Abstract
Numerous studies have supported the idea that de novo protein synthesis is critical for synaptic plasticity and normal long-term memory formation. This requirement for protein synthesis has been shown for several different types of fear memories, exists in multiple brain regions and circuits, and is necessary for different stages of memory creation and storage. However, evidence has recently begun to accumulate suggesting that protein degradation through the ubiquitin–proteasome system is an equally important regulator of memory formation. Here we review those recent findings on protein degradation and memory formation and stability and propose a model explaining how protein degradation may be contributing to various aspects of memory and synaptic plasticity. We conclude that protein degradation may be the major factor regulating many of the molecular processes that we know are important for fear memory formation and stability in the mammalian brain.
Keywords: Ubiquitin, Fear conditioning, Proteasome, Consolidation, Reconsolidation, Protein degradation
1. Introduction
Pavlovian fear memories are formed when a neutral conditional stimulus (CS) is paired with an aversive unconditional stimulus (UCS). As a result of learning this association, the CS will acquire the ability to elicit a fear response. Memories formed through Pavlovian conditioning are thought to go through several stages that allow for their creation, storage and modification. Acquisition occurs when the animal is first exposed to the stimuli and their programmed relationship and this newly formed memory will remain in a labile state for a brief period of time following the acquisition period. The memory is then transferred, through a process known as memory consolidation, into a stable state in which it is no longer susceptible to disruption (McGaugh, 2000). However, upon retrieval a once consolidated memory “destabilizes” and enters a new labile phase known as memory reconsolidation (Nader, Schafe, & LeDoux, 2000). The reconsolidation process is believed to be dynamic and allows new information to be incorporated into the existing trace, which can strengthen (Inda, Muravieva, & Alberini, 2011; Lee, 2008) or weaken (Agren et al., 2012; Clem & Huganir, 2010; Lee et al., 2008; Monfils, Cowansage, Klann, & LeDoux, 2009; Rao-Ruiz et al., 2011; Schiller et al., 2010) the original memory. Additionally, after repeated presentations of the CS without the UCS, memories can also undergo extinction. The extinction process is believed to include the formation of a new inhibitory memory that reduces responding the CS without erasing the original excitatory CS–UCS association and this is largely regulated by synaptic plasticity in the infralimbic region of the prefrontal cortex and the amygdala (Myers & Davis, 2007). Like fear memory consolidation and reconsolidation, the extinction consolidation process is a distinct phase characterized by the formation of a new inhibitory memory that requires synaptic alterations in multiple brain regions.
Pavlovian fear memories are formed and stored in multiple brain regions, and the specific regions recruited depend largely on the type of memory acquired (Helmstetter, Parsons, & Gafford, 2008). In general, many of these memories require de novo protein synthesis in the amygdala for their long-term storage. For example, infusions of protein synthesis inhibitors directly into the amygdala can disrupt contextual (Jarome, Werner, Kwapis, & Helmstetter, 2011; Parsons, Gafford, & Helmstetter, 2006), auditory delay (Jarome et al., 2012; Nader et al., 2000; Parsons Gafford, Baruch, Riedner, & Helmstetter, 2006; Parsons, Gafford, & Helmstetter, 2006; Schafe & LeDoux, 2000; Schafe, Nadel, Sullivan, Harris, & LeDoux, 1999) and trace (Kwapis, Jarome, Schiff, & Helmstetter, 2011) fear memories, conditioned taste aversion memories (Bahar, Samuel, Hazvi, & Dudai, 2003) as well as fear potentiated startle (Yeh, Mao, Lin, & Gean, 2006). Inhibiting protein synthesis in the hippocampus can disrupt the long-term storage of contextual fear memories (Bourtchouladze et al., 1998; Gafford, Parsons, & Helmstetter, 2011; Lee et al., 2008) while infusion of protein synthesis inhibitors in the auditory thalamus disrupts auditory delay fear memories (Parsons, Riedner, Gafford, & Helmstetter, 2006) and conditioned taste aversion memories are disrupted by blocking protein synthesis in the insular cortex (Moguel-Gonzalez, Gomez-Palacio-Schjetnan, & Escobar, 2008). Additionally, inhibiting the atypical PKC isoform PKMζ can disrupt long-term memory for auditory delay and contextual fear memories and fear potentiated startle in the amygdala (Kwapis, Jarome, Gilmartin, & Helmstetter, 2012; Kwapis, Jarome, Lonergan, & Helmstetter, 2009; Migues et al., 2010; Parsons & Davis, 2012; Serrano et al., 2009), aversive spatial memories in the hippocampus (Pastalkova et al., 2006), and conditioned taste aversion memories in the insular cortex (Shema, Sacktor, & Dudai, 2007; Shema et al., 2011), suggesting that fear memories are stored throughout many different brain regions.
Consistent with these results, manipulation of several signaling pathways “upstream” of protein synthesis impairs fear memory formation when applied following acquisition and stability when applied following retrieval. For example, inhibiting NMDA receptor activity impairs the long-term storage of auditory delay fear and contextual fear memories (Rodrigues, Schafe, & LeDoux, 2001) and fear potentiated startle (Walker & Davis, 2000) in the amygdala, trace and contextual fear memories in the medial prefrontal cortex (Gilmartin & Helmstetter, 2010) and hippocampus (Czemiawski, Ree, Chia, & Otto, 2012), and conditioned taste aversion memories in the insular cortex (Escobar, Alcocer, & Chao, 1998). Inhibiting transcriptional control pathways such as protein kinase A (PKA), protein kinase C, ERK/MAP kinase, CaMKII, and CREB, as well as new mRNA synthesis impairs fear memory formation following acquisition and stability following retrieval in multiple brain regions (e.g., Abel et al., 1997; Adams & Sweatt, 2002; Atkins, Selcher, Petraitis, Trzaskos, & Sweatt, 1998; Bailey, Kim, Sun, Thompson, & Helmstetter, 1999; Duvarci, Nader, & LeDoux, 2005; Kida et al., 2002; Rodrigues, Farb, Bauer, LeDoux, & Schafe, 2004; Schafe & LeDoux, 2000; Selcher, Weeber, Varga, Sweatt, & Swank, 2002; Tronson, Wiseman, Olausson, & Taylor, 2006). As a result, one current model of memory consolidation proposed by Johansen, Cain, Ostroff, and LeDoux (2011) suggests that activation of NMDA receptors during acquisition critically triggers changes in the activity of a number of intracellular signaling pathways which regulate increases in gene transcription and new protein synthesis necessary for the synaptic changes important for fear memory formation. However, this model does not account for the possibility that protein degradation may also be necessary for learning-induced synaptic plasticity.
The majority of protein turnover in eukaryotes is controlled by the ubiquitin–proteasome system (UPS), which has both proteolytic and non-proteolytic functions. The UPS is involved in a number of cellular processes, including cell-cycle progression, transcription, apoptosis and synaptic plasticity. Recently, evidence has begun accumulating suggesting that protein degradation may be a critical regulator of memory formation and stability in the mammalian brain. Here, we review the published studies on the role of protein degradation and memory and propose a model of how ubiquitin–proteasome mediated protein degradation may be regulating long-term memory storage.
2. The ubiquitin–proteasome system
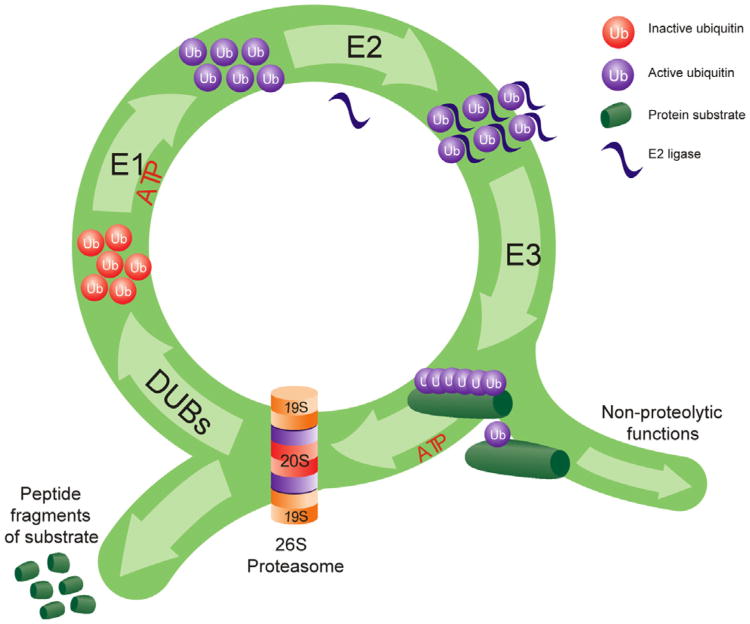
The ubiquitin–proteasome system is a complex network of ubiquitin ligases and interconnected proteasome structures that targets proteins for proteasome-dependent proteolysis. The ubiquitin–proteasome system has been reviewed extensively by others (Bingol & Schuman, 2005; Bingol & Sheng, 2011; Fioravante & Byrne, 2011; Geng, Wenzel, & Tansey, 2012; Hegde, 2010; Mabb & Ehlers, 2010; Patrick, 2006; Tai & Schuman, 2008; Yi & Ehlers, 2005), but generally proteins become targeted for degradation through the coordinated actions of many different ubiquitin ligases (Fig. 1). This occurs in a three step process where free ubiquitin is first activated by an E1 enzyme. Next, the ubiquitin modifier is conjugated to its target protein by the coordinated actions of an ubiquitin-conjugating enzyme (E2) and an ubiquitin–protein ligase (E3). There are hundreds of known E3 ligases which can target a few to many different proteins for degradation, providing a mechanism by which the UPS can control the degradation of a specific class of proteins. While proteins can acquire a single ubiquitin, thus becoming monoubiquitinated, most proteins acquire several ubiquitin tags. This process of polyubiquitination can result in a target protein being tagged by 2–7 different ubiquitin modifiers. Generally, those that acquire four or more ubiquitin modifiers will provide the maximal signal for degradation. However, this is also dependent on the type of polyubiquitin tag a target protein receives. Polyubiquitin tags form when each ubiquitin modifier links together at specific lysine residues on the first ubiquitin, and lysine-48 linkage is thought to provide the maximal signal for degradation. Once this ubiquitination process is complete, the target protein can then be recognized and degraded by the 26S proteasome complex.
Fig. 1.
The ubiquitination process. Ubiquitin protein modifiers are attached to a target protein through a series of ATP-dependent steps. Free ubiquitin is activated by an E1 ligase and conjugated to a target protein through the coordinated actions of ubiquitin E2 and E3 ligases. Short ubiquitin tags target proteins for non-proteolytic functions while longer ubiquitin tags (polyubiquitination) target the protein for degradation by the 26S proteasome complex. The proteasome removes the ubiquitin tag through deubiquitinating enzymes (DUBs) and degrades the target substrate into small peptide fragments.
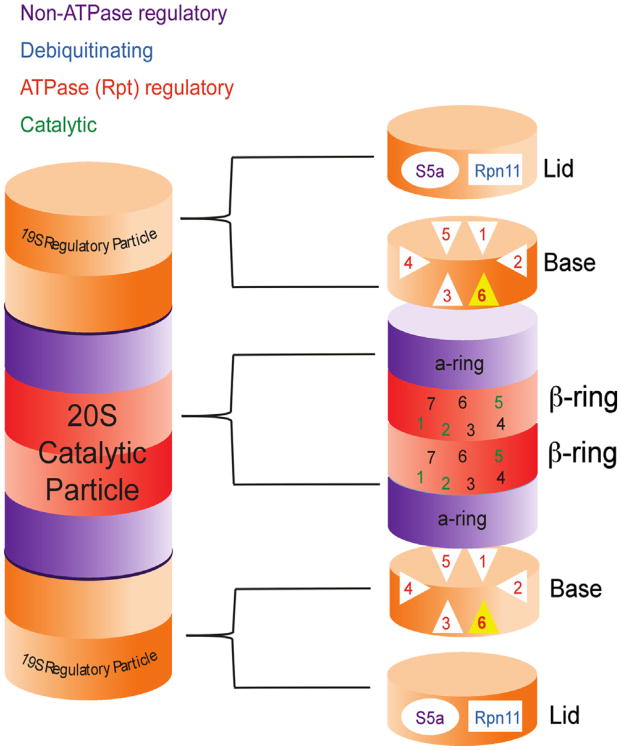
The 26S proteasome complex (Fig. 2) is a multi-subunit structure that consists of a 20S catalytic core and two 19S regulatory particles (for review, see Bedford, Paine, Sheppard, Mayer, & Roelofs, 2010). The 19S caps can be divided into lid and base complexes. The 19S lid contains deubiquitinating enzymes and the Rpn10/S5a subunit, which has been shown to bind polyubiquitinated proteins that contain the proper chain length and linkage for degradation (Wang, Young, & Walters, 2005). The base of the 19S complex contains the six ATPase subunits of the proteasome complex, known as Rpt1-6, and four non-ATPase subunits. The Rpt subunits regulate the activity of the 20S core through an ATP-dependent process and are believed to regulate protein unfolding. The catalytic core of the proteasome contains two outer rings of α subunits (1–7) and two inner rings of b subunits (1–7). The three types of proteolytic activity, chymotrypic-, trypic- and peptidylglutamyl peptide hydrolysing-like activities, are regulated by the β1, β2, and β5 subunits. The 19S proteasome recognizes polyubiquitinated proteins and shuttles them into the 20S core for degradation, making the 26S proteasome capable of recognizing, binding, deubiquitinating, unfolding and degrading polyubiquitinated proteins all within its self-compartmentalized structure.
Fig. 2.
The 26S proteasome complex. The 26S proteasome complex consists of a 20S catalytic core and two 19S regulatory particles. The regulatory particles are divided into lids and bases, The lid contains the non-ATPase Rpn11 deubiquitinating subunit and the Rpn10/S5a subunit, which captures polyubiquitinated proteins for degradation, while the base contains the only six ATPase subunits, Rpt1-6. The catalytic subunit contains two outer rings or α-subunits and two inner rings of β-subunits. The β1, β2, and β5 subunits are responsible for the catalytic activity of the proteasome.
3. The role of protein degradation in activity-dependent synaptic plasticity
A number of studies support a role for protein degradation in activity-dependent synaptic plasticity. These studies have been reviewed extensively elsewhere (e.g., Mabb & Ehlers, 2010) but some of the recent findings are discussed here. Ehlers (2003) demonstrated that chronic stimulation or inhibition of cultured hippocampal neurons resulted in dynamic changes in the protein composition of the postsynaptic density (PSD). He found that many of these changes were largely regulated by increases or decreases in polyubiquitination and subsequent proteasome-dependent degradation of proteins in the PSD. Consistent with this, inhibiting proteasome activity not only reversed many of these changes to the synaptic structure, but also affected the activity of other signaling molecules such as CREB. Furthermore, synaptic stimulation causes redistribution of proteasomes from the dendritic shaft to the spine, and the redistributed proteasomes subsequently become more active (Bingol & Schuman, 2006). This redistribution of proteasomes following synaptic excitation is largely regulated by CaMKII, which acts as a scaffold to recruit proteasomes to dendritic spines (Bingol et al., 2010). Once at the spines, CaMKII then phosphorylates the proteasome regulatory Rpt6 on Serine 120 (S120), leading to increases in proteasome activity. Consistent with this, CaMKII has been shown to target S120 in vtiro (Djakovic, Schwarz, Barylko, DeMartino, & Patrick, 2009) and increases in CaMKII phosphorylation at Thr-286 enhances phosphorylation of S120 (Djakovic et al., 2012). Transfecting cells with a phospho-dead S120 reduced the activity-dependent accumulation of proteasomes to the PSD, suggesting that phosphorylation of S120 is necessary for tethering of proteasomes to postsynaptic scaffolds, and enhancing or inhibiting S120 phosphorylation can mimic changes in synaptic strength that result from chronic stimulation or inhibition of cultured hippocampal neurons (Djakovic et al., 2012). Supporting this, the activity-dependent growth of new dendritic spines requires phosphorylation of S120 (Hamilton et al., 2012). Despite evidence that both CaMKII and PKA phosphorylate S120 (Djakovic et al., 2009; Zhang et al., 2007), this proteasome-dependent increase in spine growth was sensitive to CaMKII but not PKA inhibitors and required association of CaMKII with NMDA receptors. These results suggest that stimulation of NMDA receptors increases protein polyubiquitination and leads to increases in CaMKII phosphorylation. CaMKII then regulates redistribution of proteasomes to dendritic spines where it phosphorylates Rpt6 at Serine 120. Phosphorylation of S120 leads to increases in proteasome stability at synapses and increases in proteasome activity, which degrades the polyubiquitinated proteins at the PSD. This change in the synaptic structure then promotes the growth of new dendritic spines, though the mechanisms by which this occurs remain unknown.
Protein degradation has been also been shown to be important for long-term potentiation (LTP) and long-term depression (LTD), although some of the results are conflicting. Proteasome inhibitors have been shown to enhance or inhibit early LTP (E-LTP), while some studies have reported no effect of these same manipulations (Dong, Upadhya, Ding, Smith, & Hegde, 2008; Fonseca, Vabulas, Hartl, Bonhoeffer, & Nagerl, 2006; Karpova, Mikhavlova, Thomas, Knopfel, & Behnisch, 2006). However, these same studies showed that late LTP (L-LTP), which requires gene transcription and protein synthesis, is sensitive to proteasome inhibitors. Interestingly, simultaneously inhibiting protein degradation and protein synthesis during L-LTP rescues the loss of LTP that would normally occur by blocking either one of these mechanisms individually (Fonseca et al., 2006). This suggests that L-LTP may depend on a balance between protein synthesis and protein degradation. Additionally, inhibiting proteasome activity in the hippocampus impairs both NMDA-dependent and metabotropic glutamate receptor-dependent LTD (Colledge et al., 2003; Deng & Lei, 2007; Hou et al., 2006), though not all studies have found these effects (Citri, Soler-Llavina, Bhattacharyya, & Malenka, 2009; Mao, Lin, & Gean, 2008). As a result it remains unclear under what circumstances protein degradation is necessary for E-LTP and LTD, though it does seem to be critical for L-LTP.
Some of the earliest work implicating protein degradation in learning-dependent synaptic plasticity came from experiments examining long-term facilitation (LTF) in Aplysia. A series of experiments by Hegde, Goldberg, and Schwartz (1993) demonstrated that PKA regulatory subunits, which become dissociated from their catalytic subunits during the induction of LTF, were targeted by the UPS for degradation. Additionally, the deubiquitinating enzyme Ap-uch, which interacts with the proteasome, was induced by the same treatment that induces LTF and injection of antibodies or antisense oligonucleotides that targeted Ap-uch on the sensory-motor synapses blocked the induction of LTF (Hegde et al., 1997). A follow-up study then demonstrated that a proteasome inhibitor could indeed prevent the induction of LTF (Chain et al., 1999). These results provided the first evidence that protein degradation may be involved in memory formation, though the first evidence of this in mammals was not reported until several years later.
4. Protein degradation and memory
While numerous studies have supported a role for NMDA-receptor mediated plasticity and de novo protein synthesis in the formation and stability of long-term fear memories, only recently have researchers begun to examine the importance of ubiquitin– proteasome mediated protein degradation in memory storage. While some of the results have been conflicting, in general there is now convincing evidence that protein degradation is a critical regulator of long-term memory formation and storage in the mammalian brain. Here, we review those recent studies highlighting the requirement for protein degradation in memory consolidation, reconsolidation and extinction.
4.1. Memory consolidation
In mammals, several labs have studied the role of protein degradation in memory consolidation, reconsolidation and extinction. The first evidence that protein degradation may be involved in memory consolidation came from Lopez-Salon et al. (2001) who found that a proteasome inhibitor infused into the dorsal hippocampus impaired the consolidation of an inhibitory avoidance (IA) memory. They found that IA training lead to an increase in polyubiquitination and proteasome trypsin-like activity, and that one potential target of the proteasome was the Inhibitory Kappa B (IκB) protein, an inhibitor of the nuclear factor kappa B (NF-κB) signaling pathway. They did not find any change in the PKA regulatory subunit, suggesting that it may not be a target of the proteasome during IA memory consolidation. However, this result was challenged several years later by a study examining context fear memory consolidation in the hippocampus (Lee et al., 2008). Using a S5a-fusion protein, which can specifically recognize and bind polyubiquitinated proteins for degradation, they found that protein polyubiquitination was increased in the dorsal hippocampus following context fear conditioning, however, a proteasome inhibitor applied to the CA1 region of the hippocampus post-conditioning had no effect on long-term memory. Consistent with this, one recent study found that conditionally knocking out Cdh1, which regulates the E3 ligase anaphase promoting complex/cyclosome (APC/C), in neurons during development did not prevent mice from acquiring contextual fear conditioning or another hippocampal-dependent spatial task (Pick, Wang, Mayfield, & Klann, 2013). Interestingly, another study found that a hippocampus-dependent spatial memory task did require protein degradation in the CA3 region of the hippocampus (Artinian et al., 2008), suggesting that protein degradation may be important for the consolidation of certain memories in the hippocampus or that there may be a region-specific requirement for protein degradation in the hippocampus.
While it is unclear if protein degradation is necessary for the consolidation of contextual fear memories in the hippocampus, recent evidence suggests that it is important for the consolidation of both auditory and contextual fear memories in the amygdala (Jarome et al., 2011). In this study, both a S5a-fusion protein and an antibody which recognizes Lysine-48 polyubiquitination revealed that degradation-specific polyubiquitination was increased in the amygdala following fear conditioning. These increases were learning-specific, sensitive to NMDA receptor inhibitors, and occurred within the memory consolidation time window. During the increases in protein degradation, there were significant increases in the polyubiquitination of the synaptic scaffolding protein Shank and the RISC (RNA-induced Silencing Complex) factor MOV10, two known targets of the UPS in vitro (Banerjee, Neveu, & Kosik, 2009; Ehlers, 2003). Infusion of the proteasome inhibitor βlac (clasto-lactacystin β-lactone) into the amygdala following fear conditioning significantly impaired memory for both the auditory cue and the context on later tests. Interestingly, these impairments were similar to those produced by the protein synthesis inhibitor anisomycin and simultaneous blockade of protein degradation and protein synthesis did not rescue these impairments, suggesting that simply maintaining a balance between synthesis and degradation is not sufficient to promote fear memory formation in the amygdala. These results suggest that auditory and contextual fear memories likely require protein degradation in the amygdala for their long-term storage.
Consistent with this requirement for protein degradation in the amygdala following fear conditioning, one recent study found that conditionally knocking out Cdh1 in excitatory neurons after development lead to impairments in associative fear learning (Pick, Malumbres, & Klann, 2013). Additionally, amygdala slices prepared from these animals had impaired LTP and there were increased levels of Shank and NR2A following the induction of LTP in these slices. These results suggest that this specific E3 ligase is critical for synaptic plasticity and memory formation in the amygdala, and it may regulate these processes by promoting the activity-and-proteasome-dependent degradation of Shank and NR2A.
Conditioned taste aversion memories also have been shown to require protein degradation in the amygdala and the insula cortex for their consolidation (Rodriguez-Ortiz, Balderas, Saucedo-Alquicira, Cruz-Castaneda, & Bermudez-Rattoni, 2011) and the consolidation of long-term memory in the crab Chasmagnathus requires proteasome activity (Merlo & Romano, 2007), suggesting that inhibiting proteasome activity around the time of acquisition can impair various types of memories in both vertebrates and invertebrates. However, in some cases proteasome inhibitors can actually enhance memory. For example, proteasome inhibitors applied into the amygdala enhance fear potentiated startle (Yeh et al., 2006). Consistent with this, one recent study found that proteasome inhibitors enhance classical appetitive conditioning in the honeybee (Felsenberg, Dombrowski, & Eisenhardt, 2012). Nonetheless, these studies demonstrate that protein degradation seems to play an important role in the memory consolidation process, though whether it acts to support or constrain memory formation varies based on the learning paradigm and model system used.
4.2. Memory reconsolidation
While a number of studies have examined the role of protein degradation in memory consolidation, only a few have examined its role in the reconsolidation of the same memories. One study found that retrieval of a contextual fear memory led to increases in degradation specific polyubiquitination in the CA1 region of the dorsal hippocampus (Lee et al., 2008). Two synaptic scaffolding proteins, Shank and GKAP, were found to be heavily ubiquitinated during these increases in polyubiquitination, and decreases in total Shank and GKAP followed increases in their polyubiquitination suggesting that these proteins were being targeted for degradation by the proteasome. When the proteasome inhibitor βlac was applied to the CA1 region following memory retrieval it did not impair memory, but it did rescue the memory impairments that normally results from protein synthesis blockade. This suggested that protein degradation was involved in the initiation of the reconsolidation process by controlling memory destabilization (Ben Mamou, Gamache, & Nader, 2006), which induced the need for new protein synthesis in the hippocampus. In the absence of protein degradation the original memory trace was left in a consolidated state, unable to be modified or disrupted. Consistent with this, an elegant study by Lee (2008) demonstrated that applying a proteasome inhibitor into the hippocampus following additional learning would prevent memory strengthening without affecting the original memory trace. Additionally, similar results have been reported for the updating of contextual content (Lee, 2010). However, proteasome inhibitors infused into the hippocampus following retrieval have been shown to impair memory on a non-fearful hippocampal dependent spatial task, suggesting that not all hippocampal dependent memories require protein degradation for memory destabilization following retrieval, but rather some require it to restabilize memory during reconsolidation (Artinian et al., 2008).
Protein degradation is also involved in the reconsolidation of both auditory and contextual fear memories in the amygdala (Jarome et al., 2011). In this study retrieval of either an auditory or contextual fear memory resulted in increases in degradation-specific polyubiquitination in the amygdala. Similar to memory consolidation, these increases were NMDA-dependent and the proteasome targeted the synaptic scaffolding protein Shank and the RISC factor MOV10. Interestingly, the peak increases in protein polyubiquitination were observed at different times for auditory or contextual fear memory in the amygdala, with contextual fear memory retrieval showing an earlier peak in polyubiquitination than auditory memory retrieval. Despite this, infusions of βlac into the amygdala following memory retrieval did not impair either memory but did rescue the memory impairments that normally resulted from protein synthesis inhibition. This result suggests that protein degradation regulates memory destabilization in the amygdala, an effect that was downstream of NMDA receptor activity which has been shown to regulate memory destabilization in the amygdala (Ben Mamou et al., 2006). However, not all fear memories that require the amygdala for their acquisition destabilize in the amygdala following retrieval, as a proteasome inhibitor infused into the amygdala prior to retrieval had no effect on a conditioned taste aversion memory (Rodriguez-Ortiz et al., 2011).
Recently, a cellular model of memory reconsolidation has been proposed in Aplysia (Lee et al., 2012). Here they found that long-term sensitization of the gill- and siphon-withdrawal reflex became labile following retrieval, and that blocking protein degradation could prevent the effects of a protein synthesis inhibitor when applied following retrieval. Interestingly, they found that LTF, the cellular analog of the synaptic plasticity underlying the gill- and siphon-withdrawal reflex, also was destabilized by protein degradation and restabilized by protein synthesis following reactivation. These results provide additional support that protein degradation is a major regulator of synaptic destabilization following memory retrieval.
4.3. Extinction consolidation
The role of protein degradation in the extinction of Pavlovian fear memories has received little attention and currently very little is known about the involvement of this process in extinction memory consolidation. One study reported that infusion of a proteasome inhibitor into the CA1 region of the dorsal hippocampus following extinction training could prevent extinction consolidation for a contextual fear memory (Lee et al., 2008). Consistent with, mice that lack Cdh1 in neurons showed impaired extinction of previously consolidated auditory fear memory (Pick, Wang, Mayfield, & Klann, 2013). Another study reported that a proteasome inhibitor infused into the amygdala could prevent d-cycloserine (DCS)-induced enhancement of memory extinction, suggesting that protein degradation may be necessary for memory extinction in the amygdala (Mao et al., 2008). Additionally, proteasome inhibitors impair extinction consolidation in honeybees (Felsenberg et al., 2012). Collectively, these results suggest that protein degradation may be important for memory extinction, though more research is needed before we can gain a better understanding of the role of the UPS in memory extinction.
5. The role of protein degradation in long-term memory formation and storage
While evidence is accumulating supporting the theory that protein degradation is critical for the formation and stability of long-term fear memories, what we know very little about is how protein degradation is involved in memory consolidation and reconsolidation. However, in vitro work provides us with a possible model for how protein degradation could be directly linked to the current signaling pathways that regulate memory storage. Here we propose cellular models for memory consolidation and reconsolidation. These models are adoptions from the comprehensive working models recently proposed by Johansen et al. (2011) which did not account for the involvement of protein degradation in long-term memory storage. Thus, our models focus on the idea that protein degradation can serve as the main intermediate mechanism of learning-induced synaptic plasticity, linking upstream signaling pathways (NMDA receptors, CaMKII) to downstream processes (transcription, translation) already known to be involved in the storage of long-term memories.
5.1. Memory consolidation model
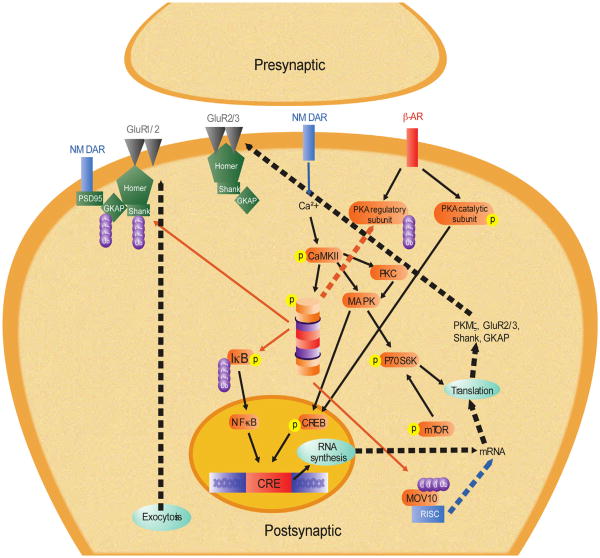
Our model for memory consolidation is presented in Fig. 3. The initiation of the consolidation process starts with activation of the NMDA receptors (NMDAR), which leads to the increases in intracellular calcium levels necessary for activation of the signal pathways which regulate memory consolidation. One of the pathways is the UPS, which we have shown to be dependent on NMDAR activity in the amygdala (Jarome et al., 2011). Indeed, numerous studies have reported that ubiquitin–proteasome function is dependent on NMDAR activity in vitro (e.g., Banerjee et al., 2009; Bingol & Schuman, 2006; Bingol et al., 2010). The likely pathway by which NMDARs regulate increases in UPS activity is two-fold. First, activation of various intracellular signaling pathways can increase the activity the of ubiquitin ligases, which leads to increased protein polyubiquitination. Consistent with this, inhibiting NMDAR in the amygdala following fear conditioning prevents increases in protein polyubiquitination (Jarome et al., 2011). Next, the catalytic activity of the proteasome is likely increased by CaMKII following fear conditioning. CaMKII has been shown to regulate proteasome translocation and activity in cultured hippocampal neurons, primarily through the phosphorylation of Rpt6-S120 (Bingol et al., 2010; Djakovic et al., 2009, 2012; Hamilton et al., 2012). This regulation of S120 by CaMKII is regulated by phosphorylation of CaM-KII at Thr-286 (CaMKII-T286). Indeed, fear conditioning increases CaMKII-T286 phosphorylation in the amygdala and inhibiting CaMKII activity impairs fear memory consolidation (Rodrigues et al., 2004). It is possible then that CaMKII regulates increases in proteasome activity following fear conditioning, and this may occur through phosphorylation of S120. Consistent with this, we have recently shown that inhibiting CaMKII, but not PKA, in the amygdala following fear conditioning reduces learning-induced increases in S120 phosphorylation and proteasome activity in the amygdala (Jarome, Kwapis, Ruenzel, & Helmstetter, Unpublished result). This indicates that the UPS is activated following fear conditioning through an NMDA–CaMKII-dependent mechanism.
Fig. 3.
Cellular model of memory consolidation. Solid lines denote hypothetical pathways based on tested mechanisms. Dotted lines denote untested, hypothetical pathways. Red lines indicate proteasome-dependent degradation of target protein. Abbreviations: β-AR, β-adrenergic receptor; Ca2+, calcium; CaMKII, calcium–calmodulin dependent protein kinase II; CREB, cAMP response element (CRE) binding protein; GluR1/2/3, glutamate AMPA receptor subunit 1, 2 and 3; GKAP, guanylate kinase-associated protein; IκB, inhibitory kappa B protein; MAPK, mitogen-activated protein kinase; MOV10, Moloney leukemia virus 10; mRNA, messenger ribonucleic acid; mTOR, mammalian target of rapamycin; NF-κB, nuclear factor kappa B; NMDA, N-methyl-d-aspartate glutamate receptor; P70S6K, P70 S6 kinase; PKA, protein kinase A; PKC, protein kinase C; PKMζ, protein kinase Mζ; PSD95, postsynaptic density protein 95; RISC, RNA-induced silencing complex; SHANK, SH3 and multiple ankyrin repeat domains protein. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Once activated, the UPS can potentially regulate a large number of downstream signaling pathways and processes. One potential process the UPS could regulate is new gene transcription. The NF-κB signaling pathway, a regulator of gene transcription, is critical for memory formation (Ahn et al., 2008; Freudenthal et al., 2005; Yeh, Lin, Lee, & Gean, 2002; Yeh et al., 2006). Activation of this pathway requires proteasome-dependent degradation of the IjB protein, which normally prevents phosphorylation and nuclear translocation of the NF-κB protein. Consistent with this, IκB has been implicated as a target of the UPS during fear memory formation (Lopez-Salon et al., 2001), suggesting that the UPS could be involved in the regulation of gene transcription. Additionally, the transcription factor CREB has been suggested as a downstream affector of proteasome activity in cultured hippocampal neurons (Ehlers, 2003). The UPS could regulate CREB phosphorylation and activation through two different mechanisms. First, the proteasome could degrade the PKA regulatory subunit, allowing persistent phosphorylation and activation of the PKA catalytic subunit (Hegde et al., 1993) which phosphorylates CREB. Second, the proteasome could target CREB repressors. Consistent with this, the proteasome targets the CREB repressor protein during the induction of LTF (Upadhya, Smith, & Hegde, 2004), though this has not yet been examined in mammals. However these are just two examples of how protein degradation could be involved in the regulation of mRNA transcription during long-term memory formation, though many more likely exist.
A number of recent studies have shown that epigenetic changes are critical for long-term memory formation (Chwang, Arthur, Schumacher, & Sweatt, 2007; Gupta et al., 2010; Kim, Levenson, Korsmeyer, Sweatt, & Schumacher, 2007; Levenson et al., 2004; Lubin & Sweatt, 2007; Miller, Campbell, & Sweatt, 2008; Miller et al., 2010). Modifications of the chromatin structure make the DNA more or less accessible to transcription factors, resulting in long-term changes in gene transcription. Normally, histone acetylation and methylation have been studied as ways of modifying histones and making DNA more or less accessible to the transcription machinery, respectively, however, histones also undergo ubiquitination and sumoylation. These modifications are analogous to methylation and acetylation, respectively, however have not yet been examined in the context of long-term memory formation. It is possible that a non-proteolytic function of the UPS during memory consolidation is ubiquitination of histones. Additionally, this may be regulated by 19S proteasome subunits Rpt4 and Rpt6 (Ezhkova & Tansey, 2004), though more research is needed to address this in more detail.
The UPS could also regulate the requirement for protein synthesis during memory formation, both through proteolytic and non-proteolytic functions. For proteolytic activity of the UPS, translation could be regulated two different ways. First, UPS-mediated increases in protein synthesis could be a consequence of UPS-mediated increases in gene transcription. Second, the UPS could target translational repressors independent of its actions of gene transcription. For example, the proteasome has been shown to target the RISC factor MOV10 in vitro (Banerjee et al., 2009). MOV10 undergoes a NMDA- and proteasome-dependent degradation process, which leads to greater protein synthesis at synapses. Interestingly, MOV10 has been shown to be a target of the proteasome during fear memory consolidation in the amygdala (Jarome et al., 2011). This suggests that protein degradation could regulate protein synthesis through both transcription-dependent and transcription-independent mechanisms. Additionally, the UPS could regulate translation through a non-proteolytic mechanism. A seminal study by Pavlopoulos et al. (2011) demonstrated that the cytoplasmic polyadenylation element-binding protein 3 (CPEB3) undergoes monoubiquitination by the E3 ligase Neuralized1. This monoubiquitination of CPEB3 lead to an increase in the synthesis of GluR1 and GluR2 and a growth of new dendritic spines in cultured hippocampal neurons. Additionally, overexpression or knockdown of Neuralized1 enhanced or impaired memory formation and synaptic plasticity, suggesting that this monoubiquitination of CPEB3 was critical for memory formation. Collectively, this suggests that the UPS could regulate protein synthesis during memory formation by both proteasome-dependent and proteasome-independent mechanisms.
The UPS could also regulate memory consolidation through its actions on the synaptic structure. Ubiquitin–proteasome mediated protein degradation has been shown to be important for activity dependent rearrangement of the PSD (Ehlers, 2003) and growth of new dendritic spines (Hamilton et al., 2012) in cultured hippocampal neurons. Following fear conditioning, there is a change in the synaptic structure and possible growth of new dendritic spines (Lai, Franke, & Gan, 2012; Ostroff, Cain, Bedont, Monfils, & LeDoux, 2010; Radley et al., 2006). It is possible then that protein degradation regulates changes in the PSD and growth of new dendritic spines. Consistent with this, the UPS targets the synaptic scaffolding protein Shank for degradation in the amygdala following fear conditioning (Jarome et al., 2011) and memory impairments resulting from knockout of a specific E3 ligase is correlated with increased Shank expression following LTP induction (Pick, Malumbres, et al., 2013). Shank is a “master” scaffold protein that has connections with receptor proteins and the actin cytoskeleton (Zheng, Seabold, Horak, & Petralia, 2011), suggesting that protein degradation could regulate changes to the synaptic structure through removal of receptor scaffolds. Additionally, the UPS could target proteins that normally negatively regulate dendritic spine growth, such as MEF2 which normally inhibits learning-dependent spine growth and synaptic plasticity (Cole et al., 2012). Thus it is possible that UPS-mediated protein degradation may be involved in structure changes to synapses following memory formation, but no direct link has yet been established in a behaving animal. Future research will need to address this in more detail.
Collectively, our model for memory consolidation suggests that new memories are formed by the coordinated activation of the UPS, which regulates transcriptional and translational control proteins to promote increases in gene transcription and protein synthesis necessary for normal memory formation. In this hypothetical model, protein polyubiquitination is increased through a NMDA-dependent mechanism and proteasome activity is increased by NMDA–CaMKII mediated phosphorylation of the 19S subunit Rpt6 at Serine-120. The proteasome then can regulate de novo gene transcription and protein translation through the removal of transcriptional and translational repressors and proteasome-dependent degradation of “master” scaffolds such as Shank could control alterations to the postsynaptic structure, allowing long-term changes in synaptic strength. Additionally, non-proteolytic ubiquitination of histone proteins could act to coordinate the histone acetylation and methylation changes that have been shown to be important in long-term memory formation, allowing long-term changes in gene expression. This model would suggest then that protein degradation could be a major regulator of memory consolidation by linking upstream signaling mechanisms to the downstream transcriptional and translational processes thought to be important in long-term memory formation.
5.2. Memory reconsolidation model
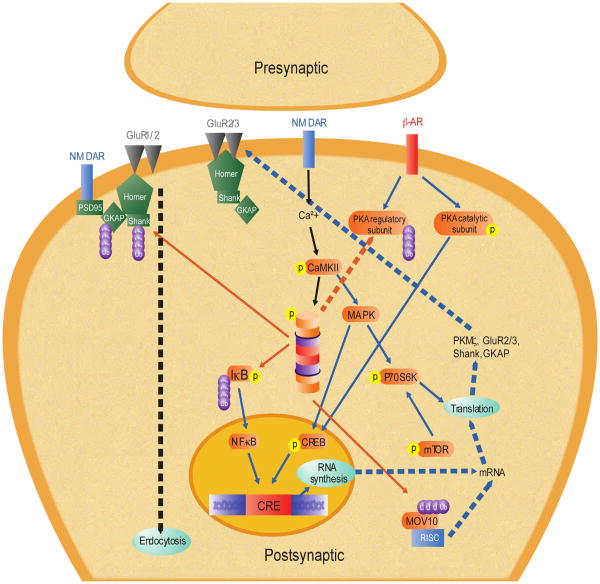
Our model for memory reconsolidation is presented in Fig. 4. As discussed in detail elsewhere, while memory reconsolidation shares some of the same molecular mechanisms as memory consolidation, reconsolidation is not a recapitulation of consolidation (Alberini, 2005; Alberini & Chen, 2012; Finnie & Nader, 2012; Nader & Hardt, 2009; Tronson & Taylor, 2007). Despite this, reconsolidation does seem to require UPS-mediated protein degradation, however, how protein degradation regulates the reconsolidation process may be both similar and different than how it regulates memory consolidation. For example, increases in protein degradation after retrieval are dependent on NMDAR receptor activity (Jarome et al., 2011). Downstream of NMDARs, it is unknown how protein degradation is regulated during the reconsolidation process, though CaMKII remains an attractive mechanism. Currently, the role of CaMKII in memory reconsolidation has not yet been examined, though it is possible that it regulates memory flexibility or lability following retrieval (Radwanska et al., 2011). Consistent with this, our lab has preliminary evidence demonstrating that CaMKII regulates proteasome activity following memory retrieval through the phosphorylation of Rpt6-S120 (Jarome et al., personal communication). Thus it is possible then that CaMKII may regulate protein degradation during memory reconsolidation through its actions on the proteasome.
Fig. 4.
Cellular model of memory reconsolidation. Solid lines denote hypothetical pathways based on tested mechanisms. Dotted lines denote untested, hypothetical pathways. Red lines indicate proteasome-dependent degradation of target protein. Black lines denote destabilization mechanisms. Blue lines denote restabilization mechanisms. Abbreviations: β-AR, β-adrenergic receptor; Ca2+, calcium; CaMKII, calcium–calmodulin dependent protein kinase II; CREB, cAMP response element (CRE) binding protein; GluR1/2/3, glutamate AMPA receptor subunit 1, 2 and 3; GKAP, guanylate kinase-associated protein; IκB, inhibitory kappa B protein; MOV10, Moloney leukemia virus 10; mRNA, messenger ribonucleic acid; mTOR, mammalian target of rapamycin; NF-κB, nuclear factor kappa B; NMDA, N-methyl-d-aspartate glutamate receptor; P70S6K, P70 S6 kinase; PKA, protein kinase A; PSD95, postsynaptic density protein 95; RISC, RNA-induced silencing complex; SHANK, SH3 and multiple ankyrin repeat domains protein. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
One of the most intriguing theories for the role of protein degradation during memory reconsolidation is that it regulates memory destabilization following retrieval. Indeed, infusion of a proteasome inhibitor after retrieval can prevent the effects of protein synthesis inhibitors (Jarome et al., 2011; Lee et al., 2008, 2012) and prevent memory updating (Lee, 2008, 2010). This suggests that protein degradation acts to induce memory destabilization following retrieval, however, it is unknown how this occurs. One possibility is that the UPS regulates structural modifications to synapses following memory retrieval (Kaang & Choi, 2012). This theory would suggest that memory destabilization is initiated by a coordinated disassembly of the postsynaptic structure, which induces the need for new protein synthesis to complete the synaptic remodeling process. Consistent with this, the proteasome has been shown to target “master” synaptic scaffolding proteins such as Shank in the amygdala (Jarome et al., 2011) and hippocampus (Lee et al., 2008) following retrieval. This suggests that protein degradation could regulate memory destabilization by controlling the activity-dependent remodeling of the postsynaptic density, though such a remodeling process has yet to be identified in vivo.
Protein degradation may also directly regulate protein synthesis during reconsolidation, possibly through similar mechanisms as those outlined in the consolidation model. For example, the UPS could regulate increases in protein synthesis by enhancing gene transcription, however, there have been a number of conflicting results for the involvement of new gene transcription in the reconsolidation process (Duvarci, Nader, & LeDoux, 2008; Kida et al., 2002; Lee, Everitt, & Thomas, 2004; Lubin & Sweatt, 2007; Parsons, Gafford, Baruch, et al., 2006; Parsons, Gafford, & Helmstetter, 2006; Parsons, Riedner, et al., 2006; Si et al., 2012; Solntseva & Nikitin, 2012). Additionally, the UPS could regulate translation directly by degrading RISC factors such as MOV10 after retrieval (Jarome et al., 2011). Regardless, the UPS likely regulates memory consolidation and reconsolidation in similar ways, though, considering the large number of potential targets, it could act as a way to differentiate the two memory storage processes.
Collectively, our model for memory reconsolidation suggests that consolidated memories are modified by the coordinated activation of the UPS, which controls the destabilization of the memory. In this hypothetical model, protein polyubiquitination is increased through a NMDA-dependent mechanism and proteasome activity is increased by NMDA–CaMKII mediated phosphorylation of the 19S subunit Rpt6 at Serine-120. The proteasome then can regulate the need for de novo protein translation by disassembling the synaptic structure through the degradation of “master” scaffolds such as Shank, allowing new long-term changes in synaptic strength. Additionally, the UPS could directly regulate increases in gene transcription and protein translation, but these would occur as a consequence of memory destabilization initiated by disassembly of the synaptic structure. This model would suggest then that protein degradation could be a major regulator of memory reconsolidation by controlling the ability of synapses to be plastic following retrieval, allowing modification of previously consolidated memories.
6. Conclusions
For decades it was proposed that protein synthesis was the critical cellular process underlying new memory formation and stability – only recently has the idea that protein degradation may be important in memory formation process begun to emerge. While some of the early results have been conflicting, there does appear to be a clear need for protein degradation in the multiple stages of memory formation and storage. Despite this, we still have a very basic understanding of how protein degradation may be involved in making and modifying memories. Here we have proposed simple working models that incorporate protein degradation into the existing cellular models of memory consolidation and reconsolidation. These models could serve as tools to help us determine not only if protein degradation is generally necessary for memory formation and stability, but also how it contributes to these processes, ultimately unraveling the mystery of how protein degradation is involved in learning and memory in the mammalian brain.
Acknowledgments
We thank Janine Kwapis for helpful comments on an earlier version of this manuscript. This work was supported by National Institute of Mental Health (NIMH) Grants R01-06558 (F.J.H.) and F31-088125 (T.J.J.).
References
- Abel T, Nguyen PV, Barad M, Deuel TA, Kandel ER, Bourtchoulazde R. Genetic demonstration of a role for PKA in the late phase of LTP and in hippocampus-based long-term memory. Cell. 1997;88:615–622. doi: 10.1016/s0092-8674(00)81904-2. [DOI] [PubMed] [Google Scholar]
- Adams JP, Sweatt JD. Molecular psychology: Roles for the ERK MAPK kinase cascade in memory. Annual Review of Pharmacology and Toxicology. 2002;42:135–163. doi: 10.1146/annurev.pharmtox.42.082701.145401. [DOI] [PubMed] [Google Scholar]
- Agren T, Engman J, Frick A, Bjorkstrand J, Larsson EM, Furmark T, et al. Disruption of reconsolidation erases a fear memory trace in the human amygdala. Science. 2012;337:1550–1552. doi: 10.1126/science.1223006. [DOI] [PubMed] [Google Scholar]
- Ahn HJ, Hernandez CM, Levenson JM, Lubin FD, Liou HC, Sweatt JD. C-Rel, an NF-kappaB family transcription factor, is required for hippocampal long-term synaptic plasticity and memory formation. Learning and Memory. 2008;15:539–549. doi: 10.1101/lm.866408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini CM. Mechanisms of memory stabilization: Are consolidation and reconsolidation similar or distinct processes? Trends in Neurosciences. 2005;28:51–56. doi: 10.1016/j.tins.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Alberini CM, Chen DY. Memory enhancement: Consolidation, reconsolidation and insulin-like growth factor 2. Trends in Neurosciences. 2012;35:274–283. doi: 10.1016/j.tins.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artinian J, McGauran AM, De Jaeger X, Mouledous L, Frances B, Roullet P. Protein degradation, as with protein synthesis, is required during not only long-term spatial memory consolidation but also reconsolidation. European Journal of Neuroscience. 2008;27:3009–3019. doi: 10.1111/j.1460-9568.2008.06262.x. [DOI] [PubMed] [Google Scholar]
- Atkins CM, Selcher JC, Petraitis JJ, Trzaskos JM, Sweatt JD. The MAPK cascade is required for mammalian associative learning. Nature Neuroscience. 1998;1:602–609. doi: 10.1038/2836. [DOI] [PubMed] [Google Scholar]
- Bahar A, Samuel A, Hazvi S, Dudai Y. The amygdalar circuit that acquires taste aversion memory differs from the circuit that extinguishes it. European Journal of Neuroscience. 2003;17:1527–1530. doi: 10.1046/j.1460-9568.2003.02551.x. [DOI] [PubMed] [Google Scholar]
- Bailey DJ, Kim JJ, Sun W, Thompson RF, Helmstetter FJ. Acquisition of fear conditioning in rats requires the synthesis of mRNA in the amygdala. Behavioral Neuroscience. 1999;113:276–282. doi: 10.1037//0735-7044.113.2.276. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Bedford L, Paine S, Sheppard PW, Mayer RJ, Roelofs J. Assembly, structure and function of the 26S proteasome. Trends in Cell Biology. 2010;20:391–401. doi: 10.1016/j.tcb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Mamou C, Gamache K, Nader K. NMDA receptors are critical for unleashing consolidated auditory fear memories. Nature Neuroscience. 2006;9:1237–1239. doi: 10.1038/nn1778. [DOI] [PubMed] [Google Scholar]
- Bingol B, Schuman EM. Synaptic protein degradation by the ubiquitin proteasome system. Current Opinion in Neurobiology. 2005;15:536–541. doi: 10.1016/j.conb.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:1144–1148. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140:567–578. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- Bingol B, Sheng M. Deconstruction for reconstruction: The role of proteolysis in neural plasticity and disease. Neuron. 2011;69:22–32. doi: 10.1016/j.neuron.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Bourtchouladze R, Abel T, Berman N, Gordon R, Lapidus K, Kandel ER. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learning and Memory. 1998;5:365–374. [PMC free article] [PubMed] [Google Scholar]
- Chain DG, Casadio A, Schacher S, Hegde AN, Valbrun M, Yamamoto N, et al. Mechanisms for generating the autonomous cAMP-dependent protein kinase required for long-term facilitation in Aplysia. Neuron. 1999;22:147–156. doi: 10.1016/s0896-6273(00)80686-8. [DOI] [PubMed] [Google Scholar]
- Chwang WB, Arthur JS, Schumacher A, Sweatt JD. The nuclear kinase mitogen-and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. Journal of Neuroscience. 2007;27:12732–12742. doi: 10.1523/JNEUROSCI.2522-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Soler-Llavina G, Bhattacharyya S, Malenka RC. N-methyl-D-aspartate receptor- and metabotropic glutamate receptor-dependent long-term depression are differentially regulated by the ubiquitin-proteasome system. European Journal of Neuroscience. 2009;30:1443–1450. doi: 10.1111/j.1460-9568.2009.06950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem RL, Huganir RL. Calcium-permeable AMPA receptor dynamics mediate fear memory erasure. Science. 2010;330:1108–1112. doi: 10.1126/science.1195298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CJ, Mercaldo V, Restivo L, Yiu AP, Sekeres MJ, Han JH, et al. MEF2 negatively regulates learning-induced structural plasticity and memory formation. Nature Neuroscience. 2012;15:1255–1264. doi: 10.1038/nn.3189. [DOI] [PubMed] [Google Scholar]
- Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, et al. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czemiawski J, Ree F, Chia C, Otto T. Dorsal versus ventral hippocampal contributions to trace and contextual conditioning: Differential effects of regionally selective NMDA receptor antagonism on acquisition and expression. Hippocampus. 2012;22:1528–1539. doi: 10.1002/hipo.20992. [DOI] [PubMed] [Google Scholar]
- Deng PY, Lei S. Long-term depression in identified stellate neurons of juvenile rat entorhinal cortex. Journal of Neurophysiology. 2007;97:727–737. doi: 10.1152/jn.01089.2006. [DOI] [PubMed] [Google Scholar]
- Djakovic SN, Schwarz LA, Barylko B, DeMartino GN, Patrick GN. Regulation of the proteasome by neuronal activity and calcium/calmodulin-dependent protein kinase II. Journal of Biological Chemistry. 2009;284:26655–26665. doi: 10.1074/jbc.M109.021956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic SN, Marquez-Lona EM, Jakawich SK, Wright R, Chu C, Sutton MA, et al. Phosphorylation of Rpt6 regulates synaptic strength in hippocampal neurons. Journal of Neuroscience. 2012;32:5126–5131. doi: 10.1523/JNEUROSCI.4427-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Upadhya SC, Ding L, Smith TK, Hegde AN. Proteasome inhibition enhances the induction and impairs the maintenance of late-phase long-term potentiation. Learning and Memory. 2008;15:335–347. doi: 10.1101/lm.984508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvarci S, Nader K, LeDoux JE. Activation of extracellular signal-regulated kinase mitogen activated protein kinase cascade in the amygdala is required for memory reconsolidation of auditory fear conditioning. European Journal of Neuroscience. 2005;21:283–289. doi: 10.1111/j.1460-9568.2004.03824.x. [DOI] [PubMed] [Google Scholar]
- Duvarci S, Nader K, LeDoux JE. De novo mRNA synthesis is required for both consolidation and reconsolidation of fear memories in the amygdala. Learning and Memory. 2008;15:747–755. doi: 10.1101/lm.1027208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Activity levels control postsynaptic composition and signaling via the ubiquitin-proteasome system. Nature Neuroscience. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Escobar ML, Alcocer I, Chao V. The NMDA receptor antagonist CPP impairs conditioned taste aversion and insular cortex long-term potentiation in vivo. Brain Research. 1998;812:246–251. doi: 10.1016/s0006-8993(98)00931-7. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Molecular Cell. 2004;13:435–442. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- Felsenberg J, Dombrowski V, Eisenhardt D. A role of protein degradation in memory consolidation after initial learning and extinction learning in the honeybee (Apis mellifera) Learning and Memory. 2012;19:470–477. doi: 10.1101/lm.026245.112. [DOI] [PubMed] [Google Scholar]
- Finnie PS, Nader K. The role of metaplasticity mechanisms in regulating memory destabilization and reconsolidation. Neuroscience and Biobehavioral Reviews. 2012;36:1667–1707. doi: 10.1016/j.neubiorev.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Fioravante D, Byrne JH. Protein degradation and memory formation. Brain Research Bulletin. 2011;85:14–20. doi: 10.1016/j.brainresbull.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nagerl UV. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron. 2006;52:239–245. doi: 10.1016/j.neuron.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Freudenthal R, Boccia MM, Acosta GB, Blake MG, Merlo E, Baratti CM, et al. NF-kappaB transcription factor is required for inhibitory avoidance long-term memory in mice. European Journal of Neuroscience. 2005;21:2845–2852. doi: 10.1111/j.1460-9568.2005.04126.x. [DOI] [PubMed] [Google Scholar]
- Gafford GM, Parsons RG, Helmstetter FJ. Consolidation and reconsolidation of contextual fear memory requires mammalian target of rapamycin-dependent translation in the dorsal hippocampus. Neuroscience. 2011;182:98–104. doi: 10.1016/j.neuroscience.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng F, Wenzel S, Tansey WP. Ubiquitin and proteasomes in transcription. Annual Review of Biochemistry. 2012;81:177–201. doi: 10.1146/annurev-biochem-052110-120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin MR, Helmstetter FJ. Trace and contextual fear conditioning require neural activity and NMDA receptor-dependent transmission in the medial prefrontal cortex. Learning and Memory. 2010;17:289–296. doi: 10.1101/lm.1597410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, et al. Histone methylation regulates memory formation. Journal of Neuroscience. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AM, Oh WC, Vega-Ramirez H, Stein IS, Hell JW, Patrick GN, et al. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron. 2012;74:1023–1030. doi: 10.1016/j.neuron.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Goldberg AL, Schwartz JH. Regulatory subunits of cAMP-dependent protein kinase are degraded after conjugation to ubiquitin: A molecular mechanism underlying long-term synaptic plasticity. Proceedings of the National Academy of Science USA. 1993;90:7436–7440. doi: 10.1073/pnas.90.16.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Inokuchi K, Pei W, Casadio A, Ghirardi M, Chain DG, et al. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell. 1997;89:115–126. doi: 10.1016/s0092-8674(00)80188-9. [DOI] [PubMed] [Google Scholar]
- Hegde AN. The ubiquitin-proteasome pathway and synaptic plasticity. Learning and Memory. 2010;17:314–327. doi: 10.1101/lm.1504010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstetter FJ, Parsons RG, Gafford GM. Macromolecular synthesis, distributed synaptic plasticity, and fear conditioning. Neurobiology of Learning and Memory. 2008;89:324–337. doi: 10.1016/j.nlm.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Inda MC, Muravieva EV, Alberini CM. Memory retrieval and the passage of time: From reconsolidation and strengthening to extinction. Journal of Neuroscience. 2011;31:1635–1643. doi: 10.1523/JNEUROSCI.4736-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Werner CT, Kwapis JL, Helmstetter FJ. Activity dependent protein degradation is critical for the formation and stability of fear memory in the amygdala. PLoS ONE. 2011;6:e24349. doi: 10.1371/journal.pone.0024349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Kwapis JL, Werner CT, Ruenzel WL, Parsons RG, Gafford GM, et al. The timing of multiple retrieval events can alter GluR1 phosphorylation and the requirement for protein synthesis in fear memory reconsolidation. Learning and Memory. 2012;19:300–306. doi: 10.1101/lm.024901.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Kwapis JL, Ruenzel WL, Helmstetter FJ. CaMKII, but not Protein Kinase A, regulates Rpt6 phosphorylation and proteasome activity during the formation of long-term memories. doi: 10.3389/fnbeh.2013.00115. Unpublished result. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen JP, Cain CK, Ostroff LE, LeDoux JE. Molecular mechanisms of fear learning and memory. Cell. 2011;147:509–524. doi: 10.1016/j.cell.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaang BK, Choi JH. Synaptic protein degradation in memory reorganization. Advances in Experimental Medicine and Biology. 2012;970:221–240. doi: 10.1007/978-3-7091-0932-8_10. [DOI] [PubMed] [Google Scholar]
- Karpova A, Mikhavlova M, Thomas U, Knopfel T, Behnisch T. Involvement of protein synthesis and degradation in long-term potentiation of Schaffer collateral CA1 synapses. Journal of Neuroscience. 2006;26:4949–4955. doi: 10.1523/JNEUROSCI.4573-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida S, Josselyn SA, Pena de Ortiz S, Kogan JH, Chevere I, Masushiqe S, et al. CREB required for the stability of new and reactivated fear memories. Nature Neuroscience. 2002;5:348–355. doi: 10.1038/nn819. [DOI] [PubMed] [Google Scholar]
- Kim SY, Levenson JM, Korsmeyer S, Sweatt JD, Schumacher A. Developmental regulation of Eed complex composition governs a switch in global histone modification in brain. Journal of Biological Chemistry. 2007;282:9962–9972. doi: 10.1074/jbc.M608722200. [DOI] [PubMed] [Google Scholar]
- Kwapis JL, Jarome TJ, Lonergan ME, Helmstetter FJ. Protein kinase Mzeta maintains fear memory in the amygdala but not in the hippocampus. Behavioral Neuroscience. 2009;123:844–850. doi: 10.1037/a0016343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwapis JL, Jarome TJ, Schiff JC, Helmstetter FJ. Memory consolidation in both trace and delay fear conditioning is disrupted by intra-amygdala infusion of the protein synthesis inhibitor anisomycin. Learning and Memory. 2011;18:728–732. doi: 10.1101/lm.023945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwapis JL, Jarome TJ, Gilmartin MR, Helmstetter FJ. Intra-amygdala infusion of the protein kinase Mzeta inhibitor ZIP disrupts foreground context fear memory. Neurobiology of Learning and Memory. 2012;98:148–153. doi: 10.1016/j.nlm.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Franke TF, Gan WB. Opposite effects of fear conditioning and extinction on dendritic spine remodeling. Nature. 2012;483:87–91. doi: 10.1038/nature10792. [DOI] [PubMed] [Google Scholar]
- Lee JL, Everitt BJ, Thomas KL. Independent cellular processes for hippocampal memory consolidation and reconsolidation. Science. 2004;304:839–843. doi: 10.1126/science.1095760. [DOI] [PubMed] [Google Scholar]
- Lee JL. Memory reconsolidation mediates the strengthening of memories by additional learning. Nature Neuroscience. 2008;11:1264–1266. doi: 10.1038/nn.2205. [DOI] [PubMed] [Google Scholar]
- Lee JL. Memory reconsolidation mediates the updating of hippocampal memory content. Frontiers in Behavioral Neuroscience. 2010;4:168. doi: 10.3389/fnbeh.2010.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Choi JH, Lee N, Lee HR, Kim JI, Yu NK, et al. Synaptic protein degradation underlies destabilization of retrieved fear memory. Science. 2008;319:1253–1256. doi: 10.1126/science.1150541. [DOI] [PubMed] [Google Scholar]
- Lee SH, Kwak C, Shim J, Kim JE, Choi SL, Kim HF, et al. A cellular model of memory reconsolidation involves reactivation-induced destabilization and restabilization at the sensorimotor synapse in Aplysia. Proceedings of the National Academy of Science USA. 2012;109:14200–14205. doi: 10.1073/pnas.1211997109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. Journal of Biological Chemistry. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Lopez-Salon M, Alonson M, Vianna MR, Mello e Souza T, Izquierdo I, Pasquini JM, et al. The ubiquitin-proteasome cascade is required mammalian long-term memory formation. European Journal of Neuroscience. 2001;14:1820–1826. doi: 10.1046/j.0953-816x.2001.01806.x. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Sweatt JD. The IkappaB kinase regulates chromatin structure during the reconsolidation of conditioned fear memories. Neuron. 2007;55:942–957. doi: 10.1016/j.neuron.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabb AM, Ehlers MD. Ubiquitination in postsynaptic function and plasticity. Annual Review of Cell and Developmental Biology. 2010;26:179–210. doi: 10.1146/annurev-cellbio-100109-104129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao SC, Lin HC, Gean PW. Augmentation of fear extinction by D-Cycloserine is blocked by proteasome inhibitors. Neuropsychopharmacology. 2008;33:3085–3095. doi: 10.1038/npp.2008.30. [DOI] [PubMed] [Google Scholar]
- McGaugh JL. Memory – A century of consolidation. Science. 2000;287:248–251. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- Merlo E, Romano A. Long-term memory consolidation depends on proteasome activity in the crab Chasmagnathus. Neuroscience. 2007;147:46–52. doi: 10.1016/j.neuroscience.2007.04.022. [DOI] [PubMed] [Google Scholar]
- Migues PV, Hardt O, Wu DC, Gamache K, Sacktor TC, Wang YT, et al. PKMzeta maintains memories by regulating GluR2-dependent AMPA receptor trafficking. Nature Neuroscience. 2010;13:630–634. doi: 10.1038/nn.2531. [DOI] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiology of Learning and Memory. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, et al. Cortical DNA methylation maintains remote memory. Nature Neuroscience. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moguel-Gonzalez M, Gomez-Palacio-Schjetnan A, Escobar ML. BDNF reverse the CTA memory deficits produced by inhibition of protein synthesis. Neurobiology of Learning and Memory. 2008;90:587–587. doi: 10.1016/j.nlm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Monfils MH, Cowansage KK, Klann E, LeDoux JE. Extinction-reconsolidation boundaries: Key to persistent attenuation of fear memories. Science. 2009;324:951–955. doi: 10.1126/science.1167975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KM, Davis M. Mechanisms of fear extinction. Molecular Psychiatry. 2007;12:120–150. doi: 10.1038/sj.mp.4001939. [DOI] [PubMed] [Google Scholar]
- Nader K, Schafe GE, LeDoux JE. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature. 2000;406:722–726. doi: 10.1038/35021052. [DOI] [PubMed] [Google Scholar]
- Nader K, Hardt O. A single strand for memory: A case for reconsolidation. Nature Reviews Neuroscience. 2009;10:224–234. doi: 10.1038/nrn2590. [DOI] [PubMed] [Google Scholar]
- Ostroff LE, Cain CK, Bedont J, Monfils MH, LeDoux JE. Fear and safety learning differentially affect synapse size and dendritic translation in the lateral amygdala. Proceedings of the National Academy of Science USA. 2010;107:9418–9423. doi: 10.1073/pnas.0913384107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons RG, Gafford GM, Helmstetter FJ. Translational control via the mammalian target of rapamycin pathway is critical for the formation and stability of long-term fear memories in amygdala neurons. Journal of Neuroscience. 2006a;26:12977–12983. doi: 10.1523/JNEUROSCI.4209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons RG, Gafford GM, Baruch DE, Riedner BA, Helmstetter FJ. Long-term stability of fear memory depends on the synthesis of protein but not mRNA in the amygdala. European Journal of Neuroscience. 2006b;23:1853–1859. doi: 10.1111/j.1460-9568.2006.04723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons RG, Riedner BA, Gafford GM, Helmstetter FJ. The formation of auditory fear memory requires the synthesis of protein and mRNA in the auditory thalamus. Neuroscience. 2006c;141:1163–1170. doi: 10.1016/j.neuroscience.2006.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons RG, Davis M. Temporary disruption of fear potentiated startle following PKMf inhibition in the amygdala. Nature Neuroscience. 2011;14:295–296. doi: 10.1038/nn.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastalkova E, Serrano P, Pinkhasova D, Wallace E, Fenton AA, Sacktor TC. Storage of spatial information by the maintenance mechanism of LTP. Science. 2006;313:1141–1144. doi: 10.1126/science.1128657. [DOI] [PubMed] [Google Scholar]
- Patrick GN. Synapse formation and plasticity: Recent insights from the perspective of the ubiquitin proteasome system. Current Opinion in Neurobiology. 2006;16:90–94. doi: 10.1016/j.conb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E, Trifilieff P, Chevaleyre V, Fioriti L, Zairis S, Pagano A, et al. Neuralized1 activated CPEB3: A function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell. 2011;147:1369–1383. doi: 10.1016/j.cell.2011.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick JE, Wang L, Mayfield JE, Klann E. Neuronal expression of the ubiquitin E3 ligase APC/C-Cdh1 during development is required for long-term potentiation, behavioral flexibility, and extinction. Neurobiology of Learning and Memory. 2013;100:25–31. doi: 10.1016/j.nlm.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick JE, Malumbres M, Klann E. The E3 ligase APCC-CDh1 is required for associative fear memory and long-term potentiation in the amygdala of adult mice. Learning and Memory. 2013;20:11–20. doi: 10.1101/lm.027383.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radley JJ, Johnson LR, Janssen WG, Martino J, Lamprecht R, Hof PR, et al. Associative Pavlovian conditioning leads to an increase in spinophilin-immunoreactive dendritic spines in the lateral amygdala. European Journal of Neuroscience. 2006;24:876–884. doi: 10.1111/j.1460-9568.2006.04962.x. [DOI] [PubMed] [Google Scholar]
- Radwanska K, Medvedev NI, Pereira GS, Engmann O, Thiede N, Moraes MF, et al. Mechanism for long-term memory formation when synaptic strengthening is impaired. Proceedings of the National Academy of Science USA. 2011;108:18471–18475. doi: 10.1073/pnas.1109680108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao-Ruiz P, Rotaru DC, van der Loo RJ, Mansvelder HD, Stiedl O, Smit AB, et al. Retrieval-specific endocytosis of GluA2-AMPARs underlies adaptive reconsolidation of contextual fear. Nature Neuroscience. 2011;14:1302–1308. doi: 10.1038/nn.2907. [DOI] [PubMed] [Google Scholar]
- Rodrigues SM, Schafe GE, LeDoux JE. Intra-amygala blockade of the NR2B subunit of the NMDA receptor disrupts the acquisition but not expression of fear conditioning. Journal of Neuroscience. 2001;21:6889–6896. doi: 10.1523/JNEUROSCI.21-17-06889.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues SM, Farb CR, Bauer EP, LeDoux JE, Schafe GE. Pavlovian fear conditioning regulates Thr286 autophosphorylation Ca2+/calmodulin dependent protein kinase II at amygdala synapses. Journal of Neuroscience. 2004;24:3281–3288. doi: 10.1523/JNEUROSCI.5303-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ortiz CJ, Balderas I, Saucedo-Alquicira F, Cruz-Castaneda P, Bermudez-Rattoni F. Long-term aversive taste memory requires insular and amygdala protein degradation. Neurobiology of Learning and Memory. 2011;95:311–315. doi: 10.1016/j.nlm.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Schafe GE, Nadel NV, Sullivan GM, Harris A, LeDoux JE. Memory consolidation for contextual and auditory fear conditioning is dependent on protein synthesis, PKA, and MAP kinase. Learning and Memory. 1999;6:97–110. [PMC free article] [PubMed] [Google Scholar]
- Schafe GE, LeDoux JE. Memory consolidation of auditory Pavlovian fear conditioning requires protein synthesis and protein kinase A in the amygdala. Journal of Neuroscience. 2000;20:RC96. doi: 10.1523/JNEUROSCI.20-18-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller D, Monfil MH, Raio CM, Johnson DC, LeDoux JE, Phelps EA. Preventing the return of fear in humans using reconsolidation update mechanisms. Nature. 2010;463:49–53. doi: 10.1038/nature08637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcher JC, Weeber EJ, Varga AW, Sweatt JD, Swank M. Protein kinase signal transduction cascades in mammalian associative conditioning. Neuroscientist. 2002;8:122–131. doi: 10.1177/107385840200800208. [DOI] [PubMed] [Google Scholar]
- Serrano P, Friedman EL, Kenney J, Taubenfeld SM, Zimmerman JM, Hanna J, et al. PKMzeta maintains spatial, instrumental, and classically conditioned long-term memories. PLoS Biology. 2009;6:2698–2706. doi: 10.1371/journal.pbio.0060318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema R, Sacktor TC, Dudai Y. Rapid erasure of long-term memory associations in the cortex by an inhibitor of PKM zeta. Science. 2007;317:951–953. doi: 10.1126/science.1144334. [DOI] [PubMed] [Google Scholar]
- Shema R, Haramati S, Ron S, Hazvi S, Chen A, Sacktor TC, et al. Enhancement of consolidated long-term memory by overexpression of protein kinase Mzeta in the neocortex. Science. 2011;331:1207–1210. doi: 10.1126/science.1200215. [DOI] [PubMed] [Google Scholar]
- Si J, Yang J, Xue L, Yang C, Luo Y, Shi H, et al. Activation of NF-κB in basolateral amygdala is required for memory reconsolidation in auditory fear conditioning. PLoS ONE. 2012;7:e43973. doi: 10.1371/journal.pone.0043973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solntseva S, Nikitin V. Conditioned food aversion reconsolidation in snails is impaired by translation inhibitors but not transcription inhibitors. Brain Research. 2012;1467:42–47. doi: 10.1016/j.brainres.2012.05.051. [DOI] [PubMed] [Google Scholar]
- Tai HC, Schuman EM. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nature Reviews Neuroscience. 2008;9:826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- Tronson NC, Wiseman SL, Olausson P, Taylor JR. Bidirectional behavioral plasticity of memory reconsolidation depends on amygdalar protein kinase A. Nature Neuroscience. 2006;9:167–169. doi: 10.1038/nn1628. [DOI] [PubMed] [Google Scholar]
- Tronson NC, Taylor JR. Molecular mechanisms of memory reconsolidation. Nature Reviews Neuroscience. 2007;8:262–275. doi: 10.1038/nrn2090. [DOI] [PubMed] [Google Scholar]
- Upadhya SC, Smith TK, Hegde AN. Ubiquitin-proteasome-mediated CREB repressor degradation during induction of long-term facilitation. Journal of Neurochemistry. 2004;91:210–219. doi: 10.1111/j.1471-4159.2004.02707.x. [DOI] [PubMed] [Google Scholar]
- Walker DL, Davis M. Involvement of NMDA receptors within the amygdala in short- versus long-term memory for fear conditioning as assessed with fear-potentiated startle. Behavioral Neuroscience. 2000;114:1019–1033. [PubMed] [Google Scholar]
- Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. Journal of Molecular Biology. 2005;348:727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Yeh SH, Lin CH, Lee CF, Gean PW. A requirement of nuclear factor-kappaB activation in fear-potentiated startle. Journal of Biological Chemistry. 2002;277:46720–46729. doi: 10.1074/jbc.M206258200. [DOI] [PubMed] [Google Scholar]
- Yeh SH, Mao SC, Lin HC, Gean PW. Synaptic expression of glutamate receptor after encoding of fear memory in the rat amygdala. Molecular Pharmacology. 2006;69:299–308. doi: 10.1124/mol.105.017194. [DOI] [PubMed] [Google Scholar]
- Yi JJ, Ehlers MD. Ubiquitin and protein turnover in synapse function. Neuron. 2005;47:629–632. doi: 10.1016/j.neuron.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Zhang F, Hu Y, Huang P, Toleman CA, Paterson AJ, Kudlow JE. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. Journal of Biological Chemistry. 2007;282:22460–22471. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]
- Zheng CY, Seabold GK, Horak M, Petralia RS. MAGUKs, synaptic development, and synaptic plasticity. Neuroscientist. 2011;17:493–512. doi: 10.1177/1073858410386384. [DOI] [PMC free article] [PubMed] [Google Scholar]