Abstract
While calcium-phosphate has been used to deliver plasmid DNA (pDNA) for decades, the method is typically characterized by low and irreproducible transfection efficiency relative to the other non-viral approaches, such as liposomes and polymers. Here we report a novel gene transfer vector comprising lipid-coated nano-calcium-phosphate (LNCP) that provides consistently efficient and satisfactory pDNA delivery. It is based on core-shell nanoparticles comprising a calcium-phosphate core and a cationic lipid shell. This method, in contrast to the solution precipitation methods used in the past, yields colloidally stable calcium-phosphate nanoparticles inside the cationic liposomes. Our results indicate that the particle size and the size distribution of the LNCP remain virtually unchanged even after 21 days of storage. Atomic force microscopy measurements reveal that the LNCP have a 5-fold higher rigidity than common cationic liposomes. The LNCP transfected pDNA 24 times greater than the naked pDNA and 10-fold greater relative to the standard calcium-phosphate precipitation preparations, suggesting that the LNCP may have potential as a novel transfection agent for gene therapy.
Keywords: Nanoparticles, Cationic liposome, Calcium-phosphate, Gene therapy
1. Introduction
Gene therapy holds a great potential for treatment of a wide variety of acquired and genetic diseases. Transfer of plasmid DNA by non-viral vectors, although generally not as efficient as viral vectors, is preferred because of safety concerns associated with viral vectors (Kim et al., 2004). Among non-viral vectors, cationic liposomes are the most frequently used transfection vectors, due to their low toxicity and weak immunogenicity, and ease of production in clinically relevant quantities (Maitani et al., 2007; Olton et al., 2007).
While the use of calcium-phosphate precipitation to transfect cultured cells has been introduced decades ago (Olton et al., 2007; Welzel et al., 2004), its poor colloidal stability hinders extensive application (Bisht et al., 2005; Luo et al., 2004). It has been shown that the control of the particle size is a critical issue in achieving satisfactory transfection efficiency (Kneuer et al., 2000; Roy et al., 2003). A small size and good colloidal stability of the calcium-phosphate nanoparticles have been achieved by liposome-directed growth of the calcium-phosphate nanoshells (Schmidt and Ostafin, 2002). However, calcium nitrate has been used in that study, which is a strong oxidizer and can induce severe cytotoxicity. In addition, the gene transfection efficiency of the liposome-calcium-phosphate hybrid nanoparticles has not been completely explored.
In the present report, we described a novel design of lipid-coated nano-calcium-phosphate (LNCP) to enhance pDNA delivery efficiency. These core-shell nanoparticles consist of a calcium-phosphate core and a cationic lipid shell. This method, in contrast to the solution precipitation methods used traditionally (Bisht et al., 2005; Olton et al., 2007; Sokolova et al., 2007), yields colloidally stable vectors, comprising calcium-phosphate nanoparticles inside cationic liposomes. The particle size and the size distribution of the LNCP remained virtually unchanged after 21 days. Atomic force microscopy (AFM) measurements revealed that the LNCP had a 5-fold higher rigidity than typical cationic liposomes, which indicated the presence of a calcium-phosphate core. The LNCP transfected pDNA 24 times more efficiently than the naked pDNA and 10-fold greater relative to the standard calcium-phosphate precipitation preparations in NIH 3T3 cells, suggesting that the LNCP can efficiently promote gene delivery.
2. Materials and methods
2.1. Material
DOPE (L-alpha-dioleoyl phosphatidylethanolamine), DOTAP (1,2-dioleoyl-3-trimethylammonium-propane) and DC-Chol (cholesteryl 3β-N-(dimethylaminoethyl)carbamate hydrochloride) were purchased from Avanti Polar Lipids Inc. Cholesterol and TPGS (d-α-Tocopheryl Polyethylene Glycol-1000 Succinate) were obtained from Fisher Scientific Inc. DDAB (dimethyldioctadecylammonium bromide), MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide), calcium acetate, and monosodium phosphate were purchased from Sigma–Aldrich Chemical Co. (St. Louis, MO, USA). The plasmid (pCMV-GFP) was purchased from Invitrogen. All other reagents were of analytical grade.
2.2. Liposomes and lipid-coated nano-calcium-phosphate (LNCP) preparation
The DDAB liposomes, composed of DDAB/Cholestrol/TPGS (59:40:1 mol/mol), were prepared by ethanol injection method (Guo and Lee, 2000; Maitani et al., 2007). Briefly, 20–50 mg/ml of individual lipid stock solutions were prepared by dissolving the lipids in ethanol. Appropriate amounts of the lipid stock solutions were mixed to achieve the desired lipid ratio. The ethanol solution was rapidly injected to 4× volume of HEPES buffer (20 mM, pH 7.4). After the ethanol injection, the liposomes were sonicated for 1–2 min by a probe sonicator and dialyzed against distilled water for 4 h.
For the preparation of the LNCP, the same lipid composition was used, while a mixture of calcium acetate and monosodium phosphate was used instead of HEPES buffer. The concentrations of calcium acetate and monosodium phosphate in the buffer were 18 and 5.4 mM, respectively. After the probe sonication, the LNCP was dialyzed against distilled water for 4 h to remove the Ca2+ and PO43− ions outside the liposomes and form nano-calcium-precipitate inside the liposomes (Chu and Liu, 2005).
Two other cationic liposome formulations, with the compositions of DOTAP/DOPE (1:1 mol/mol) (DOTAP liposome) (Guo et al., 2002) and DC-Chol/DOPE (1:2 mol/mol) (DC-Chol liposome) (Maitani et al., 2007), were also prepared by the ethanol injection method as described above.
2.3. Gel retardation assay
Gel retardation assay was used to determine the ability of the LNCP to bind DNA. LNCP/pDNA complexes were formed by mixing the pCMV-GFP plasmid DNA (0.2 μg) with the LNCP at selected lipid/DNA (L/D, w/w) ratios. After allowing 20 min for complexes formation, samples were subjected to electrophoresis on a 1.0% agarose gel containing 0.1 μg/ml ethidium bromide. A quantity of 0.1 μg of pDNA was loaded into each well.
2.4. Serum stability of LNCP/pDNA complexes
The ability of LNCP/pDNA complexes to protect DNA from serum DNase digestion was characterized by serum protection assay (Adami et al., 1998). LNCP/pDNA complexes were formed by mixing the pDNA with LNCP at selected L/D ratios, and incubating them for 20 min at room temperature. The resulting complexes were combined with equal volume of fetal bovine serum and allowed to incubate at 37 °C for 2 h. The samples were loaded onto 1.0% agarose gel containing 0.1 μg/ml ethidium bromide for gel electrophoresis. An equal amount of naked pDNA without serum incubation was loaded as a control.
2.5. Calcium determination
Inductively coupled plasma-optical emission spectrometers (ICP-OES) was used to determine the total calcium amount in the LNCP suspension. The sample preparation method has been described previously (Barth et al., 1991). Briefly, the liposome samples were digested by adding appropriate amounts of concentrated sulfuric acid, incubated in oil bath at 105 °C, and decolorized by adding 50% H2O2. Once the solution was clear, deionized water was added to the sample to bring up it to desired volume for calcium determination. Calcium concentrations were then determined by ICP-OES.
2.6. Size, zeta potential and morphology of the liposomes and LNCP
The particle size distributions of the liposomes and LNCP were examined by dynamic light scattering on a NICOMP Submicron Particle Sizer Model 370 (NICOMP, Santa Barbara, CA). All particle size data refer to volume-weighted distributions.
The zeta potentials (ζ) of DDAB liposomes and LNCP were determined in 0.1× PBS on a ZetaPALS instrument (Brookhaven Instruments Corp., Holtsville, NY).
The liposomes and LNCP were examined under an AFM. A drop of diluted liposome or LNCP suspensions was placed on a fresh mica surface and allowed to dry at room temperature. AFM imaging was performed using a Multi-Mode scanning probe microscope (Vecoo Instrument) in a tapping mode. Rectangular cantilevers with a nominal spring constant of 12–103 N/m, a resonance frequency of ~300 kHz and a tip radii curvature of ~10 nm were used in all measurements. All images were recorded in a height model and the scanning rate was 1.0 Hz.
2.7. Cell culture
NIH 3T3 cells were grown in DMEM culture medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin, at 37 °C in a humidified atmosphere containing 5% CO2.
2.8. In vitro transfection
NIH 3T3 cells were grown until 80% confluence was reached. One day before transfection, the cells were seeded in a 24-well plate (6 × 104 cells/well) with 500 μl of DMEM growth medium and incubated at 37 °C in a 5% CO2 environment. On the day of transfection, cells were incubated with the transfection agents, containing 1 μg plasmid DNA, in 200 μl serum-free DMEM growth medium for 4 h at 37 °C. The transfection medium was then removed, and the cells were incubated for a further 24 h in fresh DMEM growth medium before analysis of the GFP expression.
The experimental groups included: (1) cells transfected with the standard calcium-phosphate preparation (Sokolova et al., 2006), (2) cells transfected with pDNA/DDAB lipoplexes, (3) cells transfected with pDNA/DDAB lipoplexes mixed with Ca(Ac)2 and NaH2PO4, (4) cells transfected with pDNA/LNCP complexes, (5) cells transfected with pDNA/DOTAP lipoplexes, (6) cells transfected with pDNA/DCChol lipoplexes, (7) cells transfected with pDNA alone, (8) untreated control.
In all transfection protocols, 1 μg plasmid DNA was mixed with various amounts of trasfection vectors, followed by a 10 s vortex mixing, and then incubated for 10 min at room temperature before use. Appropriate amounts of cationic liposome and the LNCP were used to generate an L/D ratio between 2 and 12 (Guo et al., 2002).
2.9. Transfection in the presence of chloroquine
Transfection experiments were performed with or without chloroquine in order to examine the transfection mechanism of the LNCP. Transfection experiments and assays were performed generally following the transfection protocol described above, except that cells were incubated with LNCP/pDNA complexes or naked pDNA in the presence or absence of 100 μM chloroquine during transfection.
2.10. Flow cytometry and fluorescent microscopy
The cells were analyzed for GFP expression 24 h after exposure to the transfection agents. The cells were rinsed with 500 μl PBS (pH 7.4) twice and detached with trypsin-EDTA (0.25%). The cells were then centrifuged down and resuspended with 500 μl of PBS. The cell suspension was directly introduced into a Beckman Coulter EPICS XL (Beckman Coulter) to determine the level of expression of GFP. For each cell sample, a minimum of 10,000 events were collected under the LIST mode. The data were analyzed using WinMDI software. Transfection efficiencies of various formulations were reported as multiples of those of untreated control.
The presence of GFP fluorescence was visually determined by an X51 Olympus fluorescence microscope (Olympus Optical Co., Tokyo, Japan).
2.11. Cell viability assay
The cytotoxicity of different transfection agents was determined by MTT assay, as described previously (Sudimack et al., 2002). Briefly, NIH 3T3 cells were seeded in 96-well plates at 3 × 103 cells/well. The cells were treated with different trasfection reagents, diluted with DMEM growth medium to provide the same concentration in the protocols of transfection, and incubated for 24 h prior to toxicity assessment. MTT was then added to the culture medium at a concentration of 0.6 mg/ml, and the cells were incubated at 37 °C for additional 2 h. The medium was removed, the cells were dissolved in DMSO, and the absorption at 495 nm was measured as an indicator of cell viability by using an automatic plate reader. Viability was expressed as a percentage of the untreated cells control. Cytotoxicity experiments were repeated on three separate occasions.
2.12. Statistical analysis
The statistically significant differences were analyzed with Microsoft Excel 2003 software (Microsoft, Redmond, WA). Student's t-test was used to compare differences between two groups. Results were considered significant when p values were <0.05.
3. Results and discussion
3.1. Particle size, zeta potential, and stability
The particle size of various cationic liposomes and the LNCP determined by dynamic light scattering (DLS) are presented in Table 1. The average particle size of the LNCP was ~30% larger relative to the DDAB liposomes. This increase is not uncommon, because calcium ions have been known to induce liposome aggregation and/or fusion during the formation (Mortazavi et al., 2007; Yohannes et al., 2006). The zeta potentials of the DDAB liposomes and the LNCP were 52.87 ± 2.44 and 60.97 ± 1.92 mV, respectively.
Table 1.
Mean particle size of various cationic liposomes and the LNCP.
| Lipid composition | Lipid composition (mol ratio) | Mean particle size (nm) |
|---|---|---|
| DDAB liposome | DDAB/Chol/TPGS = 59:40:1 | 101.3 ± 58.1 |
| DC-Chol liposome | DC-Chol/DOPE = 1:2 | 130.9 ± 64.8 |
| DOTAP liposome | DOTAP/DOPE = 1:1 | 116.1 ± 52.0 |
| LNCP | DDAB/Chol/TPGS = 59:40:1 | 132.0 ± 71.7 |
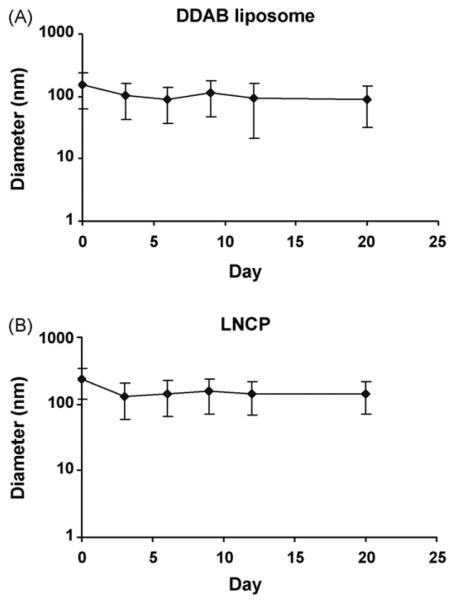
Fig. 1 illustrates the effect of storage (4 °C) on the average particle size of the DDAB liposomes and the LNCP. The mean particle size of the DDAB liposomes and the LNCP decreased during the first 3 days, then remained almost unchanged even at 21 days.
Fig. 1.
Particle size and colloidal stability. The mean diameter of (A) DDAB liposomes and (B) the LNCP remained stable for up to 3 weeks after synthesis and storage at 4 °C.
3.2. Gel retardation assays and serum stability of LNCP/pDNA complexes
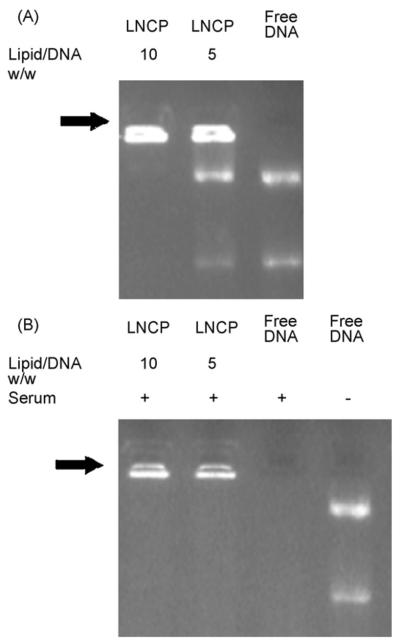
The pDNA condensing ability of the LNCP as a function of L/D ratio was examined by gel retardation assay. The L/D ratio of LNCP/pDNA complexes reflects the overall positive to negative charge balance of the complexes. The results in Fig. 2A show that L/D ratio of 10:1 results into complete gel retardation of pDNA, suggesting that the LNCP was capable of forming LNCP/pDNA complexes.
Fig. 2.
Mobility of pDNA/LNCP complexes analyzed by agarose gel electrophoresis. (A) Agarose gel electrophoresis of pDNA/LNCP complexes with ethidium bromide staining, particles formed at L/D ratios of 5, 10, and free pDNA control. (B) pDNA protection assay. Plasmid DNA alone or pDNA/LNCP complexes at different lipid/DNA ratios were exposed to 50% FBS for 2 h before gel electrophoresis analysis. Complexes were prepared at varying lipid/DNA ratio as described in the method section. Black arrow indicates the place of the wells.
Protecting the pDNA from enzymatic degradation during formulation and delivery is an important factor for a successful gene transfer. The endogenous endonucleases in serum are known to degrade uncomplexed pDNA within half an hour during in vitro incubation (Adami et al., 1998). The LNCP/pDNA complexes and naked DNA were challenged by incubation in 50% serum for 2 h. As shown in Fig. 2B, after incubation with serum, the pDNA in LNCP/pDNA complexes remained intact, whereas naked DNA was completely digested. Furthermore, LNCP/pDNA complexes still showed complete gel retardation of pDNA after serum incubation. The result also demonstrated that the LNCP/pDNA complexes exhibit good colloidal stablility in the presence of serum proteins.
3.3. Morphological characterization of DDAB liposome and the LNCP
We hypothesized that the calcium-phosphate nano-precipitate was formed during dialysis. During this process, acetic acid gradually diffused out of the liposomes. This would increase the pH inside the liposomes, resulting in the deposition of calcium-phosphate in the liposomal nucleation site (Olton et al., 2007; Schmidt and Ostafin, 2002). In order to examine the presence of calcium-phosphate nano-precipitate within the liposome coat, we assayed the calcium concentration of the LNCP suspension by ICP-OES and examined the morphology of the DDAB liposomes and the LNCP by AFM.
The presence of a calcium-phosphate core was supported by the presence of the calcium in the LNCP suspension. The ICP-OES assay showed that Ca2+ concentration in the LNCP was 0.556 mM. The liposome internal volume (5 mg/ml lipid) was estimated to be ~3% of the entire solution (Lasic et al., 1998), which meant that the theoretical Ca2+ concentration in the LNCP solution should be ~3% of the Ca2+ concentration before dialysis (18 mM). So the Ca2+ concentration in the LNCP solution could be expected to be ~0.540 mM hypothetically. The consistence between the theoretical and actual Ca2+ concentration indicate that the Ca2+ inside the LNCP particles was virtually completely retained during the dialysis process, and that the Ca2+ concentration inside the LNCP particles was approximately 18 mM. Since the lipid bilayer is impermeable for Ca2+ and PO43−, they will form a calcium-phosphate core inside the LNCP particles, when acetic acid diffuses away from the liposome.
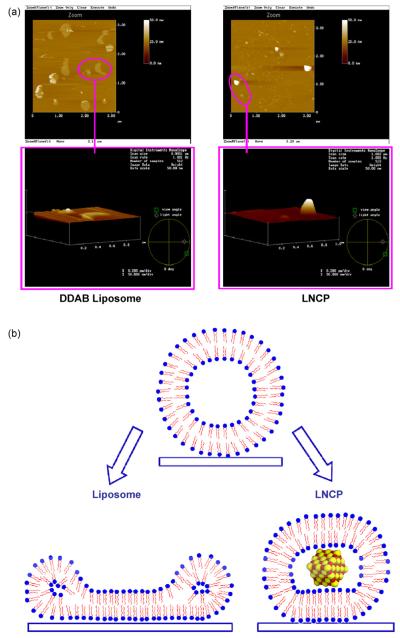
Fig. 3A shows an AFM image of adsorbed DDAB liposomes and LNCP onto the surface of mica. The maximum height of the DDAB liposome and the LNCP was estimated as 5.05 and 32.1 nm, respectively, by the depth analysis software of the AFM apparatus, while the particle sizes of the same particles were 101.3 ± 58.1 and 132.0 ± 71.7 nm, respectively, by DLS. Therefore, the rigidity (maximum height of adsorbed particles/particle size) (Nakano et al., 2008) of the DDAB liposome was calculated to be 0.050, compared to a value of 0.243 for the LNCP.
Fig. 3.
AFM images and schematic model. (A) AFM images of DDAB liposomes and LNCP adsorbed onto surface-modified mica. Scale: 9 μm2 × 50 nm. (B) Schematic model of DDAB liposomes and the LNCP adsorbed on the mica surface.
Based on an earlier model of liposomes adsorbed onto mica surface (Nakano et al., 2008), here we propose a new scheme for the behavior of LNCP adsorbed onto mica (Fig. 3B). The DDAB liposomes showed usually a flattened shape onto the mica surface, where the height of the DDAB liposomes was 5.05 nm. This was consistent with previous finding that the DDAB liposomes had low rigidity (Nakano et al., 2008). It had been shown previously that 6.0–7.5 nm is the estimated height of a single bilayer (Jass et al., 2000). It could be thus anticipated that the low height of the DDAB liposomes could be due to the liposome collapse after adsorption, so that the top and bottom bilayers merged into a single bilayer on the mica surface. On the other hand, the LNCP, which showed an ~5-fold increase in rigidity, retained its shape much better than DDAB liposomes. The increased rigidity indicates that the solid calcium-phosphate core in the LNCP is able to support the internal structure of the LNCP particles. Otherwise DDAB liposomes could be expected to exhibit higher rigidity than the LNCP, since smaller particles tend to maintain their spherical shape and rigidity better than larger ones.
3.4. Cytotoxicity evaluations
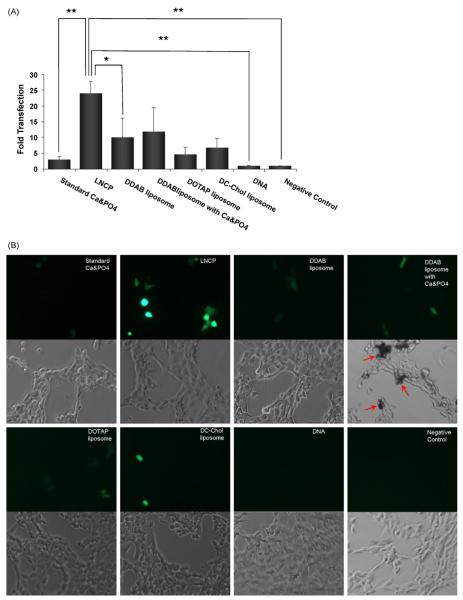
As expected, the LNCP showed minimal cytotoxicity towards cultured NIH 3T3 cells, similar to the cytotoxicity levels of other three types of cationic liposomes (Fig. 4). In contrast, standard calcium-phosphate precipitation preparations showed significant cytotoxicity. This is in accordance with a recent finding that cationic liposomes prepared by a modified ethanol injection method exhibit low toxicity when tested on HeLa cells (Maitani et al., 2007). Light microscopy experiments indicated that treatment with the LNCP did not alter the morphology of the cells (Fig. 5B), while treatment with a mixture of DDAB liposomes and calcium-phosphate resulted in significant changes of the cell morphology. This morphological change may be an indication of toxicity of the mixture of DDAB liposomes and calcium-phosphate, which was not detected by the MTT assay. The considerably lower cytotoxicity of the LNCP relative to the standard calcium-phosphate precipitation preparations can be attributed to the fact that calcium-phosphate was encapsulated within the liposomal shell of the LNCP, thereby preventing from a direct contact with the cell membrane, which may cause membrane disruption.
Fig. 4.
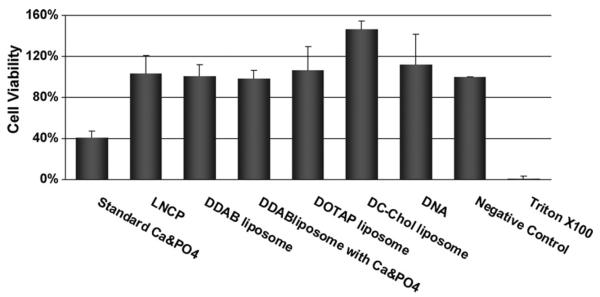
Cytotoxicity of the LNCP. Results of MTT cell viability test after removal of the transfection medium. Values represent the mean ± S.D. (n = 3).
Fig. 5.
Comparison of transfection efficiencies of gene transfer vectors. (A) Transfection efficiencies of standard calcium-phosphate precipitation preparation, LNCP, DDAB liposomes, DDAB liposomes/calcium-phosphate mixture, DC-Chol liposomes, DOTAP liposomes, naked pDNA and a control of untreated cells, measured by flow cytometry. Values represent the mean ± S.D. (n = 3). *p < 0.05; **p < 0.01. (B) Micrographs from fluorescent microscopy under fluorescence (first and third row) or phase contrast (second and fourth row) modes of cells after transfection with GFP pDNA. Red arrow indicates the changes in cellular morphology after transfection. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
3.5. Transfection efficiency of the LNCP
The LNCP and the cationic liposomes were combined with GFP plasmid DNA several different L/D ratios, to test the efficacy of gene transfer. Preliminary data indicated that at an L/D ratio of 10:1, both the LNCP and the cationic liposomes showed best transfection activity in NIH 3T3 cells (data not shown). Therefore, we decided to use the L/D ratio of 10:1 in the later experiments.
To assess the relative transfection efficiency of the LNCP, GFP gene expression was measured against standard calcium-phosphate precipitation preparation (Sokolova et al., 2006) and various liposome formulations. Cationic liposomes were prepared using three well-characterized formulations, i.e., DDAB/Cholestrol/TPGS (59:40:1 mol/mol), DOTAP/DOPE (1:1 mol/mol) and DC-Chol/DOPE (1:2 mol/mol). As shown in Fig. 5, the LNCP yielded a 24 times higher GFP expression relative to naked pDNA, and about 10-fold higher transfection than standard calcium-phosphate precipitation preparation. In addition, the DDAB liposomes alone showed a 58% lower transfection activity than the LNCP, suggesting that the calcium-phosphate core in the LNCP promotes more efficient gene delivery.
3.6. Transfection efficiency of the LNCP in the presence of chloroquine
Transfection experiments in the presence of chloroquine were performed in order to examine the transfection mechanism of the LNCP. Chloroquine (100 μM) was added to the cells along with the LNCP/pDNA complexes and the cells were further incubated for 4 h. Free pDNA was used as a control.
Chloroquine is known to cause swelling and bursting of the endosomes, and lead to the release of DNA from endosome (Kim et al., 2009). Yet, the addition of 100 μM chloroquine caused a 60% decrease in the transfection efficiency of the LNCP, as shown in Fig. 6. It has been shown that the weak base chloroquine became protonated rapidly after entering endosome (Fredericksen et al., 2002; Sonawane et al., 2003), thus neutralizing the acidic environment of endocytic vesicles. The reduced transfection efficiency of the LNCP in the presence of chloroquine suggested that the endosomal acidification is necessary for its superior transfection efficiency, and that the endosome buffering ability of the calcium-phosphate core in the LNCP may play a pivotal role in boosting its transfection activity.
Fig. 6.
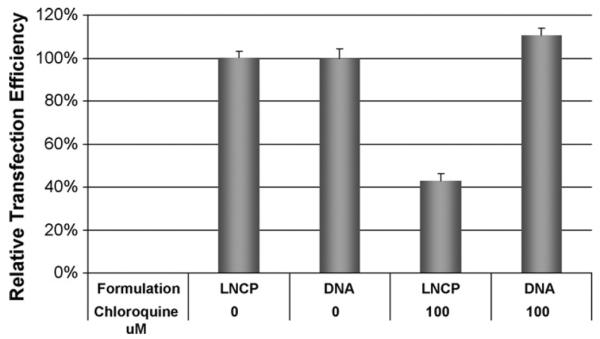
Transfection activity with or without chloroquine. Free pDNA or pDNA/LNCP complexes at L/D ratio of 10 were used to transfect 3T3 cells with or without 100 μM chloroquine. Values represent the mean ± S.D. (n = 3).
The possible reason for the low transfection efficiency of calcium-phosphate nanoparticles was the nuclease susceptibility of calcium-phosphate-DNA complexes, which inhibited the entry of these complexes into the nucleus (Lechardeur et al., 2005; Sokolova et al., 2006). In contrast, liposomal protection of the encapsulated DNA will increase the efficiency of transfection (Mortazavi et al., 2007). Fig. 5 shows that in the case of LNCP, the liposomal coat and the calcium-phosphate core exhibited a synergistic gene transfection effect. Meanwhile, DDAB liposomes and calcium-phosphate when combined only showed an additive effect in transfection efficiency. The synergistic effect of the liposomal coat and the calcium-phosphate core possibly was due to several factors. First, the cationic liposomal coat could protect the pDNA from enzymatic cleavage and promote the fusion of the LNCP with cell membranes with subsequent endocytosis. Upon entering the endosome, the “proton sponge” effect (Akinc et al., 2005) of the phosphate ion will ultimately promote the release of the pDNA from the endosome. The calcium-phosphate core is re-dissolved in the acidic endosomal environment (Li et al., 2010), and because the phosphate ions can buffer considerable amount of H+, it will cause additional pumping of protons into the endosome, together with the influx of chloride counter ions, thus increasing the osmotic pressure inside the endosomes. This would result in osmotic swelling that would lead to physical rupture of the endosomes and the escape of the pDNA from the degradative lysosomal trafficking pathway (Godbey et al., 1999). The third possible reason for the enhanced transfection efficiency of the LNCP was the high concentration of the encapsulated Ca2+ in the LNCP. It has been shown that 8 mM Ca2+ can act together with cationic lipids to destabilize the endosomal membrane following uptake (Palmer et al., 2003). The calcium concentration inside the LNCP particle in our experiments was estimated to be 18 mM, therefore this high internal calcium concentration can promote endosomal escape of pDNA after internalization. Because the high Ca2+ concentration required to enhance transfection efficiency (Palmer et al., 2003), is unlikely to be achieved systemically, the encapsulated Ca2+ in LNCP could be a very desirable means to utilize Ca2+ to promote transfection in vivo. Moreover, the rigidity of the LNCP particles, related to their stability and drug release activity, may also be a factor in improving the gene delivery efficiency (Drummond et al., 1999; Nakano et al., 2008).
4. Conclusions
Non-toxic lipid-coated nano-calcium-phosphate particles exhibiting high DNA transfection efficiency were prepared by a new and simple method. Efficient delivery of plasmid DNA was achieved in a mammalian cell culture using LNCP/pDNA complexes. By incorporating a calcium-phosphate core, the LNCP was able to deliver DNA very efficiently. The LNCP prepared by the present method are superior to the standard calcium-phosphate transfection preparation in terms of cytotoxicity. The LNCP described here seem to be useful for ex vivo and in vitro DNA transfer, particularly with respect to their ability to protect the incorporated DNA, and their superior safety and efficiency profiles.
Acknowledgements
This work was support in part by NSF Grant EEC-0425626 and NIH Grant R01 CA135243 and R21CA131832. We wish to thank Dr. JingtaoYan for assistance with the AFM. The authors would like to thank Dr. Rumiana Koynova for her helpful comments and suggestions on editing this manuscript.
References
- Adami RC, Collard WT, Gupta SA, Kwok KY, Bonadio J, Rice KG. Stability of peptide condensed plasmid DNA formulations. J. Pharm. Sci. 1998;87:678–683. doi: 10.1021/js9800477. [DOI] [PubMed] [Google Scholar]
- Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- Barth RF, Adams DM, Soloway AH, Mechetner EB, Alam F, Anisuzzaman AKM. Determination of boron in tissues and cells using direct-current plasma atomic emission-spectroscopy. Anal. Chem. 1991;63:890–893. doi: 10.1021/ac00009a010. [DOI] [PubMed] [Google Scholar]
- Bisht S, Bhakta G, Mitra S, Maitra A. pDNA loaded calcium phosphate nanoparticles: highly efficient non-viral vector for gene delivery. Int. J. Pharm. 2005;288:157–168. doi: 10.1016/j.ijpharm.2004.07.035. [DOI] [PubMed] [Google Scholar]
- Chu MQ, Liu GJ. Preparation and characterization of hydroxyapatite/liposome core-shell nanocomposites. Nanotechnology. 2005;16:1208–1212. [Google Scholar]
- Drummond DC, Meyer O, Hong K, Kirpotin DB, Papahadjopoulos D. Optimizing liposomes for delivery of chemotherapeutic agents to solid tumors. Pharmacol. Rev. 1999;51:691–743. [PubMed] [Google Scholar]
- Fredericksen BL, Wei BL, Yao J, Luo TC, Garcia JV. Inhibition of endosomal/lysosomal degradation increases the infectivity of human immunodeficiency virus. J. Virol. 2002;76:11440–11446. doi: 10.1128/JVI.76.22.11440-11446.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbey WT, Wu KK, Mikos AG. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc. Natl. Acad. Sci. U.S.A. 1999;96:5177–5181. doi: 10.1073/pnas.96.9.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Gosselin MA, Lee RJ. Characterization of a novel diolein-based LPDII vector for gene delivery. J. Control. Release. 2002;83:121–132. doi: 10.1016/s0168-3659(02)00167-0. [DOI] [PubMed] [Google Scholar]
- Guo W, Lee RJ. Efficient gene delivery using anionic liposome-complexed polyplexes (LPDII) Biosci. Rep. 2000;20:419–432. doi: 10.1023/a:1010338219401. [DOI] [PubMed] [Google Scholar]
- Jass J, Tjarnhage T, Puu G. From liposomes to supported, planar bilayer structures on hydrophilic and hydrophobic surfaces: an atomic force microscopy study. Biophys. J. 2000;79:3153–3163. doi: 10.1016/S0006-3495(00)76549-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Lee EH, Choi SH, Kim CK. In vitro and in vivo transfection efficiency of a novel ultradeformable cationic liposome. Biomaterials. 2004;25:305–313. doi: 10.1016/s0142-9612(03)00534-9. [DOI] [PubMed] [Google Scholar]
- Kim TI, Ou M, Lee M, Kim SW. Arginine-grafted bioreducible poly(disulfide amine) for gene delivery systems. Biomaterials. 2009;30:658–664. doi: 10.1016/j.biomaterials.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneuer C, Sameti M, Haltner EG, Schiestel T, Schirra H, Schmidt H, Lehr CM. Silica nanoparticles modified with aminosilanes as carriers for plasmid DNA. Int. J. Pharm. 2000;196:257–261. doi: 10.1016/s0378-5173(99)00435-4. [DOI] [PubMed] [Google Scholar]
- Lechardeur D, Verkman AS, Lukacs GL. Intracellular routing of plasmid DNA during non-viral gene transfer. Adv. Drug Deliv. Rev. 2005;57:755–767. doi: 10.1016/j.addr.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Li J, Chen YC, Tseng YC, Mozumdar S, Huang L. Biodegradable calcium phosphate nanoparticle with lipid coating for systemic siRNA delivery. J. Controlled Release. 2010;142:416–421. doi: 10.1016/j.jconrel.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasic D, Weiner N, Riaz M, Martin F. Liposomes. In: Lieberman A, Rieger M, Banker G, editors. Pharmaceutical Dosage Forms: Disperse Systems. vol 3. Marcel Dekker; New York, NY: 1998. pp. 43–86. [Google Scholar]
- Luo D, Han E, Belcheva N, Saltzman WM. A self-assembled, modular DNA delivery system mediated by silica nanoparticles. J. Control. Release. 2004;95:333–341. doi: 10.1016/j.jconrel.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Maitani Y, Igarashi S, Sato M, Hattori Y. Cationic liposome (DC-Chol/DOPE = 1:2) and a modified ethanol injection method to prepare liposomes, increased gene expression. Int. J. Pharm. 2007;342:33–39. doi: 10.1016/j.ijpharm.2007.04.035. [DOI] [PubMed] [Google Scholar]
- Mortazavi SM, Mohammadabadi MR, Khosravi-Darani K, Mozafari MR. Preparation of liposomal gene therapy vectors by a scalable method without using volatile solvents or detergents. J. Biotechnol. 2007;129:604–613. doi: 10.1016/j.jbiotec.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Nakano K, Tozuka Y, Yamamoto H, Kawashima Y, Takeuchi H. A novel method for measuring rigidity of submicron-size liposomes with atomic force microscopy. Int. J. Pharm. 2008;355:203–209. doi: 10.1016/j.ijpharm.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Olton D, Li JH, Wilson ME, Rogers T, Close J, Huang L, Kumta PN, Sfeir C. Nanostructured calcium phosphates (NanoCaPs) for non-viral gene delivery: influence of the synthesis parameters on transfection efficiency. Bio-materials. 2007;28:1267–1279. doi: 10.1016/j.biomaterials.2006.10.026. [DOI] [PubMed] [Google Scholar]
- Palmer LR, Chen T, Lam AM, Fenske DB, Wong KF, MacLachlan I, Cullis PR. Transfection properties of stabilized plasmid-lipid particles containing cationic PEG lipids. Biochim. Biophys. Acta. 2003;1611:204–216. doi: 10.1016/s0005-2736(03)00058-0. [DOI] [PubMed] [Google Scholar]
- Roy I, Mitra S, Maitra A, Mozumdar S. Calcium phosphate nanoparticles as novel non-viral vectors for targeted gene delivery. Int. J. Pharm. 2003;250:25–33. doi: 10.1016/s0378-5173(02)00452-0. [DOI] [PubMed] [Google Scholar]
- Schmidt HT, Ostafin AE. Liposome directed growth of calcium phosphate nanoshells. Adv. Mater. 2002;14:532–535. [Google Scholar]
- Sokolova V, Kovtun A, Prymak O, Meyer-Zaika W, Kubareva EA, Romanova EA, Oretskaya TS, Heumann R, Epple M. Functionalisation of calcium phosphate nanoparticles by oligonucleotides and their application for gene silencing. J. Mater. Chem. 2007;17:721–727. [Google Scholar]
- Sokolova VV, Radtke I, Heumann R, Epple M. Effective transfection of cells with multi-shell calcium phosphate-DNA nanoparticles. Biomaterials. 2006;27:3147–3153. doi: 10.1016/j.biomaterials.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Sonawane ND, Szoka FC, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J. Biol. Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- Sudimack JJ, Guo W, Tjarks W, Lee RJ. A novel pH-sensitive liposome formulation containing oleyl alcohol. Biochim. Biophys. Acta. 2002;1564:31–37. doi: 10.1016/s0005-2736(02)00399-1. [DOI] [PubMed] [Google Scholar]
- Welzel T, Radtke I, Meyer-Zaika W, Heumann R, Epple M. Transfection of cells with custom-made calcium phosphate nanoparticles coated with DNA. J. Mater. Chem. 2004;14:2213–2217. [Google Scholar]
- Yohannes G, Pystynen KH, Riekkola ML, Wiedmer SK. Stability of phospholipid vesicles studied by asymmetrical flow field-flow fractionation and capillary electrophoresis. Anal. Chim. Acta. 2006;560:50–56. [Google Scholar]