Abstract
Mitotic spindle position is controlled by interactions of cortical molecular motors with astral microtubules. In animal cells, Partner of Inscuteable (Pins) acts at the cortex to coordinate the activity of Dynein and Kinesin-73 (Khc73; Kif13B in mammals) to orient the spindle. Though the two motors move in opposite directions, their synergistic activity is required for robust Pins-mediated spindle orientation. Here we identify a physical connection between Dynein and Khc73 that mediates cooperative spindle positioning. Khc73’s motor and MBS domains link Pins to microtubule plus ends, while its stalk domain is necessary for Dynein activation and precise positioning of the spindle. A motif in the stalk domain binds, in a phospho-dependent manner, 14-3-3ζ, which dimerizes with 14-3-3ε. The 14-3-3ζ/ε heterodimer binds the Dynein adapter NudE to complete the Dynein connection. The Khc73 stalk/14-3-3/NudE pathway defines a physical connection that coordinates the activities of multiple motor proteins to precisely position the spindle.
INTRODUCTION
Positioning of the mitotic spindle is critical for biological processes that require precisely oriented cell divisions, such as cellular differentiation, embryogenesis, and organogenesis (Toyoshima and Nishida, 2007). The position of the spindle can be directed by cortical proteins that bias the division orientation in response to extrinsic or intrinsic cues (Gillies and Cabernard, 2011; Morin and Bellaiche, 2011; Siller and Doe, 2009). Cortical spindle position regulators coordinate cytoskeletal elements such as microtubule motors that generate forces on astral microtubules. Here we examine how one such cortical spindle position regulator, Partner of Inscuteable (Pins; GPR-1/2 in C. elegans; LGN or mPins in mammals), regulates spindle orientation in metazoans.
Pins is a modular protein that regulates spindle positioning in a diversity of cell types by orchestrating the control of at least two motor proteins (David et al., 2005; Du and Macara, 2004; Schaefer et al., 2000). Pins’ architecture allows for integration of upstream signals, and activation of multiple downstream pathways (Du and Macara, 2004; Siegrist and Doe, 2005; Siller et al., 2006; Wee et al., 2011). GoLoco motifs within the Pins COOH-terminus bind the heterotrimeric G-protein subunit, Gai, which is thought to direct the division orientation (Du and Macara, 2004). NH2-terminal TPR motifs (PinsTPR) bind Mud (NuMA in mammals), which in turn binds the minus-end directed microtubule motor Dynein (Kotak et al., 2012; Nipper et al., 2007; Siller et al., 2006; Wang et al., 2011). Finally, a “Linker” motif (Pinslinker) between the TPRs and GoLocos (Pinslinker) that is phosphorylated by the mitotic kinase Aurora A (Johnston et al., 2012) binds the tumor suppressor Discs large (Dlg) which recruits the kinesin Khc73 (Siegrist and Doe, 2005; Yamada et al., 2007). Thus, Pins responds to a cellular cue (Gai), by activating two microtubule motor pathways, one minus-end directed (Dynein) and one plus-end directed (Khc73).
The PinsTPR and Pinslinker pathways act synergistically to robustly position the spindle (Johnston et al., 2009; Siegrist and Doe, 2005). The Pinslinker pathway is thought to function through a microtubule capture mechanism, and can partially orient the spindle on its own. The PinsTPR pathway, on the other hand, is not sufficient for any spindle orienting activity, but generates force on astral microtubules through Dynein for precise positioning when functioning along with Pinslinker (Figure 1A). Synergy requires linkage of both pathways, as PinsTPR must be coupled in cis to Pinslinker for robust alignment of the spindle (Johnston et al., 2009).
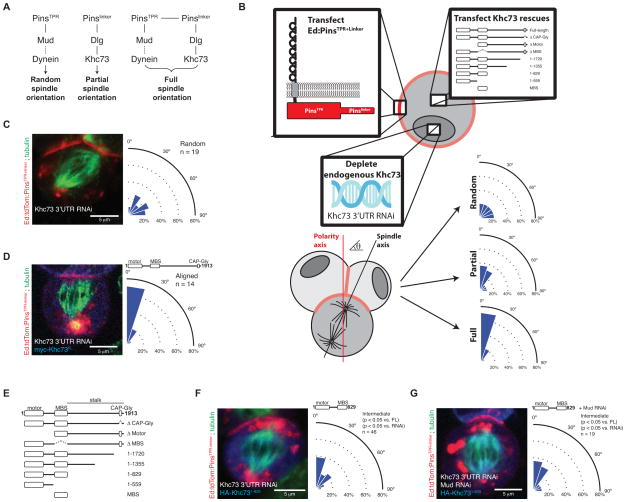
Figure 1. Determinants of Khc73-mediated spindle orientation.
(A) Pathways of Pins-mediated spindle orientation. The Pinslinker pathway alone has partial spindle orientation through the activity of Discs large (Dlg) and Khc73. The PinsTPR pathway terminates in Dynein through Mushroom body defect (Mud; NuMA in vertebrates) and is necessary for full activity but is insufficient for any activity by itself. Together, the PinsTPR and Pinslinker pathways synergize to induce completely aligned spindles.
(B) Schematic of the induced polarity spindle orientation assay in S2 cells. S2 cells are transfected with Ed:PinsTPR+linker and a Khc73 rescue construct. Transfected S2 cells are treated with RNAi targeted for the 3′UTR of Khc73, thereby depleting endogenous Khc73. Intercellular interactions sequester Ed:PinsTPR+linker to points of cell-cell contact. The spindle angle is measured relative to the polarity axis, determined by Ed:PinsTPR+linker crescent, and is categorized as a random, intermediate, or aligned phenotype based on statistical comparisons to controls.
(C) Khc73 3′UTR RNAi in an Ed:PinsTPR+linker context results in complete loss of spindle orientation. The resulting spindle angle distribution is used as the control for a random distribution. All S2 images shown are oriented such that the polarity axis dictated by the Ed:PinsTPR+linker crescent, located at the top of the cell, is vertically aligned with the image.
(D) The knockdown of endogenous Khc73 can be rescued by expression of a transgenic full-length Khc73. The resulting spindle angle distribution is used as the control for an aligned distribution. The domain architecture of Khc73 is shown.
(E) The domain architecture of Khc73 proteins analyzed for rescue of endogenous Khc73 RNAi in Ed:PinsTPR+linker spindle orientation assay. Amino acids 830-1913 define Khc73’s stalk region.
(F) Khc73’s motor and MBS domains are sufficient to yield the partial spindle orienting activity suggesting that, although PinsTPR+linker is present on the cortex, only the Pinslinker pathway is active. Statistical significance compared to full-length Khc73 (FL) and Khc73 RNAi were determined using t-tests.
(G) Only the Pinslinker pathway is active when Khc73 lacks it stalk domain. Mud RNAi (which inactivates the PinsTPR pathway) was used to show that there is no residual PinsTPR pathway activity in cells where Khc73 lacks its stalk domain. Statistical significance compared to full-length Khc73 (FL) and Khc73 RNAi were determined using t-tests. P > 0.5 vs. condition in figure 1F. See also Figure S1, Movie S1, and Movie S2.
The molecular mechanism by which the PinsTPR and Pinslinker pathways coordinate motor activity to orient the mitotic spindle remains unclear. A number of possibilities exist for the source of the synergy—the Pinslinker pathway could be required for the recruitment and activation of the PinsTPR pathway, or the Pinslinker pathway could directly activate the PinsTPR pathway through some allosteric mechanism. Alternatively, the activation of the PinsTPR pathway by the Pinslinker pathway could occur through the crosstalk of any of the downstream components of either pathway, including the two terminating microtubule motor proteins Khc73 and Dynein.
Dynein adapters and cofactors are potential mediators of crosstalk between microtubule motor pathways. Several Dynein associated proteins, including Dynactin, Lis1, and NudE, are known to interact with proteins that are not typical members of the Dynein complex (Kardon and Vale, 2009). Dynactin interacts with Kinesin-2 and Kinesin-5 (Berezuk and Schroer, 2007; Blangy et al., 1997; Deacon et al., 2003), which suggests that Dynein complex components might serve as a link between the opposing motors, at least in certain contexts. Here we sought to identify the components of two Pins-mediated spindle orientation pathways that mediate crosstalk, with the goal of identifying factors the link Dynein and the kinesin Khc73. Defining the connection between these pathways is a foundation for understanding the coordinated action of minus- and plus-end directed motors during spindle orientation.
RESULTS
The Khc73 stalk domain mediates synergy between Pins spindle orientation pathways
To understand how the PinsTPR and Pinslinker pathways cooperate to generate robust spindle positioning, we first sought to identify the components that mediate synergy. To determine the spindle positioning activities of the Pins pathways, we used the “induced polarity” spindle orientation assay that exploits the homophilic cell adhesion protein Echinoid (Ed) (Johnston et al., 2009). In this assay, Ed with its cytosolic domain replaced with PinsTPR+linker forms oligomers at sites of cell-cell contact, effectively polarizing PinsTPR+linker. Mitotic S2 cells harboring Ed:PinsTPR+linker crescents are then scored for spindle alignment relative to the induced PinsTPR+linker crescents (Figure 1B, methods). We took advantage of the observation that Pinslinker by itself has partial activity, PinsTPR has no activity, and both combined yield full activity (Figure 1A) (Johnston et al., 2009). Thus, we reasoned that inactivation of components that link the two pathways would reduce the full activity of PinsTPR+linker to that of Pinslinker alone. The induced cell polarity S2 cell system therefore allows for the rapid analysis of pathway interactions as knockdown of PinsTPR components yields a Pinslinker phenotype and knockdown of Pinslinker components results in full loss of spindle positioning activity (Figure 1A). In this manner, components that affected the connection between the two pathways, but not intrinsic Pinslinker function, could be identified with this system.
The Pinslinker components that link to PinsTPR/Mud/Dynein could be within Pins itself, the adapter Dlg, the kinesin Khc73, or some other component that has yet to be identified. A structure/function analysis of Dlg revealed that the GK domain, which couples to Khc73 (Siegrist and Doe, 2005), is sufficient for Pinslinker activity when polarized on the cell cortex (Johnston et al., 2009). Furthermore, the Dlg GK domain alone restores full Ed:PinsTPR+linker activity when endogenous Dlg is knocked down by RNAi (Johnston et al., 2009). Taken together, these observations suggest that the connection to the PinsTPR lies downstream of Dlg.
The only known Pinslinker pathway component downstream of Dlg is the kinesin Khc73. However, little is known about the functional requirements for Khc73 in spindle orientation. We performed a thorough analysis of Khc73 in the context of the PinsTPR+linker spindle orientation assay to potentially identify variants that support Pinslinker, but not PinsTPR pathway activity. This approach has the advantage that we can activate select spindle orientation pathways, unlike in vivo systems such as the Drosophila neuroblast, where spindle orientation is highly redundant (Siegrist and Doe, 2005). We used a rescue assay in which we knocked down endogenous Khc73 in S2 cells using a double-stranded RNA targeted to its 3′ UTR (Figure 1B). We confirmed the knockdown by measuring spindle orientation, where successful Khc73 depletion results in complete randomization of spindle angles (Figure 1C), the phenotype of loss of a Pinslinker pathway component. To establish the feasibility of the rescue assay, we confirmed that full PinsTPR+linker spindle orientation can be restored in the presence of Khc73 3′ UTR RNAi by expressing full length Khc73 by transient transfection (Figure 1D). The full rescue we observed in this system allowed us to test Khc73 variants lacking specific domains (Figure 1E) for their ability to mediate spindle orientation by PinsTPR+linker.
We first performed a domain deletion analysis and determined that all domains, except the CAP-Gly domain (Khc73CAP-Gly), were required for full Pins activity (Figure 1E, F, Figure S1A-D). However, while nearly all Khc73 domains are required for full (PinsTPR+linker) activity, the motor domain (Khc73motor) and MAGUK Binding Stalk (MBS) domain (Khc73MBS) together are sufficient to restore intermediate activity (statistically distinct from the Khc73 RNAi or FL rescue distributions; Figure 1F, Figure S1C, D). This result suggests that Khc73 sequences after the MBS domain are not required for intrinsic Pinslinker activity, but could mediate crosstalk between Pinslinker and PinsTPR pathways. To determine if the partial restoration of Pins activity by Khc73motor+MBS is indeed due to sole activation of the Pinslinker pathway and not attenuated activity of both PinsTPR+linker pathways, we examined whether inactivating PinsTPR (using Mud RNAi) influences Khc73motor+MBS activity. We observed that Mud knockdown reduced PinsTPR+linker activity as previously observed (Johnston et al., 2009) (Figure S1E), but had no effect on Khc73motor+MBS activity (Figure 1G). These results, taken with Khc73’s domain architecture, indicate that regions between the MBS and CAP-Gly domains mediate PinsTPR and Pinslinker pathway synergy.
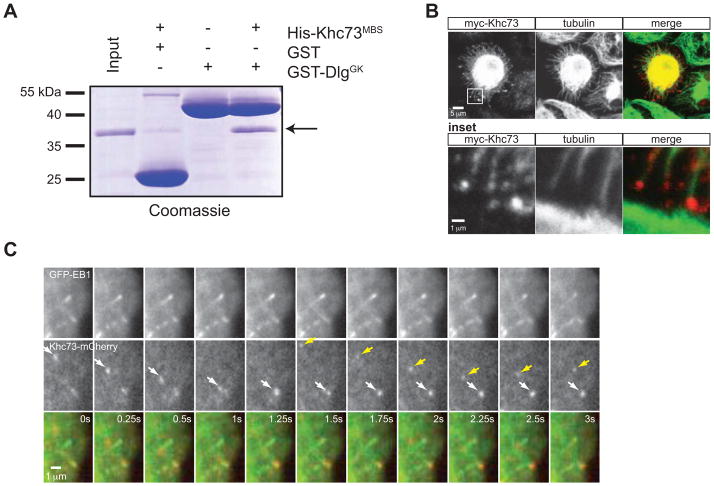
The Khc73 motor and MBS domains connect Dlg to microtubules
The Khc73motor and Khc73MBS domains are sufficient to restore Pinslinker activity, but the mechanism by which these domains position the spindle remains unclear. The simplest explanation for the function of these domains in Pins-mediated spindle orientation is a “connector” model, where Khc73 acts as a link between the cortex and microtubules through the Khc73MBS-Dlg and Khc73motor-MT interactions. To test this model, we first performed in vitro binding experiments using purified components and found that the Khc73MBS domain interacts directly with Dlg’s GK domain (Figure 2A). Khc73’s microtubule motility has been reported (Huckaba et al., 2011), and we also found that Khc73 localizes to the ends of microtubules in fixed stains (Figure 2B) and directly tracks along microtubules at an average rate of 0.76 ± 0.26 μm/sec (Figure 2C, movie S1). By simultaneously imaging with EB1-GFP, we also found that Khc73 stops at the plus ends of microtubules (Figure 2C, movie S2), as we observe a small difference between EB1’s velocity (average rate of 0.11 ± 0.08 μm/sec) and the velocity of Khc73 stopped at MT ends (average rate of 0.06 ± 0.04 μm/sec). The functional spindle orientation assay, in vitro binding assay, and immunofluorescence data support a model in which Khc73’s N-terminus acts as a direct link between the cortex (Dlg) and the spindle (astral microtubule plus ends).
Figure 2. Khc73’s motor and MBS domains provide a physical link between the cortex and microtubule plus ends.
(A) Purified Khc73’s MBS domain interacts directly with Dlg’s GK domain.
(B) Fixed interphase S2 cells expressing myc-Khc73FL (plated on ConA-coated coverslips) reveal Khc73’s localization to ends of microtobules. An expanded view of the box shown in the upper panel is shown as “inset”.
(C) Live imaging analysis by TIRF microscopy of interphase S2 cells expressing GFP-EB1 and Khc73FL-mCherry show Khc73 localizing to plus ends of microtubules (arrows show the progress of two different Khc73 signals). EB1 was used to mark plus-ends of microtubules. Cells were plated on ConA-coated chambers. Motile Khc73-mCherry travels at 0.76 ± 0.26 μm/sec, whereas “stopped” Khc73-mCherry moves at 0.06 ± 0.04 μm/sec, which varies slightly from a GFP-EB1 comet tail velocity of 0.11 ± 0.08 μm/sec. Mean ± S.D. shown for n = 5 of each. See also Figure S2.
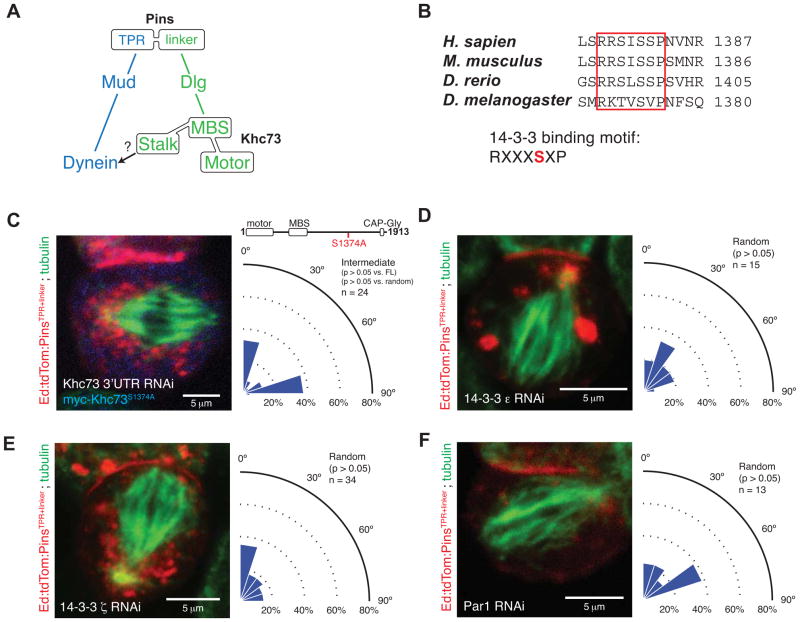
A 14-3-3 Binding Motif in the Khc73 stalk connects Pins spindle orientation pathways
Although the Khc73motor and Khc73MBS domains are sufficient to restore Pinslinker activity, the C-terminal stalk (amino acids 830-1913) is required for full activity (Figures 1F, 3A). We analyzed this region in more detail to determine how it mediates synergy between Pins spindle orientation pathways. Although the stalk region is generally uncharacterized, it does contain a conserved 14-3-3 binding motif. This motif was initially identified in the mammalian Khc73 ortholog, KIF13B, and was found to be required for association with mammalian 14-3-3β (Yoshimura et al., 2010) (Figure 3B). Furthermore, the 14-3-3 site in KIF13B was found to be phosphorylated by Par-1, a kinase implicated in the regulation of asymmetric cell division (Gonczy, 2008; Tabler et al., 2010). Finally, because 14-3-3 proteins, also known as Par-5, are known to regulate cell divisions (Benton and St Johnston, 2003; Hao et al., 2010), we hypothesized that the 14-3-3 binding motif in Khc73’s stalk region could couple the two Pins spindle orientation pathways.
Figure 3. Khc73 contains a conserved 14-3-3 binding motif that is necessary for Pins-mediated spindle positioning.
(A) Model for the role of the Khc73 stalk domain in Pins-mediated spindle orientation. An unknown factor links the stalk domain to the Mud/Dynein pathway.
(B) Khc73 contains a conserved 14-3-3 binding motif in its C-terminal stalk. The serine residue highlighted in red is phosphorylated. 14-3-3 protein interactions are typically phosphoregulated, although 14-3-3 proteins can bind to non-phosphorylated motifs.
(C) A Khc73 mutant containing a serine-to-alanine mutation in its 14-3-3 motif cannot rescue endogenous Khc73 knockdown in Pins-mediated spindle positioning. This mutant exhibits a novel intermediate activity phenotype in which the spindle is either aligned or orthogonal relative to cortical Ed-Pins. The statistical significance compared to full-length Khc73 (FL) was determined using a t-test, and the comparison to a random distribution was done using a chi-square analysis.
(D) Knockdown by RNAi of endogenous 14-3-3ε in S2 cells expressing Ed:PinsTPR+linker reduces Pins spindle-orientation activity to random. The statistical significance compared to Khc73 RNAi was determined using a t-test.
(E) Knockdown by RNAi of endogenous 14-3-3ζ in S2 cells expressing Ed:PinsTPR+linker reduces Pins spindle-orientation activity to random. The statistical significance compared to Khc73 RNAi was determined using a t-test.
(F) Knockdown by RNAi of endogenous Par-1 in S2 cells expressing Ed:PinsTPR+linker reduces Pins spindle-orientation activity to random. The statistical significance compared to Khc73 RNAi was determined using a t-test.
To determine whether the 14-3-3 binding motif in Khc73’s stalk region (RKTVSVP) is required for Pins-mediated spindle positioning, we determined if Khc73 lacking this motif could mediate spindle alignment with Ed-PinsTPR+linker crescents. We tested a variant in which the putative phosphorylation site was inactivated (S1374A; RKTVAVP) for its ability to mediate spindle orientation. Unlike wild-type Khc73, which fully rescues endogenous Khc73 RNAi, Khc73 containing the S1374A mutation is unable to fully rescue spindle alignment to Ed-PinsTPR+linker (Figure 3C). Instead, cells expressing this protein exhibit a novel, intermediate activity phenotype in which spindle angles are bimodally distributed between fully-aligned or fully-orthogonal orientations (see Discussion).
We also tested directly whether both Drosophila 14-3-3 proteins, 14-3-3ε and 14-3-3ζ, are required for Pins-mediated spindle positioning. We observed that knockdown of either paralogue by RNAi leads to near-random spindle orientation activity (Figure 3D, E), consistent with a role for these proteins in connecting the PinsTPR and Pinslinker pathways, and possibly other roles in the process. Given that the 14-3-3 binding motif in Khc73’s stalk region is required for Pins-mediated spindle positioning, we also tested the role of its putative kinase, Par-1, in Pins-mediated spindle positioning by measuring spindle orientation in cells where Par-1 had been inactivated by RNAi. Par-1 is also completely required for PinsTPR+linker–mediated spindle positioning, as these cells exhibit randomized spindle orientation (Figure 3F).
The complete loss of spindle orientation activity in cells lacking both 14-3-3 proteins and Par-1 suggest that they have other roles in spindle orientation besides connecting the PinsTPR and Pinslinker pathways. Par-1 is essential for several cellular processes including the maintenance of the dynamic state of microtubules (reviewed in (Hayashi et al., 2012), and among its diverse set of substrates are several microtubule-associated proteins including the microtubule stabilizing protein Tau (reviewed in (Drewes et al., 1997; Matenia and Mandelkow, 2009)). Also essential for several cellular functions, 14-3-3 proteins are ubiquitous adapter proteins that are involved in cell cycle control (reviewed in (Gardino and Yaffe, 2011)), microtubule maintenance (Zhou et al., 2010), and spindle assembly and regulation (De and Kline, 2013). Because of their various roles in spindle-related processes, disruption of the 14-3-3 proteins and Par-1 may have compounding effects on Pins-mediated spindle positioning, producing the random phenotype we observe.
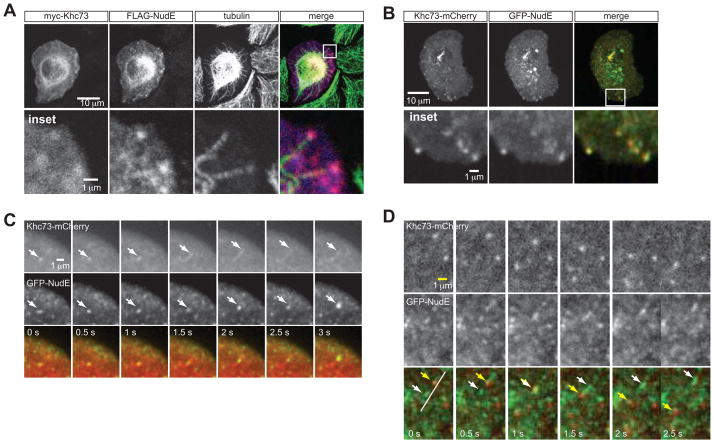
NudE is a spindle orientation factor that migrates with Khc73 along microtubules
The requirement for the 14-3-3 binding motif in Khc73’s stalk for Pins pathway synergy prompted us to examine whether the motif could be a connection node from Pinslinker to the PinsTPR pathway. Mammalian 14-3-3 proteins have several binding partners including Khc73 ortholog KIF13B (Yoshimura et al., 2010) and NudE, an essential Dynein co-factor (Johnson et al., 2010; Kardon and Vale, 2009). Mammalian 14-3-3 was shown to interact with NudE within its unstructured serine/threonine-rich C-terminus; this interaction depends on NudE phosphorylation by AuroraA (Johnson et al., 2010). Because both 14-3-3 and NudE are conserved in Drosophila, we hypothesized that NudE could be part of the bridge between the two Pins spindle orientation pathways.
We first determined the localization of NudE in S2 cells and observed that, although it is diffusely localized throughout the cell, it co-localizes with Khc73FL at the ends of microtubules in fixed cells (Figure 4A, B). TIRF live imaging analysis of NudE in S2 cells revealed that NudE is highly dynamic and when expressed with Khc73FL-mCherry, GFP-NudE can be seen to move with the motor protein in S2 cells (Figure 4C, 5A, quantified in Figure 5D, movie S3). Another population of NudE moves in the opposite direction from Khc73 along the same linear path (Figure 4D, movie S4), however, suggesting that NudE may be trafficked by Dynein moving toward microtubule minus-ends. Because Khc73 is known to interact with Rab5, an early endosome marker (Huckaba et al., 2011), we tested whether Rab5 plays a role in Pins-mediated spindle positioning and whether NudE/Khc73 co-localization was occurring at Rab5-containing vesicles. We first tested whether Rab5 is required for Pins-mediated spindle positioning, but found that Rab5 RNAi interfered with Ed:PinsTPR+linker crescent formation (supplemental information, Figure S4A, B) and also delayed the growth of S2 cells (supplemental information, Figure S4D). However, we analyzed NudE and Rab5 localization by live imaging and found that there is little overlap between these two proteins suggesting that NudE does not localize to Rab-5 containing vesicles (Figure S3A, C). We also tested the effects of Rab5 RNAi on NudE and Khc73 localization and determined that Rab5 is not required for NudE/Khc73 co-localization (Figure S3B, C). We conclude from our live imaging analysis that NudE and Khc73 co-migrate towards microtubule plus ends in a Rab5-independent manner, suggesting that NudE migrates with Khc73.
Figure 4. NudE migrates with Khc73 in S2 cell.
(A) Fixed interphase S2 cells transfected with myc-Khc73FL and FLAG-NudE preps show that NudE localizes to plus ends of microtubules which is where Khc73 localizes.
(B) Live imaging analysis of interphase S2 cells transfected with GFP-Nude and Khc73FL-mCherry shows colocalization of the two components at the periphery of the cell, presumably to microtubule plus-ends. The two can be seen moving together in S2 cells towards the periphery of the cell.
(C) Live imaging analysis of interphase S2 cells plated on a Con-A coated slide transfected with GFP-NudE and Khc73FL-mCherry. This panel demonstrates an example of the two signals co-migrating towards the plus-end (white arrows).
(D) From the same experiment shown in panel C, an example of the GFP-NudE and Khc73FL-mCherry migrating in opposite directions (white versus yellow arrows). See also Figure S3, Movie S3, and Movie S4.
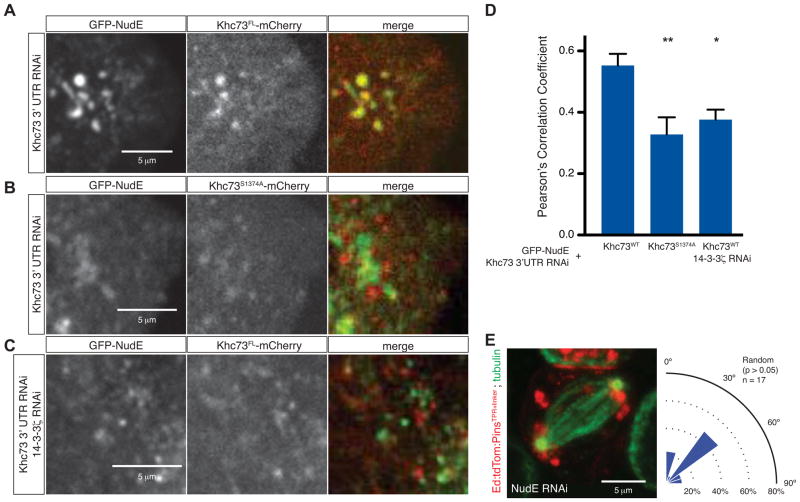
Figure 5. Khc73 and NudE co-migration requires 14-3-3.
(A) Still frame from live-imaging analysis of S2 cells transfected with GFP-NudE and wild-type Khc73FL-mCherry. Endogenous Khc73 was depleted by RNAi and S2 cells were plated on ConA-coated chambers.
(B) Still frame from live-imaging analysis of S2 cells transfected with GFP-NudE and a 14-3-3 binding motif mutant Khc73S1374A-mCherry. Endogenous Khc73 was depleted by RNAi and S2 cells were plated on ConA-coated chambers.
(C) Still frame from live-imaging analysis of S2 cells transfected with GFP-NudE and treated with an RNAi directed against 14-3-3ζ. Endogenous Khc73 was depleted by RNAi and S2 cells were plated on ConA-coated chambers.
(D) Quantification of GFP-NudE and Khc73FL-mCherry or Khc73S1374A-mCherry colocalization demonstrates that colocalization of these two components is significantly reduced in the 14-3-3ζ RNAi background and in Khc73S1374A-mCherry mutants. Asterisks denote statistical significance relative to wild-type Khc73 rescue. A t-test was used to evaluate statistical significance (*, p < 0.05 and **, p < 0.01). n=5, mean ± S.D. shown.
(E) NudE RNAi in S2 cells expressing Ed:PinsTPR+linker reduces Pins spindle orienting activity to random levels. The statistical significance compared to Khc73 RNAi was determined using a t-test. See also Figure S4.
The live-imaging analysis suggests that NudE is a Khc73 cargo and we hypothesized that NudE binding might occur via 14-3-3. We tested whether the 14-3-3 binding motif is required for NudE and Khc73 co-migration and found that the S1374A Khc73 mutant does not migrate with NudE as robustly as wild-type Khc73 (Figure 5A, B, D). Furthermore, 14-3-3ζ is required for Khc73/NudE co-localization suggesting that it is required for NudE to be a Khc73 cargo (Figure 5C, D). Finally, we tested whether NudE itself is required for Pins-mediated spindle positioning and found that knockdown of NudE by RNAi leads to loss of PinsTPR+linker activity (Figure 5E). Like the 14-3-3 and Par1 knockdowns, NudE RNAi also leads to a complete loss of spindle positioning, suggesting that it plays other roles in spindle orientation. We were surprised that NudE RNAi had such a strong phenotype, as we have found that knockdown of Dynein light chain only partially affects spindle orientation (Johnston et al., 2009). The difference in the two phenotypes indicates that NudE may have a Dynein-independent function in spindle orientation. NudE is essential in a variety of cellular activities including spindle positioning, kinetochore activity, and organelle transport (reviewed in (Kardon and Vale, 2009)). Together, these results indicate that the Khc73 14-3-3 binding site, 14-3-3ε, 14-3-3ζ, and NudE are all essential components of the PinsTPR+linker pathways. Furthermore, NudE appears to be trafficked by Khc73 via 14-3-3 proteins.
Khc73/14-3-3/NudE form a Phospho-Dependent Complex that links to the PinsTPR/Mud/Dynein Pathway
We have identified several components that appear to link the PinsTPR and Pinslinker pathways. Based on our observations, we hypothesize that Khc73’s stalk region forms a physical connection to NudE via 14-3-3 proteins. We first tested whether the 14-3-3 binding motif in Khc73’s stalk region directly interacts with the purified 14-3-3 proteins, 14-3-3ε and 14-3-3ζ, and found that 14-3-3ζ, but not 14-3-3ε, binds to Khc73’s stalk (Figure 6A). In addition to directly associating with Khc73’s stalk region, we also observed that 14-3-3ζ localizes to spindle microtubules in S2 cells (Figure 6B). Rather than interacting with Khc73, we found that 14-3-3ε associates with NudE (Figure 6C), which is consistent with findings in mammalian systems (Toyo-oka et al., 2003). 14-3-3 proteins are known to heterodimerize (Aitken et al., 2002; Liang et al., 2008), and we observed a direct interaction between 14-3-3ε and 14-3-3ζ (Figure 6D), suggesting that Pins spindle orientation pathway synergy might arise from a Khc73/14-3-3ζ/14-3-3ε/NudE complex quaternary complex. We expressed all of the components as epitope-tagged full-length fusions in HEK293 cells and performed immunoprecipitation experiments to test whether the entire complex could form. As shown in Figure 6E, we observed that 14-3-3ζ, 14-3-3ε, and NudE are all immunoprecipitated with Khc73. However, immunoprecipitation of the 14-3-3 motif mutant Khc73S1374A failed to co-immunoprecipitate any of the complex components beyond background levels (Figure 6E). We conclude from our biochemical studies that NudE binds to Khc73 through a heterodimeric complex of the 14-3-3 adapter proteins whose interaction to Khc73 may be regulated by Par-1.
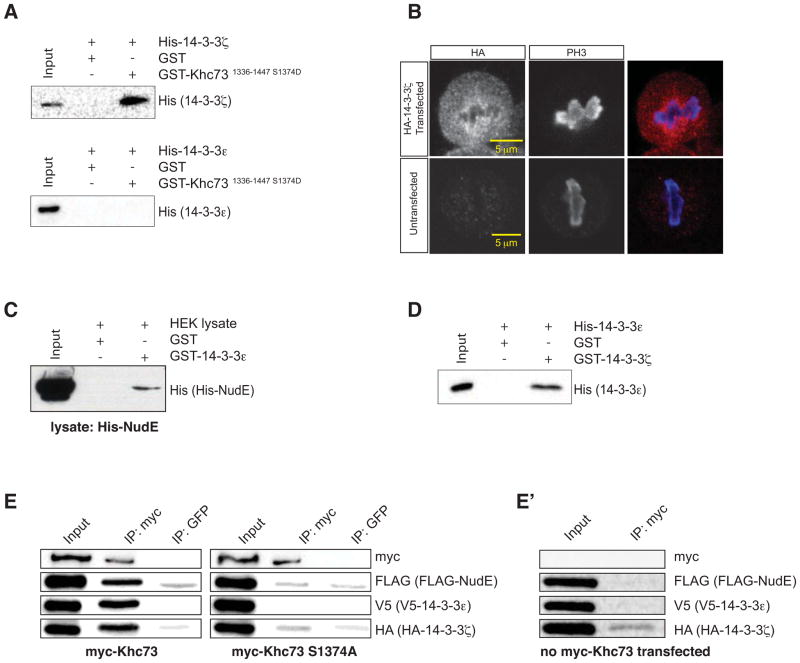
Figure 6. Khc73 forms a quaternary complex with NudE via 14-3-3 proteins.
(A) Western blot of GST-pulldown assays using purified GST alone (solid phase) and purified GST-Khc731336-1447 S1374D (solid phase) with purified His-14-3-3ζ and purified His-14-3-3ε as bait. 14-3-3ζ directly interacts with Khc73’s stalk region containing the phosphomimetic 14-3-3 binding motif (amino acids 1336-1447) whereas 14-3-3ε does not. Anti-His western blot for specificity (14-3-3 proteins are approx. same size as GST).
(B) 14-3-3ζ can been seen decorating spindle microtubules in mitotic S2 cells transfected with HA-14-3-3ζ. Untransfected S2 cells were stained for HA as a negative control.
(C) Western blot analysis of GST-pulldown assays using purified GST alone (solid phase) and purified GST-14-3-3ε (solid phase) with HEK lysate containing His-NudE. 14-3-3ε immobilized on the solid phase can pull-down His-NudE from lysate harvested from HEK293 cells transfected with His-NudE.
(D) Western blot analysis of GST-pulldown assays using purified GST (solid phase) and purified GST-14-3-3ε (solid phase) with purified His-14-3-3ζ as prey. 14-3-3ζ directly interacts with 14-3-3ε that is immobilized on the solid phase in GST-pulldown assays. Anti-His western blot for specificity (14-3-3 proteins are approx. same size as GST).
(E) HEK cells were trasfected with FLAG-NudE, V5-14-3-3ε, HA-14-3-3ζ, and either myc-Khc73FL wt or myc-Khc73FL S1374A (left and right sets of blots, respectively). Khc73 was immunoprecipitated in both cases, then probed for FLAG-NudE, V5-14-3-3ε, HA-14-3-3ζ by Western blotting for their epitope tags. Khc73FL wt could co-immunoprecipitate all components, whereas Khc73FL S1374A could not, indicating that complex formation is phosphoregulated.
(E′) A negative control showing that an anti-myc antibody fails to immunoprecipitate FLAG-NudE, V5-14-3-3ε, or HA-14-3-3ζ from HEK cells transfected with only these components.
DISCUSSION
Mitotic spindle orientation requires the coordination of several pathways that act on astral microtubules. These pathways may establish cortical-microtubule connections and generate the forces necessary for movement of this large cellular structure with metaphase spindle lengths varying from 2 μm in yeast to 60 μm of a Xenopus single-cell stage (Goshima and Scholey, 2010). The spindle orientation protein Pins has a domain that has been thought to capture microtubules (Pinslinker), and another that generates force (PinsTPR). We attempted to understand how these two pathways function together by taking advantage of an induced polarity system in cultured S2 cells in which the two pathways can be selectively activated. This system allowed for the identification of the Khc73 stalk domain as a critical element that links PinsTPR and Pinslinker pathways. This observation was used as a platform for establishing a complete physical connection between the two pathways. We have also clarified the role of 14-3-3 proteins in spindle orientation, establishing that their interaction with Pins is likely to be indirect (through Dlg and Khc73).
Khc73’s Motor and MBS Domains Facilitate Cortical Microtubule Capture
We have shown that Khc73 performs two functions in Pins-mediated spindle positioning. First, it functions in the Pinslinker pathway to mediate cortical microtubule capture through its MBS and motor domains, respectively. The N-terminal portion of Khc73 is sufficient for linker activity, which is likely occurring through a DlgGK/Khc73MBS interaction at the cortex and a microtubule/Khc73motor interaction at the spindle. This suggests that Khc73’s motor domain could function at the cortex by itself, however, Ed:Khc73motor did not have spindle positioning activity (Figure S2A), indicating that other factors could be required or the motor domain is not functional in this context (e.g. as a monomer with the coiled-coil stalk). Khc73 must therefore rely on Dlg as an adapter to target it to the cortex, which is where it can potentially function to facilitate the initial contact of astral microtubules.
Although Khc73’s MBS domain directly interacts with Dlg, Khc73 is not seen to colocalize with cortical Pins (Figure S2B), even though Dlg robustly localizes to Pins crescents (Johnston et al., 2009). Instead, the motor protein is seen distinctly at the ends of microtubules (Figure 2B), suggesting that Khc73 moves to the plus-ends where it may be poised for capture by the cortical Pinslinker/Dlg complex. Thus, Khc73’s N-terminal domains are likely to facilitate cortical microtubule capture by linking microtubule plus ends to cortical Dlg.
Khc73 Traffics the Dynein activator NudE to Microtubule Plus-Ends
In addition to facilitating cortical microtubule capture, we found that Khc73 also forms a physical connection to the PinsTPR/Mud/Dynein pathway with its stalk region that is essential for the synergistic function of the two pathways. Khc73 may activate Dynein by delivering NudE to the cortex, where Dynein is presumably localized by PinsTPR/Mud. Although it is not possible to observe the localization of Dynein in S2 cells for technical reasons, there is good evidence that it is cortically localized by way of PinsTPR/Mud. In HeLa cells, Dynein localizes to the cortex with the mammalian homolog of Mud, NuMA, along with mPins, during mitosis (Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012).
We propose that a 14-3-3 motif in Khc73’s stalk region activates an “idling” cortically-localized Dynein by cargoing it via NudE. Interestingly, although the Khc73 14-3-3 motif mutant Khc73S1374A has a distribution of spindle orientation angles that isn’t random, the distribution is bimodal such that the spindle angles are either fully aligned or orthogonal to the polarity axis. The bimodal phenotype is distinct from the Khc73motor+MBS fragment, which has a canonical intermediate distribution of spindle angles, suggesting that there may be additional regions or domains in the stalk that are contributing to the bimodal phenotype. We hypothesize that an element within Khc73’s stalk region is required for the proper application of the forces generated from by two motor proteins to properly orient the mitotic spindle. Nevertheless, our biochemical and genetic studies demonstrate that the 14-3-3 binding motif is, at the very least, required for proper Pins-mediated spindle positioning and is required for Khc73’s interaction with the 14-3-3 proteins and NudE.
A Model for coordinated motor activity in Pins-mediated spindle positioning
Pins mediates spindle positioning by coordinating two motor proteins that, as a pair, facilitate the cortical capture of microtubules and also provide pulling forces to robustly orient the mitotic spindle. We propose a model (Figure 7) in which orientation occurs through an ordered series of events, beginning with the initial polarization of Pins, followed by recruitment of Mud through its PinsTPR domain and Dlg through Pinslinker region. Cortical Mud then recruits cytoplasmic Dynein, which is not yet active and will remain inert, but poised at the cortex. Khc73 localizes to the plus-ends of microtubules where establishes cortical-microtubule contact through direct binding to Dlg, and also delivers NudE to cortical Dynein, thereby activating it. As astral microtubules enter the proximity of the Dynein complex, Dynein can generate specifically timed cortical pulling forces necessary for robust spindle positioning. Future work will be directed at dissecting the precise timing of these synergistic events that underlie differentiation and tissue architec.
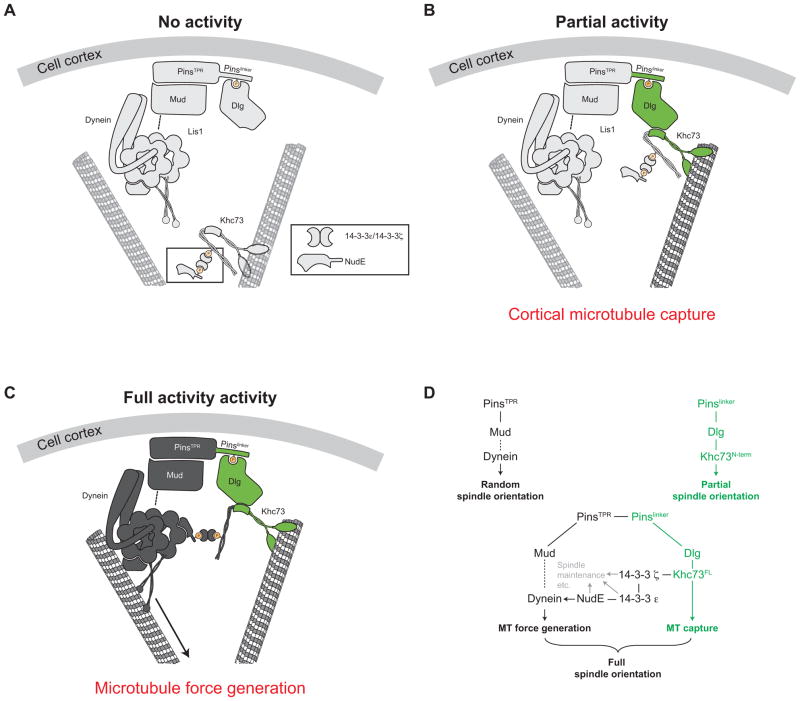
Figure 7. Model for coordinated control of Dynein and Khc7 during Pins-mediated spindle positioning.
(A) Cortical Pins recruits an “idling” Dynein through its TPR domain; the PinsTPR pathway alone is insufficient for any activity.
(B) Khc73 walks to MT plus ends where its N-terminus provides a physical link between MTs and cortical Pins; the Pinslinker pathway is sufficient for partial spindle orientation activity through cortical-microtubule capture.
(C) Khc73 transports NudE to MT plus ends, bringing NudE within proximity to activate cortical Dynein. Activated Dynein moves toward MT minus ends to generate the necessary cortical pulling required for robust spindle positioning; the PinsTPR+linker pathway is sufficient for full spindle orientation. NudE was depicted to associate with the ATPase region of cytoplasmic Dynein for clarity.
(D) Schematic representation of the genes and the functions of PinsTPR (black) and Pinslinker (green) pathway components for Pins-mediated spindle positioning. The PinsTPR pathway components 14-3-3 ε, 14-3-3 ζ, and NudE have roles outside of PinsTPR pathway function (gray) that may be essential for proper Pins-mediated spindle positioning.
EXPERIMENTAL PROCEDURES
Plasmid Construction, S2 Cell Culture, and RNAi
Ed:tdTomato:PinsTPR+linker was made in the metallothionine-promoter-based pMT-V5 vector (Invitrogen, Carlsbad, CA) by replacing the Ed cytoplasmic domain with an in-frame tdTomato and Pins (amino acids 1-466) cassette. All Khc73 fragments used for the structure/function analysis were cloned into pMT with an N-terminal myc epitope. 14-3-3ζ was cloned into pMT with an N-terminal FLAG epitope tag. Khc73:mCherry used for TIRF and spinning disk confocal live imaging was expressed from a pMT vector containing a full-length Khc73 construct cloned from S2 cells which differs slightly from the deposited sequence (amino acids 1-1913, sequence in supplemental information) with an in-frame C-terminal mCherry cassette. GFP:NudE used in TIRF and spinning disk was expressed from pMT vectors containing an N-terminal GFP cassette with either full-length Dm NudE isoform C (amino acids 1-377). The pMT-GFP:a-tubulin vector used for the live imaging experiments was a gift from Dr. Chris Q. Doe (Johnston et al., 2009). Site-directed mutagenesis was performed to generate point mutations in the all plasmids mentioned in the material and methods section.
S2 cells were grown in Schneider’s media (Sigma) supplemented with 10% fetal bovine serum and seeded at ~2 × 106 cells per well of a 6-well culture dish on the day of transfection. S2 cells were transfected with 0.4–1 μg total DNA with Effectene (QIAGEN, Germandtown, MD), and gene expression was induced 16–24 hours after transfection with the addition of 500 μM CuSO4 for 24–48 hours.
RNA intereference was performed as described in (Johnston et al., 2009) and is described in detail in the supplemental experimental procedures. Primer sequences and determination of RNAi efficiency are also detailed in the supplemental information.
Induced polarity assay and Measuring Spindle Orientation in S2 cells quantification and analysis
S2 cells expressing Ed:PinsTPR+linker were clustered by orbital shaking at 175RPM for 2 hours. Cells were then fixed and stained and mitotic S2 cells exhibiting Ed:PinsTPR+linker cortical crescents were imaged by confocal microscopy. All spindle orientation angles were quantified using ImageJ’s angle tool. Ed:PinsTPR+linker crescents consuming more than a third of the S2 cell circumference and atypical spindles were excluded from spindle orientation analysis. The polarity axis was determined by measuring a line perpendicular through the center of the Ed:PinsTPR+linker crescent and the spindle axis was measured as a line drawn through both spindle poles. To analyze significance between two independent samples, p-values were determined using students t-test, unless otherwise noted. Partial alignment phenotypes were determined as significantly different from the “random” Khc73 RNAi condition as well as the “aligned” full-length rescue condition.
Immunofluorescence Staining, Live Imaging, and TIRF Microscopy
For immunofluorescence staining of S2 cells, cells were allowed to settle onto 12mm coverslips, either coated with Con-A (0.5mg/mL) or not, for 1–2 hrs before being fixed for 20 min with 4% paraformaldehyde in PBS. Cells were washed with 0.1% saponin in PBS, and blocked for 30 min in 0.1% saponin/1% BSA in PBS. Primary antibodies diluted in blocking buffer were added to cells immobilized on coverslips and allowed to incubate overnight at 4°C. Primary antibody dilutions are as follows: 1:1000 Rabbit anti-myc (Sigma), 1:1000 mouse anti-FLAG (Sigma), 1:1000 mouse anti-HA (Sigma), and rat anti-α tubulin (AbCam). Fixed S2 cells were imaged using a Leica TCS SP2 confocal microscope with a 60X 1.4 NA immersion-oil lens using 488 Ar laser/500–530nm emission filter, 543 HeNe laser/560–620 emission filter, and 633 HeNe laser/650–750 emission filter. The refractive index of the immersion oil is 1.518.
For TIRF and spinning disk confocal live imaging, S2 cells were allowed to settle onto Conconavalin-A (0.5mg/mL)-coated 4- or 8-well chambers (Labtek) for 2 hrs. Time-lapse images were acquired on a Nikon TE2000 confocal microscope outfitted with an Andor D4-891 EM-CCD camera and a 100X 1.49 NA oil-immersion TIRF objective lens.
Colocalization quantification and Khc73/Eb1 velocity determination
Images obtained from spinning-disk microscopy of live S2 cells plated on ConA-coated coverslips expressing GFP-NudE and Khc73FL-mCherry or Khc73S1374A-mCherry were analyzed for colocalization. Colocalization analysis was performed as previously described (Oser et al., 2010). Briefly, images were processed in ImageJ using LoG 3D, a spot enhancing Laplacian of Gaussian 2D filter (Sage et al., 2005) using a sigma value of 2 or 3. The processed images were then analyzed for colocalization using ImageJ plugin JACoP (Bolte and Cordelieres, 2006). Pearson’s Correlation Coefficients were collected from five trials for each condition and mean and S.E.M. are reported.
To determine the velocities of Khc73-mCherry and GFP-EB1, we analyzed time-lapse TIRF images of S2 cells expressing Khc73-mCherry and GFP-EB1 plated on ConA-treated chambers by tracking pixels over the course of 10 successive frames (2.5 seconds).
Bacterial Protein Purification GST-Pulldown Assays
The purification of bacterially expressed proteins is detailed in the supplemental experimental procedures.
For GST-pulldown experiments, purified GST-fusion proteins were immobilized onto glutathione-agarose resin (Sigma), allowed to incubate with His-tagged prey proteins or HEK lysate for 15 min at room temperature, and followed by three washes in 50mM HEPES 7.5, 1mM DTT, 300mM NaCl (for experiments using 14-3-3 proteins and NudE)/100mM NaCl (for GST-Dlg/His-Khc73 pulldown), 0.5% tween-80, and 50mM MgCl2. Complexes were eluted off the glutathione-agarose with SDS.
For Western blotting, 1:1000 Mouse anti-His antibody in 5% low-fat milk in TBS-T. HRP-conjugated goat anti-mouse secondary antibody (Santa Cruz) and the Pico-sensitivity peroxidase chemiluminescent substrate (Pierce) was used for detection.
HEK293 Protein Expression and Co-Immunoprecipitation
For protein expression in HEK293T cells, Drosophila full-length NudE isoform C containing an N-terminal FLAG epitope tag, full length Khc73 containing an N-terminal myc epitope tag, 14-3-3ε containing an N-terminal V5 epitope tag, and 14-3-3ζ containing an N-terminal HA tag were cloned into the mammalian cell expression vector pCMV (Invitrogen). 30mL freestyle HEK293T cells were transfected with 30μg total DNA using 60ml 293fectin reagent (Invitrogen). HEK293 suspension cells were incubated at 37°C at 5% CO2 shaking at 200 rpm, and proteins were expressed for 72 hours.
For immunoprecipitation experiments, HEK 293T cells were harvested and lysed by homogenization through a 21G needle in a 5mL high-salt lysis buffer containing 50mM Tris pH 8.0, 0.5% NP-40, and 500mM NaCl. 5μg Rabbit anti-myc antibody was added to 1mL clarified lysate and incubated at 4°C for 1 hour. Complexes were precipitated by the addition of 50μl of a 50:50 slurry of Protein-G sepharose (Invitrogen) and incubated at 4°C for 1 hour. The Protein-G sepharose immune complexes were then washed three times in lysis buffer and once in 50mM Tris pH 8.0. Immunocomplexes were dissociated by the addition of 1% SDS and 100mM DTT in 50mM Tris pH 7.5.
For Western blotting, 1:1000 Rabit anti-myc (Sigma), 1:1000 mouse anti-HA (Covance), 1:1000 mouse anti-FLAG (Santa Cruz), and 1:1000 mouse anti-V5 (Santa Cruz) were used in 5% low-fat milk in TBS-T. HRP-conjugated goat anti-rabbit and HRP-conjugated goat anti-mouse (Santa Cruz) were used with the Femto-sensitivity peroxidase chemiluminescent substrate (Pierce).
Supplementary Material
Highlights.
The N-terminus of the kinesin Khc73 provides a cortical-microtubule connection
A 14-3-3 binding motif in the Khc73 stalk is necessary for robust spindle orientation
Khc73 forms a quaternary complex with 14-3-3 ε, 14-3-3ζ, and NudE
Khc73 delivers NudE to the cortex where it activates cortically-localized Dynein
Footnotes
AUTHOR CONTRIBUTIONS
M.S.L. and K.E.P. designed research; M.S.L. performed research; M.S.L. and K.E.P. analyzed data. M.S.L. and K.E.P. wrote manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aitken A, Baxter H, Dubois T, Clokie S, Mackie S, Mitchell K, Peden A, Zemlickova E. Specificity of 14-3-3 isoform dimer interactions and phosphorylation. Biochem Soc Trans. 2002;30:351–360. doi: 10.1042/bst0300351. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Berezuk MA, Schroer TA. Dynactin enhances the processivity of kinesin-2. Traffic. 2007;8:124–129. doi: 10.1111/j.1600-0854.2006.00517.x. [DOI] [PubMed] [Google Scholar]
- Blangy A, Arnaud L, Nigg EA. Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150. J Biol Chem. 1997;272:19418–19424. doi: 10.1074/jbc.272.31.19418. [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- David NB, Martin CA, Segalen M, Rosenfeld F, Schweisguth F, Bellaiche Y. Drosophila Ric-8 regulates Galphai cortical localization to promote Galphai-dependent planar orientation of the mitotic spindle during asymmetric cell division. Nat Cell Biol. 2005;7:1083–1090. doi: 10.1038/ncb1319. [DOI] [PubMed] [Google Scholar]
- De S, Kline D. Evidence for the requirement of 14-3-3eta (YWHAH) in meiotic spindle assembly during mouse oocyte maturation. BMC Dev Biol. 2013;13:10. doi: 10.1186/1471-213X-13-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon SW, Serpinskaya AS, Vaughan PS, Lopez Fanarraga M, Vernos I, Vaughan KT, Gelfand VI. Dynactin is required for bidirectional organelle transport. J Cell Biol. 2003;160:297–301. doi: 10.1083/jcb.200210066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Gardino AK, Yaffe MB. 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin Cell Dev Biol. 2011;22:688–695. doi: 10.1016/j.semcdb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies TE, Cabernard C. Cell division orientation in animals. Curr Biol. 2011;21:R599–609. doi: 10.1016/j.cub.2011.06.055. [DOI] [PubMed] [Google Scholar]
- Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Goshima G, Scholey JM. Control of mitotic spindle length. Annu Rev Cell Dev Biol. 2010;26:21–57. doi: 10.1146/annurev-cellbio-100109-104006. [DOI] [PubMed] [Google Scholar]
- Hao Y, Du Q, Chen X, Zheng Z, Balsbaugh JL, Maitra S, Shabanowitz J, Hunt DF, Macara IG. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical Pins. Curr Biol. 2010;20:1809–1818. doi: 10.1016/j.cub.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Suzuki A, Ohno S. PAR-1/MARK: a kinase essential for maintaining the dynamic state of microtubules. Cell Struct Funct. 2012;37:21–25. doi: 10.1247/csf.11038. [DOI] [PubMed] [Google Scholar]
- Huckaba TM, Gennerich A, Wilhelm JE, Chishti AH, Vale RD. Kinesin-73 is a processive motor that localizes to Rab5-containing organelles. J Biol Chem. 2011;286:7457–7467. doi: 10.1074/jbc.M110.167023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, MacKintosh C. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J. 2010;427:69–78. doi: 10.1042/BJ20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Doe CQ, Prehoda KE. Structure of an enzyme-derived phosphoprotein recognition domain. PLoS One. 2012;7:e36014. doi: 10.1371/journal.pone.0036014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Hirono K, Prehoda KE, Doe CQ. Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell. 2009;138:1150–1163. doi: 10.1016/j.cell.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Busso C, Gonczy P. Cortical dynein is critical for proper spindle positioning in human cells. J Cell Biol. 2012;199:97–110. doi: 10.1083/jcb.201203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Butterworth MB, Peters KW, Walker WH, Frizzell RA. An obligatory heterodimer of 14-3-3beta and 14-3-3epsilon is required for aldosterone regulation of the epithelial sodium channel. J Biol Chem. 2008;283:27418–27425. doi: 10.1074/jbc.M803687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matenia D, Mandelkow EM. The tau of MARK: a polarized view of the cytoskeleton. Trends Biochem Sci. 2009;34:332–342. doi: 10.1016/j.tibs.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Morin X, Bellaiche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell. 2011;21:102–119. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Nipper RW, Siller KH, Smith NR, Doe CQ, Prehoda KE. Galphai generates multiple Pins activation states to link cortical polarity and spindle orientation in Drosophila neuroblasts. Proc Natl Acad Sci U S A. 2007;104:14306–14311. doi: 10.1073/pnas.0701812104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M, Mader CC, Gil-Henn H, Magalhaes M, Bravo-Cordero JJ, Koleske AJ, Condeelis J. Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. J Cell Sci. 2010;123:3662–3673. doi: 10.1242/jcs.068163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage D, Neumann FR, Hediger F, Gasser SM, Unser M. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans Image Process. 2005;14:1372–1383. doi: 10.1109/tip.2005.852787. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Shevchenko A, Knoblich JA. A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr Biol. 2000;10:353–362. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Tabler JM, Yamanaka H, Green JB. PAR-1 promotes primary neurogenesis and asymmetric cell divisions via control of spindle orientation. Development. 2010;137:2501–2505. doi: 10.1242/dev.049833. [DOI] [PubMed] [Google Scholar]
- Toyo-oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, Ward HL, Ayala R, Tsai LH, Dobyns W, Ledbetter D, et al. 14-3-3epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat Genet. 2003;34:274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- Toyoshima F, Nishida E. Spindle orientation in animal cell mitosis: roles of integrin in the control of spindle axis. J Cell Physiol. 2007;213:407–411. doi: 10.1002/jcp.21227. [DOI] [PubMed] [Google Scholar]
- Wang C, Li S, Januschke J, Rossi F, Izumi Y, Garcia-Alvarez G, Gwee SS, Soon SB, Sidhu HK, Yu F, et al. An ana2/ctp/mud complex regulates spindle orientation in Drosophila neuroblasts. Dev Cell. 2011;21:520–533. doi: 10.1016/j.devcel.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Wee B, Johnston CA, Prehoda KE, Doe CQ. Canoe binds RanGTP to promote Pins(TPR)/Mud-mediated spindle orientation. J Cell Biol. 2011;195:369–376. doi: 10.1083/jcb.201102130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada KH, Hanada T, Chishti AH. The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin-3 motor GAKIN/KIF13B. Biochemistry. 2007;46:10039–10045. doi: 10.1021/bi701169w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y, Terabayashi T, Miki H. Par1b/MARK2 phosphorylates kinesin-like motor protein GAKIN/KIF13B to regulate axon formation. Mol Cell Biol. 2010;30:2206–2219. doi: 10.1128/MCB.01181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Kee YS, Poirier CC, Jelinek C, Osborne J, Divi S, Surcel A, Will ME, Eggert US, Muller-Taubenberger A, et al. 14-3-3 coordinates microtubules, Rac, and myosin II to control cell mechanics and cytokinesis. Curr Biol. 2010;20:1881–1889. doi: 10.1016/j.cub.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.