Abstract
Tailored biomaterials with tunable functional properties are desirable for many applications ranging from drug delivery to regenerative medicine. To improve the predictability of biopolymer materials functionality, multiple design parameters need to be considered, along with appropriate models. In this article we review the state of the art of synthesis and processing related to the design of biopolymers, with an emphasis on the integration of bottom-up computational modeling in the design process. We consider three prominent examples of well-studied biopolymer materials – elastin, silk, and collagen – and assess their hierarchical structure, intriguing functional properties and categorize existing approaches to study these materials. We find that an integrated design approach in which both experiments and computational modeling are used has rarely been applied for these materials due to difficulties in relating insights gained on different length- and time-scales. In this context, multiscale engineering offers a powerful means to accelerate the biomaterials design process for the development of tailored materials that suit the needs posed by the various applications. The combined use of experimental and computational tools has a very broad applicability not only in the field of biopolymers, but can be exploited to tailor the properties of other polymers and composite materials in general.
Keywords: Biopolymers, Material design, Elastin, Collagen, Silk, Integration
1. Introduction
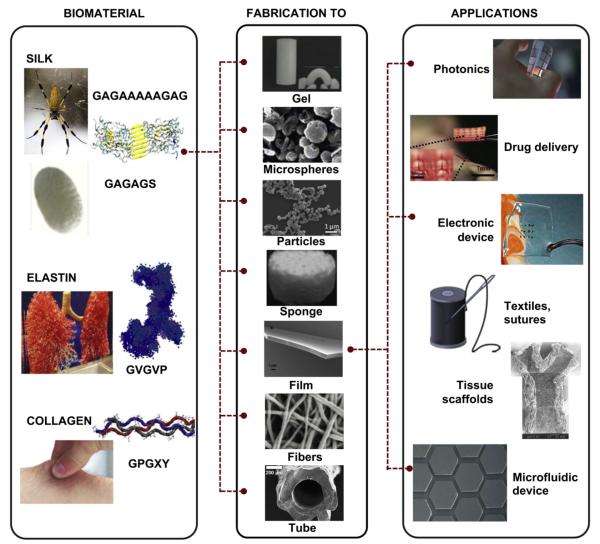
In applications such as tissue engineering, drug delivery and implantable devices, materials that meet a combination of specific medical requirements are desired. These requirements include features such as biocompatibility and biodegradability as well as tunability of critical functional properties, such as mechanical strength, resilience and toughness [1–4]. Advanced healthcare is just one example of the challenges of the 21st century that rely on advanced functional materials [5]. Biopolymers show highly diverse characteristics with respect to their shape (e.g., gels to fibers and particles) and their properties (e.g. diverse mechanical properties such as strength, elasticity, toughness or robustness), and often require transformative solutions for the design of new materials. Fig. 1 shows different shapes that can be fabricated from the biopolymers collagen, elastin and silk, and their potential applications.
Fig. 1.
Possible structures and technical applications of the materials silk, elastin and collagen. The dotted line shows an example of the versatility of silk and the multiple possible applications. Courtesy for images is as follows: Spider is from David Maiolo uploaded on 14.08.2011 on Wikipedia under the CC BY-SA 3.0 license. Silk cocoon, gel, sponge, fiber and electronic device are adapted from Omenetto and Kaplan [111]. Lungs from “jemsweb” on 06.07.2007 uploaded via flickr under the CC BY-SA 2.0 license. Elastin molecule is adapted from Baldock et al. [160]. Copyright 2011 National Academy of Sciences, USA. Collagen molecule is adapted from Gautieri et al. [161]. Microspheres are reprinted from Wang et al. [162] with permission from Elsevier. Particles are reprinted from Lammel et al. [115] with permission from Elsevier. Film is adapted from Krishnaji et al. [163]. Tube is reprinted from Lovett et al. [164] with permission from Elsevier. Photonics is from Fiorenzo Omenetto. Drug delivery is reprinted from Tsioris et al. [165]. Collagen scaffold is reprinted from Sachlos et al. [166] with permission from Elsevier. Microfluidic device is reprinted with permission from Bettinger et al. [167].
The combination of characteristics of biopolymers is often based on the distinct hierarchical levels in their structure, which leads to an increased diversification and enhancement of material properties [6–9]. Based on relatively simple building blocks, a highly controllable assembly strategy is the foundation of this structural complexity. It is of interest to tailor functional materials to match relevant requirements. The design of biological or bioinspired materials for specific applications can be achieved by tuning multiple design-variables in polymer synthesis and processing that pertain to the functional outcomes required [7–9]. Thereby, factors such as chemistry, molecular weight, processing conditions, and final post curing must be addressed to identify building blocks and recurring patterns in hierarchical structures and their subsequent links to properties. The parameter complexity is a significant factor in the serial design process that has usually been applied with little data transparency [5]. An urgent challenge to a concurrent or synergistic design is the incongruity of the different perspectives on materials (e.g. from a chemical point of view, processing, modeling, or in vivo performance). The serial design process, where a polymer is prepared, characterized, processed, and then evaluated for function, is inefficient and often results in materials lacking key functional features. Furthermore, outcomes considered as failures in this mode of discovery may be useful materials with alternative properties or instructive in new design approaches. In this context, a polymer design process, which integrates synthesis and processing, coupled to mechanistically driven bottom-up computational simulation may greatly accelerate the design process and rescue the failures to explore their full potential. However, to date, such an integrated strategy has rarely been employed in materials design, in spite of the fact that it seems to be an important and almost logical evolution of the field. An immediate consequence of such an approach would be a more rationally directed design based on model predictions. The ability to control structural features on multiple hierarchical levels via integrating computational modeling and processing at early stages of the material design could contribute toward more cost- and time-efficient design processes.
Here we review the state of the art for the design of biopolymers, and point out material design approaches based on the combination of experimental and computational studies. We believe that the exploration of the versatile parameter effects in biopolymer synthesis and processing gives the chance to build mechanistic models and to explore the design space of new materials. Thereby, we purposely omit studies that are solely focused on the structure analysis of natural biomaterials and their characterization. This review begins with an introduction to common synthesis, processing and modeling methods for biopolymers that highlights which parameters can be controlled or studied with these methods. Thereafter, we discuss the structure–property and process–property relationships of collagen, elastin and silk inspired biopolymers and investigate the approaches used to find these results. Finally, we examine the combination of experimental and computational works in the field of biopolymer design and outline challenges and opportunities.
2. Experimental, computational and theoretical methods for exploring the structure–process–property relationships
We highlight three fields related to the design process of biopolymers, which we identified to be a useful guide for the discussion: i) The field of polymer synthesis, which is chemically driven and dictates the polymer sequence chemistry that in turn has a major influence on the material structure at multiple scales. ii) The field of polymer processing and how its control of environmental conditions influences biopolymer assembly, from secondary structure to the macroscale. iii) The field of computational modeling that guides the biopolymer design process and is able to simulate structure and assembly at different length scales. We do not consider further material characterization steps for function which include in vivo or in situ studies and do not discuss materials development steps that are aimed toward scaling up and commercializing the production processes.
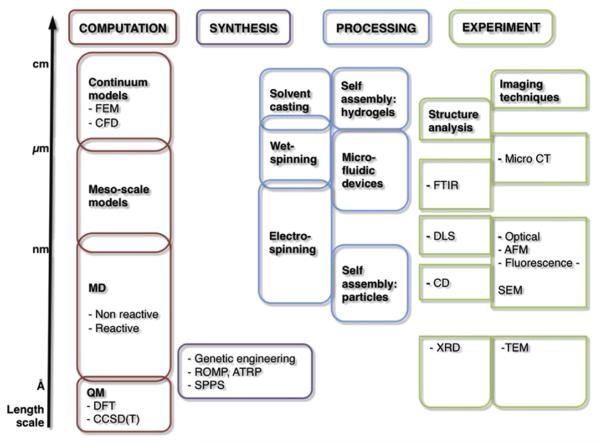
In Fig. 2 we show the design approaches of synthesis, processing and computational modeling together with relevant methods and associate them with the length scale they typically address. The synthesis of polymers is chemically driven and accesses the length scale of nanometers, while different processing methods control higher level structures and influence the final form (e.g. fibers, particles, films, gels) at the length scale of millimeters and higher. The secondary structure of peptides is influenced by both sequence and processing conditions and can therefore be tuned by synthesis and processing. Fig. 2 also illustrates how the synthesis and processing remain limited in the biopolymer design process when each is applied alone. Besides these experimental approaches we identify computational modeling as a third approach to handle biopolymer design. A brief introduction to the methods of biopolymer synthesis, processing and computational modeling is presented in the following sections, supported by Tables 1–3.
Fig. 2.
Experimental and computational methods to study the structure and properties of hierarchical biopolymers according to the length scale they address. Note how the different approaches work on distinct scales. In the bottom-up design of biopolymers all hierarchical levels must be considered. Therefore, only an integration of these approaches will allow full control of biopolymer design. Modeling could fill in the gaps in understanding processes that cannot be analyzed experimentally. Abbreviations are as follows: FEM: Finite Element Method, CFD: Computational Fluid Dynamics, MD: Molecular Dynamics, QM: Quantum Mechanics, DFT: Density Functional Theory, CCSD(T): Coupled Cluster Method, ROMP: Ring Opening Metathesis Polymerization, ATRP: Atom Transfer Radical Polymerization, SPPS: Solid Phase Peptide Synthesis, FTIR: Fourier Transform Infrared Spectroscopy, DLS: Dynamic Light Scattering, CD: Circular Dichroism Spectroscopy, XRD: X-ray Diffraction, MicroCT: Microtomography, AFM: Atomic Force Microscopy, SEM: Scanning Electron Microscopy, TEM: Transmission Electron Microscopy.
Table 1.
Overview of methods of chemical–biological hybrid polymer synthesis indicated along with outcomes.
| Synthesis | Type of polymer | Impact | Outcome/merits or demerits | Ref. |
|---|---|---|---|---|
| Bio-organic homopolymers | ||||
| Controlled radical, atom transfer radical, ring opening polymerization |
Polypeptides | Demerits-chain breaking and termination reactions, precipitation of the growing polypeptide chain at a certain molecular weight, and formation of unwanted secondary structures making it difficult to prepare homo polypeptides with defined molecular weights and low polydispersity indices |
[11–13] | |
| Catalyzed enzymatic polymerization |
Polysaccharides | Synthesized polysaccharides treated with enzyme allow for living polymerization with predictable molecular weight, Poisson molecular weight distribution |
[169,170] | |
| Sequenced bio-organic polymers | ||||
| Recombinant DNA |
Peptide polymers |
Site specific incorporation of unnatural amino acids at any position by using either chemically acylated transfer-RNA (tRNA), or engineered tRNAs and synthetases; unique codons encode for glycosylated amino acids, and amino acids with keto, azido, acetylenic and heavy atom containing side chain |
Fidelity in high molecular weight monodisperse |
[26,171,172] |
| Solid phase peptide synthesis |
Overcomes a number of difficulties related to the selectivity of chemical reactions, the insolubility of protected peptides, the optimization of the applied polymer supports concerning accessibility, diffusion properties and non-specific interactions with the peptide, as well as the difficulty to drive the step-wise reaction to quantitative conversion |
Sequence-specific incorporation of unnatural amino acids by chemical synthesis is virtually unlimited. It allows versatile modifications of the peptides (ranging from single position mutants, where one amino acid is substituted, to the synthesis of pseudo-peptides comprising of non peptidic backbones) and their chemistry |
[173,174] | |
| Ligation (native, coupling) |
Diverse selective coupling reactions, adapted from peptide ligation, were used such as formation of thioesters, oximes, thiazolidine/oxazolidine, thioether, and disulfides to attach polymers to peptides |
[175,176] | ||
| Templated polymerization |
DNA polymers | Programmable interconnections to guide structure formation processes in synthetic polymers for the preparation of materials with preconceived architectural parameters and new properties; enormous potential of oligonucleotides in biodiagnostics |
Demerits – Rapid degradation by DNAses, low solubility |
[177–179] |
| PNA polymers | Stability/tolerance to pH, ionic strengths, expensive synthesis |
Pharmacological potential to hybridize with DNA/RNA |
[180,181] | |
Table 3.
Summary of modeling methods for biopolymers at different length scales with restrictions and possibilities.
| Scale | Methods | Variables | Outcome | Limitations |
|---|---|---|---|---|
| Quantum scale |
Quantum mechanics (QM): Density Functional Theory (DFT), Coupled Cluster Method (CCSD(T)) |
Crystal symmetries | Based on band structure and density of states: Thermal properties, electrical properties and chemical reactions |
Restrictions in time scale and system size due to computational performance (below 5000 atoms and 10−14 s) |
| Molecular scale |
Molecular dynamics (MD): Energy Minimization & Equilibration, Steered molecular dynamics (SMD), Replica exchange molecular dynamics (REMD), Umbrella sampling, Well-tempered metadynamics, Reactive force-fields |
Sequence, pH, ions, force (loading conditions), velocity, temperature |
Conformation (e.g. number of H-bonds, secondary structure, free energy landscape, folding pathways), chemical reactions, mechanical properties (e.g. robustness, strength, adhesion strength) |
Restrictions in time scale and system size due to constraints in computational performance (below several hundreds of nm/ below billion atoms, time step about 10−9 s); With most of the force fields no reactions |
| Mesoscale |
Coarse-grained methods: Quasicontinuum method, Kinetic Monte-Carlo method, Coarse-grained Molecular dynamics (e.g. MARTINI force field), Dissipative Particle Dynamics (DPD) |
Structure, structural flaws and defects, hydrophilic– hydrophobic-ratio, molecular weight, thermal fluctuation |
Size effects, self-assembly, radius of gyration, hydrodynamic radius, fracture properties |
Either restrictions in size or in accuracy |
| Macroscale |
Continuum methods: Solid and CFD (FDM, FEM, FVM, BEM) MBS-Simulation |
Global structure (geometry), dynamic mechanical properties, process conditions (thermal, mechanical, electrical) |
Nonlinear structure analysis, global thermal and electrical analysis, multi-body problems |
Restriction due to homogenization of structure/properties; description via partial differential equations; Not based on first principles |
2.1. Synthesis
Simple molecular concepts from biology are a source of inspiration for polymer chemists. These concepts can be exploited by integrating bio-inspired polymer conjugates to exhibit interesting structural and functional properties beyond those of their individual components. Here, we discuss the different synthesis methods for the generation of functional chemical–biological hybrid systems that exploit the advantages of each system. Polymer synthesis is considered to be a relatively mature discipline, yet there still remains a wide gap for chemically (as opposed to genetically) engineered systems to mimic material performance of natural systems. Advancements in the field of conventional polymer science have led to the synthesis of nearly monodisperse materials with defined molecular weights, and reasonable control over architecture. However, the design of highly functional polymers unavoidably requires a high design complexity together with a detailed control of their synthesis. Biological systems with their limited repertoire of ‘monomers’ could benefit from chemical modifications to expand the functionality of natural building blocks.
Current chemical methods to synthesize polymers use both amino acids and other chemicals as monomers. These monomers can be synthesized as copolymers. In particular, the repetitive nature of block copolymers is well suited for structural assembly. There are diverse strategies for the preparation of hybrid block copolymers [10]. Some traditional methods include controlled radical polymerization, ring-opening polymerization (ROP), polymerization of macromonomers, and convergent synthesis of peptide-polymer hybrids. The most frequently used synthetic methodology to prepare homopolypetide blocks is ROP of α-amino acid-N-carboxyanhydride (α-NCA). For example, ROP of α-NCA initiated by a primary amino end-functionalized polymer was used to synthesize a series of polybutadiene-b-poly(γ-benzyl-l-glutamate) and polybutadiene-b-poly(N-hydroxypropyl-l-gluta-mine) diblock copolymers [11–13]. This technique allows gram scale synthesis, but has some major limitations. These limitations include the dependence on purity of reagents, abrupt chain termination, precipitation of growing polypeptides with increased molecular weight and unwanted secondary structures leading to branching of polypeptides, and a high polydispersity index. Polymerization is however considered to be living in nature depending on temperature, i.e., reactions other than nucleophilic ring opening chain growth are absent. Different approaches to synthesizing various polymers are briefly described in Table 1.
In order to impart structural control characteristic of biopolymers to synthetic polymers, let us first consider the factors that control structure in biopolymers. In the case of protein-based biopolymers, the primary sequence and linkages between the monomers are responsible for the formation of well-defined secondary structures (α-helices and β-sheets) by folding that later self-assemble into supramolecular structures [14]. Hence, precise control over defined properties can be achieved at different length scales by programming primary sequences of the polymers to obtain predictable hierarchical structures. Recently, conjugation of peptides to synthetic polymers has enabled new materials that expand on the properties of single component systems. For instance, peptide/biopolymer conjugates can enhance control over the nano-structure formation of the synthetic component. The biopolymer segment can reduce the toxicity and immunogenicity of materials, whereas the synthetic segment prevents enzymatic degradation or loss of function due to steric hindrance and unfolding of the conjugate systems.
Current methods to produce genetically engineered biopolymers include recombinant DNA technology that has been a major focus for preparing protein-based polymers, including silk-like block copolymers [15–17], elastin-like [18–21] and silk–elastin-like copolymers [22–24]. The major advantages of this approach include sequence-controlled programming of biopolymers with precise molecular weight using genetic templates and production of reasonable amounts of protein via bacterial expression systems that do not require harsh organic solvents for synthesis. Moreover, purification of genetically engineered proteins is performed under aqueous conditions and ambient temperature and pressure. Through careful design of genetic constructs and the use of appropriate expression systems, successful recombinant proteins have been used commercially after FDA approval, including vaccines for papillomavirus and hepatitis B, and drugs such as anti-thrombin, insulin and erythropoietin.
However, there are some disadvantages to this approach, such as the time involved in assembly of genetic constructs and inability to easily prepare biopolymers with d-amino acids, unnatural amino acids or with selective chemical modifications, most of which cannot be encoded efficiently by bacterial systems. The incorporation of unnatural amino acids has become feasible through modification of host strains or chemical ligation techniques. These techniques allow the use of unnatural amino acids to generate polymers that are otherwise difficult to prepare [25]. Native chemical ligation is an alternative and elegant strategy that allows selective coupling of peptide fragments under natural conditions to form native peptide linkages. By this approach, a peptide with a C-terminal thioester is temporarily ligated to an N-terminal cysteine residue of a second peptide through a trans-thio-esterification reaction and can be routinely used for the preparation of polypeptides over 100 amino acid residues in length.
Recently, an approach of expanding the array of unnatural amino acids was achieved by chemically acylated transfer RNA (tRNA) or engineered t-RNAs and synthetases. Methods were developed to expand the genetic code at the nucleic acid level, including 4-base codons (sequence of adjacent nucleotides in the genetic code, determining the insertion of a specific amino acid in a polypeptide chain) and unnatural base-pairs [26]. Over thirty unnatural amino acids have been genetically encoded in response to unique triplet and quadruplet codons, including fluorescent, photoreactive and redox active amino acids, glycosylated amino acids, and amino acids with keto, azido, acetylenic and heavy-atom containing side chains. This approach provides new options for genetic engineering to overcome obstacles in the incorporation of unnatural amino acids.
2.2. Processing
In addition to the diversity of biopolymer synthesis techniques, a wide variety of versatile processing techniques can be utilized to prepare substrates ranging from fibers to particles for varying applications (see Table 2). Due to the large number of polymer processing methods, we will focus on those that have been utilized to increase the control of resulting material structure or properties. These processing techniques range from electrospinning to microfluidics.
Table 2.
Summary of processing techniques sorted by the type of the microstructure they achieve. The variables can be adapted to tailor the material properties.
| Product | Technique | Variables | Application | Ref. |
|---|---|---|---|---|
| Fibers | Electrospinning | Electric field, pH, solvent, plate distance, set-up parameters, post-spin treatment |
Tissue engineering scaffolds, fiber optics, nanowires |
[27–29] |
| Wet-spinning | Solution concentration, pH, solvent, set-up parameters, post-spin treatment |
Tissue engineering scaffolds, textiles |
[30,31] | |
| Microfluidics | pH, flow rate, channel geometry, post-spin treatment |
Fiber optics, tissue engineering scaffolds, macromolecular assembly mechanisms |
[32–34, 154] |
|
| Films | Solvent casting | Solvent, concentration, casting conditions, post treatment; patterned surfaces via lithography |
Complex patterns for cell guiding |
[37,182] |
| Microfluidics | Geometry, physical constraints, flow constraints |
Cell guidance | [35] | |
| Hydrogels, 3D matrices |
Controlled assembly |
Crosslinking method, pH, concentration |
Artificial organs, drug delivery |
[39,40] |
| Self-assembly | Amphiphilic characteristics, ionic strength, pH |
ECM mimic | [41,183] | |
| Particles | Self-assembly | Amphiphilic characteristics, concentration, pH |
Drug delivery | [42,43, 184] |
| Microfluidics | Viscosity, concentration, relative flow rates, device dimensions, post fabrication treatment |
Drug delivery, ultrasound contrast agents |
[36] |
One common biopolymer fabrication technique is electrospinning, a process that utilizes a large electric field between a polymer solution and a collection plate to induce a stable jet to be ejected from the solution reservoir and travel toward the plate [27]. The result of this process is a non-woven fiber mat that is either randomly oriented or aligned depending on the collection method used. Variables in electrospinning include solution conditions (pH, concentration, solvent), device conditions (the distance between the tip and plate, strength of the electric field, nozzle dimensions) and collection methods (plate vs. rotating mandrel, speed of collection), which affect fiber diameter, fiber mechanical properties, and organization or alignment of the fiber mat [27]. A direct benefit of electrospinning is that the biopolymer material can be efficiently used, thus applicable to small amounts of polymers for exploratory studies. Fibers with diameters ranging from a few nanometers to microns can be fabricated for applications ranging from nanowires to fiber optics [28]. Additionally, electrospinning of natural polymers such as elastin, collagen and silk is useful for scaffold designs for tissue engineering [28,29].
Wet-spinning is another popular processing technique for biopolymer fibers. This technique is similar to an extrusion process where a polymer solution is forced through a nozzle of controlled dimensions into a solution bath to fabricate fibers. Fibers can be collected in a variety of ways to make 3D constructs, such as meshes, tubes and related materials. Variables in wet-spinning include polymer solution conditions (concentration, pH, solvent), nozzle dimensions, reservoir solution composition and additional post-spin treatment such as drawing. These variables affect the diameter of the fibers as well as the mechanical and surface properties [30,31].
Microfluidic processing techniques are very versatile and can be used to make fibers [32–34], gels [35] or particles [36] from biopolymers. Microfluidic fiber fabrication techniques utilize flow conditions and gradient environments [33] to control polymer assembly inside a microfluidic channel. Variables include solutions used, device geometry and dimensions, flow rates, and post-spin processing. Manipulation of these variables allows for control over parameters such as fiber diameter and mechanical properties [32]. Microfluidics has been used in the processing of biopolymer gels to constrict assembly and further advance polymer organization within the gel [35]. Microfluidic approaches to particle fabrication often utilize flow focusing of multiple solutions or phases. For example, drug encapsulation in monodisperse particles can be easily achieved via microfluidics and has significantly advanced the area of drug delivery research [36].
Biopolymer films can be fabricated via solvent casting [37] where a solution is cast onto a mold and allowed to dry, forming a film. Variables in solvent casting include the solvent used, the concentration of the biopolymer, the casting conditions (i.e. forced drying) and film post-treatment. These variables affect the surface and bulk properties of the material. Additionally, the mold can be flat or can have complicated patterning via lithography, creating a film with patterns such as alternating grooves and ridges or complex designs that can be used for cell orientation. These films are often used for tissue engineering. While lithography allows for complex patterns to be formed, casting does not allow for a large degree of control over material assembly, which can be important for many biopolymers [37].
Another important biopolymer processing technique with rather widespread utility is self-assembly, which can be utilized to fabricate hydrogels, 3D matrices and particles. Hydrogels are 3D polymer networks that swell but do not dissolve in water [38]. These materials have been used as biomaterials since the mid-1900s [39] and their use ranges from contact lenses to artificial organs, extracellular matrix (ECM) mimics and controlled/sustained drug delivery. Variables in hydrogel fabrication include physical and chemical crosslinking [40], pH, biopolymer composition (amphiphilic characteristics, ionic strength [41]) and concentration, which all affect the properties of the material, including stiffness and swelling ratio. Amphiphilic polymers that form hydrogels at low concentrations often form micelles or other particles at higher concentrations or in certain environments. Self-assembled particles are also valuable tools in drug delivery. Variables in self-assembly of particles include the amphiphilic nature of the biopolymer, concentration, and assembly conditions including pH [42,43]. These variables will determine if assembly will occur, control the size/shape of the resulting particles, and have implications in the drug delivery profile.
These processing techniques are as diverse as the materials they generate, and each variable or parameter presents an opportunity to control the assembly of the biopolymer and thus affect the structure and resulting properties. Controlled processing is one key step in the tailored design and fabrication of tunable materials.
2.3. Computational modeling
A recent white paper published by the U.S. National Science and Technology Council [5] states that computational methods are crucial to accelerate the materials design process. In the study of biopolymer properties a multiscale approach is beneficial, as many remarkable material properties of biological materials are evoked by a hierarchical architecture [6]. Computational models should therefore be able to incorporate all critical length and time scales. It is currently impossible to study materials like biopolymers with a single computational method on all scales simultaneously. For the observation and prediction of sequence–structure–property relationships, a bottom-up modeling approach can provide a rigorous basis for the design of functional materials. In Table 3 several key modeling techniques to study the structure and behavior of materials are shown. It is possible to describe the behavior of materials from fundamental laws of physics. Therefore, a model of a material based on the basic building blocks and their interactions can result in the description of material properties. Based on this idea, molecular dynamics (MD) considers atoms as point masses, and force fields between atoms are either derived from quantum mechanical modeling or defined empirically. In MD, the trajectory of a set of atoms is simulated following Newton’s equation of motion, as opposed for example to Monte Carlo methods that find the system’s equilibria according to the generation of random states. All-atom simulations are typically used to sample the energy landscape and predict the protein structure of the sequence. MD is also able to predict the folding pathway of small proteins. Ideally, in all-atom simulations the solvent is modeled explicitly; however, this approach quickly reaches its limits of accessible time and size scales. The structure of medium sized proteins is explored by either accelerated sampling methods (e.g. replica exchange molecular dynamics [44], metadynamics [45]) or by the reduction of the system complexity by using implicit solvent and/or simplified protein description [46]. Nevertheless, it remains a challenge to fully describe large proteins such as elastin or silk, based on the fact that non-equilibrium processes influence also the formation of secondary structures.
Only distinct parts of biopolymers are simulated by MD. Steered molecular dynamics is a non-equilibrium MD technique that is able to elucidate mechanisms of deformation behavior on the atomistic scale, for example unfolding or stick-slip failure. Such modeling techniques help to relate the secondary structure of proteins to their mechanical properties. For instance, the mechanisms related to the deformation behavior of α-helices and β-structures have been extensively characterized and size and rate effects were quantified [47]. The obtained data is fed into mesoscale models that represent structural features on levels of hierarchy, which are, up to now, impossible to reach by all-atomistic simulations [48,49]. These mesoscale models bridge the gap between continuum methods and MD by enabling a continuous multiscale description of materials from the atomistic to the macroscale.
A common approach to model materials on the mesoscale is the use of coarse-graining models that merge several atoms to a single bead and use simplified force fields. Hence, the degrees of freedom are reduced. Consequently, larger system sizes can be simulated on larger time scales. While MD simulations with, for example, the widely used CHARMM force field [50] are generically applicable for most proteins, coarse-grained models are often only valid for a particular material system they have specifically been trained for. Some generalized coarse-graining frameworks exist, such as the MARTINI force field [51], the OPEP force field [52], the Honeycutt–Thirumalai model [53] and the Head-Gordon model [54]. The MARTINI force field, for example, only includes four types of beads – polar, nonpolar, apolar and charged – and further subtypes of these. However, for the effective simulation of systems with high molecular weight biopolymers this coarse-graining level is not sufficient. With increasing levels of coarse-graining, i.e. increasing size and time scale, chemical details are more and more simplified or neglected.
To represent processing conditions mesoscale simulation methods incorporating hydrodynamic effects are interesting for the study of biopolymers. Two main approaches are commonly used to model the solvent. Lattice methods such as the lattice-Boltzmann method have been reviewed by Dünweg and Ladd [55] and particle-based methods such as dissipative particle dynamics and multi-particle collision dynamics have been reviewed by Noguchi et al. [56].
On larger scales, continuum methods, such as the finite element method (FEM), enable the modeling of materials such as homogenized composite structures, but face difficulties in describing the impact of hierarchical structures due to their dependence on governing partial differential equations, and often lack a mechanistic basis to the model.
3. Biopolymer design: case studies and examples
The variation of input variables and possible system configurations is a necessity that allows tweaking a material. Natural materials based on biopolymers and the remarkable performance that they offer provide an excellent starting point for further exploration of configurations, as the materials and their production processes have been highly optimized during evolution. Here, we categorize studies on collagen, silk, silk–elastin and elastin-like biopolymers that deal with the interaction and influence of design variables on structure and properties. We purposely omit studies solely focused on structure analysis. Therefore, we exclude the biomimetic design approach that aims at the understanding of how certain structure–property relationships are achieved in nature. The state of the art of this biomimetic material design approach has recently been reviewed elsewhere [57,58]. Vollrath et al. [59] state that in order to employ a biomimetic design, nature has to be (fully) understood first and subsequently these insights shall become integrated in the resulting material design. Given this, they argue that for example derivatives of spider silks attained by biomimetic approaches shall incorporate the envelope of natural spider silk properties. However, the interactions between spiders and their environment are highly complex. Most likely unknown dependencies have influenced the evolutionary design, which may not be in scope for the desired technical application [60].
Mimicking nature gives us only a single configuration in the respective parameter space (e.g., a certain set of sequence and process condition input variables leading to a specific set of output variables, such as: structure and property). For applications that require multi-functional materials with controlled properties, a fit of the material to the functionality is preferred rather than a redesign under the constraint of the intrinsic material properties. For example, some applications could take advantage of the biocompatibility and strength of spider silk, but its high extensibility could be a limitation. Moreover, the building blocks in silk, only a few distinct amino acids, could be dependent on the spider’s nutrition. Modified building blocks could be more efficient or serve to generate different functional properties of interest [60]. The observation of one configuration does not provide any information about structure–process–property relationships, while the study of more than one parameter configuration may already indicate a trend. For example Gosline et al. [61] compared data of silk from different species which spin silk with different mechanical behavior to gain insight into structure–property relationships. However, to tailor biopolymers for engineering applications, the investigation of multiple parameter configurations is useful. With more configurations investigated, the existence of parameter optima can be proven. In some cases metastable or local minima may also provide useful information for the fabrication of advanced materials.
3.1. Elastin
Elastic proteins exhibit rubber-like elasticity and are capable of undergoing large deformations without rupture. Elastin is an excellent example and one of the most extensively studied elastomeric proteins, as it serves as an efficient energy storage system. Materials with dominant elasticity must meet the following criteria: the individual components must be flexible and conformationally free, so that they can quickly correspond to stress, and they must be cross-linked to distribute the stress throughout the system. Thus, the elastic properties are influenced by the nature of the elastomeric domains, their size and the degree of cross-linking.
Elastin is an important extracellular matrix protein conferring elasticity to tissues and organs [62–64]. Native elastin isolated from animal tissues has two domains, a repeating hydrophobic domain and an alanine-rich cross-linking region [62]. It is widely accepted that the hydrophobic domain contributes to the elasticity of elastin [65,66]. Because of its remarkable elasticity [62–64] and stimuli-responsive properties [67–69], recently elastin and elastin-like polypeptides (ELPs) have been actively pursued as biomaterials for various biomedical applications [70], including protein purification [71], drug delivery [19,72,73], and tissue engineering [74–76].
3.1.1. Primary sequence and structural features of elastin
Elastin is secreted as a soluble precursor, tropoelastin, which is comprised of alternating hydrophobic domains of variable length (elastic repeats) and alanine rich, lysine containing domains that form cross-links. The hydrophobic domains are comprised of modular repeat domains of tri-, tetra- or pentapeptide repeats of (VPGXG)n, where X can be any residue other than proline [77,78], whereas the lysine–alanine regions provide functionality by formation of oxidative cross-links, fixing the structure [79]. Genetic engineering has made it possible to generate recombinant elastin mimetic protein polymers that undergo reversible, temperature dependent, hydrophobic assembly from aqueous solution in analogy to phase behavior of native elastins. The self-assembly process results in a spontaneous phase separation of protein polymers above the lower critical solution temperature (LCST) that coincides with conformational rearrangement of the local secondary structure within the pentapeptide motifs. Spectroscopic analysis has demonstrated that the pentapeptide sequence units undergo a conformational transition from a random coil to β-turns as the temperature approaches the transition temperature [69]. Substitution of the third amino acid residue of the repeat sequence (glycine to alanine) results in a change of the mechanical response of the material from elastic to plastic. Additionally, amino acid substitution of the fourth position modulates the temperature dependent phase behavior of the material [80]. These tunable properties lead to the generation of recombinant elastin-mimetic protein polymers that can serve as promising biomaterials for tissue engineering and biomedical applications. Recombinant elastin-like protein polymers provide significant opportunities to modulate material microstructure and can be processed in various forms including particles, films, gels and fiber networks. As a consequence, biological and mechanical responses of elastin-based polymers are tunable through precise primary sequence and block length.
3.1.2. Elastin-mimicking polymers
The combined effect of elasticity, durability and stimuli responsiveness (e.g. LCST behavior) of elastin makes it a promising biomimetic target. Numerous ELPs have been synthesized chemically [81–83] and by recombinant DNA synthesis [72,76] for mechanistic studies.
The characterization of elastin-like peptides started with (VPG)n, (VPGG)n and (VPGVG)n chemically synthesized to determine conformational properties. Early structural studies proposed β-turns adopted by these proteins, with both linear and cyclic polymers [84]. The regular periodicity of β-turns within these polypeptides was originally predicted to favor the formation of a putative right-handed helical structure termed a β-spiral, modeled with just under three VPGVG motifs per spiral turn [83]. Subsequent analyses determined the presence of β-turns within sequences of (GVPGV)7 and domains containing VPGVG and VAPGVG repeats, and a lack of extended secondary structures. Separately, MD simulations of a (VPGVG)18 sequence modeled using β-spiral dihedral angles predicted the instability of ideal β-spiral parameters within this sequence in solution [85,86]. Varying parameters like ionic strength and surfactants to determine thermodynamic parameters led to a conclusion that conformation was interdependent of intramolecular backbone conformations and aggregate states [69].
3.1.3. Recombinant elastin-like peptides
Recombinant elastin-like polymers have been fabricated as films, fibers and hydrogels for a variety of applications. The thermoresponsive behavior of recombinant elastin-like peptides was determined by altering lengths of diblock or triblock copolymers to demonstrate a wide range of inverse transition temperatures (temperature below which block copolymers are completely miscible) [19,21,68,87]. Thus, above the lower critical solution temperature Tt, the polymer undergoes hydrophobic assembly from a soluble, extended state, to a collapsed, aggregated state. As a consequence, ELPs undergo a reversible temperature-dependent assembly, leading to variety of morphologies. Later, a model was designed to quantify the LCST and stimuli responsiveness of ELPs using a single equation comprised of three parameters, sequence, molecular weight and concentration [68]. Incorporation of different guest residues (in position X of VPGXG) enabled a wide range of Tt as a function of chain length and concentration for a fixed composition. Thus, by tailoring chemistry with block length and guest residue, tunable and stimuli responsive ELPs could be designed and engineered to be functional. In addition, to improve control over self-assembly and cross-linking, the sequence composition of block copolymers with altering repeats of hydrophobic blocks with polylysine/cross-linking domains and hydrophilic elastin blocks were engineered [76,87]. Lysine domains cross-linked with hydroxymethylphosphine even under physiological conditions and hydrogels with different swelling ratios, microstructure and mechanical properties were obtained. In vitro studies indicated that mouse fibroblasts were successfully embedded into the hydrogels and remained viable for at least three days [76]. ELPs were physically impregnated onto expanded polytetrafluoroethylene (ePTFE) as a non-thrombogenic coating, due to the ability of ELPs to limit platelet adhesion [88].
With the advent of single molecule force spectroscopy (SFM) and atomic force microscopy (AFM), characterization of mechanical strength of ELPs has been feasible. High molecular weight elastin-like peptides that were chemically modified with cysteine residues were subjected to stretch and relaxation modes at the tip of an SFM probe [63]. Below the inversion temperature Tt, the peptides showed extension curves with perfect storage of strain energy, following the worm-like chain model of molecular elasticity. But above the LCST, there was an abrupt change in the modulus, indicating that at the single molecule level the hydrophobically collapsed structure has a significant amount of order that unfolds under stress in a similar way as expected from extended β-spiral rich random coils.
3.2. Collagen
Collagen type I and type III are the predominant constituents of most extracellular matrix material, and as such are ubiquitous and important structural biomaterials. The secondary structure of collagen, including the characteristic triple helix, is closely related to mechanical properties and cellular interactions. Additionally, the precise spatial organization of collagen fibers in vivo is closely related to cellular response as well as tissue tensile strength [35]. However, nearly all in vitro preparations of ECM involve randomly oriented collagen fibrils due to the self-assembly of soluble collagen subunits [89]. This section will discuss current methods used to study collagen in vitro related to control of sequence or composition, processing parameters, and relevant models.
3.2.1. Synthesizing collagen
Native collagens are often derived from rat tails or human placenta, and type I and III collagens are the most common types of the near 30 chemical variants of collagens reported to date. For in vitro studies, the protein is denatured into an intermediate state, fibrils, that are subsequently reassembled via a variety of techniques. While collagen substrates have been successfully prepared from these native materials, there is often difficulty in recapitulating native collagen structure and alignment, both of which are closely tied to collagen functions in vivo [27,28,37]. Non-native collagen for in vitro use can be fabricated either chemically or via recombinant DNA methods. Chemically derived collagen consists of short synthetic collagen peptides (24–63 amino acids long) that may be linked via terminal cysteines into fibrillar-like structures. However, the short length of these peptides compared to native collagens affects the scope of the nanoscale architecture and the overall mechanical properties [90–92]. Collagens derived via recombinant DNA methods are small collagen-like polymers fabricated via the tandem ligation of partial collagen regions. It is difficult to prepare recombinant full-length collagen due to the glycine–X–Y sequence repetition that promotes mismatched hybridization. Additionally there are limitations to the location, frequency, identity and combination of biologically relevant sites that can be introduced via this method [91,93,94]. Other recombinant proteins are designed to mimic collagen or contain collagen-like domains: a tailored DNA sequence is designed and built and the desired protein is expressed in a host via genetic engineering. The protein sequence can be dictated to control parameters such as mechanics and cellular responses, and then optimized for tailored applications [91,95–97].
3.2.2. Processing collagen
In addition to the control of collagen composition and sequence, an expanded degree of control can be achieved through different processing techniques and conditions. Collagen can be processed via a variety of techniques ranging from electrospinning to microfluidics (see processing section for additional details). Solution parameters such as pH [33], concentration, collagen type [27,37], and solvent affect the properties of the material fabricated. For example concentration and viscosity will affect the diameter of an electrospun fiber [98]; concentration, solvent and pH will affect the assembly of the collagen peptides into higher ordered structures, and thus affect mechanical properties as well as cell responses. Physical constraints during processing induce fibril alignment similar to that found in vivo and thus result in a more natural material [35,98,99]. On the macroscale, collection methods [27,33] and crosslinking of fibers [37,100] can affect bulk material properties, including mechanical properties, anisotropy and porosity.
3.2.3. Computational modeling of collagen
Given the hierarchical nature of collagen and the close interplay between levels of structural organization, many attempts to computationally model the relationships between sequence, structure and properties have been made. Hierarchical multiscale approaches, from atomistic to coarse-grained models, simulate native collagens and can be used to investigate deformation mechanics. These models explore relationships between the hierarchical organization of collagen and the properties, and allow for the representation of collagen fibrils on much larger and more diverse length scales than previous full atomistic models [101,102]. Additionally, there are many approaches that model defects in collagen primary sequence that are closely related to known diseases. These models relate single point mutations to structure and mechanics at multiple scales and have the potential to be expanded to a broad class of genetic disorders [103,104].
In spite of the large amount of work that has been done to study and understand collagen at all structural levels, there are still significant gaps in the overall interplay of sequence, structure, processing and properties; and especially in connecting molecular to larger scales. To our knowledge, there is no work that models the unique recombinant collagen-like proteins in the same detail that is used for modeling native collagen and collagen mutations. This is an area with great opportunities for biomedical applications.
3.3. Silk
Silk proteins can be regarded as high molecular weight amphiphilic block copolymers. The sequence is highly repetitive, where hydrophilic and hydrophobic domains alternate approximately a hundred times. Hydrophobic poly-alanine runs form β-sheet nanocrystals that are embedded in a semi-amorphous matrix consisting of glycine-rich repeats. The β-sheet content amounts to ~50% for the silkworm (Bombyx mori) cocoon silk and ~36–37% in spider dragline silk (Nephila clavipes and Nephila edulis) [105,106]. The β-sheet nanocrystals are thought to be responsible for strength, whereas the semi-amorphous phase uncoils upon stretching and triggers extensibility.
Different sorts of silk exhibit versatile and exceptional mechanical properties [107–109]. Silkworm cocoon silk fibers are strong and stiff and commonly used for textiles and wound dressings [60]. Spiders produce different types of silk that differ in properties and function. Spider dragline silk is the most studied spider silk and features a remarkable combination of strength and extensibility that leads to a superior toughness modulus, i.e. it is able to absorb massive kinetic energy before breaking [61,110]. Viscid silk of spiders, in contrary, behaves rubber-like and exhibits a low initial Young’s modulus and high extensibility [61].
Not only are the mechanical properties versatile, but silk is also not limited to the form of fibers and can be processed into a variety of shapes such as gels, films, particles, and sponges [111,112]. Applications in medicine, e.g. sutures [113], tissue scaffolds [114], and drug delivery systems [115] benefit from silk’s biocompatibility, degradability and mutability, i.e. the change of specific properties through external triggers such as pH, temperature or electric fields [116]. Recently, applications in photonics and electronics have been reported [117].
Silk has been thoroughly investigated in regard to its structure and the spinning process aiming to understand how a material that relies on weak hydrogen bonds and is processed in water at ambient conditions can outperform the mechanical properties of many synthetic materials. In the following, we describe which design variables can be influenced by synthesizing and processing silk to tune the material properties.
3.3.1. Synthesizing silk
Native silkworm silk fibers are available by large-scale cultivation of silkworms at reasonable costs and can be processed into multiple material formats as regenerated silk [111]. To achieve biocompatibility for medical applications the sericin coating needs to be removed of the fibroin core of silkworm silk (see for example [118] for protocols). Spider silk is biocompatible without further preparation, however, the commercial production through spiders itself is not feasible because of their territorial behavior and collecting silk from webs would not be profitable [119]. Therefore alternative synthesis routes are explored, including the expression in plants [120], in mammalian cells [121], in bacteria [22,122]; and recently the production of composite silkworm silk/spider silk fibers through silkworms [123].
Instead of reproducing the exact sequence of special silk types, peptides are engineered to represent main features found in silk sequences. Distinct naturally occurring repetitive motifs have been characterized as building blocks of spider silk that each fulfills different functions [111,119]. Next to the already mentioned polyalanine domain a (GA)n motif found in minor ampullate spidroin leads also to a β-sheet crystalline phase. Another motif that is found in many sorts of silks is the GGX motif, where X typically stands for tyrosine, leucine, alanine or glutamine. This motif is less understood and believed to consist mainly of little ordered β-sheet structures, 31 helices and β-turns [106,124–127]. Repeats with proline of the form of GPGXX lead to a spiral structure that is considered to evoke elasticity in operating like a spring [128]. In natural silk, nonrepetitive amino- [129,130] and carboxy-terminal domains [131] flank the motifs discussed above. The terminal domains are highly conserved over different species and contain several amino acids that are able to form salt bridges [132]. While the terminal domains are believed not to contribute to the mechanical properties of spider silk, it has been shown that they play an important role in the fiber assembly process [132,133]. Furthermore, spacers that contain charged side groups can be found [134] that also might relate to storing and fiber assembly. The design of a sequence out of these building blocks could potentially lead to a tailor-made material with specific, desired properties. However, some characteristics are difficult to reproduce. Silk’s large molecular weight of ~200–350 kDa [111] results in low yields when expressed by the commonly used bacterium Escherichia coli, so recombinantly produced silk proteins are usually in the size of 30–110 kDa [119].
3.3.2. Processing silk
In general, processing has a considerable effect on the silk fiber properties. For instance, under modified processing conditions, silkworm silk fibers can achieve properties comparable to spider silk [108]. Influential process parameters are mechanical stresses during spinning (shear and elongational flow), pH, ion-concentration, solvent type (organic or water based) and protein concentration (see Table 4). For example, phosphate ions induce protein aggregation due to the kosmotropic nature of phosphate anions and also shear triggers aggregation and alignment in the fiber assembly [135]. The high molecular weight does not only challenge synthesizing silk proteins; it is also difficult to store them without irreversible aggregation at a concentration of up to 50% (w/v) as found in spiders [136]. Ion- and pH-shifts induce a change in shape in the terminal domains. The “switching” of the terminal domains is believed to be crucial for the storage of the spidroin and may be also for chain alignment in the spinning duct [132,133,137].
Table 4.
Input-variables and the affected outputs-variables of biomaterial design studies on collagen, elastin, silk and silk–elastin. The references are ordered according to the relationships they address (compare to Table 5 and Fig. 3). For detailed information about these relationships please refer to the cited references.
| Sequence/composition | Processing | Structure | Property | Ref. |
|---|---|---|---|---|
| Collagen | ||||
| Recombinant collagen mimic |
Molecules retained triple helical assembly |
[91,96] | ||
| Length of tropocollagen molecule |
Deformation mechanics | [101] | ||
| Altered channel width | Alignment of fibrils | [35] | ||
| Langmuir blodgett conditions |
Alignment of fibrils | [99] | ||
| Concentration; shear rate | Alignment of fibrils | [98] | ||
| Point mutation (OI) | Mechanical properties | [161] | ||
| Concentration; voltage | Fiber diameter, spinnability | [27,28] | ||
| Simultaneous fibril formation and crosslinking |
Mechanics and morphology of fibers |
[100] | ||
| pH gradient; viscosity | Assembly | Fiber diameter | [33] | |
| Recombinant collagen mimic |
Monitored assembly via AFM |
Measured modulus | [95] | |
| Mutation – Alport syndrome | Tropocollagen structure | Mechanical properties | [103] | |
| Recombinant elastin-like polymers | ||||
| Different chain length | Concentration | Shift in inverse transition temperature |
[20,68] | |
| Elastin triblock copolymers |
Hydrogels | Viscoelastic and mechanical responses |
[19,21] | |
| Elastin diblock/triblock copolymers |
Physical and chemical crosslinking to hydrogels |
Rheology for gelation kinetics, swelling, microstructures, mechanical strength, biocompatibility studies |
[76,87] | |
| Temperature; ionic salts; solvent; guest residue |
Force-extension of single molecule to study hydrophobic hydrations |
[185] | ||
| Exons of recombinant elastin |
α-Helix | [186] | ||
| Single molecule | Force extension curve | [63] | ||
| Particles; temperature | Inverse transition temperature kinetics, condensation of polyplexes |
[187,188] | ||
| Temperature; pull-out | Mechanism of hydrophobic collapse in the presence of water leading to inverse transition temperature and elasticity |
[85,86] | ||
| Coating on PTFE grafts | β-Spiral structure | [88] | ||
| Temperature; pressure | Reversible conformation from random coil to β-turns/strands |
[152,153] | ||
| Heavy metal binding domains | Hydrogels | Inverse transition temperature, binding studies with heavy metals |
[18,189] | |
| Cartilage oligomeric matrix protein with different chain length |
Temperature | Secondary structure – conformational change from random coil to α-helix/β-sheets |
[190] | |
| Silk–elastin like polymers | ||||
| Varying chain length of elastin and guest residue |
pH; temperature; ionic strength; concentration |
Shift in inverse transition temperature |
[72,191–193] | |
| Temperature; ionic strength; concentration; hydrogels |
Swelling, rheology, gelation DNA release study |
[23,24] | ||
| Ratio of silk to elastin | Temperature; pH; ionic strength |
Irreversible conformational change from random coil to β-sheets |
Shift in inverse transition temperature; self assembly mechanism |
[58,194] |
| Films | Conformational change from silk I to silk II structure and formation of β-strands on methanol treatment |
Swelling, mechanical strength, biocompatibility |
[195] | |
| Fibers | Mechanical strength | [196] | ||
| Silk-like polymers | ||||
| Poly-alanine region length | β-Sheet content | [148] | ||
| Ion concentration | Secondary structure (conformation transition from random coil and/or helical structure to β-sheet) |
[197–200] | ||
| Fibroin concentration; pH; temperature; Ultrasonic treatment; shear forces |
Secondary structure | [200] | ||
| Geometric confinement | Strength and toughness | [49] | ||
| β-Sheet crystal size | Strength and toughness | [143,168] | ||
| Deformation behavior | Robustness of spider web | [146] | ||
| Reeling speed | β-Sheet crystal size; degree of orientation |
Strength and toughness | [149] | |
| Ion concentration; fibroin concentration; pH; temperature; poly (ethylene oxide) content |
Secondary structure | Gelation time; mechanical compressive properties | [150] | |
| Protein concentration; chemical crosslinking; temperature |
Secondary structure | Rheological properties | [201] | |
Out of the various material forms that are possible via different processing methods the most obvious form is silk fibers. Methods to artificially assemble fibers out of silk proteins are electro-spinning [138,139], wet-spinning [31,140,141], and spinning with a microfluidic device [32,34,142]. Silk spinning microfluidic devices allow to partly mimic the spinning process in nature via varying the aforementioned parameter such as imposing elongational flow, inducing a pH shift and exchanging ions [32,34]. Additional process variables are the flow rate and optional fiber drawing at different speeds by attaching the fiber to a spool.
3.3.3. Modeling of silk
The hierarchical structure of spider silk has been investigated with simulation on several scales, from the repeat unit of a set of dragline silk protein strands in atomistic resolution [9,127,143,144] to mesoscale models of fibrils [49,145] to the whole spider web [146] (Table 4). These studies address the relationship between structure and properties and specifically how material phenomena at the molecular scale affect material properties and ultimately the function of the spider web. We refer the reader to a recent article[147] that provides an in-depth review of these studies. Some influential design-variables in the sequence of silk have been identified and explored: the size of the β-sheet nanocrystals is controlled by the length of a repeating poly-alanine region in the peptide [148] (together with a processing design-variable, the reeling speed [149], see Table 4). Further, the experimentally observed [149] finding that geometric confinement of β-sheet nanocrystals to a few nanometers enhances toughness, stiffness and strength of spider dragline silk, could be explained by molecular mechanics studies [143].
Early studies have used a network model with an assumption of rubber-like amorphous domain to examine the influence of water content and crystal size on the stress–strain curve [145]. Group interaction modeling, which builds on mean field theory, predicted different stress–strain profiles by changing the fraction of order in the silk fiber morphology. An additional confinement effect appears at a higher level of hierarchy. Silk fibers are composed of fibril bundles, where fibrils exhibit diameters in the range of 20–150 nm. A computational mesoscale model demonstrated that a confinement to diameters of 50 ± 30 nm yields in a homogeneous deformation state in the fibril so that the nanoscale properties of a repeat unit can be scaled up to the silk fiber, despite the existence of large flaws [49].
We note from Table 5 that processing conditions have not yet been taken into account for silk by modeling, despite the fact that experimental investigations show a significant dependence on parameters such as pH, ion concentration and shear forces [34,150]. For triblock copolymers with a silk-like middle block MD simulations and replica exchange MD simulations showed a solvent depending structure change of the middle part from a β-roll in water to antiparallel β-sheets in methanol [151].
Table 5.
A categorization of studies on silk, collagen, elastin and silk-elastin, considering the approach taken and the field of biopolymer design they address. The chosen studies deal with the interaction and influence of design-variables on structure and properties (outcome-parameter). Studies solely focused on structure analysis are omitted. For a full description of the biopolymer design all fields should be incorporated (four checkmarks). Table 4 shows the investigated design-variables and outcome-parameters of the here listed references.
| Sequence | Processing | Structure | Property | Ref. |
|---|---|---|---|---|
| Elastin – experiment | ||||
|
|
|
[20,68] | |
|
|
[19,21] | ||
|
|
|
[76,87] | |
|
|
[88] | ||
|
|
[187,188] | ||
|
|
|
[18,189] | |
|
|
[63] | ||
| Elastin – computation | ||||
|
|
[85,86] | ||
|
|
[186] | ||
|
|
|
[185] | |
| Elastin – experiment and computation | ||||
|
|
[152,153](combined) | ||
| Silk-elastin – experiment | ||||
|
|
|
[72,191–193] | |
|
|
|
|
[194] |
|
|
|
[195] | |
|
|
|
[23,24] | |
| Elastin-cartilage – experiment | ||||
|
|
|
[190] | |
| Collagen – experiment | ||||
|
|
|
[91] | |
|
|
[96] | ||
|
|
|
[95] | |
|
|
[27] | ||
|
|
[28] | ||
|
|
[100] | ||
|
|
[98] | ||
|
|
[35] | ||
| Collagen – computation | ||||
|
|
|
[102] | |
|
|
|
[103] | |
|
|
[161] | ||
|
|
[101] | ||
| Collagen – experiment and computation | ||||
|
|
|
[33] | |
| Silk – computation | ||||
|
|
[148] | ||
|
|
[49,143,146,168] | ||
| Silk- experiment | ||||
|
|
[197–200] | ||
|
|
|
[149,150,201] |
4. From natural biopolymers to tailored materials using experiment and computational modeling
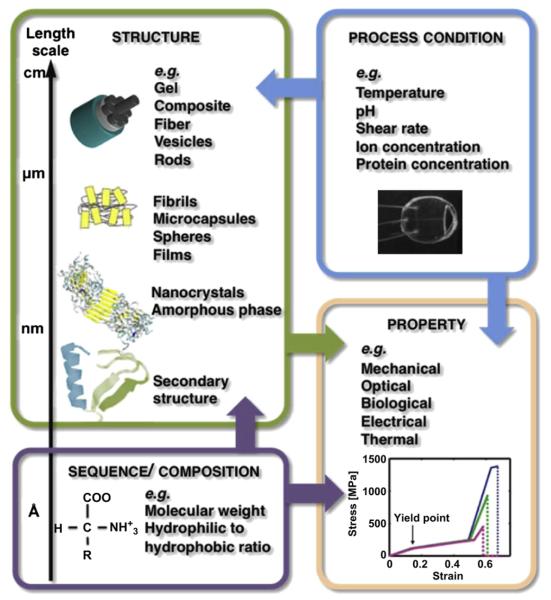
In the preceding sections we discussed methods that allow the investigation of the structure–process–property relationships at multiple scales of structure and organization, while considering a set of critical input and output variables. In biomaterials these relationships are complex as there are parameters in sequence and processing that address several hierarchical levels. This complexity challenge has mainly been addressed through a serial design process and the input parameters often rely on scientific intuition or trial and error experimentation [5]. To explore more configurations of the variety of chemistry compositions in a systematic way, the integration of polymer synthesis and processing with computational modeling may accelerate the design process to specific functional materials. For the overlap of experimental and computational works, four basic concepts can be distinguished.
4.1. Identification of mechanisms
Combining experimental and computational data achieved with similar boundary conditions results in a more complete description of material behavior. As shown in Fig. 2, the methods for experimental analysis cannot provide data for all length scales and the data is usually only available at spatially and temporally limited discrete points. In computer simulations data can be obtained throughout the system and is limited in resolution only by the model approximation and time step for MD and coarse-grained models. Furthermore, simulations can provide trajectories, which help in identifying the mechanisms of phenomena rather than empirical correlation. This leads to a basic understanding of phenomena seen in experiments. For a minimalistic elastin-like model a joint experimental and computational study (using MD) has been carried out [152,153]. Conclusions drawn out of structural and thermodynamic experimental measurements could be confirmed by MD simulations, which provide insight in hydrogen bond dynamics and the free energy profile for peptide structural transitions upon temperature changes.
4.2. Exchange of information
Computational simulations may support the analysis of experimental work as seen in the following example. In a mostly experimental study on collagen spinning in microchannels [154] a finite element method is used to describe diffusive phenomena, flow profile and pH gradient in a microchannel, which was a challenge to measure experimentally. Similarly, experimental data can provide appropriate starting conditions to massively shorten simulation runtime. The structure prediction for the atomistic model of spider dragline silk, for instance, was started using an approximated elongated configuration deduced from insight of experimental work [9].
4.3. Design of experiments and prediction of phenomena
Complex models can be used to determine the feasibility of an experiment to avoid losing time and valuable resources. A model that has been validated for one set of parameters can be adjusted or extended to facilitate educated decisions in experimental design. For example, mesoscale models predict tendencies, which are then checked more thoroughly by empirical investigations. Such models, if appropriately validated, may become more and more helpful in materials design.
4.4. Optimization by computational modeling
Applying experimentally obtained boundary conditions in the computational work allows the comparison to experimental data to validate the computer models. Once a model is validated, a set of optimized parameters can be found by simulations and finally the determined parameters are returned into a feed-back loop to the experimentalist.
5. Conclusion and outlook
We discussed various methods for the synthesis and processing of biopolymers. Synthetic polymers do not exhibit hierarchical structural organization as permeated throughout biopolymers (synthetic polymers form spheres, cylinders, etc., in contrast to the α-helices, β-sheets for proteins). Thus, it is essential to design and bridge the gap in structure–architecture–function relationships in protein-based block copolymers. A further step in the development of functional materials is the implementation and controlling of mutability, that means to change specific properties in the place and situation of interest through external triggers such as pH, temperature or electric fields [116]. Leisk et al., for example, showed an electrically mediated, reversible gel forming process of silk with potential application as a temporary glue [155]. While most synthetic or biopolymer materials allow some degree of control over composition and structure, the full potential to maximize their properties is often not exploited [8]. Silk is an example of a biopolymer where the three tiers of control (sequence, structure, process) have been well studied, but a full combination of approaches has not been realized yet. Successive studies with the integration of computational and experimental work, combined with feedback loops, may trigger an optimized material design.
For a system’s structure–function description, mathematical methods such as category theoretic data analysis may serve as a tool to store, share and relate the data and insights gained during research [156]. Recently, first analyses have described and related the structural hierarchical buildup of protein materials, social networks, and music [157,158]. Thereby, the elucidation of natural materials design principles can shape the synthesis of engineered materials. This is of interest because most biological materials are produced in moderate conditions, i.e. in aqueous solution at ambient temperature, without high energy input or extreme pH. Additionally, the starting material is often abundantly available and the product mostly degradable. Therefore, a translation of natural processes to materials engineering would yield highly sustainable and functional materials. An improved understanding of how biomaterials are structured and how they assemble with respect to environmental conditions might help in developing mechanistic models of, and eventual treatments for diseases such as Alzheimer’s [159], rheumatoid arthritis, brittle bone disease, or Ehlers-Danlos syndrome [91]. These diseases include material factors as a critical element in disease etiology and treatment, where defects such as mutations, or a change in processing conditions, lead to a loss of critical functional properties.
While polymer chemistry is considered a mature disciplinary field, there remains a gap in mimicking and extending the material performance of natural systems, with respect to structure–process–property relationships. Some examples of good integration of modeling, synthesis, and processing for biopolymers are available. In other fields, such as aerospace engineering, car design, or drug discovery, the combination of computational and experimental work is well established. We believe that this integrated approach may also boost advancements in the design of biopolymers and in polymers and composites in general.
Fig. 3.
The four main fields of study of biopolymer design: sequence, structure, process condition and properties. The examples show silk as a versatile biopolymer. The arrows show the relationships between the fields of study. Examples for studies on these relationships are shown in Table 4. Courtesy for images is as follows: Silk representation is adapted from Ref. [143]. Stress–strain plot is reprinted with permission from Ref. [168], copyright © 2011 American Chemical Society. Microfluidic device is reprinted with permission from Ref. [32], copyright © 2011 American Chemical Society.
Acknowledgments
We acknowledge support from NIH U01 EB014976 (M.J.B., D.L.K., J.Y.W.). Additional support from NSF (M.J.B., D.L.K.), DOD (AFOSR AND MURI) (M.J.B., D.L.K.), NIH R01HL72900 (J.Y.W.) and P41 EB002520 (D.L.K.), the NSF Graduate Research Fellowship Program (M.E.K.), the German National Academic Foundation (Studienstiftung des deutschen Volkes) (G.G.) and the Dr. Jürgen-Ulderup-Foundation (G.G.) is acknowledged. The sponsors did not play any role in the study design, in the collection, analysis and interpretation of data and in the writing of the report or in the decision to submit the paper for publication.
References
- [1].Burg KJL, Porter S, Kellam JF. Biomaterial developments for bone tissue engineering. Biomaterials. 2000;21:2347–59. doi: 10.1016/s0142-9612(00)00102-2. [DOI] [PubMed] [Google Scholar]
- [2].Jeong B, Bae YH, Lee DS, Kim SW. Biodegradable block copolymers as injectable drug-delivery systems. Nature. 1997;388:860–2. doi: 10.1038/42218. [DOI] [PubMed] [Google Scholar]
- [3].Qiu Y, Park K. Environment-sensitive hydrogels for drug delivery. Adv Drug Deliv Rev. 2001;53:321–39. doi: 10.1016/s0169-409x(01)00203-4. [DOI] [PubMed] [Google Scholar]
- [4].Griffith LG. Emerging design principles in biomaterials and scaffolds for tissue engineering. Ann N Y Acad Sci. 2002;961:83–95. doi: 10.1111/j.1749-6632.2002.tb03056.x. [DOI] [PubMed] [Google Scholar]
- [5].U.S. National Science and Technology Council . Materials genome initiative for global competitiveness. Executive Office of the President; Washington: 2011. [Google Scholar]
- [6].Fratzl P, Weinkamer R. Nature’s hierarchical materials. Prog Mater Sci. 2007;52:1263–334. [Google Scholar]
- [7].Buehler MJ, Yung YC. Deformation and failure of protein materials in physiologically extreme conditions and disease. Nat Mater. 2009;8:175–88. doi: 10.1038/nmat2387. [DOI] [PubMed] [Google Scholar]
- [8].Cranford SW, Buehler MJ. Shaky foundations of hierarchical biological materials. Nano Today. 2011;6:332–8. [Google Scholar]
- [9].Keten S, Buehler MJ. Nanostructure and molecular mechanics of spider dragline silk protein assemblies. J R Soc Interf. 2010;7:1709–21. doi: 10.1098/rsif.2010.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Engler AC, Bonner DK, Buss HG, Cheung EY, Hammond PT. The synthetic tuning of clickable pH responsive cationic polypeptides and block copolypeptides. Soft Matter. 2011;7:5627–37. [Google Scholar]
- [11].Billot J-P, Douy A, Gallot B. Preparation, fractionation, and structure of block copolymers polystyrene–poly(carbobenzoxy-L-lysine) and polybutadiene–poly(carbobenzoxy-L-lysine) Makromol Chem. 1977;178:1641–50. [Google Scholar]
- [12].Smeenk JM, Schon P, Otten MBJ, Speller S, Stunnenberg HG, van Hest JCM. Fibril formation by triblock copolymers of silk-like beta-sheet polypeptides and poly(ethylene glycol) Macromolecules. 2006;39:2989–97. [Google Scholar]
- [13].Vandermeulen GWM, Tziatzios C, Klok H-A. Reversible self-organization of poly(ethylene glycol)-based hybrid block copolymers mediated by a de novo four-stranded alpha-helical coiled coil motif. Macromolecules. 2003;36:4107–14. [Google Scholar]
- [14].Koehl P, Delarue M. Polar and nonpolar atomic environments in the protein core: implications for folding and binding. Proteins. 1994;20:264–78. doi: 10.1002/prot.340200307. [DOI] [PubMed] [Google Scholar]
- [15].Rabotyagova OS, Cebe P, Kaplan DL. Self-assembly of genetically engineered spider silk block copolymers. Biomacromolecules. 2009;10:229–36. doi: 10.1021/bm800930x. [DOI] [PubMed] [Google Scholar]
- [16].Rabotyagova OS, Cebe P, Kaplan DL. Role of polyalanine domains in beta-sheet formation in spider silk block copolymers. Macromol Biosci. 2010;10:49–59. doi: 10.1002/mabi.200900203. [DOI] [PubMed] [Google Scholar]
- [17].Krishnaji ST, Huang WW, Rabotyagova O, Kharlampieva E, Choi I, Naik R, et al. Thin film assembly of spider silk-like block copolymers. Langmuir. 2011;27:1000–8. doi: 10.1021/la102638j. [DOI] [PubMed] [Google Scholar]
- [18].Lao UL, Sun M, Matsumoto M, Mulchandani A, Chen W. Genetic engineering of self-assembled protein hydrogel based on elastin-like sequences with metal binding functionality. Biomacromolecules. 2007;8:3736–9. doi: 10.1021/bm700662n. [DOI] [PubMed] [Google Scholar]
- [19].Wright ER, Conticello VP. Self-assembly of block copolymers derived from elastin-mimetic polypeptide sequences. Adv Drug Deliv Rev. 2002;54:1057–73. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- [20].Lee T, Cooper A, Apkarian RP, Conticello VP. Thermo-reversible self-assembly of nanoparticles derived from elastin-mimetic polypeptides. Adv Mater. 2000;12:1105–10. [Google Scholar]
- [21].Wright ER, McMillan RA, Cooper A, Apkarian RP, Conticello VP. Thermo-plastic elastomer hydrogels via self-assembly of an elastin-mimetic triblock polypeptide. Adv Funct Mater. 2002;12:149–54. [Google Scholar]
- [22].Xia XX, Qian Z, Ki CS, Park YH, Kaplan DL, Lee SY. Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber. Proc Natl Acad Sci U S A. 2010;107:14059–63. doi: 10.1073/pnas.1003366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Haider M, Leung V, Ferrari F, Crissman J, Powell J, Cappello J, et al. Molecular engineering of silk–elastinlike polymers for matrix-mediated gene delivery: biosynthesis and characterization. Mol Pharm. 2005;2:139–50. doi: 10.1021/mp049906s. [DOI] [PubMed] [Google Scholar]
- [24].Megeed Z, Cappello J, Ghandehari H. Genetically engineered silk- elastin-like protein polymers for controlled drug delivery. Adv Drug Deliv Rev. 2002;54:1075–91. doi: 10.1016/s0169-409x(02)00063-7. [DOI] [PubMed] [Google Scholar]
- [25].Dawson P, Muir T, Clark-Lewis I, Kent S. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–9. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- [26].Link AJ, Mock ML, Tirrell DA. Non-canonical amino acids in protein engineering. Curr Opin Biotechnol. 2003;14:603–9. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- [27].Boland ED, Matthews JA, Pawlowski KJ, Simpson DG, Wnek GE, Bowlin GL. Electrospinning collagen and elastin: preliminary vascular tissue engineering. Front Biosci. 2004;9:1422–32. doi: 10.2741/1313. [DOI] [PubMed] [Google Scholar]
- [28].Buttafoco L, Kolkman NG, Engbers-Buijtenhuijs P, Poot AA, Dijkstra PJ, Vermes I, et al. Electrospinning of collagen and elastin for tissue engineering applications. Biomaterials. 2006;27:724–34. doi: 10.1016/j.biomaterials.2005.06.024. [DOI] [PubMed] [Google Scholar]
- [29].Matthews JA, Wnek GE, Simpson DG, Bowlin GL. Electrospinning of collagen nanofibers. Biomacromolecules. 2002;3:232–8. doi: 10.1021/bm015533u. [DOI] [PubMed] [Google Scholar]
- [30].Hirano S, Zhang M, Nakagawa M, Miyata T. Wet spun chitosan–collagen fibers, their chemical N-modifications, and blood compatibility. Biomaterials. 2000;21:997–1003. doi: 10.1016/s0142-9612(99)00258-6. [DOI] [PubMed] [Google Scholar]
- [31].Matsumoto K, Uejima H, Iwasaki T, Sano Y, Sumino H. Studies on regenerated protein fibers. III. Production of regenerated silk fibroin fiber by the self-dialyzing wet spinning method. J Appl Polym Sci. 1996;60:503–11. [Google Scholar]
- [32].Kinahan ME, Filippidi E, Koster S, Hu X, Evans HM, Pfohl T, et al. Tunable silk: using microfluidics to fabricate silk fibers with controllable properties. Biomacromolecules. 2011;12:1504–11. doi: 10.1021/bm1014624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Köster S, Evans HM, Wong JY, Pfohl T. An in situ study of collagen self-assembly processes. Biomacromolecules. 2008;9:199–207. doi: 10.1021/bm700973t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rammensee S, Slotta U, Scheibel T, Bausch AR. Assembly mechanism of recombinant spider silk proteins. Proc Natl Acad Sci U S A. 2008;105:6590–5. doi: 10.1073/pnas.0709246105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lee P, Lin R, Moon J, Lee LP. Microfluidic alignment of collagen fibers for in vitro cell culture. Biomed Microdevices. 2006;8:35–41. doi: 10.1007/s10544-006-6380-z. [DOI] [PubMed] [Google Scholar]
- [36].Xu Q, Hashimoto M, Dang TT, Hoare T, Kohane DS, Whitesides GM, et al. Preparation of monodisperse biodegradable polymer microparticles using a microfluidic flow-focusing device for controlled drug delivery. Small. 2009;5:1575–81. doi: 10.1002/smll.200801855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zorlutuna P, Elsheikh A, Hasirci V. Nanopatterning of collagen scaffolds improve the mechanical properties of tissue engineered vascular grafts. Biomacromolecules. 2009;10:814–21. doi: 10.1021/bm801307y. [DOI] [PubMed] [Google Scholar]
- [38].Kopecek J. Smart and genetically engineered biomaterials and drug delivery systems. Eur J Pharm Sci. 2003;20:1–16. doi: 10.1016/s0928-0987(03)00164-7. [DOI] [PubMed] [Google Scholar]
- [39].Wichterle O, Lim D. Hydrophilic gels for biological use. Nature. 1960;185:117–8. [Google Scholar]
- [40].Hennink WE, van Nostrum CF. Novel crosslinking methods to design hydrogels. Adv Drug Deliv Rev. 2002;54:13–36. doi: 10.1016/s0169-409x(01)00240-x. [DOI] [PubMed] [Google Scholar]
- [41].Kisiday J, Jin M, Kurz B, Hung H, Semino C, Zhang S, et al. Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: implications for cartilage tissue repair. Proc Natl Acad Sci U S A. 2002;99:9996–10001. doi: 10.1073/pnas.142309999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kataoka K, Harada A, Nagasaki Y. Block copolymer micelles for drug delivery: design, characterization and biological significance. Adv Drug Deliv Rev. 2001;47:113–31. doi: 10.1016/s0169-409x(00)00124-1. [DOI] [PubMed] [Google Scholar]
- [43].Vinogradov SV, Bronich TK, Kabanov AV. Nanosized cationic hydrogels for drug delivery: preparation, properties and interactions with cells. Adv Drug Deliv Rev. 2002;54:135–47. doi: 10.1016/s0169-409x(01)00245-9. [DOI] [PubMed] [Google Scholar]
- [44].Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141–51. [Google Scholar]
- [45].Bussi G, Gervasio FL, Laio A, Parrinello M. Free-energy landscape for beta hairpin folding from combined parallel tempering and metadynamics. J Am Chem Soc. 2006;128:13435–41. doi: 10.1021/ja062463w. [DOI] [PubMed] [Google Scholar]
- [46].Chebaro Y, Dong X, Laghaei R, Derreumaux P, Mousseau N. Replica exchange molecular dynamics simulations of coarse-grained proteins in implicit solvent. J Phys Chem B. 2009;113:267–74. doi: 10.1021/jp805309e. [DOI] [PubMed] [Google Scholar]
- [47].Buehler MJ, Keten S, Ackbarow T. Theoretical and computational hierarchical nanomechanics of protein materials: deformation and fracture. Prog Mater Sci. 2008;53:1101–241. [Google Scholar]
- [48].Paparcone R, Cranford S, Buehler MJ. Compressive deformation of ultralong amyloid fibrils. Acta Mech Sin. 2010;26:977–86. [Google Scholar]
- [49].Giesa T, Arslan M, Pugno NM, Buehler MJ. Nanoconfinement of spider silk fibrils begets superior strength, extensibility, and toughness. Nano Lett. 2011;11:5038–46. doi: 10.1021/nl203108t. [DOI] [PubMed] [Google Scholar]
- [50].MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- [51].Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111:7812–24. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- [52].Derreumaux P, Mousseau N. Coarse-grained protein molecular dynamics simulations. J Chem Phys. 2007:126. doi: 10.1063/1.2408414. [DOI] [PubMed] [Google Scholar]
- [53].Honeycutt JD, Thirumalai D. The nature of folded states of globular proteins. Biopolymers. 1992;32:695–709. doi: 10.1002/bip.360320610. [DOI] [PubMed] [Google Scholar]
- [54].Brown S, Fawzi NJ, Head-Gordon T. Coarse-grained sequences for protein folding and design. Proc Natl Acad Sci U S A. 2003;100:10712–7. doi: 10.1073/pnas.1931882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dünweg B, Ladd AJC. Lattice Boltzmann simulations of soft matter systems. In: Holm C, Kremer K, editors. Advanced computer simulation approaches for soft matter sciences III. Springer; Berlin/Heidelberg: 2009. pp. 89–166. [Google Scholar]
- [56].Noguchi H, Kikuchi N, Gompper G. Particle-based mesoscale hydrodynamic techniques. Europhys Lett. 2007:78. [Google Scholar]
- [57].Kushner AM, Guan Z. Modular design in natural and biomimetic soft materials. Angew Chem Int Ed. 2011;50:9026–57. doi: 10.1002/anie.201006496. [DOI] [PubMed] [Google Scholar]
- [58].Ma PX. Biomimetic materials for tissue engineering. Adv Drug Deliv Rev. 2008;60:184–98. doi: 10.1016/j.addr.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Vollrath F, Porter D, Holland C. There are many more lessons still to be learned from spider silks. Soft Matter. 2011;7:9595–600. [Google Scholar]
- [60].Kaplan DL. Silk polymers: materials science and biotechnology. American Chemical Society; Washington, D.C.: 1993. [Google Scholar]
- [61].Gosline JM, Guerette PA, Ortlepp CS, Savage KN. The mechanical design of spider silks: from fibroin sequence to mechanical function. J Exp Biol. 1999;202:3295–303. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- [62].Li B, Daggett V. Molecular basis for the extensibility of elastin. J Muscle Res Cell Motil. 2002;23:561–73. doi: 10.1023/a:1023474909980. [DOI] [PubMed] [Google Scholar]
- [63].Urry DW, Hugel T, Seitz M, Gaub HE, Sheiba L, Dea J, et al. Elastin: a representative ideal protein elastomer. Philos Trans R Soc Lond B Biol Sci. 2002;357:169–84. doi: 10.1098/rstb.2001.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gosline J, Lillie M, Carrington E, Guerette P, Ortlepp C, Savage K. Elastic proteins: biological roles and mechanical properties. Philos Trans R Soc Lond B Biol Sci. 2002;357:121–32. doi: 10.1098/rstb.2001.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Urry DW. Entropic elastic processes in protein mechanisms. I. Elastic structure due to an inverse temperature transition and elasticity due to internal chain dynamics. J Protein Chem. 1988;7:1–34. doi: 10.1007/BF01025411. [DOI] [PubMed] [Google Scholar]
- [66].Urry DW. Entropic elastic processes in protein mechanisms. II. Simple (passive) and coupled (active) development of elastic forces. J Protein Chem. 1988;7:81–114. doi: 10.1007/BF01025240. [DOI] [PubMed] [Google Scholar]
- [67].Urry DW. Physical chemistry of biological free energy transduction as demonstrated by elastic protein-based polymers. J Phys Chem B. 1997;101:11007–28. [Google Scholar]
- [68].Meyer DE, Chilkoti A. Quantification of the effects of chain length and concentration on the thermal behavior of elastin-like polypeptides. Biomacromolecules. 2004;5:846–51. doi: 10.1021/bm034215n. [DOI] [PubMed] [Google Scholar]
- [69].Yamaoka T, Tamura T, Seto Y, Tada T, Kunugi S, Tirrell DA. Mechanism for the phase transition of a genetically engineered elastin model peptide (VPGIG)40 in aqueous solution. Biomacromolecules. 2003;4:1680–5. doi: 10.1021/bm034120l. [DOI] [PubMed] [Google Scholar]
- [70].Daamen WF, Veerkamp JH, van Hest JCM, van Kuppevelt TH. Elastin as a biomaterial for tissue engineering. Biomaterials. 2007;28:4378–98. doi: 10.1016/j.biomaterials.2007.06.025. [DOI] [PubMed] [Google Scholar]
- [71].Chow D, Nunalee ML, Lim DW, Simnick AJ, Chilkoti A. Peptide-based biopolymers in biomedicine and biotechnology. Mater Sci Eng. 2008;62:125–55. doi: 10.1016/j.mser.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Dandu R, Ghandehari H. Delivery of bioactive agents from recombinant polymers. Prog Polym Sci. 2007;32:1008–30. [Google Scholar]
- [73].Wu Y, MacKay JA, McDaniel JR, Chilkoti A, Clark RL. Fabrication of elastin-like polypeptide nanoparticles for drug delivery by electrospraying. Biomacromolecules. 2009;10:19–24. doi: 10.1021/bm801033f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Martin L, Alonso M, Möller M, Rodriguez-Cabello JC, Mela P. 3D microstructuring of smart bioactive hydrogels based on recombinant elastin-like polymers. Soft Matter. 2009;5:1591–3. [Google Scholar]
- [75].Grieshaber SE, Farran AJE, Lin-Gibson S, Kiick KL, Jia X. Synthesis and characterization of elastin-mimetic hybrid polymers with multiblock, alternating molecular architecture and elastomeric properties. Macromolecules. 2009;42:2532–41. doi: 10.1021/ma802791z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Lim DW, Nettles DL, Setton LA, Chilkoti A. In situ cross-linking of elastin-like polypeptide block copolymers for tissue repair. Biomacromolecules. 2008;9 doi: 10.1021/bm7007982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Martino M, Tamburro AM. Chemical synthesis of cross-linked poly(KGGVG), an elastin-like biopolymer. Biopolymers. 2001;59:29–37. doi: 10.1002/1097-0282(200107)59:1<29::AID-BIP1003>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- [78].Cappello J, Crissman J, Dorman M, Mikolajczak M, Textor G, Marquet M, et al. Genetic engineering of structural protein polymers. Biotechnol Prog. 1990;6:198–202. doi: 10.1021/bp00003a006. [DOI] [PubMed] [Google Scholar]
- [79].Gray WR, Sanberg LB, Foster JA. Molecular model for elastin structure and function. Nature. 1973;246:461–6. doi: 10.1038/246461a0. [DOI] [PubMed] [Google Scholar]
- [80].Urry DW, Luan CH, Parker TM, Gowda DC, Prasad KU, Reid MC, et al. Temperature of polypeptide inverse temperature transition depends on mean residue hydrophobicity. J Am Chem Soc. 1991;113:4346–8. [Google Scholar]
- [81].Morelli AC, Scopa A, Tamburro AM. Spectroscopic studies on elastin-like synthetic polypeptides. Int J Biol Macromol. 1990;12:363–8. doi: 10.1016/0141-8130(90)90044-b. [DOI] [PubMed] [Google Scholar]
- [82].Martino M, Coviello A, Tamburro AM. Synthesis and structural characterization of poly(LGGVG), an elastin-like polypeptide. Int J Biol Macromol. 2000;27:59–64. doi: 10.1016/s0141-8130(99)00118-x. [DOI] [PubMed] [Google Scholar]
- [83].Okamoto K, Urry DW. Synthesis of a cross-linked polypentapeptide of tropoelastin. Biopolymers. 1976;15:2337–51. doi: 10.1002/bip.1976.360151203. [DOI] [PubMed] [Google Scholar]
- [84].Urry DW, Cunningham WD, Ohnishi T. Studies on the conformation and interactions of elastin. Proton magnetic resonance of the repeating pentapeptide. Biochemistry. 1974;13:609–16. doi: 10.1021/bi00700a032. [DOI] [PubMed] [Google Scholar]
- [85].Li B, Alonso DOV, Daggett V. The molecular basis for the inverse temperature transition of elastin. J Mol Biol. 2001;305:581–92. doi: 10.1006/jmbi.2000.4306. [DOI] [PubMed] [Google Scholar]
- [86].Li B, Alonso DOV, Daggett V. Hydrophobic hydration is an important source of elasticity in elastin-based biopolymers. J Am Chem Soc. 2001;123:11991–8. doi: 10.1021/ja010363e. [DOI] [PubMed] [Google Scholar]
- [87].Sallach RE, Cui W, Wen J, Martinez A, Conticello VP, Chaikof EL. Elastin-mimetic protein polymers capable of physical and chemical crosslinking. Biomaterials. 2009;30:409–22. doi: 10.1016/j.biomaterials.2008.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Jordan SW, Haller CA, Sallach RE, Apkarian RP, Hanson SR, Chaikof EL. The effect of a recombinant elastin-mimetic coating of an ePTFE prosthesis on acute thrombogenicity in a baboon arteriovenous shunt. Biomaterials. 2007;28:1191–7. doi: 10.1016/j.biomaterials.2006.09.048. [DOI] [PubMed] [Google Scholar]
- [89].Kadler KE, Holmes DF, Trotter JA, Chapman JA. Collagen fibril formation. Biochem J. 1996;316:1–11. doi: 10.1042/bj3160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Brodsky B, Ramshaw J. The collagen triple-helix structure. Matrix Biol. 1997;15:545–54. doi: 10.1016/s0945-053x(97)90030-5. [DOI] [PubMed] [Google Scholar]
- [91].Chan SW, Hung SP, Raman SK, Hatfield GW, Lathrop RH, Da Silva NA, et al. Recombinant human collagen and biomimetic variants using a de novo gene optimized for modular assembly. Biomacromolecules. 2010;11:1460–9. doi: 10.1021/bm100052y. [DOI] [PubMed] [Google Scholar]
- [92].Yamasakia C, Kadoyab Y, Hozumic K, Okano-Kosugid H, Asadad S, Kitagawad K, et al. A collagen-mimetic triple helical supramolecule that evokes integrin-dependent cell responses. Biomaterials. 2010;31:1925–34. doi: 10.1016/j.biomaterials.2009.10.014. [DOI] [PubMed] [Google Scholar]
- [93].Goldberg I, Salerno AJ, Patterson T, Williams JI. Cloning and expression of a collagen-analog-encoding synthetic gene in Escherichia coli. Gene. 1989;1980:305–14. doi: 10.1016/0378-1119(89)90294-1. [DOI] [PubMed] [Google Scholar]
- [94].Majsterek I, McAdams E, Adachi E, Dhume ST, Fertala A. Prospects and limitations of the rational engineering of fibrillar collagens. Protein Sci. 2003;12:2063–72. doi: 10.1110/ps.0385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Bracalello A, Santopietro V, Vassalli M, Marletta G, Del Gaudio R, Bochicchio B, et al. Design and production of a chimeric resilin-, elastin-, and collagen-like engineered polypeptide. Biomacromolecules. 2011;12:2957–65. doi: 10.1021/bm2005388. [DOI] [PubMed] [Google Scholar]
- [96].Yao J, Yanagisawa S, Asakura T. Design, expression and characterization of collagen-like proteins based on the cell adhesive and crosslinking sequences derived from native collagens. J Biochem. 2004;136:643–9. doi: 10.1093/jb/mvh172. [DOI] [PubMed] [Google Scholar]
- [97].Yeo I-S, Oh J-E, Jeong L, Lee TS, Lee SJ, Park WH, et al. Collagen-based biomimetic nanofibrous scaffolds: preparation and characterization of collagen/silk fibroin bicomponent nanofibrous structures. Biomacromolecules. 2008;9:1106–16. doi: 10.1021/bm700875a. [DOI] [PubMed] [Google Scholar]
- [98].Lanfer B, Freudenberg U, Zimmermann R, Stamov D, Korber V, Werner C. Aligned fibrillar collagen matrices obtained by shear flow deposition. Biomaterials. 2008;29:3888–95. doi: 10.1016/j.biomaterials.2008.06.016. [DOI] [PubMed] [Google Scholar]
- [99].Goffin AJ, Rajadas J, Fuller GG. Interfacial flow processing of collagen. Langmuir. 2009;26:3514–21. doi: 10.1021/la9031317. [DOI] [PubMed] [Google Scholar]
- [100].Yunoki S, Matsuda T. Simultaneous processing of fibril formation and cross-linking improves mechanical properties of collagen. Biomacromolecules. 2008;9:879–85. doi: 10.1021/bm7012058. [DOI] [PubMed] [Google Scholar]
- [101].Buehler MJ. Nature designs tough collagen: explaining the nanostructure of collagen fibrils. Proc Natl Acad Sci U S A. 2006;103:12285–90. doi: 10.1073/pnas.0603216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Gautieri A, Russo A, Vesentini S, Redaelli A, Buehler MJ. Coarse-grained model of collagen molecules using an extended MARTINI force field. J Chem Theory Comput. 2010;6:1210–8. [Google Scholar]
- [103].Srinivasan M, Uzel SG, Gautieri A, Keten S, Buehler MJ. Alport syndrome mutations in type IV tropocollagen alter molecular structure and nano-mechanical properties. J Struct Biol. 2009;168:503–10. doi: 10.1016/j.jsb.2009.08.015. [DOI] [PubMed] [Google Scholar]
- [104].Tang Y, Ballarini R, Buehler MJ, Eppell SJ. Deformation micromechanisms of collagen fibrils under uniaxial tension. J R Soc Interf. 2009;7:839–50. doi: 10.1098/rsif.2009.0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Rousseau ME, Lefevre T, Beaulieu L, Asakura T, Pezolet M. Study of protein conformation and orientation in silkworm and spider silk fibers using Raman microspectroscopy. Biomacromolecules. 2004;5:2247–57. doi: 10.1021/bm049717v. [DOI] [PubMed] [Google Scholar]
- [106].Lefevre T, Rousseau ME, Pezolet M. Protein secondary structure and orientation in silk as revealed by Raman spectromicroscopy. Biophys J. 2007;92:2885–95. doi: 10.1529/biophysj.106.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Becker N, Oroudjev E, Mutz S, Cleveland JP, Hansma PK, Hayashi CY, et al. Molecular nanosprings in spider capture-silk threads. Nat Mater. 2003;2:278–83. doi: 10.1038/nmat858. [DOI] [PubMed] [Google Scholar]
- [108].Shao ZZ, Vollrath F. Surprising strength of silkworm silk. Nature. 2002;418:741. doi: 10.1038/418741a. [DOI] [PubMed] [Google Scholar]
- [109].Vollrath F, Knight DP. Liquid crystalline spinning of spider silk. Nature. 2001;410:541–8. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- [110].Agnarsson I, Kuntner M, Blackledge TA. Bioprospecting finds the toughest biological material: extraordinary silk from a giant riverine orb spider. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0011234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Omenetto FG, Kaplan DL. New opportunities for an ancient material. Science. 2010;329:528–31. doi: 10.1126/science.1188936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Vepari C, Kaplan DL. Silk as a biomaterial. Prog Polym Sci. 2007;32:991–1007. doi: 10.1016/j.progpolymsci.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen JS, et al. Silk-based biomaterials. Biomaterials. 2003;24:401–16. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- [114].MacIntosh AC, Kearns VR, Crawford A, Hatton PV. Skeletal tissue engineering using silk biomaterials. J Tissue Eng Regen Med. 2008;2:71–80. doi: 10.1002/term.68. [DOI] [PubMed] [Google Scholar]
- [115].Lammel A, Schwab M, Hofer M, Winter G, Scheibel T. Recombinant spider silk particles as drug delivery vehicles. Biomaterials. 2011;32:2233–40. doi: 10.1016/j.biomaterials.2010.11.060. [DOI] [PubMed] [Google Scholar]
- [116].Buehler MJ. Strength in numbers. Nat Nanotechnol. 2010;5:172–4. doi: 10.1038/nnano.2010.28. [DOI] [PubMed] [Google Scholar]
- [117].Omenetto FG, Kaplan DL. A new route for silk. Nat Photonics. 2008;2:641–3. [Google Scholar]
- [118].Rockwood DN, Preda RC, Yucel T, Wang XQ, Lovett ML, Kaplan DL. Materials fabrication from Bombyx mori silk fibroin. Nat Protoc. 2011;6:1612–31. doi: 10.1038/nprot.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Eisoldt L, Smith A, Scheibel T. Decoding the secrets of spider silk. Mater Today. 2011;14:80–6. [Google Scholar]
- [120].Scheller J, Guhrs KH, Grosse F, Conrad U. Production of spider silk proteins in tobacco and potato. Nat Biotechnol. 2001;19:573–7. doi: 10.1038/89335. [DOI] [PubMed] [Google Scholar]
- [121].Lazaris A, Arcidiacono S, Huang Y, Zhou JF, Duguay F, Chretien N, et al. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science. 2002;295:472–6. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- [122].Huemmerich D, Helsen CW, Quedzuweit S, Oschmann J, Rudolph R, Scheibel T. Primary structure elements of spider dragline silks and their contribution to protein solubility. Biochemistry. 2004;43:13604–12. doi: 10.1021/bi048983q. [DOI] [PubMed] [Google Scholar]
- [123].Teule F, Miao YG, Sohn BH, Kim YS, Hull JJ, Fraser MJ, et al. Silkworms transformed with chimeric silkworm/spider silk genes spin composite silk fibers with improved mechanical properties. Proc Natl Acad Sci U S A. 2012;109:923–8. doi: 10.1073/pnas.1109420109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].van Beek JD, Hess S, Vollrath F, Meier BH. The molecular structure of spider dragline silk: folding and orientation of the protein backbone. Proc Natl Acad Sci U S A. 2002;99:10266–71. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Thiel BL, Guess KB, Viney C. Non-periodic lattice crystals in the hierarchical microstructure of spider (major ampullate) silk. Biopolymers. 1997;41:703–19. doi: 10.1002/(SICI)1097-0282(199706)41:7<703::AID-BIP1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- [126].Cunniff PM, Fossey SA, Auerbach MA, Song JW, Kaplan DL, Adams WW, et al. Mechanical and thermal properties of dragline silk from the spider Nephila clavipes. Polym Adv Technol. 1994;5:401–10. [Google Scholar]
- [127].Keten S, Buehler MJ. Atomistic model of the spider silk nanostructure. Appl Phys Lett. 2010;96 [Google Scholar]
- [128].Hayashi CY, Shipley NH, Lewis RV. Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins. Int J Biol Macromol. 1999;24:271–5. doi: 10.1016/s0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- [129].Rising A, Hjalm G, Engstrom W, Johansson J. N-terminal nonrepetitive domain common to dragline, flagelliform, and cylindriform spider silk proteins. Biomacromolecules. 2006;7:3120–4. doi: 10.1021/bm060693x. [DOI] [PubMed] [Google Scholar]
- [130].Motriuk-Smith D, Smith A, Hayashi CY, Lewis RV. Analysis of the conserved N-terminal domains in major ampullate spider silk proteins. Biomacromolecules. 2005;6:3152–9. doi: 10.1021/bm050472b. [DOI] [PubMed] [Google Scholar]
- [131].Sponner A, Vater W, Rommerskirch W, Vollrath F, Unger E, Grosse F, et al. The conserved C-termini contribute to the properties of spider silk fibroins. Biochem Biophys Res Commun. 2005;338:897–902. doi: 10.1016/j.bbrc.2005.10.048. [DOI] [PubMed] [Google Scholar]
- [132].Hagn F, Eisoldt L, Hardy JG, Vendrely C, Coles M, Scheibel T, et al. A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature. 2010;465:239–42. doi: 10.1038/nature08936. [DOI] [PubMed] [Google Scholar]
- [133].Askarieh G, Hedhammar M, Nordling K, Saenz A, Casals C, Rising A, et al. Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature. 2010;465:236–8. doi: 10.1038/nature08962. [DOI] [PubMed] [Google Scholar]
- [134].Colgin MA, Lewis RV. Spider minor ampullate silk proteins contain new repetitive sequences and highly conserved non-silk-like “spacer regions”. Protein Sci. 1998;7:667–72. doi: 10.1002/pro.5560070315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Eisoldt L, Hardy JG, Heim M, Scheibel TR. The role of salt and shear on the storage and assembly of spider silk proteins. J Struct Biol. 2010;170:413–9. doi: 10.1016/j.jsb.2009.12.027. [DOI] [PubMed] [Google Scholar]
- [136].Hijirida DH, Do KG, Michal C, Wong S, Zax D, Jelinski LW. C-13 NMR of Nephila clavipes major ampullate silk gland. Biophys J. 1996;71:3442–7. doi: 10.1016/S0006-3495(96)79539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Hagn F, Thamm C, Scheibel T, Kessler H. pH-Dependent dimerization and salt-dependent stabilization of the N-terminal domain of spider dragline silk – implications for fiber formation. Angew Chem Int Ed. 2011;50:310–3. doi: 10.1002/anie.201003795. [DOI] [PubMed] [Google Scholar]
- [138].Jin HJ, Fridrikh SV, Rutledge GC, Kaplan DL. Electrospinning Bombyx mori silk with poly(ethylene oxide) Biomacromolecules. 2002;3:1233–9. doi: 10.1021/bm025581u. [DOI] [PubMed] [Google Scholar]
- [139].Wang M, Jin HJ, Kaplan DL, Rutledge GC. Mechanical properties of electrospun silk fibers. Macromolecules. 2004;37:6856–64. [Google Scholar]
- [140].Phillips DM, Drummy LF, Naik RR, De Long HC, Fox DM, Trulove PC, et al. Regenerated silk fiber wet spinning from an ionic liquid solution. J Mater Chem. 2005;15:4206–8. [Google Scholar]
- [141].Liivak O, Blye A, Shah N, Jelinski LW. A microfabricated wet-spinning apparatus to spin fibers of silk proteins. Structure–property correlations. Macromolecules. 1998;31:2947–51. [Google Scholar]
- [142].Martel A, Burghammer M, Davies R, DiCola E, Panine P, Salmon JB, et al. A microfluidic cell for studying the formation of regenerated silk by synchrotron radiation small- and wide-angle X-ray scattering. Biomicrofluidics. 2008;2 doi: 10.1063/1.2943732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Keten S, Xu ZP, Ihle B, Buehler MJ. Nanoconfinement controls stiffness, strength and mechanical toughness of beta-sheet crystals in silk. Nat Mater. 2010;9:359–67. doi: 10.1038/nmat2704. [DOI] [PubMed] [Google Scholar]
- [144].Bratzel G, Buehler MJ. Molecular mechanics of silk nanostructures under varied mechanical loading. Biopolymers. 2011;97:408–17. doi: 10.1002/bip.21729. [DOI] [PubMed] [Google Scholar]
- [145].Termonia Y. Molecular modeling of spider silk elasticity. Macromolecules. 1994;27:7378–81. [Google Scholar]
- [146].Cranford SW, Tarakanova A, Pugno NM, Buehler MJ. Nonlinear material behaviour of spider silk yields robust webs. Nature. 2012;482:72–6. doi: 10.1038/nature10739. [DOI] [PubMed] [Google Scholar]
- [147].Tarakanova A, Buehler MJ. A materiomics approach to spider silk: protein molecules to webs. JOM. 2012;64:214–25. [Google Scholar]
- [148].Bratzel G, Buehler MJ. Sequence–structure correlations in silk: poly-Ala repeat of N. clavipes MaSp1 is naturally optimized at a critical length scale. J Mech Behav Biomed Mater. 2012;7:30–40. doi: 10.1016/j.jmbbm.2011.07.012. [DOI] [PubMed] [Google Scholar]
- [149].Du N, Liu XY, Narayanan J, Li LA, Lim MLM, Li DQ. Design of superior spider silk: from nanostructure to mechanical properties. Biophys J. 2006;91:4528–35. doi: 10.1529/biophysj.106.089144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Kim UJ, Park JY, Li CM, Jin HJ, Valluzzi R, Kaplan DL. Structure and properties of silk hydrogels. Biomacromolecules. 2004;5:786–92. doi: 10.1021/bm0345460. [DOI] [PubMed] [Google Scholar]
- [151].Schor M, Martens AA, Dewolf FA, Stuart MAC, Bolhuis PG. Prediction of solvent dependent beta-roll formation of a self-assembling silk-like protein domain. Soft Matter. 2009;5:2658–65. [Google Scholar]
- [152].Rousseau R, Schreiner E, Kohlmeyer A, Marx DD. Temperature-dependent conformational transitions and hydrogen-bond dynamics of the elastin-like octapeptide GVG(VPGVG): a molecular-dynamics study. Biophys J. 2004;86:1393–407. doi: 10.1016/S0006-3495(04)74210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Nicolini C, Ravindra R, Ludolph B, Winter R. Characterization of the temperature- and pressure-induced inverse and reentrant transition of the minimum elastin-like polypeptide GVG(VPGVG) by DSC, PPC, CD, and FT-IR spectroscopy. Biophys J. 2004;86:1385–92. doi: 10.1016/S0006-3495(04)74209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Köster S, Leach JB, Struth B, Pfohl T, Wong JY. Visualization of flow-aligned type I collagen self-assembly in tunable pH gradients. Langmuir. 2007;23:357–9. doi: 10.1021/la062473a. [DOI] [PubMed] [Google Scholar]
- [155].Leisk GG, Lo TJ, Yucel T, Lu Q, Kaplan DL. Electrogelation for protein adhesives. Adv Mater. 2010;22:711–5. doi: 10.1002/adma.200902643. [DOI] [PubMed] [Google Scholar]
- [156].Spivak DI, Kent RE. Ologs: a categorical framework for knowledge representation. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0024274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Spivak DI, Giesa T, Wood E, Buehler MJ. Category theoretic analysis of hierarchical protein materials and social networks. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0023911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Giesa T, Spivak DI, Buehler MJ. Reoccurring patterns in hierarchical protein materials and music: the power of analogies. Bionanoscience. 2011;1:153–61. [Google Scholar]
- [159].Paparcone R, Cranford SW, Buehler MJ. Self-folding and aggregation of amyloid nanofibrils. Nanoscale. 2011;3:1748–55. doi: 10.1039/c0nr00840k. [DOI] [PubMed] [Google Scholar]
- [160].Baldock C, Oberhauser AF, Ma LA, Lammie D, Siegler V, Mithieux SM, et al. Shape of tropoelastin, the highly extensible protein that controls human tissue elasticity. Proc Natl Acad Sci U S A. 2011;108:4322–7. doi: 10.1073/pnas.1014280108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Gautieri A, Uzel S, Vesentini S, Redaelli A, Buehler MJ. Molecular and mesoscale mechanisms of osteogenesis imperfecta disease in collagen fibrils. Biophys J. 2009;97:857–65. doi: 10.1016/j.bpj.2009.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Wang XQ, Wenk E, Matsumoto A, Meinel L, Li CM, Kaplan DL. Silk microspheres for encapsulation and controlled release. J Control Release. 2007;117:360–70. doi: 10.1016/j.jconrel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- [163].Krishnaji ST, Bratzel GH, Kinahan ME, Wong JY, Buehler MJ, Kaplan DL. Sequence–structure–property relationships with recombinant spider silk-like proteins. Adv Funct Mater. in press. [Google Scholar]
- [164].Lovett M, Cannizzaro C, Daheron L, Messmer B, Vunjak-Novakovic G, Kaplan DL. Silk fibroin microtubes for blood vessel engineering. Biomaterials. 2007;28:5271–9. doi: 10.1016/j.biomaterials.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Tsioris K, Raja WK, Pritchard EM, Panilaitis B, Kaplan DL, Omenetto FG. Fabrication of silk microneedles for controlled-release drug delivery. Adv Funct Mater. 2012;22:330–5. [Google Scholar]
- [166].Sachlos E, Reis N, Ainsley C, Derby B, Czernuszka JT. Novel collagen scaffolds with predefined internal morphology made by solid freeform fabrication. Biomaterials. 2003;24:1487–97. doi: 10.1016/s0142-9612(02)00528-8. [DOI] [PubMed] [Google Scholar]
- [167].Bettinger CJ, Cyr KM, Matsumoto A, Langer R, Borenstein JT, Kaplan DL. Silk fibroin microfluidic devices. Adv Mater. 2007;19:2847. doi: 10.1002/adma.200602487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Nova A, Keten S, Pugno NM, Redaelli A, Buehler MJ. Molecular and nano-structural mechanisms of deformation, strength and toughness of spider silk fibrils. Nano Lett. 2010;10:2626–34. doi: 10.1021/nl101341w. [DOI] [PubMed] [Google Scholar]
- [169].Ziegast G, Pfannemuller B. Phosphorolytic syntheses with di-, oligo- and multi-functional primers. Carbohydr Res. 1987;160:185–204. [Google Scholar]
- [170].Pfannemüller B. Living-polymerisation und enzymatische polysaccharidsynthese. Naturwissenschaften. 1975;62:231–3. doi: 10.1007/BF00603168. [DOI] [PubMed] [Google Scholar]
- [171].Liu DR, Magliery TJ, Pastrnak M, Schultz PG. Engineering a tRNA and aminoacyl-tRNA synthetase for the site-specific incorporation of unnatural amino acids into proteins in vivo. Proc Natl Acad Sci U S A. 1997;94:10092–7. doi: 10.1073/pnas.94.19.10092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].ACS J, Magliery TJ. Expanding the natural repertoire of protein structure and function. Curr Pharm Biotechnol. 2002;3:299–315. doi: 10.2174/1389201023378102. [DOI] [PubMed] [Google Scholar]
- [173].Hartmann L, Krause E, Antonietti M, Börner HG. Solid-phase supported polymer synthesis of sequence-defined, multifunctional poly(amidoamines) Biomacromolecules. 2006;7:1239–44. doi: 10.1021/bm050884k. [DOI] [PubMed] [Google Scholar]
- [174].Nielsen PE. Pseudo-peptides in drug discovery. Wiley-VCH; Weinheim: 2004. [Google Scholar]
- [175].Tam JP, Yu Q, Miao Z. Orthogonal ligation strategies for peptide and protein. Biopolymers. 1999;51:311–32. doi: 10.1002/(SICI)1097-0282(1999)51:5<311::AID-BIP2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- [176].Albericio F. Developments in peptide and amide synthesis. Curr Opin Biotechnol. 2004;8:211–21. doi: 10.1016/j.cbpa.2004.03.002. [DOI] [PubMed] [Google Scholar]
- [177].Li Z, Zhang Y, Fullhart P, Mirkin CA. Reversible and chemically programmable micelle assembly with DNA block-copolymer amphiphiles. Nano Lett. 2004;4:1055–8. [Google Scholar]
- [178].Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem Rev. 2005;105:1547–62. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- [179].Watson KJ, Park SJ, Im JH, Nguyen ST, Mirkin CA. DNA-block copolymer conjugates. J Am Chem Soc. 2001;123:5592–3. doi: 10.1021/ja0156845. [DOI] [PubMed] [Google Scholar]
- [180].Uhlmann E, Peyman A. Antisense oligonucleotides: a new therapeutic principle. Chem Rev. 1990;90:543–84. [Google Scholar]
- [181].Nielsen PE. Sequence-selective DNA recognition by synthetic ligands. Bioconjug Chem. 1991;2:1–12. doi: 10.1021/bc00007a001. [DOI] [PubMed] [Google Scholar]
- [182].Lawrence BD, Marchant JK, Pindrus MA, Omenetto FG, Kaplan DL. Silk film biomaterials for cornea tissue engineering. Biomaterials. 2009;30:1299–308. doi: 10.1016/j.biomaterials.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Hamley IW, Daniel C. From hard spheres to soft spheres: the effect of copolymer composition on the structure of micellar cubic phases formed by diblock copolymers in aqueous solution. Langmuir. 2000;16:2508–14. [Google Scholar]
- [184].Duncanson WJ, Figa MA, Hallock K, Zalipsky S, Hamilton JA, Wong JY. Targeted binding of PLA microparticles with lipid-PEG-tethered ligands. Biomaterials. 2007;28:4991–9. doi: 10.1016/j.biomaterials.2007.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Valiaev A, Dong WL, Schmidler S, Clark RL, Chilkoti A, Zauscher S. Hydration and conformational mechanics of single, end-tethered elastin-like polypeptides. J Am Chem Soc. 2008;130:10939–46. doi: 10.1021/ja800502h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Djajamuliadi J, Kagawa TF, Ohgo K, Kumashiro KK. Insights into a putative hinge region in elastin using molecular dynamics simulations. Matrix Biol. 2009;28:92–100. doi: 10.1016/j.matbio.2008.12.001. [DOI] [PubMed] [Google Scholar]
- [187].Bae Y, Buresh RA, Williamson TP, Chen T-HH, Furgeson DY. Intelligent biosynthetic nanobiomaterials for hyperthermic combination chemotherapy and thermal drug targeting of HSP90 inhibitor geldanamycin. J Control Release. 2007;122:16–23. doi: 10.1016/j.jconrel.2007.06.005. [DOI] [PubMed] [Google Scholar]
- [188].Chen T-H, Bae Y, Furgeson D. Intelligent biosynthetic nanobiomaterials (IBNs) for hyperthermic gene delivery. Pharm Res. 2008;25:683–91. doi: 10.1007/s11095-007-9382-5. [DOI] [PubMed] [Google Scholar]
- [189].Kostal J, Mulchandani A, Gropp KE, Chen W. A temperature responsive biopolymer for mercury remediation. Environ Sci Technol. 2003;37:4457–62. doi: 10.1021/es034210y. [DOI] [PubMed] [Google Scholar]
- [190].Haghpanah JS, Yuvienco C, Civay DE, Barra H, Baker PJ, Khapli S, et al. Arti-ficial protein block copolymers blocks comprising two distinct self-assembling domains. Chembiochem. 2009;10:2733–5. doi: 10.1002/cbic.200900539. [DOI] [PubMed] [Google Scholar]
- [191].Nagarsekar A, Crissman J, Crissman M, Ferrari F, Cappello J, Ghandehari H. Genetic synthesis and characterization of pH and temperature-sensitive silk–elastin-like protein block copolymers. J Biomed Mater Res. 2002;62:195–203. doi: 10.1002/jbm.10272. [DOI] [PubMed] [Google Scholar]
- [192].Hatefi A, Cappello J, Ghandehari H. Adenoviral gene delivery to solid tumors by recombinant silk–elastin-like protein polymers. Pharm Res. 2007;24:773–9. doi: 10.1007/s11095-006-9200-5. [DOI] [PubMed] [Google Scholar]
- [193].Nagarsekar A, Crissman J, Crissman M, Ferrari F, Cappello J, Ghandehari H. Genetic engineering of stimuli-sensitive silk–elastin-like protein block copolymers. Biomacromolecules. 2003;4:602–7. doi: 10.1021/bm0201082. [DOI] [PubMed] [Google Scholar]
- [194].Xia XX, Xu Q, Hu X, Qin G, Kaplan DL. Tunable self-assembly of genetically engineered silk–elastin-like protein polymers. Biomacromolecules. 2011;12:3844–50. doi: 10.1021/bm201165h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Teng W, Huang Y, Cappello J, Wu X. Optically transparent recombinant silk–elastinlike protein polymer films. J Phys Chem B. 2011;115:1608–15. doi: 10.1021/jp109764f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Qiu W, Teng W, Cappello J, Wu X. Wet-spinning of recombinant silk–elastin-like protein polymer fibers with high tensile strength and high deformability. Biomacromolecules. 2009;10:602–8. doi: 10.1021/bm801296r. [DOI] [PubMed] [Google Scholar]
- [197].Chen X, Knight DP, Shao ZZ, Vollrath F. Conformation transition in silk protein films monitored by time-resolved Fourier transform infrared spectroscopy: effect of potassium ions on Nephila spidroin films. Biochemistry. 2002;41:14944–50. doi: 10.1021/bi026550m. [DOI] [PubMed] [Google Scholar]
- [198].Peng XN, Shao ZZ, Chen X, Knight DP, Wu PY, Vollrath F. Further investigation on potassium-induced conformation transition of Nephila spidroin film with two-dimensional infrared correlation spectroscopy. Biomacromolecules. 2005;6:302–8. doi: 10.1021/bm049598j. [DOI] [PubMed] [Google Scholar]
- [199].Ruan QX, Zhou P, Hu BW, Ji D. An investigation into the effect of potassium ions on the folding of silk fibroin studied by generalized two-dimensional NMR–NMR correlation and Raman spectroscopy. FEBS J. 2008;275:219–32. doi: 10.1111/j.1742-4658.2007.06191.x. [DOI] [PubMed] [Google Scholar]
- [200].Li XG, Wu LY, Huang MR, Shao HL, Hu XC. Conformational transition and liquid crystalline state of regenerated silk fibroin in water. Biopolymers. 2008;89:497–505. doi: 10.1002/bip.20905. [DOI] [PubMed] [Google Scholar]
- [201].Schacht K, Scheibel T. Controlled hydrogel formation of a recombinant spider silk protein. Biomacromolecules. 2011;12:2488–95. doi: 10.1021/bm200154k. [DOI] [PubMed] [Google Scholar]