Abstract
The eukaryotic centromere is an essential chromatin region required for accurate segregation of sister chromatids during cell division. Centromere protein B (CENP-B) is a highly conserved protein which can bind to the 17-bp CENP-B box on the centromeric DNA. In this study, we found that CENP-B could be α-N-methylated in human cells. We also showed that the level of the α-N-methylation was stimulated in cells in response to a variety of extracellular stimuli, including increased cell density, heat shock, and arsenite treatment, though the methylation level was not altered upon metaphase arrest. We identified N-terminal RCC1 methyltransferase (NRMT) as a major enzyme required for the CENP-B methylation. Additionally, we found that chromatin-bound CENP-B was primarily trimethylated and α-N-trimethylation could enhance CENP-B’s binding to CENP-B box in cells. Our study also expands the function of protein α-N-methylation that has been known for decades and whose function remains largely unexplored.
Keywords: CENP-B, LC-MS/MS, α-N-methylation, NRMT, CENP-B box
INTRODUCTION
The centromere is a chromatin domain that is essential for proper segregation of chromosomes to daughter cells during cell division. The centromeric DNA, consisting of large arrays of repetitive DNA, is normally in a heterochromatin state. Human α-satellite DNA (alphoid DNA) is comprised of tandem repeats of about 170-bp and it clusters in the centromeric region 1, 2. In human centromere, histone H3 is replaced by a centromere-specific variant, centromere protein A (CENP-A), which is required for the assembly of other centromere/kinetochore components 3–5. In addition to CENP-A, a collection of about 16 proteins termed the constitutive centromere-associated network (CCAN) also associate with centromeric chromatin 6.
CENP-B is a highly conserved protein in mammalian cells 7. The CENP-B homologs in fission yeast are required for the formation of centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications 8. The CENP-B homologs have roles in host genome surveillance for retrotransposons and the maintenance of genome integrity 9, 10, yet CENP-B null mice are viable and display no mitotic or meiotic defects 11–13. However, CENP-B is required for the de novo assembly of centromere on input naked DNA lacking a functional centromere, but preventing undesired assembly of centromere by stimulating heterochromatin formation on human alphoid DNA integrated into ectopic sites 14. CENP-B contains a helix-loop-helix DNA-binding motif at the N-terminus, and a dimerization domain at the C-terminus 15–19. CENP-B can bind to a 17-bp DNA motif (CENP-B box) within centromeric α-satellite DNA through its N-terminal region and link the centromeric DNA to the kinetochore 14, 17, 18, 20. The CENP-B box is highly conserved in human alphoid DNA and mouse minor satellite DNA, and it is essential for de novo assembly of CENP-A and kinetochore 21.
Post-translational modification constitutes a ubiquitous mechanism to expand proteins’ structure, interactions, localization, and function 22. Among them, protein N-terminal α-methylation has been known for several decades, and it is conserved from Escherichia coli to man 23, 24. Recently several eukaryotic proteins were reported to be αN-methylated, including regulator of chromatin condensation 1 (RCC1) 25, histone H2B from different organisms 26–28, and others 29–34. N-terminal methylation of RCC1 is essential for normal bipolar spindle formation and chromosome segregation during mitosis 25; however, the biological functions of α-N-methylation for all other eukaryotic proteins have not yet been elucidated.
Recently, the first α-N-methyltransferase in human [i.e., N-terminal RCC1 methyltransferase (NRMT)] and its orthologs in yeast and Drosophila melanogaster were discovered 24, 35, 36. Among the known NRMT substrates, the initiator methionine residue was cleaved and there is a common N-terminal sequence motif of XPK (‘X” represents alanine, proline, or serine) 24. In addition, recombinant human NRMT can also methylate synthetic peptide substrates in which X in the XPK motif is a C, F, G, H, K, M, N, Q, R, or Y 37. Considering that CENP-B contains an N-terminal GPK motif after removal of the initiating methionine, we reason that this protein might also be α-N-methylated by NRMT.
In this study, we demonstrated the α-N-methylation of CENP-B in human cells and the involvement of NRMT in this methylation. We showed that the N-terminal methylation level of CENP-B could be elevated by various cellular stresses. Additionally, we found that chromatin-bound CENP-B is primarily trimethylated, and α-N-trimethylation can strengthen CENP-B’s binding to the CENP-B boxes in mouse endogenous centromeric minor satellite DNA and human synthetic alphoid DNA at the ectopic integration site.
MATERIALS AND METHODS
Cell Culture
HEK293T human embryonic kidney epithelial cells (ATCC) and CENP-B−/− mouse embryonic fibroblast cells (gift from Prof. W.R. Brinkley) were cultured in Dulbecco’s Modified Eagle Medium (DMEM, ATCC) supplemented with 10% fetal bovine serum (FBS, Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin (ATCC). Cells were maintained in a humidified atmosphere with 5% CO2 at 37°C.
Construction of Vectors
Human CENP-B gene was amplified from HEK293T cells by RT-PCR to introduce a 5′ XbaI site and a 3′ BglII site, and subcloned into a modified mammalian expression vector pRK7 in which three tandem repeats of the FLAG epitope tag (DYKDDDDK) were inserted between BamHI and EcoRI sites to produce vector for expressing C-terminally FLAG-tagged CENP-B. CENP-B-AGPK and -K4Q mutants were amplified from FLAG-tagged CENP-B plasmid using primers containing designed mutations.
Preparation of FLAG-Tagged CENP-B Protein and Mass Spectrometric Analysis
FLAG-tagged CENP-B plasmids (1.5 μg) were transfected into HEK293T cells in 6-well plates with Lipofectamine 2000 (Invitrogen). After a 48-hr incubation, the cells were harvested by using trypsin-EDTA solution (ATCC) and cellular extracts were prepared by suspending cells in CelLytic M (Sigma) lysis buffer containing protease inhibitor cocktail (Sigma). The FLAG-tagged CENP-B was isolated from the whole cell lysate by affinity purification with anti-FLAG M2 beads (Sigma), digested using Glu-C (NEB) at a protein/enzyme ratio of 20:1, and then analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
For LC–MS/MS experiments, peptide samples were automatically injected and separated by online liquid chromatography (LC) on an EASY-nLCII, and analyzed on an LTQ Orbitrap Velos mass spectrometer equipped with a nanoelectrospray ionization source (Thermo, San Jose, CA) following similar procedures as described previously 38. The separation was conducted by using a homemade trapping column (150 μm × 50 mm) and a separation column (75 μm × 120 mm), packed with ReproSil-Pur C18-AQ resin (3 μm in particle size, Dr. Maisch HPLC GmbH, Germany). Peptide samples were initially loaded onto the trapping column with a solvent mixture of 0.1% formic acid in CH3CN/H2O (2:98, v/v) at a flow rate of 4.0 μL/min. The peptides were then separated using a 120-min linear gradient of 2–40% acetonitrile in 0.1% formic acid and at a flow rate of 300 nL/min. The LTQ-Orbitrap Velos mass spectrometer was operated in the positive-ion mode, and the spray voltage was 1.8 kV. The full-scan mass spectra (m/z 350–2000) were acquired with a resolution of 60,000 at m/z 400 after accumulation to a target value of 500,000. MS/MS experiments were carried out in the pre-selected ion mode where the fragmentations of the protonated ions of the unmodified and mono-, di- or tri-methylated forms of the N-terminal peptide of CENP-B were monitored. All the MS/MS data were analyzed manually.
Cellular Stress Experiments
FLAG-tagged CENP-B plasmids were transfected into 30% (low density) or 100% (high density) confluent HEK293T cells in cell density experiments. For heat shock experiments, CENP-B plasmids were transfected into HEK293T cells at 70% confluence for 48 h and then incubated at 45°C for 1 h before harvesting. For arsenite treatment, HEK293T cells transfected with CENP-B plasmids were incubated for 24 h, and then treated with 5 μM arsenite (Sigma) for another 24 h. For colcemid treatment, CENP-B expression plasmid was transfected into HEK293T cells, and 48 h later, the cells were treated with 0.075 μg/mL colcemid (Sigma) for 2 h. After these treatments, the FLAG-tagged CENP-B proteins were extracted and analyzed by LC-MS/MS as described above.
In Vitro Methylation Assay
The NRMT-His6 plasmid, a gift from Prof. Ian G. Macara 24, were expressed in E. coli Rosetta (DE3) pLysS after growth at 37°C in Luria broth supplemented with 2% (vol/vol) ethanol to an optical density at 600 nm of approximately 0.8, followed by induction with 0.5 mM isopropylthiogalactopyranoside at room temperature overnight. The proteins were then purified by using Talon affinity resin (Clontech) following the manufacturer’s recommended procedures.
In vitro methylation assay was performed as previously described with minor modification 24, 25. Briefly, 200 ng unmodified peptide, i.e., GPKRRQLTFREK (Genemed Synthesis Inc.), was mixed with 100 μM S-adenosyl-L-methionine (S-AdoMet, Sigma), which serves as the methyl group donor, and 1 μg NRMT-His6. The reaction mixture was brought to 50 μL with the methyltransferase buffer (50 mM Tris, 50 mM potassium acetate, pH 8.0), incubated at 30°C for 1 h and subjected to LC-MS/MS analysis.
SiRNA Knockdown of NMRT for In vivo Methylation Assay
The siRNAs were purchased from Dharmacon: NRMT SMARTpool (L-008461) and siControl Non-Targeting pool (D-001210). The HEK293T cells were seeded in 6-well plates at 40–60% confluence level and transfected with approximately 100 pmol siRNAs using Lipofectamine 2000 (Invitrogen). After a 48-h incubation, 1.5 μg CENP-B expression plasmid was co-transfected into the cells together with another aliquot of siRNA using Lipofectamine 2000. The FLAG-tagged CENP-B proteins were extracted from the cells 48 h after transfection as described above.
Real-Time Quantitative RT-PCR
Total RNA was extracted from the cells 48 h after transfection with siRNA using the Total RNA Kit I (Omega), and cDNA was generated by using M-MLV reverse transcriptase (Promega) and an oligo(dT)16 primer. The siRNA knockdown efficiency of NMRT was evaluated by quantitative real-time RT-PCR using iQ SYBR Green Supermix kit (Bio-Rad) and GAPDH was used as an internal control as described elsewhere 39. Primer sequences used for real-time PCR were as follows: NRMT-S, 5′-GCCCTCCCTTCCTCTTCC-3′; NRMT-AS, 5′-CCAACCACGGCTCTACTCA-3′; GAPDH-S, 5′-TTTGTCAAGCTCATTTCCTGGTATG-3′; GAPDH-AS, 5′-TCTCTTCCTCTTGTGCTCTTGCTG-3′.
Isolation of Chromatin-Associated Proteins
Chromatin-associated proteins were isolated as previously described with minor modification 40. Briefly, cells were lysed with cytoplasmic lysis buffer (10 mM Tris-HCl, pH 8.0, 0.34 M sucrose, 3 mM CaCl2, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.5% NP-40), and intact nuclei were pelleted by centrifugation at 5000 rpm for 2 min. Nuclei were lysed with nuclear lysis buffer (20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 1 mM EDTA, 150 mM KCl, 0.1% NP-40, 1 mM DTT, 10% glycerol, protease and phosphotase inhibitors) by homogenization. The nucleoplasmic fraction was cleared by centrifugation at 14,000 rpm for 30 min. The chromatin-enriched pellet was then resuspended in a buffer containing 20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 150 mM KCl, 10% glycerol, protease and phosphotase inhibitors and 0.15 unit/μL benzonase (Sigma), and the reaction mixture was incubated on ice for 1 h. Chromatin-associated proteins were collected by spinning down the debris and collecting supernatant.
ChIP Analysis
CENP-B expression plasmid (500 ng) was transfected into 70% confluent CENP-B−/− MEF cells with a human synthetic alphoid DNA (WTR11.32Bsr) ectopic integration site (MEF CENP-B−/− Int W1) (Okada et al. 2007) in 6 well plates with FugeneHD (Promega). Two days after the transfection, the cells were trypsinized, harvested in a centrifuge tube, washed once with PBS, and fixed in 0.5% formaldehyde (Sigma) at 22°C for 10 min. The reaction was stopped by addition of glycine until its final concentration reached 100 mM. The cells were then incubated at room temperature for 5 min and washed with PBS. Soluble chromatin was prepared by sonicating the cells suspended in sonication buffer (10 mM HEPES, 1 mM EDTA, 1.5 μM aprotinin, 10 μM leupeptin, 1 mM DTT, 0.05% SDS and 40 μM MG132) to an average DNA size of 0.5–1 kb. After sonication, soluble chromatin (as input) was recovered by centrifugation, resuspended in IP buffer (55 mM HEPES, 150 mM NaCl, 1 mM EGTA, 2 mM MgCl2, 2 mM ATP, 1.5 μM aprotinin, 10 μM leupeptin, 1 mM DTT, 0.01% SDS, 1% NP-40) and immunoprecipitated using 2 μg anti-CENP-B N-ter monoclonal antibody (5E6C1) with protein G sepharose beads (GE: 17-0618) 14. DNA purified from the immunoprecipitates and the input soluble chromatin fraction were quantified by real-time PCR using the following primer sets: CH 4mer F1 and JNCRevNo for synthetic alphoid, MS 24C and MS box 1 for minor satellite DNA, MMS 24C and MMS24d for major satellite DNA and rDNA-f and rDNA-r for ribosomal DNA 14.
Western Blotting
MEF CENP-B−/− Int W1 cells transfected with each plasmid were harvested at two days after transfection. Transfected whole cell mixture was electrophoresed on Mini-PROTEAN TGX Gel (Bio-Rad). The proteins were transferred to PVDF membrane by Trans-Blot® Turbo System (Bio-Rad) and blocked with TPBS (PBS containing 0.1% Tween) containing 1% skim milk at room temperature for 30 min. The membrane was incubated with primary antibody at 4°C for overnight and with secondary antibody at room temperature for 1 h. The HRP signals were detected using Pierce ECL Western Blotting Substrate (Thermo).
Anti-CENP-B N-terminal polyclonal antibody (BN1, 0.8 μg/mL), anti-GAPDH antibody ab9482 (Abcam, 0.2 μg/ml, (HRP)-Loading Control) and anti-DDDDK tag antibody F-tag-01 ab18230 (Abcam, 1 μg/ml) were employed as primary antibodies, and horseradish peroxidase-conjugated anti-rabbit IgG and anti-mouse IgG (Nakarai Tesque, 1:2,000 dilution) were used as secondary antibodies.
RESULTS AND DISCUSSION
Identification of α-N-Methylation of CENP-B
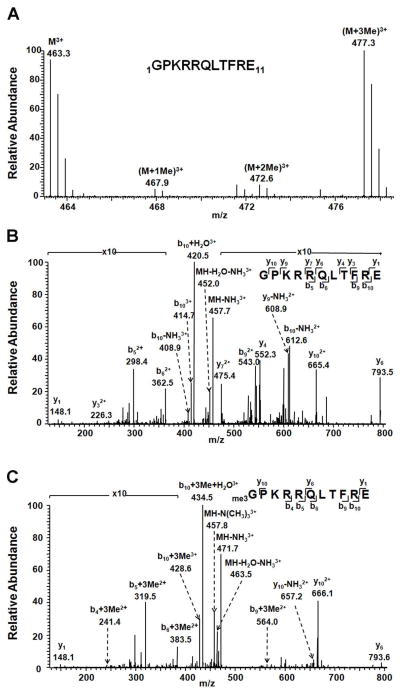
To assess whether CENP-B is α-N-methylated in human cells, we constructed plasmids allowing for the expression, in human embryonic kidney 293T (HEK293T) cells, of fusion proteins where 3 tandem repeats of the FLAG epitope tag (3×FLAG) are conjugated to the C-terminus of CENP-B. The FLAG-tagged CENP-B was isolated from the whole cell lysate by affinity purification with anti-FLAG M2 beads. Because CENP-B is rich in lysine and arginine residues on the N-terminal moiety, we employed Glu-C to digest the protein. LC-MS/MS analysis of the resulting peptide mixture revealed unambiguously the α-N-methylation of this protein (Figure 1). In this context, we observed that the N-terminus of the protein is heavily trimethylated (Figure 1A and C). In addition, our results showed that the levels of unmodified and trimethylated forms of the N-terminal peptide of CENP-B were similar (Figure 1), whereas those of the mono- and dimethylated forms were markedly lower (Supplementary Figure S1).
Figure 1.
Identification of α-N-methylation of CENP-B. (A) ESI-MS of the N-terminal peptide GPKRRQLTRE from the Glu-C digestion of C-terminally FLAG-tagged human CENP-B. (B) and (C) MS/MS of unmodified (B) and tri-methylated N-terminal peptide (C) of CENP-B.
Methylation Level of CENP-B Is Up-Regulated in Cells Under Stress Conditions
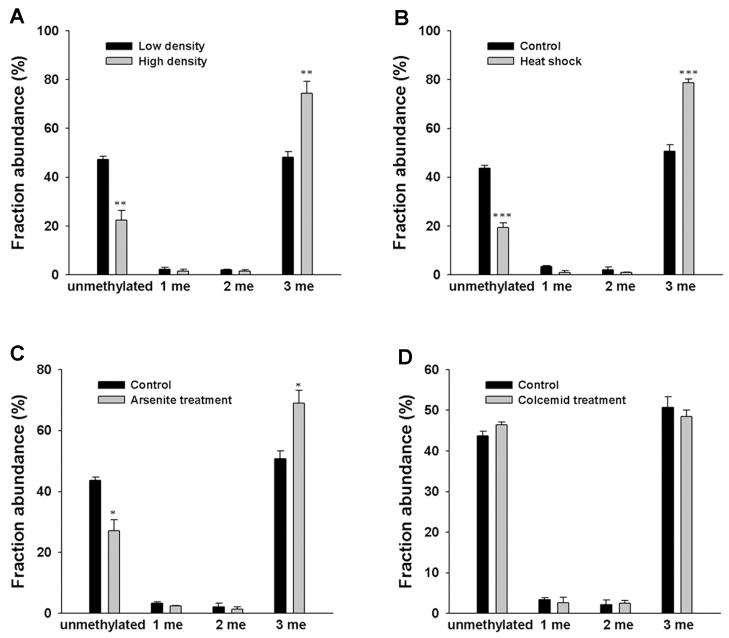
A previous study showed that the methylation of the N-terminal proline in histone H2B from Drosophila melanogaster was stimulated by several external signals from proliferative or physiological stress, such as cell density and heat-shock 36. We therefore asked whether such stress can also alter the N-terminal methylation of human CENP-B. To this end, we first examined the effect of cell density on the modification pattern of CENP-B isolated from the cultured cells expressing C-terminally FLAG-tagged CENP-B at low or high cell densities (30% and 100% confluent, respectively). We found that, when cells are seeded at high density, the level of α-N-trimethylation of CENP-B is increased, which is accompanied with a decrease in the level of the unmodified peptide (Figure 2A).
Figure 2.
N-terminal methylation levels of CENP-B were influenced by cellular stress, but not by metaphase arrest. MS analysis showing the relative abundances of different methylation forms of N-terminal peptide GPKRRQLTFRE of CENP-B isolated from cells at different cell densities (A) or upon treatment with heat shock (B), arsenite (C), or colcemid (D). Error bars represent the SEM (n=3). *, P<0.05; **, P<0.01; ***, P<0.001. The P values were calculated by using unpaired two-tailed Student’s t-test.
To assess whether the α-N-methylation of CENP-B is regulated by heat shock, we treated HEK293T cells expressing FLAG-tagged CENP-B at 45°C for 1 h and then isolated the CENP-B protein. As displayed in Figure 2B, we observed that the proportion of trimethylated form of CENP-B increased whereas the amount of unmodified CENP-B decreased after the heat shock (Figure 2B). On the grounds that arsenite treatment is known to induce the synthesis of heat-shock proteins and thus may mimic a heat-shock response 41–43, we also treated the cells with arsenite and measured the methylation level of CENP-B. It turned out that arsenite treatment again led to an increase in the N-terminal trimethylation of CENP-B in human cells (Figure 2C).
As CENP-B can form foci at centromere region in metaphase cells 14, we next examined whether the distribution of its N-terminal methylation depends on cell cycle. To this end, we treated HEK293T cells with colcemid, which inhibits spindle fiber formation and leads to the arrest of cells in the metaphase, and isolated the FLAG-tagged CENP-B from the treated cells. Our LC-MS/MS results showed that there was no significant difference in the methylation pattern of the N-terminal peptide of CENP-B isolated from control or colcemid-treated cells (Figure 2D). This result suggested that the metaphase arrest does not alter the N-terminal methylation of CENP-B.
NRMT Is a Major Enzyme Required for the α-N-Methylation of CENP-B
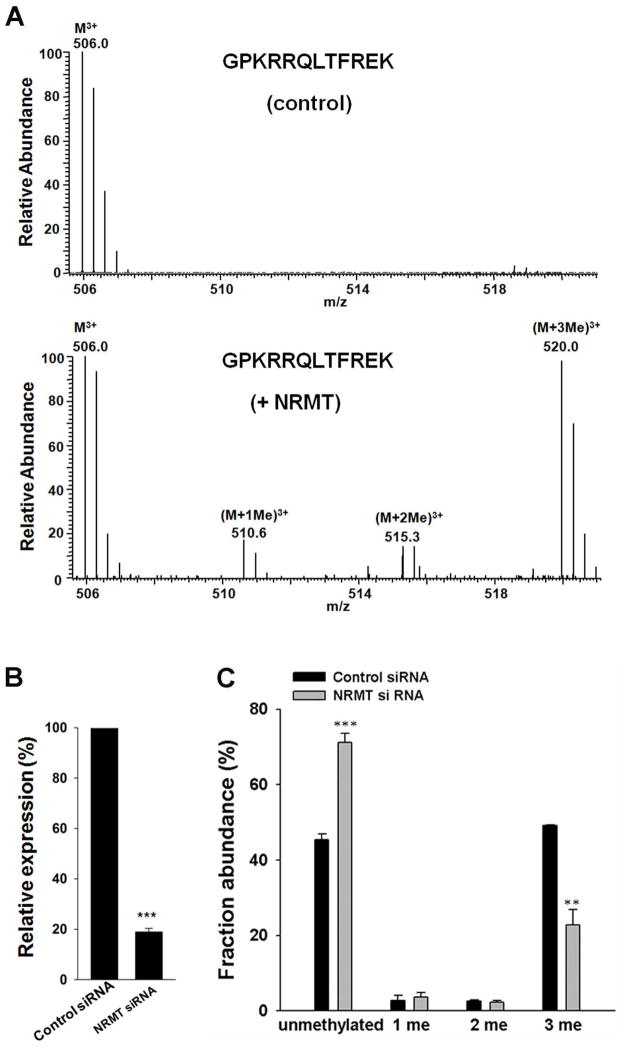
Viewing that NRMT can catalyze the α-N-methylation of RCC1 and several other human proteins carrying the conserved N-terminal XPK motif 24, 25, we asked whether this enzyme can also catalyze the α-N-methylation of CENP-B, which harbors an N-terminal GPK motif. To this end, we incubated a peptide containing the N-terminal twelve amino acids (without the initial methionine) of CENP-B with NRMT and S-adenosyl-L-methionine (S-AdoMet) in vitro. LC-MS/MS analysis revealed the presence of mono-, di-, and tri-methylated N-terminal peptides in samples with NRMT as supported by MS/MS (Figure 3A, bottom), but the absence of methylated N-terminal peptides in samples without the addition of NRMT (Figure 3A, top). These results demonstrated that NRMT can catalyze the α-N-methylation of CENP-B in vitro.
Figure 3.
NRMT can catalyze α-N-methylation of CENP-B. (A) In vitro methylation assay of CENP-B N-terminal peptide with (bottom) or without NRMT (top). (B) Relative mRNA level of the NRMT gene in HEK293T cells treated with control non-targeting or NRMT siRNA, as assessed by real-time PCR with the use of GAPDH as control. (C) Relative abundances of different methylation forms of CENP-B N-terminal peptide (2–11) in cells with control and NRMT siRNA knockdown, as determined by MS analysis. Error bars represent the SEM (n=3). **, P<0.01; ***, P<0.001. The P values were calculated by using unpaired two-tailed Student’s t-test.
We next assessed whether CENP-B is also a substrate for NRMT in cells. To this end, we knocked down the expression of NRMT in HEK293T cells by using siRNA and subsequently co-transfected the cells with NRMT siRNA and the plasmid for expressing the C-terminally FLAG-tagged CENP-B. Quantitative real-time PCR results showed that the knockdown of NRMT was highly efficient (Figure 3B). We then estimated the extent of N-terminal methylation based on the relative abundances of precursor ions of the methylated and unmodified N-terminal peptides of FLAG-tagged CENP-B isolated from HEK293T cells. Upon NRMT knockdown, there is a significant decrease in the level of α-N-methylation in CENP-B relative to that observed for CENP-B obtained from cells treated with control, non-targeting siRNA (Figure 3C). This result further supported that NRMT is an important enzyme responsible for the N-terminal methylation of CENP-B in human cells.
N-Terminal Methylation Is Required for CENP-B’s Binding to CENP-B Box
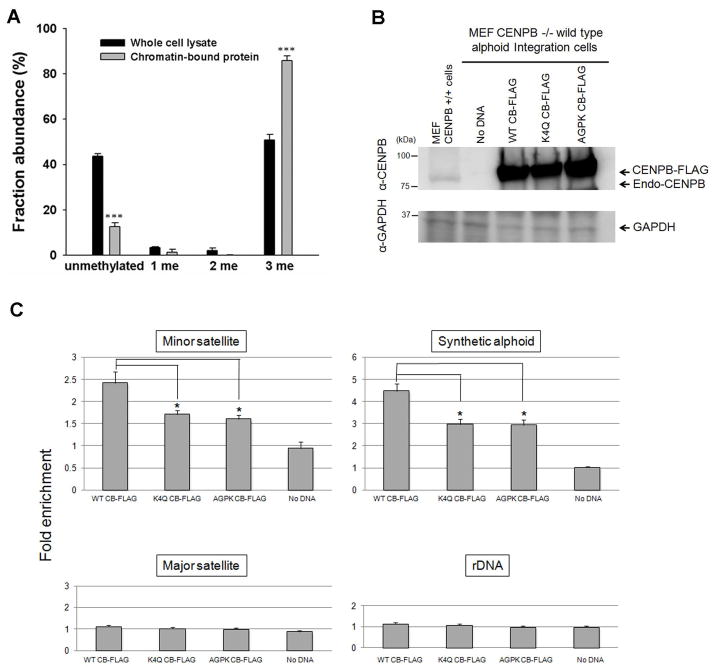
To gain additional insights into the function of the N-terminal methylation of CENP-B, we extracted chromatin-associated proteins using a previously described method 40, isolated the FLAG-tagged CENP-B using anti-FLAG M2 beads, and digested the resulting protein with Glu-C, as described above. LC-MS/MS analysis of the peptide mixture showed that the chromatin-bound CENP-B was primarily trimethylated (Figure 4A). Viewing that α-N-trimethylation introduces a permanent cation to the N-terminus of protein, we deduce that this modification facilitates CENP-B’s binding to CENP-B box on the centromeric DNA.
Figure 4.
The α-N-trimethylation of CENP-B enhances its binding to the CENP-B box. (A) MS analysis showing chromatin-bound CENP-B was more heavily trimethylated than CENP-B purified from whole cell lysate. (B) Western blotting showing the expression levels of the wild-type or mutant CENP-Bs in CENP-B−/− MEF transfected with the FLAG-tagged CENP-B plasmids as indicated. (C) ChIP and real-time PCR analysis of the mouse minor satellite DNA, integrated human alphoid DNA, mouse major satellite DNA, and mouse ribosomal DNA using anti-CENP-B antibody. Error bars represent the SEM (n=3). *, P<0.05; ***, P<0.001. The P values were calculated by using unpaired two-tailed Student’s t-test.
To investigate this further, we produced CENP-B-K4Q and CENP-B-AGPK mutants, which result in loss of the consensus motif for NRMT methylation. Indeed LC-MS/MS analysis showed that the N-terminal methylation was abrogated in these two mutants (Supplementary Figure S2). We further performed chromatin immunoprecipitation (ChIP) experiment to investigate the binding ability of wild-type and mutant CENP-B proteins to CENP-B box using CENP-B−/− MEF with a human synthetic alphoid DNA ectopic integration site (MEF CENP-B−/− Int W1) 14. In this vein, FLAG-tagged CENP-Bs were overexpressed in MEF CENP-B−/− Int W1, and the expression levels of the wild-type and the mutant CENP-Bs were similar in the transfected cells (Figure 4B). Mouse major satellite DNA and rDNA without CENP-B box were employed as controls (Figure 4C). The ChIP analysis results revealed that both the CENP-B-K4Q and -AGPK mutants can bind to the CENP-B boxes in mouse endogenous centromeric minor satellite DNA and human synthetic alphoid DNA at the ectopic integration site, but the binding of the mutant proteins toward the CENP-B boxes was significantly reduced relative to the wild-type counterpart (Figure 4C). These results provided solid evidence supporting that α-N-methylation of CENP-B can enhance its binding to the CENP-B box in the centromere.
CONCLUSIONS
Herein, we identified the α-N-methylation of CENP-B and found that chromatin-bound CENP-B was present mainly in the α-N-trimethylated form. On the grounds that trimethylation introduces a quaternary ammonium ion, a permanent cation, to the N-terminus of the protein 23, α-N-trimethylation is expected to enhance the binding of the protein to DNA through the strengthened electrostatic interaction between the protein N-terminus and phosphate groups in DNA. In agreement with this hypothesis, our ChIP assay directly demonstrated that α-N-trimethylation of CENP-B can enhance its binding to CENP-B boxes in mouse endogenous centromeric minor satellite DNA and human synthetic alphoid DNA at the ectopic integration site.
The N-terminal α-methylation level of CENP-B increased when the cells were seeded at high density. It has been previously shown that histone H2B of Drosophila melanogaster also exhibited an increased proline methylation under high cell density 36. Additionally, heat shock or arsenite treatment, which can elicit a similar cellular response as heat shock, also led to an increase in the α-N-methylation of CENP-B. As discussed above, N-terminal trimethylation of CENP-B can enhance its binding to the CENP-B box. Thus, cells may respond to these stresses by strengthening the interaction between CENP-B and centromeric DNA, thereby maintaining centromere activity.
This study revealed the α-N-methylation of CENP-B in human cells, identified NRMT as an important enzyme involved with the methylation, and characterized the biological function of this post-translational modification. Our results showed that the αN-trimethylation of CENP-B can enhance its binding to CENP-B box on human α-satellite DNA and mouse centromeric minor satellite DNA, which might play an important role in assembly, disassembly and/or maintenance of centromere activity in normal human and mouse cells. The present study also expands the biological functions of N-terminal protein methylation.
Supplementary Material
Synposis.
CENP-B is a highly conserved protein which can bind to the 17-bp CENP-B box on centromeric DNA. We found that CENP-B could be α-N-methylated in human cells by NRMT, and this methylation was stimulated in cells in response to a variety of extracellular stimuli, including increased cell density, heat shock, and arsenite treatment. Additionally, chromatin-bound CENP-B was primarily trimethylated and α-N-trimethylation could enhance CENP-B’s binding to CENP-B box in cells.
Acknowledgments
The authors would like to thank Prof. Ian G. Macara for providing NRMT-His6 vector and Prof. William R. Brinkley for MEFs derived from wild type or from CENP-B null (CENP-B −/−) embryos. This work was supported by the National Institutes of Health (R01 ES019873 to Y.W.). H. M. was supported by a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Kazusa DNA Research Institute Foundation. Z. W. was supported by an NRSA Institutional Training grant (T32 ES018827).
Footnotes
Supporting Information Available. MS and MS/MS results. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Manuelidis L. Chromosomal localization of complex and simple repeated human DNAs. Chromosoma. 1978;66(1):23–32. doi: 10.1007/BF00285813. [DOI] [PubMed] [Google Scholar]
- 2.Waye JS, Willard HF. Nucleotide sequence heterogeneity of alpha satellite repetitive DNA: a survey of alphoid sequences from different human chromosomes. Nucleic Acids Res. 1987;15(18):7549–69. doi: 10.1093/nar/15.18.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KH. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci U S A. 2000;97(3):1148–53. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan KF. A solid foundation: functional specialization of centromeric chromatin. Curr Opin Genet Dev. 2001;11(2):182–8. doi: 10.1016/s0959-437x(00)00177-5. [DOI] [PubMed] [Google Scholar]
- 5.Goshima G, Kiyomitsu T, Yoda K, Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J Cell Biol. 2003;160(1):25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120(5):425–46. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan KF, Glass CA. CENP-B is a highly conserved mammalian centromere protein with homology to the helix-loop-helix family of proteins. Chromosoma. 1991;100(6):360–70. doi: 10.1007/BF00337514. [DOI] [PubMed] [Google Scholar]
- 8.Nakagawa H, Lee JK, Hurwitz J, Allshire RC, Nakayama J, Grewal SI, Tanaka K, Murakami Y. Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 2002;16(14):1766–78. doi: 10.1101/gad.997702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cam HP, Noma K, Ebina H, Levin HL, Grewal SI. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451(7177):431–6. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- 10.Zaratiegui M, Vaughn MW, Irvine DV, Goto D, Watt S, Bahler J, Arcangioli B, Martienssen RA. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature. 2011;469(7328):112–5. doi: 10.1038/nature09608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hudson DF, Fowler KJ, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wreford NG, de Kretser DM, Cancilla MR, Howman E, Hii L, Cutts SM, Irvine DV, Choo KH. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J Cell Biol. 1998;141(2):309–19. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapoor M, Montes de Oca Luna R, Liu G, Lozano G, Cummings C, Mancini M, Ouspenski I, Brinkley BR, May GS. The cenpB gene is not essential in mice. Chromosoma. 1998;107(8):570–6. doi: 10.1007/s004120050343. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Castro AV, Shamanski FL, Meneses JJ, Lovato TL, Vogel KG, Moyzis RK, Pedersen R. Centromeric protein B null mice are viable with no apparent abnormalities. Dev Biol. 1998;201(2):135–43. doi: 10.1006/dbio.1998.9005. [DOI] [PubMed] [Google Scholar]
- 14.Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131(7):1287–300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 15.Earnshaw WC, Sullivan KF, Machlin PS, Cooke CA, Kaiser DA, Pollard TD, Rothfield NF, Cleveland DW. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J Cell Biol. 1987;104(4):817–29. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109(5):1963–73. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T. Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J Cell Biol. 1992;116(3):585–96. doi: 10.1083/jcb.116.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoda K, Kitagawa K, Masumoto H, Muro Y, Okazaki T. A human centromere protein, CENP-B, has a DNA binding domain containing four potential alpha helices at the NH2 terminus, which is separable from dimerizing activity. J Cell Biol. 1992;119(6):1413–27. doi: 10.1083/jcb.119.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pluta AF, Saitoh N, Goldberg I, Earnshaw WC. Identification of a subdomain of CENP-B that is necessary and sufficient for localization to the human centromere. J Cell Biol. 1992;116(5):1081–93. doi: 10.1083/jcb.116.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka Y, Nureki O, Kurumizaka H, Fukai S, Kawaguchi S, Ikuta M, Iwahara J, Okazaki T, Yokoyama S. Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 2001;20(23):6612–8. doi: 10.1093/emboj/20.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohzeki J, Nakano M, Okada T, Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol. 2002;159(5):765–75. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 23.Stock A, Clarke S, Clarke C, Stock J. N-terminal methylation of proteins: structure, function and specificity. FEBS Lett. 1987;220(1):8–14. doi: 10.1016/0014-5793(87)80866-9. [DOI] [PubMed] [Google Scholar]
- 24.Tooley CE, Petkowski JJ, Muratore-Schroeder TL, Balsbaugh JL, Shabanowitz J, Sabat M, Minor W, Hunt DF, Macara IG. NRMT is an alpha-N-methyltransferase that methylates RCC1 and retinoblastoma protein. Nature. 2010;466(7310):1125–8. doi: 10.1038/nature09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, Hunt DF, Macara IG. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat Cell Biol. 2007;9(5):596–U203. doi: 10.1038/ncb1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desrosiers R, Tanguay RM. Methylation of Drosophila histones at proline, lysine, and arginine residues during heat shock. J Biol Chem. 1988;263(10):4686–92. [PubMed] [Google Scholar]
- 27.Martinage A, Briand G, Van Dorsselaer A, Turner CH, Sautiere P. Primary structure of histone H2B from gonads of the starfish Asterias rubens. Identification of an N-dimethylproline residue at the amino-terminal. Eur J Biochem. 1985;147(2):351–9. doi: 10.1111/j.1432-1033.1985.tb08757.x. [DOI] [PubMed] [Google Scholar]
- 28.Xiong L, Adhvaryu KK, Selker EU, Wang Y. Mapping of lysine methylation and acetylation in core histones of Neurospora crassa. Biochemistry. 2010;49(25):5236–43. doi: 10.1021/bi1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porras-Yakushi TR, Whitelegge JP, Clarke S. A novel SET domain methyltransferase in yeast: Rkm2-dependent trimethylation of ribosomal protein L12ab at lysine 10. J Biol Chem. 2006;281(47):35835–45. doi: 10.1074/jbc.M606578200. [DOI] [PubMed] [Google Scholar]
- 30.Sadaie M, Shinmyozu K, Nakayama J. A conserved SET domain methyltransferase, Set11, modifies ribosomal protein Rpl12 in fission yeast. J Biol Chem. 2008;283(11):7185–95. doi: 10.1074/jbc.M709429200. [DOI] [PubMed] [Google Scholar]
- 31.Carroll AJ, Heazlewood JL, Ito J, Millar AH. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol Cell Proteomics. 2008;7(2):347–69. doi: 10.1074/mcp.M700052-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Meng F, Du Y, Miller LM, Patrie SM, Robinson DE, Kelleher NL. Molecular-level description of proteins from saccharomyces cerevisiae using quadrupole FT hybrid mass spectrometry for top down proteomics. Anal Chem. 2004;76(10):2852–8. doi: 10.1021/ac0354903. [DOI] [PubMed] [Google Scholar]
- 33.Smith GM, Pettigrew GW. Identification of N,N-dimethylproline as the N-terminal blocking group of Crithidia oncopelti cytochrome c557. Eur J Biochem. 1980;110(1):123–30. doi: 10.1111/j.1432-1033.1980.tb04847.x. [DOI] [PubMed] [Google Scholar]
- 34.Henry GD, Dalgarno DC, Marcus G, Scott M, Levine BA, Trayer IP. The occurrence of alpha-N-trimethylalanine as the N-terminal amino acid of some myosin light chains. FEBS Lett. 1982;144(1):11–5. doi: 10.1016/0014-5793(82)80558-9. [DOI] [PubMed] [Google Scholar]
- 35.Webb KJ, Lipson RS, Al-Hadid Q, Whitelegge JP, Clarke SG. Identification of protein N-terminal methyltransferases in yeast and humans. Biochemistry. 2010;49(25):5225–35. doi: 10.1021/bi100428x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villar-Garea A, Forne I, Vetter I, Kremmer E, Thomae A, Imhof A. Developmental regulation of N-terminal H2B methylation in Drosophila melanogaster. Nucleic Acids Res. 2012;40(4):1536–49. doi: 10.1093/nar/gkr935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petkowski JJ, Schaner Tooley CE, Anderson LC, Shumilin IA, Balsbaugh JL, Shabanowitz J, Hunt DF, Minor W, Macara IG. Substrate specificity of mammalian N-terminal alpha-amino methyltransferase NRMT. Biochemistry. 2012;51(30):5942–50. doi: 10.1021/bi300278f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang F, Dai X, Wang Y. 5-Aza-2′-deoxycytidine induced growth inhibition of leukemia cells through modulating endogenous cholesterol biosynthesis. Mol Cell Proteomics. 2012;11(7):M111 016915. doi: 10.1074/mcp.M111.016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You C, Dai X, Yuan B, Wang J, Brooks PJ, Niedernhofer LJ, Wang Y. A quantitative assay for assessing the effects of DNA lesions on transcription. Nat Chem Biol. 2012;8(10):817–22. doi: 10.1038/nchembio.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci U S A. 2008;105(25):8580–4. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashburner M, Bonner JJ. The induction of gene activity in drosophilia by heat shock. Cell. 1979;17(2):241–54. doi: 10.1016/0092-8674(79)90150-8. [DOI] [PubMed] [Google Scholar]
- 42.Johnston D, Oppermann H, Jackson J, Levinson W. Induction of four proteins in chick embryo cells by sodium arsenite. J Biol Chem. 1980;255(14):6975–80. [PubMed] [Google Scholar]
- 43.Vincent M, Tanguay RM. Different intracellular distributions of heat-shock and arsenite-induced proteins in Drosophila Kc cells. Possible relation with the phosphorylation and translocation of a major cytoskeletal protein. J Mol Biol. 1982;162(2):365–78. doi: 10.1016/0022-2836(82)90532-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.