Abstract
Pre-mRNA splicing is required for the accurate expression of virtually all human protein coding genes. However, splicing also plays important roles in coordinating subsequent steps of pre-mRNA processing such as polyadenylation and mRNA export. Here, we test the hypothesis that nuclear pre-mRNA processing influences the polyribosome association of alternative mRNA isoforms. By comparing isoform ratios in cytoplasmic and polyribosomal extracts, we determined that the alternative products of ∼30% (597/1954) of mRNA processing events are differentially partitioned between these subcellular fractions. Many of the events exhibiting isoform-specific polyribosome association are highly conserved across mammalian genomes, underscoring their possible biological importance. We find that differences in polyribosome association may be explained, at least in part by the observation that alternative splicing alters the cis-regulatory landscape of mRNAs isoforms. For example, inclusion or exclusion of upstream open reading frames (uORFs) in the 5′UTR as well as Alu-elements and microRNA target sites in the 3′UTR have a strong influence on polyribosome association of alternative mRNA isoforms. Taken together, our data demonstrate for the first time the potential link between alternative splicing and translational control of the resultant mRNA isoforms.
The coding sequences (exons) of most metazoan genes are separated by intervening sequences (introns) that must be removed from the precursor message RNA (pre-mRNA) prior to translation. Pre-mRNA splicing is an essential step in gene expression and is catalyzed by the spliceosome, a dynamic macromolecular ribonucleoprotein particle, which assembles de novo on each intron (Hoskins and Moore 2012). Pre-mRNA splicing is also a major regulatory point in the gene expression pathway (Braunschweig et al. 2013). The vast majority of multiexon genes are alternatively spliced, giving rise to a diverse but functionally enigmatic pool of transcripts (Castle et al. 2008; Pan et al. 2008; Wang et al. 2008).
In eukaryotic systems, there is a poor correlation between steady-state mRNA and protein levels (Lian et al. 2001; Griffin et al. 2002; Cox et al. 2005; Kislinger et al. 2006; Wilhelm et al. 2006; Schmidt et al. 2007; Fu et al. 2009). Although some of this discrepancy can be attributed to mRNA stability and the efficiency of translation initiation, the contribution of alternative splicing (AS) to this problem is largely ignored. Conversely, the impact of AS on mRNA diversity is indisputable and yet the underlying biological relevance for many transcript isoforms remains enigmatic. Bridging this gap between transcript diversity and protein complexity requires a closer examination of isoform-specific mRNA fates.
AS regulates the expression of human genes by at least two well-established mechanisms. First, AS expands protein diversity by generating different mRNA isoforms from protein-coding genes. Second, AS attenuates gene expression by producing substrates for nonsense-mediated decay (AS-NMD) (Ni et al. 2007; Pan et al. 2008; Wang et al. 2008). In contrast to NMD, little is known regarding how mRNA isoforms differ in their translatability. It is clear that the trans-acting factors associated with alternative mRNA isoforms can vary just as distinctly as the protein-coding capacities of the messages they encode. Many RNA binding proteins, including shuttling proteins, are implicated in multiple steps of post-transcriptional gene regulation, including mRNA export, stability, surveillance, localization, and translation (Sanford and Caceres 2004; Zhang and Krainer 2004; Zhong et al. 2009; Han et al. 2011). These factors can be transcript-associated throughout the life cycle of an mRNA and may coordinate nuclear and cytoplasmic steps of gene expression (Dreyfuss et al. 2002; Nashchekin et al. 2007; Kylberg et al. 2008). This hypothesis is supported by the observation that exonic splicing enhancers can also alter the efficiency of mRNA translation in a position-specific manner (Sanford et al. 2004; Michlewski et al. 2008; Delestienne et al. 2010). Thus, an unanticipated consequence of alternative splicing may lie within the subsequent regulation of the RNA binding proteins packaged upon each isoform: functionally distinct mRNPs with distinct cytoplasmic fates (Zhong et al. 2009; Wang et al. 2012).
Here, we test the hypothesis that alternative mRNA isoforms are subject to translational regulation. This approach combines subcellular fractionation and high throughput RNA sequencing (Frac-seq) to assay partitioning of mRNA isoforms between the cytosolic and the polyribosome-associated fractions. Our data shows that ∼30% of alternative splicing events exhibit isoform-specific polyribosome association. The sequences encompassing these events are more evolutionarily conserved compared to archetypal alternative pre-mRNA processing events, underscoring their biological importance. We also suggest that alternative splicing alters the cis-regulatory landscape of mRNA isoforms, leading to distinct cytoplasmic fates. Taken together, our results document an unexpected link between alternative pre-mRNA processing and the cytoplasmic fate of endogenous mRNA isoforms.
Results
Frac-seq reveals isoform-specific differences in polyribosome association
Polyribosome profiling is widely used to estimate the translation status of mRNAs (Masek et al. 2011). We hypothesized that alternative splicing not only expands the protein coding capacity of human genes, but also establishes patterns of isoform-specific translational control. To test this hypothesis, we propose a genomic approach comparing alternative mRNA isoform diversity in cytoplasmic and polyribosome subcellular fractions (Fig. 1A). The former sample represents the output of the nucleus, whereas the latter represents those mRNAs that are engaged in the translation machinery. We extracted only the polyribosomal peaks from each gradient thereby avoiding possible contaminating RNA from other regions of the gradient (Supplemental Fig. 1).
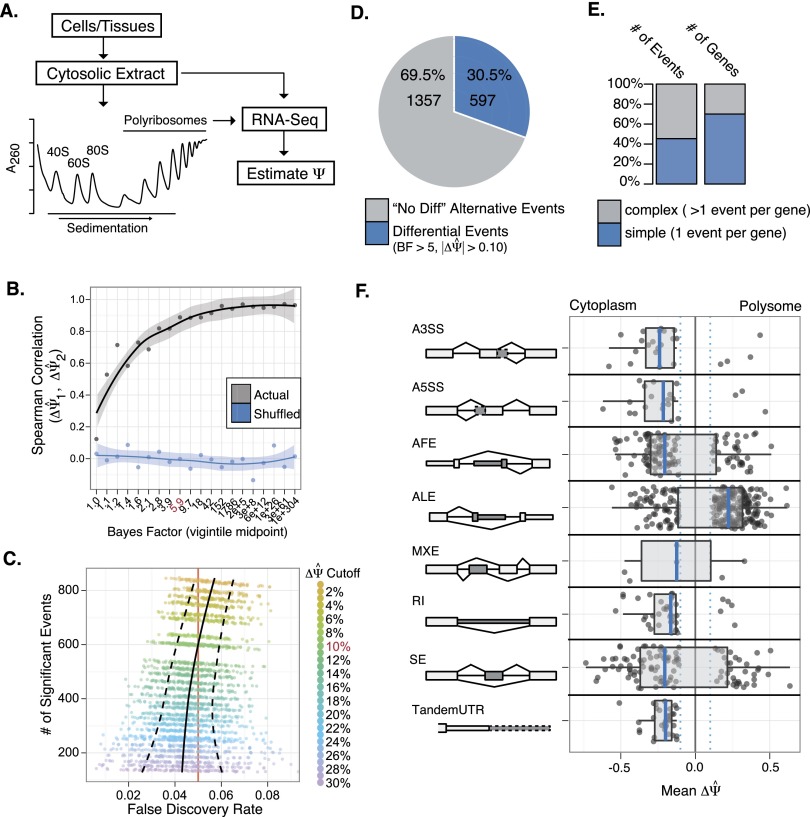
Figure 1.
Frac-seq reveals isoform-specific differences in polyribosome association. (A) Schematic diagram of the Frac-seq protocol. (B) Bayes factor threshold estimation as a function of biological reproducibility (measured via Spearman rank correlation). Local smoothing of the mean and standard error of the mean (SEM) provided as line and shading, respectively. (C) Empirical false discovery estimation (FDR) using bootstrapped (100×) label permutation of one biological replicate for each |ΔΨ| cutoff. Mean FDR and standard error are plotted as solid and dashed lines, respectively. (D) Percentage of alternative events showing significant changes in polyribosome association (blue) compared to detectable alternatively spliced events showing no difference (gray). (E) Complexity of RNA processing events per gene (columns scaled to percentage), with complex events (more than one differential event per gene) in gray, and simple (one differential event per gene) events in blue. (F) Breakdown of significant event types and their distributions with respect to ΔΨ. Schematics on the left side present the “spliced in” region as darkly shaded. For example, the skipped exon (SE) diagram presents the shaded exon that when preferentially associated with polyribosomes would show a positive value for ΔΨ.
Following paired-end sequencing of polyA+ selected RNA libraries and mapping to hg19 (Supplemental Table 1), we employed a previously developed Bayesian inference methodology to estimate percentage spliced-in (Ψ, MISO) (Katz et al. 2010) in the polyribosomal and cytoplasmic fractions. The value of Ψ was estimated for eight different types of alternative events, including skipped exons (SE), alternative 5′ splice sites (A5SS), alternative 3′splice sites (A3SS), mutually exclusive exons (MXE), retained introns (RI), alternative first exons (AFEs), alternative last exons (ALEs), and tandem 3′UTRs (alternative polyadenylation). We found that the Ψ values estimated for biological replicates are strongly correlated, suggesting a sufficient degree of biological reproducibility in the data (Supplemental Fig. 2).
To determine which events, if any, showed significant changes in Ψ upon comparison of polyribosomal and cytoplasmic fractions, we utilized the Bayes factor (BF) as a measure of statistical significance for ΔΨ (ΨPolyribosomal – ΨCytoplasm) (Fig. 1B). As a prerequisite for biological significance, we assume that the events must be biologically reproducible. Proceeding from this logic, we split the data (BF > 1 in both replicates) into vigintiles following the distribution of Bayes factor values. Plotting Spearman's rank correlation coefficient (ΔΨ replicate I vs. replicate II) for the events in each quantile produces a hyperbolic curve that approaches perfect correlation and plateaus near a Bayes factor of five (Fig. 1B). These data suggest that the Bayes factor is indeed a measure that corresponds to increasing biological reproducibility, and a cutoff of five is an appropriate threshold. To determine the rate of false discovery (FDR), we utilized the real data as a set of true positives and estimated false positives from 100× bootstrapped label permutations. Introducing |ΔΨ| consistency between replicates as a second stringency measure, we plotted this empirical FDR for each bootstrapped replicate following increasing cutoffs for ΔΨ (Fig. 1C; see Methods). These data allowed us to choose relevant criteria for significance (BF > 5, |ΔΨ| > 0.1) that maximize the number of significant events and minimize the false discovery rate to an acceptable level (FDR ≈ 5.02 ± 0.9%).
If mRNA translation is independent of the process of alternative splicing, one prediction would be that the relative fractional abundance of alternatively spliced messages would be consistent in both the experimental fractions. Using Frac-seq, we identified 597 alternative events with significant difference in Ψ between the cytoplasmic and polyribosomal fractions (Supplemental Table 2). We also identified 1357 alternative processing events (0.1 < Ψ < 0.9) that are unchanged between the two fractions (“no-diff”, |ΔΨ| < 0.1; BF < 1). Taken together, we find that 30.5% of detectable alternative processing events are differentially partitioned between the cytosolic and polyribosomal fractions (Fig. 1D). Further investigation showed that >40% of these events are the only differential event per gene (Fig. 1E, “simple events”). This observation suggests that loss or gain of sequence elements associated with single alternative events may contribute to the isoform-specific polyribosome association we detect. The 597 significant events encompass all eight distinct pre-mRNA processing classes defined in the MISO database. Furthermore, we recognize a marked preference for significant changes in Ψ affiliated with alternative last exon (ALE) and alternative first exon (AFE) events (Fig. 1F; Supplemental Fig. 3). This is consistent with roles for 5′ and 3′ UTRs (untranslated regions) in modulation of both mRNA stability and translation. Additionally, tandem UTR events (alternative polyadenylation) showed a clear bias toward increased polyribosome association for the shorter isoform. This observation is consistent with previous reports (Sandberg et al. 2008; Mayr and Bartel 2009). Collectively, these data strongly support our hypothesis that alternative pre-mRNA splicing impacts the translation of mRNA. Finally, genes associated with significant differences in isoform-specific polyrbosome association are enriched in gene ontology terms related to RNA binding and pre-mRNA splicing (Supplemental Table 4).
Validation of alternative event-specific polyribosome association by RT-PCR
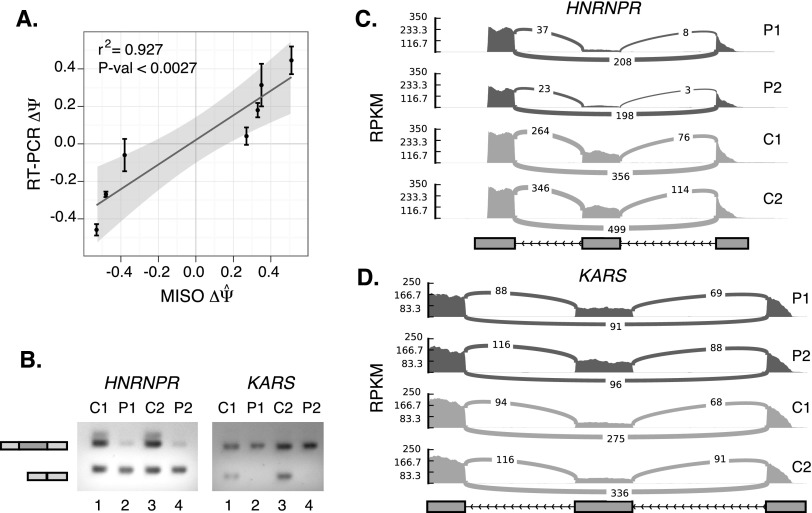
We tested seven alternative cassette exons and one alternative last exon (ALE) estimated by MISO to be differentially partitioned between the cytoplasmic and polyribosomal fractions (Supplemental Fig. 4). Qualitatively, the RT-PCR analysis for seven of the eight events was in agreement with the computational estimates (summarized in Supplemental Table 3; Supplemental Fig. 4). Figure 2A compares ΔΨ estimates from sequencing reads and RT-PCR for seven cassette exons. This provides a ∼12.5% experimental estimate of the FDR that is not unexpected given the sample size and our computational estimate. One splicing event in the MOV10 gene was clearly a false positive and another, in the SGOL1 gene, proved to be qualitatively similar to the MISO estimate but was difficult to quantify by RT-PCR (Supplemental Fig. 4). Despite these two caveats and a relatively small sample size, we observe significant positive correlation between RT-PCR and computational estimates of ΔΨ (Fig. 2A; Supplemental Table 3). Two exemplars, HNRNPR and KARS, are shown in Figure 2B and represent robust examples of event-specific polyribosome association. Computational estimates of Ψ suggest that 15%–32% of HNRNPR transcripts contain the alternative exon in the polyribosome fraction compared to 58%–66% in the cytoplasm (Fig. 2C). Alternative mRNA isoforms produced by the KARS gene show the opposite pattern of partitioning. Estimates of Ψ suggest that 46%–64% of KARS polyribosome associated transcripts contain the alternative cassette exon compared to only 23%–31% in the cytoplasm (Fig. 2D). Representative endpoint RT-PCR confirmed these results (Fig. 2B).
Figure 2.
Validation of alternative mRNA isoforms in polyribosomal and cytosolic subcellular fractions by RT-PCR. (A) Correlation of endpoint RT-PCR (in triplicate with standard error) determined ΔΨ with average MISO estimated ΔΨ. Linear regression plotted as a solid line with 95% confidence interval (shaded region). Significance P-value derived from t-distribution test using Pearson's product-moment correlation coefficient. (B) Representative agarose gels of endpoint RT-PCR for two exemplar genes, HNRNPR and KARS. C1 and C2 refer to the cytoplasmic derived replicates, whereas P1 and P2 refer to the polyribosome-bound replicates. (C,D) Sashimi plots depict RPKM coverage of RNA-seq reads used to estimate Ψ values for HNRNPR and KARS, respectively. Junction reads are annotated as connecting lines.
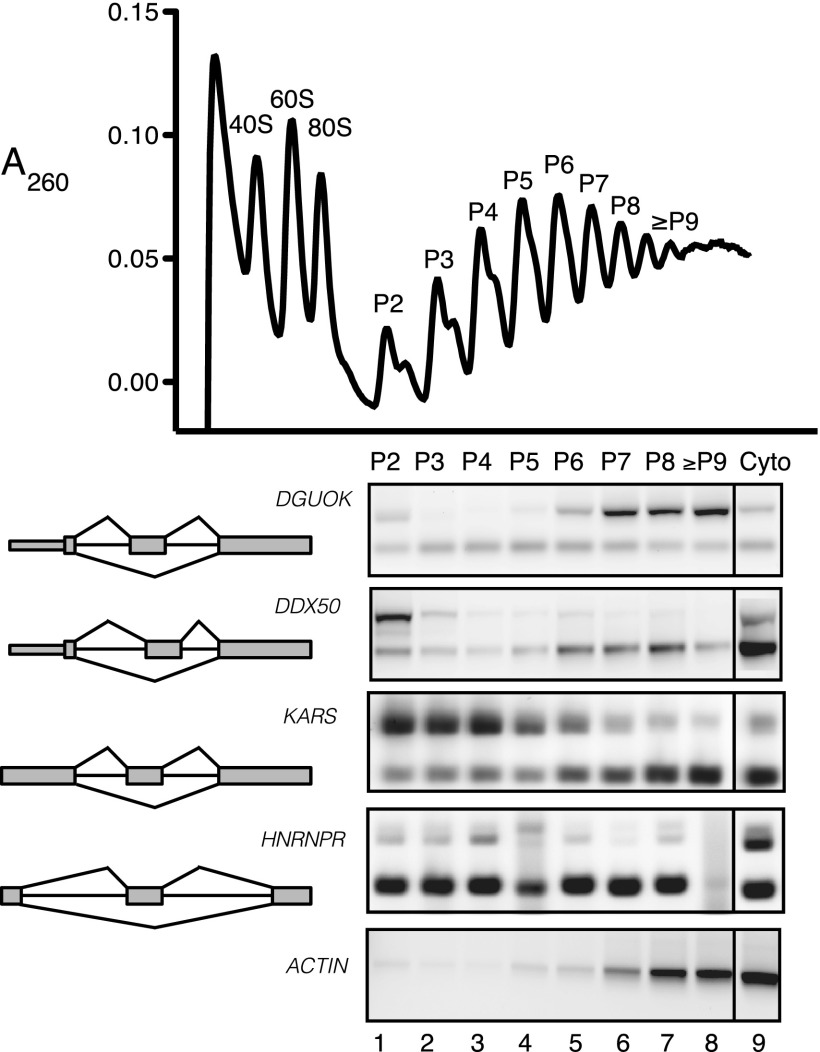
Translational control can be assayed by determining the distribution of mRNAs across individual polyribosome fractions. One caveat of Frac-seq is that the polyribosome fractions are pooled prior to sequencing. To determine if isoform-specific polyribosome association is indicative of translational control, we examined the sucrose gradient distributions of alternative mRNAs from four genes: DDX50, DGUOK, HNRNPR, and KARS. Quantification of RT-PCR analysis of skipping events in different fractions demonstrates that polyribosome association of alternative mRNA isoforms is highly variable (Supplemental Fig. 5). For example, the DGUOK isoform including a cassette exon is associated almost exclusively with the heavier polyribosome fractions (Fig. 3, lanes 5–8). In contrast, the isoform lacking this exon is distributed fairly evenly across the gradient, suggesting that the skipped isoform is translated less efficiently than the included isoform. Contrary to DGUOK, both KARS and DDX50 behave similarly with increased exon inclusion in the lighter polyribosome fractions and enrichment of the skipped isoform in the heavier fractions (Fig. 3, lanes 1–4 and 5–8, respectively). For HNRNPR, inclusion of the alternative cassette exon is enriched in the cytoplasmic fraction and is dramatically reduced in the polyribosome fractions (Fig. 3, cf. lane 9 to lanes 1–7).
Figure 3.
Distribution of alternative mRNA isoforms in ranked polyribosome complexes. (Upper panel) Absorbance profile (A260) of cytosolic extracts across 15%–45% sucrose gradient. Purified polyribosome fractions are indicated by P2 to ≥P9 corresponding to two ribosomes (P2) up to nine or more (≥P9). (Lower panels) Alternative splicing of KARS, HNRNPR, DGUOK, and DDX50 in different polyribosomal peak fractions assayed by RT-PCR and agarose gel electrophoresis. The constitutively spliced ACTB transcript is present at the bottom.
Sequences corresponding to isoform-specific polyribosome association are conserved through evolution
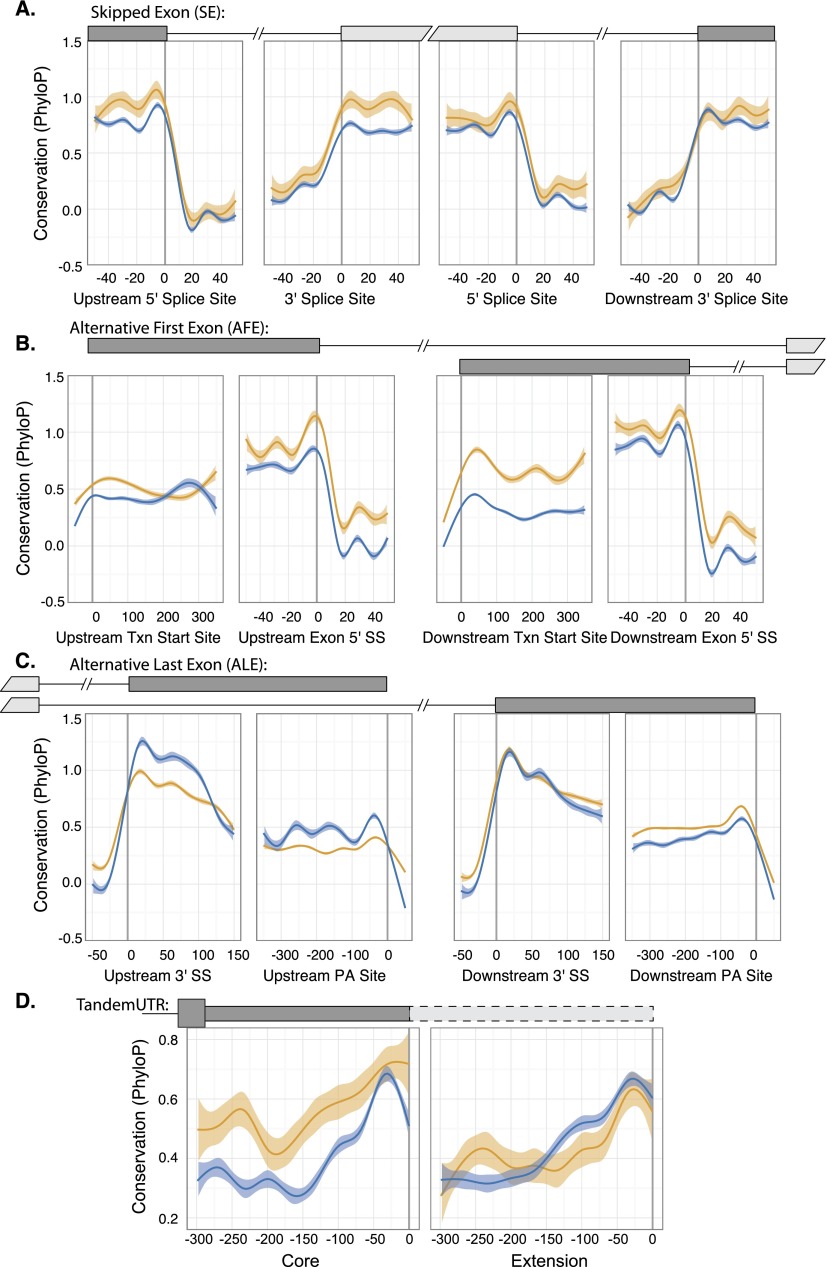
Although our data demonstrate that ∼30% of alternatively processed messages are differentially associated with polyribosomes, the mechanisms by which they do so remain elusive. Phylogenic conservation of DNA sequences often underscores their functional importance. This hallmark has been successfully applied to alternative splicing to identify putative regulatory elements in alternative exons and flanking intronic sequences (Sugnet et al. 2006; Wang et al. 2008). We compared the PhyloP conservation scores (placental mammals using a 46-way alignment) for different classes of alternative events associated with isoform-specific polyribosome association (Siepel et al. 2005; Pollard et al. 2010). As expected, cassette exons appear to be less conserved than adjacent constitutive exons, suggesting recent evolution (Gelfman et al. 2012). Surprisingly, we observe elevated conservation scores for differential cassette exons in the coding sequences flanking the upstream intron (Fig. 4A, gold and blue lines, respectively). Most notably, we detect convincing changes in conservation near the transcription start site (TSS) for the proximal alternative first exon and 250-bp internal to the upstream alternative polyadenylation (PA) site (Fig. 4B,C). In both cases, sequences enriched in the polyribosome fraction have higher conservation scores compared to those in the “no-diff” set (Fig. 4B,C, respectively). In contrast, we observe sub-background levels of conservation present in the proximal ALE and subtle increases in conservation 300 bp upstream of the PA site in the distal ALE (Fig. 4D). Taken together, our data suggest that splicing events related to strong polyribosome association show evolutionary conservation, which may reflect the biological importance and/or functionality of those sequences.
Figure 4.
Conservation of DNA sequences encompassing different classes of polyribosome associated alternative mRNA processing events. Local nonlinear smoothing using a generalized additive model shows best-fit sequence conservation (46-way PhyloP) scores of alternative events and flanking sequence. Significant events are depicted as yellow, while “no diff” events are colored blue (shaded 95% confidence interval). (A) Skipped exon events; (B) alternative first exon events; (C) alternative last exon events; and (D) alternative polyadenylation events.
Alternative pre-mRNA processing alters the cis-regulatory landscape of mRNA isoforms
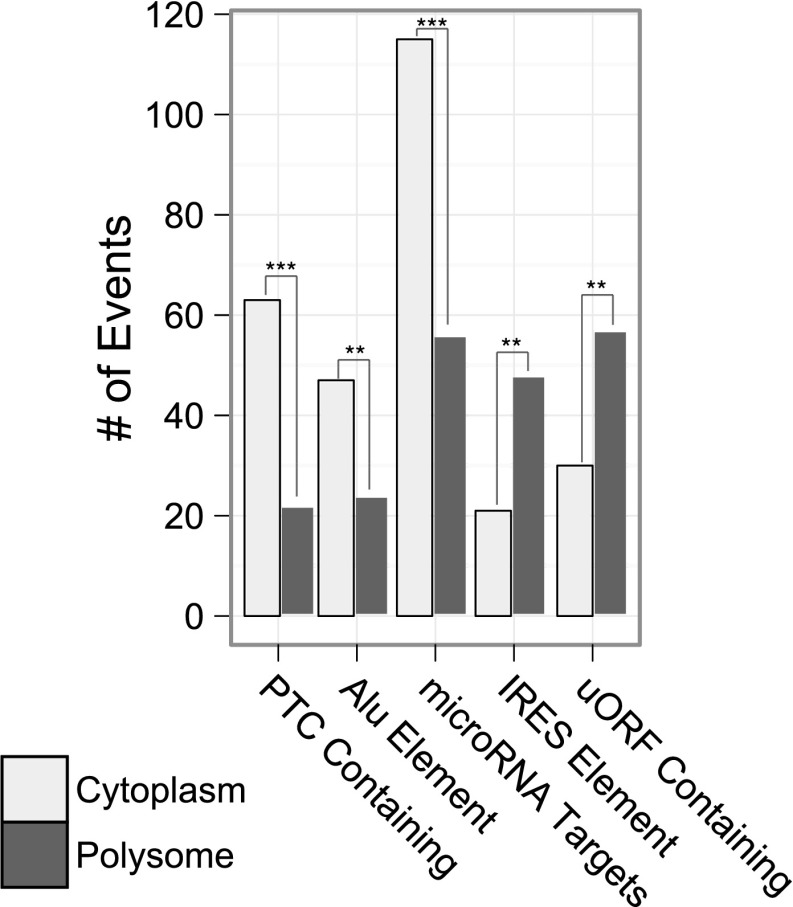
A significant fraction of alternative splicing events give rise to nonfunctional mRNAs containing premature termination codons (PTC). These potentially deleterious mRNA isoforms are highly unstable due to their efficient elimination by the nonsense-mediated mRNA decay pathway. Transcripts containing PTCs are hypothesized to be degraded in a pioneer round of mRNA translation and thus should be largely excluded from the polyribosome fraction. To identify possible NMD substrates, we searched for alternative splicing events that introduced sequences containing termination codons in all three frames. We observe that alternative events containing PTCs are depleted from the polyribosome fraction (Fig. 5). Given their prominent role in translational control and mRNA stability, we also analyzed alternative events that remodel sequences in 5′ and 3′ UTRs associated with isoform-specific polyribosome association. We exploited a list of predicted functional elements annotated in the UTR database (UTRdb) (Grillo et al. 2009) and intersected these annotations with the set of alternative events exhibiting a change in polyribosome association (see Methods). Included in this list are well-annotated 5′ and 3′ UTR specific regulatory elements such as upstream ORFs (uORFs) and internal ribosomal entry site (IRES), as well as microRNA target sites and Alu elements, respectively. These element classes are well characterized for their roles in translational control and mRNA stability (Grillo et al. 2009; Gong and Maquat 2011a,b; Ramani et al. 2011). Our analysis demonstrates that uORFs and IRES elements are associated with an increase in polyribosome association for element-harboring isoforms compared to the element-deficient isoform (P-value < 3.79 × 10−3 and <1.15 × 10−3, respectively) (Fig. 5). For alternative pre-mRNA processing events within the 3′UTR, we observe a significant decrease in polyribosome association for isoforms containing microRNA target sites or Alu elements when compared to the isoform lacking these elements (P-value < 6.42 × 10−6 and <6.34 × 10−3, respectively) (Fig. 5).
Figure 5.
Alternative pre-mRNA processing alters the cis-regulatory landscape of mRNA isoforms. Intersection of the alternative portion of each significant event with predicted cis-acting RNA elements. Events whose alternative region harbors or overlaps a cis-acting element are labeled by their direction of change (positive or negative ΔΨ, polyribosomal or cytoplasmic, respectively). Premature termination codon (PTC) containing events have at least one termination codon in all three reading frames of the alternative portion of the event. Events with uORF annotations contain the start codon of the annotated upstream open reading frame. The tendency of an annotated element-containing sequence toward the cytoplasm is colored gray, while the polyribosome is dark gray. Results of a χ2 goodness-of-fit test to an even ratio are denoted as follows: (***) <0.001; (**) <0.01; (*) <0.05.
The analysis described above suggests that remodeling of known cis-regulatory elements may contribute to event-specific patterns of polyribosome association. To identify novel cis-elements, we searched sequences associated with preferential polyribosome or cytoplasmic distribution for over-represented n-mers. It was most feasible to analyze the ALE and AFE events given the relatively small sample size of other event types. We identified several distinct hexamer sequence clusters that are over-represented in either polyribosome or cytoplasmic fractions for AFE events (Supplemental Fig. 6A). These sequences correspond primarily to GC-rich hexamers. The only enriched hexamers we identified in cytoplasmic or polyribosomal-associated ALE events resembled the cis-elements related to polyadenylation signals (Supplemental Fig. 6C).
Discussion
Here we present Frac-seq, a method to determine the enrichment or depletion of alternatively spliced mRNAs in polyribosomes. The Frac-seq method is related to the recently reported ribosome footprinting method but differs in several important ways (Ingolia et al. 2009; Ingolia 2010). First, Frac-seq is not intended to quantify the translational state of all mRNAs but rather to determine the alternatively spliced transcriptome that is associated with the translation machinery. The footprinting method produces a ribosomal silhouette within the coding sequence of each mRNA that provides an extremely accurate measure of ribosome density. However the nuclease digestion decreases the complexity of the library and lowers the resolution of different mRNA isoforms. Additionally, information pertaining to processing within UTRs is missing, as these regions are not protected from nuclease.
In eukaryotic systems, mRNA transcript levels often do not correspond with protein quantities (Lian et al. 2001; Griffin et al. 2002; Cox et al. 2005; Kislinger et al. 2006; Wilhelm et al. 2006; Schmidt et al. 2007; Fu et al. 2009). For example, in human medulloblastoma cells, there is a relatively poor correlation between steady-state protein and mRNA levels (R = 0.29) (Vogel et al. 2010). The authors of this study identified sequence features such as transcript length, secondary structure and known regulatory elements accounting for up to 67% of the variation between mRNA and protein levels (Vogel et al. 2010). However the contribution of alternative splicing (AS) to this problem is poorly understood. Vogel et al. (2010) demonstrate that coding and 3′UTR sequences explain a significant fraction of protein abundance variation. Our data largely agree with these results. In particular, we find that isoforms enriched in the polyribosome fraction frequently correspond to exon skipping events or processing events leading to distinct or shorter 3′UTRs (Fig. 1F). However, because we adopted an event-based analysis, our results do not address the relationship between isoform length and polyribosome association. Based on results from ribosome footprinting experiments in yeast, we speculate that isoforms with shorter coding regions will be translated more efficiently (Ingolia et al. 2009).
Alternative splicing plays two well-established roles in gene regulation. First, it expands protein diversity by generating different mRNA isoforms from protein-coding genes. Second, alternative splicing attenuates gene expression by producing substrates for nonsense-mediated decay (Ni et al. 2007; Pan et al. 2008; Wang et al. 2008). The work presented here suggests a third and unexpected consequence of alternative splicing—the translational control of resultant mRNA isoforms (AS-TC). Our data suggests that splicing generates transcripts with different affinities for the translational machinery. We hypothesize that this isoform-specific mRNA translation may be regulated depending on the cellular needs.
Translational control is predominantly achieved through regulation of initiation (Lackner and Bahler 2008). Cis-acting translational enhancers have been characterized in diverse species and influence either cap-dependent or independent translation (Pfeiffer et al. 2012). Eukaryotic viruses and many cellular mRNAs contain regulatory elements in 5′ UTRs such as internal ribosome entry sites and tandem oligopyrimidine elements that regulate translation initiation under a variety of cellular conditions (Jefferies et al. 1997; Jackson 2005). One possibility is that alternative splicing alters the topology of these cis-acting regulatory elements, resulting in isoform-specific fates (Sanford et al. 2004; Staudacher et al. 2011). Another possibility is that multifunctional cis-acting elements, such as the FN1 EDA splicing enhancer, may directly couple splicing decisions to translational control (Lu and Cullen 2003; Nott et al. 2004; Sanford et al. 2004; Barberan-Soler et al. 2009). There is also new evidence supporting the idea of functional microRNA targets sites within coding exons (Hausser et al. 2013). This model, if true, provides an elegant mechanism(s) for coupling alternative splicing with isoform-specific mRNA translation. An alternative model, not considered here, is that cis-elements regulating isoform-specific translation function in auto-regulatory feedback loops as described for the TYMS and DHFR mRNAs (Lin et al. 2000; Tai et al. 2004).
RNA secondary structures can also regulate translational control of mRNA isoforms (Bonnal et al. 2003; Goodarzi et al. 2012). A prime example of this type of regulation occurs during the localization of the Oskar mRNA to the posterior pole of the Drosophila oocyte. Splicing of the first intron of the Oskar pre-mRNA generates a secondary structure that is required for precise localization of the mRNA. This stem–loop structure encompasses the exon–exon junction and creates binding sites for a variety of trans-acting factors (Hachet and Ephrussi 2004; Yano et al. 2004; Ghosh et al. 2012). It is not known if similar coupled-regulatory mechanisms exist in mammalian genomes. It will be interesting to determine if local structural elements in alternatively spliced exons (both the “skipped” and “included” products) influence translation initiation.
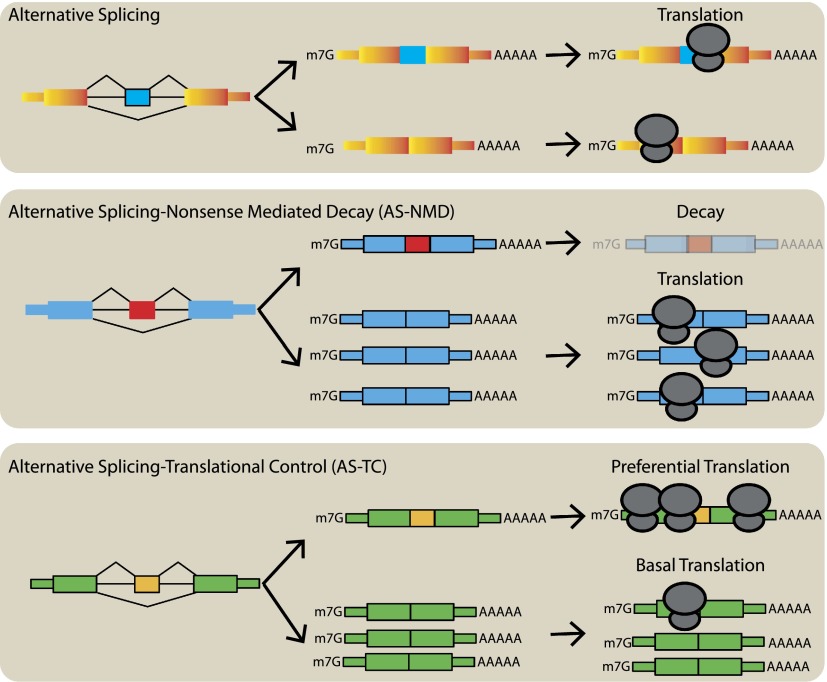
We think our data support a “coupled fate hypothesis,” suggesting that alternative splicing and/or mRNP assembly pathways in the nucleus can result in different fates for messages (Fig. 6); this may result in active translation, sequestration or mRNA decay (Dreyfuss et al. 2002). We propose that alternative splicing is not only important for expanding the protein coding potential of the human genome but also for altering the cis-regulatory composition of mRNAs leading to an unexpected level of post-transcriptional regulation in the cytoplasm.
Figure 6.
Alternative splicing generates mRNA isoforms with diverse cytoplasmic fates. Alternative splicing expands the protein coding capacity of human genes (upper panel). Alternative splicing can also be coupled to nonsense mediated decay (AS-NMD, middle panel). This mechanism can attenuate gene expression post-transcriptionally by producing unstable PTC-containing isoforms. In this paper, we present a novel mode of splicing-based gene regulation linking alternative splicing and translational control (AS-TC, lower panel) of the resultant mRNA isoforms. Our data suggest the intriguing hypothesis that alternative splicing of both coding and noncoding exons alters the cis-regulatory landscape of mRNA isoforms leading to different levels of polyribosome association.
Methods
Cell culture, fractionation, and polyribosome profiling
Human embryonic kidney (HEK) 293 T cells were cultured in complete medium (DMEM 10% FBS) at 37°C, 5% CO2. Prior to lysis, cells were treated with cycloheximide (100 μg/mL), 10 min at 37°C 5%CO2. Cells were washed three times with ice cold PBS and lysed in ice cold buffer A (0.5% NP40, 20 mM Tris HCl pH 7.5, 100 mM KCl and 10 mM MgCl2). Lysates were passed three times through a 23G needle and incubated on ice for 7 min. Extracts were then centrifuged at 10K rpm for 7 min at 4°C. The supernatant was collected as crude cytosolic extract. Cytosolic extracts were overlaid on 10%–50% sucrose gradients prepared in 20 mM Tris HCl pH 7.5, 100 mM KCl and 10 mM MgCl2 buffer (prepared using the Gradient Station, Biocomp Instruments). Gradients were then ultracentrifuged at 40K rpm for 1h 20 min at 4°C using an SW41 in a Beckman ultracentrifuge. Individual polyribosome fractions were subsequently purified using a Gradient Station (Biocomp Instruments). RNA was purified from both cytosolic and polyribosomal fractions using TRI Reagent LS (Sigma), converted to double stranded libraries using the TruSeq polyA+ kit (Illumina) and sequenced using the Illumina HiSeq2000 platform.
High-throughput data analysis
High-throughput RNA sequencing was performed at the UCSC Genome Sequencing Center, producing ∼50–100M reads per sample (Supplemental Table 3). Mapping of single-end ∼35-bp reads was performed using Bowtie and TopHat (Langmead et al. 2009; Trapnell et al. 2009) to the hg19 build of the human genome (February 2009 GRCh37, NCBI Build 37.1). Mapping parameters are listed in Supplemental Table 1. All data collection and parsing was performed with the Perl programming language and statistical analyses with the R statistical language, version 2.14.1. All external library packages used are available on CPAN or CRAN, respectively.
Estimation of alternative splicing
We used the Mixture of Isoforms (MISO) Bayesian Inference model v1.0 for quantification of alternative RNA processing events (Katz et al. 2010). The MISO algorithm counts the numbers of reads that are common to both events and the reads that are exclusive to one isoform or the other, in order to estimate the percent-spliced-in (Ψ) in a given sample. For a given RNA processing event, we calculated ΔΨ by subtracting Ψ from two different samples. We employed an “event-centric” approach, considering annotated alternative mRNA processing events with at least 20 exclusive reads and a maximum confidence interval width of 0.45 for the MISO estimated posterior distribution of percent-spliced-in (“Psi”, Ψ, Supplemental Table 5 lists all of the MISO outputs). The MISO events database was downloaded from the MISO website (http://genes.mit.edu/burgelab/miso). Orientation of Ψ vs. 1-Ψ represents the included vs. the excluded isoform for alternative 5′ and 3′ splice sites (A5SS, A3SS), skipped exons (SE), and retained introns (RI). For alternative first and last exons (AFE, ALE), Ψ and 1-Ψ refer to the proximal and distal exon sets, respectively. In the case of the analysis of functional elements in Figure 5, the orientation of AFE, ALE, and MXE events may be switched such that the functional element in question is contained within the Ψ isoform rather than 1-Ψ. Classification into 5′ UTR, CDS, and 3′ UTR events was based upon the alternative region's orientation with respect to the March 18th, 2012, update of human NCBI Consensus CDS database (NCBI Build 37.3) (ftp://ftp.ncbi.nlm.nih.gov/pub/CCDS/).
False discovery rate calculations
To empirically estimate the FDR based on biological reproducibility, we considered the filtered events common to both replicates. For each value of x in 0.01 to 0.3 increasing by 0.01, we perform 100 bootstrapped label permutations of replicate 2. For each of the 100 shuffled sets, we determined the number of events where both replicates have at least a BF > 5, and |ΔΨ| > x, and following the same direction of change. The FDR for each of the 100 permutations of each value of x was then estimated as FP/(FP + TP), which is the number of permuted events passing the criterion divided by the number of real events passing the criterion. This was plotted against the number of true positives, the number of real events passing the criterion minus the number of permuted events.
Reverse transcription-polymerase chain reaction
RNA was purified from cytoplasmic extracts and polyribosome fractions using TRI Reagent LS (Sigma) following manufacturer's instructions. cDNA synthesis was performed on each fraction using random hexamers (Applied Biosystems) following manufacturer's instructions. PCR was performed using Taq Titanium (Clontech). For cytoplasm to polyribosme comparisons, rate of inclusion was calculated using the following formula: %Inclusion= Included/(Skipped+Included) in nmol as determined using an Agilent 2001 Bioanalyzer. Statistics were done using a Mann-Whitney one-tailed t-test.
Oligos
MOV10 FOR: GGCGACTTTCAGTTTCATTTCCAC
MOV10 REV: TCTTGGCTAGCAGTGACTTGTGG
THOC5 FOR: GCTCCGGGACTGAGGTGCT
THOC5 REV: CTCGGTGTCAGATCGATTCCG
DHX9 FOR: CAGAAGAAGTGGATTTAAATGCTGG
DHX9 REV: CCCAGCTGCTTGATATAAATGGTC
KARS FOR: GGAAGATGGCGGCCGTGCA
KARS REV: CTCTTCCTCAGGACCCACAC
HNRNPR FOR: TGCTGCCGGGATCGCGTCT
HNRNPR REV: AGTCACTTTCCTTGAACTGCTGTAG
SGOL1 FOR: GCAAATTCCTCTTGAAGAAACTGAAC
SGOL1 REV: AGGGTGGGCTCCTTATAGTTCAC
NDUFS1 FOR: ATATGTTAAGGATACCTGTAAGAAAGGC
NDUFS1 REV: GGGCTTTCTCAATTTCAACAAGGC
DDX50 FOR: AATGCCTGGGAAACTCCTCT
DDX50 REV: TGGTGCCTTGACTTCCTTCT
DGUOK FOR: CTCTCCATCGAAGGCAACAT
DGUOK REV: TTTCCAAGACTTTGGGCAGT
Data access
The Frac-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE45237.
Acknowledgments
We thank Al Zahler and Demian Cazalla for thoughtful comments on the manuscript. We thank Manny Ares, David Coombs (Biocomp Instruments), Anders Krogh, and the Center for Computational Applied Transcriptomics (University of Copenhagen and Danish Council for Strategic Research) for helpful discussion. We thank Doyle Coyne for technical contributions to this work. This project was supported by grants from the NIH (GM085121) and the Ellison Medical Foundation to J.R.S. Partial funding for Open Access was provided by the UC Santa Cruz Open Access Fund.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.148585.112.
Freely available online through the Genome Research Open Access option.
References
- Barberan-Soler S, Lambert NJ, Zahler AM 2009. Global analysis of alternative splicing uncovers developmental regulation of nonsense-mediated decay in C. elegans. RNA 15: 1652–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal S, Boutonnet C, Prado-Lourenco L, Vagner S 2003. IRESdb: The Internal Ribosome Entry Site database. Nucleic Acids Res 31: 427–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ 2013. Dynamic integration of splicing within gene regulatory pathways. Cell 152: 1252–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM 2008. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet 40: 1416–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B, Kislinger T, Emili A 2005. Integrating gene and protein expression data: Pattern analysis and profile mining. Methods 35: 303–314 [DOI] [PubMed] [Google Scholar]
- Delestienne N, Wauquier C, Soin R, Dierick JF, Gueydan C, Kruys V 2010. The splicing factor ASF/SF2 is associated with TIA-1-related/TIA-1-containing ribonucleoproteic complexes and contributes to post-transcriptional repression of gene expression. FEBS J 277: 2496–2514 [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N 2002. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3: 195–205 [DOI] [PubMed] [Google Scholar]
- Fu X, Fu N, Guo S, Yan Z, Xu Y, Hu H, Menzel C, Chen W, Li Y, Zeng R, et al. 2009. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics 10: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfman S, Burstein D, Penn O, Savchenko A, Amit M, Schwartz S, Pupko T, Ast G 2012. Changes in exon-intron structure during vertebrate evolution affect the splicing pattern of exons. Genome Res 22: 35–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Marchand V, Gáspár I, Ephrussi A 2012. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat Struct Mol Biol 19: 441–449 [DOI] [PubMed] [Google Scholar]
- Gong C, Maquat LE 2011a. “Alu”strious long ncRNAs and their role in shortening mRNA half-lives. Cell Cycle 10: 1882–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE 2011b. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Najafabadi HS, Oikonomou P, Greco TM, Fish L, Salavati R, Cristea IM, Tavazoie S 2012. Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature 485: 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin TJ, Gygi SP, Ideker T, Rist B, Eng J, Hood L, Aebersold R 2002. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics 1: 323–333 [DOI] [PubMed] [Google Scholar]
- Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, et al. 2009. UTRdb and UTRsite (RELEASE 2010): A collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res 38: D75–D80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A 2004. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 428: 959–963 [DOI] [PubMed] [Google Scholar]
- Han J, Xiong J, Wang D, Fu XD 2011. Pre-mRNA splicing: Where and when in the nucleus. Trends Cell Biol 21: 336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser J, Syed AP, Bilen B, Zavolan M 2013. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res 23: 604–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Moore MJ 2012. The spliceosome: A flexible, reversible macromolecular machine. Trends Biochem Sci 37: 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT 2010. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol 470: 119–142 [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ 2005. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans 33: 1231–1241 [DOI] [PubMed] [Google Scholar]
- Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G 1997. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J 16: 3693–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, et al. 2006. Global survey of organ and organelle protein expression in mouse: Combined proteomic and transcriptomic profiling. Cell 125: 173–186 [DOI] [PubMed] [Google Scholar]
- Kylberg K, Bjorkroth B, Ivarsson B, Fomproix N, Daneholt B 2008. Close coupling between transcription and exit of mRNP from the cell nucleus. Exp Cell Res 314: 1708–1720 [DOI] [PubMed] [Google Scholar]
- Lackner DH, Bahler J 2008. Translational control of gene expression from transcripts to transcriptomes. Int Rev Cell Mol Biol 271: 199–251 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Z, Wang L, Yamaga S, Bonds W, Beazer-Barclay Y, Kluger Y, Gerstein M, Newburger PE, Berliner N, Weissman SM 2001. Genomic and proteomic analysis of the myeloid differentiation program. Blood 98: 513–524 [DOI] [PubMed] [Google Scholar]
- Lin X, Parsels LA, Voeller DM, Allegra CJ, Maley GF, Maley F, Chu E 2000. Characterization of a cis-acting regulatory element in the protein coding region of thymidylate synthase mRNA. Nucleic Acids Res 28: 1381–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cullen BR 2003. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9: 618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek T, Valasek L, Pospisek M 2011. Polysome analysis and RNA purification from sucrose gradients. Methods Mol Biol 703: 293–309 [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Sanford JR, Cáceres JF 2008. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell 30: 179–189 [DOI] [PubMed] [Google Scholar]
- Nashchekin D, Masich S, Soop T, Kukalev A, Kovrigina E, Nashchekina O, Daneholt B 2007. Two splicing isoforms of the Y-box protein ctYB-1 appear on the same mRNA molecule. FEBS J 274: 202–211 [DOI] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr 2007. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Le Hir H, Moore MJ 2004. Splicing enhances translation in mammalian cells: An additional function of the exon junction complex. Genes Dev 18: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Truman JW, Rubin GM 2012. Using translational enhancers to increase transgene expression in Drosophila. Proc Natl Acad Sci 109: 6626–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani AK, Calarco JA, Pan Q, Mavandadi S, Wang Y, Nelson AC, Lee LJ, Morris Q, Blencowe BJ, Zhen M, et al. 2011. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res 21: 342–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Caceres JF 2004. Pre-mRNA splicing: Life at the centre of the central dogma. J Cell Sci 117: 6261–6263 [DOI] [PubMed] [Google Scholar]
- Sanford JR, Gray NK, Beckmann K, Cáceres JF 2004. A novel role for shuttling SR proteins in mRNA translation. Genes Dev 18: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MW, Houseman A, Ivanov AR, Wolf DA 2007. Comparative proteomic and transcriptomic profiling of the fission yeast Schizosaccharomyces pombe. Mol Syst Biol 3: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudacher K, Baldea I, Kisselbach J, Staudacher I, Rahm AK, Schweizer PA, Becker R, Katus HA, Thomas D 2011. Alternative splicing determines mRNA translation initiation and function of human K2P10.1 K+ channels. J Physiol 589: 3709–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet CW, Srinivasan K, Clark TA, O'Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, et al. 2006. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol 2: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai N, Schmitz JC, Chen TM, Chu E 2004. Characterization of a cis-acting regulatory element in the protein-coding region of human dihydrofolate reductase mRNA. Biochem J 378: 999–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Abreu Rde S, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO 2010. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Cody NA, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, et al. 2012. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 150: 710–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm D, Huang E, Svingen T, Stanfield S, Dinnis D, Koopman P 2006. Comparative proteomic analysis to study molecular events during gonad development in mice. Genesis 44: 168–176 [DOI] [PubMed] [Google Scholar]
- Yano T, López de Quinto S, Matsui Y, Shevchenko A, Ephrussi A 2004. Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev Cell 6: 637–648 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Krainer AR 2004. Involvement of SR proteins in mRNA surveillance. Mol Cell 16: 597–607 [DOI] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD 2009. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell 35: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]