Abstract
Live imaging provides exciting opportunities to study dynamic cellular events as they occur in real time. However, such experimental procedures present several challenges. This chapter discusses some of the major considerations relating to the maintenance of live biological samples during ex vivo imaging and presents some relatively simple, low-cost solutions to keeping samples healthy.
INTRODUCTION
The development of new fluorescent probes and GFP reporter animal models, coupled with improved access to and convenience of imaging technologies, has generated a good deal of excitement in the field. One of the challenges that would-be users face is the application of these technologies to live specimens that remain viable in the imaging setup. Although some preparations permit high-resolution imaging of live cells in vivo, in many cases it is still necessary or desirable to isolate the cells and tissues so that they may be stained and subsequently observed over time. Such procedures can compromise the health of the cells and tissues. The length of time needed to make the required observations will clearly dictate the kinds of measures taken to maintain the specimens. Experimental observations made over the course of many hours require a different level of specimen maintenance than those requiring only a few minutes. Under optimal conditions, it is possible to maintain viable tissues on the stage of a microscope for many hours—even days—while collecting valuable information on dynamic events in live cells and tissues.
How can biological samples be mounted and maintained to last long enough to make meaningful observations? This chapter offers some practical advice based on the authors’ experiences imaging neurons (see for example, O’Rourke et al. 1992; Dailey and Smith 1994, 1996; Dailey et al. 1994; Marrs et al. 2001; Zha et al. 2005; Ahmed et al., 2006), astrocytes (Benediktsson et al. 2005), and microglia (Dailey and Waite 1999; Stence et al. 2001; Dailey 2002; Grossmann et al. 2002; Petersen and Dailey 2004; Kurpius et al. 2006, 2007) in rodent brain tissue slices, as well as neurons in excised chick neural retinal preparations (Marrs et al. 2006). Although the specific requirements of different samples will vary, there are some general principles to consider when attempting to maintain viable specimens in the imaging setup (Dailey et al., 2006). The emphasis here is on simple, low-cost approaches for maintaining live mammalian brain cells and tissue slices in imaging setups.
KEEPING THE SPECIMEN ALIVE IN THE IMAGING SETUP
There are several considerations for maintaining live cells and tissues in the imaging setup, including temperature, pH, gas exchange, and light exposure. The relative importance of each of these factors varies with the particular specimen, the spatial and temporal sampling requirements, and the expected length of observation.
Medium Considerations
The medium in which the specimen is mounted must support the cells and tissues for the duration of the observation. When working with cultured cells and tissues, it is necessary to consider the changes that may occur when the sample is taken from the environment of a culture incubator to ambient air. Most bicarbonate-based culture media are designed to buffer an air environment composed of ~5% CO2, but in ambient air, these media rapidly (within ~5 minutes) become alkaline. Thus, by the time the sample is mounted, the gas content and pH of the culture medium may have changed substantially. Such changes should be monitored. For determining pH changes, a pH indicator (phenol red) can be included in the chosen medium, or pH indicator strips (colorpHast; e.g., EMD Chemicals) may be used to test the medium at different stages in the mounting and imaging procedure. In the authors’ experience, a saline solution (e.g., phosphate-buffered saline) is sufficient to maintain neonatal mammalian brain tissue slices for a few hours. Longer observations usually require the more substantial nutritional support of culture media. In addition, long-term imaging of mammalian brain tissues derived from more mature animals (>postnatal day [PND]7) may require continuous perfusion of oxygenated medium.
Mounting Live Specimens for Microscopic Observation
This section describes a simple mounting technique for rodent organotypic hippocampal slice cultures grown on filter culture membranes, a popular in vitro neural tissue slice preparation (Stoppini et al 1991; Gähwiler et al. 1997). These slice cultures are readily mounted and imaged because the culture membrane provides a stable substratum that can be secured within an imaging chamber without applying pressure directly to the tissues. The authors use translucent filter membrane inserts (Falcon 3090 or 3102) containing polyethylene terephthalate, track-etched porous membranes (1-micron pore size). Typically, two to five tissue slices are cultured on a single membrane insert. Although it is possible to image samples through a translucent filter membrane using low-magnification or long-working-distance lenses, the highest resolution will be achieved by mounting samples in a way that allows direct imaging without an intervening filter membrane. The mounting technique described here (Fig. 1) is especially useful for imaging tissues through a coverslip on an inverted microscope system, but it could be adapted for an upright system in conjunction with saline or water-dipping lenses.
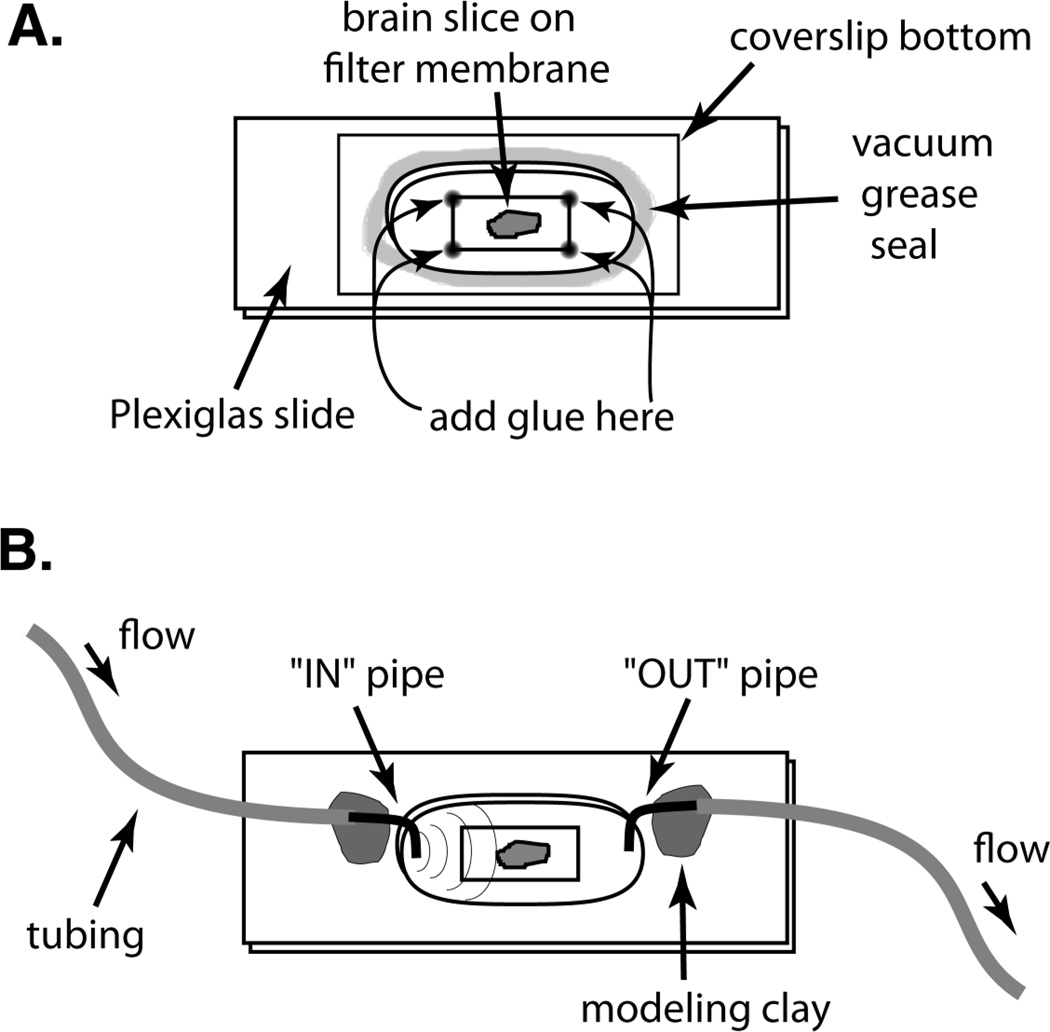
Figure 1.
A simple specimen chamber for live-cell or tissue imaging. (A) The specimen chamber is made by drilling out an elongated hole (1.5×3 cm) through a thin (3 mm thick) piece of Plexiglas. A rectangular coverslip (22×50 mm) is attached to the bottom of the Plexiglas slide with a ring of silicone vacuum grease. The Plexiglas slide and coverslip are firmly pressed together. The specimen is secured to the coverslip using a plasma/thrombin clot, netting, tape, glue, or small weight, depending on the specimen. Here, a brain slice culture growing on a filter membrane is shown attached to a coverslip via cyanoacrylate glue. A second Plexiglas slide may be added to increase the chamber volume. Then, the chamber is partially filled with medium such that the bottom of the chamber is covered. The chamber opening may be covered with a second glass coverslip or kept uncovered for perfusion. (B) Perfusate is delivered by gravity feed to one side of the open chamber and withdrawn by vacuum suction from the other side. The tips of two syringe needles are cut off, bent, and inserted into the ends of the tubing to serve as small “in” and “out” pipes. The pipes, which stick down into the chamber well, are held in place with tape or modeling clay. The “out” pipe works best if the tip of the needle is beveled, or if a small filter-paper wick is inserted into the end of the needle. This allows continuous withdrawal of perfusate, rather than a pulsed removal that can induce severe focus jumps. The position of the “out” pipe must be adjusted (raised) so as not to suck the chamber dry.
First, a membrane culture insert with attached organotypic tissue slices is removed from the culture well and the entire insert is placed upside down on a flat surface. A piece of membrane with attached slices is excised from the culture insert by making a rectangular cut through the membrane and around the slices using a razor blade or scalpel. The rectangular piece of membrane is then placed tissue side down in the center of a dry 22×50-mm glass coverslip. To minimize air pockets and bubbles, the membrane is lowered slowly onto the coverslip starting at one edge. A very small drop of cyanoacrylate glue is then applied to each of the corners of the membrane. Care is taken to avoid placing glue too near or on top of any tissue slices. The glue is allowed to dry for 10–20 sec. Then a custom-made Plexiglas slide (~3 mm thick) with an oblong hole (~1.5×3 cm) in the center is used to form a chamber around the tissue. First, a thin bead of silicone vacuum grease (e.g., Dow Corning) is laid down around the edge of the hole in the Plexiglas slide. [Note: A 3cc syringe, loaded from the back with vacuum grease, works well as a grease applicator. A 22 GA tapered polyethylene tip (Small Parts, Inc., #FFT-22-05) works well to lay down a line of grease.] The greased Plexiglas slide is then inverted and carefully positioned around the membrane rectangle and sealed to the coverslip by application of firm pressure. The vacuum grease forms a stable, waterproof seal between the Plexiglas slide and the coverslip, thus forming an open chamber containing the specimen. It is important that the piece of membrane and glue spots fit entirely inside the hole of the Plexiglas slide so that the grease ring is able to make a tight, leak-resistant seal. A second Plexiglas slide may be similarly sealed on top of the first to create a chamber with greater volume. HEPES-buffered culture medium (1–2 ml) is then added directly on top of the membrane to fill the chamber 1/2 to 2/3 full, with care to spread the medium across the entire floor of the well. After transferring the imaging chamber to the microscope stage, a second coverslip (22×40 mm or larger) is placed, without sealing, across the top of the well cavity to prevent evaporation of medium during imaging. Medium in the chamber cavity should not directly contact the top coverslip as this can increase focal drift during time-lapse experiments. Also, by not sealing the chamber top, it is possible to remove the top coverslip to add or replace the chamber medium during the course of the experiment. It is important to mount the samples swiftly to prevent evaporation and thus maintain proper salinity and pH of the medium.
For some short-term (<2 hours) time-lapse experiments, it may be possible to maintain the specimen in a closed-chamber configuration for the duration of the experiment. Larger volumes of chamber medium can support tissues for longer periods of time, although the pH of the chamber medium should be monitored. When rather large pieces (or several pieces) of tissue are confined to small volumes, the pH of the medium may significantly change (acidify) within a couple of hours as a consequence of tissue metabolism. This can cause run-down in cell behavior resulting, for example, in greatly reduced cell motility or migration. Mounting the specimen in a larger volume of medium may help, or even periodic exchange of medium may adequately support the sample for many hours (O’Rourke et al. 1992). An alternative solution for longer-term imaging experiments is to set up a continuous perfusion system. The rate of perfusion need not be very high—1–2 ml of HEPES-buffered culture medium per hour can support developing rodent brain tissue slices and slice cultures on the stage of the microscope for many hours (Dailey and Smith 1996). The challenge is to set up a perfusion system that does not leak (thus minimizing risk of damage to microscope components) and that does not induce focus jumps or drift. Drop-wise delivery of perfusate to the specimen chamber, as well as periodic sucking of medium from the chamber, can cause pressure changes that translate into focus jumps. A rather simple and inexpensive gravity-fed perfusion setup can be constructed mostly from common laboratory supplies (Fig. 2). This can be integrated into a microscope setup with an external stage heater, as shown in Figure 3. However, more elaborate chamber and perfusion systems can be constructed or purchased commercially (see Table 19.3 in Dailey et al., 2006). For a more thorough treatment of specimen chambers for live-cell imaging, see Dailey et al. (2009).
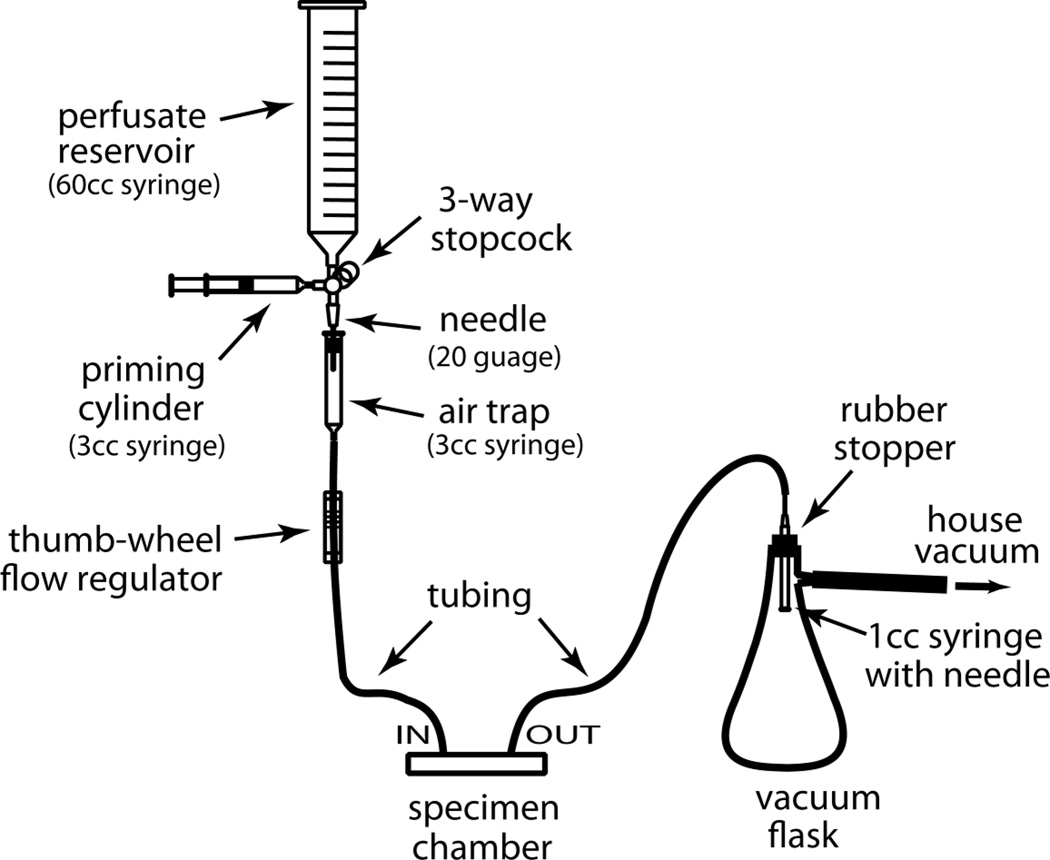
Figure 2.
A simple gravity-feed perfusion system constructed from common lab supplies. Perfusate is added to the reservoir (e.g., 60 cc syringe), and the system is primed to remove any air using a 3-cc syringe. The flow of perfusate is monitored by observing drip rates in the air trap (3-cc syringe), and the rate is controlled by a thumb-wheel flow regulator taken from a standard medical IV set. A vacuum flask serves as the waste container.
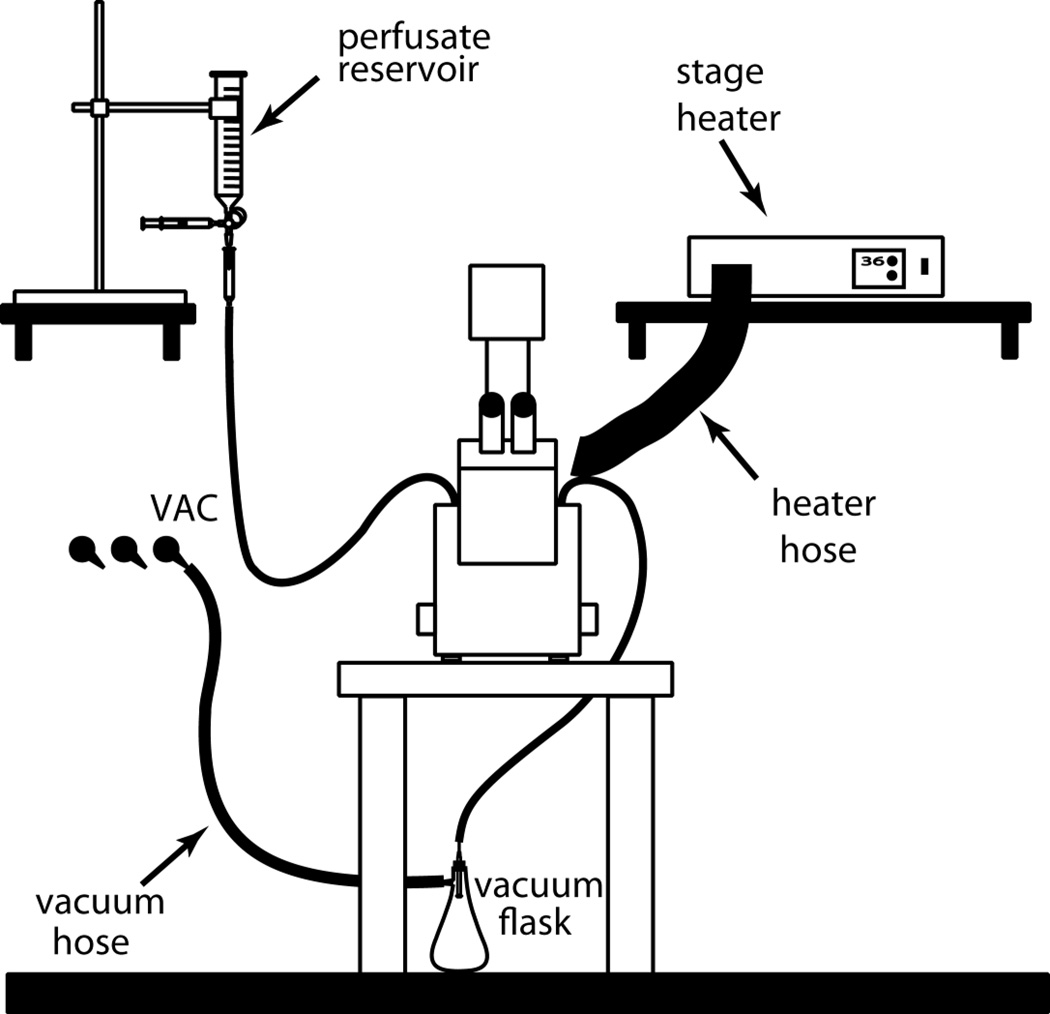
Figure 3.
A simple, integrated setup for live-cell imaging. A gravity-feed perfusion system and forced-air stage heater is shown in the context of an inverted microscope.
For mature mammalian brain tissues, it is usually necessary to provide continuous perfusion of oxygenated medium. Figure 4 shows a simple perfusion arrangement that could be used to maintain mature brain tissue slices. Whatever the exact chamber configuration, for lengthy time-lapse experiments it appears important that the imaging setup provide sufficient tissue oxygenation and removal of metabolic by-products such as CO2.
Figure 4.
Perfusion setup for imaging adult rodent brain tissue slices on an inverted microscope platform. Warm perfusate is maintained in a covered reservoir (1-liter glass bottle) sitting within a warm water bath. For perfusates requiring oxygenation, a tube (white arrowhead) is run from the gas tank (95% O2, not shown) into the reservoir perfusate, and the perfusate is continuously aerated with the gas. Perfusate is delivered to and removed from the specimen chamber using a two-channel, variable-speed pump (black arrowhead; e.g., Bioptechs) that maintains a constant rate of perfusate delivery and removal (~1 ml/min). The specimen chamber (arrow) holds ~2 ml of perfusate. Tubes running to and from the specimen chamber are taped to the mobile portion of the microscope stage to minimize drag. Used perfusate is aspirated from the chamber and pumped into a waste container. Care must be taken to avoid dripping or spilling perfusate on any microscope or electronic components. Here, a custom-made Plexiglas shield (beneath the pump) was built to prevent perfusate from leaking onto electronic components in the confocal scan head.
Temperature Considerations
Maintaining the specimen at a physiological temperature during the imaging session increases experimenter confidence that the observed phenomena truly reflect native behaviors and not artifacts of the experimental arrangement. Indeed, many dynamic cellular activities are highly sensitive to temperature. In the case of mammalian cells and tissues, it is often desirable to maintain a constant temperature of 34–36 degrees C. This can be accomplished by perfusing warmed medium through the specimen chamber, heating the stage, or by blowing warm air onto the preparation. There are very elaborate ways to accomplish this (see, e.g, Dailey et al. 2009), although it is also possible to put together a crude, inexpensive arrangement that will adequately maintain constant temperature for many hours. For example, a standard inexpensive hair dryer can be used to blow warm air onto the microscope stage. Make sure, however, that the sample is protected from evaporation, which may occur when blowing warm, nonhumidified air over the specimen chamber. Evaporation is reduced or prevented by using a closed chamber configuration, or by continuous replacement (perfusion) of the chamber medium. Blowing warm air from the objective side of the chamber (on an inverted microscope) will help reduce airflow over the chamber and will also serve to more efficiently heat the objective, which may be a significant heat sink when using oil-coupled objectives. On inverted platforms, use of (nondipping) water-or saline-immersion lenses in combination with forced-air stage heating can lead to rapid evaporation of the immersion medium and loss of the image, but this problem can be overcome by using a non-drying synthetic immersion medium that has an index of refraction (IR) matched to water (IR=1.33; R.P. Cargille Laboratories, Cedar Grove, NJ).
Unfortunately, heating the specimen can introduce further complications such as increased rate of fluorescence photobleaching, or focus drift due to thermal expansion across a chamber or stage that is heated nonuniformly. Preheating the stage and chamber can minimize temperature-induced focus drift. The medium should also be preheated to avoid the formation of bubbles within the chamber when cold medium is heated.
Collecting Images
How are images with sufficient spatial and temporal resolution collected without inducing changes in cell structure or physiology? Photodamage may occur when imaging live, fluorescently labeled specimens. The cellular consequences of photodamage can appear within seconds, and the effects of a single photodamaging event can persist for many minutes or even hours. A compromise between the desired spatial and temporal resolution, and the risk of photodamage to the sample, must be reached. This is a particularly important issue when performing extended time-lapse imaging. Increasing the incident illumination (until fluorophore saturation) can improve the signal-to-noise ratio (Rs/n) of the image and permit higher rates of image collection for a given Rs/n. However, most fluorescently stained samples cannot tolerate continuous light exposure for very long. The problem is particularly acute when attempting to sample a 3D tissue volume over time (4D imaging). It is often necessary to sacrifice some spatial and/or temporal resolution, but the experimental goals will dictate what must be sacrificed. For example, for fast physiological events (such as calcium spikes) it may be necessary to reduce exposure time and sacrifice some spatial resolution in order to collect images at the fastest rate possible. On the other hand, for relatively slow events (such as long-distance migration of cells over periods of hours), the required long imaging sessions can be achieved only at the cost of reduced temporal or spatial resolution.
These considerations raise the question of whether it is better to keep the intensity of incident illumination low while increasing exposure time, or whether it is preferable to turn up the light and reduce the exposure time. The incident illumination intensity will be a primary determinant of image quality and temporal sampling. In scanning imaging systems, image quality (Rs/n) can be improved by increasing pixel dwell time or by frame or line averaging. Unfortunately, there is no clear consensus as to whether it is better (for the specimen) to absorb less intense light over a longer period of time or to allow the sample sufficient time to “recover” between events of higher illumination intensity. In live specimens, movement of organelles and cells can “smear” the image when several frames are averaged over the period of a few seconds. This is less of an issue when imaging relatively slow events such as cell migration or axon and dendrite growth, but it is problematic when attempting to image fast physiological events such as calcium spikes, which tend to be averaged out. Most newer scanning imaging systems are capable of “line averaging” (resampling line by line) or slowing the scan rate (increasing pixel dwell time), which helps minimize the smearing of fast events while maintaining high Rs/n. Suffice it to say that the optimal imaging parameters vary with different specimens and fluorescent tags, and they need to be determined empirically for each biological and imaging system.
Photodynamic damage of the sample may be reduced somewhat by the inclusion of antioxidants such as ascorbate (vitamin C, 25–30 mM) in the chamber medium. However, the pH of the medium should be monitored because addition of significant amounts of ascorbate will acidify the medium.
KNOWING WHEN THE SPECIMEN IS UNHEALTHY
Deterioration of specimens in the imaging setup will, of course, limit the usefulness of observations. But what criteria might be used to determine the health of the specimen? The primary consideration would seem to be whether the essential features of cell morphology, physiology, and dynamic behavior change under “control” conditions during the imaging session. If abnormal morphological or physiological features are apparent in the first images, then the culturing, staining, or mounting conditions must be altered. If abnormal features appear only after the imaging session has begun, one would suspect a problem with the imaging protocol or possibly the mounting procedure.
What are the indicators of ill health? Different cells vary in their responses to different conditions. Neurons are especially sensitive to changing conditions. When stained with fluorescent membrane dyes, nerve cell processes can show abnormal features such as beading, blebbing, and triangulation of the branch points. The same cell types in different samples can show different patterns of beading and blebbing that may indicate different stress factors. Declining health can also affect the rates of cell growth and motility, usually resulting in a general decrease in activity. However, ill health is not always indicated by a decline in cellular activity. For example, compromised cells can exhibit a marked increase in Brownian-like organelle motions. Likewise, increases in physiological activity (calcium spikes) are sometimes seen when imaging cells loaded with a fluorescent calcium indicator (e.g., fluo-3).
Whatever morphological or physiological criteria are used, a critical question is whether the specimen looks and behaves in a similar way throughout the course of a control imaging session. For fluorescence imaging, it is important to remember that photodynamic damage can occur long before significant photobleaching is observed. To determine if the light load contributes to specimen deterioration, it can be helpful to compare the morphology or physiology of experimental specimens with control specimens that receive similar treatments (i.e., staining, mounting, drug application, etc.) but which are imaged only at the end of a comparable experimental period. It can be especially instructive to sacrifice a few good-looking specimens to clearly define the sequence of changes that occur when the sample is subjected to phototoxic stress. Appropriate control experiments will help identify which cell behaviors are due to the experimental treatment and which are consequences of less-than-optimal imaging conditions.
CONCLUSION
Even as imaging technologies improve, the problems encountered in live-cell imaging can be daunting. Researchers are often limited by the photosensitivity of the biological sample, especially when fluorescent probes are used. Improvements in imaging system performance have allowed higher-quality images to be acquired with greater frequency, but the limits of the biological systems under study will always be tested. Increasingly, imaging involves the simultaneous use of two or three different fluorophores in individual cells. It is necessary, therefore, to become thoroughly familiar with the limitations of both the imaging and biological systems so that sound conclusions about the observed biological phenomena can be made.
ACKNOWLEDGMENTS
Development of the techniques described herein was facilitated by grants from the National Institutes of Health (NS37159, NS43468, and AA018823), the Whitehall Foundation (#S98-6), the Roy J. Carver Charitable Trust, the American Heart Association (0950160G), and a seed grant from the American Cancer Society (#IN-122R) administered through the University of Iowa Cancer Center.
REFERENCES
- Ahmed R, Zha X-M, Green SH, Dailey ME. Synaptic activity and F-actin coordinately regulate CaMKIIα localization to dendritic postsynaptic sites in developing hippocampal slices. Molec. Cell. Neurosci. 2006;31:37–51. doi: 10.1016/j.mcn.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Benediktsson AM, Schachtele SJ, Green SH, Dailey ME. Ballistic labeling and dynamic imaging of astrocytes in organotypic hippocampal slice cultures. J. Neurosci. Methods. 2005;141:41–53. doi: 10.1016/j.jneumeth.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Dailey ME. Optical imaging of neural structure and physiology: Confocal fluorescence microscopy in live brain slices. In: Toga A, Mazziotta J, editors. Brain mapping: The methods. 2nd edition. San Diego: Elsevier; 2002. pp. 49–76. [Google Scholar]
- Dailey ME, Smith SJ. Spontaneous Ca2+ transients in developing hippocampal pyramidal cells. J. Neurobiol. 1994;25:243–251. doi: 10.1002/neu.480250305. [DOI] [PubMed] [Google Scholar]
- Dailey ME, Smith SJ. The dynamics of dendritic structure in developing hippocampal slices. J. Neurosci. 1996;16:2983–2994. doi: 10.1523/JNEUROSCI.16-09-02983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey ME, Waite M. Confocal imaging of microglial cell dynamics in hippocampal slice cultures. Methods. 1999;18:222–230. doi: 10.1006/meth.1999.0775. [DOI] [PubMed] [Google Scholar]
- Dailey ME, Buchanan J, Bergles D, Smith SJ. Mossy fiber growth and synaptogenesis in rat hippocampal slices in vitro. J. Neurosci. 1994;14:1060–1078. doi: 10.1523/JNEUROSCI.14-03-01060.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey ME, Manders E, Soll D, Terasaki M. Confocal microscopy of live cells. In: Pawley JB, editor. Handbook of Biological Confocal Microscopy. 3rd Edition. New York: Plenum; 2006. [Google Scholar]
- Dailey ME, Focht DC, Khodjakov A, Rieder CL, Spring KR, Claxton NS, Olenych SG, Griffin JD, Davidson MW. [Accessed 10/15/09];Culture Chambers for Live-Cell Imaging. 2009 http://microscopyu.com/articles/livecellimaging/culturechambers.html.
- Gähwiler BH, Capogna M, Debanne D, McKinney RA, Thompson SM. Organotypic slice cultures: A technique has come of age. Trends Neurosci. 1997;20:471–477. doi: 10.1016/s0166-2236(97)01122-3. [DOI] [PubMed] [Google Scholar]
- Grossmann R, Stence N, Carr J, Fuller L, Waite M, Dailey ME. Juxtavascular microglia migrate along brain capillaries following activation during early postnatal development. Glia. 2002;37:229–240. [PubMed] [Google Scholar]
- Kurpius D, Wilson N, Fuller L, Hoffman A, Dailey ME. Early activation, motility, and homing of neonatal microglia to injured neurons does not require protein synthesis. Glia. 2006;54:58–70. doi: 10.1002/glia.20355. [DOI] [PubMed] [Google Scholar]
- Kurpius D, Nolley EP, Dailey ME. Purines induce directed migration and rapid homing of microglia to injured pyramidal neurons in developing hippocampus. Glia. 2007;55:873–884. doi: 10.1002/glia.20509. [DOI] [PubMed] [Google Scholar]
- Marrs G, Green SH, Dailey ME. Rapid formation and remodeling of postsynaptic densities in developing dendrites. Nat. Neurosci. 2001;4:1006–1013. doi: 10.1038/nn717. [DOI] [PubMed] [Google Scholar]
- Marrs G, Honda T, Fuller L, Thangavel R, Balsamo J, Lilien J, Dailey ME, Arregui C. Dendritic arbors of developing retinal ganglion cells are stabilized by β1-integrins. Molec. Cell. Neurosci. 2006;32:230–241. doi: 10.1016/j.mcn.2006.04.005. [DOI] [PubMed] [Google Scholar]
- O’Rourke NA, Dailey ME, Smith SJ, McConnell SK. Diverse migratory pathways in the developing cerebral cortex. Science. 1992;258:299–302. doi: 10.1126/science.1411527. [DOI] [PubMed] [Google Scholar]
- Petersen MA, Dailey ME. Diverse microglial motility behaviors during clearance of dead cells in hippocampal slices. Glia. 2004;46:195–206. doi: 10.1002/glia.10362. [DOI] [PubMed] [Google Scholar]
- Stence N, Waite M, Dailey ME. Dynamics of microglial activation: A confocal time-lapse analysis in hippocampal slices. Glia. 2001;33:256–266. [PubMed] [Google Scholar]
- Stoppini L, Buchs P-A, Muller D. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Zha X-M, Green SH, Dailey ME. Regulation of hippocampal synapse remodeling by epileptiform activity. Molec. Cell. Neurosci. 2005;29(4):494–506. doi: 10.1016/j.mcn.2005.04.007. [DOI] [PubMed] [Google Scholar]