Abstract
Objectives. We examined depression within a multidimensional framework consisting of genetic, environmental, and sociobehavioral factors and, using machine learning algorithms, explored interactions among these factors that might better explain the etiology of depressive symptoms.
Methods. We measured current depressive symptoms using the Center for Epidemiologic Studies Depression Scale (n = 6378 participants in the Wisconsin Longitudinal Study). Genetic factors were 78 single nucleotide polymorphisms (SNPs); environmental factors—13 stressful life events (SLEs), plus a composite proportion of SLEs index; and sociobehavioral factors—18 personality, intelligence, and other health or behavioral measures. We performed traditional SNP associations via logistic regression likelihood ratio testing and explored interactions with support vector machines and Bayesian networks.
Results. After correction for multiple testing, we found no significant single genotypic associations with depressive symptoms. Machine learning algorithms showed no evidence of interactions. Naïve Bayes produced the best models in both subsets and included only environmental and sociobehavioral factors.
Conclusions. We found no single or interactive associations with genetic factors and depressive symptoms. Various environmental and sociobehavioral factors were more predictive of depressive symptoms, yet their impacts were independent of one another. A genome-wide analysis of genetic alterations using machine learning methodologies will provide a framework for identifying genetic–environmental–sociobehavioral interactions in depressive symptoms.
Depression is a widespread mental disorder associated with a host of undesirable health, social, and economic outcomes. One in 6 Americans is diagnosed with depression in his or her lifetime.1 The etiology of depression is complex and heterogeneous2; although many environmental factors such as prolonged stress and traumatic life events have important ties with depression, so too do gender and many genetic and epigenetic factors. Recent research has focused largely on the ties between genetics and depression. Twin studies have estimated that genetic factors typically account for 37% of the risk for developing major depression.3
Most research into the genetics of depression has focused on single nucleotide polymorphisms (SNPs) of common variants and variable-number tandem repeat regions. Recent advances in genotyping have produced numerous high-throughput studies that have assessed SNPs on the genome-wide level, making them commonly known as genomewide association studies (GWAS). Despite the rise in genotyping capacity, only 1 variant—SLC6A15 (neutral amino acid transporter) rs1545843—of a total of 8 separate GWAS on depression has been found to associate with depression at the level of genome-wide significance.4,5 In a recent mega-analysis of GWAS data5—the largest genetic study of depression to date—no SNPs reached the level of genome-wide significance. Because of the relatively higher prevalence but lower heritability of depression, extraordinarily large sample sizes (n = 100 000) may be necessary to detect SNPs at genome-wide significant levels.5
Besides the recent trend toward GWAS, the majority of depression genetics work has relied on candidate gene studies. A 2008 article by Lopez-Leon et al.6 reviewed all prior candidate gene articles covering 393 polymorphisms in 102 different genes, and meta-analyses were carried out on 22 of these polymorphisms. Of the 22 variants examined in meta-analyses, 6 showed significant associations with depression: apolipoprotein E (ApoE) e2 (odds ratio [OR] = 0.51; 95% confidence interval [CI] = 0.27, 0.97), guanine nucleotide-binding protein (GNB3) rs5443 (C825T; OR = 1.38; 95% CI = 1.13, 1.69), methylenetetrahydrofolate reductase (MTHFR) rs1801133 (C677T; OR = 1.2), dopamine transporter (DAT1) 40–base pair variable-number tandem repeat (OR = 2.06; 95% CI = 1.25, 3.40), serotonin transporter–linked polymorphic region (5-HTTLPR; OR = 1.11; 95% CI = 1.04, 1.19), and dopamine receptor D4 (DRD4) 48–base pair variable-number tandem repeat (OR = 1.73; 95% CI = 1.29, 2.32).6 On the basis of these results and the results from GWAS, one sees that single main-factor SNP associations contribute only a small effect size that does not explain all of the recognized genetic contributions to depression.
Given the lack of predictive power among individual genetic alterations together with the heterogeneous etiology of depression, we and others have begun exploring gene–gene (G×G) interactions in depression.7 Other studies have reported epistatic effects with depression among glycogen synthase kinase 3β rs6782799, brain-derived neurotrophic factor (BDNF) rs7124442, and BDNF Val66Met (rs6265)8; between BDNF Val66Met (rs6265) and 4 SNPs in neurotrophic tyrosine kinase receptor 29; between serotonin-transporter-linked polymorphic region (5-HTTLPR) short allele and a chromosome 4 gene10; between 5-HTTLPR short allele and solute carrier family 6, member 4 (SLC6A4) rs14070011; and between serotonin receptor 1A (HTR1A) rs6295 (C[-1019]G) and SNPs in phospholysine phosphohistidine inorganic pyrophosphate phosphatase.12
A way to further model the heterogeneity of depression is to explore gene–environment (G×E) interactions. Caspi et al.13 first reported on the interaction between the 5-HTTLPR short allele and early life stress and depression in 2003. Since then, numerous other studies (at least 148 publications14) have examined the same interaction, and some have replicated the findings, others did not, and some even found opposite trends (see Table 1 in Karg et al.14). Meta-analysis of 54 of these studies found strong evidence supporting the existence of the interaction.14 Other recent studies have examined different polymorphisms for interactions with early life stress in depression. Bradley et al.15 and others16,17 have shown interactions between multiple SNPs in corticotropin-releasing hormone type 1 receptor and early life stress for depression. Further G×E interactions involving genetic alterations in stress hormone–related genes are reviewed in Heim and Binder,18 including the genes FK506 binding protein 5,19,20 glucocorticoid receptor,21 oxytocin receptor,22 serotonin receptor 3A,23 and dopamine receptor D2 (DRD2).24 In addition, Heim and Binder reviewed studies examining higher order G×G×E interactions associated with depression.25–28 Kim et al. and Wichers et al. also reported G×G×E interactions with depression.29,30
TABLE 1—
Comparison of Machine Learning Algorithm AUCs for G×E Data and G×E×SB Data: Wisconsin Longitudinal Study, 1957–2005
| G×E Data |
G×E×SB Data |
|||
| Model | No. | AUC ROC (95% CI) | No. | AUC ROC (95% CI) |
| Linear SVM | 0.497 (0.475, 0.520) | 0.539 (0.513, 0.564) | ||
| Gaussian SVM | 0.537 (0.516, 0.557) | 0.624 (0.600, 0.649) | ||
| Naïve Bayes | 3 | 0.568 (0.546, 0.589)a | 13 | 0.691 (0.671, 0.712)b |
| TAN Bayes | 3 | 0.561 (0.546, 0.576) | 11 | 0.673 (0.651, 0.695) |
Note. AUC = area under curve; CI = confidence interval; E = environmental; G = genetic; ROC = receiver operating characteristic; SB = sociobehavioral; SVM = support vector machine; TAN = tree-augmented naïve.
Top-performing G×E model.
Top-performing model.
With its rich covariate composition and longitudinal nature, the Wisconsin Longitudinal Study (WLS; http://www.ssc.wisc.edu/wlsresearch) is ideal for examining interactive correlates associated with a heterogeneous disorder such as depression. Relevant WLS covariates include genetic factors (78 SNPs), environmental factors (stressful life events [SLEs]), and a multitude of sociobehavioral factors. Previously, we examined G×G interactions associated with self-reported lifetime depression (i.e., depression experienced at any stage of life).7 Because the human genetic code is thought to be constant over time, lifetime depression was an appropriate measure for studying genetic associations. However, the time dependence of environmental variables requires them to be considered in a contributory-to-depression context. That is to say, environmental variables must precede the onset of depressive symptoms. For this reason, in this study we chose to examine current depression (as assessed with the Center for Epidemiologic Studies Depression Scale) as our outcome measure so that previous life events would act in a contributory fashion to depression.
Given that integrated G×E risk factor studies of complex diseases tend to produce data sets containing many covariates relative to smaller sample sizes, traditional multiple regression methods are not optimal for seeking out interactions.31 Machine learning algorithms are a useful alternative; we have previously explored G×G interactions using the tree-based algorithm called recursive partitioning.7 In this study, we performed analyses using 2 different machine learning algorithms—support vector machines (SVMs) and Bayesian networks—that we chose because of their ability to detect interactions among factors in large, complex data sets. SVMs have previously been used to search for G×G interactions in breast cancer,32 chronic fatigue syndrome,33 and glaucoma,34 and Bayesian networks have been used to search for G×G interactions in stroke,35 Alzheimer’s disease,36 chronic fatigue syndrome,33 and asthma and eczema.37
Our goals were (1) to explore genetic, environmental, and sociobehavioral interactions that might better explain the etiology of depressive symptoms and (2) to determine the utility of SVMs and Bayesian networks to identify these interactions. Preliminary testing found no single SNPs to associate with depressive symptoms after correcting for multiple testing. Naïve Bayes—a linear form of Bayesian network that assumes risk factors contribute independently to depressive symptom status—was our top-performing model, indicating that there were no interactions of notable magnitude or predictability. When considering only genetic and environmental factors, our model consisted of gender and 2 environmental factors: Being female, experiencing spousal physical abuse, and proportion of stressful life events experienced were all associated with higher odds for depressive symptoms. Secondary analyses showed that adding various sociobehavioral traits (personality, IQ, years of educational attainment, marriage, and body mass index) to our models significantly improved the prediction of depressive symptoms. Nevertheless, we again did not find evidence for interactions.
METHODS
We collected data from the WLS, a random sample originally consisting of 10 317 men and women who graduated from Wisconsin high schools in 1957. Later, in 1977, the WLS also began interviewing 1 randomly selected sibling of each graduate, when possible. The cohort consists almost entirely of non-Hispanic White people whose average level of educational attainment was 1.5 years of post–high- school education at the time of interview in 2004–2005. Participants’ ages ranged from 35 to 90 years, and 83% of participants were aged between 60 and 70 years. Additional characteristics of the WLS cohort may be found in detail elsewhere.38 Our main measure of depression, assessed in 2004–2005, was based on the Center for Epidemiologic Studies Depression Scale. The scale is a 20-question self-report of depression symptomatology, with questions having the form “On how many days during the past week did you feel/think… ?” We converted scores to match the traditional scale, on which 0 days was scored 0, 1 to 2 days was scored 1, 3 to 4 days was scored 2, and 5 to 7 days was scored 3 (scoring of positive items was reversed). Depressive symptoms were defined as being present in those participants having a score of 16 or higher.39
Environmental Variables
Environmental variables consisted of 13 potentially SLEs, reported in 2004–2005. Participants were asked whether they had ever experienced any of the following: death of a close friend, parental drug abuse, sibling physical abuse, life-threatening natural disaster, serving in war, witnessing severe injury or death, spousal (or partner) physical abuse, divorce of children, life-threatening illness or accident of child, adult child moving back into home, increased responsibility for care of grandchildren, aging parent or in-law moving into home, and placing an aging spouse, in-law, or parent in a nursing home. Last, we created an index for the proportion of SLEs experienced (proportion of SLEs index), ranging from 0 to 1 (0 = “experienced no events among those answered,” 1 = “experienced all events among those answered”).
Sociobehavioral Variables
In our secondary analysis, we also examined an additional 18 sociobehavioral variables. We obtained IQ, measured during junior year of high school with the Henmon-Nelson Test of Mental Ability,40 and rank in high school class from Wisconsin high schools’ administrative records. We rescaled IQ to the conventional metric with a mean of 100 and a standard deviation of 15. We also ascertained total years of educational attainment in 2004–2005 (< 12 = did not graduate high school, 12 = high school graduate, 16 = college graduate, > 16 = higher degree, etc.).
We took data on marital status—married and not married (including separated, divorced, widowed, or never married)—from the 1992–1993 survey and number of biological children and whether a participant had a deceased child from the 2004–2005 survey.
We collected personality measures from the 1992–1993 survey using a 5-factor model of personality, which includes extraversion, openness, neuroticism, conscientiousness, and agreeableness.41 We used a 29-item subset of the 54-item Big Five Inventory for our 5-factor model (5 questions for neuroticism, 6 questions for each of the other 4 personality measures).42 Questions were scored on a 6-point scale (1 = agree strongly, 6 = disagree strongly) and summed (we reverse-scored items where appropriate).
We determined body mass index from self-reported height and weight information from the 1992–1993 survey. Heart attack, diabetes, cancer, and stroke information came from the 2004–2005 survey, on which we asked, “Has a doctor ever told you that you had/have… ?” For women, we also examined 2 menopausal-related measures—years of life since last menstruation and hormone replacement therapy ever-users—measured in 2004–2005.
Genotype Variables
The 7101 participants (4569 graduates and 2532 siblings) provided saliva samples in Oragene DNA sample collection kits from which DNA was extracted and genotyped for 78 SNPs.7 Previous depression studies have examined 15 of these SNPs: serotonin transporter (SLC6A4) rs2553343 and rs8076005,44 HTR1A rs878567,45 HTR2A rs631446 and rs7997012,47 HTR2C rs6318,48 catechol-O-methyltransferase (COMT) rs4680,49 cholinergic receptor, muscarinic 2 (CHRM2) rs206117450 and rs8191992,51 DRD2 rs180049752 and rs6277,53 BDNF rs626554,55 and rs908867,56 and APOE ɛ4 rs429358 and rs7412.57 We also examined 11 additional candidate SNPs that are located within genes thought to be associated with depression58 but that to our knowledge have not previously been assessed. We selected the 52 remaining SNPs on the basis of their candidacy for associating with various age-related conditions and diseases. Genotyping was performed by KBioscience (Hoddesdon, UK) using a homogeneous fluorescent resonance energy transfer technology coupled to competitive allele-specific polymerase chain reaction. All SNP genotypes described in our results were in Hardy-Weinberg equilibrium, and their frequencies matched those reported in the literature for European samples. We excluded from our analyses participants who were missing more than 10% genotype data (133 individuals).
SNP Analysis
We performed single SNP analysis using logistic regression likelihood ratio tests (df = 2). Each SNP was represented as 2 indicator variables, v1 and v2, with homozygous major allele carriers being the reference level. Specifically, participants homozygous for the major allele were coded 0 0, heterozygous carriers were coded 1 0, and minor allele homozygous carriers were coded 0 1. Note that our indicator variables do not represent the number of minor alleles. To account for multiple testing, we recorded likelihood ratio test P values and then used them to calculate q values.59 For subsequent multifactorial machine learning analyses, we coded SNPs in genotype form as 3 nominal values.
Machine Learning Algorithms
We implemented analysis via multiple classification algorithms, using the freely available data-mining software Weka, version 3.6.6 (Machine Learning Group, University of Waikato, Hamilton, New Zealand).60 For each algorithm, we used 10-fold cross-validation to estimate prediction error. A fold is created by modeling only 90% of the data (the training data) and then running the remaining 10% of data (the test data) through the training model. Independent fold runs are performed 10 times, such that each data point is used exactly once as a test case; misclassifications are recorded for each fold (resulting in confidence values) and pooled to give overall prediction of error rates for the final model.
We evaluated the performance of the different classification techniques using receiver operating characteristic (ROC) curves. On the basis of positive (having depressive symptoms) and negative (not having depressive symptoms) outcome classes, we defined true positive rate (also known as sensitivity or recall) as the proportion of actual positive cases that are correctly predicted as positive and false positive rate (also known as 1 − specificity) as the proportion of actual negative cases that are incorrectly predicted as positive. A given classifier will predict cases as positive if the confidence (test value) is above a given threshold. By varying the threshold, the true and false positive rates change in value, resulting in ROC points that can be linearly connected into a curve. ROC curves are displayed by plotting the true positive rate on the vertical axis and the false positive rate on the horizontal axis. We used area under curve (AUC), defined as the area between the ROC curve and the horizontal axis, as a measure for classification model comparison. To assess statistical significance when comparing 2 classification models by AUC, we used a 2-sided paired-samples t test. The paired-samples values used in the test are the AUC values computed on the 10 cross-validation test sets for each model.
Support vector machines.
SVMs61 belong to the family of generalized linear models. We used SVMs with both linear and Gaussian kernels. The linear version is essentially an (n − 1)-dimensional hyperplane that separates the instances of the 2 classes (i.e., depressive symptoms and non-depressive symptoms) in the n-dimensional feature space. The hyperplane maximizes the margin with the closest training instances, with the goal of lowering the model’s generalization error. The closest training instances become known as the support vectors because they are responsible for fixing the position and orientation of the hyperplane. When the training data are not linearly separable, SVMs rely on soft margins,62 which seek to minimize the sum of the distances to the training instances that are incorrectly classified by the hyperplane in addition to maximizing the margin.
A Gaussian (or radial base function) SVM follows the same idea as the linear SVM, except a nonlinear curve is used to separate classes. A nonlinear separator has the potential to increase predictability, especially when interactions are present in the data. We used the radial basis function to transform our data into a higher dimensional space, where it was then possible to apply a hyperplane separator as before. Thus, we create a hypersurface in a higher dimensional space than our model, but the model can be used to classify without needing to characterize the space. A disadvantage of using Gaussian SVMs is comprehensibility—it can be difficult to understand the model because it classifies participants as a function of their similarities to other participants, and the learning algorithm assigns weights to the other participants. Linear SVMs do not share this disadvantage.
In Weka, we selected CVParameterSelection as our classifier, with the SMO function running the PolyKernel for our linear SVM. For the Gaussian SVM, we used the RBFKernel. Using CVParameterSelection tunes the relevant SVM parameters in a methodologically sound manner. Specifically, on each fold of cross-validation for evaluation, CVParameterSelection uses an internal cross-validation run, using only the training data for the current evaluation fold, to find the optimal parameter settings on the basis of the training data alone, not the test data. All other parameters were set to default, except we chose to fit logistic models to the outputs.
Bayesian networks.
In general, a Bayesian network treats features in the data as random variables and represents them as nodes in a directed acyclic graph. If the Bayesian network has a directed arc (represented as a line with an arrow) from a node A to another node B, then A is said to be a “parent” of B. Each node can take 1 of multiple continuous or nominal values (“states”), and the Bayesian network associates a probability distribution over the possible states a node can take that is conditional on the states of the parent nodes. For instance, the depression node has 2 states that represent the outcome of interest (depressive symptoms or not); if the Bayesian network has a tree structure, and this node is the root (no incoming arcs), then it will store the prior probability of these states (the prevalence of depressive symptoms). The remaining nodes in the model represent the attributes (risk factors). Directed arcs in the network encode dependence relationships among variables. Although in the past investigators have typically used preexisting knowledge about the probabilistic relationships among variables to build a Bayesian network, we learned the structure and probabilities from our existing data set.63
The first Bayesian network we used is known as naïve Bayes,64 in which each attribute node is assigned exactly 1 parent node, which must be the root node. Thus, naïve Bayes assumes (perhaps “naively”) that all risk factors are conditionally independent given the outcome of interest (the class); intuitively, this means that all risk factors in the model contribute independently to the probability of the outcome of interest. This assumption reduces the computational cost involved with classifying and in practice often works surprisingly well. Naïve Bayes predicts a class (depressive symptoms or not) with the rule 
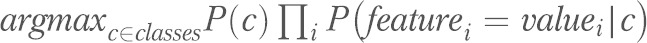 . Note that
. Note that 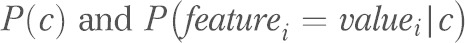 are learned from the training data. Any non-nominal (continuous) features were discretized before running naïve Bayes.
are learned from the training data. Any non-nominal (continuous) features were discretized before running naïve Bayes.
Our second Bayesian network, tree-augmented naïve (TAN) Bayes,65 shares a similar structure to naïve Bayes in that the root node serves as a parent to each attribute node. However, TAN also allows each attribute to have at most 1 other parent node, as long as no directed cycles are created. This keeps TAN a directed acyclic graph (as required for a Bayesian network). In other words, TAN allows for some dependencies between attributes. To decide which attribute-to-attribute arcs to include in the augmented network, the algorithm initially makes a complete graph between all attributes (besides depressive symptoms), in which the weight of each edge is given as the conditional mutual information between those 2 attributes. A maximum-weight spanning tree is constructed over this graph, and the edges that appear in the spanning tree are added to the network.
In Weka, we selected Bayesian networks under classifier AttributeSelectedClassifier. We used the information gain, or mutual information, of attributes with the class and the Ranker method to identify the top attributes, and we selected final models on the basis of the largest ROC area. Analogously to CVParameterSelection tuning with SVMs, when combing feature selection with cross-validation for algorithm evaluation, we were careful to repeat feature selection on every fold of cross-validation, using only the training data for that fold. This avoids the overly optimistic accuracy estimates that arise from a single feature selection run on the entire data set.
We explored the use of other Weka classifiers including J48 and CART decision trees as well as random forests. None of these methods performed nearly as well as SVMs or Bayesian networks and so were excluded.
RESULTS
Among those 6968 participants with less than 10% missing genotype data, we found survey information on depressive symptoms for 6378 (3047 men and 3331 women). We identified 936 participants (558 women and 372 men) with depressive symptoms. Initial single SNP logistic regression likelihood ratio tests identified 9 SNPs associated with depressive symptoms below the P = .05 level, but after correcting for multiple testing, none remained significant. Broken down by gender, we found 8 SNPs in women and 5 SNPs in men that reached the .05 level, but again none remained significant after correcting for multiple testing.
To determine the presence of G×G and G×E interactions, we initially compared the performance of linear and Gaussian SVMs and then selected a final model after comparing SVMs with naïve and TAN Bayes. See Table 1 for performance of our machine learning models. The Gaussian SVM outperformed the linear SVM, but the difference was not significant, suggesting that there were likely no interactions present in our data. Naïve and TAN Bayes both significantly outperformed the linear SVM, but they did not outperform one another or the Gaussian SVM. We chose naïve Bayes (attribute size n = 3) as our final model in the interest of parsimony and because it showed the largest AUC ROC. The model contained only environmental risk factors (partner physical abuse and proportion of SLEs index) and gender. See Table 2 for odds ratios for the attributes of this model, listed in rank order. Experiencing physical abuse by a spouse or partner was associated with increased odds of 112%. Overall, each additional SLE experienced was associated with a 9% increase in odds for depressive symptoms. Being female was associated with a 45% increase in odds for depressive symptoms.
TABLE 2—
Odds of Depressive Symptoms for Attributes from Final Model for G×E Data: Wisconsin Longitudinal Study, 1957–2005
| Attribute | Year Measured | Value | LR OR (95% CI) |
| Spouse or partner physical abuse | 2004–2005 | Yes–no | 2.12 (1.70, 2.63) |
| Proportion of SLEs index | 2004–2005 | Continuous | 3.06 (1.93, 4.86) |
| Female | … | Yes–no | 1.45 (1.26, 1.67) |
Note. AUC = area under curve; CI = confidence interval; E =environmental; G =genetic; LR = logistic regression; OR = odds ratio; ROC = receiver operating characteristic; SLEs = stressful life events. Depressive symptoms status was measured in 2004–2005. The final model was naïve Bayes classifier (AUC ROC = 0.568). Attributes (n = 3) are listed in ranked order.
In the interest of comparison with our machine learning results, we also performed logistic regression models for every single SNP × single SLE interaction. After correction for multiple testing, we found no interactions below the Q = 0.05 level. We found 1 potential interaction, between HTR1A rs878567 and the proportion of SLEs index, which reached a marginal level of significance (Q = 0.061; P ≤ .001). Participants with a C/C genotype for HTR1A rs878567 had 18% higher odds of having depressive symptoms for each additional SLE experienced than those with a T/T genotype.
From our AUC ROC values, our best model, naïve Bayes, would accurately discriminate between a randomly selected depressive symptom individual and a randomly selected non- depressive symptom individual 56.8% of the time, which is 6.8% more often than by random chance (i.e., 50%). Thus, to enhance the predictability of our models, we examined additional sociobehavioral measures (Table 1). Once again, naïve Bayes (attribute size n = 13) produced our top-performing model, suggesting there were also no G×E×sociobehavioral (SB) interactions in our data. Nevertheless, the AUC ROC value showed that inclusion of sociobehavioral measures increased predictability; because the model was able to accurately classify 19.1% more often than by random chance, we designated it our overall best model. See Table 3 for odds ratios for the attributes of the model, listed in rank order (note that this model retained spouse or partner physical abuse, proportion of SLEs index, and gender). A 1-point increase in neuroticism score and 1-point decreases in openness, conscientiousness, extraversion, and agreeableness scores were associated with increased odds of depressive symptoms of 21%, 11%, 10%, 8%, and 10%, respectively. Each additional point increase in IQ score and each additional year of educational attainment was associated with a 2% and 9% decrease in odds of depressive symptoms, respectively. Experiencing physical abuse by a sibling was associated with increased odds of depressive symptoms of 115%. Being married was associated with a 39% decrease in odds for depressive symptoms. Each additional point increase in body mass index score was associated with a 3% increase in odds for depressive symptoms.
TABLE 3—
Odds of Depressive Symptoms for Attributes From Final Model for G×E×SB Data: Wisconsin Longitudinal Study, 1957–2005
| Attribute | Year Measured | Value | LR OR (95% CI) |
| Neuroticism score | 1992–1993 | Continuous | 1.21 (1.18, 1.24) |
| Openness score | 1992–1993 | Continuous | 0.89 (0.87, 0.92) |
| High school IQ | Varies | Continuous | 0.98 (0.98, 0.99) |
| Years educational attainment | 2004–2005 | Continuous | 0.91 (0.88, 0.94) |
| Conscientiousness score | 1992–1993 | Continuous | 0.90 (0.87, 0.92) |
| Spouse or partner physical abuse | 2004–2005 | Yes–no | 2.12 (1.70, 2.63) |
| Extraversion score | 1992–1993 | Continuous | 0.92 (0.89, 0.94) |
| Proportion of SLEs index | 2004–2005 | Continuous | 3.06 (1.93, 4.86) |
| Agreeableness score | 1992–1993 | Continuous | 0.90 (0.87, 0.93) |
| Female | … | Yes–no | 1.45 (1.26, 1.67) |
| Married | 1992–1993 | Yes–no | 0.61 (0.51, 0.73) |
| Sibling physical abuse | 2004–2005 | Yes–no | 2.15 (1.60, 2.90) |
| Body mass index | 1992–1993 | Continuous | 1.03 (1.02, 1.05) |
Note. AUC = area under curve; CI =confidence interval; E = environmental; G = genetic; LR =logistic regression; OR =odds ratio; ROC = receiver operating characteristic; SB = sociobehavioral; SLEs = stressful life events. Depressive symptoms status was measured in 2004–2005. The final model was naïve Bayes classifier (AUC ROC = 0.691). Attributes (n = 13) are listed in ranked order.
DISCUSSION
We found no significant associations in the WLS between depressive symptoms and single genetic variants. Previous studies reported significant single SNP associations between depression and ApoE4 (in women without Alzheimer’s disease),57 BDNF rs6265,54,55 CHRM2 rs8191992 (in women),51 COMT rs4680 (in men),49 and SLC6A4 rs8076005.44
In the WLS, we found marginal associations between only 2 of these SNPs and depressive symptoms, the COMT SNP in women (P = .021) and the BDNF SNP in men (P = .036). In the study by Åberg et al.,49 depression prevalence for the COMT rs4680 genotype in men (n = 1049) was 12.3%, 12.5%, and 5.9% for A/A, A/G, and G/G, respectively, indicative of an association between the Met (A) allele and depression (P = .009). When we compare this trend for men in the Åberg et al. study with the prevalence values for WLS women (n = 3260; A/A = 13.9%, A/G = 17.4%, and G/G = 18.7%), we see a dissimilar (opposite) trend. The trend in WLS men (n = 2985; A/A = 11.9%, A/G = 12.3%, and G/G = 12.7%) was also dissimilar from that of Åberg et al.49 Other studies also have demonstrated no association between COMT rs4680 and depression.66,67 In the BDNF studies by both Hwang et al.54 (n = 281; prevalence by genotype: G/G = 29.3%, G/A = 38.0%, and A/A = 56.7%; case–control G allele frequency = 46.8% vs 61.4%) and Taylor et al.55 (n = 245; prevalence by genotype: G/G = 67.9%, G/A = 79.4%, and A/A = 90.9%; case–control G allele frequency = 61.2% vs 75.5%), the BDNF rs6265 Met (A) allele was associated with higher prevalence of geriatric depression (men and women combined), which was seen in a dose–response fashion. In the WLS (men and women combined, n = 6317; prevalence: G/G = 15.1%, G/A = 12.8%, and A/A = 18.0%; case–control G allele frequency = 81.8% vs 80.8%), we see that, as in the studies by Hwang et al. and Taylor et al., prevalence is highest among A/A carriers and lowest among G/G carriers. However, WLS case–control allele frequencies were virtually identical, and we did not find a dose–response relationship for the A allele.
We were unable to replicate the findings of these 3 previous studies reporting single genetic associations for COMT and BDNF SNPs and depression. Not only did we not find statistical significance between these SNPs and depressive symptoms in the WLS, but the allele effects also did not show the same pattern as those in these previous studies. Another recent analysis of the WLS also failed to replicate previous candidate gene studies dealing with general intelligence.68 Comparison of our results with those of other studies assessing depression is, however, complicated by the differing scales used to assess depression (see Table 4 for comparison among the studies). For comparative purposes, clinical diagnosis would ideally provide the most robust measure for depression, although it is not always possible to obtain this type of diagnosis, especially in larger cohort studies. Thus, although our findings did not support those of previous studies, we cannot rule out that this was because different depression measures were used.
TABLE 4—
Measures for Depression Used in Studies Reporting SNP Associations Under Discussion
| SNP | Depression Measure |
| ApoE4 | Geriatric Depression Scale57 |
| BDNF rs6265 | Hamilton Rating Scale for Depression54 |
| Clinical diagnosis of unipolar major depression55 | |
| CHRM2 rs8191992 | Structured Clinical Interview for DSM-III-R51 |
| COMT rs4680 | Major Depression Inventory (DSM-IV diagnosis)49 |
| SLC6A4 rs8076005 | Beck Depression Inventory II44 |
| WLS (above SNPs) | Center for Epidemiologic Studies Depression Scale |
Note. DSM-III-R = Diagnostic and Statistical Manual of Mental Disorders, Revised Third Edition; DSM-IV = Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition; SNP = single nucleotide polymorphism.
Nevertheless, the message to be taken from our single SNP association results is that for a disease of complex origin such as depression, it is highly unlikely that single (common) genetic variants will be found that can explain etiology with anything more than nominal predictability.
Beyond Single Associations
Our main focus in this study was to explore potential G×G, E×E, and G×E interactions associated with depressive symptoms through the use of modern machine learning methods. These types of methods have become popular in recent medical genetics research because of their ability to efficiently explore diseases showing potential nonlinear causation.69
As such, we first used SVMs as a tool for determining whether the WLS contained any interactions between genetic and environmental variables. The Gaussian SVM did not significantly outperform the linear SVM (Table 1), which is evidence that no interactions occurred. Additionally, after comparing the SVMs with Bayesian networks, we saw that naïve Bayes was our top-performing model, which provided further evidence that there were no G×E interactions, because naïve Bayes by definition assumes that all risk factors contribute independently to depressive symptom status. Environmental variables (partner physical abuse, proportion of SLEs experienced) and gender were the attributes selected by this model. Single logistic regression (Table 2) supported these findings: that ever experiencing physical abuse from a spouse or partner, experiencing more SLEs in general, and being female were all associated with higher odds of depressive symptoms.
To confirm our finding of a lack of interaction, we decided to perform traditional logistic regression analysis for every pairwise G×E interaction model. No interactions remained significant below the Q = 0.05 level, supporting our machine learning results. Nevertheless, we found 1 marginally significant interaction between HTR1A rs878567 and the proportion of SLEs experienced index, in which homozygous C carriers had higher odds for depressive symptoms because they experienced more SLEs than homozygous T carriers. This interaction provided only a small additional predictive utility (OR = 1.18) and was likely too weak to be detected by machine learning algorithms. Nevertheless, Brezo et al.70 reported a significant G×E interaction involving the same SNP (rs878567) and the same risk allele. They found that C allele carriers who experienced childhood physical abuse showed higher risk for mood disorders (including major depression and bipolar disorder) than T allele carriers. Further testing of this G×E interaction appears warranted.
Next, because our machine learning models could only classify depressive symptoms 6.8% more often than by the flip of a coin, we considered a variety of additional sociobehavioral variables in our analysis. Naïve Bayes was once again our top-performing model, suggesting a lack of any interactions among genetic, environmental, and sociobehavioral variables. Including sociobehavioral variables increased our ability to classify depressive symptoms to 19.1% more often than by the flip of a coin, making this our overall top-performing model. The top 5 ranked risk factors were all sociobehavioral variables: neuroticism, openness, IQ, educational attainment, and conscientiousness. However, the model also contained gender and environmental variables—partner physical abuse and proportion of SLEs index—from our first naïve Bayes model. Thus, our results support that depressive symptoms is a disorder of heterogeneous origin and sociobehavioral variables seem to be the most predictive.
As has been shown in previous research (for major depression), we found that higher levels of neuroticism and lower levels of openness, conscientiousness, extraversion, and agreeableness were all associated with higher odds of depressive symptoms.71,72 We also found that lower high school IQ73 and lower overall educational attainment74 were both associated with higher depressive symptom odds.
Our results also showed that experiencing physical abuse from a sibling or a spouse or partner were both associated with higher odds of depressive symptoms. Coker et al.75 (and many additional studies referenced therein) showed that physical violence by a spouse or partner was associated with significant mental health consequences. Al-Modallal et al.76 found a general positive association in women between experiencing physical abuse (as a child or adult) and depressive symptoms in adulthood. In terms of overall SLEs, we found that a higher number of events experienced was associated with higher odds of depressive symptoms. A review by Kessler77 covered the topic of SLEs and depression in great detail, and although the relationship is complex, he concluded that, in general, experiencing SLEs predicts subsequent depression.
Next, we found that being female was associated with a 45% increase in odds of depressive symptoms. It is well established that women report depression at a higher rate than men.78,79 Being married as opposed to not married (12 years before assessment of depressive symptoms) decreased odds for current depressive symptoms by almost half, which is similar to the trend reported by Gutiérrez-Lobos et al.80 between current marital status and depression. Last, we found that higher body mass index was associated with increased odds for current depressive symptoms. Recent studies have reported that obesity increases risk for depression.81,82
We note that although our final models showed heterogeneous origins for depressive symptoms, they contained no genetic attributes as risk factors, which may suggest that depressive symptoms is conditionally independent of this study’s genetic information, given the sociobehavioral features. In other words, any information held in our SNPs was already manifesting in the sociobehavioral variables. Another possibility is that our selection of SNPs had no pertinent connections with the etiology of depressive symptoms. This is not to say that SNPs are not important; future, more extensive GWAS will provide a more complete determination of which polymorphisms may interact with one another or with environmental and sociobehavioral factors to modulate risk of depressive symptoms. The methodology we used in this study is the perfect tool with which to analyze high-dimensional genotypic data sets for G×E×SB interactions. Additionally, the WLS is an especially suitable cohort for this type of study because of its rich covariate nature.
Limitations
The predictive value of our genetic data is limited because of user bias in selection of SNPs. Other genetic variants that we did not examine in this study may have provided information about crucial interactions involved with depressive symptoms. One such variant, for example, would be 5-HTTLPR, a highly studied repeat in recent depression work. The WLS has social factor homogeneities, such as participants being almost exclusively non-Hispanic White people with middle- to upper-middle-class backgrounds. Another limiting factor is that besides our genotype data (and high school rank and IQ scores), our analyses relied exclusively on self-reported data for all predictors. Moreover, we should point out that the initial onset of depressive symptoms may have preceded the occurrence of SLEs. Nevertheless, all life events may contribute to current depressive symptoms status in some manner, whether they are the initial cause or a perpetuating factor. We note that although we have reported G×G interactions associated with lifetime depression (Composite International Diagnostic Interview Short Form) in the WLS,7 in this study we examined a different measure of depression (current depressive symptoms; Center for Epidemiologic Studies Depression Scale), and we found no interactions. Future directions for research will involve using these same machine learning methodologies on more complete genetic information, such as from genome-wide SNP or sequencing data, with which we will be able to explore all genetic interaction possibilities rather than a limited subset of variants.
Acknowledgments
This research used data from the Wisconsin Longitudinal Study (WLS) of the University of Wisconsin–Madison. Since 1991, the WLS has been supported principally by the National Institute on Aging (AG-9775, AG-21079, and AG-033285), with additional support from the Vilas Estate Trust, the National Science Foundation, the Spencer Foundation, and the Graduate School of the University of Wisconsin–Madison. A public use file of data from the Wisconsin Longitudinal Study is available from the WLS, University of Wisconsin–Madison, 1180 Observatory Drive, Madison, WI 53706, and at http://www.ssc.wisc.edu/wlsresearch/data. This material is the result of work supported with resources at the William S. Middleton Memorial Veterans Hospital, Madison, WI.
Note. The opinions expressed herein are those of the authors. The contents do not represent the views of the Department of Veterans Affairs or the US government. This article is Geriatrics Research, Education and Clinical Center VA paper 2013–17.
Human Participant Protection
Ethics approval was provided by the social sciences institutional review board, University of Wisconsin–Madison.
References
- 1.Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62(6):593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 2.Goltser-Dubner T, Galili-Weisstub E, Segman RH. Genetics of unipolar major depressive disorder. Isr J Psychiatry Relat Sci. 2010;47(1):72–82. [PubMed] [Google Scholar]
- 3.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157(10):1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 4.Kohli MA, Lucae S, Saemann Philipp G et al. The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron. 2011;70(2):252–265. doi: 10.1016/j.neuron.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18(4):497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.López-León S, Janssens AC, González-Zuloeta Ladd AM et al. Meta-analyses of genetic studies on major depressive disorder. Mol Psychiatry. 2008;13(8):772–785. doi: 10.1038/sj.mp.4002088. [DOI] [PubMed] [Google Scholar]
- 7.Roetker NS, Yonker JA, Lee C et al. Multigene interactions and the prediction of depression in the Wisconsin Longitudinal Study. BMJ Open. 2012;2(4):e000944. doi: 10.1136/bmjopen-2012-000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang K, Yang C, Xu Y et al. Genetic association of the interaction between the BDNF and GSK3B genes and major depressive disorder in a Chinese population. J Neural Transm. 2010;117(3):393–401. doi: 10.1007/s00702-009-0360-4. [DOI] [PubMed] [Google Scholar]
- 9.Lin E, Hong CJ, Hwang JP et al. Gene-gene interactions of the brain-derived neurotrophic-factor and neurotrophic tyrosine kinase receptor 2 genes in geriatric depression. Rejuvenation Res. 2009;12(6):387–393. doi: 10.1089/rej.2009.0871. [DOI] [PubMed] [Google Scholar]
- 10.Neff CD, Abkevich V, Potter J, Riley R, Shattuck D, Katz DA. Evidence for epistasis between SLC6A4 and a chromosome 4 gene as risk factors in major depression. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(1):321–322. doi: 10.1002/ajmg.b.30979. [DOI] [PubMed] [Google Scholar]
- 11.Lazary J, Lazary A, Gonda X et al. New evidence for the association of the serotonin transporter gene (SLC6A4) haplotypes, threatening life events, and depressive phenotype. Biol Psychiatry. 2008;64(6):498–504. doi: 10.1016/j.biopsych.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 12.Neff CD, Abkevich V, Packer JCL et al. Evidence for HTR1A and LHPP as interacting genetic risk factors in major depression. Mol Psychiatry. 2009;14(6):621–630. doi: 10.1038/mp.2008.8. [DOI] [PubMed] [Google Scholar]
- 13.Caspi A, Sugden K, Moffitt TE et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301(5631):386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 14.Karg K, Burmeister M, Shedden K, Sen S. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Arch Gen Psychiatry. 2011;68(5):444–454. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradley RG, Binder EB, Epstein MP et al. Influence of child abuse on adult depression: moderation by the corticotropin-releasing hormone receptor gene. Arch Gen Psychiatry. 2008;65(2):190–200. doi: 10.1001/archgenpsychiatry.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polanczyk G, Caspi A, Williams B et al. Protective effect of CRHR1 gene variants on the development of adult depression following childhood maltreatment: replication and extension. Arch Gen Psychiatry. 2009;66(9):978–985. doi: 10.1001/archgenpsychiatry.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grabe HJ, Schwahn C, Appel K et al. Childhood maltreatment, the corticotropin-releasing hormone receptor gene and adult depression in the general population. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(8):1483–1493. doi: 10.1002/ajmg.b.31131. [DOI] [PubMed] [Google Scholar]
- 18.Heim C, Binder EB. Current research trends in early life stress and depression: review of human studies on sensitive periods, gene-environment interactions, and epigenetics. Exp Neurol. 2012;233(1):102–111. doi: 10.1016/j.expneurol.2011.10.032. [DOI] [PubMed] [Google Scholar]
- 19.Luijk MP, Velders FP, Tharner A et al. FKBP5 and resistant attachment predict cortisol reactivity in infants: gene–environment interaction. Psychoneuroendocrinology. 2010;35(10):1454–1461. doi: 10.1016/j.psyneuen.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Zimmermann P, Bruckl T, Pfister H et al. The interplay of variations in the FKBP5 gene and adverse life events in predicting the first onset of depression during a ten-year follow-up. Pharmacopsychiatry. 2009;42(5):249. [Google Scholar]
- 21.Bet PM, Penninx BW, Bochdanovits Z et al. Glucocorticoid receptor gene polymorphisms and childhood adversity are associated with depression: new evidence for a gene–environment interaction. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(5):660–669. doi: 10.1002/ajmg.b.30886. [DOI] [PubMed] [Google Scholar]
- 22.Thompson RJ, Parker KJ, Hallmayer JF, Waugh CE, Gotlib IH. Oxytocin receptor gene polymorphism (rs2254298) interacts with familial risk for psychopathology to predict symptoms of depression and anxiety in adolescent girls. Psychoneuroendocrinology. 2011;36(1):144–147. doi: 10.1016/j.psyneuen.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gatt JM, Nemeroff CB, Schofield PR et al. Early life stress combined with serotonin 3A receptor and brain-derived neurotrophic factor valine 66 to methionine genotypes impacts emotional brain and arousal correlates of risk for depression. Biol Psychiatry. 2010;68(9):818–824. doi: 10.1016/j.biopsych.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 24.Hayden EP, Klein DN, Dougherty LR et al. The dopamine D2 receptor gene and depressive and anxious symptoms in childhood: associations and evidence for gene-environment correlation and gene-environment interaction. Psychiatr Genet. 2010;20(6):304–310. doi: 10.1097/YPG.0b013e32833adccb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguilera M, Arias B, Wichers M et al. Early adversity and 5-HTT/BDNF genes: new evidence of gene-environment interactions on depressive symptoms in a general population. Psychol Med. 2009;39(9):1425–1432. doi: 10.1017/S0033291709005248. [DOI] [PubMed] [Google Scholar]
- 26.Conway CC, Hammen C, Brennan PA, Lind PA, Najman JM. Interaction of chronic stress with serotonin transporter and catechol-O-methyltransferase polymorphisms in predicting youth depression. Depress Anxiety. 2010;27(8):737–745. doi: 10.1002/da.20715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaufman J, Yang B-Z, Douglas-Palumberi H et al. Brain-derived neurotrophic factor–5-HTTLPR gene interactions and environmental modifiers of depression in children. Biol Psychiatry. 2006;59(8):673–680. doi: 10.1016/j.biopsych.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 28.Ressler KJ, Bradley B, Mercer KB et al. Polymorphisms in CRHR1 and the serotonin transporter loci: gene x gene x environment interactions on depressive symptoms. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(3):812–824. doi: 10.1002/ajmg.b.31052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J-M, Stewart R, Kim S-W et al. Interactions between life stressors and susceptibility genes (5-HTTLPR and BDNF) on depression in Korean elders. Biol Psychiatry. 2007;62(5):423–428. doi: 10.1016/j.biopsych.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 30.Wichers M, Kenis G, Jacobs N et al. The BDNF Val66Met × 5-HTTLPR × child adversity interaction and depressive symptoms: an attempt at replication. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(1):120–123. doi: 10.1002/ajmg.b.30576. [DOI] [PubMed] [Google Scholar]
- 31.Szymczak S, Biernacka JM, Cordell HJ et al. Machine learning in genome-wide association studies. Genet Epidemiol. 2009;33(suppl 1):S51–S57. doi: 10.1002/gepi.20473. [DOI] [PubMed] [Google Scholar]
- 32.Listgarten J, Damaraju S, Poulin B et al. Predictive models for breast cancer susceptibility from multiple single nucleotide polymorphisms. Clin Cancer Res. 2004;10(8):2725–2737. doi: 10.1158/1078-0432.ccr-1115-03. [DOI] [PubMed] [Google Scholar]
- 33.Huang L-C, Hsu S-Y, Lin E. A comparison of classification methods for predicting chronic fatigue syndrome based on genetic data. J Transl Med. 2009;7(1):81. doi: 10.1186/1479-5876-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhuo Z, Jiang L, Chee Keong K . Buenos Aires, Argentina: August 31, 2010. Learning in glaucoma genetic risk assessment. Paper presented at: 32nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society. [Google Scholar]
- 35.Sebastiani P, Ramoni MF, Nolan V, Baldwin CT, Steinberg MH. Genetic dissection and prognostic modeling of overt stroke in sickle cell anemia. Nat Genet. 2005;37(4):435–440. doi: 10.1038/ng1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang X, Barmada MM, Visweswaran S. Identifying genetic interactions in genome-wide data using Bayesian networks. Genet Epidemiol. 2010;34(6):575–581. doi: 10.1002/gepi.20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma S, Poon A, Himes BE et al. Association of variants in innate immune genes with asthma and eczema. Pediatr Allergy Immunol. 2012;23(4):315–323. doi: 10.1111/j.1399-3038.2011.01243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sewell WH. As We Age: The Wisconsin Longitudinal Study, 1957-2001. Madison, WI: Center for Demography and Ecology, University of Wisconsin–Madison; 2001. [Google Scholar]
- 39.Weissman MM, Sholomskas D, Pottenger M, Prusoff BA, Locke BZ. Assessing depressive symptoms in five psychiatric populations: a validation study. Am J Epidemiol. 1977;106(3):203–214. doi: 10.1093/oxfordjournals.aje.a112455. [DOI] [PubMed] [Google Scholar]
- 40.Lamke TA, Nelson MJ. Henmon-Nelson Tests of Mental Ability, Rev. Ed. and Earlier Editions: Grades 3-6, 6-9, 9-12, 13-17. Boston, MA: Houghton Mifflin; 1929. [Google Scholar]
- 41.McCrae RR, Costa PT. Personality in Adulthood: A Five-Factor Theory Perspective. New York, NY: Guilford Press; 2003. [Google Scholar]
- 42.Flynn KE, Smith MA. Personality and health care decision-making style. J Gerontol B Psychol Sci Soc Sci. 2007;62(5):P261–P267. doi: 10.1093/geronb/62.5.p261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin E, Chen PS, Chang HH et al. Interaction of serotonin-related genes affects short-term antidepressant response in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33(7):1167–1172. doi: 10.1016/j.pnpbp.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 44.Su S, Zhao J, Bremner JD et al. Serotonin transporter gene, depressive symptoms, and interleukin-6. Circ Cardiovasc Genet. 2009;2(6):614–620. doi: 10.1161/CIRCGENETICS.109.870386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kishi T, Tsunoka T, Ikeda M et al. Serotonin 1A receptor gene and major depressive disorder: an association study and meta-analysis. J Hum Genet. 2009;54(11):629–633. doi: 10.1038/jhg.2009.84. [DOI] [PubMed] [Google Scholar]
- 46.Minov C, Baghai TC, Schüle C et al. Serotonin-2A-receptor and -transporter polymorphisms: lack of association in patients with major depression. Neurosci Lett. 2001;303(2):119–122. doi: 10.1016/s0304-3940(01)01704-9. [DOI] [PubMed] [Google Scholar]
- 47.Illi A, Setala-Soikkeli E, Viikki M et al. 5-HTR1A, 5-HTR2A, 5-HTR6, TPH1 and TPH2 polymorphisms and major depression. Neuroreport. 2009;20(12):1125–1128. doi: 10.1097/WNR.0b013e32832eb708. [DOI] [PubMed] [Google Scholar]
- 48.Brummett BH, Kuhn CM, Boyle SH, Babyak MA, Siegler IC, Williams RB. Cortisol responses to emotional stress in men: association with a functional polymorphism in the 5HTR2C gene. Biol Psychol. 2012;89(1):94–98. doi: 10.1016/j.biopsycho.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Åberg E, Fandino-Losada A, Sjoholm LK, Forsell Y, Lavebratt C. The functional Val158Met polymorphism in catechol-O-methyltransferase (COMT) is associated with depression and motivation in men from a Swedish population-based study. J Affect Disord. 2011;129(1-3):158–166. doi: 10.1016/j.jad.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 50.Cohen-Woods S, Gaysina D, Craddock N et al. Depression case control (DeCC) study fails to support involvement of the muscarinic acetylcholine receptor M2 (CHRM2) gene in recurrent major depressive disorder. Hum Mol Genet. 2009;18(8):1504–1509. doi: 10.1093/hmg/ddp051. [DOI] [PubMed] [Google Scholar]
- 51.Comings DE, Wu S, Rostamkhani M, McGue M, Iacono WG, MacMurray JP. Association of the muscarinic cholinergic 2 receptor (CHRM2) gene with major depression in women. Am J Med Genet. 2002;114(5):527–529. doi: 10.1002/ajmg.10406. [DOI] [PubMed] [Google Scholar]
- 52.Elovainio M, Jokela M, Kivimäki M et al. Genetic variants in the DRD2 gene moderate the relationship between stressful life events and depressive symptoms in adults: cardiovascular risk in young Finns study. Psychosom Med. 2007;69(5):391–395. doi: 10.1097/psy.0b013e31806bf365. [DOI] [PubMed] [Google Scholar]
- 53.Huuhka K, Anttila S, Huuhka M et al. Dopamine 2 receptor C957T and catechol-O-methyltransferase Val158Met polymorphisms are associated with treatment response in electroconvulsive therapy. Neurosci Lett. 2008;448(1):79–83. doi: 10.1016/j.neulet.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 54.Hwang J-P, Tsai S-J, Hong C-J, Yang C-H, Lirng J-F, Yang Y- M. The Val66Met polymorphism of the brain-derived neurotrophic-factor gene is associated with geriatric depression. Neurobiol Aging. 2006;27(12):1834–1837. doi: 10.1016/j.neurobiolaging.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 55.Taylor WD, Zuchner S, McQuoid DR, Steffens DC, Speer MC, Krishnan KR. Allelic differences in the brain-derived neurotrophic factor Val66Met polymorphism in late-life depression. Am J Geriatr Psychiatry. 2007;15(10):850–857. doi: 10.1097/JGP.0b013e318050c9d5. [DOI] [PubMed] [Google Scholar]
- 56.Kocabas NA, Antonijevic I, Faghel C et al. Brain-derived neurotrophic factor gene polymorphisms: influence on treatment response phenotypes of major depressive disorder. Int Clin Psychopharmacol. 2011;26(1):1–10. doi: 10.1097/yic.0b013e32833d18f8. [DOI] [PubMed] [Google Scholar]
- 57.Slifer MA, Martin ER, Gilbert JR, Haines JL, Pericak-Vance MA. Resolving the relationship between ApolipoproteinE and depression. Neurosci Lett. 2009;455(2):116–119. doi: 10.1016/j.neulet.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raymer KA, Waters RF, Price CR. Proposed multigenic composite inheritance in major depression. Med Hypotheses. 2005;65(1):158–172. doi: 10.1016/j.mehy.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 59.Storey JD, Taylor JE, Siegmund D. Strong control, conservative point estimation and simultaneous conservative consistency of false discovery Rates: a unified approach. J R Stat Soc Series B Stat Methodol. 2004;66(1):187–205. [Google Scholar]
- 60.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software. An update. SIGKDD Explor. 2009;11(1):10–18. [Google Scholar]
- 61.Vapnik VN. Statistical learning theory. In: Haykin S, editor. Adaptive and Learning Systems for Signal Processing, Communications, and Control. New York, NY: Wiley; 1998. pp. 736–757. [Google Scholar]
- 62.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20(3):273–297. [Google Scholar]
- 63.Burnside ES, Davis J, Chhatwal J et al. Probabilistic computer model developed from clinical data in national mammography database format to classify mammographic findings 1. Radiology. 2009;251(3):663–672. doi: 10.1148/radiol.2513081346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Domingos P, Pazzani M. On the optimality of the simple Bayesian classifier under zero-one loss. Mach Learn. 1997;29(2-3):103–130. [Google Scholar]
- 65.Friedman N, Geiger D, Goldszmidt M. Bayesian network classifiers. Mach Learn. 1997;29(2-3):131–163. [Google Scholar]
- 66.Illi A, Setala-Soikkeli E, Kampman O et al. Catechol-O-methyltransferase val108/158met genotype, major depressive disorder and response to selective serotonin reuptake inhibitors in major depressive disorder. Psychiatry Res. 2010;176(1):85–87. doi: 10.1016/j.psychres.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 67.Wray NR, James MR, Dumenil T et al. Association study of candidate variants of COMT with neuroticism, anxiety and depression. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(7):1314–1318. doi: 10.1002/ajmg.b.30744. [DOI] [PubMed] [Google Scholar]
- 68.Chabris CF, Hebert BM, Benjamin DJ et al. Most reported genetic associations with general intelligence are probably false positives. Psychol Sci. 2012;23(11):1314–1323. doi: 10.1177/0956797611435528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cordell HJ. Detecting gene–gene interactions that underlie human diseases. Nat Rev Genet. 2009;10(6):392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brezo J, Bureau A, Merette C et al. Differences and similarities in the serotonergic diathesis for suicide attempts and mood disorders: a 22-year longitudinal gene-environment study. Mol Psychiatry. 2010;15(8):831–843. doi: 10.1038/mp.2009.19. [DOI] [PubMed] [Google Scholar]
- 71.Chien L-L, Ko H-C, Wu JY- W. The five-factor model of personality and depressive symptoms: one-year follow-up. Pers Individ Dif. 2007;43(5):1013–1023. [Google Scholar]
- 72.Weber K, Giannakopoulos P, Bacchetta J-P et al. Personality traits are associated with acute major depression across the age spectrum. Aging Ment Health. 2012;16(4):472–480. doi: 10.1080/13607863.2011.630375. [DOI] [PubMed] [Google Scholar]
- 73.Koenen KC, Moffitt TE, Roberts AL et al. Childhood IQ and adult mental disorders: a test of the cognitive reserve hypothesis. Am J Psychiatry. 2009;166(1):50–57. doi: 10.1176/appi.ajp.2008.08030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bjelland I, Krokstad S, Mykletun A, Dahl AA, Tell GS, Tambs K. Does a higher educational level protect against anxiety and depression? The HUNT study. Soc Sci Med. 2008;66(6):1334–1345. doi: 10.1016/j.socscimed.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 75.Coker AL, Davis KE, Arias I et al. Physical and mental health effects of intimate partner violence for men and women. Am J Prev Med. 2002;23(4):260–268. doi: 10.1016/s0749-3797(02)00514-7. [DOI] [PubMed] [Google Scholar]
- 76.Al-Modallal H, Peden A, Anderson D. Impact of physical abuse on adulthood depressive symptoms among women. Issues Ment Health Nurs. 2008;29(3):299–314. doi: 10.1080/01612840701869791. [DOI] [PubMed] [Google Scholar]
- 77.Kessler RC. The effects of stressful life events on depression. Annu Rev Psychol. 1997;48(1):191–214. doi: 10.1146/annurev.psych.48.1.191. [DOI] [PubMed] [Google Scholar]
- 78.Kessler RC, McGonagle KA, Swartz M, Blazer DG, Nelson CB. Sex and depression in the National Comorbidity Survey I: lifetime prevalence, chronicity and recurrence. J Affect Disord. 1993;29(2-3):85–96. doi: 10.1016/0165-0327(93)90026-g. [DOI] [PubMed] [Google Scholar]
- 79.Weissman MM, Bland RC, Canino GJ et al. Cross-national epidemiology of major depression and bipolar disorder. JAMA. 1996;276(4):293–299. [PubMed] [Google Scholar]
- 80.Gutiérrez-Lobos K, Wölfl G, Scherer M, Anderer P, Schmidl-Mohl B. The gender gap in depression reconsidered: the influence of marital and employment status on the female/male ratio of treated incidence rates. Soc Psychiatry Psychiatr Epidemiol. 2000;35(5):202–210. doi: 10.1007/s001270050229. [DOI] [PubMed] [Google Scholar]
- 81.Luppino FS, de Wit LM, Bouvy PF et al. Overweight, obesity, and depression: a systematic review and meta-analysis of longitudinal studies. Arch Gen Psychiatry. 2010;67(3):220–229. doi: 10.1001/archgenpsychiatry.2010.2. [DOI] [PubMed] [Google Scholar]
- 82.Faith MS, Butryn M, Wadden TA, Fabricatore A, Nguyen AM, Heymsfield SB. Evidence for prospective associations among depression and obesity in population-based studies. Obes Rev. 2011;12(5):e438–e453. doi: 10.1111/j.1467-789X.2010.00843.x. [DOI] [PubMed] [Google Scholar]


