Abstract
Our body habitat-associated microbial communities are of intense research interest because of their influence on human health. Because many studies of the microbiota are based on the same bacterial 16S ribosomal RNA (rRNA) gene target, they can, in principle, be compared to determine the relative importance of different disease/physiologic/developmental states. However, differences in experimental protocols used may produce variation that outweighs biological differences. By comparing 16S rRNA gene sequences generated from diverse studies of the human microbiota using the QIIME database, we found that variation in composition of the microbiota across different body sites was consistently larger than technical variability across studies. However, samples from different studies of the Western adult fecal microbiota generally clustered by study, and the 16S rRNA target region, DNA extraction technique, and sequencing platform produced systematic biases in observed diversity that could obscure biologically meaningful compositional differences. In contrast, systematic compositional differences in the fecal microbiota that occurred with age and between Western and more agrarian cultures were great enough to outweigh technical variation. Furthermore, individuals with ileal Crohn's disease and in their third trimester of pregnancy often resembled infants from different studies more than controls from the same study, indicating parallel compositional attributes of these distinct developmental/physiological/disease states. Together, these results show that cross-study comparisons of human microbiota are valuable when the studied parameter has a large effect size, but studies of more subtle effects on the human microbiota require carefully selected control populations and standardized protocols.
Targeting our indigenous human microbial communities (microbiota) to prevent or treat disease is difficult due to their complexity, as well as their intra- and interpersonal variations (Lozupone et al. 2012b). Major efforts are underway to understand the predominant factors that shape the human gut microbiota and the inter-relationships between the organismal composition of the microbiota, its pool of microbial genes (microbiome), their expressed functions, and host physiologic and disease phenotypes. In the case of the gut, which contains the largest collection of microbes, these factors and interrelationships include diet (Muegge et al. 2011; Wu et al. 2011; Yatsunenko et al. 2012), host genetic and familial relationships (Turnbaugh et al. 2009; Hansen et al. 2011; Yatsunenko et al. 2012), varying cultural traditions and geography (De Filippo et al. 2010; Hehemann et al. 2010; Yatsunenko et al. 2012; Zupancic et al. 2012), age (Palmer et al. 2007; Biagi et al. 2010; Koenig et al. 2011; O'Sullivan et al. 2011; Yatsunenko et al. 2012), pregnancy (Koren et al. 2012), route of delivery (Huurre et al. 2008; Dominguez-Bello et al. 2010), obesity, metabolic syndrome, and type II diabetes (Ley et al. 2005; Turnbaugh et al. 2009; Qin et al. 2010; Graessler et al. 2012; Vrieze et al. 2012), cardiovascular disease (Wang et al. 2011), disturbances produced by antibiotics (Jakobsson et al. 2010; Dethlefsen and Relman 2011) including Clostridium difficile colitis (Chang et al. 2008; Khoruts et al. 2010; Gough et al. 2011), and other forms of inflammatory bowel diseases (Willing et al. 2010).
Bacteria dominate our various microbial communities. The composition of these communities is typically evaluated by targeting the bacterial 16S rRNA gene as a phylogenetic marker. Trends in community-level diversity differences can be interrogated by computing the amount of diversity that is shared between samples (β-diversity), followed by clustering using an unsupervised multivariate statistical technique such as Principal Coordinates Analysis (PCoA). These techniques sometimes reveal clear associations between subject characteristics and overall diversity. Strong drivers of gut microbial community relatedness include age (Koenig et al. 2011; Yatsunenko et al. 2012), culture/geography (Yatsunenko et al. 2012), inflammatory bowel disease (IBD) (Willing et al. 2010), and kinship (Dicksved et al. 2008; Turnbaugh et al. 2009; Yatsunenko et al. 2012).
Because different studies of the human microbiota often use the same 16S rRNA gene target, studies performed by different research groups can in principle be compared, and parallels among different disease, physiological, or developmental states discovered. Such comparative analyses have yielded key insights when applied to 16S rRNA gene libraries generated by different laboratories focusing on a number of environmental habitats; for example, these comparisons have revealed that salinity is an important factor structuring the bacterial diversity in free-living communities (Lozupone and Knight 2007) and that bacterial communities in the vertebrate gut are highly divergent from free-living communities (Ley et al. 2008). Analysis of sequences from different studies also allows comparisons to relevant control populations. For example, pregnant women differ in the composition of their gut microbiota between the first and third trimester (Koren et al. 2013), and third but not first trimester composition was shown to be distinctive from nonpregnant adults by comparison to the healthy reference data set sequenced by the NIH-sponsored Human Microbiome Project (HMP) (The Human Microbiome Project Consortium 2012).
Particularly in comparisons restricted to a specific type of sample (e.g., only from human fecal samples), technical differences in experimental protocols between laboratories, including the manner in which samples are obtained and stored, DNA extraction methods, the selection of PCR primers for generating amplicons from bacterial 16S rRNA genes, the region of the 16S rRNA gene targeted for PCR, and the instruments used to determine the nucleotide sequences of these amplicons, might all produce variability that could outweigh biological differences (Mao et al. 2012). Here, we conducted meta-analyses to identify overall patterns that drive differences in the human microbiota and to ascertain the degree to which technical variability between studies impacts observed diversity.
Results
Differences between human body sites are greater than those produced by technical variation
We compared sequences generated from different regions of the 16S rRNA gene by using a reference mapping protocol for Operational Taxonomic Unit (OTU) assignment, in which sequences from different regions of the 16S rRNA gene will map to the same full-length reference sequence if they are from the same species (see Methods). In short, we picked OTUs on the February 4, 2011 Greengenes database, which is composed of all available near full-length 16S rRNA gene sequences in GenBank, at a 97% threshold using UCLUST (Edgar 2010), and then assigned 16S rRNA fragments to a reference sequence if they were within a 97% threshold. A 97% similarity threshold is typically used to denote bacterial species. Although many different techniques are available for identifying relationships between the overall microbiota compositions in different samples once OTU counts per sample have been calculated, we chose to use unweighted UniFrac and PCoA because of its successful application in previous meta-analyses (Lozupone and Knight 2007; Ley et al. 2008; Lozupone et al. 2012a), and also after verifying that unweighted UniFrac performed well compared with other beta diversity measures in clustering human microbiome samples by body site (see Supplemental Materials). UniFrac evaluates the distance between two samples based on the degree to which the 16S rRNA sequences are from unique versus shared phylogenetic lineages (Lozupone and Knight 2005).
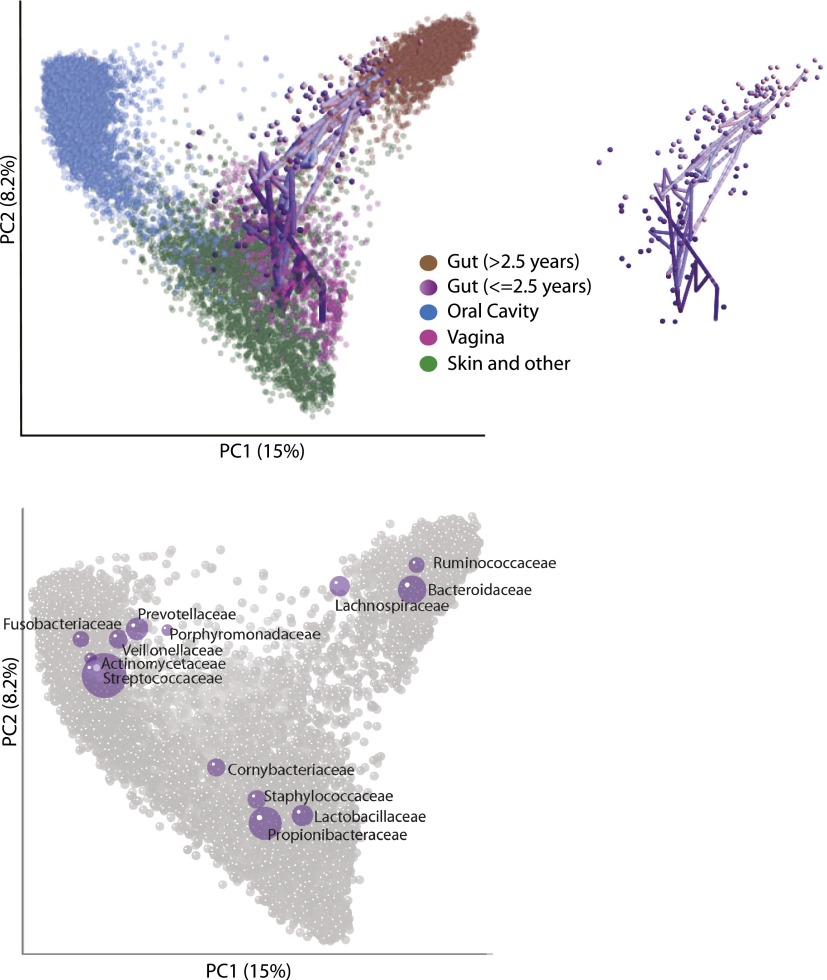
When combining data from 12 different published studies of the human microbiota (Table 1), including three studies of multiple individuals that sequenced multiple body sites from the same person over time (HMP, Costello_whole_body, and Dense_timeseries; see Table 1), the samples clustered primarily by body site rather than by study (Fig. 1). This was the case despite considerable differences in experimental protocols across the studies, including different DNA extraction methods, sequencing platforms, primers, and regions of 16S rRNA targeted (Table 1). One notable exception to the broad clustering by body site was that samples from the gut of infants generally clustered with samples from the adult vagina or skin rather than the adult gut (Fig. 1). This was observed for all four studies that collected fecal samples from infants and age information for study participants (Table 1; US_infant_timeseries, Global_gut, Italy/Burkina Faso, Newborns_and_mothers) (Fig. 1). Furthermore, fecal samples collected from a single USA infant in US_infant_timeseries (Table 1) progressed from the adult vaginal region of the PCoA plot to the adult fecal region over the first 2½ yr of life (Fig. 1).
Table 1.
Figure 1.
Unweighted UniFrac PCoA plot illustrating that samples from the human microbiome cluster primarily by body site. Each point represents a sample from one of the studies detailed in Table 1. Samples were classified broadly as from the Gut (mostly feces but also colon, ileum, and rectum), vagina, oral cavity (e.g., saliva, tongue, cheek), and skin and other (diverse skin sites, hair, nostril, and urine). Gut samples from individuals older than 2½ yr are colored brown and from individuals ages 0 to 2½ yr are colored across a dark purple (0 yr) to light purple (2½ yr) spectrum. Samples from one infant sampled repeatedly over the first 2½ yr of life are joined together with a purple line with a decreasingly dark hue with age. The infant samples are also shown in the inset. The most abundant bacterial families are superimposed on the same PCoA plot in the lower panel in purple. The size of the sphere representing a taxon is proportional to the mean relative abundance of the taxon across all samples.
One way to explore taxonomic driving factors of patterns in a PCoA analysis is to produce biplots, where bacterial taxa are plotted in the same PCoA space based on the weighted average of the PCoA coordinates of all samples, where the weights are the relative abundances of the taxon in the samples. By plotting bacterial families in this way (Fig. 1) we show that clustering by body site is driven by taxa that have previously been shown to characterize the different body sites, such as an abundance of Ruminococcaceae, Bacteroidaceae, and Lachnospiraceae in the adult gut, Lactobacillaceae in the vagina, Propionibacteraceae/Staphylococcaceae on the skin, and Streptococcaceae/Prevotellaceae in the mouth (Costello et al. 2009; The Human Microbiome Project Consortium 2012).
Age and geography/culture drive major clustering patterns across studies of the gut (stool) microbiota
We found strong clustering of the gut microbiota by study when comparing fecal samples alone, suggesting that technical differences between laboratories cause significant differences in the observed diversity. However, some host factors produced sufficiently large and characteristic changes in the gut microbiota to drive global clustering patterns even when combining studies that used diverse protocols. Age was especially important, with a progression toward an adult-like state over the first 3 yr of life, explaining the first principal coordinate when three studies with age gradients were combined (Fig. 2A). These studies assessed the fecal microbiota in individuals between the ages of 0 and 83 yr living in North America (USA), Africa (Malawi), and in South America (Amazonas State of Venezuela) (Global_gut) (Table 1), children aged 0–6 yr from Europe and Africa (Italy/Burkina Faso), and a single infant in the USA sampled repeatedly from age 0 to 2.3 yr and its mother (US_infant_timeseries). The effect of age transcended any differences introduced by experimental protocol in these particular studies, including different sequencing platforms (454 and Illumina) or PCR primers (e.g., those targeting the V2 and V4 hypervariable regions of the bacterial 16S rRNA gene) (Table 1).
Figure 2.
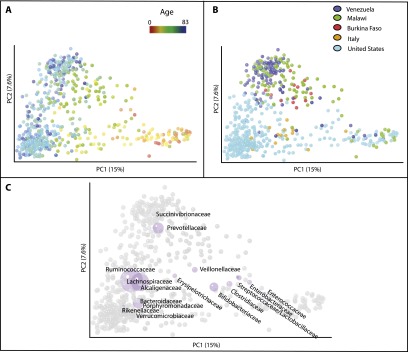
Unweighted UniFrac PCoA plots illustrating the relative degree to which age, cultural/geographic stratification, systematic differences in the collection of samples, and sequencing method affect the observed diversity of the gut microbiota. (A–C) Data from three different studies with age gradients from culturally diverse populations (Global_gut, US_infant_timeseries, and Italy/Burkina Faso) (Table 1). Points are colored by age gradient in A or by county in B. C plots the most abundant bacterial families as a weighted average of the coordinates of all samples in purple, where the weights are the relative abundances of the taxon in the samples. The size of the sphere representing a taxon is proportional to the mean relative abundance of the taxon across all samples.
The observed age gradient across these three studies was associated with a transition from communities enriched in Enterococcaceae, Enterobacteraceae, Streptococcaceae, Lactobacillaceae, Clostridiaceae, and Bifidobacteraceae in early age, followed by a progression to communities enriched in Lachnospiraceae, Ruminococcaceae, Bacteroidaceae, and Prevotellaceae (among others) in adults (Fig. 2C). This result is consistent with taxa previously reported to differ during development of the human fecal microbiota in early life (Stark and Lee 1982; Palmer et al. 2007; Dominguez-Bello et al. 2010; Koenig et al. 2011; Yatsunenko et al. 2012).
Characteristic differences between “Western” and agrarian cultures were also large enough to outweigh study effects when combining these three studies. The fecal microbiota of USA children from two different studies (US_infant_timeseries, Global_gut) clustered along the second principal coordinate with children from Italy from another study (Italy/Burkina Faso) (Fig. 2B; Table 1). Similarly, samples from the Malawian and Amerindian children from Global_gut clustered with children from Burkina Faso from Italy/Burkina Faso at the opposite end of the same axis (Fig. 2B). Taxa biplot analysis indicated that this difference was associated with an enrichment of Prevotellaceae in the adults from agrarian cultures and Bacteroidaceae in adults from Western cultures (Fig. 2C). This observation is consistent with the independent reports of enrichment of Prevotella in fecal samples from individuals living in non-Western societies in two of the studies included in this analysis (De Filippo et al. 2010; Yatsunenko et al. 2012).
Samples from adults in Western populations cluster by study
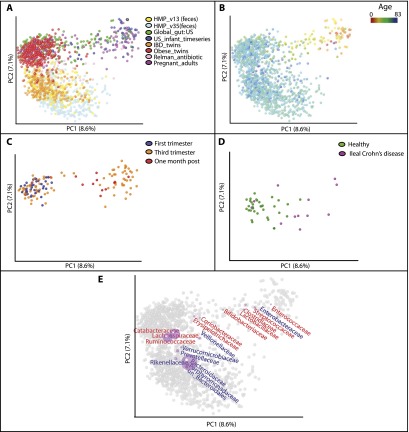
In contrast to the striking difference in microbiota across the age gradient and between Western and non-Western societies, the fecal samples from Western adults clustered primarily by study (Fig. 3A). Although study-based clustering dominated many different data sets, we illustrate this phenomenon here with the following seven studies of individuals living in the USA or Sweden (Table 1): (1) a reference population of healthy US adults characterized by the HMP (using primers targeting the V3–5 region of bacterial 16S rRNA genes and the V1–3 region for a subset of the samples; HMP_V13, HMP_V15); (2) individuals with IBD and healthy controls (IBD_twins); (3) three healthy adults whose microbiota was sampled before, during, and after two short periods of voluntary consumption of the antibiotic ciprofloxacin (Antibiotic_timeseries); (4) obese and lean mono- and dizygotic twin pairs and their mothers (Obese_twins); (5) healthy individuals who were 14–66 yr old from Global_gut; (6) healthy US individuals who were 14–66 yr old from a study comparing microbiota within and between families (Family_study); and (7) healthy US adults sampled at four different timepoints (Healthy_whole_body) (Table 1). The strength of study-driven clustering was unexpected given that some of the individual studies had identified factors that were driving community differences, such as antibiotic administration (Dethlefsen and Relman 2011). In Figure 3B, which clusters samples from the study of twins discordant for IBD only (IBD_twins), several individuals with ileal Crohn's disease deviate strongly from healthy controls (Fig. 3B), and yet their deviation from healthy people sequenced in other studies is clearly confounded by study effects (Fig. 3A).
Figure 3.
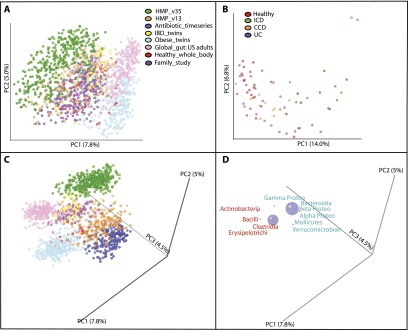
Unweighted UniFrac PCoA plots illustrating a strong study effect when comparing fecal samples of Western adults. (A) Studies conducted with Western adult populations. (B) Clustering of the fecal samples from IBD_twins (Table 1) colored by disease state. (ICD) Ileal Crohn's disease, (CCD) Colonic Crohn's Disease, (UC) Ulcerative Colitis. (C,D) Same as in A but with the axes rotated to maximize clustering by study. D shows just the bacterial orders as a weighted average of the coordinates of all samples, where the weights are the relative abundances of the taxon in the samples. The size of the sphere representing a taxon is proportional to the mean relative abundance of the taxon across all samples. Gram-positive bacterial orders are labeled in red text and Gram-negative in blue.
Although many different experimental parameters could have an effect, these results reinforce the well-known point that choice of PCR primers used for amplification of different regions of the bacterial 16S rRNA gene is important; HMP samples that were subjected to the same extraction and storage protocols, but amplified with different primers cluster separately. Plotting the bacterial orders in the same plot, and rotating the plot of the first three PC axes to maximize study-based clustering (Fig. 3 C,D) showed that the studies that targeted the V1–3/V2 region of rRNA (Antibiotic_timeseries, HMP_V13, Healthy_whole_body, Family_study, and Obese_twins) tended to have an enrichment in Erysipelotrichi and Verrucomicrobia and a depletion in Actinobacteria and Gamma proteobacteria compared with studies that targeted the V3–5/V4 region of rRNA (HMP_V35 and Global_gut). Consistent with a depletion of Actinobacteria in the V2 studies, Antibiotic_timeseries, Obese_twins, Healthy_whole_body, and Family_study used the 27F forward PCR primer (Table 1), which has previously been shown to have three primer mismatches and poor amplification of the Bifidobacteriales group, the dominant type of Actinobacteria in the human gut (Frank et al. 2008; van den Bogert et al. 2011).
It is also clear, however, that primer choice is not the only factor in study-based clustering. For instance, the two included studies from the laboratory of Dr. Jeffrey Gordon (Obese_twins and Global_gut) cluster closer to each other than to other studies even though they used PCR primers that targeted different regions of 16S rRNA (V2 and V4) and sequenced using different platforms (454 GS FLX [Roche] and Illumina HiSeq 2000). Biplot analysis indicates that this separation is associated with an enrichment of Gram-positive and depletion of Gram-negative bacteria in the Gordon lab studies (Fig. 3 C,D). This finding may be due to differences in the DNA extraction protocol utilized at the different sites (i.e., a Phenol:Chloroform:Isoamyl alcohol-based technique [P:C:I] by the Gordon lab and primarily the MO BIO PowerSoil kit by the others) (Table 1), suggesting that the (P:C:I) technique, which includes a bead-beating step, may be more efficient at extracting DNA from Gram-positive bacteria. Cell wall architecture correlates with this study bias independently of phylogeny; there is an enrichment in Gram-positive Erysipelotrichi in the Gordon lab data sets and depletion of the phylogenetically related Mollicutes, which have lost an ancestral Gram-positive cell-wall trait during the course of genome reduction (Bove 1993).
The sequencing platform also appeared to be a driver of study-based clustering. Healthy adult fecal samples from Healthy_whole_body and Family_study clustered apart even though they used the same PCR/sequencing primers and DNA extraction protocol (Table 1). Samples from Family_study, which sequenced using the Illumina HiSeq platform, clustered closer to Global_gut (the one other study that used this sequencing platform) than did samples from Healthy_whole_body, which used 454 GS FLX (Fig. 3C). Clustering by a sequencing platform is consistent with reports of GC-related bias in data sequenced on the Illumina platform (Ratan et al. 2013).
Comparison of infant development to physiological and disease states in adults
Although study effects dominated the clustering pattern of studies of Western adults by adding samples from US infants (including samples from a single infant sampled continuously over the first 2.3 yr of life) (Koenig et al. 2011) and the US infants from a survey conducted across age and cultures (Yatsunenko et al. 2012), we found that an age gradient was again observable over PC1 (Fig. 4B), although systematic differences across studies were still observable on PC2 (Fig. 4A). These systematic biases were again related to the research group who conducted the studies, with reports from the lab of one of the co-authors of this article (Jeffrey Gordon) (Obese_twins, Global_gut) and the lab of Ruth Ley (Pregnant_adults, US_infant_timeseries) having high values for PC2, and the remaining studies low. This separation was again associated with an enrichment of Gram-positive bacteria in the Ley and Gordon labs (Fig. 4E).
Figure 4.
Unweighted UniFrac PCoA plots illustrating the relationship between the bacterial diversity in fecal samples from different disease/physiologic states in adults and the infant microbiome. (A) Compares samples from same studies as in Figure 3A, but with the US_infant_timeseries, the US infants and children from Global_gut, and adults from a study of pregnancy (Pregnant_adults) (Table 1) added in addition. (A–D) The same plot, except that different subsets of the samples are shown or are colored differently. (A) Points colored by study. (B) Points colored by an age gradient. The samples from Pregnant_adults are not shown because the age of study participants was not available. (C) Samples from pregnant women in their first and third trimesters and 1 mo post-delivery (from Pregnant_adults) (Table 1) (D) Healthy individuals and individuals with ileal Crohn's disease from IBD_twins. (E) Bacterial families are plotted as a weighted average of the coordinates of all samples where the weights are the relative abundances of the taxon in the samples (purple circles). Gram-negative bacterial orders are in blue text, Gram-positive in red.
With the addition of US infants to the analysis, parallels between infant development and physiological and disease states become apparent. Specifically, a large subset of pregnant women in their third trimester, but not their first trimester, cluster with infants rather than adults (Fig. 4C); this phenotype generally persisted 1 mo after delivery. Samples from individuals with ileal Crohn's disease also resembled infant samples to a greater degree than healthy control samples from the same study (Fig. 4D).
We again added bacterial families to the same PCoA plot based on their average relative abundance across the samples to explore which are driving factors of community-level variation along these PC axes. Essentially, the same pattern of taxa turnover is shown as in the age gradient in Figure 2, with a transition from communities enriched in Enterococcaceae, Enterobacteraceae, Streptococcaceae, and Lactobacillaceae in early age, followed by a progression to communities enriched in Lachnospiraceae, Ruminococcaceae, and Bacteroidaceae (Fig. 4F). The enrichment of these infant-associated taxa in individuals with ileal Crohn's disease and in third trimester pregnancy is consistent with analyses of taxa associated with these states reported in the original publications. For instance, individuals with ileal Crohn's disease had significantly greater Enterobacteraceae and Lactobacillaceae (infant-associated) in their fecal microbiomes and significantly less Lachnospiraceae and unclassified Bacteroidales (adult-associated) compared with healthy controls (Willing et al. 2010). Similarly, OTUs within the Enterobacteraceae, Streptococcaceae, and Enterococcaceae families (infant-associated) were enriched, and several OTUs within the Lachnospiraceae and Ruminococcaceae families (adult-associated) were depleted in third versus first trimester pregnant women (Koren et al. 2012).
Overall, this clustering pattern suggests that the gut environment in infants, in late pregnancy, and with ileal Crohn's disease may share biological or physiological characteristics that result in the selection of some of the same types of bacteria. Although these characteristics are not fully understood, the parallel between ileal Crohn's disease, which is characterized by high inflammation in the gut and third trimester pregnancy is interesting in light of the finding that stool samples from pregnant women in their third trimester had significantly higher levels of the proinflammatory cytokines IFNG, IL2, IL6, and TNF compared with first trimester, and inflammatory markers were increased in gnotobiotic mice into which a third but not first trimester gut microbiota had been transferred (Koren et al. 2012).
An additional way to gain insight into the driving factors of parallel microbiota changes is to determine the shared biological properties of the diverse bacteria that associate independently with infants and with disease or with late pregnancy. For instance, particular taxa in the Lachnospiraceae group such as Roseburia and Eubacterium have decreased relative abundance in infants, in individuals with ileal Crohn's disease (Willing et al. 2010) and in late pregnancy (Koren et al. 2012). Within the Clostridiales, however, there are also species such as Clostridium bolteae that show an opposite pattern, thriving in infants and with various disturbances and having relatively low prevalence in the healthy adult gut (Lozupone et al. 2012a). Genomic comparisons of these species revealed that infant/disturbance-adapted taxa in the Lachnospiraceae had a selection for genes predicted to confer resistance to osmotic and oxidative stress as well as distinctive metabolic capabilities (Lozupone et al. 2012a).
Discussion
Our results demonstrate that differences in the human microbiota across body sites are sufficiently large that samples from each site cluster together even when experimental protocols differ substantially. For comparisons within gut (fecal) samples, we found that compositional differences associated with age and culture/geography also were greater than those driven by the experimental protocols used in the particular studies combined here, which included differences in the sequencing platform, the region of 16S rRNA targeted, and the DNA extraction technique used. However, the strong clustering by study in fecal samples from Western adults indicates that differences in experimental protocol, including choice of PCR primers/16S rRNA region targeted, DNA extraction protocol, and sequencing platform can be associated with significant differences in the observed diversity. Experimental protocols must thus be carefully standardized for studies conducted within populations and age groups, especially when the effects of a biological parameter on the (gut) microbiota are expected to be subtle. As more data sets with different combinations of PCR primers, 16S rRNA region, DNA extraction protocol, and sequencing platform are made available, it should be possible to develop a better understanding of which techniques produce particular types of bias in the resulting data and the optimal techniques for minimizing these biases.
The large effect of age on the human gut microbiota shown here is consistent with studies that have detailed dramatic changes in the gut community composition over the first 3 yr of life (Palmer et al. 2007; Koenig et al. 2011; Yatsunenko et al. 2012). Studies of the early assembly of the gut microbiota, whether longitudinal (Koenig et al. 2011) or cross-sectional within the USA (Palmer et al. 2007) or across countries representing distinct cultural traditions (Yatsunenko et al. 2012), indicate that the infant microbiota undergoes a successional trajectory until a relatively stable adult-like configuration is established. Our results further show that the early gut microbiota more closely resembles other body sites, including the vagina and skin, than the fecal microbiota of Western adults. This is consistent with studies of the effects of delivery mode on the microbiota of newborns, which showed that the vaginally delivered babies had microbiota resembling the mother's vagina across multiple body sites and babies born by C-section had an initial microbiota resembling skin (Dominguez-Bello et al. 2010). However, given that the progression of the infant gut microbiota to one resembling an adult occurs slowly over the first 1–3 yr of life, during which time exposures to gut microbes occur, the trajectory from infant adult gut microbiota is likely not solely due to a replacement of initial colonizers. Another possible contributor is shared biological or physiological characteristics between the vagina and infant gut, such as more similar environmental stressors, which results in the selection of some of the same types of bacteria.
The separation of the gut microbiota of Western cultures (USA) (Koenig et al. 2011; Yatsunenko et al. 2012) and Italy (De Filippo et al. 2010) from the gut microbiota of individuals in the developing world (Burkina Faso) (De Filippo et al. 2010), Malawi (Yatsunenko et al. 2012), and Venezuela (Yatsunenko et al. 2012), supports the conclusion that strong and consistent differences exist between the gut microbiota of individuals in Western compared with agrarian cultures. The clustering of fecal samples from individuals from Burkina Faso, Malawi, and Venezuela is consistent with independent reports of enrichment of Prevotella in fecal samples from individuals living in non-Western societies compared with Italy (De Filippo et al. 2010) or the US (Yatsunenko et al. 2012). The finding that Prevotella could predict community-wide diversity patterns is not surprising given the observation that a trade-off between Prevotella and Bacteroides are associated with overall stratification of diversity both within Western populations and in cross-cultural comparisons (Arumugam et al. 2011; Wu et al. 2011; Yatsunenko et al. 2012; Koren et al. 2013).
In this work, sequences from studies that targeted different non-overlapping regions of the 16S rRNA gene were related via a reference-sequence mapping protocol, in which sequences were only considered if they were related to near full-length sequences in the Greengenes database (McDonald et al. 2012). This protocol allows for comparison of sequences generated from different regions of the 16S rRNA gene, because sequences from different regions from the same microbial species will map to the same full-length reference sequence. However, it limits the analysis to taxa related to those that have been observed before in studies that generated long sequence reads, and these studies have become less common recently, since next generation sequencing produces only short reads. Because the human body is among the most intensely sampled microbial habitats on Earth, a high percentage (76.77%–94.95%) of raw sequences from each study were successfully assigned to reference sequences at a 97% similarity threshold (Table 1), and re-analysis of the same data using these reference-mapping techniques readily reproduced published results. The continued expansion of the full-length 16S rRNA sequences from a diversity of environments will make the reference-sequence mapping protocol more powerful, particularly for habitats whose microbial communities are diverse and not deeply surveyed. Using the MIMARKS standards (Minimum Information about a MARKer gene Sequence) (Yilmaz et al. 2011) and the QIIME database (http://www.microbio.me/qiime), comparative analysis of large numbers of samples processed by many different researchers should help determine factors that structure microbial communities in a variety of systems, and further investigation of which experimental parameters produce the largest biases in microbial diversity estimates.
Methods
The studies included in these meta-analyses, which all used 16S rRNA gene sequencing to survey the human microbiota, are detailed in Table 1. PCR-generated amplicons from bacterial 16S rRNA genes were sequenced using a diversity of sequencing platforms across the studies (Illumina HiSeq 2000, 454 Titanium or standard FLX chemistries [Roche]) and primers that targeted different regions of the 16S rRNA gene (Table 1). Raw sequences from each study were processed and quality filtered using the default parameters of QIIME version 1.5.0 (Caporaso et al. 2010). Specifically, for Titanium and FLX (pyrosequencing) data we excluded sequences that were not between 200 and 1000 nucleotides in length, had greater than six ambiguous bases, had homopolymer runs longer than 6 nucleotides, had mismatches in the primer, or that could not be assigned to a sample using the barcode. For Illumina HiSeq data, we truncated the reads after runs of more than three consecutive low-quality base calls and excluded reads with <0.75 of the original read length after truncation. We excluded reads with ambiguous bases after quality trimming. Because 454 Titanium chemistry yields longer read lengths, we trimmed all reads generated with this chemistry to the length achieved with standard FLX chemistry.
We grouped the sequences for all studies into “species-level” operational taxonomic units (OTUs) by using a reference mapping protocol. Specifically, the QIIME database uses a reference data set that is derived from all near full-length 16S rRNA sequences that were available on February 4, 2011 in the Greengenes database (McDonald et al. 2012). Closely related/redundant sequences were removed from this set by selecting OTUs using UCLUST (http://www.drive5.com/usearch/) and a minimum pairwise nucleotide sequence identity threshold between reads of 97% over the full-length sequence. Sequences from the studies included in this meta-analysis were all compared with this same nonredundant reference set and assigned to a reference sequence using the UCLUST reference protocol if they were within a 97% identity threshold. Sequences that did not have ≥97% identity to any of the reference sequences in the Greengenes database were not assigned to OTUs and thus not considered further in these analyses. The average percent of assigned sequences across samples for each study is summarized in Table 1. Samples that did not have at least 100 sequences after quality filtering and OTU assignment (and thus were excluded from all of the PCoA analyses) were also excluded from these calculations.
This reference mapping protocol for OTU assignment allows comparison of sequences from studies that used primers that targeted heterogeneous regions of the 16S rRNA gene, because sequences from the same bacteria will match the same reference sequence regardless of the length or region of the 16S rRNA gene targeted, although in practice a greater degree of similarity will be required if the targeted region of 16S rRNA is particularly variable.
We then performed an unweighted UniFrac analyses on tables of OTU counts. UniFrac performs a pairwise comparison of all communities in a data set, defining the overall degree of phylogenetic similarity between any two communities based on the degree of branch length they share on a bacterial tree of life (Lozupone and Knight 2005). Since sampling depth can impact UniFrac values, and thus clustering patterns (Lozupone et al. 2011), 1000 sequences were randomly selected from each sample before performing the PCoA analyses, except for the analysis in Figure 2 in which 100 sequences per sample were used because of low-sequence counts in the Italy/ Burkina Faso study after filtering out low-quality reads. OTUs were selected and studies collated using the QIIME database (http://www.microbio.me/qiime/). All analyses were carried out using QIIME 1.5 (Caporaso et al. 2010) or in the QIIME database web interface, including the taxa biplots, which can be generated with the make_3d_plots.py script of QIIME. Taxa summaries at the family and order level were performed using the RDP classifier trained on the February 4, 2011 Greengenes 97% reference data set using QIIME.
The 16S rRNA data and all metadata used to conduct these analyses were from previously published studies and are available for download and analysis in the publicly accessible QIIME database (http://www.microbio.me/qiime/).
Acknowledgments
C.L. was supported by the National Institutes of Health (K01DK090285). This work was supported in part by the National Institutes of Health, the Crohns and Colitis Foundation of America, and the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.151803.112.
Freely available online through the Genome Research Open Access option.
References
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. 2011. Enterotypes of the human gut microbiome. Nature 473: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, Nikkila J, Monti D, Satokari R, Franceschi C, et al. 2010. Through ageing, and beyond: Gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE 5: e10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bove JM 1993. Molecular features of mollicutes. Clin Infect Dis (Suppl 1) 17: S10–S31 [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, et al. 2011. Moving pictures of the human microbiome. Genome Biol 12: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB 2008. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile–associated diarrhea. J Infect Dis 197: 435–438 [DOI] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R 2009. Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci 107: 14691–14696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci (Suppl 1) 108: 4554–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicksved J, Halfvarson J, Rosenquist M, Jarnerot G, Tysk C, Apajalahti J, Engstrand L, Jansson JK 2008. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J 2: 716–727 [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci 107: 11971–11975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461 [DOI] [PubMed] [Google Scholar]
- Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ 2008. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol 74: 2461–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough E, Shaikh H, Manges AR 2011. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis 53: 994–1002 [DOI] [PubMed] [Google Scholar]
- Graessler J, Qin Y, Zhong H, Zhang J, Licinio J, Wong ML, Xu A, Chavakis T, Bornstein AB, Ehrhart-Bornstein M, et al. 2012. Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: Correlation with inflammatory and metabolic parameters. Pharmacogenomics J doi: 10.1038/tpj.2012.43 [DOI] [PubMed] [Google Scholar]
- Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, et al. 2011. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci (Suppl 1) 108: 4599–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G 2010. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464: 908–912 [DOI] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huurre A, Kalliomaki M, Rautava S, Rinne M, Salminen S, Isolauri E 2008. Mode of delivery—effects on gut microbiota and humoral immunity. Neonatology 93: 236–240 [DOI] [PubMed] [Google Scholar]
- Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L 2010. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE 5: e9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ 2010. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol 44: 354–360 [DOI] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci (Suppl 1) 108: 4578–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Kling Backhed H, Gonzalez A, Werner JJ, Angenent LT, Knight R, et al. 2012. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150: 470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE 2013. A guide to enterotypes across the human body: Meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol 9: e1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci 102: 11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI 2008. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6: 776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R 2005. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R 2007. Global patterns in bacterial diversity. Proc Natl Acad Sci 104: 11436–11440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R 2011. UniFrac: An effective distance metric for microbial community comparison. ISME J 5: 169–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Faust K, Raes J, Faith JJ, Frank DN, Zaneveld J, Gordon JI, Knight R 2012a. Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles of human gut symbionts. Genome Res 22: 1974–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R 2012b. Diversity, stability and resilience of the human gut microbiota. Nature 489: 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao DP, Zhou Q, Chen CY, Quan ZX 2012. Coverage evaluation of universal bacterial primers using the metagenomic datasets. BMC Microbiol 12: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332: 970–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan O, Coakley M, Lakshminarayanan B, Claesson MJ, Stanton C, O'Toole PW, Ross RP 2011. Correlation of rRNA gene amplicon pyrosequencing and bacterial culture for microbial compositional analysis of faecal samples from elderly Irish subjects. J Appl Microbiol 111: 467–473 [DOI] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO 2007. Development of the human infant intestinal microbiota. PLoS Biol 5: e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratan A, Miller W, Guillory J, Stinson J, Seshagiri S, Schuster SC 2013. Comparison of sequencing platforms for single nucleotide variant calls in a human sample. PLoS ONE 8: e55089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, Caporaso JG, Knights D, Clemente JC, Nakielny S, et al. 2013. Cohabiting family members share microbiota with one another and with their dogs. eLife 2: e00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark PL, Lee A 1982. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the 1st year of life. J Med Microbiol 15: 189–203 [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. 2009. A core gut microbiome in obese and lean twins. Nature 457: 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bogert B, de Vos WM, Zoetendal EG, Kleerebezem M 2011. Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples. Appl Environ Microbiol 77: 2071–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, et al. 2012. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143: 913–916 [DOI] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, et al. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, Jarnerot G, Tysk C, Jansson JK, Engstrand L 2010. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139: 1844–1854 [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. 2012. Human gut microbiome viewed across age and geography. Nature 486: 222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz P, Kottmann R, Field D, Knight R, Cole JR, Amaral-Zettler L, Gilbert JA, Karsch-Mizrachi I, Johnston A, Cochrane G, et al. 2011. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat Biotechnol 29: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupancic ML, Cantarel BL, Liu Z, Drabek EF, Ryan KA, Cirimotich S, Jones C, Knight R, Walters WA, Knights D, et al. 2012. Analysis of the gut microbiota in the old order Amish and its relation to the metabolic syndrome. PLoS ONE 7: e43052. [DOI] [PMC free article] [PubMed] [Google Scholar]