SUMMARY
Toll-like receptor-9 (TLR9) is largely responsible for discriminating self from pathogenic DNA. However, association of host DNA with autoantibodies activates TLR9, inducing the pathogenic secretion of type I interferons (IFNs) from plasmacytoid dendritic cells (pDCs). Here, we found that in response to DNA-containing immune complexes (DNA-IC), but not to soluble ligands, IFN-α production depended upon the convergence of the phagocytic and autophagic pathways, a process called microtubule-associated protein 1A/1B-light chain 3 (LC3)-associated phagocytosis (LAP). LAP was required for TLR9 trafficking into a specialized interferon signaling compartment by a mechanism that involved autophagy-related proteins, but not the conventional autophagic preinitiation complex, or adaptor protein-3 (AP-3). Our findings unveil a new role for nonconventional autophagy in inflammation and provide one mechanism by which anti-DNA autoantibodies, such as those found in several autoimmune disorders, bypass the controls that normally restrict the apportionment of pathogenic DNA and TLR9 to the interferon signaling compartment.
INTRODUCTION
Pattern recognition receptors, including Toll-like receptors (TLRs), recognize conserved microbial features as a strategy of pathogen detection and play a central role in innate and adaptive immunity (Akira et al., 2001). The cellular mechanisms of innate DNA sensing are emerging, but it is well established that microbial nucleic acids trigger the induction of type I interferons (IFNs) (Barbalat et al., 2011), such as IFN-α, and that this represents a key host defense strategy to limit the replication of invading microorganisms (Stetson and Medzhitov, 2006). However, under some circumstances, recognition of host DNA has been linked to autoimmunity, highlighting the importance of the mechanisms that ensure the distinction between self and foreign DNA (Gilliet et al., 2008) in the type I IFN response.
Unabated activation of TLR9 in plasmacytoid dendritic cells (pDCs) has been associated with autoimmune disorders such as systemic lupus erythematosus (SLE) (Gilliet et al., 2008). A hallmark of such diseases is the production of IFN-α in response to large DNA-containing immune complexes (DNA-IC) that are engulfed by phagocytosis. Phagocytes internalize DNA-IC using cell surface Fcγ-receptors (FcγR) (Means et al., 2005). FcγR initiates a tyrosine phosphorylation cascade that activates phosphatidylinositol 3-kinase (PI3K), and results in dynamic and rapid reorganization of the actin cytoskeleton, which is essential for phagocytic uptake (Jaumouillé and Grinstein, 2011), compartment maturation (Yates et al., 2005; Trivedi et al., 2006), and the secretion of proinflammatory cytokines (Abrahams et al., 2000). How DNA-IC trigger TLR9 is poorly understood, but some of the mechanisms leading to TLR9 activation have been described using agonistic CpG-containing phosphorothioate-modified oligodeoxynucleotides (CpG-ODN) (Klinman, 2004) that are internalized by endocytosis. CpG-ODN actively signal in intracellular compartments (Ahmad-Nejad et al., 2002) as they are inactive when immobilized (Manzel and Macfarlane, 1999), and lipofection enhances the stimulatory activity of CpG-ODN (Gursel et al., 2001). Unc-93 homolog B (UNC93B) physically interacts with the transmembrane domain of TLR9 and mediates TLR9 transport from the endoplasmic reticulum (ER) to the endosome. Endosomal maturation is required for TLR9 stimulation (Häcker et al., 1998) where its ectodomain is cleaved by cathepsins (Park et al., 2008). Upon ligand recognition, recruitment of the adaptor protein myeloid differentiation primary response gene 88 (MyD88) initiates the secretion of nuclear factor κB (NF-κB)-dependent inflammatory cytokines (Kawai and Akira, 2007) and interferon regulatory factor 7 (IRF7)-dependent type I IFNs (Honda et al., 2006). Although these two pathways are triggered by the same adaptor protein, it is apparent that TLR9 trafficking through the endolysosomal pathway further shapes signaling outcome, as differential cytokine production results from targeting TLR9 ligands to specific compartments (Sasai et al., 2010). Although largely unknown, the mechanisms associated with TLR9 compartmentalization are starting to emerge. Activation of type I IFN genes is initiated by TNF receptor-associated factor 3 (TRAF3) in a mature compartment. Trafficking of TLR9 to this interferon signaling compartment (ISC) is mediated by the clathrin adaptor protein-3 (AP-3); however, this requirement can be bypassed if TRAF3 is forced onto early endosome compartments (Sasai et al., 2010).
Autophagy, a self-digestion pathway, has been linked to innate immunity and its activation plays a role in the degradation of a wide range of intracellular pathogens, including bacteria and viruses (Levine et al., 2011). Autophagy is engaged by immune cells to isolate cytosolic replicating viruses in the lumen of autophagosomes and deliver their genetic material to intracellular TLRs for recognition. However, there are additional situations where the autophagic machinery is involved in host defense: proteins that participate in the formation of the autophagosome can be directly recruited to the phagosome after engulfment of particles associated with ligands for TLR2, 4 or 6 (Sanjuan et al., 2007) or in response to T cell immunoglobulin mucin-4 (TIM-4) binding to phosphatidylserine (Martinez et al., 2011). As opposed to conventional autophagy, this process appears to occur without the formation of a double membrane, and it promotes a more rapid maturation of the phagosome. Similarly to conventional autophagy, the recruitment of microtubule-associated protein (MAP) 1A/1B-light chain 3 (LC3) to phagosomes is preceded by class III PI3K activity and involves the autophagy proteins 5 and 7 (ATG5 and ATG7). However, it proceeds in the absence of ULK1 (Martinez et al., 2011) and FIP200 (Florey et al., 2011), two components of the autophagosome preinitiation complex. To distinguish this process from conventional autophagy, we refer to it as LC3-associated phagocytosis (LAP) (Sanjuan et al., 2009).
The results presented here demonstrate that IFN-α secretion depends on this convergence of the phagocytic and autophagic pathway, LAP, in response to DNA-IC. Collectively, these data provide a mechanism by which inflammatory disease is triggered by host-DNA bound to autoantibodies.
RESULTS
DNA-IC Elicits TLR9-Dependent IFN-α Secretion
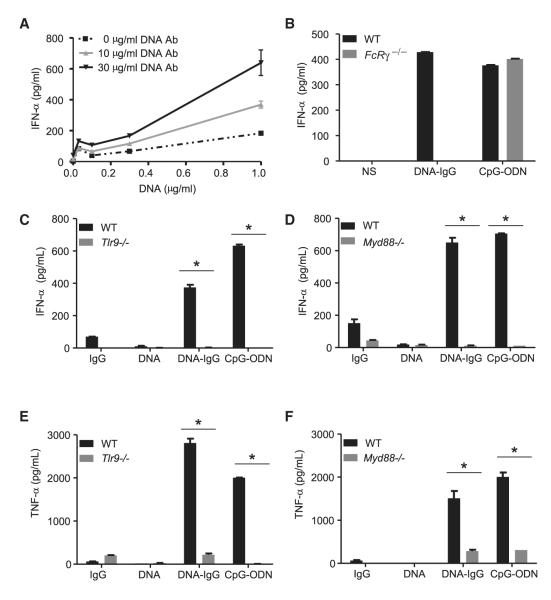
The presence of anti-nuclear antibodies (ANA) in serum is used to diagnose autoimmune disorders such as SLE. These autoantibodies form DNA-IC that trigger type I IFN production in pDCs (Means et al., 2005), a response that normally is elicited by pathogenic nucleic acids. In order to study the interferon response engaged by such complexes, we formed in vitro DNA-IC by combining monoclonal DNA antibodies with CG50, a plasmid optimized to activate TLR9 (Viglianti et al., 2003). DNA antibodies greatly enhanced the formation of IFN-α in murine pDCs (Figure 1A) and were also required for the secretion of IFN-α in response to mammalian DNA by human peripheral blood mononuclear cells (PBMCs) (see Figure S1A available online).
Figure 1. DNA-IgG-Mediated Production of IFN-α Requires Both FcγR and TLR9.
(A) Bone-marrow-derived pDCs from WT mice were stimulated with increasing concentrations of CG50 plasmid DNA (DNA) in presence of different concentrations of the DNA antibody E11 (DNA Ab). Production of IFN-α was measured by ELISA after 24 hr.
(B) Bone-marrow-derived pDCs from WT and Fcer1g (FcRγ −/−) mice were unstimulated (NS) or stimulated with DNA-IC formed by combining CG50 plasmid DNA and DNA antibody E11 (DNA-IgG). We used 5 μM of ODN1585 (CpG-ODN) as positive control. Secretion of IFN-α was measured by ELISA 24 hr after pDC stimulation. Data is representative of three independent experiments.
(C–F) Fetal liver-derived pDCs from WT, Tlr9−/− (C and E) or Myd88−/− (D and F) mice were stimulated with DNA antibody E11 (IgG), CG50 plasmid DNA (DNA), or DNA-IgG. ODN1585 (CpG-ODN) was used as a positive control. IFN-α and TNF-α in supernatants were measured by ELISA 24 hr after pDC stimulation. Data are presented as mean ± SD of three independent experiments. (*p < 0.05).
DNA-IC sensing strictly depends on FcγR. Mouse pDCs lacking the common γ-chain of FcγR (FcRγ) did not secrete IFN-α in response to DNA-IC (Figure 1B). Similarly, inhibition of FcγR with blocking antibodies (Figure S1B) or with aggregated IgG (Figure S1C) inhibited IFN-α triggered by DNA-IC in human PBMCs, suggesting an interplay between FcγR and TLR9 for recognizing DNA-IC. However, synthetic CpG-containing phosphorothioate oligodeoxynucleotides (ODN) induced IFN-α in FcRγ−/− cells (Figure 1B), indicating that activation of FcγR is an important step in the induction of interferon in response to DNA-IC, but not to CpG-ODN.
The DNA-IC found in lupus patients trigger interferon responses through TLR9 (Means et al., 2005). Similarly, IFN-α production by human PBMCs in response to our DNA-IC were reduced in the presence of antagonistic ODN (Figure S1D) or chloroquine (Figure S1E), both of which inhibit TLR9 activation (Trieu et al., 2006; Macfarlane and Manzel, 1998). We further investigated our model of in vitro formed DNA-IC in fetal liver-derived murine pDCs, a cell type that in our knowledge has not previously been used to investigate TLR9 responses and that was used throughout the current study. Neither DNA antibodies nor DNA alone were capable of inducing significant type I IFN responses in fetal liver-derived pDCs, whereas DNA-IC were potently active (Figure 1C). Ablation of TLR9 (Figure 1C) or the adaptor protein MyD88 (Figure 1D) completely blocked the secretion of IFN-α in response to DNA-IC. Because ODN induce TLR9-dependent tumor necrosis factor (TNF)-α secretion in an NF-κB-dependent manner (Kawai and Akira, 2007), we investigated whether DNA-IC triggered TNF-α production in fetal liver-derived pDCs. DNA-IC, but not antibody or DNA alone, induced a robust secretion of TNF-α (Figure 1E) in pDCs. Cells deficient for TLR9 (Figure 1E) or MyD88 (Figure 1F) did not secrete TNF-α in response to DNA-IC (or CpG-ODN), indicating that the TLR9-MyD88 axis triggers both IRF7-dependent and NF-κB-dependent responses upon sensing DNA-IC.
IgG Component of DNA-IC Enhances Both TLR9 and UNC93B Trafficking to Phagosomes
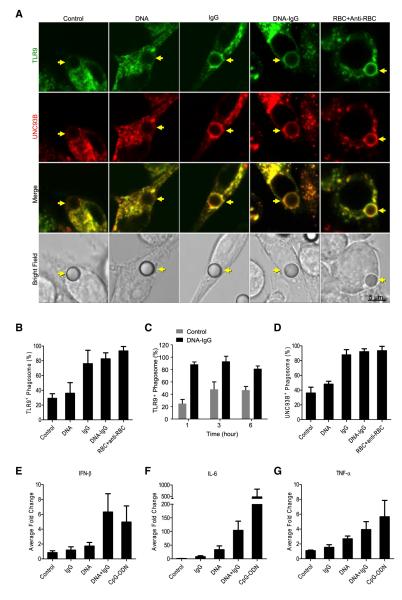
FcγR and TLR9 are localized in different compartments in resting cells, with FcγR exposed at the cell surface (Nimmerjahn and Ravetch, 2006) and TLR9 localized intracellularly (Brinkmann et al., 2007). We hypothesized that TLR9 mobilizes to engulfed particles in order to be activated. We prepared beads coated with DNA, IgG, or both DNA and IgG (DNA-IgG) and examined the intracellular localization of TLR9 after beads were engulfed by phagocytosis in mouse macrophages expressing a TLR9-GFP. Beads adsorbed to IgG or the combination of DNA and IgG resulted in an enhanced recruitment of TLR9 to phagosomes containing the engulfed particles (Figures 2A and 2B), lasting up to 6 hr (Figure 2C). UNC93B is required for TLR9 trafficking from the ER to the endosome (Brinkmann et al., 2007) and we observed that this protein also accompanied TLR9 to the phagosome (Figures 2A and 2D). Our data indicate that DNA is not required for the enhancement of TLR9 and UNC93B translocation to phagosomes by DNA-IgG complexes, and this was further confirmed with antibody-opsonized erythrocytes, which lack nuclear DNA (Figures 2A, 2B, and 2D).
Figure 2. Intracellular TLR9 and UNC93B Are Specifically Recruited to IgG-Opsonized Phagosomes.
(A) Mouse macrophages expressing TLR9-GFP and UNC93B-mCherry were allowed to ingest uncoated polystyrene beads (Control) or polystyrene beads coated with double-stranded DNA (DNA), anti-DNA IgGs (IgG), or a combination of both (DNA-IgG). RBC opsonized with RBC antibodies (RBC+Anti-RBC) were used as control. Engulfed beads were followed by time lapse confocal microscopy and representative frames taken 30 min after bead internalization are shown. Yellow arrows point to internalized beads. Scale bar represents 5 μm.
(B and C) The percentage of phagosomes positive for TLR9 recruitment were quantified for three independent experiments (n ≥ 75 phagosomes per group) on cells treated as described in (A). Data are presented as mean ± SD. TLR9 recruitment was also quantified in (C) for phagosomes containing DNA-IgG at 1, 3, and 6 hr after bead phagocytosis for three independent experiments (n ≥ 75 phagosomes per group).
(D) The percentage of phagosomes positive for UNC93B recruitment were quantified for three independent experiments (n ≥ 75 phagosomes per group). Data are presented as mean ± SD.
(E–G) Mouse macrophages were stimulated with beads as described in (A). We used 3 μM ODN 1826 (CpG-ODN) as a positive control. IFN-β, IL-6, and TNF-α mRNA expression was assessed by quantitative RT-PCR 4 hr after stimulation with beads. Results are presented as mean ± SD of three independent experiments.
As previously reported (Latz et al., 2004), we detected CpG-ODN in TLR9 positive compartments (Figure S2A). This is consistent with the idea that free CpG-ODN accumulates in endosomal compartments by nonspecific endocytosis (Häcker et al., 1998) where it directly binds TLR9. However, immobilization of agonistic CpG-ODN on beads appeared to force this ligand to enter the cell through phagocytosis, which did not trigger a mobilization of TLR9 (Figures S2A and S2B), UNC93B (Figure S2A, and S2C), or the activation of the interferon pathway (Figure S2D). Further, DNA-IgG coated beads triggered interferon production (Figure 2E) and also induced more IL-6 than DNA-or IgG-coated beads (Figure 2F). TNF-α induction was also modestly enhanced by DNA-IgG (Figure 2G). Together, our results indicate that TLR9 recruitment to the phagosome is an important step for the recognition of DNA-IC.
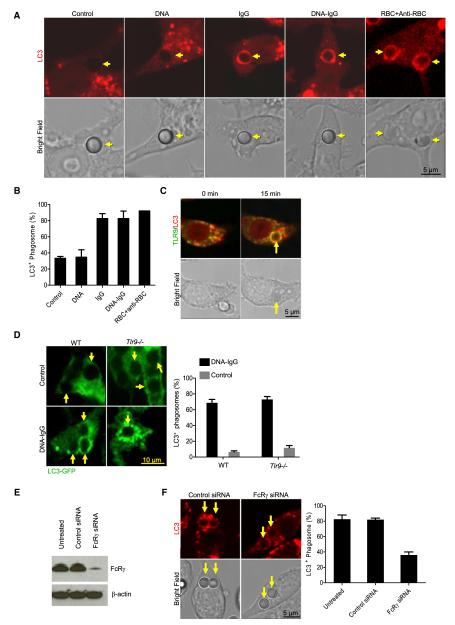
Our confocal studies indicate that TLR9 recruitment is enhanced by the IgG component of DNA-IC and that TLR9 was not efficiently mobilized to uncoated beads. Supporting this conclusion, we were able to detect a dramatic increase in the recruitment of TLR9 to the phagosome by immunoblotting using the phagosome fraction from cells fed with DNA-IgG coated beads, compared to DNA-coated beads (Figure S2E). To further investigate FcγR-mediated mobilization of TLR9 to phagosomes, we adsorbed beads to IgG fragments. Beads bound to IgG or Fc fragments induced an increase in TLR9 trafficking to the phagosome compared to control beads, whereas those coated with F(ab)2 fragments did not (Figures S2F and S2G). The same effect was seen for UNC93B (Figures S2F and S2H). FcγR signaling in mouse macrophages is largely initiated by a tyrosine kinase cascade (Ravetch and Bolland, 2001). Ablation of FcRγ expression in macrophages using small interfering RNA (siRNA) silencing (Figure S2I) reduced the recruitment of TLR9 and UNC93B to beads coated with IgG (Figures S2J and S2K).
Together, our results indicate that the engagement of FcγRs during engulfment of DNA-IC is responsible for the enhanced mobilization of both TLR9 and UNC93B to phagosome membranes in a DNA-independent manner.
DNA-IC Induce LC3-Associated Phagocytosis
Recently, we uncovered a process in which components of the autophagy pathway are recruited to the membrane of the phagosome. This process, LAP (Sanjuan et al., 2009), is engaged upon phagocytosis of particles containing some TLR ligands (those for TLR2, TLR4, or TLR6) (Sanjuan et al., 2007) or phosphatidylserine on liposomes or dying cells (Martinez et al., 2011; Florey et al., 2011) and functions in phagosome maturation. We therefore asked whether engulfment of DNA-IC can trigger the recruitment of LC3 to the phagosome. To this end, we employed the macrophage cell line RAW 264.7, expressing fluorescent LC3-mCherry, and monitored engulfment of beads. We observed recruitment of LC3 to phagosomes containing beads coupled with DNA-IgG, but not uncoated or DNA-coated beads (Figures 3A and 3B). Bead adsorption to IgG was sufficient to elicit LC3 recruitment to phagosomes (Huang et al., 2009) (Figures 3A and 3B). Similarly, engulfment of red blood cells (RBC) opsonized with RBC antibody-induced recruitment of LC3 (Figures 3A and 3B). Phagosomes containing IgG recruited both LC3 and TLR9, and these two proteins colocalized in the phagosome compartment (Figure 3C). LC3 mobilization did not depend on TLR9 signaling as it mobilized normally to phagosomes in TLR9-deficient pDCs (Figure 3D). However, siRNA suppression of FcRγ (Figure 3E) interfered with the recruitment of LC3 to phagosomes (Figure 3F), indicating that LC3 mobilization to the phagosome depends on FcγR signaling.
Figure 3. LC3 Colocalizes with TLR9 in IgG-Opsonized Phagosomes.
(A) Raw 264.7 cells expressing LC3-mCherry were allowed to ingest uncoated polystyrene beads (Control) or polystyrene beads coated with double-stranded DNA (DNA), anti-DNA IgGs (IgG), or a combination of both (DNA-IgG). RBC opsonized with RBC antibodies (RBC+Anti-RBC) were used as control. Engulfed beads were followed by time lapse confocal microscopy and representative frames taken 15 min after bead internalization are shown. Yellow arrows point to internalized beads. Scale bar represents 5 μm.
(B) The percentage of phagosomes that were positive for LC3 upon particle ingestion were quantified for three independent experiments (n ≥ 75 phagosomes per group). Data are presented as mean ± SD.
(C) Mouse macrophages expressing TLR9-GFP (green) and LC3-mCherry (red) were exposed to IgG-opsonized RBCs and engulfed particles were visualized by time lapse confocal microscopy. Representative frames from cells before and 15 min after particle internalization are shown. Yellow arrows point to internalized particles. Scale bar represents 5 μm.
(D) Bone marrow-derived pDCs from WT and Tlr9−/− mice expressing LC3-GFP were allowed to ingest uncoated polystyrene beads (Control) or polystyrene beads coated with a combination of CG50 plasmid DNA and DNA antibody E11 (DNA-IgG). Engulfed beads were followed by time-lapse confocal microscopy and representative frames taken 30 min after bead internalization are shown. Yellow arrows point to internalized beads. The percentage of phagosomes that were positive for LC3 upon bead ingestion were quantified for three independent experiments (n ≥ 75 phagosomes per group). Data are presented as mean ± SD. Scale bar represents 10 μm.
(E and F) Raw 264.7 cells expressing LC3-mCherry were treated with siRNA oligonucleotides targeting FcRγ. Nontargeting siRNA was used as control. FcRγ expression was assessed by immunoblotting 48 hr after the cells were treated with siRNA (E). siRNA-treated cells were allowed to ingest polystyrene beads coated with DNA-IgG. Engulfed beads were followed by time lapse confocal microscopy and representative frames taken 15 min after bead internalization are shown. Yellow arrows point to internalized beads. The percentage of phagosomes that were positive for LC3 upon bead ingestion were quantified for three independent experiments (n ≥ 75 phagosomes per group). Data are presented as mean ± SD. Scale bar represents 5 μm.
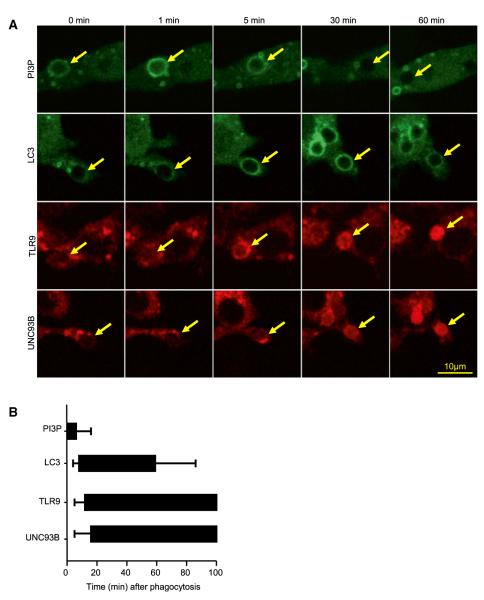
Using live cell imaging, we noted that the recruitment of LC3, TLR9, and UNC93B proceeded with similar, rapid kinetics (Figures 4A and 4B). As observed for LAP under other conditions (Sanjuan et al., 2007), PI3P, the product of PI3-kinases, was formed at the phagosome membrane as detected with a PX-domain probe (Figures 4A and 4B). Both LAP (Sanjuan et al., 2007) and TLR9 signaling (Kuo et al., 2006) require PI3-kinase activity. Consistent with this, we found that the PI3-kinase inhibitor Wortmannin prevented recruitment of LC3, TLR9, and UNC93B to phagosomes in macrophages (Figure S3A) and the secretion of IFN-α (Figure S3B) and TNF-α (Figure S3C) in response to DNA-IC in human pDCs.
Figure 4. PI3K Activity Precedes LC3 and TLR9 Recruitment to the Phagosome.
(A) RAW 264.7 cells expressing PX-GFP, LC3-GFP, TLR9-mCherry, or UNC93B-mCherry were fed with IgG-opsonized RBCs. Internalization was followed by acquiring time-lapse confocal images every 1 min for a period 2 hr and representative images are shown. Yellow arrows point to internalized particles. Scale bar represents 10 μm.
(B) Translocation of PX, LC3, TLR9, and UNC93B was quantified by measuring the time and duration of recruitment for each of the proteins represented to individual phagosomes. Phagosomes were followed for 100 min. Data are presented as mean ± SD (n = 25 phagosomes per group).
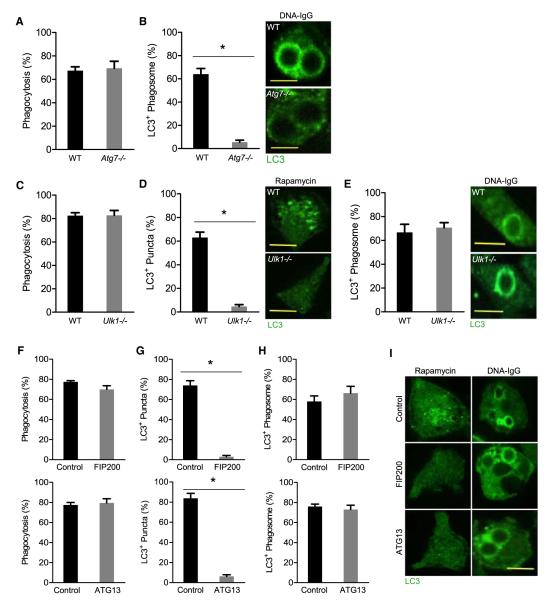
Although LAP requires Beclin-1, ATG7, and ATG5 (Sanjuan et al., 2007; Martinez et al., 2011), it is distinct from conventional autophagy in that it involves only the single phagosome membrane rather than the double membrane of the autophagosome. We therefore generated pDCs from fetal livers carrying an LC3-GFP transgene (Kuma and Mizushima, 2008) with or without deletion of Atg7. Although wild-type (WT) and Atg7−/− pDCs equivalently engulfed beads coated with DNA-IC (Figure 5A), no recruitment of LC3 to phagosomes was observed in Atg7−/− pDCs (Figure 5B).
Figure 5. Phagosome Recruitment of LC3 Requires ATG7 but Not the Autophagy Initiation Complex.
(A and B) Fetal liver-derived pDCs from WT and Atg7−/− mice were fed with beads coated with CG50 plasmid DNA and the DNA antibody E11 (DNA-IgG). Bead phagocytosis and LC3 recruitment at the phagosomes were visualized by time-lapse confocal microscopy. The percentage of cells that were positive for bead phagocytosis (A) and the percentage of phagosomes that were positive for LC3 upon particle ingestion (B) were quantified for three independent experiments. (n = 25 phagosomes per group). Scale bars represent 5 μm.
(C–E) Fetal liver-derived pDCs from WT and Ulk1−/− mice were treated with rapamycin (200 nM) or fed with beads coated with DNA-IgG as in (A). The percentage of cells that were positive for bead phagocytosis (C), LC3 puncta formation (D), and the percentage of phagosomes that were positive for LC3 upon particle ingestion (E) were quantified for three independent experiments (n = 25 phagosomes per group). Scale bars represent 5 μm.
(F–I) RAW 264.7 cells expressing LC3-GFP were transfected with nontargeting (control), FIP200, or ATG13 siRNA oligonucleotides. At 24 hr after transfection, cells were treated with 200 nM rapamycin or fed DNA-IgG coated beads as in (A). Bead phagocytosis and LC3 recruitment at the phagosomes were visualized by time-lapse confocal microscopy. The percentage of cells that were positive for bead phagocytosis (F), LC3 puncta formation (G and I), and the percentage of phagosomes that were positive for LC3 upon particle ingestion (H and I) were quantified for three independent experiments (n ≥ 50 cells per / group). Data are presented as mean ± SD (*p value < 0.05). Scale bars represent 5 μm.
Unlike conventional autophagy, LAP appears to proceed independently of ULK1 (Martinez et al., 2011) or FIP200 (Florey et al., 2011), two proteins of the autophagic preinitiation complex. We therefore examined pDCs from Ulk1−/− mice carrying LC3-GFP. ULK1 deficiency was not associated with defects in the rate of particle internalization (Figure 5C) but eliminated rapamycin-induced autophagic LC3 puncta formation (Figure 5D). However, LC3 recruitment to phagosomes containing DNA-IC-coupled beads was equivalently observed in both WT and Ulk1−/− cells (Figure 5E).
We then extended these observations to RAW 264.7 cells expressing LC3-GFP. Silencing of Fip200 or Atg13 with siRNA did not impact the rate of particle internalization (Figure 5F), whereas the formation of LC3-positive autophagosomes in response to rapamycin was dramatically reduced (Figures 5G and 5I). Nevertheless, LC3 recruitment to phagosomes containing DNA-IC-coated beads was unaffected (Figures 5H and 5I). Therefore, whereas ATG7 is required for FcγR-mediated LAP, the elements of the autophagy preinitiation complex, ULK1, FIP200, and ATG13 appear to be dispensable.
IFN-α Secretion in Response to DNA-IC Requires LAP
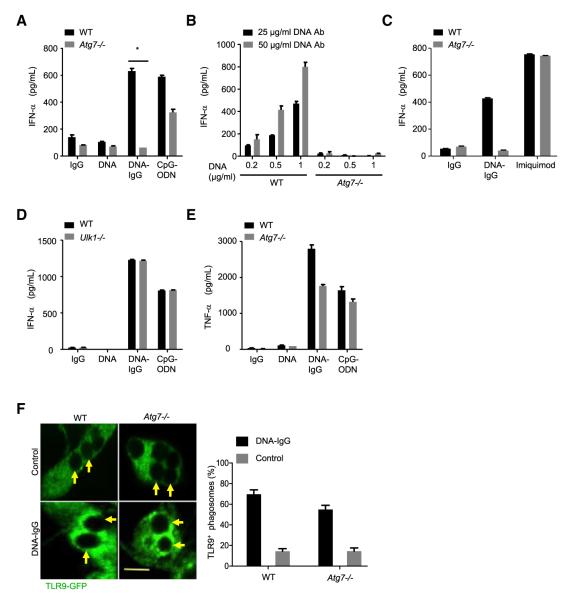
We then asked whether LAP was involved in signaling upon TLR9 ligation. Expression of TLR9, MyD88, and IRF7 messenger RNA (mRNA) and protein were similar in WT and Atg7−/− fetal liver-derived pDCs (Figures S4A–S4D). However, Atg7−/− pDCs were strikingly deficient in their IFN-α response to DNA-IC, and IFN-α secretion in response to CpG-ODN was also partially reduced (Figure 6A). This defect was observed over a range of concentrations of DNA-IC (Figure 6B) but was not observed upon treatment with imiquimod (Figure 6C), a cell-permeable TLR7 ligand that, as CpG-ODN, does not require phagocytosis for entry. In contrast, WT and Ulk1−/− pDCs, capable of engaging LAP (Figure 5E) but not conventional autophagy (Figure 5D), showed nearly identical IFN-α responses to both CpG-ODN and DNA-IC (Figure 6D). Although LAP was required for the secretion of IFN-α, production of TNF-α was only moderately reduced in response to DNA-IC and CpG-ODN in Atg7−/− pDCs (Figure 6E; Figures S4E and S4F). TLR9 translocation to phagosomes was not compromised in Atg7−/− pDCs (Figure 6F). This was further confirmed by silencing Atg5 using siRNA in RAW 264.7 cells, where TLR9 and UNC93B mobilization was sustained (Figure S4G), whereas LC3 recruitment was strongly reduced in the absence of ATG5 (Figure S4H). Therefore, neither LAP nor conventional autophagy is involved in TLR9 trafficking to phagosomes, but LAP is necessary for IFN-α secretion in response to DNA-IC.
Figure 6. LAP Is Required for IFN-α Secretion in Response to DNA-IgG.
(A) Fetal liver-derived pDCs from WT and Atg7−/− mice were incubated with DNA antibody E11 (IgG), CG50 plasmid DNA (DNA) or the immune complexes that result from combining both (DNA-IgG). We used 5 μM of ODN 1585 (CpG-ODN) as a control. IFN-α in supernatants was measured by ELISA 24 hr after stimulation. Representative data from at least three independent experiments are presented. Data are presented as mean ± SD (*p value < 0.05).
(B) IFN-α secretion in Atg7−/− pDCs was further evaluated by using DNA-IgG that were formed using increasing concentrations of CG50 plasmid DNA (DNA) combined with either 25 or 50 μg/ml of DNA antibody (IgG).
(C) Fetal liver-derived pDCs from WT and Atg7−/− mice were stimulated with the TLR7 ligand imiquimod (5 μg/ml) and also with DNA-IgG as described in (A).
(D) Fetal liver-derived pDCs from WT and Ulk1−/− mice were incubated with DNA-IgG as in (A). ODN 1585 (CpG-ODN) was used as a control.
(E) Fetal liver-derived pDCs from WT and Atg7−/− mice were incubated with DNA-IgG as in (A). TNF-α in supernatants was measured by ELISA 24 hr after stimulation. Representative data from at least three independent experiments are presented. Data are presented as mean ± SD.
(F) Fetal liver-derived pDCs from WT and Atg7−/− mice transiently expressing TLR9-GFP were allowed to ingest uncoated polystyrene beads (Control) or polystyrene beads coated with DNA-IgG. Engulfed beads were followed by time-lapse confocal microscopy and representative frames taken 30 min after bead internalization are shown. Yellow arrows point to internalized beads. The percentage of phagosomes that were positive for TLR9 upon bead ingestion was quantified for three independent experiments (n ≥ 75 phagosomes per group). Data are presented as mean ± SD. Scale bar represents 5 μm.
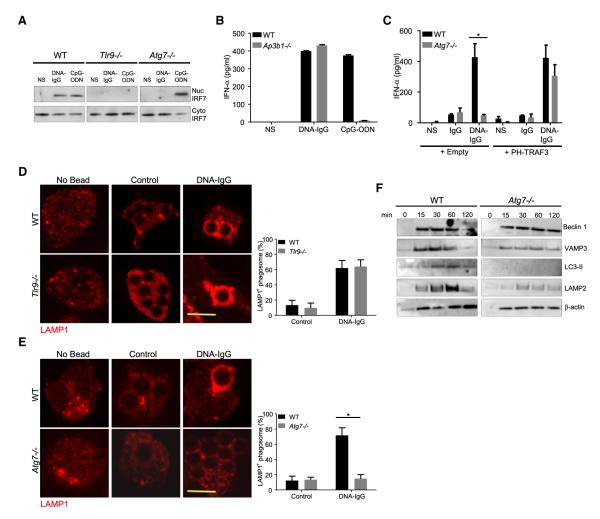
TLR9 localization has been associated with differential cytokine production. Some TLR9 ligands activate inflammatory cytokines upon entering the endosomal system, but IRF7 activation requires TLR9 trafficking into a specialized IRF7-signaling compartment (Sasai et al., 2010). We therefore examined the activation of IRF7 in pDCs in response to DNA-IC. In WT pDCs, free CpG-ODN and DNA-IC both induced nuclear accumulation of IRF7, which was not observed in pDCs lacking TLR9 (Figure 7A). In contrast, Atg7−/− cells showed robust nuclear translocation of IRF7 in response to free CpG-ODN but not to DNA-IC, indicating that LAP is required for initiating the IFN pathway in pDCs engulfing DNA-IC.
Figure 7. IRF7 Activation Requires the Formation of an Interferon Signaling Compartment.
(A) Fetal liver-derived pDCs from WT, Tlr9−/−, or Atg7−/− mice were left untreated (NS) or were treated with a combination of CG50 plasmid DNA and DNA antibody E11 (DNA-IgG) or with 5 μM ODN 1585 (CpG-ODN) for 6 hr. The presence of IRF7 in nuclear and cytoplasmic extracts was assessed by immunoblotting. Data presented are representative of three independent experiments.
(B) Bone marrow-derived pDCs from WT and Ap3b1−/− mice were incubated for 24 hr with DNA-IgGs or ODN 1585 (CpG-ODN) as in (A). IFN-α in supernatant was measured by ELISA. Representative data from at least two independent experiments are presented. Data are presented as mean ± SD.
(C) Fetal liver-derived pDCs from WT and Atg7−/− mice transiently expressing PH-TRAF3 or an empty vector were incubated for 24 hr with DNA-IgG as in (A). No treatment (−) or DNA antibody E11 alone (IgG) were used as control. IFN-α in supernatant was measured by ELISA. Representative data from at least two independent experiments are presented. Data are presented as mean ± SD.
(D and E) Fetal liver-derived pDCs were fed with uncoated beads (Control) or DNA-IgG-coated beads for 4 hr. Cells were then fixed and LAMP1 was detected by immunofluorescence. Confocal images for TLR9−/− cells (D) and Atg7−/− (E) pDCs were obtained and representative images shown. The percentage of phagosomes that were positive for LAMP1 upon bead ingestion was quantified for three independent experiments (n ≥ 50 phagosomes per group). Data are presented as mean ± SD (*p value < 0.05). Scale bars represent 5 μm.
(F) Immunoblotting analysis of phagosome proteins. Fetal liver-derived pDCs from WT and Atg7−/− mice were allowed to phagocytose latex beads coated with DNA-IgG for the indicated time. Phagosomes were purified using sucrose gradient as described in experimental procedures. Phagosome proteins were solubilized in SDS-PAGE and blotted with the indicated antibodies. The results presented are representative of three independent experiments.
AP-3 is required for the TLR9 response to CpG-ODN, functioning in the formation of this IRF7 signaling compartment (Sasai et al., 2010). As expected, bone-marrow-derived pDC from Ap3b1−/− mice were defective in the interferon response to CpG-ODN, but remarkably, IFN-α secretion in response to DNA-IC was normal in Ap3b1−/− pDCs (Figure 7B), indicating that LAP, but not AP-3, may be required for the formation of the specialized IFN-signaling compartment in response to DNA-IC. To test this, we targeted TRAF3 to early compartments using a chimeric molecule in which a PI(3,5)P2-binding plextrin homology (PH) domain of centaurin β2 is fused to the N-terminal end of TRAF3 (PH-TRAF3) (Sasai et al., 2010), a strategy that had been shown to bypass the requirement for AP-3 in the response to CpG-ODN (Sasai et al., 2010). Ectopic expression of PH-TRAF3 was sufficient to reconstitute TLR9 signaling for IFN induction in fetal liver derived Atg7−/− pDC (Figure 7C).
TLR9 trafficking into the specialized IRF7-signaling compartment is associated with late-endolysosomal proteins (Sasai et al., 2010).We therefore examined the recruitment of the lysosomal marker LAMP1 to phagosomes containing DNA-IC-coated beads. Phagosomes in macrophages constitutively acquire lysosomal markers (Huang et al., 2009). However, we found that recruitment of LAMP1 in pDCs was regulated by the cargo, as particles covered with DNA-IC mobilized LAMP1 to the phagosome but not control particles (Figure 7D). The recruitment of LAMP1 to such phagosomes in pDCs was independent of TLR9 (Figure 7D) or MyD88 (Figure S5A) but required ATG7 (Figure 7E). Beclin-1, a binding partner of class III PI3K, is activated upstream of ATG7 (Kang et al., 2011). ATG7-deficient cells effectively recruited Beclin-1 but failed to recruit LC3 (Figure 7F). The vesicle-associated membrane protein 3 (VAMP3), an early maturation marker, was found on phagosomes from WT and Atg7−/− pDC (Figure 7F), but strikingly, Atg7−/− phagosomes failed to progress into mature compartments as they failed to recruit LAMP1 (Figure 7E), LAMP2 (Figure 7F), or acidify (Figure S5B) in pDCs. Together, our data indicates that LAP is required for the progression of the TLR9 compartment into a mature LAMP1+ phagosome.
Collectively, these results suggest that IFN-α production in response to DNA-IC is initiated in a mature IRF7-signaling compartment and that ATG7 is required for the TLR9+ phagosome to progress into that compartment. The mature phagosome is formed in response to FcγR engagement and does not require TLR9 signaling. The presence of native DNA can then signal IRF7 to engage IFN-α production. We conclude that such signaling requires LAP to facilitate the formation of the IRF-7 signaling compartment.
DISCUSSION
Recognition of DNA appears to be a mechanism by which cells identify invading pathogens, but there is ample evidence that autoantibodies that bind directly or indirectly to host DNA can induce IFN-responses and cause autoimmunity (Gilliet et al., 2008). Here we have provided evidence that IFN-α secretion depends on a convergence of the phagocytic and autophagic pathways in response to DNA-IC. FcγR-mediated internalization of DNA-IC induced an enhanced recruitment of TLR9 to the phagosome. Upon recruitment, TLR9 triggered the secretion of the inflammatory cytokine TNF-α. However the activation of the transcription factor IRF7 and subsequent IFN-α secretion required phagosome progression into a late compartment that was positive for LAMP1 and LAMP2 in pDCs. This compartment was generated upon recruitment of the autophagy protein LC3, dependent on ATG7. Because neither LC3 recruitment nor IFN-α secretion required components of the classical autophagy preinitiation complex, we propose that LAP is required for IFN-α secretion in response to DNA-IC.
DNA is a shared feature of both host and pathogens, and thus safeguards must be in place to avoid TLR9 response to self-DNA. Surface expression of transgenic cleaved TLR9 on hematopoietic stem cells results in lethal autoinflammatory disease in mice (Mouchess et al., 2011), highlighting the importance of concealing TLR9 in intracellular membranes. We have provided evidence that TLR9 trafficking inside the cell is also strictly regulated, because receptor mobilization to phagosomes was enhanced by the phagosome content. Beads coated with DNA plus IgG strongly colocalized with TLR9, a phenomenon also observed in cells incubated with human lupus autoantibody-DNA complexes that are associated with disease pathogenesis (Means et al., 2005). We found that signals triggering TLR9 mobilization did not emanate from nucleic acids but rather from the IgG in the immune complexes. This trafficking is likely to be mediated by PI3K because TLR9 mobilization was impaired upon pharmacological inhibition of this lipid kinase. We noticed that FcγR activation also mobilized UNC93B, a protein responsible for TLR9 transport from the ER to endosomes (Kim et al., 2008), and it is therefore likely that it also escorts the receptor to phagosome membranes. Phagosomes containing Asperguillus fumigatus conidia also recruit TLR9 (Kasperkovitz et al., 2010), suggesting that there are other receptors associated with microbial defense that trigger TLR9 trafficking. It is therefore apparent that there are active mechanisms that limit the presence of TLR9 to those phagosomes most likely to contain pathogens in order to prevent aberrant activation, a protection against inflammatory responses that is circumvented by the formation of DNA-IC.
Autophagy has previously been linked to IFN-responses in pDCs. The recognition of single-stranded RNA in cells infected with viruses such as vesicular stomatitis virus (VSV) or Sendai virus (SV) requires transport of cytosolic viral replication intermediates into the lysosome by the process of autophagy (Lee et al., 2007). Atg5−/− cells fail to encapsulate viral RNA in autophagosomes, which was required for viral nucleic acid sensing, and infected Atg5−/− cells do not secrete cytokines, including IFN-α (Lee et al., 2007). Thus, in addition to LAP, pDCs may use the conventional autophagy pathway to deliver some ligands to nucleic acid sensors, although the role of the preinitiation complex (ULK1, FIP200, ATG13) has not been examined in this process.
While LAP is required for the IFN-α response to DNA-IC, autophagy deficiency did not block all responses to these immune complexes. Atg7−/− pDCs failed to produce IFN-α, but the secretion of TNF-α was only moderately reduced, indicating that LC3-phagosome recruitment was associated with shaping TLR9 signaling but not with ligand delivery to the receptor. Further supporting this conclusion, TLR9 was found at the phagosome in Atg7−/− pDCs and in macrophages where ATG5 was silenced with siRNA.
Signals emanating from TLR9 are determined by the compartment where the receptor is activated; however, the nature of those compartments remains controversial. Compartmentalization of IRF7-dependent responses into a LAMP2+ compartment capable of recruiting TRAF3 has previously been reported for CpG-ODN. Although NF-κB responses remain intact, AP-3 deficient cells failed to traffic TLR9 into LAMP2+ mature compartments, which impedes TRAF3 recruitment, and resulted in a profound defect in CpG-ODN-induced IFN-α production. However, AP-3 deficient cells showed normal IFN-α secretion when treated with DNA-IC, indicating that AP-3 is not part of the machinery used to sense this ligand. Compartment maturation in response to DNA-ICs was not regulated by TLR9 signaling as pDCs from Tlr9−/− and Myd88−/− mice recruited LAMP1 to phagosomes containing DNA-IC. Failure to induce LAP in Atg7−/− pDCs impeded LAMP1 and LAMP2 recruitment to the phagosome and this abrogated IRF7 activation. Targeting TRAF3 to early compartments restored secretion of IFN-α in response to DNA-ICs. Thus we conclude that FcγR bypass the need of AP-3 by providing a signal that recruits LAP to the phagosome, which in turn leads to phagosome progression into a LAMP+ compartment, and ultimately IRF7-dependent IFN-α secretion in response to DNA-ICs.
We noted that Atg7−/− pDCs produced IFN-α in response to CpG-ODN, although the response was decreased compared to WT pDCs. This observation is further supported by a previous report where the deletion of Atg5 in pDCs reduced but did not prevent IFN-responses (Lee et al., 2007). We also observed that nuclear translocation of IRF7 was defective in Atg7−/− pDCs when treated with DNA-IC, but not with free CpG-ODN. Thus our data suggest that LAP is required for the activation of IRF7 in phagosomes but may not be the only mechanism employed by pDCs to traffic endosomal ligands such as CpG-ODN to the specialized IRF7-signaling compartment.
The anti-inflammatory functions of autophagy have been reported. Autophagy deficiency has been linked to exacerbated intestinal inflammation in a mouse model of induced-acute colitis (Saitoh et al., 2008), and polymorphisms in autophagy genes are strongly associated to Crohn’s disease susceptibility (Hampe et al., 2007). The role of LAP versus conventional autophagy in such inflammatory responses has not been fully explored. Macrophages deficient for ATG7, but not ULK-1 (Martinez et al., 2011), secrete higher amounts of inflammatory cytokines than WT macrophages in response to dying cells. The requirement for LAP to induce a DNA-IC-mediated IFN-α response in pDCs implies that the pathway we described here could be involved in the pathogenesis of diseases such as SLE, where type I IFNs secreted in response to DNA-IC contributes to disease progression. Supporting this conclusion, genome-wide association studies have found that single nucleotide polymorphisms in ATG5 are associated with increased susceptibility to SLE (Harley et al., 2008). It is then conceivable that pharmacological interventions designed to treat SLE might benefit from incorporating agents that target the LAP machinery involved in sensing of nucleic acid immune complexes.
EXPERIMENTAL PROCEDURES
Mice
Mice were housed pathogen-free and experiments were done in accordance with the guidelines of the Institutional Animal Care and Use Committee of Med-Immune or the Committee for Animal Care at the St. Jude Children’s Research Institute. Ap3b1−/− and C57BL/6 mice were obtained from Jackson Laboratories. Tlr9−/− and Myd88−/− mice were kindly provided by Shizuo Akira (Osaka University, Japan). To generate Atg7−/− mice, male Atg7flox/flox mice (Komatsu et al., 2005) were bred to females EIIaCre mice (Williams-Simons and Westphal, 1999). Atg7−/− and Atg7+/+ littermates were used to generate E14 fetal liver stem cells for pDC generation. Ulk1−/− mice were kindly provided by Mondira Kundu (St. Jude Children’s Research Hospital). Fcer1g mice, deficient for the common gamma chain (FcRγ) of FcγR, have been previously described (Park et al., 1998) and were a generous gift from Jeffery Ravetch.
Cells
Human peripheral blood mononuclear cells (PBMCs) were isolated from fresh peripheral venous blood from healthy humans in accordance with the guidelines of the Institutional Review Board of MedImmune, using CPT tubes (BD Biosciences). EasySep Human pDC Enrichment kit (STEMCELL Technologies) was used to isolate human pDCs, and the enriched fraction was consistently ≥90%. Mouse macrophages expressing TLR9-GFP were generated as previously described (Hornung et al., 2008). Briefly, TLR9-deficient macrophages were immortalized from bone marrow of Tlr9−/− mice using the J2 recombinant retrovirus (carrying v-myc and v-raf oncogenes). Bone marrow cells were infected with J2 virus after 3 days of culture in L929 conditioned medium. Immortalized macrophages were then retrovirally transduced with mouse TLR9-GFP, UNC93B-mCherry, and/or LC3-mCherry. Jurkat and Raw 264.7 cells were obtained from ATCC. RAW 264.7 cells were retrovirally transduced with PX-GFP, LC3-mCherry, LC3-GFP, TLR9-mCherry, and/or UNC93B-mCherry.
Phagocytosis Assays
Polystyrene beads (Spherotech) were resuspended in PBS and incubated at 4°C for 16 hr with 10 ug/ml of thymic double-stranded DNA (dsDNA) (Trevigen), 50 ug/ml of anti-dsDNA antibody (clone MAB030, Millipore), 1.0 μg/ml CG50 DNA, 50 ug/ml of anti-DNA clone E11, 10 μM ODN2216, 50 μg/ml mouse IgG, 50 μg/ml mouse F(ab)2, or 50 μg/ml mouse Fc, as indicated in figure legend. Polystyrene beads precoated with IgG (Spherotech) were also used. Sheep erythrocytes were obtained from Bioreclamation and were used either uncoated or opsonized with 50 μg/ml sheep RBC antibody (Rockland Immunochemicals). RAW 264.7 cells were cultured in the presence of IFN-γ (30 ng/ml) for 16 hr prior to internalization experiments.
Cell Imaging
Confocal microscopy on live cells was performed with either a Leica TCS SP5 confocal system consisting of a Leica DMI6000 B inverted microscope (Leica Microsystems) or a Marianas SDC imaging system (Intelligent Imaging Innovations/3i) (Carl Zeiss MicroImaging). Temperature was maintained at 37°C with 5% CO2 using an environmental control chamber (Life Imaging Services and Solent Scientific). Images were acquired at time points noted in the figure and were analyzed using the LAS AF version 2.2.1 Leica Application Suite software (Leica Microsystems) or the SlideBook 4.2 software (3i).
Phagosome Isolation
Raw 267.4 cells expressing TLR9-GFP were incubated with smooth magnetic beads (Spherotech) coated with either 1 μg/ml CG50 plasmid DNA alone or with both CG50 and 50 μg/ml E11 DNA antibodies. Cells were transferred into HEPES-Sucrose Buffer pH 7.2 supplemented with Complete Mini protease inhibitor tablet (Roche) and DTT. Cells were homogenized using a Dounce homogenizer and phagosomes were isolated using DynaMag magnets (Life Technologies). Phagosomal proteins were extracted in RIPA lysis buffer and used for immunoblot analysis. Phagosomes from mouse pDCs were obtained as previously described (Desjardins et al., 1994). Briefly, after culture of pDCs with CG50 plasmid DNA plus E11 antibody-coated latex beads, the cells were washed in cold PBS, pelleted, resuspended in 1 ml of homogenization buffer (250 mM sucrose, 3 mM imidazole, pH 7.4), and homogenized on ice in a Dounce homogenizer. Phagosomes were then isolated by flotation on a sucrose step gradient during centrifugation for 1 hr at 100,000 g at 4°C. Latex-bead phagosomal fraction was then collected from the interface of the 10% and 25% sucrose solutions and resuspended in RIPA buffer for protein immunoblot analysis.
Statistical Analysis
Unpaired, two-tailed Student’s t test was used to determine the statistical differences between the data sets. p ≤ 0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of Health grant AI40646. We thank Jacques Moisan for manuscript critical review and David Dilillo for his help with Fcer1g mice. The following authors are or were full-time employees of MedImmune: M.A.S, J.H., J.M.R., A.J.C., J.T., L.C., P.M. and R.K. MedImmune is developing anti-interferon therapies for autoimmune diseases.
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes five figures and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2012.09.014.
REFERENCES
- Abrahams VM, Cambridge G, Lydyard PM, Edwards JC. Induction of tumor necrosis factor alpha production by adhered human monocytes: a key role for Fcgamma receptor type IIIa in rheumatoid arthritis. Arthritis Rheum. 2000;43:608–616. doi: 10.1002/1529-0131(200003)43:3<608::AID-ANR18>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Ahmad-Nejad P, Häcker H, Rutz M, Bauer S, Vabulas RM, Wagner H. Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. Eur. J. Immunol. 2002;32:1958–1968. doi: 10.1002/1521-4141(200207)32:7<1958::AID-IMMU1958>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J. Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins M, Huber LA, Parton RG, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J. Cell Biol. 1994;124:677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat. Cell Biol. 2011;13:1335–1343. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- Gursel I, Gursel M, Ishii KJ, Klinman DM. Sterically stabilized cationic liposomes improve the uptake and immunostimulatory activity of CpG oligonucleotides. J. Immunol. 2001;167:3324–3328. doi: 10.4049/jimmunol.167.6.3324. [DOI] [PubMed] [Google Scholar]
- Häcker H, Mischak H, Miethke T, Liptay S, Schmid R, Sparwasser T, Heeg K, Lipford GB, Wagner H. CpG-DNA-specific activation of antigen-presenting cells requires stress kinase activity and is preceded by non-specific endocytosis and endosomal maturation. EMBO J. 1998;17:6230–6240. doi: 10.1093/emboj/17.21.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, et al. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, Glogauer M, Grinstein S, Brumell JH. Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl. Acad. Sci. USA. 2009;106:6226–6231. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaumouillé V, Grinstein S. Receptor mobility, the cytoskeleton, and particle binding during phagocytosis. Curr. Opin. Cell Biol. 2011;23:22–29. doi: 10.1016/j.ceb.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasperkovitz PV, Cardenas ML, Vyas JM. TLR9 is actively recruited to Aspergillus fumigatus phagosomes and requires the N-terminal proteolytic cleavage domain for proper intracellular trafficking. J. Immunol. 2010;185:7614–7622. doi: 10.4049/jimmunol.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol. Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- Klinman DM. Immunotherapeutic uses of CpG oligodeoxynucleotides. Nat. Rev. Immunol. 2004;4:249–258. doi: 10.1038/nri1329. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Mizushima N. Chromosomal mapping of the GFP-LC3 transgene in GFP-LC3 mice. Autophagy. 2008;4:61–62. doi: 10.4161/auto.4846. [DOI] [PubMed] [Google Scholar]
- Kuo CC, Lin WT, Liang CM, Liang SM. Class I and III phosphatidylinositol 3′-kinase play distinct roles in TLR signaling pathway. J. Immunol. 2006;176:5943–5949. doi: 10.4049/jimmunol.176.10.5943. [DOI] [PubMed] [Google Scholar]
- Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane DE, Manzel L. Antagonism of immunostimulatory CpG-oligodeoxynucleotides by quinacrine, chloroquine, and structurally related compounds. J. Immunol. 1998;160:1122–1131. [PubMed] [Google Scholar]
- Manzel L, Macfarlane DE. Lack of immune stimulation by immobilized CpG-oligodeoxynucleotide. Antisense Nucleic Acid Drug Dev. 1999;9:459–464. doi: 10.1089/oli.1.1999.9.459. [DOI] [PubMed] [Google Scholar]
- Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. USA. 2011;108:17396–17401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Means TK, Latz E, Hayashi F, Murali MR, Golenbock DT, Luster AD. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. J. Clin. Invest. 2005;115:407–417. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchess ML, Arpaia N, Souza G, Barbalat R, Ewald SE, Lau L, Barton GM. Transmembrane mutations in Toll-like receptor 9 bypass the requirement for ectodomain proteolysis and induce fatal inflammation. Immunity. 2011;35:721–732. doi: 10.1016/j.immuni.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV. Fcgamma receptors: old friends and new family members. Immunity. 2006;24:19–28. doi: 10.1016/j.immuni.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Park SY, Ueda S, Ohno H, Hamano Y, Tanaka M, Shiratori T, Yamazaki T, Arase H, Arase N, Karasawa A, et al. Resistance of Fc receptor-deficient mice to fatal glomerulonephritis. J. Clin. Invest. 1998;102:1229–1238. doi: 10.1172/JCI3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B, Brinkmann MM, Spooner E, Lee CC, Kim YM, Ploegh HL. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat. Immunol. 2008;9:1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravetch JV, Bolland S. IgG Fc receptors. Annu. Rev. Immunol. 2001;19:275–290. doi: 10.1146/annurev.immunol.19.1.275. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- Sanjuan MA, Milasta S, Green DR. Toll-like receptor signaling in the lysosomal pathways. Immunol. Rev. 2009;227:203–220. doi: 10.1111/j.1600-065X.2008.00732.x. [DOI] [PubMed] [Google Scholar]
- Sasai M, Linehan MM, Iwasaki A. Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science. 2010;329:1530–1534. doi: 10.1126/science.1187029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Trieu A, Roberts TL, Dunn JA, Sweet MJ, Stacey KJ. DNA motifs suppressing TLR9 responses. Crit. Rev. Immunol. 2006;26:527–544. doi: 10.1615/critrevimmunol.v26.i6.50. [DOI] [PubMed] [Google Scholar]
- Trivedi V, Zhang SC, Castoreno AB, Stockinger W, Shieh EC, Vyas JM, Frickel EM, Nohturfft A. Immunoglobulin G signaling activates lysosome/phagosome docking. Proc. Natl. Acad. Sci. USA. 2006;103:18226–18231. doi: 10.1073/pnas.0609182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viglianti GA, Lau CM, Hanley TM, Miko BA, Shlomchik MJ, Marshak-Rothstein A. Activation of autoreactive B cells by CpG dsDNA. Immunity. 2003;19:837–847. doi: 10.1016/s1074-7613(03)00323-6. [DOI] [PubMed] [Google Scholar]
- Williams-Simons L, Westphal H. EIIaCre — utility of a general deleter strain. Transgenic Res. 1999;8:53–54. doi: 10.1023/a:1008994831937. [DOI] [PubMed] [Google Scholar]
- Yates RM, Hermetter A, Russell DG. The kinetics of phagosome maturation as a function of phagosome/lysosome fusion and acquisition of hydrolytic activity. Traffic. 2005;6:413–420. doi: 10.1111/j.1600-0854.2005.00284.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.