Abstract
Phytochemicals from diet and herbal medicines are under intensive investigation for their potential use as chemopreventive agents to block and suppress carcinogenesis. Chemical diversity of phytochemicals, together with complex metabolic interactions between phytochemicals and biological system, can overwhelm the capacity of traditional analytical platforms, and thus pose major challenges in studying chemopreventive phytochemicals. Recent progresses in metabolomics have transformed it to become a robust systems biology tool, suitable for examining both chemical and biochemical events that contribute to the cancer prevention activities of plant preparations or their bioactive components. This review aims to discuss the technical platform of metabolomics and its existing and potential applications in chemoprevention research, including identifying bioactive phytochemicals in plant extracts, monitoring phytochemical exposure in humans, elucidating biotransformation pathways of phytochemicals, and characterizing the effects of phytochemicals on endogenous metabolism and cancer metabolism.
KEY WORDS: chemoprevention, metabolism, metabolomics, phytochemical
INTRODUCTION
Cancer is a major cause of disease-related mortalities over the world. In the USA, nearly one in four deaths is due to cancer (1). Therefore, developing potent therapeutic and preventive agents with low toxicity has become a focus of cancer research in the past decades. Poor prognosis and high cost are commonly associated with cancer therapy targeting the late-stage carcinoma. In contrast, by intervening with the early stages of carcinogenesis, cancer prevention can be more effective in both prognosis and cost, especially for human subjects who are in high risk of cancer based on their genetic background, health status, and life style. Feasibility of cancer prevention is supported by epidemiological and experimental evidences showing that the risk of cancer could be reduced at the initiating and progressing stages (2). Among various approaches to abrogate and control the risk factors of carcinogenesis, chemoprevention is under intensive investigation due to its potential efficacy and feasibility. In definition, chemoprevention is a type of pharmacological intervention that uses synthetic or natural compounds to prevent, inhibit, or reverse the development of invasive malignancy caused by carcinogenesis (3,4). An ideal chemopreventive agent is expected to be nontoxic and effective at low doses, easily administered, and relatively inexpensive. Compared to synthetic compounds, phytochemicals are presumed to be a safer and more accessible source of chemopreventive agents owing to their presence in human diet, herbal medicines, and supplements (5). This perception is partially based on the inverse correlation between fruit and vegetable consumption and cancer incidence observed in numerous epidemiological and animal studies (6–9) as well as the widespread usage of herbal medicines in human history (10). It has been shown the diets high in fruits and vegetables (>400 g/day) reduce at least 20% of total cancer incidence and lead to even more reduction of gastrointestinal cancer (11). Protective effects of these botanical products against carcinogenesis have been largely attributed to their bioactive phytochemical components, such as indole-3-carbinol in cruciferous vegetables and polyphenols on green tea (12–14). Therefore, major scientific efforts in chemoprevention are devoted to identifying bioactive phytochemicals from fruits, vegetables, and herbs and to characterizing underlying mechanisms of their anti-carcinogenic activities.
Because of complex interactions between phytochemical and biological system, the systems biology tools, including genomics, transcriptomics, and proteomics, have been widely adopted to understand biological events contributing to the cancer prevention activities of plant extracts and their bioactive components (15). Omics-based studies have yielded novel insights on the impacts of phytochemical exposures at gene and protein levels, such as intracellular targets and signaling pathways of chemoprevention. However, gene and protein analyses are unable to define the chemical identities and properties of chemopreventive phytochemicals in plant extracts as well as the metabolic interactions between phytochemicals and biological system. Traditionally, chemical and metabolic events in chemoprevention are examined with a trial-and-error approach, which usually starts with a hypothesis and followed by tedious targeted chemical or metabolite analysis. In recent years, development of metabolomics, especially untargeted metabolomics, offers a new powerful tool for investigating chemical and metabolic events in chemoprevention.
Estimated number of metabolites in human metabolome ranges from thousands to tens of thousands (16). Since metabolites are end products of gene expression and enzyme activities of organisms, analyzing metabolites can reveal the consequences of altered gene expression, protein expression, and signaling pathways. Thus, metabolomics is a better platform for defining the metabolic status of a biological system than genomics, transcriptomics, and proteomics (17,18). Another advantage of metabolomics is that metabolic profiling can be achieved by analyzing biofluids samples instead of using tissue samples that are required for genomic, transcriptomic, and proteomic analyses. This property of metabolomics increases the possibility of carrying out large-scale research in a noninvasive manner (19). Starting with its initial applications in examining inborn metabolic errors, chemical-induced toxicity, and functional nutrigenomics (20–22), metabolomics has been widely adopted in many fields of biomedical research due to its capacity for comprehensive metabolite analysis. Dependent on instruments and experiment design, metabolomic profile can be generated through targeted analysis of metabolites associated with a particular metabolic pathway (23,24) or global analysis of all detectable metabolites (15).
Untargeted metabolomics has been widely used in identifying new natural product and dissecting natural product biosynthesis pathways in plants and microbiota, including anti-cancer phytochemicals (25). However, the application of metabolomics in discovering novel chemopreventive phytochemicals remains largely unexplored, even though the rationales of using metabolomics to study other bioactive natural products are also applicable for studying chemopreventive phytochemicals. Similarly, metabolomics has been widely adopted for investigating the interactions between xenobiotics and biological system, including xenobiotic metabolism and xenobiotic-induced metabolic changes in animals and humans (26,27), but not yet for studying the biotransformation and metabolic effects of chemopreventive phytochemicals. Because of the analytical capacity of metabolomics and the importance of chemical and metabolite analysis in studying chemopreventive phytochemicals in vitro and in vivo, this review aims to provide a brief introduction to the technical platform of metabolomics, and then discuss its potential applications in identifying bioactive phytochemicals in plant extracts, monitoring phytochemical exposure in humans, elucidating biotransformation pathways of phytochemicals, and characterizing the effects of phytochemicals on endogenous metabolism and cancer metabolism.
TECHNICAL PLATFORM OF METABOLOMICS
The capacity of metabolomics for identifying bioactive phytochemicals and revealing the interactions between phytochemicals and biological system originates from its sophisticated technical platform. As nuclear magnetic resonance (NMR) and mass spectrometry (MS) are two most widely used detection methods in metabolomic analysis, sample acquisition and preparation methods adopted in metabolomics-based studies are intended to facilitate data acquisition by NMR and MS instruments, while the approaches used in data analysis aim to maximize the values of metabolomic data from NMR and MS analysis (Fig. 1).
Fig. 1.
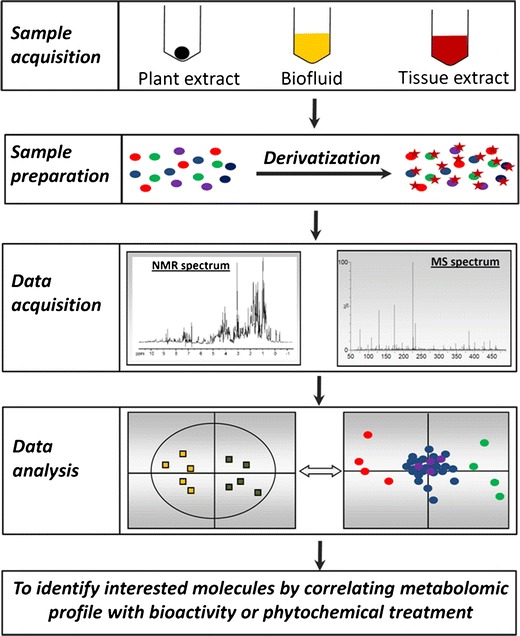
The work flow of untargeted metabolomics. Samples from diverse sources need to be processed appropriately to make them compatible with MS- and NMR-based metabolomic analysis. Chemical derivatization can be performed to facilitate the chromatographic separation of metabolites and increase the sensitivity of metabolite detection in LC-MS and GC-MS systems. Spectral data acquired by MS and NMR need to be deconvoluted to a data matrix compatible to multivariate data analysis. Subsequently, a multivariate model can be established in which the scores plot illustrates the principal or latent components of the model as well as sample classification while the loadings plot presents the contribution of each ion to sample classification in the model
Sample Acquisition and Preparation
Depending on the aims of metabolomic investigations, samples from plants, animals and humans, including plant extracts, biofluids (serum, cerebrospinal fluid), waste (urine and feces), tissue and cell extracts, can be chosen for identifying bioactive phytochemicals in vitro or examining the exposure, metabolic fate, and metabolic impact of chemopreventive phytochemicals in vivo (Fig. 1). Whether it contains interested phytochemicals or metabolites is the most important consideration in sample acquisition, while confounding factors, such as age, gender, chronobiological effects, animal species and environmental factors should also be considered in experimental design. In general, a power analysis should be conducted to ensure that a sufficient number of samples is included and the data can be statistically validated. Furthermore, to maintain the integrity of chemical composition in acquired samples, experimental techniques and storage conditions, including snap freezing in liquid nitrogen, freeze clamping, or quenching in preservation solution, should be optimized to avoid or minimize the formation of new chemical species or degradation of existing metabolites during and after sample collection (28).
Making samples compatible with analytical platform with minimal loss in sample preparation is essential for the success of metabolomic analysis, especially for MS-based metabolomics. For metabolomic analysis of phytochemical-mediated chemoprevention, ideal sample preparation process should be able to efficiently extract small-molecule phytochemicals and metabolites from plant, animal, and human materials, and also remove incompatible matrices (macromolecules and salts) simultaneously. Widely used extraction and preparation techniques include liquid–liquid extraction, solid-phase extraction, supercritical fluid extraction, microwave-assisted extraction, protein precipitation, and dialysis. Their applications in sample preparation are largely determined by chemical and physical properties of samples (29). In addition, extraction efficiency can be monitored by adding internal standards, such as stable isotope-labeled metabolite, prior to extraction.
In MS-based analysis, the barriers for detecting phytochemicals or metabolites in the MS systems are not just their concentrations in samples, but also their non-optimal performance in chromatography and MS systems, such as poor retention in liquid chromatography (LC) column and insufficient ionization in MS (30). To enhance the chromatographic and spectroscopic performance of these metabolites, one effective approach is to conduct chemical derivatization (Fig. 1). Chemical derivatization has been widely used in gas chromatography (GC)-MS analysis to improve separation, detectability and sensitivity of metabolite detection. The application of chemical derivatization in LC-MS analysis has greatly expanded in the recent years (31). In general, derivatization reactions are designed based on the functional groups, such as amino, carboxyl, carbonyl, and hydroxyl moieties in the metabolites. Increased hydrophobicity and chargeability are two desired effects of chemical derivatization (32). For example, amino acids are commonly derivatized by dansyl chloride (33). The detection of organic acids is enhanced through esterification of carboxyl group with amines, hydrazines, or alcohol, while detection of aldehydes and ketones is assisted by the formation of Schiff bases after derivatization reactions (34,35).
Analytical Platform of Metabolomics
A variety of detection methods have been adopted for metabolite analysis in metabolomics, including electrochemical array (36), infrared spectroscopy (37), NMR, and MS. Among these platforms, MS and NMR (mainly 1H-NMR) are the most widely used ones. Under electromagnetic field, NMR measures the resonant frequency of nuclei, while MS determines the mass-to-charge ratio (m/z) of ions. Compared to MS, NMR is non-destructive in nature and capable of providing more structural information, but less sensitive for detecting low-abundance metabolites. Another advantage of NMR is that chromatographic separation is not required for NMR analysis even though LC-NMR technology has become a choice for conducting high-resolution analysis of complex chemical and metabolite mixture (38). In contrast, majority of MS-based metabolomics requires a separation process, such as GC, LC, and capillary electrophoresis (CE), prior to sample introduction and mass detection in MS system. GC is an excellent platform for the metabolites that are volatile or could become volatile through derivatization while CE is suitable for separating polar compounds. However, the strength of GC and CE in separating these specific classes of chemicals also limits their application in analyzing other classes of chemicals in plant extracts, animal and human samples. At present, LC is the most commonly used platform in MS-based metabolomics owing to its good compatibility with the metabolites in biological system. High-performance liquid chromatography (HPLC) used to be the predominant LC technology used in chemical analysis. However, development of ultra-performance liquid chromatography (UPLC) or ultra-high-pressure liquid chromatography (UHPLC) that uses smaller particles, faster flow rate, and higher pressure than HPLC has greatly improved the chromatographic resolution and reduced the running time in the LC system (39). After eluting from GC, LC, or CE, analytes need to become ionized before they can be detected by mass detectors in MS-based chemical analysis. Electron-impact ionization, a hard-ionization method, is widely used in GC-MS analysis to establish the fragmentation pattern of derivatized analytes. In contract, soft-ionization methods, such as electrospray ionization and atmospheric pressure chemical ionization, are commonly used in LC-MS analysis to reduce the fragmentation in the ionization source and facilitate the detection of parent molecules since additional fragmentation of parent molecules can be achieved inside the mass detector. For mass detection, the selection of mass detector is largely based on the nature of MS-based metabolomics. In general, triple-quadrupole and ion-trap mass spectrometers are better platforms for quantitative analysis in targeted metabolomic analysis owing to their sensitivity (40), while time-of-flight, Orbitrap, or Fourier transform ion cyclotron resonance mass spectrometers are more suitable for untargeted metabolomic analysis because of their high resolution to acquire accurate mass for elemental composition analysis and their high capacity to detect multiple ions simultaneously for comprehensive metabolite profiling (41–43).
Data Analysis
NMR data comprise chemical shift and signal intensity, while LC-MS and GC-MS data comprise retention time (RT), mass-to-charge ratio (m/z), and signal intensity. To conduct untargeted metabolomic analysis, deconvolution is required to convert these data into a data matrix suitable for multivariate data analysis (MDA). Data analysis for NMR-based metabolomics was reviewed in detail previously (44). Here, we use LC-MS-based metabolomics as the example. After LC-MS analysis, chromatographic and spectral data need to be properly processed before being used in MDA. The procedure includes data condensation and reduction by centroiding and deisotoping mass spectra, chromatographic alignment for peak identification, and filtering for removing noise or isotope signals (45). To reduce the influence of systematic and sample biases, MS data can be normalized by either the parameters of whole dataset (such as total ion count or median ion count) or the intensities of single or multiple internal standards (such as creatinine in the case of urine) (46). After all these procedures, a multivariate data matrix containing information about sample identities, ion identities (RT and m/z values) and normalized ion intensities can be generated. This dataset can be either directly used for MDA, or be further statistically transformed and scaled according to the properties of data and the purpose of MDA analysis. To identify latent components or principal components (PC) in a complex dataset, data are projected onto a new coordinate system based on pattern recognition algorithm (47). Thereafter, a model containing one or multiple PCs can be established to represent a large portion of examined dataset (Fig. 1). In contrast to other statistical techniques, such as t test and ANOVA, an established MDA model and its PCs can be presented in the scores plot, in which sample–PC and sample–sample relationships can be visualized. In LC-MS-based metabolomics, the spatial distance between two samples in the scores plot reflects their differences in chemical composition. When a clear sample clustering is observed in the scores plot, the contribution of individual ions to PCs and to the group separation can be further examined in the loadings plot, in which the relationships between ions and PCs are depicted. Two major categories of MDA methods, unsupervised and supervised MDA, have been widely used in metabolomic data analysis. In unsupervised MDA, sample classification is unknown or intentionally blinded to the analytical software, while in supervised MDA this information is provided to the software for the purpose of model construction. The most popular unsupervised method is principal components analysis (PCA). Because of its indiscriminate nature, the markers identified in a robust PCA model can usually be validated. Supervised MDA encompasses many methods, including partial least squares (PLS), orthogonal partial least squares (OPLS), and partial least squares-discriminant analysis (PLS-DA) (48). The selection of supervised MDA method is determined by the data properties and the aim of MDA analysis. In metabolomic analysis of chemopreventive phytochemicals, when a clear separation of different plant preparations or different treatments is observed in the scores plot, phytochemicals related to chemopreventive activity or metabolites related to chemopreventive phytochemicals can be conveniently identified in the loadings plot through their correlation with the PCs that separate sample groups (Fig. 1). Subsequently, chemical identities of bioactive phytochemicals and endogenous metabolites can be determined by accurate mass measurement, elemental composition analysis, MS/MS fragmentation and searches of metabolite and spectral databases, such as Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/), Human Metabolome Database (http://www.hmdb.ca/), Lipid Maps (http://www.lipidmaps.org/), METLIN database (http://metlin.scripps.edu/), BioCyc (http://biocyc.org/), Spectral Database for organic compounds (http://sdbs.riodb.aist.go.jp). Recent development in bioinformatics has further facilitated metabolite annotation in metabolomics studies (49,50).
METABOLOMIC INVESTIGATION OF CHEMOPREVENTIVE PHYTOCHEMICALS
Phytochemical-elicited chemoprevention is a beneficial consequence of dynamic interactions between phytochemicals in vegetables, fruits, and herbal medicine and the biological systems within human body and cancer cells. The complexities of plant metabolome and biological system pose challenges in identifying chemopreventive phytochemicals in plant extracts and understanding the chemopreventive phytochemicals on humans and animals. Metabolomics offers an analytical platform to tackle these challenges in chemoprevention research. Based on the aim and nature of metabolomic analysis, metabolomic investigation of chemopreventive phytochemicals can be exploratory, such as the identification of metabolites and biomarkers, or hypothesis-driven, such as the investigation of enzymes and metabolic pathways.
Metabolomics-Based Identification of Bioactive Phytochemicals
Identification of bioactive phytochemicals usually starts with the bioassay-based screening of crude plant extracts, followed by the bioactivity-guided fractionation and purification of individual phytochemicals. Two types of high-throughput bioassay platforms are available for screening plant extracts or candidate bioactive phytochemicals for their activities in cancer prevention and therapy. One is cell-based platform for determining antiproliferative effects and cytotoxicity against cancer cells, such as the US National Cancer Institute (NCI)’s 60 human tumor cell lines (51). The other is a more mechanism-based approach, in which well-characterized signaling molecules (such as p53, Ras, tyrosine kinase) or enzymes (such as histone deacetylase, DT-diapharose) are chosen as the targets of screening assays (52–54). After bioassays, efficacy of interested plant extracts and phytochemicals can be further validated by animal experiments and human trials. As the large-scale in vitro bioassays have been commonly used for screening anti-cancer compounds (55), the feasibility and effectiveness of bioassay platforms are no longer the major limiting factor for identifying bioactive phytochemicals. Instead, the need to obtain pure compounds from crude plant extracts for bioassays is more challenging due to the chemical properties of plant extracts and the mechanisms of their bioactivity. Firstly, complex phytochemical composition of plant extract makes bioactivity-guided purification processes inefficient and time-consuming, especially when bioactive phytochemical is an unknown compound and also exist as a minor component in the extract. Secondly, if bioactivities of plant extract are the synergistic effects of phytochemicals, testing pure phytochemicals in bioassays might not be able to fully reveal their bioactivities. To tackle these challenges, new research strategies are needed for identifying bioactive phytochemicals, including chemopreventive phytochemicals, from plant extracts (56).
Without exhaustive fractionation and purification, NMR and MS-based metabolomics is capable of defining chemical composition of multiple plant extracts and identifying major differences among them through PCA modeling. In addition, with PLS or PLS-DA analysis, the data from NMR or MS analysis (as X variables) can be processed together with the data from bioassay (as Y responses) to establish the correlation between specific phytochemicals in plant extract and different response observed in bioassay (Fig. 2) (57). Using this approach, pure phytochemicals are not required prior to bioactivity assessment. Instead, after the interested phytochemicals are identified through the metabolomics-based correlation analysis, purification and structural analysis can be followed to determine the structure of bioactive phytochemicals, and a new round of bioassay can be conducted to validate the bioactivities of identified phytochemicals. In practice, in order to utilize this approach for identifying bioactive phytochemicals, plant extracts originated from different locations, genetic background, treatments, or extraction methods are required to establish the correlations between bioactivities and specific compounds in chemical extracts (58).
Fig. 2.
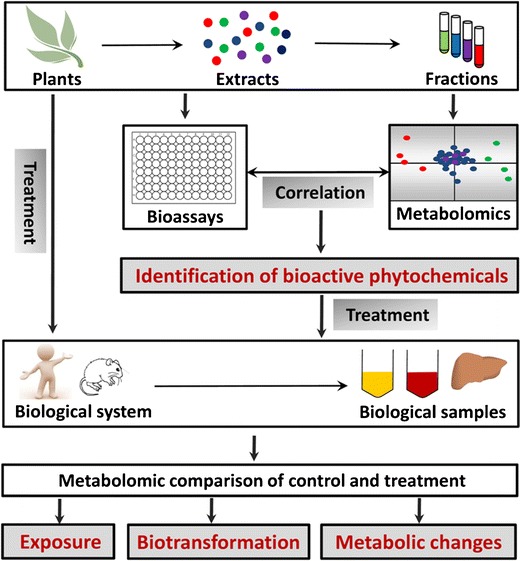
Major applications of metabolomics in studying chemopreventive phytochemicals in vitro and in vivo. Metabolomics-based phytochemical profiling and in vitro bioassays of chemopreventive activities may establish the correlation between specific phytochemicals in plant extracts and different bioactivity of plant preparations, leading to the identification of bioactive phytochemicals. Depending on the experimental design, metabolomic analysis of biological samples from the treatments of phytochemicals or plant extracts can be utilized to evaluate phytochemical exposure, elucidate biotransformation pathways of phytochemicals, and characterize the effects of phytochemicals on endogenous metabolism and cancer metabolism
The effectiveness of this metabolomics approach in identifying bioactive phytochemicals has been illustrated by several recent studies on herbal medicines (59,60). For example, Galphimia glauca, a Mexican herb, has been used for treating central nervous disorders. Mouse bioassay experiment revealed that G. glauca samples collected from six locations possess different anxiolytic and sedative activity (61). 1H NMR metabolomics of G. glauca extracts was conducted to identify the bioactive components responsible for its neuropharmacological activities. After the compositional differences among six extracts were defined by PCA analysis, galphimines and 1,3,4,5-tetra-O-galloylquinic acid were found to be the phytochemicals contributing to the separation of G. glauca extracts in the PCA model. Subsequent PLS-DA analysis established a strong correlation between the neuropharmacological activities and two identified compounds, suggesting they are the bioactive phytochemicals in G. glauca.
To our knowledge, metabolomic comparison of plant extracts with different chemopreventive activities has not been applied to identify novel chemopreventive phytochemicals. Nevertheless, since bioassay models are widely available for screening cancer prevention activities of plant preparations (62,63), the utilization of this metabolomics-based approach in chemoprevention is highly executable. It is possible that metabolomics-based identification of bioactive phytochemicals, together with high-throughput bioassays and more efficient purification of bioactive phytochemicals, such as the HPLC-MS-guided online semi-prep micro-fractionation (60), will lead to the discovery of new chemopreventive agents in near future.
Metabolomics-Based Evaluation of Phytochemical Exposure
Monitoring and measuring exposure of plant extracts and phytochemicals are indispensable components in the epidemiological surveys that evaluate correlation of plant extracts and phytochemicals with cancer incidence, as well as in the human intervention trials that examine cancer prevention effects of plant extracts and phytochemicals. Challenges in assessing their exposure level in humans are multifaceted. Diaries and questionnaires are the main sources of phytochemical exposure information in human studies. However, unpredictability of human behavior and errors in human memory affect the reliability of this type of exposure information. Furthermore, compositions and quantities of phytochemicals in plant extracts are highly variable due to both external and intrinsic factors (64). Therefore, consumption of food and herb products that are presumed to contain interested phytochemicals cannot be equivalent to definite exposure of phytochemicals or the exposure of desired amount of phytochemicals. Overall, inaccurate exposure assessment makes it difficult to establish the correlations between phytochemicals/plant extracts and health benefits.
To address these challenges, chemical analyses of phytochemicals or specific biomarkers in biological samples are commonly required to validate the exposure of interested phytochemicals or plant products in human subjects. Metabolomics, as a highly effective tool for chemical and metabolite profiling, can facilitate these analyses through its capacity in discriminating the samples undergone different phytochemical exposure (65). For example, many dietary polyphenols are considered as chemopreventive compounds due to their antioxidant and antiproliferative activities. Through targeted metabolomic analysis, polyphenol content in human urine was evaluated after the consumption of polyphenol-rich beverages in a nutrition survey study (66). The results showed that the levels of chlorogenic acid, gallic acid, epicatechin, naringenin, and hesperetin in urine can function as exposure biomarkers in epidemiological studies to assess the intake of coffee, wine, tea, cocoa and citrus juices in populations (66). This metabolomics-based approach has also been applied to evaluate the exposure of traditional Chinese medicines. Metabolomic analysis of rabbit serum samples from dosing Dangguibuxue decoction, a tonic mixture comprising of huangqi (Radix astragali) and danggui (Radix angelicae sinensis), led to the detection of flavonoids, phthalides, and triterpene saponins as the exposure biomarkers of Dangguibuxue decoction (67).
Metabolomics-Based Examination of Phytochemical Biotransformation
Metabolism of phytochemicals, including chemopreventive phytochemicals, could serve as a double-edged sword for the bioactivities of phytochemicals. On one hand, it can facilitate elimination and excretion. On the other hand, it may also cause bioactivation and toxicity. Therefore, studying the biotransformation of phytochemical in vivo is essential for understanding and predicting the biological effects of phytochemicals, including chemopreventive activities. Radiotracing is very effective technique to determine the metabolic fate of an exogenous compound in vivo due to its sensitivity and quantitative nature in metabolite analysis. However, application of radiotracing in studying phytochemical biotransformation is limited by concerns of environmental and health hazards as well as effort and cost in radiolabeled phytochemical synthesis.
Metabolomics-guided metabolite profiling provides a robust platform to examine phytochemical biotransformation (26). A straightforward approach is to conduct a metabolomics comparison between the control and phytochemical treatments. Diverse biological samples, including urine, serum, feces, tissue extracts, can be used for this purpose. Since phytochemical and its metabolites only appear in the samples from the phytochemical treatment, separation of two treatment groups in the multivariate model is expected to be contributed by phytochemical and its metabolites (Fig. 3). Using this approach, novel metabolites of phytochemicals as well as therapeutic agents and toxicants have been identified, and novel metabolic routes have been characterized (68–70). Noscapine, an alkaloid from opium, is a promising anti-tumor agent. Metabolomics-based comparison of urine and fecal samples from the control and noscapine-treated mice led to the identification of multiple novel noscapine metabolites originated from oxidation, demethylation, and glucuronidation reactions, and thus defined the metabolic pathways of noscapine in vivo (71).
Fig. 3.
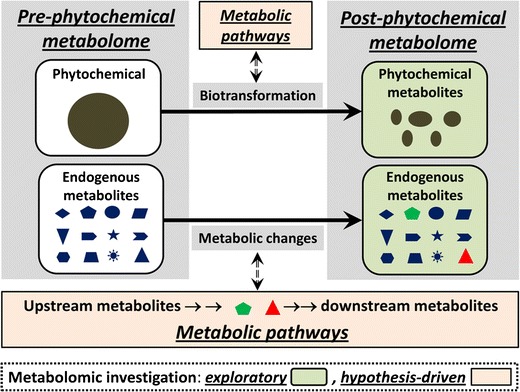
Utilization of metabolomics in both exploratory and hypothesis-driven investigation of phytochemical-related metabolic events. Metabolic events in phytochemical treatment include phytochemical biotransformation and phytochemical-induced metabolic changes. Metabolomics is not only able to identify the metabolites generated or affected by these events (exploratory investigation), but also capable of revealing the mechanisms underlying these events when combined with other experimental models and biochemical analyses on upstream/downstream metabolic reactions and regulatory pathways (hypothesis-driven investigation)
It should be noted that the exposure of phytochemicals and other exogenous compounds can also significantly affect endogenous metabolism, leading to quantitative changes in endogenous metabolite. This situation can interfere with the detection of phytochemical metabolites in metabolomic analysis since endogenous metabolites increased by phytochemical treatment also contribute to the separation of two sample groups in the multivariate model. In this case, a combination of metabolomics analysis and the use of stable isotope-labeled phytochemicals could facilitate the detection of phytochemical metabolites. To our knowledge, the stable isotope-based metabolomics has not been used in studying phytochemical metabolism, but the capacity of this method for identifying novel metabolites was demonstrated in two recent studies on ethanol and acetaminophen, in which N-acetyl taurine, a novel ethanol metabolite, and multiple novel metabolites of acetaminophen were identified through metabolomic comparisons of urine samples from mice fed unlabeled and deuterated ethanol or acetaminophen (72,73).
Besides identifying novel metabolites, metabolomics can also carry out hypothesis-driven investigation on the roles of metabolizing enzymes in biotransformation when appropriate experiment models are used, such as transgenic mice containing modified metabolizing enzymes (74). For instance, the role of CYP1A2 enzyme in the biotransformation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), a common procarcinogen in the human diet, was examined by a metabolomics comparison of urine samples from the wild-type, Cyp1a2-null, and CYP1A2-humanized mice treated with PhIP (75). Genotype-dependent separation of three groups of PhIP-treated animals demonstrated the importance of CYP1A2 in PhIP metabolism, and also revealed the interspecies differences between human and mouse as well as the potential role of other cytochrome P450 enzymes in PhIP metabolism.
Metabolomics Investigation of Phytochemical-Induced Metabolic Effects
Studying the metabolic events associated with phytochemical exposure can provide mechanistic insights on phytochemical-elicited chemopreventive activities since nutrient metabolism, antioxidant metabolism and cancer metabolism are closely related to anti-carcinogenic activities of blocking and suppressing chemopreventive agents. Nutrient and antioxidant metabolism, as the supplier of antioxidants and detoxifying enzymes, determines the capacity of endogenous detoxification system to handle oxidative stress and reactive metabolites, which are the main targets of blocking chemopreventive agents. As for cancer metabolism, altered energy and nutrient metabolism is now recognized as a major driving force behind dysregulated proliferation of cancer cells (76). Compared to normal quiescent cells, cancer cells have greater needs for energy and nutrients to match the faster rate of cell proliferation and increased biomass. Cancer cells possess more active nutrient uptake (glucose and glutamine) for anabolic biosynthesis (lipid, protein and nucleotide) and more robust glycolytic activity for ATP and lactate production than normal cells (77). These altered metabolic processes in cancer cells could be the targets of suppressing chemopreventive agents (78), and the intermediates and end products of these metabolic activities could function as metabolite biomarkers of cancer development (79). Furthermore, besides their influences on the metabolic activities directly related to carcinogenesis, chemopreventive phytochemicals, especially after high-dose or repeated exposure, might affect other physiological functions and metabolic pathways, resulting in beneficial or toxic effects in humans and animals (80–82). These needs for comprehensive metabolic profiling warrant metabolomics to be a useful tool for studying chemopreventive phytochemicals.
Both targeted and untargeted metabolomics could be conducted to examine the metabolic changes induced by chemopreventive phytochemicals. Targeted metabolomics is usually highly subjective due to its typical focus on suspected metabolites, and is commonly guided by observed changes in genes and proteins. In contrast, untargeted metabolomics could provide guidance for mechanistic investigations at the enzyme, protein, and gene levels, especially when transcriptomics and proteomic analyses are also performed (83). Sample source is important for obtaining meaningful information on phytochemical-induced metabolic changes. Biofluids (serum and urine) and tissue extracts are suitable for probing general metabolic effects of chemopreventive phytochemicals since each of them contains distinctive metabolite originated from different metabolic activities. Accordingly, immortalized cancer cells, neoplastic tissues and tumors are appropriate samples for determining the impacts on cancer metabolism. Due to the structural diversity of endogenous metabolites, selecting analytical platform that can effectively detect interested metabolites is crucial for metabolomic analysis. For example, LC-MS-based lipidomics has clear advantages in detecting complex lipid species, which makes it an ideal tool for the simultaneous examination of diverse lipid species (84,85).
Metabolomics has been applied to examine the influences of chemopreventive phytochemicals, such as isoflavones and tea polyphenols, on nutrient and antioxidant metabolism in humans (86,87). NMR-based metabolomics of urine samples from a dietary intervention study showed that a diet rich in soy isoflavones, especially unconjugated soy isoflavone, had significant effects on several metabolic pathways associated with osmolyte fluctuation and energy metabolism (86). Similarly, another separate metabolomics study revealed that the consumption of green tea significantly increased the levels of several intermediate metabolites in energy metabolites, including citrate, pyruvate and oxaloacetate, in healthy volunteers (87). Besides profiling the changes in endogenous metabolism, metabolomics is also capable of detecting phytochemicals-induced changes in gut microflora metabolism, which is a major contributor to the metabolome in serum and urine (88). For example, the increase of bacteria metabolites, hippuric acid and trimethylamine-N-oxide, in human subjects were observed after the metabolomic analysis of urine samples from consuming chamomile tea and soy isoflavones, respectively (86,89).
Metabolomics has also been applied to examine the influences of chemopreventive phytochemicals on cancer metabolism. NMR-based metabolomics was able to reveal the dose-dependent metabolic changes in breast cancer cells after the treatment of curcumin, an active chemopreventive compound in turmeric rhizome (90). The influence of curcumin on redox balance was demonstrated by the observations of increased glutathione level at low dose of curcumin and decreased glutathione level at high dose, suggesting that glutathione biosynthesis was upregulated at low dose while the consumption of glutathione elevated at high dose. In addition, the effects of curcumin treatment on lipid metabolism, including accumulation of polyunsaturated fatty acids and decrease of glyerophospholipids, were also observed (90).
CONCLUSIONS
The capacity of metabolomics to measure numerous chemicals simultaneously and detect subtle differences among sample groups makes it a powerful discovery tool in many scientific fields. Compared to traditional reductionistic approach, metabolomics provide a holistic alternative to tackle the challenges in identifying novel chemopreventive chemicals from plant extracts and studying complex interactions between chemopreventive phytochemicals and biological system (91). Despite its analytical capacity, the utilization of metabolomics in chemoprevention research, such as the four major applications discussed in the review (Fig. 2), is still in its infancy. For example, majority of metabolomics studies on the metabolic effects of chemopreventive phytochemicals remain in the observational level, even though the changes in individual metabolites or a cluster of metabolites in specific metabolic pathways provide opportunities for further biochemical investigations of underlying mechanisms, such as upstream and downstream metabolic reactions as well as regulatory signaling events (Fig. 3). Considering the newly established importance of cancer metabolism in carcinogenesis and the great need for potent chemopreventive phytochemicals, metabolomics has great promise in both exploratory and hypothesis-driven chemoprevention research.
ACKNOWLEDGEMENTS
Research projects in Dr. Chi Chen’s lab were supported in part by an Agricultural Experiment Station project MIN-18-082 from the United States Department of Agriculture (USDA). We thank all the members in Dr. Chi Chen’s lab for their help in preparing this manuscript.
REFERENCES
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Umar A, Dunn BK, Greenwald P. Future directions in cancer prevention. Nat Rev Cancer. 2012;12:835–848. doi: 10.1038/nrc3397. [DOI] [PubMed] [Google Scholar]
- 3.Wattenberg LW. Chemoprophylaxis of carcinogenesis: a review. Cancer Res. 1966;26:1520–1526. [PubMed] [Google Scholar]
- 4.Wattenberg LW. Effects of dietary constituents on the metabolism of chemical carcinogens. Cancer Res. 1975;35:3326–3331. [PubMed] [Google Scholar]
- 5.Collett NP, Amin ARMR, Bayraktar S, Pezzuto JM, Shin DM, Khuri FR, et al. Cancer prevention with natural compounds. Semin Oncol. 2010;37:258–281. doi: 10.1053/j.seminoncol.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 6.Steinmetz KA, Potter JD. Vegetables, fruit, and cancer prevention: a review. J Am Diet Assoc. 1996;96:1027–1039. doi: 10.1016/S0002-8223(96)00273-8. [DOI] [PubMed] [Google Scholar]
- 7.Reddy L, Odhav B, Bhoola KD. Natural products for cancer prevention: a global perspective. Pharmacol Ther. 2003;99:1–13. doi: 10.1016/S0163-7258(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 8.Benetou V, Orfanos P, Lagiou P, Trichopoulos D, Boffetta P, Trichopoulou A. Vegetables and fruits in relation to cancer risk: evidence from the Greek epic cohort study. Cancer Epidemiol Biomarkers Prev. 2008;17:387–392. doi: 10.1158/1055-9965.EPI-07-2665. [DOI] [PubMed] [Google Scholar]
- 9.Freedman ND, Park Y, Subar AF, Hollenbeck AR, Leitzmann MF, Schatzkin A, et al. Fruit and vegetable intake and head and neck cancer risk in a large United States prospective cohort study. Int J Cancer. 2008;122:2330–2336. doi: 10.1002/ijc.23319. [DOI] [PubMed] [Google Scholar]
- 10.Wachtel-Galor S, Benzie IFF. (2011) Herbal medicine: An introduction to its history, usage, regulation, current trends, and research needs. in Herbal medicine: Biomolecular and clinical aspects (Benzie, I. F. F., and Wachtel-Galor, S. eds.), 2nd Ed., Boca Raton (FL). pp [PubMed]
- 11.Glade MJ. Food, nutrition, and the prevention of cancer: a global perspective. American institute for cancer research/world cancer research fund, American Institute for Cancer Research, 1997. Nutrition. 1999;15:523–526. doi: 10.1016/S0899-9007(99)00021-0. [DOI] [PubMed] [Google Scholar]
- 12.Chen C, Kong ANT. Dietary cancer-chemopreventive compounds: from signaling and gene expression to pharmacological effects. Trends Pharmacol Sci. 2005;26:318–326. doi: 10.1016/j.tips.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Yang CS, Wang X, Lu G, Picinich SC. Cancer prevention by tea: animal studies, molecular mechanisms and human relevance. Nat Rev Cancer. 2009;9:429–439. doi: 10.1038/nrc2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad A, Sakr WA, Rahman KM. Anticancer properties of indole compounds: mechanism of apoptosis induction and role in chemotherapy. Curr Drug Targets. 2010;11:652–66. [DOI] [PubMed]
- 15.Urich-Merzenich G, Zeitler H, Jobst D, Panek D, Vetter H, Wagner H. Application of the “-omic-” technologies in phytomedicine. Phytomedicine. 2007;14:70–82. doi: 10.1016/j.phymed.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. Hmdb 3.0—the human metabolome database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sumner LW, Mendes P, Dixon RA. Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry. 2003;62:817–836. doi: 10.1016/S0031-9422(02)00708-2. [DOI] [PubMed] [Google Scholar]
- 18.Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat Rev Cancer. 2004;4:551–561. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 19.Di Leo A, Claudino W, Colangiuli D, Bessi S, Pestrin M, Biganzoli L. New strategies to identify molecular markers predicting chemotherapy activity and toxicity in breast cancer. Ann Oncol. 2007;18(Suppl 12):xii8–xii14. doi: 10.1093/annonc/mdm533. [DOI] [PubMed] [Google Scholar]
- 20.Kuhara T. Noninvasive human metabolome analysis for differential diagnosis of inborn errors of metabolism. J Chromatogr B Anal Technol Biomed Life Sci. 2007;855:42–50. doi: 10.1016/j.jchromb.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 21.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 22.Bino RJ, Hall RD, Fiehn O, Kopka J, Saito K, Draper J, et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 2004;9:418–425. doi: 10.1016/j.tplants.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Ryan D, Robards K. Metabolomics: the greatest omics of them all? Anal Chem. 2006;78:7954–7958. doi: 10.1021/ac0614341. [DOI] [PubMed] [Google Scholar]
- 24.Dunn WB, Bailey NJC, Johnson HE. Measuring the metabolome: current analytical technologies. Analyst. 2005;130:606–625. doi: 10.1039/b418288j. [DOI] [PubMed] [Google Scholar]
- 25.Kersten RD, Dorrestein PC. Secondary metabolomics: natural products mass spectrometry goes global. ACS Chem Biol. 2009;4:599–601. doi: 10.1021/cb900187p. [DOI] [PubMed] [Google Scholar]
- 26.Chen C, Gonzalez FJ, Idle JR. LC-MS-based metabolomics in drug metabolism. Drug Metab Rev. 2007;39:581–597. doi: 10.1080/03602530701497804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Kim S. LC-MS-based metabolomics of xenobiotic-induced toxicities. Comput Struct Biotechnol J. 2013;4:e20130108. doi: 10.5936/csbj.201301008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villas-Boas SG, Mas S, Akesson M, Smedsgaard J, Nielsen J. Mass spectrometry in metabolome analysis. Mass Spectrom Rev. 2005;24:613–646. doi: 10.1002/mas.20032. [DOI] [PubMed] [Google Scholar]
- 29.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou B, Xiao JF, Tuli L, Ressom HW. LC-MS-based metabolomics. Mol Biosyst. 2012;8:470–481. doi: 10.1039/c1mb05350g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halket JM, Waterman D, Przyborowska AM, Patel RK, Fraser PD, Bramley PM. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J Exp Bot. 2005;56:219–243. doi: 10.1093/jxb/eri069. [DOI] [PubMed] [Google Scholar]
- 32.Santa T. Derivatization reagents in liquid chromatography/electrospray ionization tandem mass spectrometry. Biomed Chromatogr. 2011;25:1–10. doi: 10.1002/bmc.1548. [DOI] [PubMed] [Google Scholar]
- 33.Jia S, Kang YP, Park JH, Lee J, Kwon SW. Simultaneous determination of 23 amino acids and 7 biogenic amines in fermented food samples by liquid chromatography/quadrupole time-of-flight mass spectrometry. J Chromatogr A. 2011;1218:9174–9182. doi: 10.1016/j.chroma.2011.10.040. [DOI] [PubMed] [Google Scholar]
- 34.Xu F, Zou L, Liu Y, Zhang Z, Ong CN. Enhancement of the capabilities of liquid chromatography-mass spectrometry with derivatization: general principles and applications. Mass Spectrom Rev. 2011;30:1143–1172. doi: 10.1002/mas.20316. [DOI] [PubMed] [Google Scholar]
- 35.Gao S, Zhang ZP, Karnes HT. Sensitivity enhancement in liquid chromatography/atmospheric pressure ionization mass spectrometry using derivatization and mobile phase additives. J Chromatogr B Anal Technol Biomed Life Sci. 2005;825:98–110. doi: 10.1016/j.jchromb.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 36.Gamache PH, Meyer DF, Granger MC, Acworth IN. Metabolomic applications of electrochemistry/mass spectrometry. J Am Soc Mass Spectrom. 2004;15:1717–1726. doi: 10.1016/j.jasms.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 37.Schattka B, Alexander M, Ying SL, Man A, Shaw RA. Metabolic fingerprinting of biofluids by infrared spectroscopy: modeling and optimization of flow rates for laminar fluid diffusion interface sample preconditioning. Anal Chem. 2011;83:555–562. doi: 10.1021/ac102338n. [DOI] [PubMed] [Google Scholar]
- 38.Wolfender JL, Queiroz EF, Hostettmann K. Phytochemistry in the microgram domain—a LC-NMR perspective. Magn Reson Chem. 2005;43:697–709. doi: 10.1002/mrc.1631. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Sun H, Zhang A, Wang P, Han Y. Ultra-performance liquid chromatography coupled to mass spectrometry as a sensitive and powerful technology for metabolomic studies. J Sep Sci. 2011;34:3451–3459. doi: 10.1002/jssc.201100333. [DOI] [PubMed] [Google Scholar]
- 40.Hopfgartner G, Varesio E, Tschappat V, Grivet C, Bourgogne E, Leuthold LA. Triple quadrupole linear ion trap mass spectrometer for the analysis of small molecules and macromolecules. J Mass Spectrom. 2004;39:845–855. doi: 10.1002/jms.659. [DOI] [PubMed] [Google Scholar]
- 41.Hu QZ, Noll RJ, Li HY, Makarov A, Hardman M, Cooks RG. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 42.Allwood JW, Parker D, Beckmann M, Draper J, Goodacre R. Fourier transform ion cyclotron resonance mass spectrometry for plant metabolite profiling and metabolite identification. Methods Mol Biol. 2012;860:157–176. doi: 10.1007/978-1-61779-594-7_11. [DOI] [PubMed] [Google Scholar]
- 43.Chernushevich IV, Loboda AV, Thomson BA. An introduction to quadrupole-time-of-flight mass spectrometry. J Mass Spectrom. 2001;36:849–865. doi: 10.1002/jms.207. [DOI] [PubMed] [Google Scholar]
- 44.Smolinska A, Blanchet L, Buydens LM, Wijmenga SS. NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: a review. Anal Chim Acta. 2012;750:82–97. doi: 10.1016/j.aca.2012.05.049. [DOI] [PubMed] [Google Scholar]
- 45.Katajamaa M, Oresic M. Data processing for mass spectrometry-based metabolomics. J Chromatogr A. 2007;1158:318–328. doi: 10.1016/j.chroma.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 46.Sysi-Aho M, Katajamaa M, Yetukuri L, and Oresic M. Normalization method for metabolomics data using optimal selection of multiple internal standards. Bmc Bioinformatics. 2007;8. [DOI] [PMC free article] [PubMed]
- 47.Schlotterbeck G, Ross A, Dieterle F, Senn H. Metabolic profiling technologies for biomarker discovery in biomedicine and drug development. Pharmacogenomics. 2006;7:1055–1075. doi: 10.2217/14622416.7.7.1055. [DOI] [PubMed] [Google Scholar]
- 48.Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469–479. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
- 49.Iijima Y, Nakamura Y, Ogata Y, Tanaka K, Sakurai N, Suda K, et al. Metabolite annotations based on the integration of mass spectral information. Plant J. 2008;54:949–962. doi: 10.1111/j.1365-313X.2008.03434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kind T, Fiehn O. Metabolomic database annotations via query of elemental compositions: mass accuracy is insufficient even at less than 1 ppm. BMC Bioinforma. 2006;7:234. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 52.Gibbs JB. Mechanism-based target identification and drug discovery in cancer research. Science. 2000;287:1969–1973. doi: 10.1126/science.287.5460.1969. [DOI] [PubMed] [Google Scholar]
- 53.Holbeck SL. Update on NCI in vitro drug screen utilities. Eur J Cancer. 2004;40:785–793. doi: 10.1016/j.ejca.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 54.Kinghorn AD, Farnsworth NR, Soejarto DD, Cordell GA, Swanson SM, Pezzuto JM, et al. Novel strategies for the discovery of plant-derived anticancer agents. Pharm Biol. 2003;41:53–67. doi: 10.1080/1388020039051744. [DOI] [Google Scholar]
- 55.Damia G, D'Incalci M. Contemporary pre-clinical development of anticancer agents—what are the optimal preclinical models? Eur J Cancer. 2009;45:2768–2781. doi: 10.1016/j.ejca.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 56.Rochfort S. Metabolomics reviewed: a new “Omics” platform technology for systems biology and implications for natural products research. J Nat Prod. 2005;68:1813–1820. doi: 10.1021/np050255w. [DOI] [PubMed] [Google Scholar]
- 57.Wold S, Sjostrom M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst. 2001;58:109–130. doi: 10.1016/S0169-7439(01)00155-1. [DOI] [Google Scholar]
- 58.Yuliana ND, Khatib A, Choi YH, Verpoorte R. Metabolomics for bioactivity assessment of natural products. Phytother Res. 2011;25:157–69. [DOI] [PubMed]
- 59.Yuliana ND, Khatib A, Verpoorte R, Choi YH. Comprehensive extraction method integrated with NMR metabolomics: a new bioactivity screening method for plants, adenosine a1 receptor binding compounds in Orthosiphon stamineus Benth. Anal Chem. 2011;83:6902–6906. doi: 10.1021/ac201458n. [DOI] [PubMed] [Google Scholar]
- 60.Wolfender JL, Queiroz EF. New approaches for studying the chemical diversity of natural resources and the bioactivity of their constituents. Chimia (Aarau) 2012;66:324–329. doi: 10.2533/chimia.2012.324. [DOI] [PubMed] [Google Scholar]
- 61.Cardoso-Taketa AT, Pereda-Miranda R, Choi YH, Verpoorte R, Villarreal ML. Metabolic profiling of the mexican anxiolytic and sedative plant Golphimia glauca using nuclear magnetic resonance spectroscopy and multivariate data analysis. Planta Med. 2008;74:1295–1301. doi: 10.1055/s-2008-1074583. [DOI] [PubMed] [Google Scholar]
- 62.Brunelle JK, Zhang B. Apoptosis assays for quantifying the bioactivity of anticancer drug products. Drug Resist Updat. 2010;13:172–179. doi: 10.1016/j.drup.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 63.Lieberman MM, Patterson GML, Moore RE. In vitro bioassays for anticancer drug screening: effects of cell concentration and other assay parameters on growth inhibitory activity. Cancer Lett. 2001;173:21–29. doi: 10.1016/S0304-3835(01)00681-4. [DOI] [PubMed] [Google Scholar]
- 64.Holst B, Williamson G. Nutrients and phytochemicals: from bioavailability to bioefficacy beyond antioxidants. Curr Opin Biotechnol. 2008;19:73–82. doi: 10.1016/j.copbio.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 65.Kussmann M, Rezzi S, Daniel H. Profiling techniques in nutrition and health research. Curr Opin Biotechnol. 2008;19:83–99. doi: 10.1016/j.copbio.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 66.Ito H, Gonthier MP, Manach C, Morand C, Mennen L, Remesy C, et al. Polyphenol levels in human urine after intake of six different polyphenol-rich beverages. Br J Nutr. 2005;94:500–509. doi: 10.1079/BJN20051522. [DOI] [PubMed] [Google Scholar]
- 67.Wang P, Liang Y, Zhou N, Chen B, Yi L, Yu Y, et al. Screening and analysis of the multiple absorbed bioactive components and metabolites of dangguibuxue decoction by the metabolic fingerprinting technique and liquid chromatography/diode-array detection mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:99–106. doi: 10.1002/rcm.2816. [DOI] [PubMed] [Google Scholar]
- 68.Chen C, Meng L, Ma X, Krausz KW, Pommier Y, Idle JR, et al. Urinary metabolite profiling reveals CYP1A2-mediated metabolism of NSC686288 (aminoflavone) J Pharmacol Exp Ther. 2006;318:1330–1342. doi: 10.1124/jpet.106.105213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giri S, Idle JR, Chen C, Zabriskie TM, Krausz KW, Gonzalez FJ. A metabolomic approach to the metabolism of the areca nut alkaloids arecoline and arecaidine in the mouse. Chem Res Toxicol. 2006;19:818–827. doi: 10.1021/tx0600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao D, Shi X, Wang L, Gosnell BA, Chen C. Characterization of differential cocaine metabolism in mouse and rat through metabolomics-guided metabolite profiling. Drug Metab Dispos. 2013;41:79–88. doi: 10.1124/dmd.112.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang ZZ, Krausz KW, Li F, Cheng J, Tanaka N, Gonzalez FJ. Metabolic map and bioactivation of the anti-tumour drug noscapine. Br J Pharmacol. 2012;167:1271–86. [DOI] [PMC free article] [PubMed]
- 72.Chen C, Krausz KW, Idle JR, Gonzalez FJ. Identification of novel toxicity-associated metabolites by metabolomics and mass isotopomer analysis of acetaminophen metabolism in wild-type and cyp2e1-null mice. J Biol Chem. 2008;283:4543–4559. doi: 10.1074/jbc.M706299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi X, Yao D, Chen C. Identification of n-acetyltaurine as a novel metabolite of ethanol through metabolomics-guided biochemical analysis. J Biol Chem. 2012;287:6336–6349. doi: 10.1074/jbc.M111.312199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang XL, Gonzalez FJ, Yu AM. Drug-metabolizing enzyme, transporter, and nuclear receptor genetically modified mouse models. Drug Metab Rev. 2011;43:27–40. doi: 10.3109/03602532.2010.512294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen C, Ma X, Malfatti MA, Krausz KW, Kimura S, Felton JS, et al. A comprehensive investigation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PHiP) metabolism in the mouse using a multivariate data analysis approach. Chem Res Toxicol. 2007;20:531–542. doi: 10.1021/tx600320w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 77.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arakaki AK, Skolnick J, McDonald JF. Marker metabolites can be therapeutic targets as well. Nature. 2008;456:443. doi: 10.1038/456443c. [DOI] [PubMed] [Google Scholar]
- 79.Serkova NJ, Glunde K. Metabolomics of cancer. Methods Mol Biol. 2009;520:273–295. doi: 10.1007/978-1-60327-811-9_20. [DOI] [PubMed] [Google Scholar]
- 80.Scalbert A, Knasmuller S. Genomic effects of phytochemicals and their implication in the maintenance of health. Br J Nutr. 2008;99(E Suppl 1):ES1–ES2. doi: 10.1017/S0007114508965740. [DOI] [PubMed] [Google Scholar]
- 81.Spencer JP. Flavonoids: modulators of brain function? Br J Nutr. 2008;99(E Suppl 1):ES60–ES77. doi: 10.1017/S0007114508965776. [DOI] [PubMed] [Google Scholar]
- 82.Fardet A, Llorach R, Martin JF, Besson C, Lyan B, Pujos-Guillot E, et al. A liquid chromatography-quadrupole time-of-flight (LC-QToF)-based metabolomic approach reveals new metabolic effects of catechin in rats fed high-fat diets. J Proteome Res. 2008;7:2388–2398. doi: 10.1021/pr800034h. [DOI] [PubMed] [Google Scholar]
- 83.Robertson DG, Watkins PB, Reily MD. Metabolomics in toxicology: preclinical and clinical applications. Toxicol Sci. 2011;120(Suppl 1):S146–S170. doi: 10.1093/toxsci/kfq358. [DOI] [PubMed] [Google Scholar]
- 84.Griffin JL, Nicholls AW. Metabolomics as a functional genomic tool for understanding lipid dysfunction in diabetes, obesity and related disorders. Pharmacogenomics. 2006;7:1095–1107. doi: 10.2217/14622416.7.7.1095. [DOI] [PubMed] [Google Scholar]
- 85.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 86.Solanky KS, Bailey NJ, Beckwith-Hall BM, Bingham S, Davis A, Holmes E, et al. Biofluid 1H NMR-based metabonomic techniques in nutrition research—metabolic effects of dietary isoflavones in humans. J Nutr Biochem. 2005;16:236–244. doi: 10.1016/j.jnutbio.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 87.Van Dorsten FA, Daykin CA, Mulder TPJ, Van Duynhoven JPM. Metabonomics approach to determine metabolic differences between green tea and black tea consumption. J Agric Food Chem. 2006;54:6929–6938. doi: 10.1021/jf061016x. [DOI] [PubMed] [Google Scholar]
- 88.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Y, Tang H, Nicholson JK, Hylands PJ, Sampson J, Holmes E. A metabonomic strategy for the detection of the metabolic effects of chamomile (Matricaria recutita L.) ingestion. J Agric Food Chem. 2005;53:191–196. doi: 10.1021/jf0403282. [DOI] [PubMed] [Google Scholar]
- 90.Bayet-Robert M, and Morvan D. Metabolomics reveals metabolic targets and biphasic responses in breast cancer cells treated by curcumin alone and in association with docetaxel. PLoS One. 2013;8:e57971. [DOI] [PMC free article] [PubMed]
- 91.Verpoorte R, Choi YH, Kim HK. Ethnopharmacology and systems biology: a perfect holistic match. J Ethnopharmacol. 2005;100:53–56. doi: 10.1016/j.jep.2005.05.033. [DOI] [PubMed] [Google Scholar]


