential (The zeta potentials of the resulting 37-nm CPT-PGA⊂SNPs and 104-nm CPT-PGA⊂SNPs are −11 ± 0.7 and −4 ± 0.2 mV, respectively.), drug encapsulation efficiency (The drug encapsulation efficiency for 37-nm and 104-nm CPT-PGA⊂SNPs are 90 ± 3% and 95 ± 2%, respectively.), stability (The results in Fig. 2A indicate that the resulting CPT-PGA⊂SNPs exhibit good stability in PBS solution under a physiological ionic strength up to 6 days.), drug release kinetics (The accumulative release of free CPT from CPT-PGA⊂SNPs was quantified by HPLC. The data in Fig. 2B point out that CPT-PGA⊂SNPs release 20% of CPT after 6 days without any associated burst release.) and in vitro viability treatments (The IC50 values, the concentration at which 50% inhibition of cellular growth occurs, were around 100 nM for free CPT and approximately 400 nM for CPT-PGA⊂SNPs and CPT-PGA (Fig. 2C). There were negligible difference among CPT-PGA and the two other sizes of CPT-PGA⊂SNPs, which is probably due to the slow release of the drug from polymer (Fig. 2B). Note that the cells treated with PGA⊂SNPs present high viability, indicating low cytotoxicity attributed to our SNPs.), of the resulting CPT-PGA⊂SNPs were also carried out and the details can be found in EXPERIMENTAL SECTION.
Scheme 1.
Schematic representations of the self-assembly synthetic method for the production of CPT-grafted PGA encapsulated Supramolecular NanoParticles (CPT-PGA⊂SNPs) from the respective molecular building blocks and CPT-PGA (camptothecin-grafted poly(l-glutamic acid)).
Fig. 1.
Two different sizes of CPT-PGA⊂SNPs were obtained by altering the mixing ratio between two building blocks (Ad-PAMAM and CD-PEI). (A) The characterizations of the resulting CPT-PGA⊂SNPs. (B and C) TEM images of 37-nm and 104-nm CPT-PGA⊂SNPs, respectively. Scale bar = 100 nm.
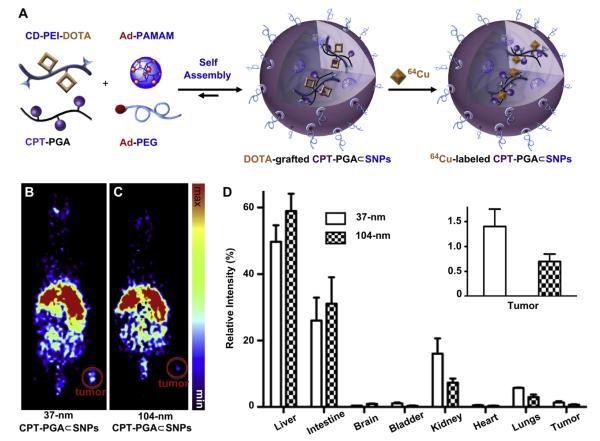
Different sizes of CPT-PGA⊂SNPs will have distinctive biodistribution patterns and varying therapeutic performances due to their differential EPR effects [12–14]. Instead of directly performing the in vivo efficacy studies of all of the two sizes of CPT-PGA⊂SNPs, which would require a significant quantity of CPT-PGA⊂SNPs and a large cohort of animals, we utilized micro-PET imaging technology to understand the biodistribution properties of the 37-nm and 104-nm CPT-PGA⊂SNPs. We anticipated that the resulting biodistribution data would correlate the sizes of CPT-PGA⊂SNPs with their delivery performances. Thus, the specific size of drug-incorporated NPs that exhibit preferential accumulation in the tumor can be identified for subsequent pre-clinical studies. The radio-labeling of SNP vectors and subsequent pre-clinical micro-PET imaging in mice have been previously demonstrated by our group [22]. Similarly, 64Cu-labeled CPT-PGA⊂SNPs (with the sizes of 37 and 104 nm) can be prepared (Fig. 3A) by incorporating CD-PEI-DOTA as one of the precursors (along with CD-PEI, Ad-PAMAM, Ad-PEG and CPT-PGA), followed by ratiometric mixing and labeling by 64Cu (see the Experimental Section). The 37-nm and 104-nm 64Cu-labeled CPT-PGA⊂SNPs were instantaneously injected into LLC tumor-bearing C57Bl/6 mice via tail vein intravenous (i.v.) administration. The respective biodistribution data (Fig. 3B–D) were acquired by micro-PET imaging technology 24 h post injection. Static micro-PET images of the two sizes of 64Cu-labeled CPT-PGA⊂SNPs (Fig. 3B and C, respectively) revealed that 37-nm 64Cu-labeled CPT-PGA⊂SNPs exhibited significantly enhanced tumor accumulation. Semi-quantitative biodistribution data (Fig. 3D and insert therein) of 64Cu-labeled CPT-PGA⊂SNPs in major organs were obtained by taking the average signal per voxel in the organ of interest and multiplying that value by the total volume of the organ via Amide software (see Experimental Section). The PET imaging-based biodistribution studies revealed that the 37-nm 64Cu-labeled CPT-PGA⊂SNPs exhibited 100% more accumulation in the tumor than that observed for the 104-nm 64Cu-labeled CPT-PGA⊂SNPs. We suggest that the preferential accumulation of 37-nm 64Cu-labeled CPT-PGA⊂SNPs in xenografted LLC tumors CPT-PGA⊂SNPs is due to their higher blood retention concentrations (see Supporting Information Fig. S3) that facilitated the EPR effect [12]-mediated accumulation of the SNPs in tumors.
Fig. 3.
(A) Schematic illustration of the incorporation of a radioisotope, i.e., 64Cu, into SNPs, resulting in 64Cu-labeled CPT-PGA⊂SNPs. Static micro-PET images of C57Bl/6 mice bearing Lewis lung carcinoma (LLC) tumor at 24 h post injection of (B) 37-nm and (C) 104-nm 64Cu-labeled CPT-PGA⊂SNPs. Tumor volumes are approximately 245 mm3. (D) In vivo biodistribution of 37-nm and 104-nm 64Cu-labeled CPT-PGA⊂SNPs after 24 h intravenous injection. 37-nm 64Cu-labeled CPT-PGA⊂SNPs had higher tumor-specific uptake than 104-nm 64Cu-labeled CPT-PGA⊂SNPs. The error bar corresponds to the standard deviation calculated by the Amide software.
Given the preferential tumor accumulation of the 37-nm 64Cu-labeled CPT-PGA⊂SNPs as compared to the larger SNPs, we next performed tumor reduction/inhibition studies using the 37-nm CPT-PGA⊂SNPs in C57/BL6 mice bearing subcutaneously implanted LLC tumors on the right flank. After the tumors developed to approximately 60–90 mm3, we performed comparative efficacy studies by dividing animals into four groups (N = 5) in a way to minimize weight and tumor size differences. The following regimens were administrated three times via i.v. injection on day 1, 6 and 11 including PBS, PGA⊂SNPs (equivalent to the amount of SNPs in the CPT-PGA⊂SNP group), 37-nm CPT-PGA⊂SNPs at 13.6 mg CPT equivalent/kg, while another group received a single dose of emulsified CPT at 13.6 mg CPT/kg through intraperitoneal injection (i.p.). CPT is very insoluble in aqueous solution and is acutely lethal when given to mice at such a high dose via i.v. due to embolization induced by the particulate matter in the drug suspension [31].Fig. 4A depicts the median tumor size for each group as a function of time, and clearly shows that the 37-nm CPT-PGA⊂SNPs had the best anti-tumor efficacy among all groups tested. As compared with PGA⊂SNPs (drug-free vectors), 37-nm CPT-PGA⊂SNPs started to show statistically significant efficacy in delaying tumor growth from day 6 (*p < 0.05 at day 6 and 8, Student’s t-test) and even higher significant inhibition effect at day 10 and 12 (**p < 0.01 at day 10 and 12). However, there was no statistical significance between the data points of PGA⊂SNPs group and PBS group indicating that the efficacy of the 37-nm CPT-PGA⊂SNPs was ascribed to the released CPT. The 37-nm CPT-PGA⊂SNPs also significantly outperformed CPT in this study with statistical significance from day 6 to day 12 (*p < 0.05). It is noteworthy that one of the five mice treated with 37-nm CPT-PGA⊂SNPs experienced complete tumor reduction on day 10. The final mean tumor burden of the 37-nm CPT-PGA⊂SNPs group was 265.5 ± 101.3 mm3 (mean ± SEM), which was smaller than any other group (*p < 0.05). On the other hand, no obvious body weight loss was observed for the group treated with the 37-nm CPT-PGA⊂SNPs at the tested dosage throughout the study (Fig. 4B), indicating there is no acute toxicity of the 37-nm CPT-PGA⊂SNPs. As a comparison, the CPT group showed emerging toxicity after a single dose with around 9% body weight loss at day 5 (data not shown). The results demonstrate that 37-nm CPT-PGA⊂SNPs group is the most efficacious group among all the groups with reduced toxicity compared to free drug. Presumably, the reason for the enhanced efficacy of 37-nm CPT-PGA⊂SNPs may be the preferential tumor accumulation due to EPR effect as demonstrated. After the SNPs reached the tumor tissues, the sustained release of CPT from CPT-PGA⊂SNPs (Fig. 2B) was ascribed to the extended inhibition on tumor growth.
4. Conclusion
We have successfully demonstrated a rational developmental approach that leverages the advantages of (i) a self-assembly synthetic method for preparation of SNP vectors and (ii) PET imaging technology to generate 37-nm CPT-PGA⊂SNPs with optimal therapeutic performance in tumor xenografted mouse model. The integration of the two technologies can lead to a breakthrough in the development of a new generation of cancer therapeutic agents. We foresee our work providing a general strategy in the search of optimal therapeutic agents among the diversity of NP-based therapeutic agents by utilizing imaging technology as a fast and early checkpoint prior to performing large-scale pre-clinical studies. In principle, this work can also be adopted for discovery of other NP-based vectors for delivery of various payloads (e.g., DNAs, RNAs, proteins or their combinations).
Supplementary Material
Acknowledgments
This research was supported by NIH R21 1R21EB008419 (HRT) and NIH 1R21CA152627 (JC).
Footnotes
Appendix. Supplementary material Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.biomaterials.2011.10.044.
References
- [1].Davis ME, Chen Z, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7(9):771–82. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- [2].Petros RA, DeSimone JM. Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov. 2010;9(8):615–27. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- [3].De M, Ghosh PS, Rotello VM. Applications of nanoparticles in biology. Adv Mater. 2008;20(22):4225–41. [Google Scholar]
- [4].Bae Y, Kataoka K. Intelligent polymeric micelles from functional poly(ethylene glycol)-poly(amino acid) block copolymers. Adv Drug Deliv Rev. 2009;61(10):768–84. doi: 10.1016/j.addr.2009.04.016. [DOI] [PubMed] [Google Scholar]
- [5].Wiradharma N, Zhang Y, Venkataraman S, Hedrick JL, Yang YY. Self-assembled polymer nanostructures for delivery of anticancer therapeutics. Nano Today. 4(4):302–317. [Google Scholar]
- [6].Duncan B, Kim C, Rotello VM. Gold nanoparticle platforms as drug and bio-macromolecule delivery systems. J Control Release. 2010;148(1):122–7. doi: 10.1016/j.jconrel.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Meng HA, Liong M, Xia TA, Li ZX, Ji ZX, Zink JI, et al. Engineered design of mesoporous silica nanoparticles to deliver doxorubicin and p-glycoprotein siRNA to overcome drug resistance in a cancer cell line. ACS Nano. 2010;4(8):4539–50. doi: 10.1021/nn100690m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nguyen HN, Wey SP, Juang JH, Sonaje K, Ho YC, Chuang EY, et al. The glucose-lowering potential of exendin-4 orally delivered via a pH-sensitive nano-particle vehicle and effects on subsequent insulin secretion in vivo. Biomaterials. 2011;32(10):2673–82. doi: 10.1016/j.biomaterials.2010.12.044. [DOI] [PubMed] [Google Scholar]
- [9].Sherlock SP, Tabakman SM, Xie LM, Dai HJ. Photothermally enhanced drug delivery by ultrasmall multifunctional FeCo/graphitic shell nanocrystals. ACS Nano. 2011;5(2):1505–12. doi: 10.1021/nn103415x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chow EK, Zhang X-Q, Chen M, Lam R, Robinson E, Huang H, et al. Nano-diamond therapeutic delivery agents mediate enhanced chemoresistant tumor treatment. Sci Transl Med. 2011;3:73. doi: 10.1126/scitranslmed.3001713. [DOI] [PubMed] [Google Scholar]
- [11].Peer D, Karp JM, Hong S, FaroKHzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat Nanotechnol. 2007;2(12):751–60. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- [12].Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J Control Release. 2000;65(1e2):271–84. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- [13].Weissig V, Whiteman KR, Torchilin VP. Accumulation of protein-loaded long-circulating micelles and liposomes in subcutaneous Lewis lung carcinoma in mice. Pharm Res-Dordr. 1998;15(10):1552–6. doi: 10.1023/a:1011951016118. [DOI] [PubMed] [Google Scholar]
- [14].Decuzzi P, Godin B, Tanaka T, Lee SY, Chiappini C, Liu X, et al. Size and shape effects in the biodistribution of intravascularly injected particles. J Control Release. 2010;141(3):320–7. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- [15].Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WCW. Mediating tumor targeting efficiency of nanoparticles through design. Nano Lett. 2009;9(5):1909–15. doi: 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]
- [16].Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer-chemotherapy – mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46(12):6387–92. [PubMed] [Google Scholar]
- [17].Goodman TT, Olive PL, Pun SH. Increased nanoparticle penetration in collagenase-treated multicellular spheroids. Int J Nanomedicine. 2007;2:265. [PMC free article] [PubMed] [Google Scholar]
- [18].Wang H, Liu K, Chen K-J, Lu Y, Wang S, Lin WY, et al. A rapid pathway toward a superb gene delivery system: programming structural and functional diversity into a supramolecular nanoparticle library. ACS Nano. 2010;4(10):6235–43. doi: 10.1021/nn101908e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang H, Chen K-J, Wang S, Ohashi M, Kamei K-I, Sun J, et al. A small library of DNA-encapsulated supramolecular nanoparticles for targeted gene delivery. Chem Commun. 2010;45:1851. doi: 10.1039/b923711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu Y, Wang H, Kamei K, Yan M, Chen K-J, Yuan Q, et al. Delivery of intact transcription factor using self-assembled supramolecular nanoparticles. Angew Chem Int Ed. 2011;50:3058. doi: 10.1002/anie.201005740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang S, Chen K-J, Wu T-H, Wang H, Lin W-Y, Ohashi M, et al. Photothermal effects of supramolecularly assembled gold nanoparticles for the targeted treatment of cancer cells. Angew Chem Int Ed. 2010;49(22):3777–81. doi: 10.1002/anie.201000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang H, Wang ST, Su H, Chen K-J, Armijo AL, Lin WY, et al. A supramolecular approach for preparation of size-controlled nanoparticles. Angew Chem Int Ed. 2009;48(24):4344–8. doi: 10.1002/anie.200900063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen K-J, Wolahan MS, Wang H, Hsu C-H, Chang H-W, Durazo A, et al. A small MRI contrast agent library of gadolinium(III)-encapsulated supramolecular nanoparticles for improved relaxivity and sensitivity. Biomaterials. 2011;32(8):2160–5. doi: 10.1016/j.biomaterials.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Phelps ME. Positron emission tomography provides molecular imaging of biological processes. Proc Nat Acad Sci. 2000;97(16):9226–33. doi: 10.1073/pnas.97.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Czernin J, Phelps ME. Positron emission tomography scanning: current and future applications. Annu Rev Med. 2002;53:89–112. doi: 10.1146/annurev.med.53.082901.104028. [DOI] [PubMed] [Google Scholar]
- [26].Phelps ME. PET: molecular imaging and its biological applications. Springer; New York: 2004. [Google Scholar]
- [27].Li SD, Huang L. Pharmacokinetics and biodistribution of nanoparticles. Mol Pharmaceut. 2008;5(4):496–504. doi: 10.1021/mp800049w. [DOI] [PubMed] [Google Scholar]
- [28].Lu H, Cheng JJ. Hexamethyldisilazane-mediated controlled polymerization of alpha-amino acid N-carboxyanhydrides. J Am Chem Soc. 2007;129(46):14114–5. doi: 10.1021/ja074961q. [DOI] [PubMed] [Google Scholar]
- [29].Lu H, Wang J, Bai Y, Lang J, Liu S, Lin Y, et al. Ionic polypeptides with unusual helical stability. Nat Commun. 2011;2:206. doi: 10.1038/ncomms1209. [DOI] [PubMed] [Google Scholar]
- [30].Bhatt RL, de Vries P, Tulinsky J, Bellamy G, Baker B, Singer JW, et al. Synthesis and in vivo antitumor activity of poly(L-glutamic acid) conjugates of 20(S)-camptothecin. J Med Chem. 2003;46(1):190–3. doi: 10.1021/jm020022r. [DOI] [PubMed] [Google Scholar]
- [31].Cheng J, Khin KT, Davis ME. Antitumor activity of beta-cyclodextrin polymer - camptothecin conjugates. Mol Pharmaceut. 2004;1(3):183–93. doi: 10.1021/mp049966y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.