Abstract
Recent analyses have discovered broad alterations in the expression of human genes across different social environments. The emerging field of social genomics has begun to identify the types of genes sensitive to social regulation, the biological signaling pathways mediating these effects, and the genetic polymorphisms that modify their individual impact. The human genome appears to have evolved specific “social programs” to adapt molecular physiology to the changing patterns of threat and opportunity ancestrally associated with changing social conditions. In the context of the immune system, this programming now fosters many of the diseases that dominate public health. The embedding of individual genomes within a broader metagenomic network provides a framework for integrating molecular, physiologic, and social perspectives on human health.
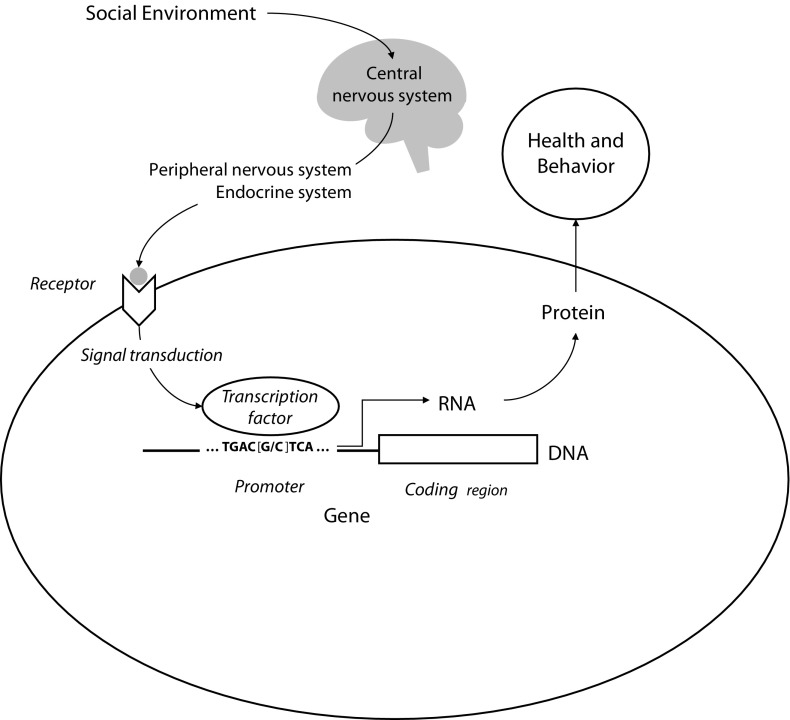
The conceptual relationship between genes and the social world has shifted significantly during the past 20 years. As genes have come to be understood in concrete molecular terms, rather than as abstract heritability constructs, it has become clear that social factors can play a significant role in regulating the activity of the human genome. DNA encodes the potential for cellular behavior, but that potential is only realized if the gene is expressed—if its DNA is transcribed into RNA (Figure 1). RNA and its translated proteins are what mediate cellular behaviors such as movement, metabolism, and biochemical response to external stimuli (e.g., neurotransmission or immune response). Absent their expression in the form of RNA, DNA genes have no effect on health or behavioral phenotypes. The development of DNA microarray and high-throughput RNA sequencing technologies now allows researchers to survey the expression of all human genes simultaneously and map the specific subset of genes that are active in a given cell at a given point in time—the RNA “transcriptome.”1 “Functional genomics” studies surveying RNA transcriptomes have shown that cells are highly selective about which genes they express, and humans’ DNA encodes a great deal more genetic potential than is actually realized in RNA. Even more striking has been the discovery that the social world outside one’s body can markedly influence these gene expression profiles.
FIGURE 1—
Social signal transduction.
Note. Socioenvironmental conditions regulate human gene expression by activating central nervous system processes that subsequently influence hormone and neurotransmitter activity in the periphery of the body. Peripheral signaling molecules interact with cellular receptors to activate transcription factors, which bind to characteristic DNA motifs in gene promoters to initiate (or repress) gene expression. Only genes that are transcribed into RNA actually have an impact on health and behavioral phenotypes. Individual differences in promoter DNA sequences (e.g., the [G/C] polymorphism shown here) can affect the binding of transcription factors and thereby influence genomic sensitivity to socioenvironmental conditions.
This article reviews the emerging field of human social genomics, including its recent scientific development, some developing themes regarding the number and nature of “socially sensitive” genes, and emerging data on the psychological, neural, and endocrine signaling pathways that mediate social influences on gene expression. The presentation also considers some evolutionary theories regarding the teleology of such “social signal transduction” and the implications of these dynamics for environmental programming of human development and life-span health trajectories. The role of gene polymorphisms (genetics) in modulating individual genomic sensitivity to socioenvironmental influences is considered, as are implications of social genomic relationships for public health and policy, including optimal intervention strategies, new opportunities for integrating social genomics into epidemiology, and implications of a public health perspective for understanding how individual human genomes cross-regulate one another in the context of social networks (i.e., social regulation of the human “metagenome,” or the collective system of individual human genomes). Social genomics research provides a concrete molecular framework for understanding the long-observed relationship between social conditions and the distribution of human health and disease.2–4
SOCIAL REGULATION OF GENE EXPRESSION
The possibility that social factors might regulate gene expression first emerged in the context of studies analyzing the effects of stress and social isolation on viral gene expression (e.g., in herpes simplex viruses,5–11 HIV-1,12–15 Epstein-Barr virus,6,16 cytomegalovirus,6,17 and the Kaposi’s sarcoma–associated human herpesvirus 818). Viruses are little more than small packages of 10 to 100 genes that hijack the protein production machinery of their host cells to make more copies of themselves. As obligate parasites of human host cells, human viruses have evolved within a microenvironment structured by our own genome. If social factors can regulate the expression of viral genes, our own complement of approximately 21 000 genes is likely to be regulated in significant ways as well.19
One of the first studies analyzing the influence of social factors on the human transcriptome compared gene expression profiles in peripheral blood leukocytes from healthy older adults who differed in the extent to which they felt socially connected to others.20 Among the 22 283 transcripts assayed, 209 showed systematically different levels of expression in people who consistently reported feeling lonely and distant from others over the course of 4 years (Figure 2). These effects did not involve a random smattering of all human genes but instead had a focal impact on 3 functionally related groups of genes, or “gene programs.” Genes supporting the early “accelerator” phase of the immune response—inflammation—were selectively up-regulated. Down-regulated were genes involved in innate antiviral responses (particularly type I interferons) and genes involved in the production of specific antibody isotypes by B lymphocytes (particularly immunoglobulin G). This complementary up-regulation of pro-inflammatory genes and down-regulation of antiviral and antibody-related genes provided a molecular framework for understanding the previously puzzling epidemiological observations that social isolation is associated with diseases that involve both up-regulated immune function (inflammation-related diseases such as heart disease, neurodegenerative diseases, and some types of cancer) and down-regulated immune function (reduced responses to vaccines and viral infections in particular). This specific proinflammatory/anti-antiviral shift in the basal leukocyte transcriptome showed that social adversity is not broadly immunosuppressive, as had previously been hypothesized, but instead selectively suppresses some groups of immune-response genes (e.g., type I interferons and specific immunoglobulin genes) while simultaneously activating others (e.g., proinflammatory cytokines).
FIGURE 2—
Social regulation of gene expression in human immune cells.
Note. Red = high expression; black = intermediate expression; green = low expression. Expression of 22 283 human gene transcripts was assayed in approximately 10 million blood leukocytes sampled from each of 14 older adults who showed consistent differences over 4 years in their level of subjective social isolation. Two hundred nine gene transcripts showed differences of 30% or more in average expression level in leukocytes from 6 people experiencing chronic social isolation versus 8 people experiencing consistent social integration. In this heat plot, each row represents data from 1 of the 14 study participants, each column contains expression values for 1 of the 209 differentially active genes, and the coloring of each cell represents the relative level of that gene’s expression in a given participant’s leukocyte sample.
Source. Adapted from Cole et al.20
A similar pattern of pro-inflammatory/anti-antiviral transcriptome skewing has since been observed in leukocytes sampled from people exposed to a diverse array of adverse life circumstances such as imminent bereavement,21 traumatic stress,22 social isolation,23 low socioeconomic status (SES),24–26 and cancer diagnosis.27 Similar dynamics have also been observed in experimental animal models of social instability, low social rank, and repeated social defeat.28–30 The mammalian immune system appears to have developed a conserved transcriptional response to adversity (CTRA) that induces a pro-inflammatory/anti-antiviral skew in the circulating leukocyte transcriptome whenever environmental conditions are experienced as threatening, stressful, or uncertain for an extended period of time.30 Although different types of social adversity can activate a common CTRA, their transcriptional effects are by no means identical because each context generally activates some distinctive transcriptional responses as well.31
Laboratory gene regulation analyses have suggested that the common transcriptional components of the CTRA likely stem from the fact that diverse types of social risk factors can induce common neural and hormonal stress responses.30–32 For example, catecholamine neurotransmitters released during fight-or-flight stress responses can directly modulate the transcription of several key master regulator genes that orchestrate the activity of broad sets of inflammatory and antiviral genes (e.g., IL1B, IL6, and IFNB).14,28,30,33 Randomized controlled studies have also shown that stress-reducing interventions can reverse CTRA-related transcriptional dynamics to down-regulate pro-inflammatory genes and up-regulate genes involved in type I interferon responses.27,34 These stress-induced changes in immune-cell gene expression provide a molecular framework for understanding why diverse types of social adversity come to be associated with a common set of diseases ranging from asthma and viral infections to cancer and cardiovascular disease.32
Transcriptome profiling of other tissues and organs has shown that social influences can penetrate remarkably deeply into the body. Adverse social conditions have been linked to gene expression alterations in the central nervous system,35,36 peripheral organs such as the lymph nodes and spleen,14,28 and diseased tissues such as ovarian carcinomas, prostate cancers, and ischemic brain injuries.37–39 Given the much smaller number of social genomics studies targeting solid tissues and the relative difficulty in ascertaining the functional significance of specific transcriptional alterations outside the well-charted territories of the immune response, it is not yet clear what basic gene programs are being activated in these other tissue contexts (e.g., are they defense responses analogous to the leukocyte CTRA?). However, the widespread penetrance of social conditions into gene regulatory dynamics in diverse tissue sites raises the question of how such external social stimuli are physically transduced into biochemical dynamics that can proximally regulate gene transcription within the nuclei of diverse cell types distributed widely throughout the body. New insights into this question have come from bioinformatic analyses of social signal transduction.
SOCIAL SIGNAL TRANSDUCTION
Biologists have traditionally construed signal transduction as the biochemical processes that translate extracellular signals, such as hormones or neurotransmitters, into changes in gene expression through the activation of protein transcription factors that bind to DNA and flag it for transcription into RNA (Figure 1). Social signal transduction extends this analysis to include the upstream neural dynamics that translate social conditions into systemically distributed signaling molecules (e.g., release of norepinephrine during fight-or-flight stress responses) and to include the specific downstream gene modules that are activated by a given transcription factor. For example, when norepinephrine is released from the sympathetic nervous system during fight-or-flight stress responses, cells bearing β-adrenergic receptors translate that signal into activation of the transcription factor cyclic 3′-5′ adenosine monophosphate response element-binding protein (CREB).40 Activated CREB proteins can up-regulate the transcription of hundreds of cellular genes.41 Which genes can be activated by CREB is determined by the nucleotide sequence of the gene’s promoter—the stretch of DNA lying upstream of the coding region of the gene that is transcribed into RNA. For example, CREB binds to the nucleotide motif TGACGTCA, whereas the microbe-responsive transcription factor nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) targets the motif GGGACTTTCC. These 2 transcription factors are activated by different receptor-mediated signal transduction pathways, providing distinct molecular channels through which specific extracellular signaling molecules and, by extension, their specific upstream environmental triggers, can regulate intracellular genomic response. The distribution of transcription factor-binding motifs across humans’ approximately 21 000 gene promoters constitutes a “wiring diagram” that maps specific types of environmental processes (e.g., infection vs a fight-or-flight stress response) onto a specific pattern of genome-wide transcriptional response (e.g., CREB vs NF-κB target genes). In that sense, each transcription factor can be said to represent some type of evolutionarily significant characteristic of the environment outside the cell (e.g., CREB = threat or stress, NF-κB = microbe or damaged cell), and the distribution of specific transcription factor-binding DNA motifs across the promoters of humans’ approximately 21 000 genes can be understood as an evolved “wisdom of the genome” regarding which genes should be activated to optimally adapt to that environment.
Biological signal transduction research has generally emphasized the role of the physicochemical or microbial stimuli in transcription factor activation, but studies of social signal transduction have suggested that subjective psychological interpretations of the external environment can also play a significant role in regulating gene expression profiles.30,31,42 For example, activation of the leukocyte CTRA is often more strongly linked to subjective perceptions of the environment than it is to objective environmental conditions,20,21,23,26,30,42 and CTRA transcriptome skewing can be reversed by psychological interventions that target those subjective psychological experiences.27,34 In studies of social connection, for example, the subjective experience of loneliness is associated with twice as many differentially expressed genes as is the objective frequency of social contacts,20,23 and psychological interventions that reduce subjective loneliness are associated with concomitant reductions in pro-inflammatory gene expression.34 In women with early-stage breast cancer, CTRA transcriptional profiles are also more strongly associated with the subjective degree of life threat women experience than with objective measures of disease severity such as tumor grade or stage, and cognitive–behavioral stress management interventions can reverse that threat-related transcriptome skewing.27 In children with asthma, SES-related perceptions of the social world as hostile or threatening are more strongly linked to leukocyte transcriptional alterations than are objective measures of SES such as household income.26 Objective features of the environment such as the number of interpersonal contacts are, of course, associated with variations in gene expression.20,23 However, subjective social experience also plays a significant role, and the effects of subjective and objective conditions are often transduced into gene expression via different molecular signaling pathways.
The combination of genome-wide transcriptional profiling with promoter-based bioinformatic analyses has greatly accelerated the identification of the specific transcription factors that translate subjective social experience into the activation of specific gene programs.31 This approach first identifies the subset of genes that is differentially expressed in response to an environmental risk factor (e.g., social isolation) and then scans the promoters of those differentially expressed genes for transcription factor–binding motifs that are substantially overrepresented relative to their prevalence across the genome as a whole and might thus reveal which specific transcription factors induced the observed transcriptional alterations.43 For example, the subset of genes up-regulated in tissues from people experiencing significant social adversity often show a higher prevalence of CREB–target promoter sequences than is found across the population of all human genes,20,38 implying that CREB may have played a role in activating that specific gene program. That inference is consistent with CREB’s known role in mediating the gene transcriptional effects of β-adrenergic receptor signaling in response to catecholamines produced during fight-or-flight stress responses.40 In the context of the leukocyte CTRA, promoter-based bioinformatics analyses have repeatedly implicated increased NF-κB transcription factor activity in the pro-inflammatory gene responses and decreased signaling by interferon regulatory factor family transcription factors in the decreased antiviral gene component.30,31,42
Promoter-based bioinformatics have also revealed some more surprising differences between the hormonal signals sent by the brain and the transcriptional signals heard by the human genome. In studies of chronic social isolation, impending bereavement, posttraumatic stress disorder, and low SES, promoter bioinformatics have indicated decreased activity of the anti-inflammatory glucocorticoid receptor (GR) in association with the leukocyte CTRA.20–22,25 Under normal circumstances, activation of the GR by cortisol from the hypothalamic–pituitary–adrenal (HPA) axis would both stimulate the expression of anti-inflammatory GR target genes and cross-inhibit the pro-inflammatory NF-κB transcription factors. However, in people experiencing chronic stress, both of those dynamics appear to be blunted, resulting in a net pro-inflammatory skew in the leukocyte transcriptome.30 None of these studies found decreases in HPA axis output of cortisol that might explain the reduced levels of GR activity. Instead, the explanation appears to involve a stress-induced reduction in the GR’s sensitivity to cortisol—rendering the leukocyte transcriptome partially deaf to the HPA axis’s request to down-regulate pro-inflammatory genes via glucocorticoid output.20,21 A similar glucocorticoid desensitization dynamic has been observed in mice repeatedly exposed to social stress.44–46 In addition to clarifying the molecular origin of the leukocyte CTRA, these findings also highlight a broader possibility that measuring blood levels of hormones, neurotransmitters, and other extracellular signaling molecules may miss some important receptor-level influences on the transcriptional mediators of health and disease. Transcriptome-based bioinformatic assessment of social signal transduction provides an integrated measure of both pre- and postreceptor dynamics at the level that matters most for the molecular biology of disease—gene expression.
Epigenetic dynamics provide another pathway by which social environments might potentially regulate gene expression.47,48 Epigenetic influences involve biochemical modifications of DNA such as methylation or histone protein engagement that block gene transcription without altering a gene’s DNA sequence.49 Research with experimental animal models has linked favorable social conditions (e.g., maternal licking of rat pups, high social rank in monkeys) to altered patterns of DNA methylation and gene expression in immune cells29 and brain structures such as the hippocampus.35,47 Correlational human studies have documented associations between DNA methylation profiles and socioenvironmental risk factors such as low SES and childhood stress exposure.50–52 Although environmental factors clearly influence epigenetic dynamics in human immune cells,53 much remains to be learned about the signaling pathways that mediate such dynamics and their functional role in social regulation of human gene expression.54
EVOLUTION OF SOCIAL PROGRAMMING
As social genomics studies map the particular gene programs that are empirically sensitive to social conditions, new theoretical analyses are emerging to explain why such connections may have evolved in the first place (i.e., the teleological basis for social programming of the human genome). In the context of the leukocyte CTRA, for example, social adversity redeploys the leukocyte’s basal transcriptional resources away from antiviral defenses and toward pro-inflammatory gene products that protect the body against bacterial infections. This shift in the leukocyte’s basal transcriptional stance may have been adaptive under Pleistocene hunter–gatherer conditions in which the social ecology outside the body played a major role in shaping the pathogen ecology within the body.
Homo sapiens is a distinctively social organism,55 and its highly social life history strategy has conferred substantial adaptive advantages56 at the price of increased vulnerability to socially transmitted infectious diseases.57 Viral infections, for example, are predominately transmitted through extended periods of close social contact,57,58 so it would be highly adaptive for an intrinsically social organism to evolve a strong antiviral bias as its default immune response bias. However, when the social world turns hostile and individuals either are isolated or confront conspecific aggression (i.e., feel threatened), the risk of wound-mediated bacterial infection increases dramatically, and it would be adaptive to temporarily redeploy leukocyte transcriptional resources toward inflammatory defenses against bacterial infection by linking pro-inflammatory gene expression to β-adrenergic fight-or-flight signaling.23,30
Such social programming of immune response biases may well have been adaptive during humans’ hunter–gatherer prehistory, but in the context of more complex and unstable contemporary social systems the connection of experienced threat, stress, or uncertainty to pro-inflammatory/anti-antiviral transcriptional skewing primes the human immune system to promote inflammation-related cardiovascular, metabolic, neurodegenerative, and neoplastic diseases while leaving it relatively unresponsive to viral infections. Similar social programming is likely to have evolved for other adaptively significant cell populations, such as the nervous and reproductive systems, and awaits more extensive social genomics studies to define both its genomic scope and its teleological rationales.
ENVIRONMENTAL EMBEDDING IN DEVELOPMENT
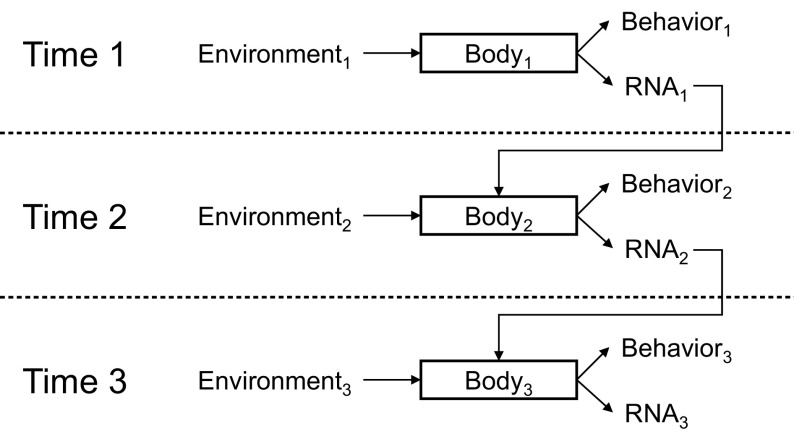
To the extent that external social conditions affect gene transcription at one point in time, the persistence of its protein products and the feedback-rich regulatory architecture of gene expression can propagate such influences over time to generate a persisting molecular “memory” of previous environmental conditions (i.e., embedding environmental influences into the molecular development of the individual).47,59–61 Compared with other biochemical response systems such as protein phosphorylation, neural and muscular activation, or ion flux, gene transcription occurs slowly (up-regulating over 0.5–2 hours) and yields proteins that can persist for weeks or years afterward (the average half-life of a human protein is about 80 days). Some gene expression dynamics are also recursively stimulated by their own products and can thus self-propagate over time once initiated. Many pro-inflammatory cytokines, for example, activate the same signal transduction pathways that trigger their initial transcriptional activation in response to cell damage or microbes and can thus self-propagate over time. A second level of extrinsic feedback can occur when the molecular changes induced by one environmental exposure affect the types of environments the individual gravitates toward in the future or the nature of the individual’s biological or behavioral responses to subsequent environmental exposures (Figure 3). One way this occurs is when social signal transduction modulates the expression of genes, which themselves play a role in mediating social signal transduction (e.g., genes encoding signaling molecules, receptors, and transcription factors). The RNA “output” from one round of social signal transduction becomes an “input” into subsequent rounds and thereby modifies the input–output relationship between subsequent environmental exposures and subsequent gene expression responses. As a result of the long intrinsic duration of gene expression effects and their capacity to self-propagate, environmental exposures that occur early in life can become embedded in an individual’s developmental trajectory.14,24,25,60,62,63 Such dynamics are hypothesized to stretch back as far as the fetal environment and its role in shaping biological development and subsequent adult vulnerability to disease (i.e., the fetal programming hypothesis).64
FIGURE 3—
RNA as a molecular medium of recursive development.
Note. Social conditions at one point in time (Environment1) are transduced into changes in behavior (Behavior1) and gene expression (RNA1) via central nervous system perceptual processes that trigger systemic neural and endocrine responses (mediated by Body1). Those RNA transcriptional dynamics may alter molecular characteristics of cells involved in environmental perception or response, resulting in a functionally altered Body2. Body2 may respond differently to a given environmental challenge than would the previous Body1, resulting in different behavioral (Behavior2) and RNA transcriptional responses (RNA2). The persisting effect of RNA transcriptional dynamics on cellular protein and functional characteristics provides a molecular framework for understanding how socioenvironmental conditions in the past may continue to affect current behavior and health and how those historical conditions interact with current environments to shape one’s future trajectories (e.g., Body3, Behavior3, RNA3). Because gene transcription serves as both a cause of social behavior (by shaping Body) and a consequence of social behavior (a product of environment × body), RNA constitutes the physical medium for a recursive developmental trajectory that integrates genetic characteristics and historical–environmental regulators to understand individual biological and behavioral responses to current environmental conditions.
One health-relevant example of socioenvironmental embedding involves the ability of chronic social stress to up-regulate transcription of the NGF gene and thereby enhance the growth of sympathetic nerve fibers in the lymph node tissues that structure the development of immune responses.14 Expressed in terms of the system outlined in Figure 3, NGF-induced up-regulation of lymph node innervation at Time1 can persist for weeks, providing a denser neural network through which subsequent social stress at Time2 can distribute norepinephrine to lymph node–resident immune cells. These arborized neural fibers also produce more NGF and thus perpetuate their own arborization. The increased norepinephrine release from these neural fibers inhibits transcription of the IFNB gene, which would otherwise play a key role in initiating antiviral responses.14,33,65 As a result, the immune system responds less effectively to a new viral exposure (e.g., Time2 or Time3) than it would have if the individual had experienced a more favorable social history at Time1. The remodeling of lymph node sympathetic innervation in response to social stress–induced NGF produces a chronic activation of CTRA transcriptional dynamics and thus undermines future antiviral responses. As such, the individual’s response to a viral infection encountered today is shaped both by the nature of that virus and by the transcriptome’s “memory” of previous environmental conditions encountered over the individual’s life history.
Socioenvironmental conditions can also regulate the molecular composition of central nervous system cells and thereby alter psychological and behavioral responses to future environments.36,60 Because the molecular composition of one’s cells constitutes the physical machinery by which one perceives and responds to the surrounding world (“Body” in Figure 3), and that molecular composition is itself subject to remodeling by socioenvironmental influences, gene expression constitutes both a cause and a consequence of behavior. RNA can be construed as the physical medium of a recursive developmental system in which social, behavioral, and health outcomes at one point in time also constitute inputs that shape one’s future responses to the environment (e.g., as in Heckman’s66 model of human capability development, which analyzes how capacities developed at Time1 have an impact on one’s ability to capitalize on environmental opportunities at Time2). Viewed from another perspective, the evolution of the RNA transcriptomes within the body provides a kind of molecular record of an individual body’s cumulative adaptation to the history of environmental exposures that it has encountered, in much the same way as the evolution of a species’ DNA genome records the history of its adaptation to the environmental exposures it has encountered over the course of its evolutionary history.
THE NEW GENETICS
The growing ability to trace social signal transduction to the molecular level is also providing new opportunities to understand and predict gene × environment interactions through computational modeling of their molecular underpinnings. One approach uses promoter-based bioinformatics to identify socially responsive transcription factors as outlined earlier (i.e., the biochemical representation of the “environment”) and then scans the promoter of each human gene to identify known genetic polymorphisms that might alter the binding of an environmentally responsive transcription factor (i.e., a regulatory polymorphism).28 One recent analysis first identified the GATA1 transcription factor as a mediator of fight-or-flight stress responses and then scanned predicted GATA1 target genes for polymorphisms that might affect GATA1 binding (Figure 3). A G/C substitution 174 bases upstream of the transcription start site for the human IL6 gene was identified as potentially inhibiting GATA1 binding and thereby disconnecting this key pro-inflammatory gene from socioenvironmental regulation.28 Laboratory biochemical analyses confirmed that the −174C allele of the IL6 promoter showed reduced transcriptional responsiveness to β-adrenergic receptor activation of GATA1, and in vivo molecular epidemiology confirmed that people bearing the GATA1-insensitive −174C allele were protected against the increased risk of inflammation-related mortality associated with significant life adversity.28 Maximal expression of IL6 required both an environmentally sensitive genotype (IL6 −174G) and its functional activation by an adverse environment (sympathetic nervous system–β-adrenergic receptor–GATA1 signaling). The IL6 regulatory polymorphism blocks the capacity of adverse environmental conditions to activate the expression of this key disease-related gene and thus renders carriers less vulnerable to socioenvironmentally mediated health risks.
Computational discovery of the social adversity × IL6 −174G/C interaction helped clarify several outstanding questions regarding genetic influences on health at the population level. Discovery that the IL6 gene requires an environmental releaser to manifest its effects clarified the basis for the incomplete penetrance of IL6 polymorphism into disease phenotypes (i.e., clarified the nature of genetic influence) and provided a genetic mechanism for individual variation in health sensitivity to adverse environments (i.e., clarified the nature of environmental influence). Identification of the specific biochemical signaling pathway conveying environmental adversity into gene expression dynamics also suggested new strategies for mitigating their jointly produced health risks (e.g., pharmacologic blockade of β-adrenergic receptor signaling).28,46,67,68 In integrating the molecular biology of gene structure (DNA), the environmental control of gene expression (RNA), and the social biology of individual behavior and survival, the IL6 regulatory polymorphism exemplifies a new “environmentally conscious” conception of genetics in which cellular and organismic behaviors constitute the fundamental units of evolutionary selection, and genes and environments depend mutually on one another to shape those behaviors by structuring humans’ brains and bodies.
LIMITATIONS AND OPPORTUNITES
The first generation of social genomics studies has opened new vistas on the connection between the human genome and its social environment, but a great deal remains to be clarified, and the existing literature needs to strengthen in several ways. Because of the substantial expense and technical demands of early microarray assays, first-generation social genomics studies involved small cross-sectional analyses with limited assessment of the socioenvironmental confounders, rendering the causal relationships unclear. As second-generation technologies have lowered cost and technical burden, studies of larger samples and experimental studies have become available.23,25,29,69 Results of these second-generation studies have broadly replicated the pattern of results from first-generation studies (e.g., compare social isolation CTRA dynamics in Cole et al.20 at n = 14 with Cole et al.23 at n = 93 or rural- and urban-related differences in Idaghdour et al.70 at n = 46 with Idaghdour et al.69 at n = 194). The surprising precision of genomic analyses in small samples stems in part from the statistical advantages of treating thousands of individual genes as multiple noisy indicators of shared higher order “themes” regarding common biochemical functions, transcription factor targets, and cellular origins of gene expression.31
Second-generation studies have also included randomized intervention studies showing that adverse social conditions can causally activate the CTRA in animal models28,29,63 and that psychologically targeted interventions can causally reduce the CTRA in human clinical studies.27,34,71 However, a great need remains for large-scale longitudinal studies involving broader assessment of the social environment throughout the life course,61 as well as large-scale intervention studies to more decisively define the causal effects of social and psychological processes; identify their mediating neural, endocrine, and transcription factor pathways; and test candidate health-protective interventions.30 There is also a great need to expand the range of tissues studied beyond the convenient pool of circulating leukocytes to encompass a broader array of health-relevant organs as well as the nervous and endocrine systems that play a central role in mediating human biological adaptation to the social environment. Given these limitations, the substantive themes summarized in this review should be considered researchers’ best available understanding, but an understanding that will surely undergo substantial revision as the empirical literature deepens over time.
IMPLICATIONS FOR PUBLIC HEALTH
Social regulation of human gene expression implies that many aspects of individual health actually constitute a form of public health in the sense that they emerge as properties of an interconnected system of human beings. Some of one’s genes operate differently depending on the presence of other people and their (subjectively perceived) implications for one’s own fitness outcomes such as survival and reproduction. As a result, some of the regulatory architecture of the human genome lies outside of the cell in the constraints and affordances present in the social ecology and in people’s subjective perceptions and interpretations of those ecologies. From this perspective, individual genomes constitute elements of a broader human metagenomic network (i.e., an interconnected system of related genomes) in which some gene regulatory dynamics represent emergent properties of the system as a whole.72 Public health can thus be understood as a metagenomic dynamic in which fast-evolving cultural systems interact with slowly evolving (but very long-memoried) human genomes and more rapidly evolving pathogen genomes to produce a pattern of RNA transcriptional responses across a network of elements whose individual properties vary as a function of both genetic and environmental polymorphism.73–75
This conception of public health raises a host of new conceptual questions, such as, Which types of genes are subject to network-level regulation? How are network transcriptional dynamics affected by individual genetic characteristics, by historical–developmental influences, or by network structural characteristics such as linkage patterns, community blocks, and individual linkage characteristics such as centrality, density, or redundancy? Which parts of the central nervous system transduce social signals into gene expression changes? What role does human culture play in metagenomic dynamics and individual social signal transduction?73,76 How has a socially networked human genome shaped the development of human social systems and gene–culture coevolution?76 Do individual transcriptional alterations affect network structure (e.g., via behavioral or biological homophily or heterophily)? How do physicochemical or microbial features of the environment interact with human social systems to regulate metagenomic systems?73–75 Are these physical environmental influences transmitted through different networks and transduction pathways than are subjective or symbolic social influences? How do positive, supportive, or playful social interactions influence human gene expression (e.g., do they simply abate the adversity-related dynamics, as recently suggested for the leukocyte CTRA,27,34 or does there exist a distinct set of prosocial genes involved in the positive neurobiological effects of social interaction77)? As the next generation of social genomics research begins to address these questions, the integration of social network analyses with individual social signal transduction and the evolved social programming of the human genome will open up an array of new opportunities for synthesizing molecular, organismic, and population-level analyses into a coherent overall understanding of human health.
In addition to these conceptual advances, new technological developments in gene expression profiling now offer new opportunities to integrate genomics-based perspectives into large-scale field epidemiology. First- and second-generation social genomics studies relied on laboratory-centered research paradigms involving venipuncture blood samples and technically intensive, time-sensitive RNA extraction procedures. However, new developments in RNA stabilization chemistry and enzymatic amplification of small RNA samples now allow genome-wide transcriptome profiling from more field-friendly sampling modes such as saliva collection tubes, finger-stick dried blood spots, and venipuncture blood samples that can be mailed or stored for months before processing. These technical innovations should allow widespread and economical collection of transcriptome data from epidemiological-scale samples (i.e., n = 1000–10 000) collected in their natural environments. Coupled with ongoing 10- to 100-fold reductions in the cost of transcriptome profiling and the emergence of automated data analytic and bioinformatic interpretation systems, these developments should allow public health research to begin routinely integrating the deep physiological, evolutionary, and molecular genetic perspectives that were formerly the province of basic laboratory research into mainstream epidemiological analyses of human host resistance and disease distribution. The new substantive insights that emerge from these field studies of the human genome will also greatly enrich laboratory and clinical studies by more clearly mapping the basic functional relationships between human social conditions and the activity of individual gene programs.
As studies more definitively link specific gene expression profiles to disease vulnerability, field-based transcriptome profiling may also provide a new form of molecular surveillance that could potentially identify both overt disease states and host vulnerability conditions that have not yet been converted into disease (i.e., up-regulated inflammatory signaling or impaired antiviral gene expression, as in the CTRA). Such a molecular window into the body could help guide public health interventions and social policies to more proactively address the general host resistance factors that seem to precipitate multiple diseases32 rather than responding reactively to specific diseases only after they clinically emerge. It might be possible, for example, to use a CTRA profile as an indicator of generalized host resistance or vulnerability (i.e., a latent liability to disease) that is assessed in parallel with realized disease to help gauge the toxicity of various social or geographic environments or the success of public policies and interventions. In combination with the conceptual advances of a network-level metagenomic approach to human health, researchers’ growing technical capacity to gauge host resistance at a molecular level before the onset of disease and within the normal social ecology will help accelerate the ongoing transformation of public health from a disease-reactive model to a more proactive and health-centered approach that also accounts for human vitality and physiological resilience.
Acknowledgments
Preparation of this article was supported by the National Institutes of Health (grants CA116778, AG033590, and AG028748).
Human Participant Protection
Human participant protection was not required because no human research participants were involved in the preparation of this article.
References
- 1.Djebali S, Davis CA, Merkel A et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Virchow R. Report on the typhus outbreak of Upper Silesia. Social Medicine. 2006;1(1):11–27. [Google Scholar]
- 3.Cassel J. The contribution of the social environment to host resistance. Am J Epidemiol. 1976;104(2):107–123. doi: 10.1093/oxfordjournals.aje.a112281. [DOI] [PubMed] [Google Scholar]
- 4.Berkman LF, Kawachi I. Social Epidemiology. New York, NY: Oxford University Press; 2000. [Google Scholar]
- 5.Rasmussen AF, Marsh JT, Brill NQ. Increases susceptibility to herpes simplex in mice subjected to avoidance-learning stress or restraint. Proc Soc Exp Biol Med. 1957;96(1):183–189. doi: 10.3181/00379727-96-23426. [DOI] [PubMed] [Google Scholar]
- 6.Glaser R, Kiecolt-Glaser JK, Speicher CE, Holliday JE. Stress, loneliness, and changes in herpesvirus latency. J Behav Med. 1985;8(3):249–260. doi: 10.1007/BF00870312. [DOI] [PubMed] [Google Scholar]
- 7.Kupfer SR, Summers WC. Identification of a glucocorticoid-responsive element in Epstein-Barr virus. J Virol. 1990;64(5):1984–1990. doi: 10.1128/jvi.64.5.1984-1990.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leib DA, Nadeau KC, Rundle SA, Schaffer PA. The promoter of the latency-associated transcripts of herpes simplex virus type 1 contains a functional cAMP-response element: role of the latency-associated transcripts and cAMP in reactivation of viral latency. Proc Natl Acad Sci U S A. 1991;88(1):48–52. doi: 10.1073/pnas.88.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuster C, Chasserot-Golaz S, Urier G, Beck G, Sergeant A. Evidence for a functional glucocorticoid responsive element in the Epstein-Barr virus genome. Mol Endocrinol. 1991;5(2):267–272. doi: 10.1210/mend-5-2-267. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins FJ, Baum A. Stress and reactivation of latent herpes simplex virus: a fusion of behavioral medicine and molecular biology. Ann Behav Med. 1995;17(2):116–123. doi: 10.1007/BF02895060. [DOI] [PubMed] [Google Scholar]
- 11.Padgett DA, Sheridan JF, Dorne J, Berntson GG, Candelora J, Glaser R. Social stress and the reactivation of latent herpes simplex virus type 1. Proc Natl Acad Sci U S A. 1998;95(12):7231–7235. doi: 10.1073/pnas.95.12.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole SW, Kemeny ME, Taylor SE, Visscher BR, Fahey JL. Accelerated course of human immunodeficiency virus infection in gay men who conceal their homosexual identity. Psychosom Med. 1996;58(3):219–231. doi: 10.1097/00006842-199605000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Cole SW, Kemeny ME, Taylor SE. Social identity and physical health: accelerated HIV progression in rejection-sensitive gay men. J Pers Soc Psychol. 1997;72(2):320–335. doi: 10.1037//0022-3514.72.2.320. [DOI] [PubMed] [Google Scholar]
- 14.Sloan EK, Capitanio JP, Tarara RP, Mendoza SP, Mason WA, Cole SW. Social stress enhances sympathetic innervation of primate lymph nodes: mechanisms and implications for viral pathogenesis. J Neurosci. 2007;27(33):8857–8865. doi: 10.1523/JNEUROSCI.1247-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capitanio JP, Mendoza SP, Lerche NW, Mason WA. Social stress results in altered glucocorticoid regulation and shorter survival in simian acquired immune deficiency syndrome. Proc Natl Acad Sci U S A. 1998;95(8):4714–4719. doi: 10.1073/pnas.95.8.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang EV, Webster Marketon JI, Chen M, Lo KW, Kim SJ, Glaser R. Glucocorticoids activate Epstein Barr virus lytic replication through the upregulation of immediate early BZLF1 gene expression. Brain Behav Immun. 2010;24(7):1089–1096. doi: 10.1016/j.bbi.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prösch S, Wendt CEC, Reinke P et al. A novel link between stress and human cytomegalovirus (HCMV) infection: sympathetic hyperactivity stimulates HCMV activation. Virology. 2000;272(2):357–365. doi: 10.1006/viro.2000.0367. [DOI] [PubMed] [Google Scholar]
- 18.Chang M, Brown H, Collado-Hidalgo A et al. Beta-adrenoreceptors reactivate Kaposi’s sarcoma-associated herpesvirus lytic replication via PKA-dependent control of viral RTA. J Virol. 2005;79(21):13538–13547. doi: 10.1128/JVI.79.21.13538-13547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunham I, Kundaje A, Aldred SF et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole SW, Hawkley LC, Arevalo JM, Sung CY, Rose RM, Cacioppo JT. Social regulation of gene expression in human leukocytes. Genome Biol. 2007;8(9):R189. doi: 10.1186/gb-2007-8-9-r189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller GE, Chen E, Sze J et al. A functional genomic fingerprint of chronic stress in humans: blunted glucocorticoid and increased NF-kappaB signaling. Biol Psychiatry. 2008;64(4):266–272. doi: 10.1016/j.biopsych.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Donovan A, Sun B, Cole S et al. Transcriptional control of monocyte gene expression in post-traumatic stress disorder. Dis Markers. 2011;30(2–3):123–132. doi: 10.3233/DMA-2011-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci U S A. 2011;108(7):3080–3085. doi: 10.1073/pnas.1014218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen E, Miller GE, Kobor MS, Cole SW. Maternal warmth buffers the effects of low early-life socioeconomic status on pro-inflammatory signaling in adulthood. Mol Psychiatry. 2011;16(7):729–737. doi: 10.1038/mp.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller GE, Chen E, Fok AK et al. Low early-life social class leaves a biological residue manifested by decreased glucocorticoid and increased proinflammatory signaling. Proc Natl Acad Sci U S A. 2009;106(34):14716–14721. doi: 10.1073/pnas.0902971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen E, Miller GE, Walker HA, Arevalo JM, Sung CY, Cole SW. Genome-wide transcriptional profiling linked to social class in asthma. Thorax. 2009;64(1):38–43. doi: 10.1136/thx.2007.095091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antoni MH, Lutgendorf SK, Blomberg B et al. Transcriptional modulation of human leukocytes by cognitive-behavioral stress management in women undergoing treatment for breast cancer. Biol Psychiatry. 2012;71(4):366–372. doi: 10.1016/j.biopsych.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole SW, Arevalo JM, Takahashi R et al. Computational identification of gene-social environment interaction at the human IL6 locus. Proc Natl Acad Sci U S A. 2010;107(12):5681–5686. doi: 10.1073/pnas.0911515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tung J, Barreiro LB, Johnson ZP et al. Social environment is associated with gene regulatory variation in the rhesus macaque immune system. Proc Natl Acad Sci U S A. 2012;109(17):6490–6495. doi: 10.1073/pnas.1202734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irwin MR, Cole SW. Reciprocal regulation of the neural and innate immune systems. Nat Rev Immunol. 2011;11(9):625–632. doi: 10.1038/nri3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole SW. Elevating the perspective on human stress genomics. Psychoneuroendocrinology. 2010;35(7):955–962. doi: 10.1016/j.psyneuen.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller G, Chen E, Cole SW. Health psychology: developing biologically plausible models linking the social world and physical health. Annu Rev Psychol. 2009;60:501–524. doi: 10.1146/annurev.psych.60.110707.163551. [DOI] [PubMed] [Google Scholar]
- 33.Collado-Hidalgo A, Sung C, Cole S. Adrenergic inhibition of innate anti-viral response: PKA blockade of type I interferon gene transcription mediates catecholamine support for HIV-1 replication. Brain Behav Immun. 2006;20(6):552–563. doi: 10.1016/j.bbi.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Creswell JD, Irwin MR, Burklund LJ et al. Mindfulness-based stress reduction training reduces loneliness and pro-inflammatory gene expression in older adults: a small randomized controlled trial. Brain Behav Immun. 2012;26(7):1095–1101. doi: 10.1016/j.bbi.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci U S A. 2006;103(9):3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karssen AM, Her S, Li JZ et al. Stress-induced changes in primate prefrontal profiles of gene expression. Mol Psychiatry. 2007;12(12):1089–1102. doi: 10.1038/sj.mp.4002095. [DOI] [PubMed] [Google Scholar]
- 37.Ornish D, Magbanua MJ, Weidner G et al. Changes in prostate gene expression in men undergoing an intensive nutrition and lifestyle intervention. Proc Natl Acad Sci U S A. 2008;105(24):8369–8374. doi: 10.1073/pnas.0803080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lutgendorf SK, Degeest K, Sung CY et al. Depression, social support, and beta-adrenergic transcription control in human ovarian cancer. Brain Behav Immun. 2009;23(2):176–183. doi: 10.1016/j.bbi.2008.04.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karelina K, Norman GJ, Zhang N, Morris JS, Peng H, DeVries AC. Social isolation alters neuroinflammatory response to stroke. Proc Natl Acad Sci U S A. 2009;106(14):5895–5900. doi: 10.1073/pnas.0810737106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanders VM, Straub RH. Norepinephrine, the beta-adrenergic receptor, and immunity. Brain Behav Immun. 2002;16(4):290–332. doi: 10.1006/brbi.2001.0639. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Odom DT, Koo SH et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102(12):4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole SW. Social regulation of human gene expression. Curr Dir Psychol Sci. 2009;18(3):132–137. doi: 10.1111/j.1467-8721.2009.01623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole SW, Yan W, Galic Z, Arevalo J, Zack JA. Expression-based monitoring of transcription factor activity: the TELiS database. Bioinformatics. 2005;21(6):803–810. doi: 10.1093/bioinformatics/bti038. [DOI] [PubMed] [Google Scholar]
- 44.Stark JL, Avitsur R, Padgett DA, Sheridan JF. Social stress induces glucocorticoid resistance in macrophages. Am J Physiol Regul Integr Comp Physiol. 2001;280(6):R1799–R1805. doi: 10.1152/ajpregu.2001.280.6.R1799. [DOI] [PubMed] [Google Scholar]
- 45.Avitsur R, Powell N, Padgett DA, Sheridan JF. Social interactions, stress, and immunity. Immunol Allergy Clin North Am. 2009;29(2):285–293. doi: 10.1016/j.iac.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Hanke ML, Powell ND, Stiner LM, Bailey MT, Sheridan JF. Beta adrenergic blockade decreases the immunomodulatory effects of social disruption stress. Brain Behav Immun. 2012;26(7):1150–1159. doi: 10.1016/j.bbi.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meaney MJ, Szyf M, Seckl JR. Epigenetic mechanisms of perinatal programming of hypothalamic-pituitary-adrenal function and health. Trends Mol Med. 2007;13(7):269–277. doi: 10.1016/j.molmed.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 48.Hunter RG. Epigenetic effects of stress and corticosteroids in the brain. Front Cell Neurosci. 2012;6:18. doi: 10.3389/fncel.2012.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riddihough G, Zahn LM. Epigenetics. What is epigenetics? Introduction. Science. 2010;330(6004):611. doi: 10.1126/science.330.6004.611. [DOI] [PubMed] [Google Scholar]
- 50.Borghol N, Suderman M, McArdle W et al. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol. 2012;41(1):62–74. doi: 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Essex MJ, Thomas Boyce W, Hertzman C et al. Epigenetic vestiges of early developmental adversity: childhood stress exposure and DNA methylation in adolescence. Child Dev. 2013;84(1):58–75. doi: 10.1111/j.1467-8624.2011.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naumova OY, Lee M, Koposov R, Szyf M, Dozier M, Grigorenko EL. Differential patterns of whole-genome DNA methylation in institutionalized children and children raised by their biological parents. Dev Psychopathol. 2012;24(1):143–155. doi: 10.1017/S0954579411000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fraga MF, Ballestar E, Paz MF et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller G. Epigenetics. The seductive allure of behavioral epigenetics. Science. 2010;329(5987):24–27. doi: 10.1126/science.329.5987.24. [DOI] [PubMed] [Google Scholar]
- 55.Cacioppo JT, Hawkley LC. Perceived social isolation and cognition. Trends Cogn Sci. 2009;13(10):447–454. doi: 10.1016/j.tics.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamilton WD. The genetical evolution of social behaviour. II. J Theor Biol. 1964;7(1):17–52. doi: 10.1016/0022-5193(64)90039-6. [DOI] [PubMed] [Google Scholar]
- 57.Nunn CL, Altizer S. Oxford, England: Oxford University Press; 2006. Infectious Diseases in Primates: Behavior, Ecology and Evolution. [Google Scholar]
- 58.Knipe DM, Howley PM, Griffin DE . Fields Virology. 5th ed. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 59.Hertzman C. The biological embedding of early experience and its effects on health in adulthood. Ann N Y Acad Sci. 1999;896:85–95. doi: 10.1111/j.1749-6632.1999.tb08107.x. [DOI] [PubMed] [Google Scholar]
- 60.Zhang TY, Bagot R, Parent C et al. Maternal programming of defensive responses through sustained effects on gene expression. Biol Psychol. 2006;73(1):72–89. doi: 10.1016/j.biopsycho.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 61.Shanahan MJ, Hofer SM. Molecular genetics, aging, and well-being: sensitive period, accumulation, and pathway models. In: Binstock RH, George LK, editors. Handbook of Aging and Social Sciences. 7th ed. New York, NY: Elsevier; 2011. pp. 135–147. [Google Scholar]
- 62.Nakamura T, Walker AK, Sominsky L, Allen T, Rosengren S, Hodgson DM. Maternal separation in early life impairs tumor immunity in adulthood in the F344 rat. Stress. 2011;14(3):335–343. doi: 10.3109/10253890.2010.548014. [DOI] [PubMed] [Google Scholar]
- 63.Cole SW, Arevalo JM, Ruggerio AM, Heckman JJ, Suomi S. Transcriptional modulation of the developing immune system by early life social adversity. Proc Natl Acad Sci U S A. 2012;109(50):20578–20583. doi: 10.1073/pnas.1218253109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barker DJ. The developmental origins of adult disease. J Am Coll Nutr. 2004;23(6, suppl):588S–595S. doi: 10.1080/07315724.2004.10719428. [DOI] [PubMed] [Google Scholar]
- 65.Sloan EK, Tarara RP, Capitanio JP, Cole SW. Enhanced replication of simian immunodeficiency virus adjacent to catecholaminergic varicosities in primate lymph nodes. J Virol. 2006;80(9):4326–4335. doi: 10.1128/JVI.80.9.4326-4335.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heckman JJ. The economics, technology, and neuroscience of human capability formation. Proc Natl Acad Sci U S A. 2007;104(33):13250–13255. doi: 10.1073/pnas.0701362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sloan EK, Priceman SJ, Cox BF et al. The sympathetic nervous system induces a metastatic switch in primary breast cancer. Cancer Res. 2010;70(18):7042–7052. doi: 10.1158/0008-5472.CAN-10-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wohleb ES, Hanke ML, Corona AW et al. Beta-adrenergic receptor antagonism prevents anxiety-like behavior and microglial reactivity induced by repeated social defeat. J Neurosci. 2011;31(17):6277–6288. doi: 10.1523/JNEUROSCI.0450-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Idaghdour Y, Czika W, Shianna KV et al. Geographical genomics of human leukocyte gene expression variation in southern Morocco. Nat Genet. 2010;42(1):62–67. doi: 10.1038/ng.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Idaghdour Y, Storey JD, Jadallah SJ, Gibson G. A genome-wide gene expression signature of environmental geography in leukocytes of Moroccan Amazighs. PLoS Genet. 2008;4(4):e1000052. doi: 10.1371/journal.pgen.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Black DS, Cole SW, Irwin MR et al. Yogic meditation reverses NF-kappaB and IRF-related transcriptome dynamics in leukocytes of family dementia caregivers in a randomized controlled trial. Psychoneuroendocrinology. 2013;38(3):348–355. doi: 10.1016/j.psyneuen.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kauffman S. The Origins of Order: Self-Organization and Selection in Evolution. Oxford, UK: Oxford University Press; 1993. [Google Scholar]
- 73.McDade TW. Life history theory and the immune system: steps toward a human ecological immunology. Am J Phys Anthropol. 2003;122(suppl 37):100–125. doi: 10.1002/ajpa.10398. [DOI] [PubMed] [Google Scholar]
- 74.Gibson G. The environmental contribution to gene expression profiles. Nat Rev Genet. 2008;9(8):575–581. doi: 10.1038/nrg2383. [DOI] [PubMed] [Google Scholar]
- 75.Rappaport SM, Smith MT. Epidemiology. Environment and disease risks. Science. 2010;330(6003):460–461. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Richerson PJ, Boyd R, Henrich J. Colloquium paper: gene-culture coevolution in the age of genomics. Proc Natl Acad Sci U S A. 2010;107(suppl 2):8985–8992. doi: 10.1073/pnas.0914631107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Panksepp J. Affective Neuroscience: The Foundations of Human and Animal Emotions. New York, NY: Oxford University Press; 1998. [Google Scholar]