Abstract
Generation of research quality, clinically relevant cell types in vitro from human pluripotent stem cells (hPSCs) requires detailed understanding of the equivalent human cell types. Here we analyzed 134 human embryonic and fetal samples from 6–20 developmental weeks and identified the stages in which cKIT+ primordial germ cells (PGCs), the precursors of gametes, undergo whole genome epigenetic reprogramming with global depletion of 5mC, H3K27me3, H2A.Z and the time where imprint erasure is initiated and 5hmC is present. Using five alternate in vitro differentiation strategies combined with single-cell microfluidic analysis and a bona fide human cKIT+ PGC signature, we show the stage of cKIT+ PGC formation in the first 16 days of differentiation. Taken together, our study creates a resource of human germ line ontogeny that is essential for future studies aimed at in vitro differentiation and unveiling mechanisms necessary to pass human DNA from one generation to the next.
The foundation of human health at a cellular and molecular level is built upon accurate lineage differentiation during embryonic and fetal life. In recent years, a major barrier to study human development was overcome through the generation of human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and human induced pluripotent stem (hIPS) cells which can be used to differentiate to embryonic and fetal cell types. However, a major caveat for using hPSCs as a surrogate model for human fetal development is the dearth of studies that provide accurate human-specific details to validate, guide and quality control differentiation in vitro.
All adult human cells are created from four major embryonic lineages, ectoderm, mesoderm, endoderm and the germ line. The first three lineages contribute a variety of cell types to multiple organs. In contrast, the germ line has one purpose that is to generate gametes, which function solely to pass DNA from one generation to the next. There is considerable interest in generating germ line from hPSCs cells, as they could serve as a potential stem cell-based intervention for infertility1,2, or a model to understand the genetic-basis of human infertility3. However, before this can be achieved, the major landmarks of human germ line development during embryonic and fetal life must be characterized.
Human germ line development begins with the formation of primordial germ cells (PGCs) that express the tyrosine kinase receptor cKIT4–10. Very little is known about the developmental progression (ontogeny) of cKIT+ PGCs, however based on the mouse model, it is clear that PGCs must undergo whole genome epigenetic reprogramming in order to remove cytosine methylation from imprinted genes and restore totipotency11–14. Given the fundamental role of epigenetic reprogramming in the germ line, it is essential to characterize reprogramming in human PGCs since mouse and human genomes are separated by ~170 million years and diverse strategies may have evolved to execute it. Once the major molecular landmarks of human PGC reprogramming are known, we propose that this information will be critical to assessing PGC differentiation and reprogramming in vitro, or identifying bottlenecks that must be overcome to generate a functional germ line from hPSCs.
By evaluating 134 human embryonic and fetal gonadal samples from 6–20 developmental weeks, we provide the first comprehensive transcriptional and epigenetic roadmap of human cKIT+ PGCs in testes and ovaries and pinpoint the timing of major epigenetic events including whole genome reprogramming and initiation of imprint erasure. Using the endogenous human cKIT+ PGCs as a reference, we can now more accurately interpret the identity of the cKIT+ subpopulation of PGCs acquired with in vitro differentiation from hESCs. Our results clearly demonstrate that single cell analysis at both RNA and protein level is critical to defining PGC identity in vitro, and indisputably shows that established hESC lines are not equivalent to human PGCs.
RESULTS
cKIT positive PGCs undergo molecular progression with fetal development
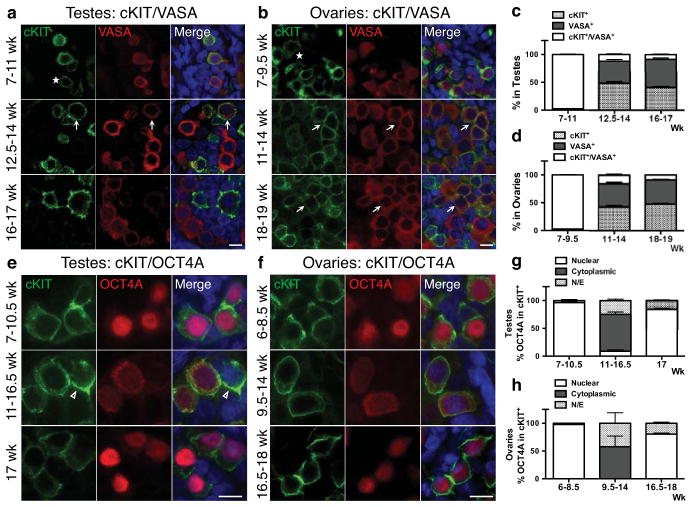
Temporal and spatial expression of cKIT in fetal testes and ovaries from 7–19 weeks of development was evaluated by immunofluorescence together with the evolutionarily conserved germ cell marker VASA. All testes samples procured had characteristic seminiferous cords by histology indicating that sex determination had been initiated15 (Supplementary Fig. S1). We identified cKIT on the surface of all VASA positive cells in testes from 7–11 weeks and ovaries from 7–9.5 weeks (Fig. 1a,b, and Supplementary Fig. S2a). However, from 12.5 weeks in testes, and 11 weeks in ovaries, cKIT and VASA protein expression becomes uncoupled, with only 10% of cKIT+ cells co-expressing VASA (arrows in Fig. 1a,b, quantified in Fig. 1c,d and Supplementary Fig. S2b). Upon uncoupling, the ratio of single cKIT+ to single VASA+ cells was 1:1. We also evaluated SSEA1, and found that although PGCs are SSEA1+ in fetal testes at the “common PGC progenitor stage” and after cKIT/VASA uncoupling, SSEA1 alone is not specific for the human germ line since it was also expressed on cKIT and VASA negative cells (not germ cells) (Supplementary Fig. S3a,b).
Fig. 1.
The dynamics of cKIT, OCT4A and VASA expression in the fetal gonad. (a,b) Representative immunofluorescence images of cKIT with VASA at the developmental weeks indicated. Asterisks indicate cKIT dim cells. (a) Shown is a 10-week testis for 7–11wk (n=5), a 13.5-week testis for 12.5–14wk (n=3) and a 16-week testis for 16–17wk (n=2). (b) Shown is a 7-week ovary for 7–9.5wk (n=4), an 11-week ovary for 11–14wk (n=3), and an 18-week ovary for 18–19wk (n=2). (c,d) Quantification of cKIT+, VASA+ and cKIT+/VASA+ cells (arrows in a,b). (c) In testes, 9 optic fields were counted at 7–11wk (n=5), 9 optic fields at 12.5–14wk (n=3) and 7 optic fields at 16–17wk (n=2). (d) In ovaries, 7 optic fields were counted at 7–9.5wk (n=4), 8 optic fields at 11–14wk (n=3) and 7 optic fields at 18–19wk (n=3). (e,f) Representative immunofluorescence images of cKIT with OCT4A at the developmental weeks indicated. (e) Shown is a 10-week testis for 7–10.5wk (n=5), a 13.5-week testis for 11–16.5wk (n=4) and a 17-week testis (n=1). (f) Shown is an 8-week for 6–8.5wk (n=3), an 11-week for 9.5–14wk (n=3) and an 18-week for 16.5–18wk (n=2). (g,h) Quantification of nuclear or cytoplasmic localization of OCT4A in cKIT+ cells. (g) In testes, 6 optic fields were counted at 7–10.5wk (n=5), 8 optic fields at 11–16.5wk (n=4) and 6 optic fields at 17wk (n=1). (h) In ovaries, 6 optic fields were counted at 7–8.5wk (n=3), 8 optic fields at 9.5–14wk (n=3) and 9 optic fields at 16.5–18wk (n=2). For immunofluorescence, nuclei were counterstained with DAPI (blue) and scale bars represent 10 um. All data are expressed as mean ± SEM. Abbreviations: wk= week, N/E= Not Expressed.
To assess the stem cell identity of cKIT+ gonadal cells we examined the germ/stem cell enriched protein OCT4A using antibodies against the N-terminal region that discriminates OCT4A from the splice variant OCT4B16,17 (Fig. 1e,f). OCT4A localized to the nucleus of cKIT+ cells from 7–10.5 weeks in testes and 6–8.5 weeks in ovaries (just prior to VASA repression). Similarly, expression of the pluripotency marker TRA-1-81 highly correlated with nuclear OCT4A in both sexes (Supplementary Fig. S3c,d). After this time, our data indicates that OCT4A+ cells become a subpopulation of cKIT+, and the majority of cKIT+ PGCs localize OCT4A protein to the cytoplasm. In addition a number of cKIT+ cells no longer express OCT4A (Fig. 1g,h). At 17 weeks in fetal testes and from 16.5 weeks in fetal ovaries OCT4A is again identified in the nucleus of a large fraction of cKIT+ cells (Fig. 1g,h). Therefore, using cKIT, OCT4A and VASA expression, we propose a common PGC progenitor stage in humans that lasts to 11 weeks in testes and 9.5 weeks in ovaries. Thereafter two major populations are established in males and females, the cKIT+ population that expresses OCT4A in most cells, and the single VASA+ cells.
To isolate individual cKIT+ cells, we performed fluorescence-activated cell sorting (FACS) of 49 testes and 42 ovaries from 8–20 developmental weeks (Table S1), using the gating strategy shown in Figures 2a,b. Applying this sorting strategy on a 15.5-week testis, in combination with quantitative (q) qRT-PCR, we verified that germ line identity was specifically enriched in the cKIT bright fraction compared to cKIT dim or single SSEA1 expressing cells (Supplementary Fig. S4a–c). We speculate that the cKIT dim gate is a heterogeneous mixture of PGCs and non-PGCs given that all germ line genes including VASA are reduced relative to the cKIT bright fraction. Therefore, to avoid potential contamination with gonadal somatic cells in down-stream applications, we excluded cKIT dim cells from all future FACS. Using the cKIT bright gate (which we call cKIT+) we sorted an average of 2.83% cKIT+ cells per testis at 8–11 weeks and 2.45% cKIT+ cells per ovary at 8–9.5 weeks. Then at 11.1–20 weeks we sorted an average of 0.9% cKIT+ cells from individual testes and 4.75% cKIT+ cells per ovary at 9.6–16.5 weeks (Fig. 2a,b). The absolute number of cKIT+ cells sorted from an individual testis was as low as 150 for a 20-week sample, with the majority of samples yielding 2,500–3,000 cKIT+ cells per testis. Similarly, fetal ovaries yielded on average 4,500–5,000 cells per ovary, ranging from as low as 276 cells for an 8-week ovary to 30,000 cells, for a 16.5-week ovary. This range in absolute numbers most likely reflects variability in sample quality (intact gonads verses fragments) and viability (which ranged from 14.4%–64.7%). However, the variability in the percentage of cKIT+ cells in the ovary did not correlate with overall sample viability, and instead we speculate that this variability was due to the presence of small amounts of attached non-gonadal tissue that varied from sample to sample.
Fig. 2.
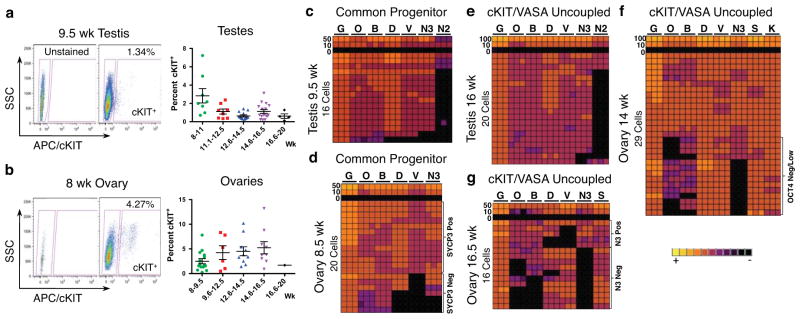
Molecular characterization of cKIT+ PGCs from 7–20 developmental weeks. (a,b) Gating strategy for sorting cKIT+ cells with an APC conjugated anti-human cKIT primary antibody against side scatter (SSC). (a) Shown is a 9.5-week testis and (b) an 8-week ovary. Also shown is the percent of cKIT+ cells sorted from the live fraction of testes in (a) and ovaries in (b) at 8–20 developmental weeks (wk). Each data point represents a single sample (biological replicate). All data are represented as mean ± SEM. (c–g) Heat map of GAPDH (G), OCT4 (O), BLIMP1 (B), DAZL (D), VASA (V), NANOS3 (N3), cKIT (K) NANOS2 (N2) and SYCP3 (S) in triplicate (columns) in 100, 50, 10, 0 or single sorted cKIT+ cells (rows).
To determine the molecular identity of cKIT+ PGCs, we performed single cell analysis with five PGC signature genes including OCT4, BLIMP1, DAZL, VASA and NANOS3 using FACS, followed by microfluidic qRT-PCR at the common progenitor and cKIT/VASA uncoupled stage (Fig. 2c–g). We also confirmed expression of cKIT in individual cells (Fig. 2f). Our single cell approach was first validated in HEK 293 cells (Supplementary Fig. S4e–h). At the common PGC progenitor stage, 14/16 cKIT+ cells in the testis, and 13/20 cKIT+ cells in the ovary coordinately expressed the five PGC signature genes (Fig. 2c,d). However, in the ovary 7/20 cKIT+ cells did not express VASA and/or DAZL at this stage and instead were OCT4/BLIMP1 double positive (O/B) or OCT4/BLIMP1/NANOS3 triple positive (O/B/N3). In testes, NANOS2 expression was also evaluated and found in <20% of cKIT+ cells in the common progenitor and this was maintained upon cKIT/VASA uncoupling in the cKIT+ cell (Fig. 2c,e). In the ovary during the uncoupled stage when OCT4A is either in the cytoplasm or no longer expressed, NANOS3 mRNA is also no longer expressed in a fraction of cells, and these NANOS3 negative cells correlated with no or low levels of OCT4 and BLIMP1 (Fig. 2f). At 16.5 weeks, when OCT4A is again localized to the nucleus or not expressed, ovarian cKIT+ cells become even more heterogeneous, most notably involving loss of NANOS3, OCT4 and BLIMP1 mRNA in some cells, with DAZL and VASA being absent in others (Fig. 2g). SYCP3 was used to indicate meiotic potential, and was expressed in every cell at 14 and 16.5 weeks (Fig. 2f,g). Furthermore, at 16.5 weeks SYCP3 and VASA mRNA expression levels were significantly enriched in the NANOS3 negative population (Supplementary Fig. S4d). Despite SYCP3 mRNA expression in every cell at a single cell level, on the protein level, only VASA+ cells are immunopositive for SYCP3 in the fetal ovary from 14 weeks and not cKIT+ (Supplementary Fig. S3e), indicating that VASA+ cells are the first to acquire meiotic potential.
Loss of 5mC from imprinted DMRs is locus specific and occurs weeks after global 5mC depletion
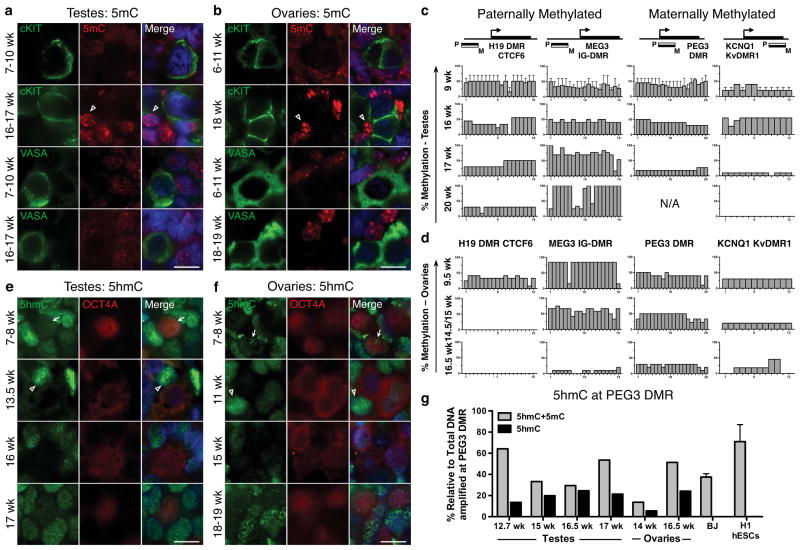
By immunofluorescence, 5-methyl cytosine (5mC) was below the level of detection at all stages of PGC development compared to somatic cells (open arrowheads on Fig. 3a,b). To evaluate cytosine methylation at differentially methylated regions (DMRs) of imprinting control centers, we used Bisulfite Sequencing (BS) followed by PCR (BS-PCR) on cKIT+ sorted PGCs (Fig. 3c,d). We evaluated two paternally methylated DMRs, H19 and MEG3 and two maternally methylated DMRs, PEG3 and KCNQ1. Primers were first verified using the BJ primary fibroblast cell line and H1 hESCs (Supplementary Fig. S5a). For the paternally methylated H19 and MEG3 DMRs, we observed CpG methylation at all developmental ages in male cKIT+ PGCs. In contrast, maternally methylated DMRs in the testis displayed a sharp reduction in CpG methylation between 16–17 weeks, and for KCNQ1 methylation was completely lost in the one 20-week sample consented to our study. Analysis of the ovary revealed a significant reduction of CpG methylation by 16.5 weeks at all loci. At paternally methylated DMRs, erasure was complete by 14.5–15 weeks for H19, and near complete by 16.5 weeks for MEG3.
Fig. 3.
Global loss of 5mC precedes loss of 5hmC (a,b) Representative immunofluorescence images shown of 5mC with cKIT or with VASA in (a) testes and (b) ovaries at the developmental weeks indicated. Open arrowheads indicate 5mC signal in somatic cells. (a) Shown is a 10-week testis for 7–10wk (n=3) and a 17-week testis for 16–17wk (n=3). (b) Shown is an 8-week ovary for 6–11wks (n=3) and an 18-week ovary for 18–19wk (n=2). (c,d) BS-PCR analysis of H19, MEG3, PEG3 and KCNQ1 in cKIT+ PGCs sorted from (c) testes at 9wk (n=2) and at 16, 17 and 20 weeks, and (d) ovaries at 9.5, 14.5, 15 and 16.5 weeks. (e,f) Representative immunofluorescence images of 5hmC with OCT4A in (e) testes and (f) ovaries at the developmental stages indicated in weeks. Arrows indicate 5hmC signal in PGCs, open arrowheads indicate 5hmC signal in somatic cells. (e) Shown is an 8-week testis for 7–8wk (n=2), a 13.5-week testis (n=1) a 16-week testis (n=2) and a 17-week testis (n=1). (f) Shown is an 8-week ovary for 7–8wk (n=2), an 11-week ovary, (n=1), 15-week ovary (n=1) and an 18-week ovary for 18–19 (n=2). (g) CGRA of the PEG3 DMR showing the percent of total methylation (5mC+5hmC) or 5hmC alone, relative to total amplified DNA (uncut) at the PEG3 DMR. DNA from BJ fibroblast and H1 hESCs were used as a negative control (for each, n=2 biological replicates). For immunofluorescence analysis nuclei were counterstained with DAPI (blue). Scale bars represent 10 um. All data are represented as mean ± SEM. Abbreviations: N/A= Not Amplified, wk= week.
5hmC is the major methylation species in the common PGC progenitor and is localized to PEG3 DMR prior to demethylation
Immunofluorescence for 5hmC, the oxidized derivative of 5mC revealed robust nuclear staining in somatic cells at all time points (open arrowheads on Fig. 3e,f), similar to previous reports in the mouse18. However in the germ line 5hmC expression is dynamic, exhibiting punctate nuclear staining in the common PGC progenitor stage (arrows on Fig. 3e,f), which is lost in OCT4A+ PGCs in the testis from 13.5–16 weeks (Fig. 3e). Enrichment of 5hmC is again detected in some OCT4A+ PGCs by 17 weeks (Fig. 3e). In fetal ovaries 5hmC is heterogeneous at 11–19 weeks, being enriched in some but not all OCT4A+ cells (Fig. 3f).
Bisulfite conversion does not distinguish between 5mC and 5hmC, therefore we used combined glycosylation restriction analysis (CGRA) at the PEG3 DMR to identify whether 5hmC is enriched at this imprinted locus in PGCs relative to somatic cells or hESCs (Fig. 3g). We identified 5ghmC at all stages of PGC development in both sexes at the PEG3 DMR. In contrast 5ghmC was not enriched at the PEG3 locus in BJ and H1 cells.
H3K27me3 and H2A.Z are enriched in common PGC progenitor cells
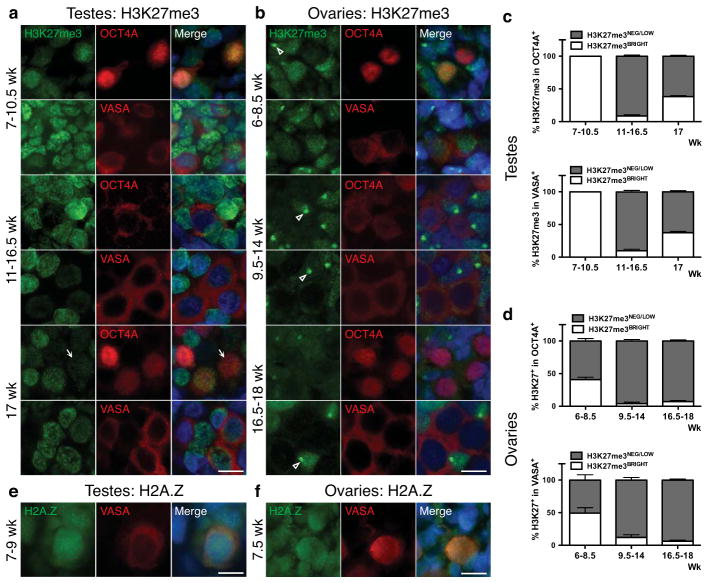
In the mouse, gonadal epigenetic reprogramming of PGCs and imprint erasure occurs from e11.5-e12.5 coincident with global changes in chromatin, including a transient loss of H3K27me3 and a permanent loss of the histone variant H2A.Z12. In humans, using immunofluorescence we show that H3K27me3 is enriched in the nucleus of common PGC progenitors in testes from 7–10.5 weeks (Fig. 4a,c). However, at 11 weeks, the endpoint of the common progenitor stage, H3K27me3 is at or below the level of detection in the majority of OCT4A+ and VASA+ PGCs (Fig. 4a,c). Interestingly at 17 weeks in testes, H3K27me3 is again observed the nucleus of ~38% OCT4A+ and VASA+ PGCs. In ovaries, H3K27me3 is absent in 50–60% of common PGC progenitors at 6–8.5 weeks (Fig. 4b,d), after which all PGCs are negative for H3K27me3 (Fig. 4b,d). Similarly, H2A.Z is enriched in the nucleus of common progenitor stage PGCs at 7–9 weeks in the testis and 7.5 weeks in the ovary (Fig. 4e,f). However, at the end of the common progenitor stage all PGCs become devoid of H2A.Z until around 17 weeks when H2A.Z reappears in the nucleus of a few VASA+ cells in both sexes (Supplementary Fig. S5b,c).
Fig. 4.
Epigenetic reprogramming of H3K27me3 and H2A.Z occurs in the common PGC progenitor. (a,b) Representative immunofluorescence images of H3K27me3 with OCT4A or VASA in (a) testes from 7–17 weeks, and (b) ovaries from 6–18 weeks. (a) Shown is a 10.5-week testis for 7–10.5wk (n=3), a 16.5-week testis for 11–16wk (n=7) and a 17-week testis (n=1). Arrow indicates PGC nucleus with H3K27me3Low levels at 17wk relative to the intensity of staining in the somatic neighbors in the same section (b) Shown is a 7-week for 6–8.5wk (n=3), an 11-week for 9.5–14wk (n=4), and an 18-week for 16.5–18wk (n=3). Open arrowhead indicates strong H3K27me3 accumulation that is indicative of X chromosome inactivation42. (c,d) Quantification of H3K27me3 in OCT4A+ or VASA+ germ cells in (c) testes and (d) ovaries, at the developmental ages indicated. (c) In testes for quantification in OCT4A+, 6 optic fields were counted at 7–10.5wk (n=3), 14 optic fields at 11–16.5wk (n=7) and 6 optic fields at 17wk (n=1). For quantification in VASA+, 6 optic fields were counted at 7–10.5wk (n=3), 10 optic fields at 11–16.5wk (n=7) and 6 optic fields at 17wk (n=1). (d) In ovaries, for quantification in OCT4A+, 6 optic fields were counted at 6–8.5wk (n=3), 10 optic fields at 9.5–14wk (n=4) and 8 optic fields at 16.5–18wk (n=3). For quantification in VASA+, 4 optic fields were counted at 6–8.5wk (n=3), 10 optic fields at 9.5–14wk (n=4) and 7 optic fields at 16.5–18wk (n=3). (e,f) Representative immunofluorescence images of H2A.Z with VASA in testes (e) from 7–9 weeks, and ovary (f) at 7.5 weeks. (e) Shown is a 9-week testis for 7–9wk (n=2) and (f) at 7.5-week ovary (n=2). For immunofluorescence, nuclei were counterstained with DAPI (blue). Scale bars represent 10 um. Data are represented as mean ± SEM. Abreviations: wk= weeks.
RNA-Seq reveals that cKIT+ PGCs are transcriptionally distinct from hESCs
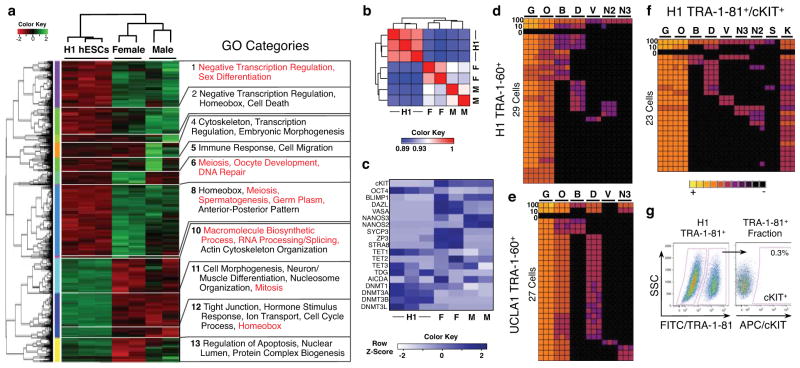
To generate a comprehensive portrait of cKIT+ PGCs in the fetal testis and ovary, we performed RNA-Sequencing (RNA-Seq) of cKIT+ PGCs sorted at 16–16.5 weeks from fetal testes (n=2), fetal ovaries (n=2) and H1 hESCs sorted with the pluripotent marker TRA-1-60 (n=3). At this developmental time point, male cKIT+ PGCs are initiating imprint erasure, whereas in females some imprinted loci show near complete demethylation (H19 and MEG3). A heatmap of the 5,455 differentially expressed genes in at least one of three pair-wise comparisons is shown in Figure 5a. Pearson Correlation Coefficient analysis showed strong correlations between biological replicates in each group (Fig. 5b). Gene Ontology (GO) analysis of the 13 differentially expressed gene clusters revealed that male and female cKIT+ PGCs are enriched in GO terms including negative transcription regulation, sex differentiation, and in females, meiosis and germ plasm when compared to hESCs. In contrast, hESCs are enriched in GO terms related to macromolecule biosynthetic processing, RNA processing/splicing and mitosis (Fig. 5a). Comparing testicular and ovarian cKIT+ PGCs revealed 433 differentially expressed genes, with GO terms such as meiosis, oocyte development and DNA repair. In females this included enrichment in DAZL, VASA, ZP3 and STRA8, and in males NANOS2 and NANOS3 (Fig. 5c).
Fig. 5.
RNA-Seq reveals the transcriptional identity of cKIT+ PGCs. Single cell analysis of hESCs shows stochastic expression of germ line genes. (a) Heat map of 5,455 differentially expressed genes (p value < 0.05) in at least one of three comparisons (male cKIT+ vs H1 hESCs; female cKIT+ vs H1 hESCs; male cKIT+ vs. female cKIT+). The enriched GO terms in the 13 resulting clusters are shown. (b) Heat map of Pearson Correlation Coefficient scores between hESCs and cKIT+ male and female PGCs. (c) Heat map of FKPM values for selected genes in hESCs and cKIT+ male and female PGCs. Abbreviations: M= Male, F= Female. (d,e) Heat map of GAPDH (G), OCT4 (O), BLIMP1 (B), DAZL (D), VASA (V), NANOS3 (N3) and NANOS2 (N2) for H1 hESCs in triplicate (columns) in 100, 10, 0 or single TRA-1-60+ cells (rows) sorted from H1 (d) and UCLA1 (e) hESCs. (f) Heat map as in d and e plus cKIT (K) for TRA-1-81+/cKIT+ H1 hESCs. (g) Gating strategy to sort TRA-1-81+/cKIT+ cells from the H1 hESC line. cKIT+ cells are gated from the TRA-1-81+ fraction, using a FITC secondary antibody against side scatter (SSC).
Given that 5hmC was detected at 16–16.5 weeks by either CGRA and/or immunofluorescence in cKIT+ PGCs, we also examined expression of Ten Eleven Translocation (TET) genes, which are responsible for converting 5mC to 5hmC19–21 (Fig. 5c). All three TET family members (TET1-3) are expressed by male and female PGCs, with a significant enrichment of TET2, and reduced expression of TET1 relative to H1 hESCs. We also evaluated the DNA Methyltransferases (DNMT) DNMT1, DNMT3A, DNMT3B and DNMT3L. Male but not female PGCs exhibited a significant decrease in DNMT1 relative to H1 hESCs. Furthermore, all PGC samples had reduced expression of DNMT3A and DNMT3B relative to H1. AICDA (also known as AID) and TDG were expressed at variable levels in H1 and also in PGCs of both sexes.
cKIT/TRA-1-81 positive PGCs generated in vitro correspond to immature pre-gonadal PGCs
We next sought to generate cKIT+ PGCs in vitro from H1 (XY) and UCLA1 (XX) hESCs22,23. In our sorting strategy we incorporated TRA-1-81, as the second marker with cKIT, based on the high OCT4/TRA-1-81 correlation in the human gonad prior to 10 developmental weeks (Supplementary Fig. S3d). Given that undifferentiated hESCs expressed detectable levels of germ line genes by RNA-Seq similar to previously reported 24,25, we performed single cell analysis of sorted TRA-1-60+ hESCs (Fig. 5d,e) and TRA-1-81+/cKIT+ hESCs (Fig 5f,g). As expected, analysis of 100-pooled undifferentiated hESCs resulted in identification of 5/5 or 3/4 PGC signature genes together with OCT4. However interrogation at a single cell level revealed that the major PGC determinant BLIMP126 was rarely expressed, and the majority of PGC signature genes were seldom co-expressed in single cells regardless of sorting strategy.
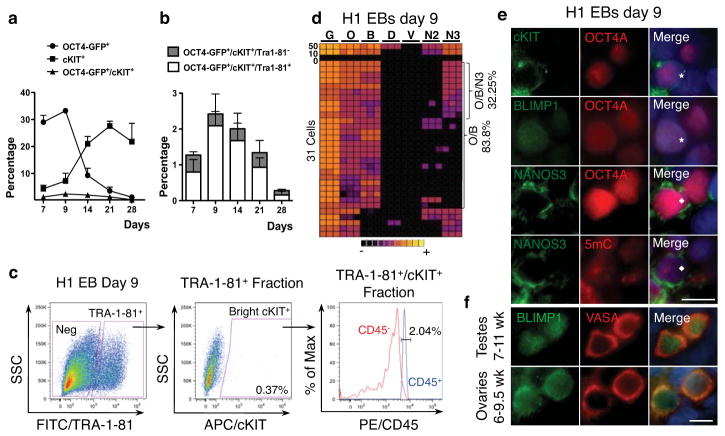
Next, PGC differentiation was evaluated for up to 16 days by serum-induced differentiation of hESCs; 1) as embryoid bodies (EBs) with and 2) without BMP4 addition, 3) adherent monolayer differentiation on growth factor reduced matrigel (GFR M/G), 4) differentiation on human fetal gonadal stromal cells (hFGSC), and 5) a combination of EB differentiation, followed by plating EBs on hFGSCs (Fig. 6 and 7). First we verified that TRA-1-81 faithfully reports OCT4 expression upon hESC differentiation for 28 days by flow cytometry using the H1 OCT4-GFP line created by homologous recombination27 (Fig. 6a,b). Using the gating strategy shown in Fig 6c (H1 day 9 EBs), we sorted the brightest TRA-1-81+/cKIT+ cells upon in vitro differentiation averaging 0.3% of the live population, with no significant difference in percentage positive cells when comparing differentiation strategy or length of time in differentiation. Our data show that >97% of TRA-1-81+/cKIT+ differentiated cells are negative for CD45, excluding the possibility of contamination with cKIT+ hematopoietic progenitors28 (Fig. 6c). Single cell analysis of cKIT+/TRA-1-81+ cells sorted from EBs at day 9 revealed a significant increase in the proportion of cells expressing BLIMP1 (Fig 6b). We show that the identity of putative PGCs was heterogeneous being either O/B double positive, or O/B/N3 triple positive with no co-expression of DAZL or VASA (Fig. 6d and Fig. 7c).
Fig. 6.
In vitro hESC differentiation generates rare germ line progenitors that are cKIT/TRA-1-81 positive. (a) Percentage of OCT4-GFP+, cKIT+ and total OCT4-GFP+/cKIT+ generated upon adherent differentiation for the indicated time points. (b) Percentage of TRA-1-81+ and TRA-1-81− cells within the OCT4-GFP+/cKIT+ population generated by adherent differentiation of H1 OCT4-GFP for the indicated time points. TRA-1-81 is co-expressed by the majority of cKIT/OCT4-GFP double positive cells upon hESC differentiation. (c) Gating strategy to sort cKIT+/TRA-1-81+ cells (shown are H1-EBs differentiated for 9 days). cKIT+ cells are gated from the TRA-1-81+ fraction. Flow cytometry for CD45 in the TRA-1-81+/cKIT+ cells reveals <3% contamination by CD45 positive cells. (d) Heat map of GAPDH (G), OCT4 (O), BLIMP1 (B), DAZL (D), VASA (V), NANOS2 (N2) and NANOS3 (N3), in triplicate (columns) in 50, 10, 0 or single cells (rows) for TRA-1-81+/cKIT+ sorted from H1 day 9 EBs. (e) Immunofluorescence of cKIT, BLIMP1 and NANOS3 with OCT4A, and of NANOS3 with 5mC on EBs differentiated for 9 days from H1 hESCs. Staining is performed on adjacent sections and asterisk and diamond indicate the same cell. (f) Representative immunofluorescence images of BLIMP1 with VASA in testes from 7–10.5 weeks (n=3), shown is a 10.5-week testis and ovaries from 6–8 weeks (n=3), shown is an 8-week ovary. Nuclei were counterstained with DAPI (blue). Scale bars represent 10 um. Abbreviations: wk= week. All data are mean ± SEM.
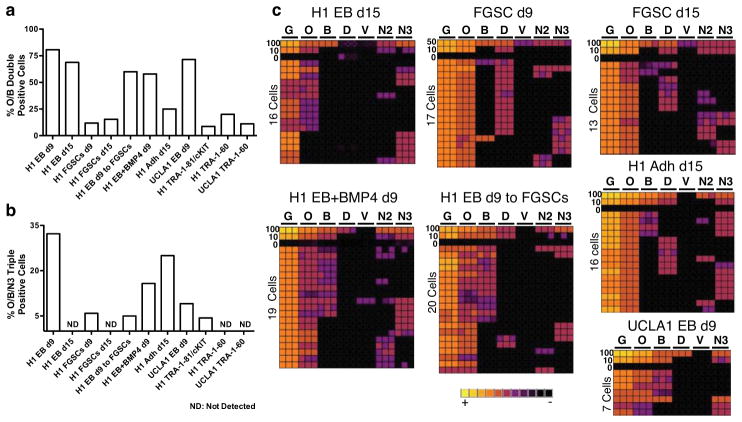
Fig. 7.
In vitro PGC differentiation from hESCs using five alternate differentiation techniques. (a,b) Comparison of (a) the percent of O/B double positive single cells, and (b) O/B/N3 triple positive within the TRA-1-81+/cKIT+ sorted fraction from five alternate differentiation techniques. (c) Heat map of GAPDH (G), OCT4 (O), BLIMP1 (B), DAZL (D), VASA (V), NANOS3 (N3) and NANOS2 (N2) for H1 in triplicate (columns) in 100, 50, 10, 0 or single TRA-1-81+/cKIT+ cells (rows) sorted from H1 and UCLA1 hESCs using five alternate differentiation strategies.
Although rare O/B and O/B/N3 single cells were identified in the undifferentiated state (Fig 5d–f), differentiation resulted in a clear enrichment for both O/B and O/B/N3 cell types. In particular, EB differentiation for 9 days yielded 9-fold enrichment in O/B and 7.5-fold enrichment in O/B/N3 cells relative to cKIT+/TRA-1-81+ self-renewing hESCs (quantified in Fig. 7a,b). Immunofluorescence of day 9 EBs verified that cKIT+ cells co-expressed OCT4A, exhibited nuclear localization of BLIMP1 and expressed NANOS3 in the cytoplasm similar to what is observed in cKIT+ PGCs from the human fetus (Fig. 6e,f and Supplementary Fig. S3c). Immunofluorescence also revealed that all NANOS3/OCT4A+ cells in the day 9 EB were positive for 5mC (Fig. 6i). Taken together, our data suggests that TRA-1-81+/cKIT+/O/B/N3 cells in EBs correspond to human PGCs prior to gonadal colonization and loss of 5mC.
Using PGC differentiation in EBs for 9 days as a comparison, we show that O/B/N3 putative PGCs are transient being lost by day 15 of EB formation (Fig. 7b,c). Transferring day 9 EBs to hFGSCs for an additional 7 days (16 days total) was consistent with an increase in survival and/or differentiation of O/B/N3 triple positive cells, however DAZL or VASA RNA were not induced in the O/B/N3 population (Fig. 7c). Sustaining the O/B/N3 triple positive population within the TRA-1-81+/cKIT+ fraction for 15 days was also achieved using adherent differentiation on GFR M/G (Fig. 7c).
DISCUSSION
By analyzing 134 human embryonic and fetal samples from 6–20 developmental weeks and in vitro PGC differentiation from hESCs we propose the following roadmap of human germ line development (Fig. 8). Our data reveal that the first 16 days of hESC differentiation in vitro, either as EBs or as monolayers creates a cKIT+/TRA-1-81+/OCT4A+ PGC population equivalent to a pre-gonadal, PGC with 5mC. After reprogramming 1 (denoted by the global depletion of 5mC from the genome followed by enrichment of H3K27me311), we speculate that DAZL and VASA are next expressed giving rise to the cKIT+/OCT4A+/VASA+ common gonadal PGC progenitors which then embark on reprogramming 2 and uncoupling of cKIT expression from VASA. This uncoupling of germ cell-expressed genes into separate populations was previously reported in second trimester testes and ovaries for OCT4 and VASA protein29. Our data is in agreement with cKIT being on the surface of OCT4+/VASA negative cells in the fetal gonad29. Furthermore, the SYCP3 staining described here also supports the hypothesis that single VASA+ germ cells in the ovary are first to enter the ovarian reserve in fetal life29.
Fig. 8.
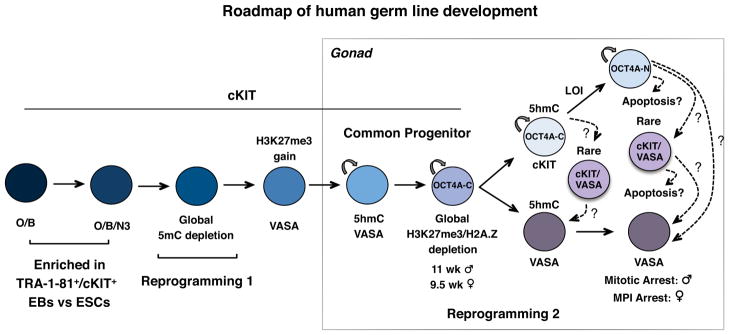
Summarized roadmap of human germ line development. Reprogramming 1, occurs prior to 6–7 developmental weeks and is characterized by global loss of 5mC from PGC DNA. Reprogramming 2 begins in the common PGC progenitor stage after acquisition of H3K27me3 (10.5 weeks in testes and 8.5 weeks in ovaries) and involves global loss of H3K27me3 and H2A.Z followed by imprint erasure in cKIT+ PGCs more than 1 month later. In vitro hESC differentiation using TRA-1-81+/cKIT+ sorting generates a rare cKIT+ PGC population that is O/B double positive or O/B/N3 triple positive and corresponds to newly specified PGCs, prior to reprogramming 1. Abbreviations: O/B = OCT4/BLIMP1, O/B/N3 = OCT4/BLIMP1/NANOS3, LOI= Loss Of Imprinting, MPI= Meiotic Prophase I.
Our results show that reprogramming 2 in the cKIT+ lineage follows a protracted series of events that are similar but not identical to the mouse. This begins with the relatively stable wholesale epigenetic loss of H3K27me3 and H2A.Z in the common progenitor followed by either loss of OCT4A or expression in the cytoplasm. Traditionally cytoplasmic localization of OCT4 is due to expression of the OCT4B splice variant16. Here we used an antibody that discriminates OCT4A from OCT4B17 suggesting that OCT4A in PGCs either translocates to the cytoplasm, or is attenuated there possibly for degradation. The significance of cytoplasmic OCT4A is unknown, but is notably coincident with major global epigenetic changes.
A major event in reprogramming 2 is the erasure of cytosine methylation from DMRs of imprinted genes, which in mice is hypothesized to be active13. In the current study we show that 5hmC and the TET enzymes are dynamically expressed by cKIT+ human PGCs, as well as additional molecular candidates that could actively modify 5mC/5hmC or remove modified 5hmC from the genome including, AICDA and TDG14,19–21,30,31. In hESCs and fibroblasts where cytosine methylation at imprinted DMRs is stably inherited, 5hmC is not detected at PEG3 DMR. In contrast in cKIT+ PGCs, where the fate of this locus is demethylation, 5hmC is enriched. Despite this tantalizing correlation, future studies are needed to determine the role of TETs and 5hmC in imprint erasure. One observation from our reference map is that imprint erasure occurs over weeks and in a locus specific manner. This relatively long window for imprint erasure in cKIT+ PGCs stands in contrast to the mouse where erasure at many imprinted loci occur within 24 hours32. Therefore, our data support the idea that removal of 5mC from imprinted DMRs in humans may involve diverse, locus-specific and time-dependent strategies.
Using undifferentiated hESCs we show that germ line genes are expressed in a stochastic manner in the undifferentiated state, with only rare undifferentiated hESCs expressing the major PGC determinant BLIMP126. However, with in vitro differentiation using cKIT with TRA-1-81 we enriched for BLIMP1 expression in cKIT+ cells, called O/B cells that we speculate represent the first lineage restricted PGCs equivalent to e6.25 in mice33,34. Differentiation also results in enrichment of O/B/N3 triple positive cKIT+ cells that we speculate represent the next stage in PGC development after O/B and prior to 5mC loss in reprogramming 1. O/B/N3 cells were never observed in single hESCs sorted for TRA-1-60 and were found in less than 5% of TRA-1-80+/cKIT+ sorted undifferentiated hESCs. The fact that at a single cell level VASA and DAZL were never co-expressed with O/B/N3 in TRA-1-81+/cKIT+ cells with differentiation does not refute previous findings using DAZL and VASA as markers to define germ line identity4,24,35–41. On the contrary, we propose that sorting for TRA-1-81+/cKIT+ specifically enriches for newly specified germ line cells prior to DAZL and VASA expression and that acquisition of this immature cell can be achieved regardless of differentiation strategy.
In conclusion, we show that accurate interpretation of in vitro differentiation requires not only a detailed understanding of the human counterpart, but also analysis at a single cell level to confirm molecular identity and rule out stochastic gene expression. In the long term for the field to move forward, functional assays to determine human germ line quality are urgently required. One possibility is using nonhuman primate hPSCs where transplantation of in vitro derived germ cells is ethically possible. Alternatively, methods for culturing endogenous PGCs in a format that promotes self-renewal and/or differentiation to gonocytes, gonia and meiotic cells rather than EGCs is also needed. The human germ cell lineage is particularly challenging to study due to lack of functional assays to test germ cell identity and quality, therefore the generation of robust molecular maps as described here are the first steps to unveiling this important lineage.
Supplementary Material
Acknowledgments
The authors would like to thank Angela Chen, MD and the UCLA Translational Pathology Core Laboratory and the UCLA Gene and Cellular Core Laboratory for some of the gonadal samples used in this study. We also thank Joseph Hargan-Calvopina, Marisabel Oliveros-Etter and Silvia Diaz-Perez for critical reading of the manuscript, Felicia Codrea and Jessica Scholes for FACS and Steven Peckman from the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research for critical assistance with human subject and embryonic stem cell review. This work was supported primarily by fund number 1R01HD058047 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD) (ATC), as well as the Iris Cantor-UCLA Women’s Health Pilot Project (ATC) and 1P01GM081621 from NIGMS. The Laboratory of Developmental Biology, University of Washington, Seattle is supported by NIH Award Number 5R24HD000836 from the NICHD. Human fetal tissue requests can be made to: bdrl@u.washington.edu.
Footnotes
AUTHOR CONTRIBUTIONS
S.G. designed and performed the experiments, analyzed data and wrote the manuscript; Z.L. performed flow analyses, RNA and DNA extraction for some gonadal samples, J.J.V. performed the single cell analysis of female ovary at 14 weeks; K.X.Z. and M.P. performed the RNA-Sequencing data analysis; A.C. provided gonadal samples used in this study; A.T.C. designed the experiments, analyzed data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no conflict of interest.
References
- 1.Ohinata Y, et al. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–84. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi K, et al. Offspring from Oocytes Derived from in Vitro Primordial Germ Cell-Like Cells in Mice. Science. 2012 Oct 4; doi: 10.1126/science.1226889. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 3.Reijo R, et al. Mouse autosomal homolog of DAZ, a candidate male sterility gene in humans, is expressed in male germ cells before and after puberty. Genomics. 1996;35:346–352. doi: 10.1006/geno.1996.0366. [DOI] [PubMed] [Google Scholar]
- 4.Park TS, et al. Derivation of primordial germ cells from human embryonic and induced pluripotent stem cells is significantly improved by co-culture with human fetal gonadal cells. Stem Cells. 2009;27:783–795. doi: 10.1002/stem.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaskell TL, Esnal A, Robinson LLL, Anderson RA, Saunders PTK. Immunohistochemical profiling of germ cells within the human fetal testis: identification of three subpopulations. Biol Reprod. 2004;71:2012–2021. doi: 10.1095/biolreprod.104.028381. [DOI] [PubMed] [Google Scholar]
- 6.Robinson L, Gaskell T, Saunders P, Anderson R. Germ cell specific expression of c-kit in the human fetal gonad. Molecular human reproduction. 2001;7:845. doi: 10.1093/molehr/7.9.845. [DOI] [PubMed] [Google Scholar]
- 7.Høyer PE, Byskov AG, Møllgård K. Stem cell factor and c-Kit in human primordial germ cells and fetal ovaries. Molecular and cellular endocrinology. 2005;234:1–10. doi: 10.1016/j.mce.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Pauls K. Spatial expression of germ cell markers during maturation of human fetal male gonads: an immunohistochemical study. Hum Reprod. 2005;21:397–404. doi: 10.1093/humrep/dei325. [DOI] [PubMed] [Google Scholar]
- 9.Kerr C, Hill C, Blumenthal P, Gearhart J. Expression of pluripotent stem cell markers in the human fetal ovary. Human reproduction. 2008;23:589. doi: 10.1093/humrep/dem411. [DOI] [PubMed] [Google Scholar]
- 10.Kerr CL, Hill CM, Blumenthal PD, Gearhart JD. Expression of pluripotent stem cell markers in the human fetal testis. Stem Cells. 2008;26:412–421. doi: 10.1634/stemcells.2007-0605. [DOI] [PubMed] [Google Scholar]
- 11.Seki Y, et al. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 12.Hajkova P, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajkova P, et al. Genome-Wide Reprogramming in the Mouse Germ Line Entails the Base Excision Repair Pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinclair AH, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 16.Atlasi Y, Mowla SJ, Ziaee SAM, Gokhale PJ, Andrews PW. OCT4 Spliced Variants Are Differentially Expressed in Human Pluripotent and Nonpluripotent Cells. Stem Cells. 2008;26:3068–3074. doi: 10.1634/stemcells.2008-0530. [DOI] [PubMed] [Google Scholar]
- 17.Warthemann R, Eildermann K, Debowski K, Behr R. False-positive antibody signals for the pluripotency factor OCT4A (POU5F1) in testis-derived cells may lead to erroneous data and misinterpretations. Molecular human reproduction. 2012 doi: 10.1093/molehr/gas032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruzov A, et al. Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res. 2011;21:1332–1342. doi: 10.1038/cr.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koh KP, et al. Tet1 and Tet2 Regulate 5-Hydroxymethylcytosine Production and Cell Lineage Specification in Mouse Embryonic Stem Cells. Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson JA. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 23.Diaz Perez SV, et al. Derivation of new human embryonic stem cell lines reveals rapid epigenetic progression in vitro that can be prevented by chemical modification of chromatin. Human Molecular Genetics. 2012;21:751–764. doi: 10.1093/hmg/ddr506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark AT. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Human Molecular Genetics. 2004;13:727–739. doi: 10.1093/hmg/ddh088. [DOI] [PubMed] [Google Scholar]
- 25.Zwaka TP. A germ cell origin of embryonic stem cells? Development. 2005;132:227–233. doi: 10.1242/dev.01586. [DOI] [PubMed] [Google Scholar]
- 26.Bao S, et al. The Germ Cell Determinant Blimp1 Is Not Required for Derivation of Pluripotent Stem Cells. Cell Stem Cell. 2012;11:110–117. doi: 10.1016/j.stem.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamei K, et al. Microfluidic image cytometry for quantitative single-cell profiling of human pluripotent stem cells in chemically defined conditions. Lab Chip. 2010;10:1113. doi: 10.1039/b922884e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briddell R, et al. Further Phenotypic Characterization and Isolation of Human Hematopoietic Progenitor Cells Using a Monoclonal-Antibody to the C-Kit Receptor. Blood. 1992;79:3159–3167. [PubMed] [Google Scholar]
- 29.Anderson R, Fulton N, Cowan G, Coutts S, Saunders P. Conserved and divergent patterns of expression of DAZL, VASA and OCT 4 in the germ cells of the human fetal ovary and testis. BMC Dev Biol. 2007;7:136. doi: 10.1186/1471-213X-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhutani N, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maiti A, Drohat AC. Thymine DNA Glycosylase Can Rapidly Excise 5-Formylcytosine and 5-Carboxylcytosine: Potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hajkova P, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 33.Ohinata Y, Sano M, Shigeta M, Yamanaka K, Saitou M. A comprehensive, non-invasive visualization of primordial germ cell development in mice by the Prdm1-mVenus and Dppa3-ECFP double transgenic reporter. Reproduction. 2008;136:503–514. doi: 10.1530/REP-08-0053. [DOI] [PubMed] [Google Scholar]
- 34.Kurimoto K, et al. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 2008;22:1617–1635. doi: 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kee K, Gonsalves JM, Clark AT, Pera RAR. Bone morphogenetic proteins induce germ cell differentiation from human embryonic stem cells. Stem Cells Dev. 2006;15:831–837. doi: 10.1089/scd.2006.15.831. [DOI] [PubMed] [Google Scholar]
- 36.Kee K, Angeles VT, Flores M, Nguyen HN, Pera RAR. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aflatoonian B, et al. In vitro post-meiotic germ cell development from human embryonic stem cells. Hum Reprod. 2009;24:3150–3159. doi: 10.1093/humrep/dep334. [DOI] [PubMed] [Google Scholar]
- 38.Tilgner K, et al. Expression of GFP under the control of the RNA helicase VASA permits fluorescence-activated cell sorting isolation of human primordial germ cells. Stem Cells. 2010;28:84–92. doi: 10.1002/stem.263. [DOI] [PubMed] [Google Scholar]
- 39.Panula S, et al. Human germ cell differentiation from fetal-and adult-derived induced pluripotent stem cells. Human Molecular Genetics. 2011;20:752–762. doi: 10.1093/hmg/ddq520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medrano JV, Ramathal C, Nguyen HN, Simon C, Reijo Pera RA. Divergent RNA-binding Proteins, DAZL and VASA, Induce Meiotic Progression in Human Germ Cells Derived in Vitro. Stem Cells. 2012;30:441–451. doi: 10.1002/stem.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chuang CY, et al. Meiotic Competent Human Germ Cell-like Cells Derived from Human Embryonic Stem Cells Induced by BMP4/WNT3A Signaling and OCT4/EpCAM (Epithelial Cell Adhesion Molecule) Selection. J Biol Chem. 2012;287:14389–14401. doi: 10.1074/jbc.M111.338434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plath K. Role of Histone H3 Lysine 27 Methylation in X Inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 43.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 44.Vincent JJ, et al. Single Cell Analysis Facilitates Staging of Blimp1-Dependent Primordial Germ Cells Derived from Mouse Embryonic Stem Cells. PLoS ONE. 2011;6:e28960. doi: 10.1371/journal.pone.0028960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boissonnas C, et al. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. European Journal of Human Genetics. 2009;18:73–80. doi: 10.1038/ejhg.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geuns E, Hilven P, Van Steirteghem A, Liebaers I, De Rycke M. Methylation analysis of KvDMR1 in human oocytes. Journal of Medical Genetics. 2006;44:144–147. doi: 10.1136/jmg.2006.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kagami M, et al. The IG-DMR and the MEG3-DMR at Human Chromosome 14q32.2: Hierarchical Interaction and Distinct Functional Properties as Imprinting Control Centers. PLoS Genet. 2010;6:e1000992. doi: 10.1371/journal.pgen.1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zechner U, et al. Quantitative methylation analysis of developmentally important genes in human pregnancy losses after ART and spontaneous conception. Molecular human reproduction. 2010;16:704–713. doi: 10.1093/molehr/gap107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.