Abstract
Ca2+-triggered neurotransmitter release depends on the formation of SNARE complexes that bring the synaptic vesicle and plasma membranes together, on the Ca2+ sensor synaptotagmin-1, and on complexins, which play active and inhibitory roles. Release of the complexin inhibitory activity by binding of synaptotagmin-1 to the SNARE complex, causing complexin displacement, was proposed to trigger exocytosis. However, the validity of this model was questioned based on the observation of simultaneous binding of complexin-I and a fragment containing the synaptotagmin-1 C2 domains (C2AB) to membrane-anchored SNARE complex. Using diverse biophysical techniques, here we show that C2AB and complexin-I do not bind to each other but can indeed bind simultaneously to the SNARE complex in solution. Hence, the SNARE complex contains separate binding sites for both proteins. However, total internal reflection fluorescence microscopy experiments show that C2AB can displace a complexin-I fragment containing its central SNARE-binding helix and an inhibitory helix (Cpx26–83) from membrane-anchored SNARE complex under equilibrium conditions. Interestingly, full-length complexin-I binds more tightly to membrane-anchored SNARE complex than Cpx26–83, and is not displaced by C2AB. These results show that interactions of N- and/or C-terminal sequences of complexin-I with the SNARE complex and/or phospholipids increase the affinity of complexin-I for the SNARE complex, hindering dissociation induced by C2AB. We propose a model whereby binding of synaptotagmin-1 to the SNARE complex directly or indirectly causes a rearrangement of the complexin-I inhibitory helix without inducing complexin-I dissociation, thus relieving the inhibitory activity and enabling cooperation between synaptotagmin-1 and complexin-I in triggering release.
Keywords: Neurotransmitter release, synaptic vesicle fusion, Ca2+ triggering, protein-protein interactions, protein-membrane interactions
Introduction
Neurotransmitter release by Ca2+-triggered synaptic vesicle exocytosis is key for communication between neurons. The sophisticated protein machinery that controls release includes the soluble N-ethylmaleimide sensitive factor attachment protein receptors (SNAREs) synaptobrevin, syntaxin-1 and SNAP-25 among its central components1–4. The three SNAREs form a tight four-helix bundle called the SNARE complex through their SNARE motifs5–7, which brings the membranes together8 and is key for membrane fusion. The acute Ca2+-dependence of release is conferred by the Ca2+ sensor synaptotagmin-19, which is believed to function in a tight interplay with the SNAREs and complexins10–13. The two C2 domains that form most of the cytoplasmic region of synaptotagmin-1 (the C2A and C2B domains) bind three and two Ca2+ ions, respectively, through loops at the top of β-sandwich structures14–17. These loops also mediate Ca2+-dependent phospholipid binding18, 19 and mutations that increase or decrease the Ca2+-dependence of this activity lead to parallel effects on the Ca2+-dependence of neurotransmitter release9, 20, demonstrating the Ca2+-sensing role of synaptotagmin-1 in release and the key functional importance of Ca2+/phospholipid binding. The function of synaptotagmin-1 is also likely to rely on its interactions with the SNARE complex21, 22 and on its ability to bind simultaneously to two membranes in a Ca2+-dependent manner, thus cooperating with the SNAREs in bringing the membranes into close proximity23, 24. The synaptotagmin-1 C2B domain is crucial for both of these activities22, 23, which correlates with the critical role of Ca2+-binding to the C2B domain for neurotransmitter release25.
Complexins are soluble proteins that bind tightly to the SNARE complex26 and play both activating and inhibitory roles in neurotransmitter release. Thus, deletion or knockdown of complexins I-III (the three major neuronal isoforms) in mice27–29, or of invertebrate complexin30–32, impairs Ca2+-triggered neurotransmitter release while at the same time increases spontaneous release in several of these systems28, 30–32 [note however that decreased spontaneous release was observed in complexin-I-III triple KO neurons29]. Dual roles have also been observed for complexins in reconstitution studies, although often only an active or an inhibitory function is manifested depending on the system11, 12, 33–35. Structural studies revealed that isolated complexin-I forms a dynamic central helix but lacks a tertiary structure, and that the central helix provides the major binding site for the SNARE complex, interacting in an antiparallel manner with the syntaxin-1 and synaptobrevin SNARE motifs; this central helix is preceded by an accessory helix that is close to but does not contact the SNARE complex36–38 (Fig. 1a). Mutagenesis showed that binding of the central helix to the SNARE complex is essential for the active function of complexin, while the accessory helix plays an inhibitory role and the N-terminal region is important to release this inhibition, likely through a weak interaction with the SNARE complex28, 39, 40. Both inhibitory and active roles have been suggested for the C-terminal region41–43, and the active role was proposed to involve phospholipid binding44.
Figure 1.
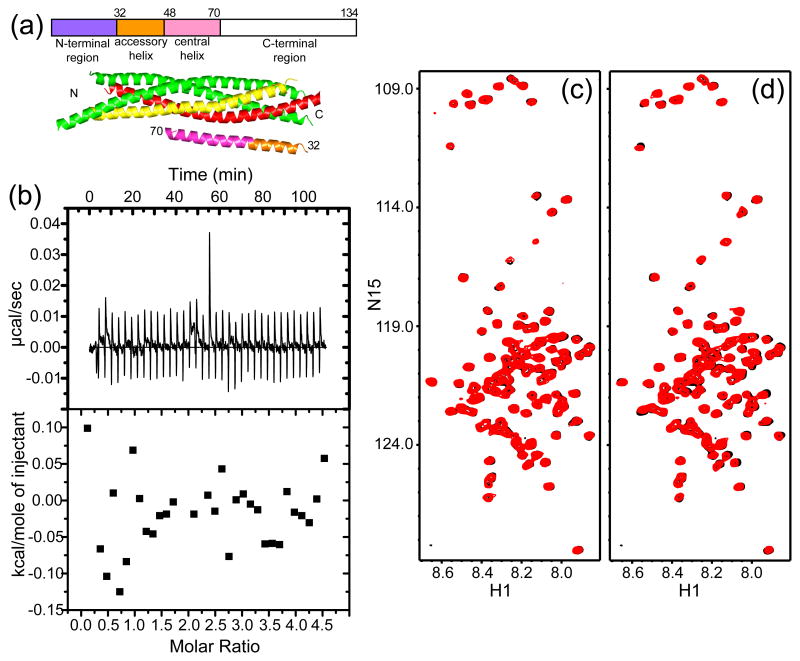
Complexin-I does not bind to the synaptotagmin-1 C2AB fragment. (a) Domain diagram of complexin-I with residue numbers above, and ribbon diagram of the Cpx26-83/SNARE complex37 with SNAP-25 in green, syntaxin-1 in yellow, synaptobrevin in red and Cpx26-83 in orange (accessory helix) and pink (central helix). The N- and C-termini of the region of Cpx26-83 that was observable are labeled with residue numbers, and the N- and C-termini of the SNAREs are indicated by N and C, respectively. (b) ITC analysis of complexin-I binding to the C2AB fragment in 1 mM Ca2+. Complexin-I (150 μM) was titrated into the C2AB fragment (10 μM). (c,d) 1H-15N HSQC spectra of 8 μM 15N-labeled complexin-I in the absence (black) and presence (red) of 10 μM C2AB fragment and 1 mM EDTA (c) or 1 mM Ca2+ (d).
Despite these advances, the mechanisms underlying the active and inhibitory functions of complexins remain unclear. Functional, reconstitution, biochemical and biophysical data led to a widespread model of Ca2+-triggered neurotransmitter release whereby complexin clamps release before Ca2+ influx and synaptotagmin-1 releases this inhibition10–13, and it was proposed that the complexin-clamped state provides a substrate for synaptotagmin-1 to trigger release synchronously10. This model was supported in part by the observation of competition between complexin-I and synaptotagmin-1 for binding to the SNARE complex using both soluble and membrane-anchored SNARE complex10, 22. However, other studies suggested that synaptotagmin-1 and complexin-I can bind simultaneously to the SNARE complex on membranes11, 45, leading some authors to the conclude that synaptotagmin-1 does not relieve a complexin-induced fusion clamp45. Clarifying these apparently contradictory data has thus become critical to understand the mechanism of neurotransmitter release.
With this purpose, we have analyzed the interplay between complexin-I and synaptotagmin-1 binding to the SNARE complex using a variety of biochemical and biophysical techniques. Our data show that complexin-I and a synaptotagmin-1 fragment spanning its two C2 domains (C2AB fragment) do not bind to each other but can bind simultaneously to soluble SNARE complex, which therefore contains separate binding sites for the complexin-I and the C2AB fragment. Intriguingly, total internal reflection fluorescence microscopy data show that full-length complexin-I binds more tightly to membrane-anchored SNARE complex than a complexin-1 fragment containing the accessory and central helices (Cpx26-83), and that the C2AB fragment can displace Cpx26-83 from membrane-anchored SNARE complex but cannot displace full-length complexin-I. These results support the notion that Ca2+-triggering of neurotransmitter release depends on a subtle interplay between interactions of synaptotagmin-1 and complexins with the SNAREs and the membrane(s).
Results
Analysis of complexin-I/synaptotagmin-1 interactions
While interactions of the SNAREs with complexins and synaptotagmin-1 have been extensively analyzed2, 4, 46, much less is known about complexin-I/synaptotagmin-1 interactions, and such interactions could be critical to couple the functions of these proteins. One study reported complexin binding to synaptotagmin-1, but the assays used relied in inmunoblotting for detection47. Since it is important to verify that protein-protein interactions can be detected by biophysical methods with purified proteins48, we tested whether the synaptotagmin-1 C2AB fragment binds to full-length complexin-I in the presence of 1 mM Ca2+ using isothermal titration calorimetry (ITC). However, no binding was observed (Fig. 1b). Because it is plausible that binding might not be detected if the associated enthalpy is small, we turned to 1H-15N heteronuclear single quantum coherence (HSQC) spectra, as these two-dimensional NMR spectra provide protein fingerprints that are highly sensitive to even very weak protein interactions48. However, 1H-15N HSQC of uniformly 15N-labeled complexin-1 in the absence or presence of 1 mM Ca2+ exhibited practically no perturbations upon addition of C2AB fragment. Hence, these results show that complexin-I does not bind to the synaptotagmin-1 C2AB fragment under the conditions of our experiments, although we cannot rule that sequences outside the synaptotagmin-1 C2 domains bind to complexin-I.
Simultaneous binding of synaptotagmin-1 and complexin-I to soluble SNARE complex
Protein solubility is an important factor to consider in the analysis of interactions of the SNARE complex with complexin-I and synaptotagmin-1. Thus, SNARE complexes formed with syntaxin-1 fragments that span its entire SNARE motif (residues 191–259) have a high tendency to aggregate49, as manifested for instance by strong broadening in NMR spectra37, by crystallization in a trimeric form7, or by precipitation under some conditions (see below). However, deletion of a few residues at the C-terminus of the syntaxin-1 SNARE motif greatly enhances the solubility of the SNARE complex and hence we normally use SNARE complex formed with a fragment spanning residues 191–253 of syntaxin-137, as we did in most of the experiments described below unless otherwise specified. Binding of the resulting SNARE complex to the synaptotagmin-1 C2AB fragment that we commonly use and spans its two C2 domains (residues 140–421) is relatively weak in the absence of Ca2+ but is readily detectable at the protein concentrations we use for 1H-15N HSQC spectra (e.g. 40 μM22). In the presence of Ca2+, the binding affinity is strongly enhanced50. Unfortunately, while the Ca2+-bound C2AB fragment is highly soluble23, 51, it has a high tendency to aggregate and precipitate when mixed with the SNARE complex even at concentrations of C2AB fragment and SNARE complex on the 10 μM range22, 52. We searched for synaptotagmin-1 fragments that alleviate this problem and we found that the solubility of the C2AB fragment/SNARE complex assembly can be somewhat improved by using a C2AB fragment that is extended by 9 residues at the N-terminus (thus including residues 131–421; referred below to as eC2AB fragment for extended C2AB fragment). Although the eC2AB fragment exhibits considerable precipitation with the SNARE complex at 20 μM protein concentrations in 1 mM Ca2+, which still hinders analysis of their interactions by NMR spectroscopy, no precipitation occurs at 10 μM protein concentrations. Hence, we used this fragment to analyze the interplay between synaptotagmin-1- and complexin-1-binding to the SNARE complex in solution.
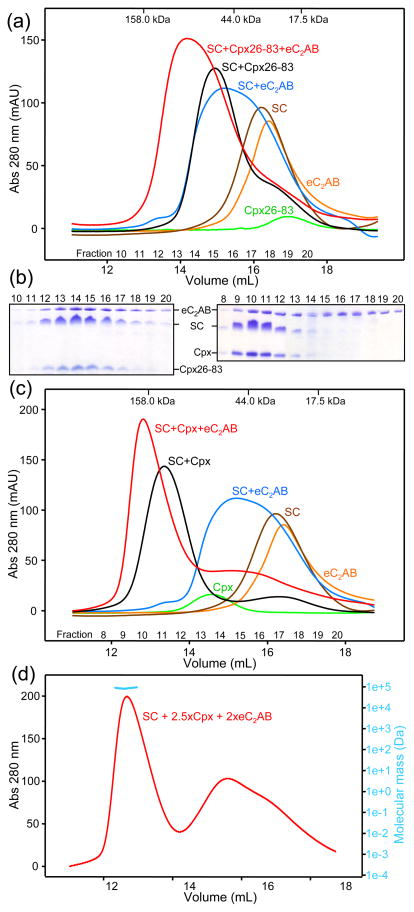
For this purpose, we first used size exclusion chromatography and analyzed binding of the SNARE complex to the eC2AB fragment and to Cpx26-83 (the complexin-I fragment that spans its accessory and central helices, which we used previously in competition assays on membranes22). These experiments were performed in the presence of 1 mM Ca2+ to increase the affinity of the eC2AB fragment for the SNARE and hence facility their co-elution in gel filtration. Indeed, addition of the eC2AB fragment caused a marked shift in the elution of the SNARE complex to smaller volumes and separate addition of Cpx26-83 caused a similar shift (Fig. 2a), revealing binding of both protein fragments to the SNARE complex individually. Importantly, addition of the Cpx26-83 and the eC2AB fragment together caused a further shift of the SNARE complex to smaller volumes, and analysis of the fractions from the chromatography by SDS/PAGE and Coomassie blue staining showed that Cpx26-83, the eC2AB fragment and the SNARE complex co-eluted under the major peak (Figs. 2a and 2b, left panel). These results unambiguously demonstrate that these complexin-I and synaptotagmin-1 fragments can bind simultaneously to the SNARE complex.
Figure 2.
The synaptotagmin-1 eC2AB fragment and complexin-I can bind simultaneously to the SNARE complex. (a,c) Gel filtration profiles of Cpx26-83 (a) or complexin-I (Cpx) (c), eC2AB fragment and soluble SNARE complex (SC) alone or in different combinations. Elution volumes of molecular weight markers are indicated at the top, and the numbers of collected fractions at the bottom. The concentrations of all proteins in the injected samples were 15 μM. (b) SDS-PAGE analysis of fractions from the chromatograms shown in panels (a) and (c) that contained mixtures of eC2AB fragment, SNARE complex and Cpx26-83 (left) or complexin-I (right). Proteins were detected with coomassie blue staining. (d) Gel filtration profile of a sample that contained 16 μM SNARE complex, 41 μM complexin-I and 32 μM eC2AB fragment. The MALS data acquired on the central part of the peak corresponding to the ternary eC2AB/SNARE complex/complexin-I assembly are shown in blue above the UV absorption profile.
We performed analogous experiments with full-length complexin-I instead of Cpx26-83 and obtained similar results (Figs. 2c and 2b, right panel). Note that in this case complexin-I or any of its complexes elute at considerably smaller volumes than those expected from their molecular weights because of the unfolded nature of much of the complexin-1 sequence53. This feature increases the separation between the elution volumes of the eC2AB fragment and the complexin-I/SNARE complex/eC2AB fragment assembly (compared to the analogous experiment performed with Cpx26-83), thus favoring dissociation of the eC2AB fragment from the complexin-I/SNARE complex during chromatography. Hence, it is not surprising that a fraction of the eC2AB fragment eluted at volumes close to that corresponding to its monomeric form in the presence of complexin-I and the SNARE complex (Fig. 2b, right panel). However, the shift of the elution volume of the complexin-I/SNARE complex upon addition of the eC2AB fragment (compare red and black curves in Fig. 2c) and the co-elution of much of the eC2AB fragment with complexin-1 and the SNARE complex (Fig. 2b, right panel) show again that both proteins can bind simultaneously to the SNARE complex.
Complexin-1 binds to the SNARE complex with high nanomolar affinity (see below) and 1:1 sotichiometry36. We attempted to determine the stoichiometry the eC2AB/SNARE complex assembly by multiangle light scattering (MALS) analysis of gel filtration chromatograms of samples with different eC2AB/SNARE complex ratios but we were unable to obtain conclusive data, which we attribute to several reasons. On one hand, partial dissociation occurs during gel filtration when the protein concentrations in the injected samples are on the 15 μM range, which underlies the broad gel filtration profile observed (Fig. 2c, blue curve) and is consistent with the affinity of the C2AB fragment for the SNARE complex (Kd estimated to be in the 1 μM range52). On the other hand, lower concentrations lead to further dissociation, and higher concentrations lead to precipitation. Such precipitation most likely arises because of similar factors to those that underlie precipitation of the C2AB fragment with the SNARE complex, which we recently showed to exhibit a primary binding mode, and one or more, weaker binding modes that promote precipitation52. Interestingly, we observed that the presence of complexin-I decreased the tendency of the eC2AB fragment and the SNARE complex to precipitate in the presence of Ca2+, which allowed us to analyze the gel filtration profile of a sample containing 16 μM SNARE complex with an excess of eC2AB fragment (32 μM) and complexin-I (41 μM) to favor formation of the ternary eC2AB/SNARE complex/complexin-I assembly. Analysis of the peak corresponding to the ternary assembly by MALS (Fig. 2d) yielded a molecular mass of 92 ± 7 kDa, which is comparable to that expected for an assembly with a 1:1:1 eC2AB/SNARE complex/complexin-I stoichiometry (calculated mass = 81.067 kDa). These finding suggest that the presence of complexin-I on the SNARE complex hinders the secondary eC2AB/SNARE complex binding mode(s), but a more systematic analysis will be required to test this notion. Note in addition that complexin-I also hinders precipitation of the SNARE complex itself (see below).
Complexin-1 binding to the SNARE complex in solution is not perturbed by eC2AB
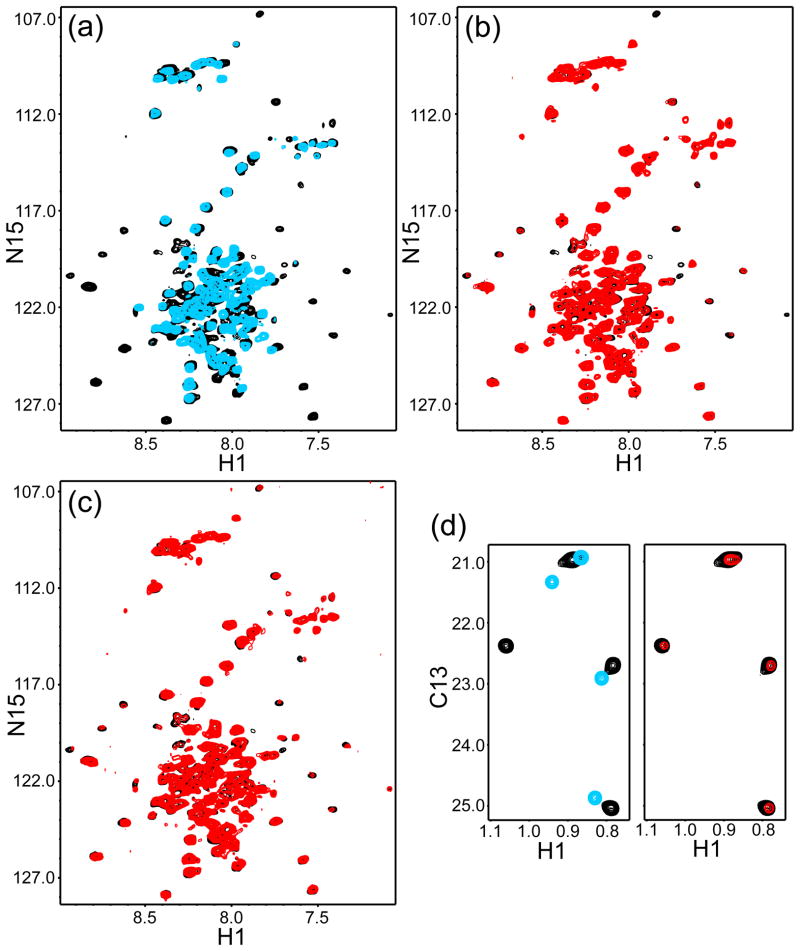
To explore whether binding of the eC2AB fragment influences the binding of complexin-I to the SNARE complex, we analyzed transverse relaxation optimized spectroscopy (TROSY)-enhanced 1H-15N HSQC spectra of uniformly 2H,15N complexin-I. The spectrum of the isolated 2H,15N complexin-I exhibited poor dispersion because of the lack of tertiary structure (Fig. 3a, blue contours), as described previously36. Binding to the SNARE complex caused dispersion of a subset of the compexin-I cross-peaks corresponding to its SNARE complex-binding region (Fig. 3a, black contours), as observed previously for Cpx26-8337. Addition of eC2AB fragment to the 2H,15N complexin-I/SNARE complex in the absence or presence of Ca2+ did not perturb the poorly dispersed cross-peaks corresponding to unstructured regions, but caused marked broadening of the well-resolved cross-peaks from the structured sequences of 2H,15N complexin-I without inducing significant shifts (Figs. 3b,c, compare black and red contours; note that some slight shifts were observed but they correspond to weak cross-peaks with poor signal-to-noise ratios). These results confirm the conclusion drawn from the gel filtration data that the eC2AB fragment binds to the complexin-I/SNARE complex, and indicate that the eC2AB fragment binds to the SNARE complex without perturbing complexin-I or its binding mode with the SNARE complex.
Figure 3.
Binding of the synaptotagmin-1 C2AB fragment does not perturb the binding mode of complexin-1 on the SNARE complex. (a) 1H-15N TROSY-HSQC spectra of 40 μM 2H,15N-labeled complexin-I in 1 mM EDTA and in the absence (blue contours) or presence (black contours) of 60 μM SNARE complex. (b,c) 1H-15N TROSY-HSQC spectra of the same sample containing 40 μM 2H,15N-labeled complexin-I and 60 μM SNARE complex in 1 mM EDTA (black contours), and of samples that contained 33 μM 2H,15N-labeled complexin-I, 50 μM SNARE complex and 50 μM eC2AB fragment in 1 mM EDTA (panel b, red contours) or 12 μM 2H,15N-labeled complexin-I, 18 μM SNARE complex and 18 μM eC2AB fragment in 1 mM Ca2+ (panel c, red contours). (d) 1H-13C HMQC spectra of 30 μM 2H-LV-13CH3-Cpx26-83 (blue contours), 15 μM 2H-LV-13CH3-Cpx26-83 plus 18 μM SNARE complex (black contours) and 10 μM 2H-LV-13CH3-Cpx26-83 plus 12 μM SNARE complex and 15 μM eC2AB fragment in 1 mM Ca2+ (red contours). In (b–d), contour levels were adjusted to correct for differences in the concentrations of 2H,15N-labeled complexin-I or 2H-LV-13CH3-Cpx26-83, and in the number of scans.
Analysis of the methyl region of 1H-13C heteronuclear multiple quantum coherence (HMQC) spectra of full-length complexin-I to further study the interplay with the eC2AB fragment for binding to the SNARE complex is complicated by the strong overlap of its multiple methyl resonances. However, Cpx26-83 contains only one leucine residue (L41 in the accessory helix) and one valine residue (V61 in the central helix) and, consequently, perdeuterated Cpx26-83 specifically 1H,13C-labeled at Leu,Val methyl groups (2H-LV-13CH3-Cpx26-83) yields high quality 1H-13C HMQC spectra that are readily interpretable. SNARE complex binding caused defined shifts on the four methyl cross-peaks of 2H-LV-13CH3-Cpx26-83 (Fig. 3d; compare blue and black cross-peaks), and addition of eC2AB fragment caused broadening of the cross-peaks from the 2H-LV-13CH3-Cpx26-83/SNARE complex without inducing significant shifts (Fig. 3d; compare red and black cross-peaks). These results further support the notion that the eC2AB fragment binds to the complexin-I/SNARE complex but does not contact complexin-I.
We also tested whether presence of the eC2AB fragment affects the affinity of Cpx26-83 or complexin-I for the SNARE complex using ITC. These experiments were performed in the presence of 1 mM Ca2+ to enhance the affinity of the eC2AB fragment for the SNARE complex, thus increasing the likelihood that the eC2AB fragment might perturb the complexin/SNARE complex affinity, but using sufficiently low SNARE complex and eC2AB fragment concentrations to minimize their aggregation together. The Kds of Cpx26-83 and complexin-I for the SNARE complex measured in the absence of eC2AB fragment were 25.3 ± 6.8 nM and 9.7 ± 2.0 nM, respectively (Figs. 4a,b). These results suggest that full-length complexin-I binds to the SNARE complex with somewhat higher affinity than Cpx26-83, perhaps because of the participation of the complexin-I N-terminus in binding40. However, it is unclear whether this difference is significant because there were some systematic deviations of the data from an ideal 1:1 binding model at low SNARE complex molar ratios for the Cpx26-83 experiments. The source of this behavior is unclear but, regardless of this issue, we obtained very similar results in the presence of eC2AB fragment, with Kds of 20.4 ± 6.3 nM and 11.3 ± 4.7 nM for the interaction of the SNARE complex with Cpx26-83 and complexin-I, respectively. Together with the gel filtration and NMR data, these results show that complexin-I and the C2AB fragment have independent binding sites for the SNARE complex used in these studies, which was formed with the syntaxin-1(191–253) fragment.
Figure 4.
The eC2AB fragment does not alter the affinity of complexin-I for the SNARE complex. Cpx26-83 (a,c) or complexin-I (b,d) (100 μM) were titrated into 10 μM soluble SNARE complex in presence of 1 mM Ca2+ and the absence (a,b) or presence (c,d) of 10 μM eC2AB fragment.
Complexin-I increases the solubility of the SNARE complex
As explained above, the presence of complexin-I decreases the tendency of the eC2AB fragment and the SNARE complex to precipitate in the presence of Ca2+. Since this precipitation appears to arise from the existence of multiple types of electrostatic interactions between the synaptotagmin-1 C2 domains and the SNARE complex52, and is likely favored by the tendency of the SNARE complex itself to self-associate (see discussion), we hypothesized that complexin-I might hinder aggregation of the SNARE complex. To test this hypothesis, we used a SNARE complex formed with a non-truncated SNARE motif of syntaxin-1 (residues 191–259), which, as explained above, has a much higher tendency to aggregate than the SNARE complex containing syntaxin-1(191–253). Intriguingly, when we assembled the non-truncated SNARE complex at 10 μM concentration and concentrated the sample 10-fold, a large percent of the complex precipitated (Fig. 5, lanes 1 and 2), but such precipitation was completely prevented if we included 12 μM complexin-I during the assembly reaction or after the assembly was completed (Fig. 5, lanes 3–6). This is an important result to consider when interpreting the available data on competition between complexins and synaptotagmin-1 because the disruption of SNARE complex aggregation caused by complexins could provide an indirect mechanism that hinders synaptotagmin-1 binding to SNARE complex oligomers. Such mechanism could reconcile the clear observation of simultaneous binding of complexin-I and the eC2AB fragment to the SNARE complex in our experiments (Figs. 2 and 3) with the competition observed in pulldown assays performed with non-truncated SNARE complex10 (see discussion).
Figure 5.
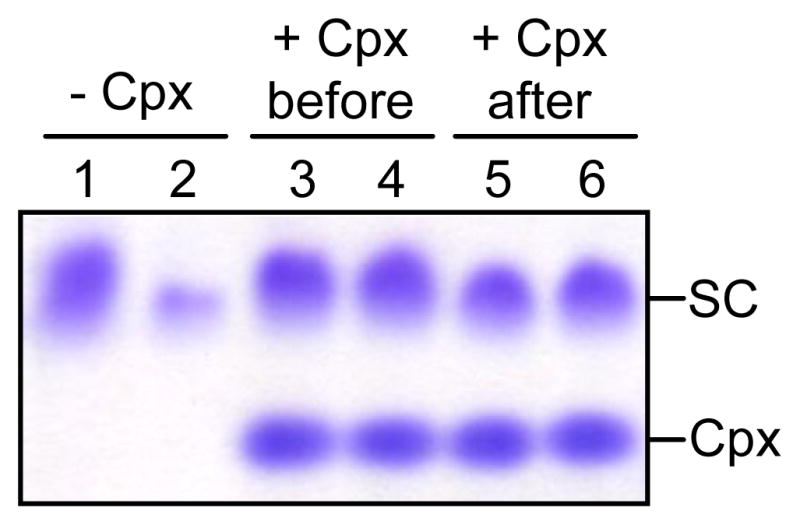
Complexin-I increases the solubility of the SNARE complex. SNARE complexes containing non-truncated syntaxin-1 SNARE motif (residues 191–259) were assembled in the absence or presence of complexin-I. For one of the samples, complexin-I was added after assembly. Samples were concentrated 10-fold and diluted 10-fold. Aliquots of the samples before (lanes 1, 3, and 5) or after (lanes 2, 4 and 6) the concetration-dilution procedure were analyzed by SDS-PAGE and commossie blue staining. The position of complexin-I (Cpx) and SNARE complex (SC) in the gels is indicated.
Subtle interplay between complexin-I and synaptotagmin-1 on membrane-anchored SNARE complex
The lack of competition observed in our solution experiments (Figs. 2 and 3) also contrasts with our finding that the C2AB fragment displaces the Cpx26-83 fragment labeled with a BODIPY- FL fluorescent probe from membrane anchored SNARE complex. This result was obtained using a confocal microscope to detect BODIPY-FL-Cpx26-83 bound to SNARE complex anchored to supported bilayers that were deposited on glass slides within microchannels, and monitoring the fluorescence as a function of added C2AB fragment10, 22, 24. Conversely, no such competition was observed in liposome floatation assays using either Cpx28-83 or full-length complexin-I45. To shed light on this apparent contradiction, we performed competition assays in supported bilayers desposited within microchannels using a similar setup to that we employed previously10, 22, 24 but detecting fluorescently-labeled complexin-I fragments with total internal reflection fluorescence (TIRF) microscopy because this approach offers an important technical advantage.
With the use of a confocal microscope, the fluorescence of BODIPY-FL-Cpx26-83 bound to the membrane-anchored SNARE complex after injecting the fluorescent protein into the microchannel could not be measured directly because the fluorescent protein remaining in solution yielded too much background fluorescence; hence, a wash with buffer was required before measurement of the fluorescence intensity remaining on the membrane10, 22. In contrast, the small volume of the detectable region above the glass in TIRF microscopy greatly reduces the background fluorescence from the fluorescent protein that remains in solution. Indeed, in initial experiments we found that the fluorescence intensity observed by TIRF after injecting 50 nM BODIPY-FL-Cpx26-83 into microchannels that contained plain phospholipid bilayers (without anchored SNARE complex) was less than 1% of the fluorescence observed after injecting the same solution into microchannels containing membrane-anchored SNARE complex (formed with a syntaxin-1 fragment spanning residues 183–288, which include the full SNARE motif and transmembrane sequence). Such low background fluorescence from the protein remaining in solution is expected based on the concentration of BODIPY-FL-Cpx26-83 and the SNARE complex-to-lipid ratio used (1:1000), and assuming that most membrane-bound SNARE complexes bound to one BODIPY-FL-Cpx26-83 molecule. Hence, the background fluorescence from the protein remaining in solution could be considered negligible (well within the error of our measurements), and the TIRF approach allowed us direct observation of the competition between the C2AB fragment and fluorescently-labeled complexin fragments under equilibrium conditions, without the requirement of washing steps. Moreover, we could monitor the effects of washing steps to compare our data with the results obtained previously with a confocal microscope.
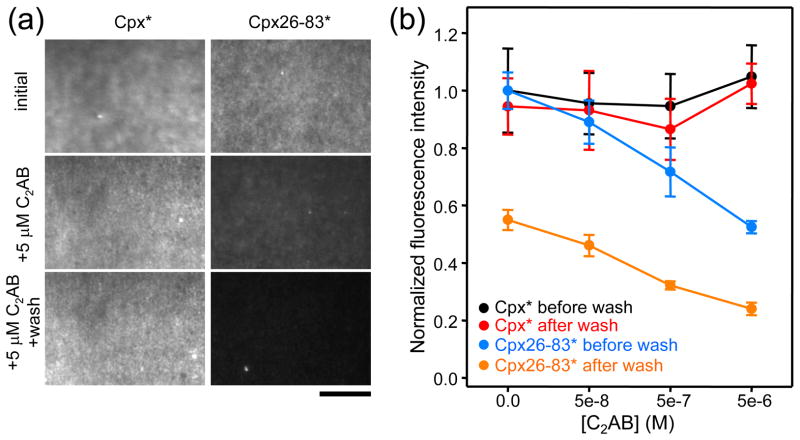
Since in the initial experiments we observed considerable photobleaching of the BODIPY-FL probe, further studies were performed with Cpx26-83 labeled with Alexa488 (below referred to as Cpx26-83*), which exhibited less photobleaching under our experimental setup. In addition, we performed parallel, side-by-side experiments with full-length complexin-1 also labeled with Alexa488 (complexin-I*) to directly compare the results obtained with both proteins. For each set of experiments, we first injected 50 nM complexin-I* or Cpx26-83* without C2AB fragment into a microchannel, washed with buffer, and then performed additional, consecutive injections of 50 nM complexin-I* or Cpx26-83* together with increasing concentrations of C2AB fragment, including a washing step with buffer alone between each injection of proteins. All samples contained 1 mM Ca2+. TIRF images were obtained in each step to measure the fluorescence on the membrane, and all the data were normalized to the fluorescence observed after the initial injection of complexin-I* or Cpx26-83*. Representative examples of the images obtained are shown in Fig. 6a, and Fig. 6b shows plots of the fluorescence measured as a function of C2AB fragment concentration before and after the washing step.
Figure 6.
The synaptotagmin-1 C2AB fragment displaces Cpx26-83 but not complexin-I from membrane-anchored SNARE complexes. (a) Representative TIRF images of supported bilayers within microchannels that contained membrane-anchored SNARE complexes formed with syntaxin-1(183–288). Images were taken after adding 50 nM complexin-I* (Cpx*) or Cpx26-83* alone or with 5 μM C2AB fragment; for the later, images were taken before and after washing. Scale bar: 10 μm. (b) Plots of normalized fluorescence intensities measured in supported bilayers as in panel (a) with different C2AB fragment concentrations before and after washing, as indicated. Fluorescence intensities from six images taken under each condition were averaged. Error bars represent standard deviations.
For complexin-I*, no substantial decrease in fluorescence intensity was observed neither after the washing steps nor with inclusion of C2AB fragment, even up to 5 μM. These results show that complexin-I* binds to the membrane-anchored SNARE complex with very high affinity and that the C2AB fragment does not displaced complexin-I* from the SNARE complex under these conditions. For the Cpx26-83* fragment, the initial fluorescence was comparable to that observed for complexin-I*, showing that the Cpx26-83* fragment also binds to the membrane-anchored SNARE complex with high affinity. However, the washing step in the absence of C2AB fragment led to a 45% reduction in fluorescence for the Cpx26-83* fragment. Thus, complexin-I has a higher affinity for the membrane-anchored SNARE complex, which may arise from additional interactions with the SNAREs mediated by the complexin-I N-terminus40 and/or from binding of the complexin-I C-terminus to the phospholipids44. We also found that inclusion of increasing concentrations of the C2AB fragment led to progressive decreases of fluorescence for the Cpx26-83* fragment both before and after the washes. The decrease in fluorescence observed after the washes exhibited a similar dependence on the C2AB fragment concentration as that observed in our previous experiments with a confocal microscope10, 22, 24, showing the overall consistency of the results obtained with this type of approach. Importantly, the progressive decreases in fluorescence observed before the washing steps show that the C2AB fragment can favor the dissociation of Cpx26-83* from membrane- anchored SNARE complex under equilibrium conditions and hence confirm that some form of competition exists between the C2AB fragment and parts of the complexin-1 sequence for binding to the SNARE complex on a membrane. This competition may be direct or indirect (see below) but may be critical to trigger neurotransmitter release, and the finding that such competition is not observed for complexin-I suggests that additional interactions involving its N- and/or C-terminal sequences compensate for the effects of C2AB fragment binding to the SNARE complex that favor dissociation of Cpx26-83*.
Discussion
Understanding how interactions among the SNAREs, synaptotagmin-1 and complexins are coordinated remains one the most crucial enigmas that need to be solved in order to elucidate the mechanism of neurotransmitter release. A convergence of results led to a widespread model whereby complexins inhibit release and Ca2+-bound synaptotagmin-1 releases this inhibition to trigger release10–13. This model was supported by data showing competition between synaptotagmin-1 and complexin-I for binding to the SNARE complex in solution and on membranes10, 22, but other results argued against such competition11, 45, leading some authors to question the model45. The experiments presented here were designed to shed light onto the apparent contradictions emerging for these studies and to help solving the complex puzzle formed by the multiple interactions that govern exocytosis. Together with previous studies, our results establish four pieces of this puzzle: i) N- and C-terminal sequences of complexin-I markedly strengthen binding to membrane-anchored complex; ii) synaptotagmin-1 and complexin-I can bind simultaneously to the SNARE complex in solution; iii) synaptotagmin-1 favors dissociation of a complexin-I fragment spanning its accessory and central helices (Cpx26-83) from membrane-anchored SNARE complex; and iv) such dissociation is hindered by N- and/or C-terminal sequences of complexin-I. Altogether, these results support the notion that a delicate interplay between interactions of synaptotagmin-1 and complexins with the SNAREs and the membranes govern Ca2+-triggering of synaptic vesicle fusion.
Neurotransmitter release depends not only on the SNAREs, synaptotagmin-1 and complexin-1 but also on several additional key proteins1–4, all together forming a sophisticated and complex protein machinery that mediates very fast membrane fusion after Ca2+ influx into a presynaptic terminal (in the 100 μs time scale54). The primed state that is formed before Ca2+ influx is likely metastable and involves a balance between multiple interactions among these proteins and of some of these proteins with the membranes. These features suggest that simple concepts often used to describe molecular mechanisms, such as ‘protein x is an inhibitor’ or ‘two proteins can bind simultaneously to a target’, may not be adequate to accurately describe the mechanism of release at or near atomic resolution. Indeed, dual active and inhibitory roles have been proposed not only for complexins but also for other critical proteins involved in release such as synaptotagmin-155, Munc18–1 and syntaxin-156, 57. Moreover, an increasing number of weak interactions of the components of the release machinery are being identified (e.g. refs. 40, 58–60). These interactions might be crucial to lower energy barriers and cause conformational rearrangements that lead to membrane fusion60, 61, and might be altered by other interactions without an overt effect in overall binding as detected in some binding assays. These considerations are important to interpret and attempt to reconcile the available data on the interplay between interactions of synaptotagmin-1 and complexin-1 with the SNARE complex.
A key result that appears to be contradictory to multiple data is the observation of competition between the C2AB fragment and the Cpx26-83* fragment for binding to SNARE complex anchored to a planar bilayer in our TIRF experiments (Fig. 6). Thus, no such competition with Cpx26-83 was observed in co-floatation assays with liposome-anchored SNARE complex45, or with complexin-I in co-floatation assays11, 45 and in our TIRF studies (Fig. 6), or with Cpx26-83 and complexin-I in our solution studies (Figs. 2–4). Moreover, a model of the synaptotagmin-1/SNARE complex built from single molecule fluorescence spectroscopy data suggests that the binding site of synaptotagmin-1 on the SNARE complex does not overlap with the Cpx26-83-binding site62. Differences in membrane curvature between our supported bilayers and liposomes were proposed to provide a potential reason for the observation of competition between the C2AB fragment and Cpx26-83 in our microchannels experiments10, 22 and the lack of it in the co-floatation assays45. However, this explanation seems unlikely because membrane curvature presumably increases the affinity of the C2AB fragment for membranes63. We favor the view that differences between the results obtained in the two systems may simply arise from technical reasons, including for instance the distinct times scales involved and the large differences in the concentrations of reagents used. Furthermore, the lack of quantification in the co-floatation assays45 limits the conclusiveness of the data and the potential to detect subtle effects, particularly at the saturating protein concentrations that were used. Conversely, the requirement for a washing step before observation10 was a limitation of the microchannels assays using a confocal microscope. The TIRF approach used here overcomes this problem by allowing direct measurement under equilibrium conditions and analysis of the effects of the washes.
The displacement of Cpx26-83* by the C2AB fragment observed by TIRF before and after washing (Fig. 6), and the consistency of these data with the results obtained with a similar setup and a confocal microscopy in three separate studies10, 22, 24, leave little doubt that there is indeed some form of competition between the C2AB fragment and Cpx26-83* for binding to membrane-anchored SNARE complex. Note however that such competition may not necessarily be direct and hence may not contradict the lack of competition observed in solution. Thus, even if the C2AB fragment and Cpx26-83 can bind simultaneously to separate sites on the membrane- anchored SNARE complex, consistent with the solution data, the simultaneous binding of the C2AB fragment to the SNARE complex and the membrane could change the overall orientation of the SNARE complex with respect to the membrane and force steric clashes of the accessory helix from the bound Cpx26-83* fragment with the membrane. Alternatively, since more than one region of the SNARE complex have been implicated in synaptotagmin-1 binding to the SNARE complex (see ref. 22), it is plausible that the major binding site for synaptotagmin-1 in solution is different from the binding site that predominates in a membrane environment, and that this second site involves at least partial competition with Cpx26-83*. This second possibility is supported by the finding that mutations in the C-terminus of SNAP-25 hinder the displacement of Cpx26-83* from membrane-anchored SNARE complex by the C2AB fragment, as binding of the C2AB fragment to this C-terminal region of SNAP-25 could result in steric clashes with the Cpx26-83* accessory helix22. Note also in this context that a recent study showed that the juxtamembrane sequence of syntaxin-1, which is present in our microchannels assays but not in our solution studies, mediates binding of synaptotagmin-1 to syntaxin-1 clusters on membranes; however, as pointed out by the authors, this effect is indirect, arising from the enrichment of PIP2 in these clusters and the affinity of synaptotagmin-1 for PIP2 64.
Both of these models (invoking steric hindrance due to the membrane or to a switch in binding site) envision that the putative steric clashes are small and that the C2AB fragment does not interfere directly with binding of the central helix of complexin-I to the SNARE complex. Thus, according to both models, the C2AB fragment and Cpx26-83* might be able to bind simultaneously to the SNARE complex, but the resulting clashes could increase the dissociation rate of Cpx26-83* from the SNARE complex. Our TIRF data showing that the C2AB fragment can displace Cpx26-83* but not complexin-I from membrane-anchored SNARE complex reveal that complexin-I dissociation is prevented or at least hindered by interactions involving the N- and C-terminal sequences of complexin-1, which are not present in Cpx26-83* and have been implicated in interactions with the SNARE complex40 and phospholipids44, respectively. Our TIRF assays also show that such interactions also stabilize binding of complexin-I to membrane-anchored SNARE complex even in the absence of C2AB fragment, as the partial release of Cpx26-83* by washing is not observed for complexin-I (Fig. 6). These observations suggest that binding of the C2AB fragment to membrane-anchored complexin-I/SNARE complex may cause a conformational rearrangement in complexin-I analogous to that caused on Cpx26-83, but without leading to complexin-I dissociation because of the additional interactions. Importantly, such rearrangement might represent a crucial step to relieve the inhibition caused by complexin-I and enable cooperation between synaptotagmin-1 and complexin-I to trigger neurotransmitter release.
The complexity of analyzing the interactions involved in this system is also illustrated by our finding that the C2AB fragment and complexin-I can bind simultaneously to the SNARE complex in solution (Figs. 2–4) and yet complexin-I released soluble SNARE complex from immobilized GST-C2AB fragment in pulldown assays10. The reason for this contradiction is unclear, but we note that the SNARE complex used in the pulldown assays contained the full SNARE motif of syntaxin-1 and has a much higher tendency to aggregate than the soluble SNARE complex used in our experiments37. Since ongoing structural studies with paramagnetic probes indicate the existence of multiple weak binding sites on the SNARE complex and the C2AB fragment (our unpublished data), and multivalency underlies the high tendency of the SNARE complex and the C2AB fragment to precipitate together52, it is plausible that aggregation of the SNARE complex used in the pulldown assays increased its affinity for the C2AB fragment. Our finding that complexin-I binding strongly hinders aggregation of the SNARE complex (Fig. 5) suggests that inclusion of complexin-I in the pulldown assays impaired the aggregation of the SNARE complex, thus decreasing its affinity for the C2AB fragment indirectly.
Much research will be required to distinguish between the different possibilities discussed above, but the results described here clearly establish that synaptotagmin-1 indeed influences the interactions of complexins with the membrane-anchored SNARE complex, and suggest that complexin N- and/or C-terminal sequences play an important role(s) in this subtle interplay. The continued development of biophysical approaches to obtain structural information of proteins on membranes (e.g. refs. 62, 65) will be critical to further understand this interplay.
Materials and Methods
Recombinant proteins
Expression vectors and protocols for expression and purification of the following protein fragments in E. coli were described previously: rat synaptotagmin-1 C2AB fragment residues 140–421 (C2AB fragment), rat syntaxin 1A residues 191–253, 191–259 and 183–288, rat synaptobrevin 2 residues 29–93, human SNAP-25 residues 11–82 and 141–203, rat complexin-I full-length (residues 2–134), and rat complexin-I residues 26–83 in WT or V61C mutant versions10, 22, 23, 36, 37, 51, 66. A construct to express rat synaptotagmin-1 residues 131–421 (eC2AB fragment) was made by standard recombinant DNA techniques using custom designed primers, and the protein was expressed in E. coli and purified as described for the C2AB fragment23. Uniform 2H,15N-labeling was performed by growing E. coli BL21(DE3) in minimal medium made with D2O as the solvent and using 15NH4Cl as the sole nitrogen source. Perdeuteration with specifically 1H,13C-labeling at Leu,Val methyl groups was accomplished as described60. Labeling of full-length complexin-I (which contains one native cysteine, C105) and the complexin-I V61C mutant fragment with BODIPY-FL or Alexa488 was performed as described10, 23.
SNARE complex assembly
Unless otherwise indicated, SNARE complexes were formed with SNAP-25 (11–82), SNAP-25 (141–203), syntaxin-1(191–253) and synaptobrevin(29–93). In some cases, we used syntaxin-1(191–259) or syntaxin-1(183–288) as indicated. Complex assembly was accomplished by incubating a mixture of the purified fragments overnight, and removing remaining unassembled fragments by concentration-dilution cycles with a 30 kDa cuttoff37.
Isothermal titration calorimetry
ITC experiments were performed using a VP-ITC system (MicroCal) at 20°C in 25 mM HEPES pH 7.5, 125 mM NaCl, 0.3 mM TCEP (buffer A) containing 1 mM Ca2+. Complexin-I (150 μM) was titrated into the chamber containing C2AB fragment (10 μM). Complexin-I and Cpx26-83 (100 μM) were titrated into 10 μM soluble SNARE complex in the absence or presence of 10 μM eC2AB fragment. All proteins were purified by gel filtration on a Superdex 75 column (Amersham Biosciences) and dialyzed in the same buffer before the experiments. The data were fitted with a nonlinear least squares routine using a single-site binding model with Origin for ITC v.5.0 (Microcal). The Kd values reported constitute averages of three experiments performed under the same conditions, and the errors correspond to the standard deviations.
Gel filtration assays
Binding experiments by gel filtration were performed using a Superdex S200(10/300GL) (GE Healthcare). Buffer A with 1 mM Ca2+ was used for elution and to dissolve all samples. The appropriate proteins were mixed to a final concentration of 15 μM (1 mL total volume) and injected into the column. Fractions were analyzed by SDS-PAGE and coomassie blue staining. MALS analysis was performed using a miniDAWN Treos detector (Wyatt Technology, Santa Barbara, CA).
NMR spectroscopy
1H-15N HSQC spectra were acquired on a Varian INOVA600 spectrometer equipped with a room temperature triple resonance probe, 1H-15N TROSY-HSQC spectra were acquired on a Varian INOVA800 spectrometer equipped with a cold probe, and 1H-13C HMQC spectra were acquired on a Varian INOVA600 spectrometer equipped with a cold probe. All spectra were acquired at 25 °C using 20 mM HEPES pH 7.1, 120 mM NaCl, 0.3 mM TCEP with 5% D O (for 1H-15N HSQC and TROSY-HSQC) or 100% D2O (for 1H-13C HMQC) as the solvent, and 1 mM EDTA or 1 mM Ca2+ as needed. Sample concentrations are indicated in the figure legends. The data were processed with NMRPipe67 and analyzed with NMRView68.
SNARE complex solubility tests
Three samples containing 10 μM each of SNAP-25 (11–82), SNAP-25 (141–203), syntaxin-1(191–259) and synaptobrevin(29–93) were incubated at room temperature for five hours. Complexin-I (12 μM) was added to one of the samples before incubation and to another sample after incubation. After taking an aliquot for analysis, each sample was concentrated 10-fold, diluted 10-fold, span down on a microfuge to remove precipitated material. Initial aliquots and final samples were analyzed by SDS-PAGE and coomassie blue staining.
Complexin displacement assays monitored by TIRF
Purified syntaxin 1A(183–288), SNAP-25 (11–82), SNAP-25 (141–203) and synaptobrevin(29–93) were used to assemble SNARE complexes in the presence of 1% β-OG. Liposomes (100 nm diameter) containing 1,2-dioleoyl-sn-glycero-3-[phospho-L-serine] (DOPS) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (Avanti Polar Lipids) 15:85 were prepared by extrusion through 0.8 μm polycarbonate membranes, and SNARE complexes were reconstituted into these preformed liposomes in reconstitution buffer (25 mM HEPES pH7.4, 100 mM KCl, 0.3mM TCEP) as described66 but with a 1:1000 protein:lipid ratio. We formed microfluidic channels by conformally attaching a glass coverslip to a poly(dimethylsiloxane) stamp presenting microscopic grooves (two parallel grooves per stamp)22. Each microchannel was 200- m wide, ~140- μm high, and 2-cm long. Lanes of supported bilayers containing SNARE complexes were formed within the microfluidic channels by allowing the SNARE complex-containing proteoliposomes (1 mM lipids) to fuse with the glass within the microchannels. Samples (6 μL) containing 50 nM V61C mutant complexin-I or Cpx26-83 labeled with Alexa488 in reconstitution buffer including 1 mM Ca2+ were loaded into the parallel microchannels, washed with buffer (6 μL) and identical samples were loaded again to ensure saturation of the membrane-anchored SNARE complexes with complexin-I* or Cpx26-83*. Microchannels were then washed again with of reconstitution buffer containing 1 mM Ca2+ and samples containing 50 nM complexin-I* or Cpx26-83* plus increasing concentrations of C2AB fragment (50, 500 and 5000 nM) in reconstitution buffer with 1 mM Ca2+ were successively loaded with a washing step in between. TIRF images of the supported bilayers were obtained on a Zeiss Axiovert 200M microscope with a Plan-Apochromat 100x 1.46 NA objective and Slidebook software (Intelligent Imaging). Alexa488 dyes were excited with a Saphire HP 488 nm laser from Coherent, and emission was detected through a Brightline FITC 3540B dichroic and emission filter (Semrock) using a QuantEM camera (Roper Scientific). Six TIRF images of each microchannels were taken at each step and the average fluorescence in each image was quantified with Image J (NIH, MD). The six averaged values were then used to calculate average fluorescence intensities and standard deviations, all normalized by the fluorescence of the first sample after loading 50 nM complexin-I* or Cpx26-83*. Fig. 6 shows data for representative experiments. Three additional experiments with 50 nM complexin-I* or Cpx26-83* were performed, yielding comparable results.
The functional interplay between synaptotagmin and complexin is unclear
Complexin N- and C-terminal sequences strengthen SNARE complex-binding on membranes
Synaptotagmin displaces a core complexin fragment from the SNARE complex on membranes
Synaptotagmin does not displace complexin from the SNARE complex on membranes
There is a delicate interplay between synaptotagmin and complexin for SNARE binding
Acknowledgments
We thank Yilun Sun for expert technical assistance on protein expression and purification, Kate Lubby-Phelps for help with the use of the TIRF microscope, and Thomas Südhof for fruitful discussions. Parts of this research were supported by faculty startup to R.P.-C. from the New Jersey Institute of Technology. This work was supported by grant I-1304 from the Welch Foundation and grant NS40944 from the NIH (to J.R.).
Abbreviations used
- HSQC
heteronuclear single quantum correlation
- NMR
nuclear magnetic resonance
- MALS
multiangle light scattering
- PIP2
phosphatidylinositol-4,5-bisphosphate
- SNARE
soluble N-ethylmaleimide sensitive factor attachment protein receptor
- TIRF
total internal reflection fluorescence
- TROSY
transverse relaxation optimized spectroscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Rizo J, Sudhof TC. The Membrane Fusion Enigma: SNAREs, Sec1/Munc18 Proteins, and Their Accomplices-Guilty as Charged? Annu Rev Cell Dev Biol. 2012;28:279–308. doi: 10.1146/annurev-cellbio-101011-155818. [DOI] [PubMed] [Google Scholar]
- 2.Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorensen JB. Conflicting views on the membrane fusion machinery and the fusion pore. Annu Rev Cell Dev Biol. 2009;25:513–537. doi: 10.1146/annurev.cellbio.24.110707.175239. [DOI] [PubMed] [Google Scholar]
- 4.Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Q Rev Biophys. 2005:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 5.Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 6.Poirier MA, Xiao W, Macosko JC, Chan C, Shin YK, Bennett MK. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat Struct Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- 7.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 8.Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Chacon R, Konigstorfer A, Gerber SH, Garcia J, Matos MF, Stevens CF, Brose N, Rizo J, Rosenmund C, Sudhof TC. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 10.Tang J, Maximov A, Shin OH, Dai H, Rizo J, Sudhof TC. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 11.Schaub JR, Lu X, Doneske B, Shin YK, McNew JA. Hemifusion arrest by complexin is relieved by Ca2+-synaptotagmin I. Nat Struct Mol Biol. 2006;13:748–750. doi: 10.1038/nsmb1124. [DOI] [PubMed] [Google Scholar]
- 12.Giraudo CG, Eng WS, Melia TJ, Rothman JE. A clamping mechanism involved in SNARE-dependent exocytosis. Science. 2006;313:676–680. doi: 10.1126/science.1129450. [DOI] [PubMed] [Google Scholar]
- 13.Roggero CM, De Blas GA, Dai H, Tomes CN, Rizo J, Mayorga LS. Complexin/synaptotagmin interplay controls acrosomal exocytosis. J Biol Chem. 2007;282:26335–26343. doi: 10.1074/jbc.M700854200. [DOI] [PubMed] [Google Scholar]
- 14.Sutton RB, Davletov BA, Berghuis AM, Sudhof TC, Sprang SR. Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell. 1995;80:929–938. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 15.Ubach J, Zhang X, Shao X, Sudhof TC, Rizo J. Ca2+ binding to synaptotagmin: how many Ca2+ ions bind to the tip of a C2-domain? EMBO J. 1998;17:3921–3930. doi: 10.1093/emboj/17.14.3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao X, Fernandez I, Sudhof TC, Rizo J. Solution structures of the Ca2+-free and Ca2+-bound C2A domain of synaptotagmin I: does Ca2+ induce a conformational change? Biochemistry. 1998;37:16106–16115. doi: 10.1021/bi981789h. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez I, Arac D, Ubach J, Gerber SH, Shin O, Gao Y, Anderson RG, Sudhof TC, Rizo J. Three-dimensional structure of the synaptotagmin 1 c(2)b-domain. Synaptotagmin 1 as a phospholipid binding machine. Neuron. 2001;32:1057–1069. doi: 10.1016/s0896-6273(01)00548-7. [DOI] [PubMed] [Google Scholar]
- 18.Chapman ER, Davis AF. Direct interaction of a Ca2+-binding loop of synaptotagmin with lipid bilayers. J Biol Chem. 1998;273:13995–14001. doi: 10.1074/jbc.273.22.13995. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Rizo J, Sudhof TC. Mechanism of phospholipid binding by the C2A-domain of synaptotagmin I. Biochemistry. 1998;37:12395–12403. doi: 10.1021/bi9807512. [DOI] [PubMed] [Google Scholar]
- 20.Rhee JS, Li LY, Shin OH, Rah JC, Rizo J, Sudhof TC, Rosenmund C. Augmenting neurotransmitter release by enhancing the apparent Ca2+ affinity of synaptotagmin 1. Proc Natl Acad Sci U S A. 2005;102:18664–18669. doi: 10.1073/pnas.0509153102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhalla A, Chicka MC, Tucker WC, Chapman ER. Ca(2+)-synaptotagmin directly regulates t-SNARE function during reconstituted membrane fusion. Nat Struct Mol Biol. 2006;13:323–330. doi: 10.1038/nsmb1076. [DOI] [PubMed] [Google Scholar]
- 22.Dai H, Shen N, Arac D, Rizo J. A Quaternary SNARE-Synaptotagmin-Ca(2+)-Phospholipid Complex in Neurotransmitter Release. J Mol Biol. 2007;367:848–863. doi: 10.1016/j.jmb.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arac D, Chen X, Khant HA, Ubach J, Ludtke SJ, Kikkawa M, Johnson AE, Chiu W, Sudhof TC, Rizo J. Close membrane-membrane proximity induced by Ca(2+)-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat Struct Mol Biol. 2006;13:209–217. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]
- 24.Xue M, Ma C, Craig TK, Rosenmund C, Rizo J. The Janus-faced nature of the C(2)B domain is fundamental for synaptotagmin-1 function. Nat Struct Mol Biol. 2008;15:1160–1168. doi: 10.1038/nsmb.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackler JM, Drummond JA, Loewen CA, Robinson IM, Reist NE. The C(2)B Ca(2+)-binding motif of synaptotagmin is required for synaptic transmission in vivo. Nature. 2002;418:340–344. doi: 10.1038/nature00846. [DOI] [PubMed] [Google Scholar]
- 26.McMahon HT, Missler M, Li C, Sudhof TC. Complexins: cytosolic proteins that regulate SNAP receptor function. Cell. 1995;83:111–119. doi: 10.1016/0092-8674(95)90239-2. [DOI] [PubMed] [Google Scholar]
- 27.Reim K, Mansour M, Varoqueaux F, McMahon HT, Sudhof TC, Brose N, Rosenmund C. Complexins regulate a late step in Ca2+-dependent neurotransmitter release. Cell. 2001;104:71–81. doi: 10.1016/s0092-8674(01)00192-1. [DOI] [PubMed] [Google Scholar]
- 28.Maximov A, Tang J, Yang X, Pang ZP, Sudhof TC. Complexin controls the force transfer from SNARE complexes to membranes in fusion. Science. 2009;323:516–521. doi: 10.1126/science.1166505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue M, Stradomska A, Chen H, Brose N, Zhang W, Rosenmund C, Reim K. Complexins facilitate neurotransmitter release at excitatory and inhibitory synapses in mammalian central nervous system. Proc Natl Acad Sci U S A. 2008;105:7875–7880. doi: 10.1073/pnas.0803012105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huntwork S, Littleton JT. A complexin fusion clamp regulates spontaneous neurotransmitter release and synaptic growth. Nat Neurosci. 2007;10:1235–1237. doi: 10.1038/nn1980. [DOI] [PubMed] [Google Scholar]
- 31.Hobson RJ, Liu Q, Watanabe S, Jorgensen EM. Complexin Maintains Vesicles in the Primed State in C. elegans. Curr Biol. 2011;21:106–113. doi: 10.1016/j.cub.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin JA, Hu Z, Fenz KM, Fernandez J, Dittman JS. Complexin has opposite effects on two modes of synaptic vesicle fusion. Curr Biol. 2011;21:97–105. doi: 10.1016/j.cub.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon TY, Lu X, Diao J, Lee SM, Ha T, Shin YK. Complexin and Ca2+ stimulate SNARE-mediated membrane fusion. Nat Struct Mol Biol. 2008;15:707–713. doi: 10.1038/nsmb.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diao J, Grob P, Cipriano DJ, Kyoung M, Zhang Y, Shah S, Nguyen A, Padolina M, Srivastava A, Vrljic M, Shah A, Nogales E, Chu S, Brunger AT. Synaptic proteins promote calcium-triggered fast transition from point contact to full fusion. elife. 2012;1:e00109. doi: 10.7554/eLife.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malsam J, Parisotto D, Bharat TA, Scheutzow A, Krause JM, Briggs JA, Sollner TH. Complexin arrests a pool of docked vesicles for fast Ca2+-dependent release. EMBO J. 2012;31:3270–3281. doi: 10.1038/emboj.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pabst S, Hazzard JW, Antonin W, Sudhof TC, Jahn R, Rizo J, Fasshauer D. Selective interaction of complexin with the neuronal SNARE complex. Determination of the binding regions. J Biol Chem. 2000;275:19808–19818. doi: 10.1074/jbc.M002571200. [DOI] [PubMed] [Google Scholar]
- 37.Chen X, Tomchick DR, Kovrigin E, Arac D, Machius M, Sudhof TC, Rizo J. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 38.Bracher A, Kadlec J, Betz H, Weissenhorn W. X-ray structure of a neuronal complexin-SNARE complex from squid. J Biol Chem. 2002;277:26517–26523. doi: 10.1074/jbc.M203460200. [DOI] [PubMed] [Google Scholar]
- 39.Xue M, Reim K, Chen X, Chao HT, Deng H, Rizo J, Brose N, Rosenmund C. Distinct domains of complexin I differentially regulate neurotransmitter release. Nat Struct Mol Biol. 2007;14:949–958. doi: 10.1038/nsmb1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue M, Craig TK, Xu J, Chao HT, Rizo J, Rosenmund C. Binding of the complexin N terminus to the SNARE complex potentiates synaptic-vesicle fusogenicity. Nat Struct Mol Biol. 2010;17:568–575. doi: 10.1038/nsmb.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue M, Lin YQ, Pan H, Reim K, Deng H, Bellen HJ, Rosenmund C. Tilting the balance between facilitatory and inhibitory functions of mammalian and Drosophila Complexins orchestrates synaptic vesicle exocytosis. Neuron. 2009;64:367–380. doi: 10.1016/j.neuron.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malsam J, Seiler F, Schollmeier Y, Rusu P, Krause JM, Sollner TH. The carboxy-terminal domain of complexin I stimulates liposome fusion. Proc Natl Acad Sci U S A. 2009;106:2001–2006. doi: 10.1073/pnas.0812813106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaeser-Woo YJ, Yang X, Sudhof TC. C-terminal complexin sequence is selectively required for clamping and priming but not for Ca2+ triggering of synaptic exocytosis. J Neurosci. 2012;32:2877–2885. doi: 10.1523/JNEUROSCI.3360-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seiler F, Malsam J, Krause JM, Sollner TH. A role of complexin-lipid interactions in membrane fusion. FEBS Lett. 2009;583:2343–2348. doi: 10.1016/j.febslet.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chicka MC, Chapman ER. Concurrent binding of complexin and synaptotagmin to liposome-embedded SNARE complexes. Biochemistry. 2009;48:657–659. doi: 10.1021/bi801962d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat Struct Mol Biol. 2008;15:665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokumaru H, Shimizu-Okabe C, Abe T. Direct interaction of SNARE complex binding protein synaphin/complexin with calcium sensor synaptotagmin 1. Brain Cell Biol. 2008;36:173–189. doi: 10.1007/s11068-008-9032-9. [DOI] [PubMed] [Google Scholar]
- 48.Rizo J, Rosen MK, Gardner KH. Enlightening molecular mechanisms through study of protein interactions. J Mol Cell Biol. 2012;4:270–283. doi: 10.1093/jmcb/mjs036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Margittai M, Fasshauer D, Pabst S, Jahn R, Langen R. Homo- and heterooligomeric SNARE complexes studied by site-directed spin labeling. J Biol Chem. 2001;276:13169–13177. doi: 10.1074/jbc.M010653200. [DOI] [PubMed] [Google Scholar]
- 50.Arac D, Murphy T, Rizo J. Facile detection of protein-protein interactions by one-dimensional NMR spectroscopy. Biochemistry. 2003;42:2774–2780. doi: 10.1021/bi0272050. [DOI] [PubMed] [Google Scholar]
- 51.Ubach J, Lao Y, Fernandez I, Arac D, Sudhof TC, Rizo J. The C2B domain of synaptotagmin I is a Ca2+-binding module. Biochemistry. 2001;40:5854–5860. doi: 10.1021/bi010340c. [DOI] [PubMed] [Google Scholar]
- 52.Zhou A, Brewer KD, Rizo J. Analysis of SNARE complex/Synaptotagmin-1 Interactions by One-dimensional NMR Spectroscopy. Biochemistry. 2013;52:3446–3456. doi: 10.1021/bi400230u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deak F, Xu Y, Chang WP, Dulubova I, Khvotchev M, Liu X, Sudhof TC, Rizo J. Munc18–1 binding to the neuronal SNARE complex controls synaptic vesicle priming. J Cell Biol. 2009;184:751–764. doi: 10.1083/jcb.200812026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 55.Shao X, Li C, Fernandez I, Zhang X, Sudhof TC, Rizo J. Synaptotagmin-syntaxin interaction: the C2 domain as a Ca2+-dependent electrostatic switch. Neuron. 1997;18:133–142. doi: 10.1016/s0896-6273(01)80052-0. [DOI] [PubMed] [Google Scholar]
- 56.Wu MN, Littleton JT, Bhat MA, Prokop A, Bellen HJ. ROP, the Drosophila Sec1 homolog, interacts with syntaxin and regulates neurotransmitter release in a dosage-dependent manner. EMBO J. 1998;17:127–139. doi: 10.1093/emboj/17.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Sudhof TC, Rizo J. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guan R, Dai H, Rizo J. Binding of the Munc13–1 MUN Domain to Membrane-Anchored SNARE Complexes. Biochemistry. 2008;47:1474–1481. doi: 10.1021/bi702345m. [DOI] [PubMed] [Google Scholar]
- 59.Xu Y, Su L, Rizo J. Binding of Munc18–1 to Synaptobrevin and to the SNARE Four-Helix Bundle. Biochemistry. 2010;49:1568–1576. doi: 10.1021/bi9021878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma C, Li W, Xu Y, Rizo J. Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat Struct Mol Biol. 2011;18:542–549. doi: 10.1038/nsmb.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339:421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi UB, Strop P, Vrljic M, Chu S, Brunger AT, Weninger KR. Single-molecule FRET-derived model of the synaptotagmin 1-SNARE fusion complex. Nat Struct Mol Biol. 2010;17:318–324. doi: 10.1038/nsmb.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hui E, Johnson CP, Yao J, Dunning FM, Chapman ER. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca(2+)-regulated fusion. Cell. 2009;138:709–721. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Honigmann A, van den BG, Iraheta E, Risselada HJ, Milovanovic D, Mueller V, Mullar S, Diederichsen U, Fasshauer D, Grubmuller H, Hell SW, Eggeling C, Kuhnel K, Jahn R. Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat Struct Mol Biol. 2013;20:679–686. doi: 10.1038/nsmb.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brewer KD, Li W, Horne BE, Rizo J. Reluctance to membrane binding enables accessibility of the synaptobrevin SNARE motif for SNARE complex formation. Proc Natl Acad Sci U S A. 2011;108:12723–12728. doi: 10.1073/pnas.1105128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen X, Arac D, Wang TM, Gilpin CJ, Zimmerberg J, Rizo J. SNARE-Mediated Lipid Mixing Depends on the Physical State of the Vesicles. Biophys J. 2006;90:2062–2074. doi: 10.1529/biophysj.105.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - A Multidimensional Spectral Processing System Based on Unix Pipes. Journal of Biomolecular Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 68.Johnson BA, Blevins RA. Nmr View - A Computer-Program for the Visualization and Analysis of Nmr Data. Journal of Biomolecular Nmr. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]