Abstract
The exact positions of nucleosomes along genomic DNA can influence many aspects of chromosome function, yet existing methods for mapping nucleosomes do not provide the necessary single base pair accuracy to determine these positions. Here we develop and apply a new approach for direct mapping of nucleosome centers based on chemical modification of engineered histones. The resulting map locates nucleosome positions genome-wide in unprecedented detail and accuracy. It reveals novel aspects of the in vivo nucleosome organization that are linked to transcription factor binding, RNA polymerase pausing, and the higher order structure of the chromatin fiber.
Nucleosomes distort and occlude the genomic DNA from access to most DNA-binding proteins, and their exact positions affect the structure of the chromatin fiber1. Even single base pair shifts of the position of a nucleosome can change chromatin configurations2 and protein binding kinetics3, 4, 5. A single base pair resolution map of nucleosome positions is necessary to fully understand a wide range of biological processes including RNA polymerase activity6, 7, 8, transcription factor binding kinetics4, 5, 9, 10, DNA replication11, 12, centromere structure13, 14, 15, and gene splicing16, 17, 18.
The most commonly used method for nucleosome mapping relies on treatment of chromatin with micrococcal nuclease (MNase) to digest DNA sequences not protected by the histone core. The nucleosome positions are indirectly inferred by the centers of undigested DNA sequence fragments. While this method has provided valuable insights into our understanding of nucleosomes, it is imprecise due to large variability of read lengths, transient unwrapping of nucleosomes, MNase sequence preferences and interference of other DNA binding proteins19, 20, 21, 22, 23. Thus, a different approach is required to measure nucleosome locations with greater accuracy.
A chemical approach to mapping nucleosomes
We developed a new, genome-wide mapping approach that directly determines nucleosome center positions with single base pair resolution. It derives from the earlier work by Flaus and Richmond in which localized hydroxyl radicals were used to map reconstituted mononucleosomes center positions 24, 25. In S. cerevisiae we introduced a unique cysteine into histone H4 at position 47 (H4S47C) to covalently attach a sulfhydryl-reactive, copper-chelating label, N-(1,10-Phenanthrolin-5-yl)iodoacetamide. Importantly, this cysteine mutation positions the label in close proximity to the DNA backbone and at sites symmetrically flanking the nucleosome center (Fig. 1a). Log-phase yeast was harvested and permeabilized immediately 26,27,28,29. The label was subsequently introduced to the cells where it reacts with the cysteines present in histone H4. Addition of copper and hydrogen peroxide catalyzes the formation of short-lived hydroxyl radicals that react with and cleave the DNA backbone (Methods). This strategy leads to a highly specific cleavage of the DNA backbone at sites that symmetrically flank the nucleosome center24, 25. On an agarose gel, a DNA banding pattern forms only when the reaction includes the sulfhydryl-reactive label, copper, H2O2, and the H4S47C mutant yeast, where the smallest fragment corresponds to DNAs between the centers of neighboring nucleosomes (Fig 1b).
Figure 1. Mapping nucleosome centers by site-specific chemical cleavage.

a, Nucleosome structure, highlighting histone H4 (green) and residue serine 47(pink), which is mutated to a cysteine where the sulfhydryl-reactive label covalently binds. b, Ethidium bromide stained agarose gel showing the chemical mapping results in a DNA banding pattern, which occurs only when the reaction includes (indicated by “+”) the sulfhydryl-reactive label, copper, H2O2, and the H4S47C mutant yeast. The cartoons adjacent to the agarose gel illustrate mapping successive nucleosomes’ centers produced the banding pattern. c, Locations of dominant hydroxyl radical cleavages relative to the nucleosome center (base pair 0).
We purified and sequenced the smallest DNA band from the agarose gel (Fig. 1b) in six independent experiments (four with single-end and two with paired-end parallel sequencing), and produced a map of 105 million cleavages on each DNA strand (Figs. 1c, 2a). The cleavage peaks on opposite DNA strands show two major correlations: a separation by either +2 or -5 nucleotides (with the 5’ to 3’ direction defined as positive) (Fig. 2b). This pattern is expected if cleavage occurs mainly at positions -1 (primary site) and +6 (secondary site) relative to the nucleosome center, similar to that described by the Richmond group for reconstituted mononucleosomes24, 25(Methods). The dominance and sharpness of these peaks reveal that the chemical maps contain high-resolution nucleosome positioning information.
Figure 2. Raw data, defined nucleosomes and pairwise comparison of nucleosome maps.
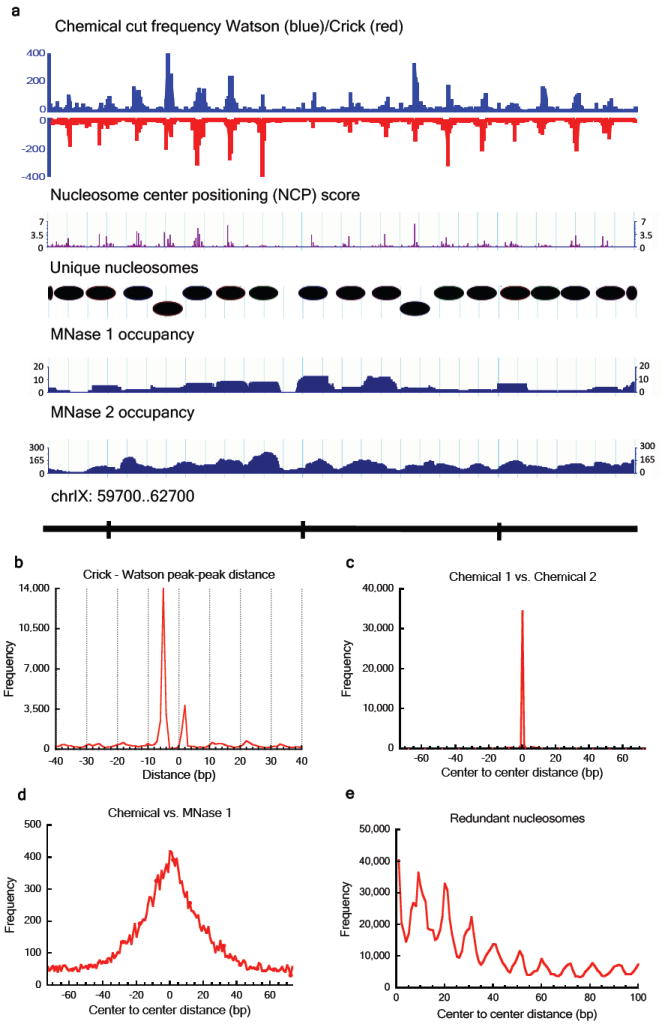
a, Screenshot of raw data, nucleosome center positioning score (NCP score), defined unique nucleosomes and two MNase maps (MNase 1 30 and MNase 2 31). b, Crick – Watson cleavage peak-peak distances showing two dominant distances: +2 and -5 nucleotides. c, Distances between the nearest nucleosome centers in two independent chemical maps and d, in the combined chemical map vs. the MNase 1 map. e. Distance between nucleosome centers in the redundant map showing preferential spacing of multiples of ~10 bp.
We defined the nucleosome centers by identifying the characteristic pattern of chemical cleavages genome-wide (Supplementary Methods). We first trained a single-template model and then a four-template model to represent the average cleavage pattern at eight nucleotide positions centered around the primary and secondary sites (Supplementary Table 1). The four-template model accounts for four major distinct cleavage patterns associated with nucleotide composition at positions -3/+3 and outperforms the single-template model (Supplementary Figs. 1a,b,c). The templates were built into a Bayesian deconvolution algorithm to calculate the nucleosome center positioning score (NCP score) at every genomic location. A larger NCP score means more cleavages observed in positions that conform to primary and secondary sites configuration, thus indicating more well positioned nucleosomes. Nucleosome centers were defined based on the magnitude of the NCP score to noise ratio (Fig. 2a, Supplementary Fig. 1d).
Comparing two independent chemical mapping experiments, the resulting nucleosome positions are extremely consistent (Fig. 2c). Hence we combined the 6-independent datasets and produced a final unique map of 67,517 nucleosomes (Supplementary Methods), allowing two neighboring nucleosomes to overlap by no more than 40 bp (occupying 79.9% of the genome, Supplementary Table 2); and a redundant map of 355,923 nucleosomes (allowing nucleosomes to overlap arbitrarily) by including all positions whose NCP score/noise ratio exceeded the smallest ratio value from the unique map (Supplementary Table 3). The chemical map shows general accordance with published MNase maps (MNase 1 30, and MNase 2 31) of nucleosome positions, establishing that the chemical mapping technique does map nucleosome centers (Figs. 2a,d, Supplementary Figs. 2a,b). All analyses (unless indicated) presented in the main text use the unique set of nucleosomes. Important conclusions drawn from the unique map were confirmed using the redundant map, and results are presented separately in Supplementary Fig. 12.
The current chemical data exhibit unbalanced cleavage patterns between the two strands at nucleosomes flanking long linkers including nucleosome free regions (NFRs) upstream of transcript start sites (TSSs) (Supplementary Figs. 3a, b). As only the densest portion of the lowest molecular weight DNA fragment (~125-200bp) was isolated from the agarose gel, DNA fragments spanning longer linkers were selected against (Fig. 1b). We confirmed this by isolating and sequencing DNA fragments of higher molecular weight (~200-500bp) produced from one chemical mapping experiment (Methods). The resulting data recovered the expected symmetric cleavage pattern at nucleosomes flanking the NFRs (Supplementary Fig. 3b), and confirmed that failure to map nucleosomes due to consecutive long linkers is not a concern in the current data (Supplementary Fig. 3c). The cleavage asymmetry is not problematic for defining nucleosome centers, as one strand can still give a local maximum positioning score in our Bayesian deconvolution approach.
Nucleosome overlap and base composition
The chemical map reveals detail of competing preferential nucleosome positions genome-wide. The redundant map suggests that in a population of cells nucleosomes often maintain multiple overlapping positions, creating closely clustered cleavage sites (Supplementary Fig. 4a). These sites were deconvoluted to yield locations and relative abundance of overlapping nucleosomes. Strikingly, overlapping nucleosomes are predominantly spaced by integral multiples of the DNA helical repeat (~10 bp) (Fig. 2e, Supplementary Fig. 4b). This result accords with ~10 bp periodic cleavage endpoints in MNase maps32, 33 and clarifies that MNase cleavage periodicities reflect multiple nucleosome positions instead of an artifact of the MNase itself32. This overlapping positioning of nucleosomes could be a consequence of the population or dynamic average, or even the co-existence of overlapping nucleosomes on the same DNA molecule34. By shifting in 10 bp steps (one helical turn) nucleosomes can maintain the local chromatin 3-dimentional structure, while sampling multiple positions.
Nucleosomes in vivo are enriched for particular DNA sequence motifs at particular points along the nucleosomal DNA, most notably ~10 bp periodic occurrences of AA/AT/TA/TT dinucleotides that are favored when the DNA backbone (minor groove) faces inward toward the histone core, and CC/CG/GC/GG dinucleotides when the DNA backbone faces outward27, 35, 36. These signals are seen in the chemical map with a period of 10.3 bp, and are far stronger than what was seen from MNase maps (Figs. 3a,b, Supplementary Fig. 5a), suggesting that imprecision in the MNase maps degrades the sequence alignment. The ~0.2 bp difference between the dinucleotide periodicity and the classical helix repeat length (i.e. 10.5 bp37) implies that DNAs likely over-stretch by 1-2 bp to accommodate the packing constraints of nucleosome structures in the chromatin fiber1. The strength of dinucleotide signal increases with the NCP score to noise ratio, suggesting that stronger dinucleotide sequence features are associated with positioning of more stable nucleosomes (Fig. 3a).
Figure 3. Nucleosome sequence preferences.

a. Position-dependent frequencies of AA, AT, TA, or TT dinucleotides in the chemical map, by quartiles of NCP score/noise ratio. b, Comparison of dinucleotide frequency between the chemical map and the MNase 1 map. The chemical map shows significantly stronger AA/AT/TA/TT dinucleotide signals suggesting an improved center alignment of nucleosomal DNA. c, 5-mer preferences of nucleosomes (left) and linker DNA regions (right) in chemical map compared to the MNase 1 map, colored by numbers of A or T nucleotides.
These dinucleotide signals are average features of the aggregated nucleosomes in the unique set, and also of the overlapping nucleosomes (Supplementary Fig. 5b). The unique nucleosomes show an average of 40-50% occurrence rate of AA/TT/TA/AT at peak positions while lacking a dominant AA/TT/AT/TA bias at the edges of the nucleosomes observed in MNase maps (Fig. 3b)19, 21. The preferences of nucleotide 5-mers from the chemical map agree well with those measured by MNase, including the strong negative preference of nucleosomes for A/T-rich 5-mers38, 39. However, both the nucleosome and linker regions are less-strongly biased against A/T 5-mers in the chemical map than in the MNase maps (Fig. 3c), indicating that MNase preferentially degrades A/T-rich nucleosomes.
Intriguingly we noted a high frequency of occurrences (~0.6) of an A at position -3, and its symmetry-related T at position +3 relative to the nucleosome center (Supplementary Figs. 6, 7). This high frequency has been observed in previous in vitro nucleosome positioning studies, with a frequency of 0.5 at these two locations40. Nucleosomes lacking both A(-3) and T(+3) (10% of the unique set) exhibit similar but slightly weaker dinucleotide preference pattern compared to those having one or two of these nucleotides (Supplementary Fig. 5c). However we cannot exclude the possibility that chemical map might present a minor bias favoring an A(-3) or a T(+3) on top of the true preference of these nucleotides.
Global features of nucleosome map
The high-resolution chemical map allows for more accurate conclusions about global nucleosome features. We defined the nucleosome occupancy score at any given location as the total NCP scores in the +/-73 bp region after correction for cleavage asymmetry on the two strands and controlling for signal/noise ratio as done in the redundant map (Supplementary Methods). The chemical map reproduces the well-known strong depletion of nucleosomes immediately upstream of the transcription start site (TSS)41 exposing the binding sites of regulatory proteins, with ordered nucleosomes extending downstream over the protein coding sequence, and much weaker ordering upstream of the TSS (Fig. 4a). The average occupancy curve within 1000 bp of TSS is highly consistent with that from the MNase map (r = 0.98, Fig. 4a), suggesting the chemical map can provide a reasonable measure of nucleosome occupancy. Substantial nucleosome depletion is shown at the ends of the open reading frames (ORFs) (Fig. 4a), implying that nucleosome depletion at 3’ gene ends20 is not an artifact of MNase digestion, but a real phenomenon. For tRNA genes, which are highly transcribed by RNA polymerase III and have their promoters inside the gene bodies42, the upstream nucleosomes are strongly ordered while the gene body regions are nucleosome-depleted (Fig. 4a). No particular ordering of nucleosomes was observed relative to exon-intron or intron-exon boundaries (Supplementary Fig. 8a). We also observed strong ordering of nucleosomes on both sides of origins of DNA replication (ARS regions), and at centromere start (start of CDEI) and end sites (end of CDEIII) (Supplementary Figs. 8a,b). For the latter, the nucleosome centers are highly localized at multiple specific positions as shown by the sharp spikes of NCP score. Thus, the locations of the unique centromeric nucleosomes appear to be positioned relative to the exact locations of other key centromere-specific DNA binding proteins.
Figure 4. Genome-wide features of nucleosome postions.

a, Genome-wide average nucleosome occupancy across all sites in the chemical map plotted relative to transcription start sites (TSSs, 3017 in total41), open reading frame (ORF) ends (center), and tRNA start sites (right). b, AA/AT/TT/TA dinucleotide frequency within nucleosomes for nucleosome classes +1, +4 and +6 from the chemical map and the MNase 2 map.
A major unresolved problem concerns the role of DNA sequence features in positioning nucleosomes in protein coding regions. Nucleosomes can be classified, with +1 referring to a nucleosome covering the start of the coding sequence, and +2…+n for each successive nucleosome downstream. Nucleosomes of all classes (not only evident in the -1 and +1 class43) show strong AA/AT/TA/TT pattern with periodicity ranging from 10.20 to 10.26 bp, at which the amplitude of transform differs by no more than 5% between any two classes (Fig. 4b, Supplementary Fig. 9, Methods). Separate analysis using MNase data exhibit a similar but weaker periodic pattern, comparable across different classes (Fig. 4b, Supplementary Figure 9,). This suggests that DNA sequence features play an important role in detailed positioning of nucleosomes. A recent in vitro study44 postulates an ATP-facilitated positioning, suggesting that chromatin complexes may use ATP to override DNA-intrinsic positioning and drive directional packaging of nucleosomes against a 5’ barrier at the 5’ ends of genes and to a lesser extent at the 3’ ends. Future studies are needed to understand the exact relationship of DNA sequence features and ATP in nucleosome positioning in vivo.
Chromatin structure and nucleosome spacing
The length of free DNA between two neighboring nucleosomes (linker length) is a crucial determinant in the structure and compaction of the chromatin fiber and is directly related to the regulation of DNA associated cellular processes2, 45, 46. The linker length distribution has a median of 23 bp (for linkers <=100 bp) and exhibits a non-random pattern with a set of strong peaks at lengths equal to 3, 15, 24, 34, and 44 bp, differing by multiples of the DNA helical repeat (Fig. 5a, Supplementary Fig. 10). If we add 1 or 2 bp uniformly to each linker length to account for over-stretching of nucleosome DNAs, the linker length will follow a form of 10.41n+4.6 bp or 10.47n+5.5 bp (for integer n) respectively by Fourier analysis (for linker length<=60 bp). The actual over-stretching varies across nucleosomes, which may alter details of the linker length distribution.
Figure 5. Nucleosome spacing and higher order chromatin structures.

a, Linker length distribution for all ~67,000 unique nucleosomes (black), for the top 50% nucleosomes with highest NCP score/noise ratio (red) and for ~45,000 MNase 2 unique nucleosomes (blue). b, AA/AT/TA/TT frequency in the genomic DNA flanking the nucleosomes. c, Fourier transform of AA/AT/TA/TT signals in the unique nucleosomes (red), in downstream flanking regions (black), and the mean (blue) and 95th percentile (green) of transform in the flanking regions after permuting the phase offset between the flanking region and the mapped nucleosome (Methods).
The non-random spacing of nucleosomes is also explicitly displayed in the genomic DNA sequence. The periodicity of AA/AT/TA/TT motifs extends from the aligned unique nucleosomes to flanking regions beyond one nucleosome repeat length (Fig. 5b) with a period of 10.26 bp and a phase offset that accords with nucleosome spacing of ~10n + 5 bp, confirming the direct analyses above and previous findings47 Fig. 5c, Methods). This suggests that given a nucleosome somewhere in the genome, the next nucleosome down the chain likely starts at approximately the opposite face of the double helical DNA, creating an open zig-zag intrinsic chromatin structure, consistent with the two-start architecture of the 30nm fiber48.
Nucleosomes and DNA binding proteins
We also examined the spatial relationships between nucleosomes and other high-resolution genomic features critical to genetic regulation. Functional transcription factor (TF) binding sites, defined as sites that are conserved and occupied in vivo49, were frequently contained inside mapped nucleosomes (Figs. 6a,b top, Supplementary Fig. 11a). This observation signifies that in a population of cells at any moment, some cells lack a nucleosome and have a bound factor, while other cells lack the bound TF and have the nucleosome instead or some cells might have TF bound to the edge of the nucleosomes while the latter are partially unwrapped. More remarkably, within the nucleosome, TF sites are strongly enriched at highly specific sites where the nucleosome sequence preferences best match those of the TF, and thus they can be predicted, given a position weight matrix for the TF. Similarly, for the Hermes transposase, another sequence-specific DNA binding protein, the transposon integration sites occur periodically through the nucleosome (Fig. 6b, Supplementary Fig. 11b) at locations predictable by the transposase sequence specificity, demonstrating a type of multiplexing of genomic information.
Figure 6. High-resolution nucleosome center positions relative to genomic features.

a, Frequency of functional Abf1 sites (left) and the average motif score calculated using position weight matrix for Abf1 (right) relative to nucleosome centers. b, Frequency of all even length functional TF binding sites (top) and Hermes transposon integration sites (bottom) relative to the nucleosome centers. c, Frequencies of RNA polymerase II pause sites mapped inside nucleosomes +2 to +4 relative to the nucleosome centers, for wildtype yeast cells (top) and dst1- Δ yeast cells (lacks polymerase backtracking ability; bottom).
Nucleosomes also influence elongation by RNA polymerase II (RNAP), with a recent genome-wide analysis showing enhanced pausing by RNAP predominantly within the first half of the nucleosome8. The chemical map reveals enhanced pause sites across the full length of nucleosomes, suggesting that DNA downstream of the RNAP can remain wrapped even after RNAP has progressed far inside the nucleosome. However, we cannot exclude the possibility that this observation is complicated by the population average of nucleosomes from different locations within clusters. Notably, the RNAP pause sites do not occur at random locations in the nucleosome, rather, they occur periodically, at the DNA helical repeat (Fig. 6c, Supplementary Fig. 11b), indicating that RNAP stalls preferentially at specific locations relative to the nucleosome rotational position. The locations at which RNAP pauses are best reflected in a dst1-Δ strain that lacks the ability for RNAP to backtrack. Strikingly, in this mutant, RNAP pause sites again appear periodically, but at locations that are systematically displaced by ~5 bp, i.e., by half of a DNA helical turn relative to the wildtype pause sites (Supplementary Fig. 11c). In the dst1-Δ strain, the RNAP has the greatest probability of pausing at locations where the DNA backbone faces out, away from the histones. This suggests that RNAP pauses at sites where further forward motion requires rotation around the DNA backbone, creating a steric clash between the polymerase and the histones. Conversely, the backtracking RNAP is most likely to pause at locations in the nucleosome at which the backbone faces inward toward the histones (with maximal steric clash between histone and RNAP). In this situation, at the initiation of transcription, the first few forward steps will reduce this clash, as the polymerase rotates away from the histone surface.
Conclusions and prospects
We developed a new method for mapping nucleosome centers in yeast with unprecedented consistency and accuracy. The resulting map shows significantly stronger dinucleotide signals in nucleosome DNAs than MNase maps. These signals are present and comparable in nucleosomes spanning entire protein coding regions, demonstrating an important role of the DNA sequence features in detailed positioning and spacing of nucleosomes genome-wide. The observed linker length distribution suggests that genome prefers an organized local chromatin structure. The exact locations of nucleosomes are associated with diverse biological phenomena, including the exact locations of TF binding sites, transposon integration sites, and RNAP pausing. We anticipate that this map will shed additional light on many other genetic processes, including replication, recombination, mutation, repair, and genome evolution, because of the intimate relationship between nucleosome locations and these diverse processes.
Methods Summary
We genetically engineered S. cerevisiae to contain a cysteine at position 47 in histone H4. Cells grown to mid-log phase were harvested, permeabilized and labeled with N(1,10 phenanthroline- 5-yl) iodoacetamide. The label covalently bound to the cysteine and allowed for copper chelation. Copper chloride, mercaptoproprionic acid and hydrogen peroxide were added sequentially creating hydroxyl radicals that cleaved the nucleosomal DNA at sites flanking the center. After the mapping reaction, the genomic DNA was purified from the cells and ran on an agarose gel. The shortest molecular weight DNA fragment (~150-200bp) was purified and prepared for highthroughput parallel sequencing.
Supplementary Material
Acknowledgments
We would like to acknowledge the life and achievements of J.W. who passed away during the preparation of this manuscript. J.W. was our dedicated leader who passionately and enthusiastically guided this project. We thank Drs. R. Holmgren, A. Matouschek, B. Meyer, R. Phillips, E. Segal and O. Uhlenbeck, for helpful comments and discussions. We are grateful to Northwestern University’s Genomic Core for all sequencing completed for this project. The work was supported by NIH Grants T32GM00806 (K.B.), R01GM058617 (J.W.), R01GM075313 (J.P.W.) and U54CA143869 (J.W.).
Footnotes
Contributions
K.B. did all experimental work. X. L. and J.P.W. developed the algorithm and performed the analyses. K.B., J.P.W. and J.W. wrote the paper. J.W directed the project.
Sequence data are deposited to NCBI GEO data base with accession #GSE36063.
References
- 1.Richmond, Davey The structure of DNA in the nucleosome core. Nature. 2003;423(6936):145–50. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 2.Koslover, et al. Local geometry and elasticity in compact chromatin structure. Biophys J. 2010;99(12):3941–50. doi: 10.1016/j.bpj.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li, Widom Nucleosomes facilitate their own invasion. Nat Struct Mol Biol. 2004;11(8):763–9. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 4.Raveh-Sadka, et al. Incorporating nucleosomes into thermodynamic models of transcription regulation. Genome Res. 2009;19(8):1480–96. doi: 10.1101/gr.088260.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao, et al. Occlusion of regulatory sequences by promoter nucleosomes in vivo. PLoS ONE. 2011;6(3):e17521. doi: 10.1371/journal.pone.0017521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, et al. The role of chromatin during transcription. Cell. 2007;128(4):707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Petesch, Lis Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134(1):74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Churchman, Weissman Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469(7330):368–73. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen-Hughes, Workman Experimental analysis of chromatin function in transcription control. Crit Rev Eukaryot Gene Expr. 1994;4(4):403–41. [PubMed] [Google Scholar]
- 10.Li, et al. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12(1):46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 11.Lipford, Bell Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell. 2001;7(1):21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- 12.Dorn, Cook Nucleosomes in the neighborhood: new roles for chromatin modifications in replication origin control. Epigenetics. 2011;6(5):552–9. doi: 10.4161/epi.6.5.15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachiwana, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476(7359):232–5. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 14.Cole, et al. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc Natl Acad Sci USA. 2011;108(31):12687–92. doi: 10.1073/pnas.1104978108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalal, et al. Structure, dynamics, and evolution of centromeric nucleosomes. Proc Natl Acad Sci USA. 2007;104(41):15974–81. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beckmann, Trifonov Splice junctions follow a 205-base ladder. Proc Natl Acad Sci USA. 1991;88(6):2380–3. doi: 10.1073/pnas.88.6.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz, et al. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16(9):990–5. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 18.Tilgner, et al. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16(9):996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 19.Dingwall, et al. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981;9(12):2659–73. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan, et al. Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3’-end formation. Proc Natl Acad Sci USA. 2010;107(42):17945–50. doi: 10.1073/pnas.1012674107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hörz, Altenburger Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981;9(12):2643–58. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung, et al. The effect of micrococcal nuclease digestion on nucleosome positioning data. PLoS ONE. 2010;5(12):e15754. doi: 10.1371/journal.pone.0015754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, et al. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009;16(8):847–52. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flaus, et al. Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc Natl Acad Sci USA. 1996;93(4):1370–5. doi: 10.1073/pnas.93.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flaus, Richmond Base-pair resolution mapping of nucleosome positions using site-directed hydroxy radicals. Meth Enzymol. 1999;304:251–63. doi: 10.1016/s0076-6879(99)04015-x. [DOI] [PubMed] [Google Scholar]
- 26.Yager, et al. Salt-induced release of DNA from nucleosome core particle. Biochemistry. 1989;28:2271–2281. doi: 10.1021/bi00431a045. [DOI] [PubMed] [Google Scholar]
- 27.Widom Role of DNA sequence in nucleosome stability and dynamics. Q Rev Biophys. 2001;34(3):269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- 28.Thåström, et al. Nucleosomal locations of dominant DNA sequence motifs for histone–DNA interactions and nucleosome positioning. J Mol Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 29.Teif, Rippe Predicting nucleosomes positions on the DNA: combining intrinsic sequence preferences and remodeler activities. Predicting nucleosome positions on the DNA. Nucleic Acid Research. 2009;37(17):5642–5655. doi: 10.1093/nar/gkp610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Field, et al. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4(11):e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Floer, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141(3):407–18. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cockell, et al. Location of the primary sites of micrococcal nuclease cleavage on the nucleosome core. J Mol Biol. 1983;170(2):423–46. doi: 10.1016/s0022-2836(83)80156-9. [DOI] [PubMed] [Google Scholar]
- 33.Albert, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446(7135):572–6. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 34.Engeholm, et al. Nucleosomes can invade DNA territories occupied by their neighbors. Nat Struct Mol Biol. 2009;16(2):151–158. doi: 10.1038/nsmb.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, Widom Improved alignment of nucleosome DNA sequences using a mixture model. Nucleic Acids Res. 2005;33(21):6743–55. doi: 10.1093/nar/gki977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segal, et al. A genomic code for nucleosome positioning. Nature. 2006;442(7104):772–8. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Travers, Klug The bending of DNA in nucleosomes and its wider implications. Philos Trans R Soc Lond, B, Biol Sci. 1987;317(1187):537–61. doi: 10.1098/rstb.1987.0080. [DOI] [PubMed] [Google Scholar]
- 38.Segal, Widom What controls nucleosome positions? Trends Genet. 2009;25(8):335–43. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal, Widom Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19(1):65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thåström, et al. Measurement of histone-DNA interaction free energy in nucleosomes. Methods. 2004;33(1):33–44. doi: 10.1016/j.ymeth.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 41.David, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA. 2006;103(14):5320–5. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schramm, Hernandez Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 43.Mavrich, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18(7):1073–83. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, et al. A packaging mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 2011;332:977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lohr, Van Holde Organization of spacer DNA in chromatin. Proc Natl Acad Sci USA. 1979;76(12):6326–30. doi: 10.1073/pnas.76.12.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Routh, et al. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci USA. 2008;105(26):8872–7. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, et al. Preferentially quantized linker DNA lengths in Saccharomyces cerevisiae. PLoS Comput Biol. 2008;4(9) doi: 10.1371/journal.pcbi.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dorigo, et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306(5701):1571–3. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 49.MacIsaac, et al. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


