Abstract
Translational research is needed to leverage discoveries from the frontiers of genome science to improve public health. So far, public health researchers have largely ignored genetic discoveries, and geneticists have ignored important aspects of population health science. This mutual neglect should end. In this article, we discuss 3 areas where public health researchers can help to advance translation: (1) risk assessment: investigate genetic profiles as components in composite risk assessments; (2) targeted intervention: conduct life-course longitudinal studies to understand when genetic risks manifest in development and whether intervention during sensitive periods can have lasting effects; and (3) improved understanding of environmental causation: collaborate with geneticists on gene–environment interaction research. We illustrate with examples from our own research on obesity and smoking.
With the completion of the Human Genome Project, discoveries linking variations in the DNA sequence with common health conditions have come thick and fast. Translational science that applies these discoveries to reveal novel drug targets, refine treatments, or prospectively identify individuals at risk has progressed at a slower pace.1,2 This slow progress reflects appropriately high standards for the clinical application of genetic discoveries.3 It also reflects the fact that a large portion of the translational research community has remained on the sidelines of genome science. In the early days of genetic discovery research, it was appropriate for social, behavioral, and health scientists doing public health research to take a “wait and see” approach to genetic discoveries. That time is now past. Personalized medicine is bringing the genome into the clinic, and direct-to-consumer genetic testing is bringing it into the community.4,5 Biologists are working hard to elucidate molecular pathways that link genetic discoveries with disease. Parallel efforts are needed in the fields of life-course epidemiology, health behavior and health education, and health services research to understand the developmental and behavioral pathways from genetic discoveries to disease and to identify opportunities for cost-effective intervention.
Public health research is uniquely positioned to contribute to the translation of genetic discoveries because of its population approach to health science. Population-representative samples, prospective longitudinal designs, and measurements of environmental context are critical to understanding how genetic risks manifest across time (e.g., development and aging) and space (e.g., policy and environmental risk strata). In turn, this population health science understanding of genetic risk can inform disease etiology and help to refine both individual- and population-level intervention strategies.
In this article, we address 3 areas where public health researchers can help to advance translation: (1) risk assessment, (2) targeted intervention, and (3) improved understanding of environmental causes of disease. We discuss the progress of discovery science as it relates to each of these applications, and we explain how public health research can contribute to translation. We also present example cases from own work on obesity and smoking, and we outline new research directions.
RISK ASSESSMENT
The most immediate translational application of genome science to improve public health is likely to come in population screening for rare, genetically determined diseases that can be prevented or mitigated with timely intervention.6 A promising example case is Lynch syndrome, in which a mutation in a single gene confers a greater than 80% risk of colon cancer, and for which treatments are available that can drastically reduce morbidity.7 Health services research is needed to determine how genetic screening for these types of diseases can be best deployed to meet ethical and cost-effectiveness criteria. For common diseases, however, this type of screening is less feasible.8
Etiological theory and empirical evidence indicate that large numbers of environmental and genetic factors contribute to common diseases. Given such complexity, diagnosis by genetic testing is an unrealistic prospect, even under the condition that all disease causing genetic variation is known.9,10 Nevertheless, genetics can provide probabilistic information about risk. Current genetic discoveries may already furnish enough information to make incremental improvements in clinical risk assessments of adults. For example, in the cases of heart disease and type 2 diabetes, panels of genetic risk markers may improve risk classification over and above existing risk indexes, although not all studies show this.11–16 More uncertain is whether these incremental improvements in risk assessment can change medical decision-making in ways that improve patient outcomes.17
Thus, a fair criticism of translational research investigating genetics for risk assessment is that known genetic risks for common health conditions offer too little information to be interesting to the clinician. An equally fair rejoinder is that known genetic risks furnish about as much information as many other risk factors currently considered in the clinic (e.g., C-reactive protein in heart disease).11,18 Faced with these options, the translational genetics researcher must make a choice: to be on the side of the angels, promoting a cost-effective medical practice that considers only as much information as needed for sound decision-making; or to lean on a tradition of a cost insensitive but intellectually curious medical practice interested in any and all information that can bring prognosis incrementally closer to truth. Quite responsibly, leaders in genetics have argued forcefully for the first option.17 But the choice is a false one. Risk assessment comprises a broad array of applications. Too little translational research has been conducted to dismiss the value of genetics for any or all of them. In the following section, we outline 2 possibilities for the application of genetics in risk assessment that have yet to be vetted through translational research.
Genetics may provide a window into clinical heterogeneity: genetic information may be useful in understanding differences in the timing of onset, rate of progression, persistence, comorbidity, and response to treatment. The last of these has received the most attention, with a recent example of success coming from the Genome-Wide Association Study (GWAS) of the treatment response to carbamazepine, a drug used in the management of epilepsy, trigeminal neuralgia, and bipolar disorder. GWAS uncovered a novel marker of risk for carbamazepine-induced hypersensitivity reactions among individuals of Northern European ancestry (the HLA-A*3101 allele).19 Consequences of these hypersensitivity reactions range from a painful rash to death. Personalization of treatment via genetic testing could reduce adverse events and possibly save lives.20 Further research is needed in this area, and progress depends on the construction of large-scale databases that link genomic data with detailed longitudinal data on the course and treatment of illness. Such efforts are ongoing,21,22 and aggressive analysis of these databases should be a priority for decision science and cost-effectiveness research into where and how integration of genetic information can improve the quality of health care.
Genetics can contribute to composite risk assessments that identify high- and low-risk segments of the population. In the near term, applications of genetics in individual-level risk assessment may be limited. At the population level, genetics can help to identify groups susceptible to developing a particular health problem. Genetics are well suited to providing this kind of background information because the DNA sequence is fixed at conception and can be reliably and noninvasively assayed at any point in the life course. Background genetic risk information can, in turn, inform investigations of other risk factors or of prevention approaches. A critical first question to ask in this application is whether genetic discoveries actually provide new information.
Example Cases From Our Research on Obesity and Smoking
In our own work on obesity and smoking, we sought to address the question of whether genetic discoveries could provide new information about risk by looking at genetic risk profiles in comparison with family history risk profiles. Family histories are measures of inherited risks that are considered to have clinical value.23,24 Like the DNA sequence, family histories can be easily and noninvasively assessed before risk exposures accrue or symptoms manifest. These parallels make family history a useful benchmark for evaluating the potential utility of genetic information. Therefore, we asked the following questions. (1) How do the magnitudes of risks predicted using genetic information compare with the magnitudes of risks predicted using family history? (2) Do genetics provide new information about risk beyond what can be gleaned from a family history?
Measuring genetic risk: from genetic discoveries to genetic risk scores.
To answer these questions, we began by developing polygenic risk profiles for obesity and smoking. Obesity and smoking are complex conditions; they are influenced by large numbers of genetic variants. Rather than being present or absent, genetic risks for obesity and for smoking are distributed along a continuum. Some people carry more risk variants in their genomes and others fewer. One way to summarize this continuum of genetic risk is to build a multi-locus profile of genetic risk, a “genetic risk score” (GRS).25,26 GRSs summarize risk information from variants across the genome. The hypothesis in a GRS analysis is that individuals with higher GRSs will be more likely to develop disease. Whole-genome methods of deriving GRSs are under development.27 In our own work, we used a more conservative approach; we built our GRSs from variants with replicated evidence of association in GWAS.28 This approach has been used extensively in GRS research on heart disease and type 2 diabetes.11,12,16,29 These more conservative GRSs may measure only the tip of the genetic iceberg.30 Therefore, we view the predictive power of our scores as representing a lower bound for what may be possible with genomic information.
Genetic risk scores versus family history scores.
We then turned to the Dunedin Multidisciplinary Health and Development Study (hereafter the “Dunedin Study”), a 4-decade longitudinal study of a representative 1972–1973 birth cohort (n = 1037) that included a DNA databank and detailed information about cohort members’ family histories. We used the DNA bank to create GRSs for obesity and smoking, and then constructed family history scores against which to compare them.
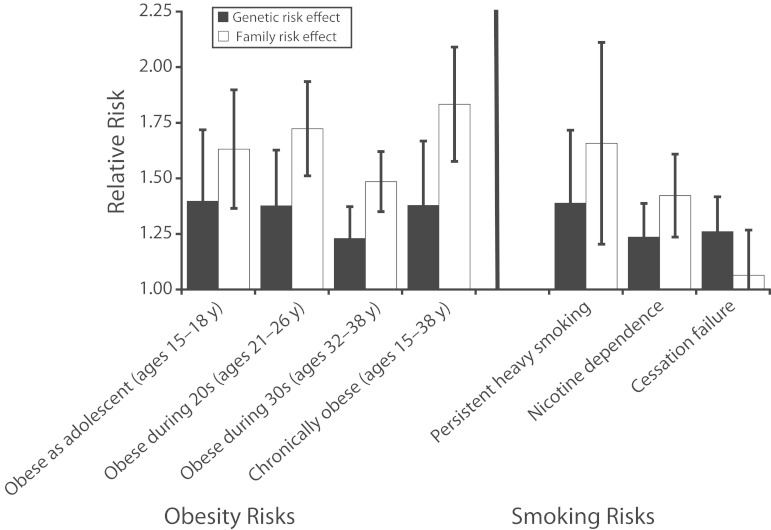
Our results revealed that, surprisingly, the GRSs were not strongly related to the corresponding family histories (see the box on the next page). In addition, effect sizes for genetic- and family history–based risk assessments were similar, although family history tended to provide somewhat more information (Figure 1). The GRSs provided information about risk that was independent of and additive to information that could be obtained from a family history.28,36 This same pattern of findings—that GWAS-discovered genetic risks are about as informative as family history and contain information not available in a family history—has also been reported for heart disease and type 2 diabetes.11,12 Thus, GWAS-discovered genetic risks appear to capture substantive and novel risk information.
FIGURE 1—
Risks for obesity and smoking associated with genetic and family history-based risk scores: Dunedin Multidisciplinary Health and Development Study, 1972–2013.
Note. Effect sizes are shown as increments in the relative risk of the outcome associated with a 1 SD increase in the risk score. The obesity family history risk score was based on parental body mass index. The smoking family history risk score was based on the smoking behavior of cohort members’ parents, siblings, and grandparents. Error bars reflect 95% confidence intervals. Chronic obesity was defined as being obese at ≥ 3 of the 6 assessments between ages 15 and 38 years. Persistent heavy smoking was defined as smoking 20 or more cigarettes/day at ≥ 3 of the 6 assessments between ages 15 and 38 years. Cessation failure was defined for cohort members who smoked daily during their 30s as being unable to quit for at least 1 year through the time of the age 38-year assessment.
Genetic Risk Scores and Family History Scores for Obesity and Smoking Capture Different Information: 3 explanations
| The genetic risks scores (GRSs) and family history scores for obesity and smoking explained only small fractions of the population variance in the traits studied (1%–4%). Family studies suggest that 50% or more of population variation in body mass index and smoking may be attributed to genetic factors.31,32 Therefore, the lack of overlap in the few percentages of variance explained by each of the genetic risk and family history scores is not implausible. Nevertheless, it was somewhat unexpected that genetic discoveries would be only weakly related to family histories (the Pearson correlations were r = 0.12 for the obesity GRS and family history score and r < 0.01 for the smoking GRS and family history score). There are 3 reasons that GRSs and family history scores might show so little overlap, and these reasons relate to differences in the types of information captured by the scores. |
| 1. GWAS-derived GRSs capture common genetic risks, whereas family history scores capture rare genetic risks or environmental risks shared by family members. GWAS measures only common genetic variation (most GWASs examine only variants with a frequency of 5% or more in the general population). However, most genetic variation is “rare” (occurring in < 1% of individuals).33 Family histories include all genetic variation in addition to environments that may be shared by family members. |
| 2. GRSs capture the additive effects of common variants, whereas family history scores capture risks arising from interactions among genetic variants (epistasis) or interactions between genetic variants and environments shared by family members. Because family members share large portions of their genomes along with many environmental exposures, family histories can capture a wide array of complex interactions among and between genes and environments. |
| 3. GRSs capture only those common variants with the largest effects, whereas family history scores capture the influence of very large numbers of variants with very small individual effects. The “infinitesimal model”34 of trait heritability posits that very large numbers of common genetic variants, each with very small effects, contribute to common diseases.35 Under this infinitesimal model, many causal common variants will escape detection in GWAS unless discovery samples grow into the millions and beyond. To the extent that family history scores reflect the combined influence of many thousands of common variants with infinitesimal effect sizes, they may not overlap with GRSs composed of a handful of common variants with effect sizes large enough to be detected in GWAS. |
Note. GRS = genetic risk score; GWAS = Genome-Wide Associated Study.
Questions for Future Research
Discovery research has identified genetic risk factors. It is already clear that these risk factors are not powerful enough on their own to provide clinically actionable information. The next steps are to determine if genetics can be combined with other risk information to improve risk assessment. Research is needed to answer 2 questions:
Do genetic risks measured by whole-genome GRSs overlap with family history? Whole-genome scores designed to capture additive “infinitesimal” genetic influences show some promise as a risk assessment technology.37 Establishing whether such genome-wide scores provide different information from family history will be important to gauging their clinical value. A rigorous way to address this question is through the use of registry data from Northern European countries. Many individuals in these registries have been genotyped for GWAS. Therefore, it is possible to calculate whole-genome scores. Similarly, linkages with data from national health systems make it possible to generate family histories. If these 2 sources of risk information were correlated, doing so could provide information about the degree to which whole-genome and family history risk scores provide independent or overlapping information about risk.
Should we conduct genetic risk assessments of individuals who do not have a family history of disease? Currently, governments and health care organizations are using family history information to determine whether genetic testing for certain diseases is warranted. Our results suggest that genetic information is equally informative regardless of family history. Therefore, research is needed to determine under what conditions and for which diseases a positive family history is an appropriate criterion for genetic testing. Further research can also help to clarify how genetic screening compares with family history to inform its use by individuals who cannot access their family’s history of disease.
Ultimately, to evaluate the application of a GRS in a clinical risk assessment, the risk information captured by genetics must be operationalized at the individual level. Population-based measures of relative risk can be misleading as to clinical significance.38–40 Many approaches to measuring the individual-level predictiveness of a risk factor have been proposed in the biostatistics literature,41,42 several of which are well-suited to evaluating a GRS.43 As clinical databases begin to incorporate genetic information, these methods can be applied to determine how genetics compares with other sources of risk information and whether it may be cost effective to include genetics in composite risk assessments applied in clinical settings.
TARGETED INTERVENTION
Genetic discoveries could help to target interventions by answering 3 questions. (1) With whom should we intervene? Genetic discoveries could identify individuals who will develop disease or respond to treatment. (2) When should we intervene? Genetic discoveries could identify sensitive or critical periods in pathogenesis when intervention could be most effective. (3) How should we intervene? Genetic discoveries can inform the selection of intervention targets.
The capacity of genetic discoveries to help to answer the “who” question remains uncertain (see the Risk Assessment section). However, there is clear opportunity for genetics to help answer questions of “when” and “how.” Theory-free discovery studies (e.g., GWAS and newer next generation sequencing approaches) survey the full spectrum of variation in the genome to identify variants that stand out as characteristic of patients with disease. This approach can leapfrog current biology to make discoveries that provide insight into new mechanisms of disease pathogenesis and refine understanding of known mechanisms.44 A prominent example is the discovery of variants in the gene FTO that predispose to obesity.45
Bottom-Up and Top-Down Approaches to Translational Research
One way to move from discovering a variant to understanding when and how it manifests to cause disease is to work from the bottom up, by tracing the path from variation in the DNA sequence to differences in RNA transcription, subsequent protein production, and onwards up through disease pathogenesis to identify a process or a molecule that can be targeted for intervention.46 The bottom-up model has received most of the attention in efforts to develop genomic medicine. In an early example of success through this approach, a research team at Stanford applied multiple “omics” analyses to repeated blood draws from a single individual, and among other discoveries, were able to detect the onset of diabetes in an individual who standard medical approaches classified as healthy.47 Less attention has been paid to complementary research that works from the top down, by relating genetic discoveries to individual differences in behavior and health states and then working backward to devise interventions that can mitigate genetic risk for disease.48 In this article, we focus on the top-down approach because it is where we think the social, behavioral, and health scientists currently sitting on the sidelines of translational genome science research can make the largest contribution.
Top-Down Research Can Support Translation of Genetic Discoveries
Currently, the dominant approaches to genetic discovery utilize theory-free data mining research designs (e.g., GWAS).49–51 These discovery approaches have been extraordinarily productive,52 but are highly data intensive. As a result, most discovery samples consist of adults drawn from a mix of cohort and case-control studies, and include only a single cross-sectional measurement of a health outcome. The goal of these studies is to identify unambiguous “signals” that a genetic variant is associated with a health outcome. Further research is then required to understand what these signals mean. Top-down research is needed to validate genetic risk effects in population-representative cohorts and locate when in the life course genetic risks manifest. Going one step further, top-down research can build on existing knowledge of etiology to test theory-driven hypotheses about how genetic risks give rise to disease.53
Example Cases From Our Research on Obesity and Smoking
Life-course epidemiology has shown that the roots of many common chronic health conditions lie in the earliest stages of life.54,55 And there is growing evidence that even the best-replicated genetic associations with disease are developmentally dynamic, meaning they change as individuals age.56–58 Yet life-course epidemiology has, for the most part, ignored genetics and genetics has ignored the life course. A life-course approach can help to map the pathway from discovery to translation by identifying when in development genetic risks manifest. The goal of this approach is to identify windows in development when it may be possible to intervene to mitigate genetic risks by disrupting the progression from early developmental phenotypes to mature disease.
For obesity, rapid growth during gestation, infancy, and early childhood are well-documented developmental risk factors.59,60 We asked (1) whether genetic risks discovered in studies of adults were also related to these developmental phenotypes, and (2) whether these early developmental phenotypes served as critical mediators of genetic risk. This second question sought to address whether intervention to disrupt or mitigate early manifestations of genetic risk might serve to prevent disease later in life. In the Dunedin cohort, genetic risks for obesity predicted rapid growth in early life, beginning after birth (genetic risk did not predict birth weight, but did predict weight gain in the first 3 years of life and earlier adiposity rebound). In turn, these developmental phenotypes of rapid early growth accounted for genetic risk for adult obesity.28
For smoking, initiation early in life and rapid progression from initiation to heavy use during adolescence are well-documented developmental risk factors.61,62 We therefore used a parallel approach to our obesity research to investigate genetic influences on the development of smoking problems across the life course. In the Dunedin cohort, genetic risks accelerated individuals’ progress from smoking initiation to heavy smoking (genetic risks did not predict the initiation of smoking, but did predict the likelihood of converting to daily smoking and progressing to smoking 20 or more cigarettes per day during adolescence). In turn, these developmental phenotypes of rapid progression from initiation to heavy use accounted for genetic risk for adult smoking problems, including persistent heavy smoking, nicotine dependence, and cessation failure.36
Collectively, these findings suggest that intervention to regulate growth in early life and to prevent teenagers from progressing to regular, heavy smoking may mitigate genetic risks and ultimately prevent mature disease.
Questions for Future Research
The approach we took in our research on obesity and smoking could be replicated for many other health problems (e.g., cardiometabolic diseases), which are also known to have their origins in early life. In addition to locating the influences of genetic risk in early development, it is also important to follow-up genetic influences later in the life course. Two priority research questions are:
Do interventions that target early developmental manifestations of genetic risk have enduring effects across the life course? To answer this question, genetics must be integrated into follow-up studies of early intervention trials. Adult follow-up of early educational interventions, such as the Abecedarian63,64 and Perry-Preschool65,66 projects, illustrate the value of this approach. To return to genetics and health, our research suggests that too rapid growth in early childhood is critical in linking genetic risk with adult obesity. Therefore, we would hypothesize that genetic risks for obesity will have weaker effects in children who received interventions to promote healthy eating and exercise early in life. In parallel, our research suggests that rapid acceleration of cigarette consumption during the teenage years is critical in linking genetic risk with adult smoking problems. Therefore, we would hypothesize that genetic risks for smoking should have weaker effects in individuals who, as teens, were exposed to environments that limited their cigarette consumption (e.g., strict monitoring or prohibitively high tobacco taxes).
Can mitigating genetic risks reduce morbidity and early mortality? A first step in answering this question is to ask whether genetic risks relate to clinical endpoints. For example, recent research points to the existence of a “benign” subtype of obesity that is uncoupled from risk for heart disease and type 2 diabetes.67,68 Important next steps are to test whether and how genetic risks are related to early mortality. Ideal settings for investigations of genetic associations with mortality include longitudinal cohort studies such as the Atherosclerosis Risk in Communities Study and the Health and Retirement Study. Regarding how genetic risks contribute to early mortality, several approaches can be taken. One approach is to analyze cause of death data to identify which specific causes account for earlier mortality in individuals at higher genetic risk. Another approach is to examine how genetic risks relate to health behavior and health behavior change.69,70 Important questions include whether individuals at higher genetic risk have more difficulty adopting healthier lifestyle practices following health shocks, whether they respond differently to health behavior change interventions, and whether they respond differently to policy changes (e.g., increases in tobacco taxes)?
UNDERSTANDING ENVIRONMENTAL CAUSATION
Environmental risks affect individuals differently. Genetics offers an opportunity to understand this heterogeneity. Interaction between genes and environments (G×E) is one of the more contentious areas of genetics research, but also the most important for public health translation of genome science.71 There are 2 reasons G×E is critical to public health translation of genome science. First, G×E is the guiding principal of personalized medicine (in which treatment is the “E”). Second, G×E may hold a key to understanding the etiology of conditions like asthma and obesity, which, although highly heritable, have dramatically increased in prevalence in recent years. In addition, G×E is an area where many public health researchers currently sitting out the genomics revolution have much to contribute—as experts in the environment. Although G×E is relevant to virtually every public health research topic, we focus our attention in this section on the issue of health inequalities.
Health inequalities have received little attention in genetics, probably attributable in part to the lingering stain of the eugenics movement. Correspondingly, genetic discoveries have been largely ignored in health inequalities research. There is reason to end this mutual neglect. Health inequalities have not been adequately explained by differences in discrete environmental or behavioral risks.72 Nor is there strong evidence that programmatic interventions to address “root causes,” such as access to health services or financial resources, or the quality of the built environment adequately ameliorate health inequalities.73 This raises questions about existing models of causation in which environmental risks are presumed to contribute additively to disease in more or less the same way across the population.
Genetic Discoveries and Socioeconomic Gradients in Health
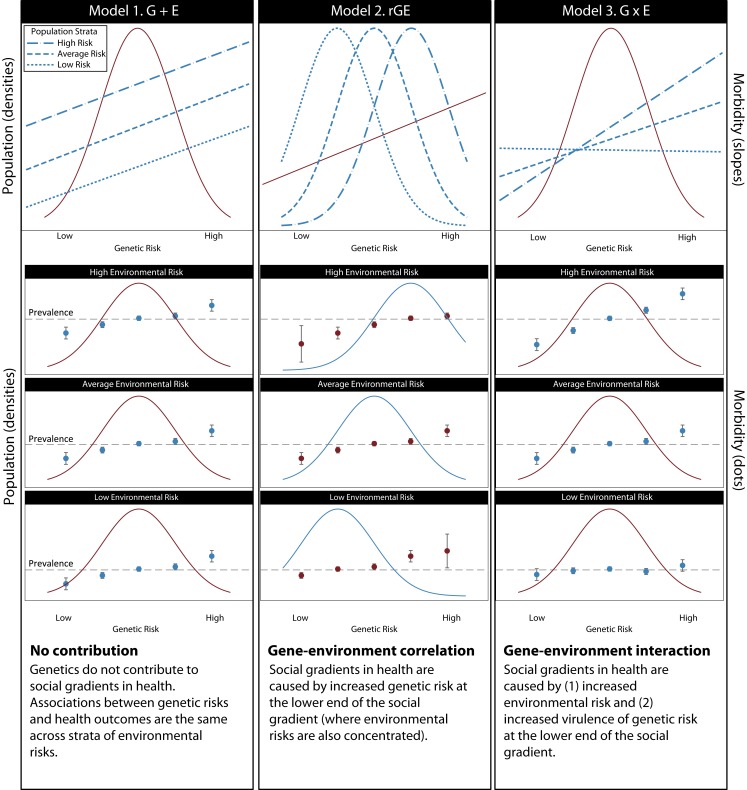
It is clear that ill health and poverty are correlated.74 It also clear that causality flows in both directions75; ill health is impoverishing, and poverty is unhealthy. Thus, there are 3 models76,77 that describe how genetic risks might contribute to socioeconomic gradients in health (Figure 2).
FIGURE 2—
Models of genetic contribution to social gradients in health.
Note. The top row of the figure shows schematic graphs of the 3 models of gene–environment interplay. Rows 2–4 show how each of these models plays out within high, average, and low environmental risk strata. Under model 1 (gene plus environment [G+E]), genetic risk effects and genetic risk distributions are the same across strata of environmental risk; the social gradient in health arises purely from differences in environmental risk. Under model 2 (gene-environment correlation [rGE]), genetic risk effects are the same across strata of environmental risk. However, the population stratum exposed to the highest environmental risk also carries the highest genetic risk and the population stratum exposed to the lowest environmental risk carries the lowest genetic risk. Under model 3 (gene-environment interaction [G×E]) genetic risk effects are stronger when environmental risk is high and weaker when environmental risk is low. The distribution of genetic risk is the same across strata of environmental risk.
No contribution: genetic and environmental risks are additive. Genetic risks are equally distributed across the social gradient. Social gradients in health are entirely caused by environmental factors (Figure 2, left column). We view this as the null hypothesis.
Gene-environment correlation (rGE): genetic risks are not equally distributed across the social gradient. Differences in health outcomes arise from the higher burden of genetic risk at the lower end of the social gradient (Figure 2, middle column).
Gene-environment interaction (G×E): genetic risks are equally distributed across the social gradient. Differences in health outcomes arise from exacerbating effects of environmental risks concentrated at the lower end of the social gradient and also from mitigating or protective effects of environmental assets concentrated at the upper end of the social gradient (Figure 2, right column).
Example Cases From Our Research on Obesity and Smoking
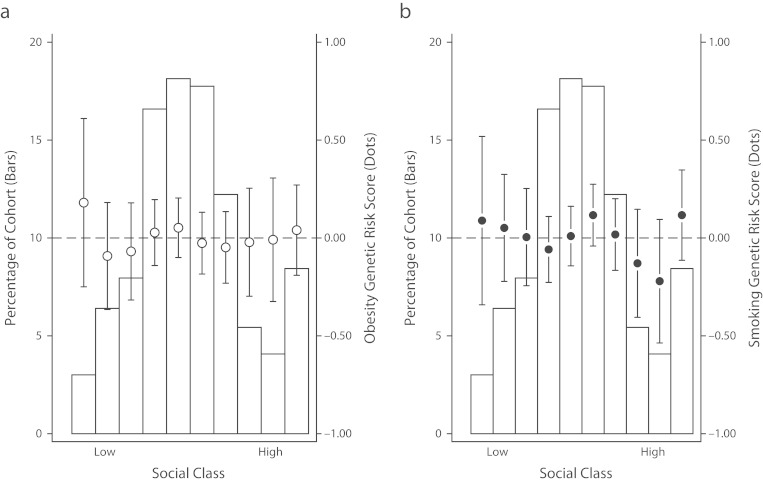
As a preliminary attempt to provide some data on this question and to illustrate how this question might be studied, we asked whether genetic risks measured by our GRSs were correlated with social class. Obesity and smoking showed strong social gradients in the Dunedin cohort, consistent with previous research.78,79 rGE would predict that children born into lower social class households would have a higher burden of genetic risk for obesity and for smoking. We found no evidence to support this rGE model. Children’s genetic risks for obesity and for smoking were unrelated to their social class (Pearson correlation r < 0.01 for both GRSs; Figure 3). We replicated this result in the ARIC cohort by comparing obesity GRSs across strata of education (average obesity GRSs for the different educational strata were all within one 20th of 1 standard deviation of the cohort-wide means; P > .2 for all comparisons). Thus, to the extent that genetic risks contribute to socioeconomic gradients in obesity or smoking, they must do so in interaction with genetic risks.
FIGURE 3—
Genetic risk by social class in the Dunedin Cohort for (a) obesity and (b) smoking: Dunedin Multidisciplinary Health and Development Study, 1972–2013.
Note. Graphs depict the distribution of social class (bars) and average genetic risk scores for obesity and smoking (dots) and associated 95% confidence intervals. Social class was defined from parents’ occupational attainment.80 Genetic risk scores were standardized to have mean of zero and standard deviation of one. The dashed gray line shows average genetic risk in the population. Under the gene-environment correlation (rGE) model of genetic contribution to social gradients in health, genetic risk should be higher for children of lower social class (dots would trend downwards from left to right). The data in this figure show that Dunedin cohort members’ genetic risks for obesity and smoking were not associated with their parents’ occupational attainment, i.e. there was no evidence of gene-environment correlation (Pearson correlations r < 0.01 for both).
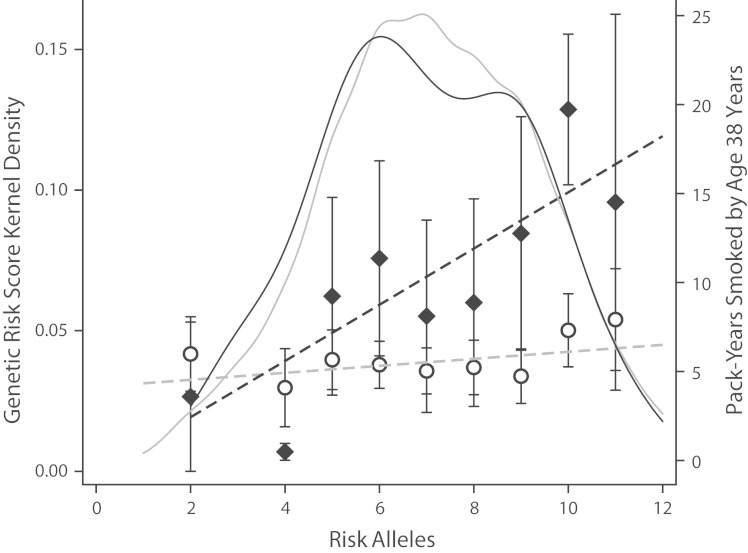
G×E predicts that genetic risk effects will be stronger in lower socioeconomic strata because protective environments that mitigate genetic risks are less prevalent, and risk environments that exacerbate genetic risks are more prevalent. To test this hypothesis, it is first necessary to identify specific environmental exposures that show social gradients and that moderate genetic risks. A number of candidate environments showing social gradients are already under investigation for interaction with genetic risks for obesity and smoking. For example, in the case of obesity, there is evidence that genetic risks can be mitigated by physical activity and exacerbated by consumption of sugary beverages.81,82 In the case of smoking, there is evidence that genetic risks can be exacerbated by peer smoking behavior83 and also by a childhood history of maltreatment.84 In the Dunedin Study, the best characterized of these exposures is childhood maltreatment,85 which shows a strong socioeconomic gradient. Therefore, we tested whether children who were maltreated showed a heightened genetic risk for smoking. Consistent with our hypothesis and in replication of a previous finding,84 children with a history of maltreatment were much more vulnerable to genetic risk for smoking; for maltreated children, the correlation between the smoking GRS and lifetime cigarette consumption was nearly 3 times as strong as for children with no history of maltreatment (r = 0.32, P = .006 for maltreated children compared with r = 0.11, P = .052 for non-maltreated children, P for difference = 0.008; Figure 4). Thus, there is initial evidence to support the hypothesis that G×E contributes to social gradients in health.
FIGURE 4—
Associations between genetic risk and lifetime cigarette consumption in cohort members with a history of childhood maltreatment and no history of childhood maltreatment: Dunedin Multidisciplinary Health and Development Study, 1972–2013.
The scatter plots and regression lines show the association between genetic risk and lifetime cigarette consumption in pack-years by age 38 years for cohort members with a history of child maltreatment (black line, diamond plot) and for cohort members with no childhood maltreatment history (gray line, circle plot). These plots show that the association between genetic risk and smoking is stronger in cohort members who were maltreated as children (i.e., there is gene-environment interaction [G×E]). The kernel density plots in the background of the graph show the distribution of genetic risk in the 2 groups (dark gray for the maltreated cohort members, light gray for the non-maltreated cohort members). These plots show that the distribution of genetic risk is similar regardless of maltreatment history (i.e., there is no rGE).
Questions for Future Research
Replication of our preliminary findings using larger datasets and more comprehensive genome-wide risk scores27 would bolster the conclusion that rGE does not contribute to social gradients in obesity and smoking, whereas G×E does. There are at least 2 other ways in which public health researchers can lead the way in investigating how genetics can improve our understanding of environmental causes of disease.
Public health researchers with expertise in environmental risks can help design studies to hunt for genes. Two G×E-informed gene-hunting approaches seem promising. One approach involves explicitly modeling G×E in discovery analyses86–88 that use large, nationally representative samples (e.g., Add Health and the Health and Retirement Survey). A second G×E-informed gene hunting approach is to conduct discovery analyses in samples selected for particular environmental exposures.89
Public health researchers can incorporate genetic information into intervention trials. A key limitation to observational G×E research is that the environmental exposure studied may not be causal in the G×E effect. Instead, the effect may be driven by unmeasured environments correlated with the exposure of interest. For example, there is evidence that physical activity attenuates the effect of variants in the gene FTO on risk for obesity.90 However, randomized trials are needed to establish that the reduction in the genetic effect is truly attributable to physical activity and not to correlated behaviors or characteristics. Randomized trials of interventions to promote weight loss through physical activity can be used to investigate this G×E. In successful trials, in which the treatment group becomes more active, researchers can ask “Is the genetic effect weaker in the treatment group as compared to the control group?” and the related question “Is the treatment effect larger in participants at higher genetic risk?”
Incorporating genetic information may be of value even when trials are unsuccessful because genetic risks may influence response to treatment.91 For example, sleep deprivation is associated with the development of obesity.92 A recent study of children found that reduced sleep predicted obesity only among those already at increased genetic risk.93 If replicated, this result suggests that genetic risk for obesity represents an important feature of a population when considering the efficacy of sleep-focused obesity interventions. In the case of smoking, there is evidence that multiple genetic variants associated with smoking also influence the effectiveness of nicotine replacement therapies.94–96 Re-analyses of failed trials that incorporate genetic risk information may provide insight into ways to “personalize” treatment.
Ultimately, findings from such genetically informed re-analyses would need to be replicated in trials designed specifically to test the effectiveness of “personalization” on the basis of genetic profile. Nevertheless, data from completed trials represent a promising resource for G×E studies.
CONCLUSIONS
The field of genetic epidemiology is fast moving and contentious. Discovery methods and the data they investigate are continuously evolving. Despite tremendous progress over the past decade, many observers are disappointed with the yield of discovery research to date.97,98 The genetics community’s response has focused on the need for bigger samples and improved technology for measuring genomes.1,99 For many in public health research, this might seem like a good argument for remaining on the sidelines. We would argue that, by contrast, this is a time to ask fresh questions. However genetic discoveries are made and whatever the nature of their molecular signatures may be, the fundamentals of population health science—representative samples, longitudinal measurements of health states, and detailed information about environmental context—are also fundamental to understanding genetic influences on health, and therefore, an opportunity for public health researchers to get in the game.
Acknowledgments
The Dunedin Multidisciplinary Health and Development Research Unit is supported by the New Zealand Health Research Council. This research received support from National Institute on Aging (grant AG032282), National Institute of Child Health and Human Development (grant HD061298) and UK Medical Research Council (grant G0601483), and additional support was provided by the Jacobs Foundation. D. W. Belsky was supported in part by fellowships from the Agency for Healthcare Research and Quality (1R36HS020524-01) and the National Institute on Aging (T32-AG000029). The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by the National Heart, Lung, and Blood Institute (contracts HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C, and grants R01HL087641, R01HL59367, and R01HL086694); the National Human Genome Research Institute (contract U01HG004402); and the National Institutes of Health (contract HHSN268200625226C). Infrastructure was partly supported by grant UL1RR025005, a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research. ARIC data (phs000090.v1.p1) were obtained from dbGaP.
We thank the Dunedin Study members, their families, Dunedin Unit Director, Richie Poulton, Unit research staff, and study founder, Phil Silva. We also thank the staff and participants of the ARIC study for their important contributions.
Human Participant Protection
The study protocol was approved by the institutional ethical review boards of the participating universities. Study members gave informed consent before participating.
References
- 1.Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470(7333):204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 2.Manolio TA, Green ED. Genomics reaches the clinic: from basic discoveries to clinical impact. Cell. 2011;147(1):14–16. doi: 10.1016/j.cell.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Rogowski WH, Grosse SD, Khoury MJ. Challenges of translating genetic tests into clinical and public health practice. Nat Rev Genet. 2009;10(7):489–495. doi: 10.1038/nrg2606. [DOI] [PubMed] [Google Scholar]
- 4.Bloss CS, Darst BF, Topol EJ, Schork NJ. Direct-to-consumer personalized genomic testing. Hum Mol Genet. 2011;20(R2):R132–R141. doi: 10.1093/hmg/ddr349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttmacher AE, McGuire AL, Ponder B, Stefansson K. Personalized genomic information: preparing for the future of genetic medicine. Nat Rev Genet. 2010;11(2):161–165. doi: 10.1038/nrg2735. [DOI] [PubMed] [Google Scholar]
- 6.Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med. 2011;13(6):499–504. doi: 10.1097/GIM.0b013e318220aaba. [DOI] [PubMed] [Google Scholar]
- 7.Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med. 2009;11(1):35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: genomic profiling to assess cardiovascular risk to improve cardiovascular health. Genet Med. 2010;12(12):839–843. doi: 10.1097/GIM.0b013e3181f872c0. [DOI] [PubMed] [Google Scholar]
- 9.Khoury MJ, McCabe LL, McCabe ERB. Population screening in the age of genomic medicine. N Engl J Med. 2003;348(1):50–58. doi: 10.1056/NEJMra013182. [DOI] [PubMed] [Google Scholar]
- 10.Roberts NJ, Vogelstein JT, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. The predictive capacity of personal genome sequencing. Sci Transl Med. 2012;4(133) doi: 10.1126/scitranslmed.3003380. 133ra58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kathiresan S, Melander O, Anevski D et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358(12):1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 12.Meigs JB, Shrader P, Sullivan LM et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359(21):2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyssenko V, Jonsson A, Almgren P et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359(21):2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 14.Paynter NP, Chasman DI, Pare G et al. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA. 2010;303(7):631–637. doi: 10.1001/jama.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talmud PJ, Hingorani AD, Cooper JA et al. Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. BMJ. 2010;340(1) doi: 10.1136/bmj.b4838. b4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ripatti S, Tikkanen E, Orho-Melander M et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376(9750):1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363(2):166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 18.Helfand M, Buckley DI, Freeman M et al. Emerging risk factors for coronary heart disease: a summary of systematic reviews conducted for the US Preventive Services Task Force. Ann Intern Med. 2009;151(7):496–507. doi: 10.7326/0003-4819-151-7-200910060-00010. [DOI] [PubMed] [Google Scholar]
- 19.McCormack M, Alfirevic A, Bourgeois S et al. HLA-A*3101 and carbamazepine-induced hypersensitivity reactions in Europeans. N Engl J Med. 2011;364(12):1134–1143. doi: 10.1056/NEJMoa1013297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auffray C, Caulfield T, Khoury MJ, Lupski JR, Schwab M, Veenstra T. Looking back at genomic medicine in 2011. Genome Med. 2012;4(1):9. doi: 10.1186/gm308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Human Genome Research Institute. eMERGE project. Available at: http://www.genome.gov/27540473. Accessed July 25, 2013.
- 22.Permanente Kaiser. Research program on genes, environment, and health. Available at: http://www.dor.kaiser.org/external/DORExternal/rpgeh/index.aspx. Accessed July 25, 2013.
- 23.Yoon PW, Scheuner MT, Peterson-Oehlke KL, Gwinn M, Faucett A, Khoury MJ. Can family history be used as a tool for public health and preventive medicine? Genet Med. 2002;4(4):304–310. doi: 10.1097/00125817-200207000-00009. [DOI] [PubMed] [Google Scholar]
- 24.Olney RS, Yoon PW. Role of family medical history information in pediatric primary care and public health: introduction. Pediatrics. 2007;120(suppl 2):S57–S59. doi: 10.1542/peds.2007-1010C. [DOI] [PubMed] [Google Scholar]
- 25.Morrison AC, Bare LA, Chambless LE et al. Prediction of coronary heart disease risk using a genetic risk score: the Atherosclerosis Risk in Communities study. Am J Epidemiol. 2007;166(1):28–35. doi: 10.1093/aje/kwm060. [DOI] [PubMed] [Google Scholar]
- 26.Plomin R, Haworth CMA, Davis OSP. Common disorders are quantitative traits. Nat Rev Genet. 2009;10(12):872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 27.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17(10):1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belsky DW, Moffitt TE, Houts R et al. Polygenic risk, rapid childhood growth, and the development of obesity: evidence from a 4-decade longitudinal study. Arch Pediatr Adolesc Med. 2012;166(6):515–521. doi: 10.1001/archpediatrics.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paynter N, Chasman DI, Pare G et al. Prospective evaluation of a comprehensive genetic risk score for future cardiovascular events among 19,313 initially healthy women: the Women’s Genome Health Study. Circulation. 2009;120(18):S607. [Google Scholar]
- 30.Yang J, Manolio TA, Pasquale LR et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43(6):519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang WJ, Kelly T, He J. Genetic epidemiology of obesity. Epidemiol Rev. 2007;29(1):49–61. doi: 10.1093/epirev/mxm004. [DOI] [PubMed] [Google Scholar]
- 32.Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6(7):521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- 33.Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336(6082):740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2012;13(2):135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era–concepts and misconceptions. Nat Rev Genet. 2008;9(4):255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 36.Belsky DW, Moffitt TE, Baker TB et al. Polygenic risk accelerates the developmental progression to heavy, persistent smoking and nicotine dependence: evidence from a 4-decade longitudinal study. JAMA Psychiatry. 2013;70(5):534–542. doi: 10.1001/jamapsychiatry.2013.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purcell SM, Wray NR, Stone JL et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wald NJ, Hackshaw AK, Frost CD. When can a risk factor be used as a worthwhile screening test? BMJ. 1999;319(7224):1562–1565. doi: 10.1136/bmj.319.7224.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. Am J Epidemiol. 2004;159(9):882–890. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- 40.Rockhill B. Theorizing about causes at the individual level while estimating effects at the population level: implications for prevention. Epidemiology. 2005;16(1):124–129. doi: 10.1097/01.ede.0000147111.46244.41. [DOI] [PubMed] [Google Scholar]
- 41.Huang Y, Pepe MS, Feng Z. Evaluating the predictiveness of a continuous marker. Biometrics. 2007;63(4):1181–1188. doi: 10.1111/j.1541-0420.2007.00814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pencina MJ, D’Agostino RB, Sr, D’Agostino RB, Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat. Med. 2008;27(2):157–172. doi: 10.1002/sim.2929. discussion 207–212. [DOI] [PubMed] [Google Scholar]
- 43.Belsky DW, Moffitt TE, Sugden K et al. Development and evaluation of a genetic risk score for obesity. Biodemogr Social Biol. 2013;59(1):87–104. doi: 10.1080/19485565.2013.774628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hindorff LA, Sethupathy P, Junkins HA et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frayling TM, Timpson NJ, Weedon MN et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plomin R, McGuffin P. Psychopathology in the postgenomic era. Annu Rev Psychol. 2003;54(1):205–228. doi: 10.1146/annurev.psych.54.101601.145108. [DOI] [PubMed] [Google Scholar]
- 47.Chen R, Mias GI, Li-Pook-Than J et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148(6):1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weissman MM, Brown AS, Talati A. Translational epidemiology in psychiatry: linking population to clinical and basic sciences. Arch Gen Psychiatry. 2011;68(6):600–608. doi: 10.1001/archgenpsychiatry.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11(6):415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 50.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12(5):363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ioannidis JP, Khoury MJ. Improving validation practices in “omics” research. Science. 2011;334(6060):1230–1232. doi: 10.1126/science.1211811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hindorff LA, Junkins HA, Mehta JP, Manolio TA. A catalog of published genome-wide association studies. Available at: http://www.genome.gov/gwastudies. Accessed April 30, 2010.
- 53.Gelernter J. Developmental perspective on the role of genes in smoking risk. Biol Psychiatry. 2011;69(7):616–617. doi: 10.1016/j.biopsych.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ben-Shlomo Y, Kuh D. A life course approach to chronic disease epidemiology: conceptual models, empirical challenges and interdisciplinary perspectives. Int J Epidemiol. 2002;31(2):285–293. [PubMed] [Google Scholar]
- 55.Ben-Shlomo Y. Rising to the challenges and opportunities of life course epidemiology. Int J Epidemiol. 2007;36(3):481–483. doi: 10.1093/ije/dym116. [DOI] [PubMed] [Google Scholar]
- 56.Nichols LM, Masdeu JC, Mattay VS et al. Interactive effect of apolipoproteine genotype and age on hippocampal activation during memory processing in healthy adults. Arch Gen Psychiatry. 2012;69(8):804–813. doi: 10.1001/archgenpsychiatry.2011.1893. [DOI] [PubMed] [Google Scholar]
- 57.Hartz SM, Short SE, Saccone NL et al. Increased genetic vulnerability to smoking at CHRNA5 in early-onset smokers. Arch Gen Psychiatry. 2012;69(8):854–860. doi: 10.1001/archgenpsychiatry.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss RB, Baker TB, Cannon DS et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4(7) doi: 10.1371/journal.pgen.1000125. e1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev. 2005;85(2):571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- 60.Frongillo EA, Lampl M. Early identification of children at risk of developing obesity. Arch Pediatr Adolesc Med. 2011;165(11):1043–1044. doi: 10.1001/archpediatrics.2011.193. [DOI] [PubMed] [Google Scholar]
- 61.Jackson KM, Sher KJ, Rose RJ, Kapiro J. Trajectories of Tobacco Use from Adolescence to Adulthood: Are the Most Informative Phenotypes Tobacco Specific? Bethesda, MD: National Cancer Institute; 2009. [Google Scholar]
- 62.Bierut LJ. Genetic vulnerability and susceptibility to substance dependence. Neuron. 2011;69(4):618–627. doi: 10.1016/j.neuron.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muennig P, Robertson D, Johnson G, Campbell F, Pungello EP, Neidell M. The effect of an early education program on adult health: the Carolina Abecedarian Project randomized controlled trial. Am J Public Health. 2011;101(3):512–516. doi: 10.2105/AJPH.2010.200063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campbell FA, Pungello EP, Burchinal M et al. Adult outcomes as a function of an early childhood educational program: an Abecedarian Project follow-up. Dev Psychol. 2012;48(4):1033–1043. doi: 10.1037/a0026644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Belfield CR, Nores M, Barnett S, Schweinhart L. The high/scope Perry preschool program: cost-benefit analysis using data from the age-40 followup. J Hum Resour. 2006;41(1):162–190. [Google Scholar]
- 66.Heckman JJ, Moon SH, Pinto R, Savelyev PA, Yavitz A. The rate of return to the HighScope Perry Preschool Program. J Public Econ. 2010;94(1-2):114–128. doi: 10.1016/j.jpubeco.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karelis AD. Metabolically healthy but obese individuals. Lancet. 2008;372(9646):1281–1283. doi: 10.1016/S0140-6736(08)61531-7. [DOI] [PubMed] [Google Scholar]
- 68.Stefan N, Kantartzis K, Machann J et al. Identification and characterization of metabolically benign obesity in humans. Arch Intern Med. 2008;168(15):1609–1616. doi: 10.1001/archinte.168.15.1609. [DOI] [PubMed] [Google Scholar]
- 69.McBride CM, Bryan AD, Bray MS, Swan GE, Green ED. Health behavior change: can genomics improve behavioral adherence? Am J Public Health. 2012;102(3):401–405. doi: 10.2105/AJPH.2011.300513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bryan AD, Hutchison KE. The role of genomics in health behavior change: challenges and opportunities. Public Health Genomics. 2012;15(3-4):139–145. doi: 10.1159/000335226. [DOI] [PubMed] [Google Scholar]
- 71.Boffetta P, Winn DM, Ioannidis JP et al. Recommendations and proposed guidelines for assessing the cumulative evidence on joint effects of genes and environments on cancer occurrence in humans. Int J Epidemiol. 2012;41(3):686–704. doi: 10.1093/ije/dys010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Link BG, Phelan J. Social conditions as fundamental causes of disease. J Health Soc Behav. 1995;35(extra issue):80–94. [PubMed] [Google Scholar]
- 73.Bambra C, Gibson M, Sowden A, Wright K, Whitehead M, Petticrew M. Tackling the wider social determinants of health and health inequalities: evidence from systematic reviews. J Epidemiol Community Health. 2010;64(4):284–291. doi: 10.1136/jech.2008.082743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marmot M. Social determinants of health inequalities. Lancet. 2005;365(9464):1099–1104. doi: 10.1016/S0140-6736(05)71146-6. [DOI] [PubMed] [Google Scholar]
- 75.Bloom DE, Canning D. Policy forum: public health. The health and wealth of nations. Science. 2000;287(5456):1207–1209. doi: 10.1126/science.287.5456.1207. [DOI] [PubMed] [Google Scholar]
- 76.Shanahan MJ, Hofer SM. Social context in gene-environment interactions: retrospect and prospect. J Gerontol B Psychol Sci Soc Sci. 2005;60(special issue 1):65–76. doi: 10.1093/geronb/60.special_issue_1.65. [DOI] [PubMed] [Google Scholar]
- 77.Boardman JD, Blalock CL, Pampel FC, Hatemi PK, Heath AC, Eaves LJ. Population composition, public policy, and the genetics of smoking. Demography. 2011;48(4):1517–1533. doi: 10.1007/s13524-011-0057-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barbeau EM, Krieger N, Soobader MJ. Working class matters: socioeconomic disadvantage, race/ethnicity, gender, and smoking in NHIS 2000. Am J Public Health. 2004;94(2):269–278. doi: 10.2105/ajph.94.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Drewnowski A. Obesity, diets, and social inequalities. Nutr Rev. 2009;67(suppl 1):S36–S39. doi: 10.1111/j.1753-4887.2009.00157.x. [DOI] [PubMed] [Google Scholar]
- 80.Poulton R, Caspi A, Milne BJ et al. Association between children’s experience of socioeconomic disadvantage and adult health: a life-course study. Lancet. 2002;360(9346):1640–1645. doi: 10.1016/S0140-6736(02)11602-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li S, Zhao JH, Luan J et al. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study. PLoS Med. 2010;7(8) doi: 10.1371/journal.pmed.1000332. e1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qi Q, Chu AY, Kang JH et al. Sugar-sweetened beverages and genetic risk of obesity. N Engl J Med. 2012;367(15):1387–1396. doi: 10.1056/NEJMoa1203039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Johnson EO, Chen LS, Breslau N et al. Peer smoking and the nicotinic receptor genes: an examination of genetic and environmental risks for nicotine dependence. Addiction. 2010;105(11):2014–2022. doi: 10.1111/j.1360-0443.2010.03074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xie P, Kranzler HR, Zhang H et al. Childhood adversity increases risk for nicotine dependence and interacts with α5 nicotinic acetylcholine receptor genotype specifically in males. Neuropsychopharmacology. 2012;37(3):669–676. doi: 10.1038/npp.2011.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Caspi A, McClay J, Moffitt TE et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297(5582):851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- 86.Thomas D. Gene-environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11(4):259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Khoury MJ, Wacholder S. Invited commentary: from genome-wide association studies to gene-environment-wide interaction studies–challenges and opportunities. Am J Epidemiol. 2009;169(2):227–230. doi: 10.1093/aje/kwn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cornelis MC, Tchetgen EJ, Liang L et al. Gene-environment interactions in genome-wide association studies: a comparative study of tests applied to empirical studies of type 2 diabetes. Am J Epidemiol. 2012;175(3):191–202. doi: 10.1093/aje/kwr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moffitt TE, Caspi A, Rutter M. Strategy for investigating interactions between measured genes and measured environments. Arch Gen Psychiatry. 2005;62(5):473–481. doi: 10.1001/archpsyc.62.5.473. [DOI] [PubMed] [Google Scholar]
- 90.Kilpeläinen TO, Qi L, Brage S et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8(11) doi: 10.1371/journal.pmed.1001116. e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van Ijzendoorn MH, Bakermans-Kranenburg MJ, Belsky J et al. Gene-by-environment experiments: a new approach to finding the missing heritability. Nat Rev Genet. 2011;12(12):881. doi: 10.1038/nrg2764-c1. [DOI] [PubMed] [Google Scholar]
- 92.Patel SR. Reduced sleep as an obesity risk factor. Obes Rev. 2009;10(suppl 2):61–68. doi: 10.1111/j.1467-789X.2009.00664.x. [DOI] [PubMed] [Google Scholar]
- 93.Prats-Puig A, Grau-Cabrera P, Riera-Perez E et al. Variations in the obesity genes FTO, TMEM18 and NRXN3 influence the vulnerability of children to weight gain induced by short sleep duration. Int J Obes (Lond) 2013;37(2):182–187. doi: 10.1038/ijo.2012.27. [DOI] [PubMed] [Google Scholar]
- 94.Rose JE, Behm FM, Drgon T, Johnson C, Uhl GR. Personalized smoking cessation: interactions between nicotine dose, dependence and quit-success genotype score. Mol Med. 2010;16(7-8):247–253. doi: 10.2119/molmed.2009.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.King DP, Paciga S, Pickering E et al. Smoking cessation pharmacogenetics: analysis of varenicline and bupropion in placebo-controlled clinical trials. Neuropsychopharmacology. 2012;37(3):641–650. doi: 10.1038/npp.2011.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen LS, Baker TB, Piper ME et al. Interplay of genetic risk factors (CHRNA5-CHRNA3-CHRNB4) and cessation treatments in smoking cessation success. Am J Psychiatry. 2012;169(7):735–742. doi: 10.1176/appi.ajp.2012.11101545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Evans JP, Burke W, Khoury M. The rules remain the same for genomic medicine: the case against “reverse genetic exceptionalism.”. Genet Med. 2010;12(6):342–343. doi: 10.1097/GIM.0b013e3181deb308. [DOI] [PubMed] [Google Scholar]
- 98.Evans JP, Meslin EM, Marteau TM, Caulfield Genomics T. Deflating the genomic bubble. Science. 2011;331(6019):861–862. doi: 10.1126/science.1198039. [DOI] [PubMed] [Google Scholar]
- 99.Sullivan P. Don’t give up on GWAS. Mol Psychiatry. 2012;17(1):2–3. doi: 10.1038/mp.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]