Abstract
In mammals, genome-wide epigenetic reprogramming systems exist in primordial germ cells and zygotes. These reprogramming systems play crucial roles in regulating genome functions during critical stages of embryonic development, and they confer the stability of gene expression during mammalian development. The frequent unexpected loss of progeny from somatic cell nuclear transfer (SCNT) is an ongoing problem. In this study, we used six cloned bovines (named NT-1 to NT-6), which were created by ear fibroblast nuclear transfer and displayed short life spans with multiple organ defects, as an experimental model. We focus here on three imprinted genes (IGF2, H19, and XIST) and four satellite loci (Satellite I, Satellite II, Art2, and VNTR) to investigate their methylation changes. The results revealed that aberrant methylation frequently occurred in the analyzed imprinted genes, but not in the satellite loci, of the cloned bovines. After the bovine fibroblast cells were treated with the 5-aza-2(′)-deoxycytidine (5-Aza-dc) demethylation agent, the methylation percentages of the XIST and H19 putative differentially methylated region (DMR) were significantly decreased (XIST, p<0.01; H19, p<0.05) followed by an increase in their mRNA expression levels (p<0.01). Furthermore, we found that five short-lived cloned bovines (NT-1 to NT-5) exhibited more severe aberrant methylation changes in the three imprinted genes examined than the little longer-lived clone (NT-6) compared with wild-type (WT) cows. Our data suggest that the reprogramming of the methylation-controlled regions between the imprinted genes and satellite loci are differences and may be involved with additional mechanisms that need further elucidation.
Introduction
Somatic cell nuclear transfer (SCNT) has been successfully used to generate genetically identical offspring and can be used to create superior agricultural animals or in medical therapy. However, SCNT animals usually suffer from developmental abnormalities and a low survival rate (Wilmut et al., 1997). Although recent work has demonstrated success in cloning various animal species, many embryonic and growth defects appear in the offspring. Genomic imprinting is a parental-origin expression phenomenon in the mammalian genome (Hitchins and Moore, 2002; Surani et al., 1986). Imprinted genes regulate many aspects of development, such as the growth of the embryo and the placenta (Wolffe and Matzke, 1999). The uniparental expression patterns of imprinted genes are regulated by the differentially methylated regions (DMRs). However, during the SCNT process, the aberrant methylation of the DMRs of imprinted genes often appeared in these cloned animals. Furthermore, the genome-wide methylation of the satellite loci was also altered after SCNT (Kang et al., 2001). The inappropriate reprogramming of the DMR from SCNT is highly correlated with these abnormalities in cloned animals. For example, cloned bovine usually exhibited large offspring syndrome (LOS), which is caused by the dysregulation of IGF2R (Spadafora and Geraci, 1975).
X inactive-specific transcript (XIST) is thought to be important for triggering X chromosome inactivation (XCI) in one X allele of the female genome. Additionally, the transcript level of XIST is responsible for maintaining normal development in the female embryo. DNA hypomethylation of the XIST DMR, which results in biallelic expression, causes death in cloned fetuses (Dindot et al., 2004; Xue et al., 2002). H19 is a maternally expressed gene that is silenced on the paternal allele and is expressed in various mesenchymal and epithelial tissues (Zhang and Tycko, 1992). H19 encodes a noncoding RNA transcript that is spliced and polyadenylated but has no open reading frames. H19 and IGF2 are both regulated by their common cis-acting elements (Sasaki et al., 2000). In some Beckwith–Wiedemann syndrome (BWS) patients, IGF2 is expressed biallelically, resulting in overgrowth syndrome (Weksberg et al., 1993).
The global DNA methylation and histone acetylation are presented in repetitive loci, such as a sequence of the higher repeat satellite I region (Satellite I), a sequence of the higher repeat satellite II region (Satellite II), a sequence near a variable number tandem repeat (VNTR), and a part of the euchromatic repeat sequence of antiretroviral treatment (Art2). In the previous study, the global DNA methylation and histone acetylation both showed aberrant patterns in cloned buffalo embryos (Suteevun et al., 2006). However, the maintenance of methylation was observed in the Satellite I locus during the preimplantation development of normal bovine embryos (Kang et al., 2005). VNTR and Art2 satellites in cloned bovine fetuses showed higher methylation levels than in in vitro–fertilized fetuses (Kang et al., 2001). We expect that during developmental stages in SCNT clones or normal individuals, the reprogramming process may play an important role to maintain the genome-wide methylation system.
In this study, we compared the methylation status between imprinted genes (XIST, IGF2, and H19) and four satellite loci (Satellite I, Satellite II, VNTR, and Art2) in the short life-span cloned bovines.
Materials and Methods
Identification of putative DMR of the bovine genes IGF2, XIST, and H19
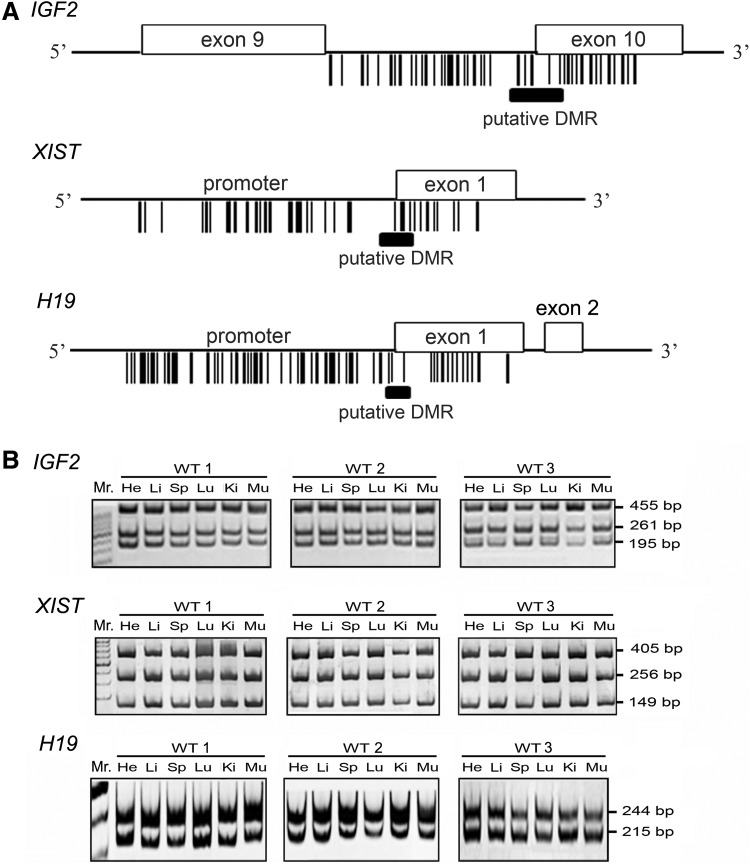
According to previous reports, the putative DMR of IGF2 was predicted to be located within the exon 9–10, and the putative DMR of XIST was predicted to be located within the promoter/exon 1 region (Fig. 1) (Liu et al., 2008; Zhang et al., 2004). A previous study identified that the sperm-specific methylation pattern of H19 within the CpG-rich region of exon 1; therefore, we predicted the putative DMR of H19 to be located in this region (Gebert et al., 2006). To identify the potential DMR of the imprinted genes within the selected regions, we used the online software MethPrimer (http://www.urogene.org/ methprimer) with restrictive conditions (GC content >50.0%, CpG observed/expected >0.6) to plot the distribution of CpG islands (Fig. 1A).
FIG. 1.
The identification of the putative DMR regions of three bovine imprinted genes using density plotting of CpG sites. (A) The defined IGF2 DMR is located in intron 9/exon 10. The defined XIST and H19 DMRs are located in the promoter/exon 1 region. (Open box) gene exon region; (solid bar) putative DMR. The vertical line indicates the CpG sites. (B) The normal methylation patterns of the three imprinted genes in several tissues of three wild-type (WT) bovines. The methylation status of IGF2, XIST, and H19 was assayed by COBRA. He, heart; Li, liver; Sp, spleen; Lu, lung; Ki, kidney; Mu, muscle.
Sample collection from cloned and control bovines
The sex of the six cloned bovines and three control wild-type (WT) bovines was female, and the species of the enucleated oocytes, somatic cell donors, and recipient bovines were all derived from the Holstein breed. The animal used in this study was approved by the Institutional Animal Care and Use Committee (IACUC), National Pingtung University of Science and Technology, Taiwan (Approval No. 98021). Various tissues that had the same genetic background as the wild-type control bovine used for the DNA and RNA extractions were collected immediately from the six cloned bovines after their death (Table 1). The tissues were snap-frozen in liquid nitrogen and stored at −80°C until they were used.
Table 1.
Summary of the Life Span and Organ Defects of the Individual Cloned Bovines
| Cloned bovine | Life span | Defective organs |
|---|---|---|
| NT-1 | <1 hour | Heart, liver, lung, thyroid |
| NT-2 | 5 hours | Heart, liver, kidney |
| NT-3 | <1 hour | Heart, lung, kidney, brain, ovary |
| NT-4 | 1 day | Liver, kidney, thyroid |
| NT-5 | <1 hour | Heart, liver, lung, kidney, thyroid, brain, ovary |
| NT-6 | 191 days | Heart, lung, kidney, muscle |
Cell culture and treatment with the demethylation agent 5-aza-2 ′-deoxycytidine
The bovine fibroblast cells were grown in Dulbecco's modified Eagle's medium (DMEM; GIBCO-BRL, Gaithersburg, MD, USA) that was supplemented with 10% fetal bovine serum (FBS; GIBCO-BRL) and 100 units·mL−1 penicillin-streptomycin (HyClone Laboratories, Losan, UT, USA). The cells were incubated at 37°C in a humidified incubator with 5% CO2. A total of 2.3×106 cells (5th passage) were seeded onto a 6-cm dish. After 24 h, the cells were treated with 5-aza-2′-deoxycytidine (5-Aza-dC; Sigma, St. Louis, MO, USA) at the following concentrations for 5 days: 0 (control), 0.5, and 1.5 μM (Chen et al., 2003; Mohana et al., 2006; Wang et al., 2011). The cultured cells were then harvested for DNA and RNA extraction.
Isolation of genomic DNA
High-quality genomic DNA was extracted using a standard Proteinase K/SDS method, as previously described (Chen et al., 1994; Shen et al., 2012). Briefly, 300 mg of tissue was homogenized and resuspended in 630 μL of lysis buffer (50 mM Tris-HCl, 100 mM NaCl, 100 mM EDTA, pH 8.0), 70 μL of 10% sodium dodecyl sulfate (SDS), 10 μL of RNase A (10 mg·m L−1; Promega, Madison, WI, USA), and 35 μL proteinase K (10 mg·mL−1; Amersco Inc., Cleveland, OH, USA). The tissue digestion was performed at 55°C for 16 h and extracted twice using an equal volume of phenol/chloroform (1:1). An addition of 1.5×volumes of 100% ethanol to the supernatant was used to precipitate the DNA, and the genomic DNA pellet was washed twice with 70% ethanol. The concentration of each DNA sample was calculated using a Nanodrop 2000 spectrophotometer (Thermo Scientific Inc., Bremen, Germany).
DNA methylation analysis by COBRA
For amplification of the repetitive elements and the putative DMR regions of the satellite genes (Satellite I, Satellite II, VNTR, and Art2) and imprinted genes (H19, IGF2, and XIST) in the bovine genome, respectively, PCR reactions were performed using 2 μL of bisulfite-converted genomic DNA as a template (Yang et al., 2003). The primer sets and the COBRA-PCR (Shen et al., 2012) annealing temperature are listed in Table S1 (Supplementary Data are available at www.liebertpub.com/cell).
The PCR products were purified using phenol/chloroform extraction followed by ethanol precipitation. The DNA was resuspended in 8.5 μL of distilled deionized water. The purified PCR products were then digested with 10 units of the appropriate restriction enzyme (New England Biolabs, Beverly, MA, USA) as follows: BstUI for H19, IGF2, and Satellite I at 60°C; HpyCH4IV for XIST at 37°C; AciI for Satellite I and VNTR at 37°C; and TaqI for Art2 at 65°C. The products were electrophoresed on a 6% native acrylamide gel, stained with 200 g·mL−1 ethidium bromide (EtBr), visualized, and quantified on a Kodak 1D imager (Kodak, Japan) (Chen et al., 2008a; Hung et al., 2010).
The definition of hypermethylation or hypomethylation (±10% compared to the average of wild-type tissues from three control bovines) was followed as we previously reported in an analysis of the methylation changes of cloned swine (Shen et al., 2012). The methylation percentages of the examined three imprinted genes and four satellite loci were based on the quantitative COBRA data.
Bisulfite sequencing
For quantification of the methylation status of each CpG site in the DMR regions, the primers were either newly designed or adapted from previous reports (Kang et al., 2001; Li et al., 2005; Lin et al., 2008; Liu et al., 2008; Yang et al., 2005) and are listed in Table S1. The PCR procedures were performed in a total volume of 25 μL for each satellite or imprinted locus. The individual PCR products were purified using phenol/chloroform extraction followed by ethanol precipitation and then cloned into a pGEM T-Easy vector (Promega). The plasmid DNA was isolated using a Mini-M™ Plasmid Purification Kit (Viogene, Taipei, Taiwan) and sequenced using the BigDye™ fluorescent-labeling chain terminator system with an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster, CA, USA).
RNA isolation and quantitative real-time RT-PCR
The total RNA was isolated from homogenized tissues using TRIzol™ (Invitrogen, Carlsbad, CA, USA) as previously described (Tung et al., 2011). One microgram of total RNA was treated with 10 U of RNase-free DNase I (Invitrogen) to degrade any contaminating DNA. The DNase reaction was terminated with heat inactivation at 65°C. First-strand cDNA synthesis was performed using 2 μg of RNA with oligo(dT) primers and Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega) in a total volume of 25 μL (Tsai et al., 2010). Quantitative RT-PCR was conducted using specific sets of primers (Table S1) for each analyzed gene. The mRNA expression levels of each gene in cloned and WT bovine genomes were normalized to the level of GAPDH mRNA and detected by SYBR Green (Sigma). To detect the low expression level of H19, we used a high-resolution melt (HRM) dye (Invitrogen). All real-time RT-PCR reactions were performed using a Rotor-Gene™ 6000 (Qiagen, Valencia, CA, USA) (Chen et al., 2008b).
Histopathological analysis
The organs or tissues were collected from the six cloned bovines immediately after their death. After being fixed with paraformaldehyde and embedded in optimal-cutting-temperature compound (Tissue-Tek; Sakura), the tissues were frozen for microdissection (Yen et al., 2009). For histopathological analysis, tissue slides were stained with Hematoxylin & Eosin and subjected to observation by microscopy and examination by two independent veterinary pathologists.
Statistical analysis
The methylation and expression differences in the imprinted genes were calculated using Student's t-test. The differences between two means were considered significant if p<0.05 (*) or p<0.01 (**). The disruption of the methylation percentage of various tissues in each cloned bovine was shown in a box plot (Gao et al., 2011). The box plot showed the outlier, median, Q1, Q3, IQR (Q3–Q1), the largest value, and the smallest value of the methylation status presenting in various tissues of each cloned individual. Q indicates the quartile number.
Results
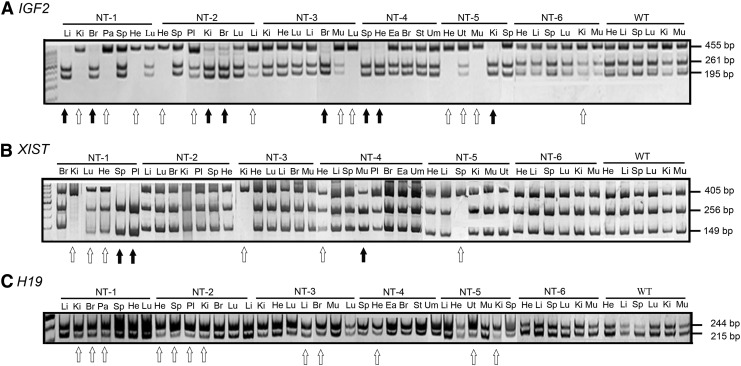
Aberrant methylation profiles of the IGF2, XIST, and H19 imprinted genes in the cloned bovines
The three putative DMRs of IGF2, XIST, and H19 were analyzed as depicted in Figure 1A. The methylation status of these three imprinted genes in three wild-type (WT-1, WT-2, WT-3) control bovine genomes was determined using COBRA, and data showed the perfect imprinting patterns in different analyzed tissues of the three control bovines (Fig. 1B). The putative DMRs of IGF2 and XIST in the cloned bovine genomes showed either hypermethylated (indicated by a black arrow) or hypomethylated (indicated by an open arrow) phenomenon compared to the WT control genomes (Fig. 2A, B). In contrast, the putative DMR of H19 in the cloned bovine genomes was simply a hypomethylated change compared to the WT control genome (Fig. 2C). The putative DMR of IGF2 showed extremely hypermethylated and hypomethylated changes in the most of brain and heart, respectively. The putative DMR of XIST showed extremely hypomethylated phenomenon in some of the kidney tissues.
FIG. 2.
COBRA of the DNA methylation status of three imprinted genes (IGF2, XIST, and H19) in different organs of wild-type (WT) and cloned bovines (NT-1 to NT-6). (A) The methylation pattern of IGF2 (exon 10). (B) The methylation pattern of XIST (exon 1). (C) The methylation pattern of H19 (exon 1). Hypermethylated and hypomethylated statuses are marked as solid arrows and hollow arrows, respectively, and compared with the corresponding loci in the average methylation percentage of control group from three WT bovines. Li, liver; Ki, kidney; Br, brain; Sp, spleen; He, heart; Lu, lung; Pl, placenta; Mu, muscle; Ea, ear; St, stomach; Um, umbilicus; Ut, uterus.
Confirmation of methylation changes in the putative DMR sequences of the imprinted genes
To acquire more detailed information on the methylation status of CpG sites, we performed bisulfite sequencing to determine the complete methylation pattern of the CpG sites. The cytosine methylation profile of the putative DMR of IGF2 exhibited a nearly complete methylation (95.7%) in the NT-1 brain and a mosaic methylation pattern in the NT-4 brain (62.5%). On the contrary, the same putative IGF2 DMR in the NT-2 heart showed a hypomethylation pattern (21.1%; Fig. 3A). The percentage of DNA methylation of the putative DMR of XIST was 2.4% in the NT-3 kidney, 50.1% in the NT-4 spleen, and 70.5% in the NT-1 spleen (Fig. 3B). XIST was hypermethylated in the NT-1 spleen but was hypomethylated in the NT-3 kidney compared with WT heart tissue (50%). The DNA methylation status of the H19 DMR showed normal imprinting patterns of methylation, with 53.3% in NT-2 muscle, 50.8% in NT-1 lung, 50.8% in NT-2 ear, and 57.8% in WT muscle; particularly, the distribution of methylation in these CpG sites showed intermittent spots (Fig. 3C). The maintenance of inherited imprinting marks on the imprinted H19 gene suggests that DNA methylation is relative stable in this region. The methylation changes of putative DMRs of IGF2 and XIST showed extremely wide variation.
FIG. 3.
The bisulfite sequencing results of IGF2, XIST, and H19 DMR regions in different organs of wild-type (WT) and cloned bovines. (A) The methylation profiles of 28 CpG sites within the putative DMR region of IGF2. (B) The methylation profiles of 17 CpG sites within the putative DMR region of XIST. (C) The methylation profiles of 12 CpG sites within the putative DMR region of H19. Scores for the percent methylation (%) in each DMR region were obtained from 10 sequencing results of individual PCR colony clones. The open and closed circles represent the unmethylated and methylated CpG sites, respectively. The score of WT group was represented as an average methylation percentage from three wild-type bovine.
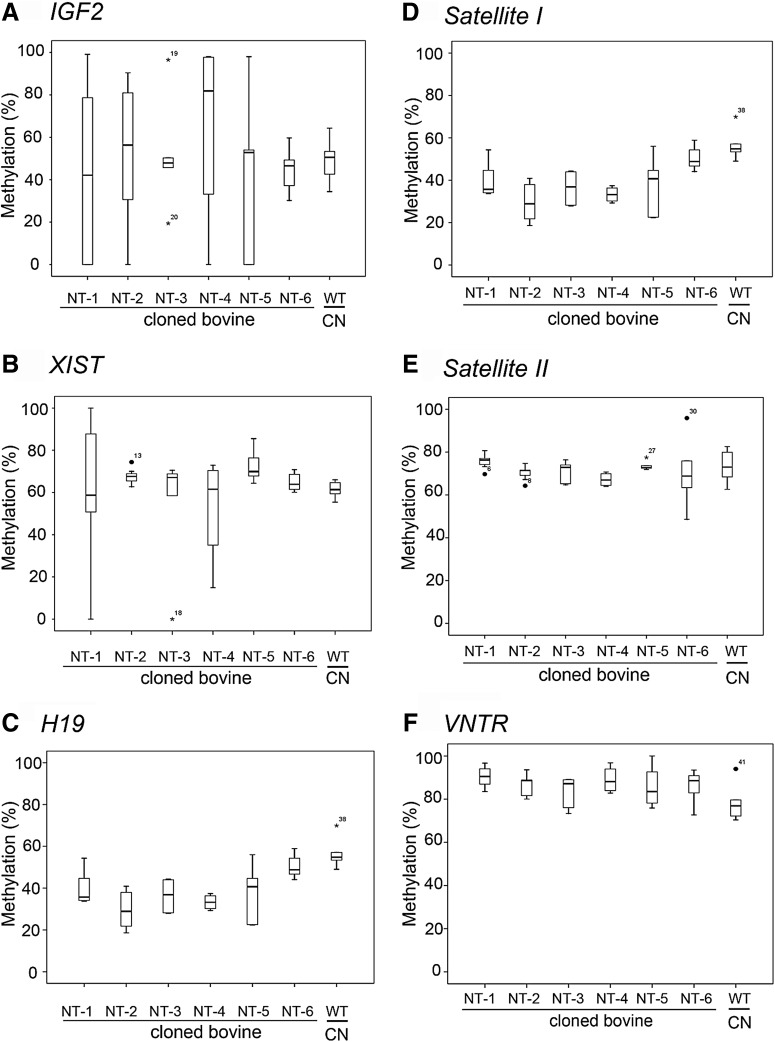
Distribution of methylation levels on imprinted and satellite genes in various tissues of each individual cloned bovine
The methylation profiles of the individual cloned bovines showed differential changes in methylation percentages in various tissues. The cloned bovines NT-1, NT-2, NT-4, and NT-5 all showed higher dispersal of methylation percentages on the IGF2 DMR region compared with the WT control (Fig. 4A). The XIST putative DMR regions of NT-1 and NT-4 (Fig. 4B) and also the H19 putative DMR region of NT-5 (Fig. 4C) displayed higher dispersal of methylation percentages than their WT control. In contrast, the methylation status of these three imprinted genes in NT-6 was more similar to the WT control (Fig. 4A–C).
FIG. 4.
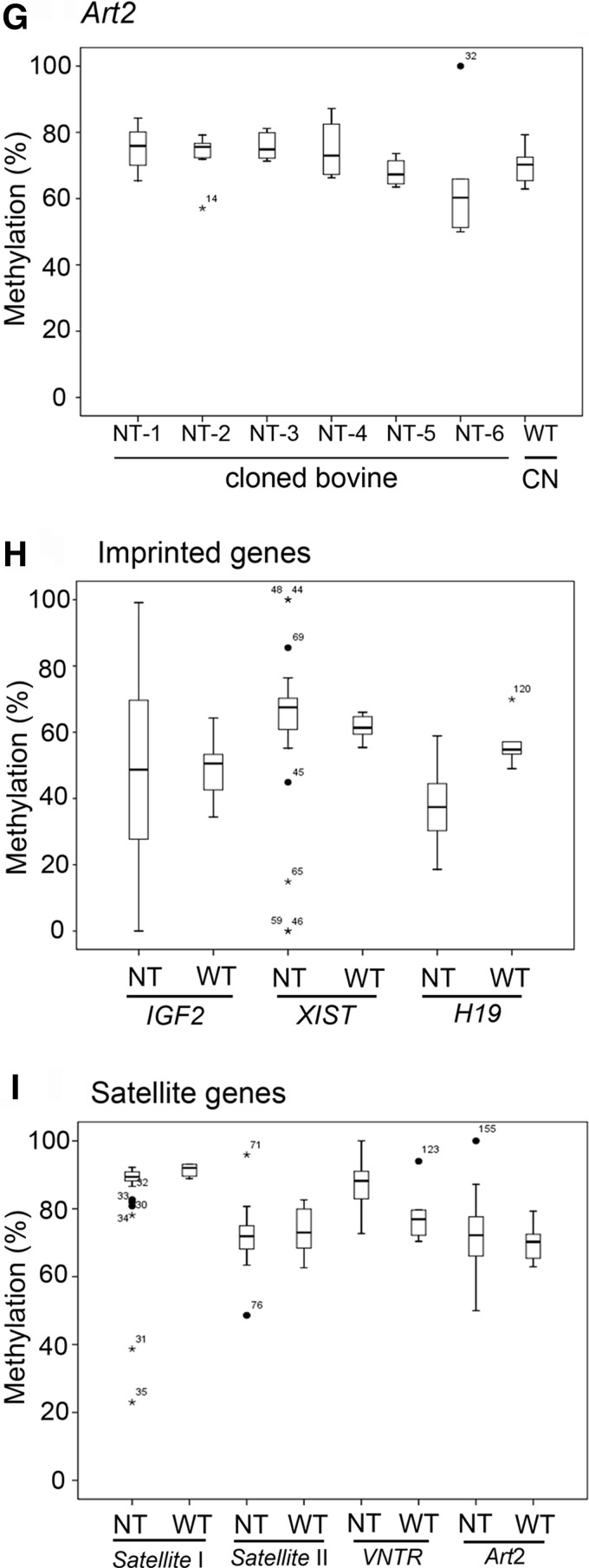
The methylation distribution of imprinted genes and satellite loci form various tissues in each cloned individual. The percent methylation for each of the three imprinted genes showed by box plot: (A) IGF2, (B) XIST, and (C) H19. Each dot represents the percent methylation within each tissue in the cloned bovine. The distribution of the percent of methylation of the satellite loci is shown for (D) Satellite I, (E) Satellite II, (F) VNTR, (G) Art2, (H) imprinted genes, and (I) Satellite genes. The horizontal bar indicates the median ratio of methylation for a cloned individual. CN, control normal bovines (n=3). (*) outer fence >3×interquartile range (IQR)[; (•) inner fence (>1×IQR). The number labeled in outer or inner fences is a far out value in the box plot.
Four satellite loci were used to investigate the methylation statuses of repetitive sequences to determine if the genome-wide methylation was altered by SCNT. Quantitative COBRA was performed, and the data showed that four satellite loci, Satellite I, Satellite II, VNTR, and Art2, displayed no noticeable differences in the methylation percentages of various tissues between the cloned bovines and the WT control genomes (Fig. 4D–G). Overall, the methylation statuses of imprinted genes showed more discordant patterns than the satellite loci in cloned bovines (Fig. 4H, I). Interestingly, we found that five short-lived cloned bovines (NT-1 to NT-5) exhibited more severe aberrant methylation changes in examined three imprinted genes (ranging from 27.8% to 72.2%) than the little longer-lived clone (NT-6; 0.06%) compared with WT cows (Table 2).
Table 2.
Summary of Methylation Change Percentages of the Imprinted Genes in the Total Examined Tissues of Six Cloned Bovines
| |
Methylation changes (%) |
|||
|---|---|---|---|---|
| Cloned bovine individual | IGF2 | XIST | H19 | Total |
| NT-1 | 5/6a | 5/6 | 3/6 | 13/18 (72.2 %) |
| NT-2 | 5/6 | 0/6 | 4/6 | 9/18 (50.0 %) |
| NT-3 | 4/6 | 1/6 | 2/6 | 7/18 (38.9 %) |
| NT-4 | 2/6 | 2/6 | 1/6 | 5/18 (27.8 %) |
| NT-5 | 4/6 | 1/6 | 2/6 | 7/18 (38.9 %) |
| NT-6 | 1/6 | 0/6 | 0/6 | 1/18 (0.06 %) |
| Total | 21/36 (58.3 %) | 9/36 (25.0 %) | 12/36 (33.3 %) | 42/108 (38.9 %) |
Number of aberrant methylation patterns in a total of six examined tissues of each cloned bovine.
The demethylation treatment of bovine ear fibroblast cells is correlated with RNA restoration
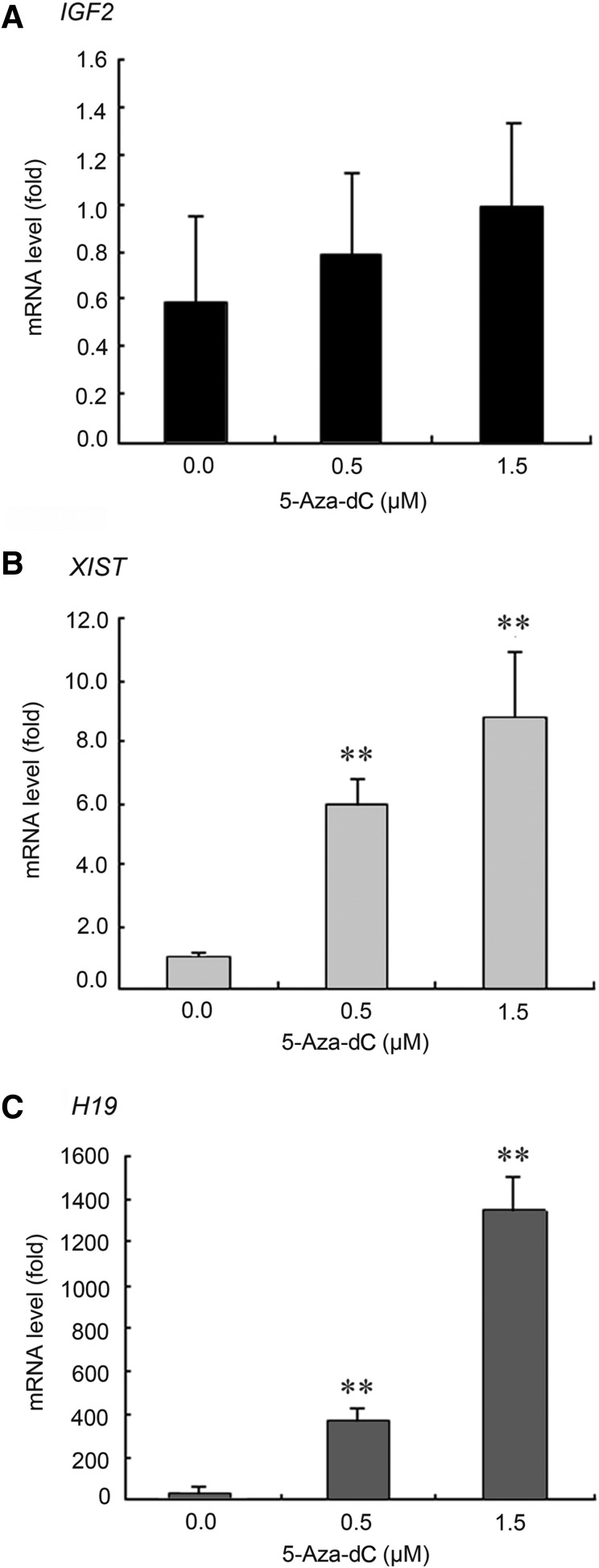
To determine if the expression of IGF2, XIST, and H19 genes is affected by DNA methylation changes in their putative DMRs, we administered different doses of 5-Aza-dC (0, 0.5, and 1.5 μM) to bovine ear fibroblast cells. The methylation percentages of these imprinted genes and satellite loci were significantly decreased after treatment with 0.5- and 1.5-μM doses of 5-Aza-dC (Table 3). Additionally, XIST and H19 exhibited a significant dose-dependent increase in mRNA levels due to 5-Aza-dC treatment (Fig. 5).
Table 3.
Methylation Percentages of the Imprinted Genes and Satellite Loci in Bovine Ear Fibroblast Cells after Treatment with Different Concentrations of 5-Aza-dC
| |
5-Aza-dC concentration |
||
|---|---|---|---|
| Gene | 0 μM | 0.5 μM | 1.5 μM |
| IGF2 | 42.7±1.18a | 36.2±1.81* | 37.1±2.00* |
| XIST | 54.5±1.11 | 46.1±1.20** | 42.9±0.26** |
| H19 | 50.2±2.16 | 22.7±1.15** | 15.2±1.68** |
| Satellite I | 55.5±0.48 | 43.9±0.66** | 45.6±0.31* |
| Satellite II | 71.8±1.42 | 24.2±2.80** | 20.2±1.42** |
| VNTR | 72.1±0.28 | 55.6±1.04** | 62.8±11.05* |
| Art2 | 68.5±0.85 | 11.9±1.18** | 9.4±0.69** |
Methylation percentage was showed as mean±standard deviation (SD) from three individual experiments.
p<0.05; **p<0.01.
FIG. 5.
The RNA expression levels of the imprinted genes in bovine fibroblast cells after 5-Aza-dC treatment. Real-time RT-PCR validation of IGF2 (A), XIST (B), and H19 (C) mRNA expression levels in bovine ear fibroblast cells after treatment with 0 μM, 0.5 μM, and 1.5 μM concentrations of 5-Aza-dC. (*) p<0.05; (**) p<0.01 compared with the untreated (0 μM) control group. All of the data were represented as means±standard deviation (SD) from a three individual repeats.
Organ defects and histopathological analyses of the cloned bovine NT-5
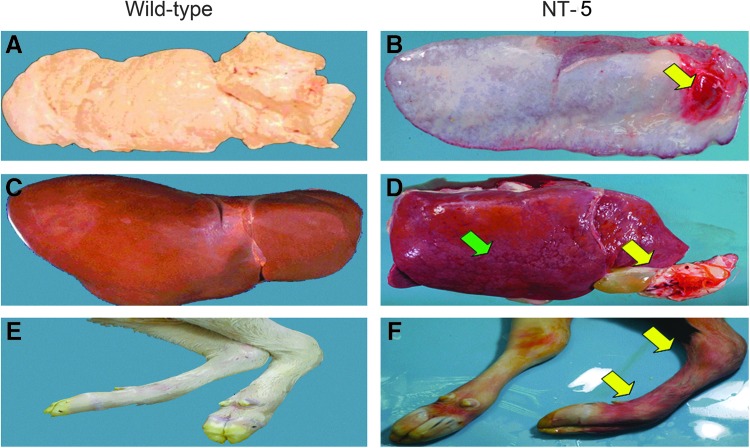
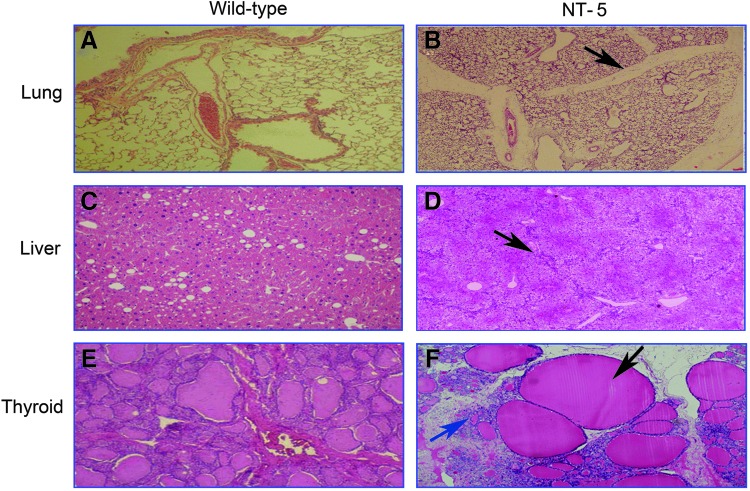
With the exception of NT-6 (191-day life span), most of the cloned bovines used in this study (NT-1 to NT-5) died less than 24 h after birth primarily due to serious defects arising from organ or tissue dysfunction (Table 1). Pathological analysis of the NT-5 cloned bovine revealed bleeding in the spleen (Fig. 6B), a cyst in the gallbladder of liver (Fig. 6D), and a twisted-limb skeleton (Fig. 6F). Furthermore, histological analyses of the NT-5 tissue sections revealed an undifferentiated bronchial and interlobular airway of the lung (Fig. 7B), fibrosis in the hepatic lobules (Fig. 7D), and agenesia (indicated by the blue arrow) or overgrowth (black arrow) of the thyroid tissues (Fig. 7F).
FIG. 6.
Clinicopathological images of the wild-type (WT) bovine and cloned bovine NT-5. Photographs of the pancreas (A), liver (C), and leg (E) indicate normal organ morphology in the WT bovine. (B) Hemorrhagic pancreatitis is indicated by the yellow arrow in the cloned bovine NT-5. (D) An abnormal liver with fibrosis (green arrow) and a cyst in the gallbladder of liver (yellow arrow) are present in the cloned bovine NT-5. (F) The twisted front limb bone (yellow arrow) is shown in the NT-5 skeleton.
FIG. 7.
Histopathological images of the wild-type (WT) bovine and the cloned bovine NT-5. The normal lung section of WT (A) is compared with the hypotrophic airway of NT-5 (B). The normal liver morphology of WT (C) is compared with the undifferentiated hepatic lobule and fibrosis of NT-5 (D). The normal morphology of the thyroid gland in the WT bovine (E) is compared with various developmental disorders of the thyroid gland, including hypertrophic thyroid follicles (black arrow) and agenesia (blue arrow), in the NT-5 bovine (F).
Discussion
The COBRA data showed the aberrant methylation patterns of the IGF2, XIST, and H19 imprinted genes in six cloned bovine genomes. The putative DMRs of IGF2 and XIST frequently exhibited aberrant methylation—either hypomethylation or hypermethylation—in the various tissue samples of the cloned bovines compared to the WT bovines. We also observed a slight change in the methylation levels of the putative DMR of H19 to a hypomethylated pattern (Fig. 2). These results indicate that the methylation change in the putative DMR of IGF2 is likely caused by an abnormal epigenetic reprogramming process rather than through co-regulation with the H19 DMR. The loss of imprinting of the putative XIST DMR results in defective placental growth, as mentioned previously (Xue et al., 2002). Hypomethylation of the XIST DMR leads to biallelic expression and random X-chromosome inactivation, which has also been reported in the previous studies (Dindot et al., 2004; Xue et al., 2002). Specifically, IGF2 controls the growth of the fetus, whereas XIST is involved with placental growth. Our data indicate that there is an aberrant methylation pattern after SCNT that is caused by incomplete epigenetic reprogramming.
IGF2 and H19 are well-known imprinted genes that are expressed on the paternal and maternal alleles, respectively. When the DMR of the H19 paternal allele is highly methylated, the enhancer element will prefer the paternal expression of IGF2; otherwise, H19 expression has a silencing effect on the paternal allele. This cis regulation of the enhancer allows for precise control of IGF2 and H19 expression during embryonic development (Zhang and Tycko, 1992). In this study, IGF2 and H19 were overexpressed, especially the H19 gene, in many tissues of the NT-5 cloned bovine (Fig. S1). The aberrant expression of H19 may cause the misregulation of genes that depend on H19 transcript for proper expression (Gabory et al., 2009; Yang et al., 2005). It has been suggested that the SCNT technique, which uses differentiated somatic cells for donor nuclei that are then transferred into an enucleated oocyte, may disturb the normal reprogramming processes during embryonic development and thus leads to a loss of the appropriate gene regulation. In cloned mice, H19 and IGF2 are usually downregulated in various tissues (Yang et al., 2005). In contrast, cloned bovines frequently exhibit LOS, IGF2 overexpression, and a high neonatal death rate (Farin et al., 2004; Young and Fairburn, 2000). Hypomethylation of the putative DMR between IGF2 and H19 has also been observed in cloned bovines that die immediately after birth (Cezar et al., 2003). In this study, we found that the cloned bovine NT-5 exhibited IGF2 overexpression (Fig. S1). The highly frequent methylation changes of IGF2 DMR were observed in the short-lived cloned bovine tissues (NT-1 to NT-5), but not in the little longer-lived NT-6 cloned bovine (Fig. 2 and Table 2).
In mammals, XIST controls X chromosome inactivation in the female genome (Hendrich et al., 1997). However, a completely hypermethylated or hypomethylated XIST DMR was frequently observed in the cloned bovine genomes that were analyzed in this study (Figs. 2 and 3), which is consistent with a previous study (Liu et al., 2008). We speculate that complete hypomethylation of the XIST DMR in the NT-3 kidney may cause biallelic gene expression, whereas complete hypermethylation in the NT-1 spleen may cause the silencing of both alleles. The loss of control of X chromosome inactivation results in an abnormal regulation of X-linked genes during fetal and placental development (Dindot et al., 2004; Xue et al., 2002). In the previous study, the X-linked gene monoamine oxidase type A (MAOA) showed an aberrant expression pattern in cloned bovines due to a loss of allele-specific expression of XIST (Xue et al., 2002). Furthermore, the hypomethylation of the putative XIST DMR has been suggested to affect the expression of X-linked genes.
The genome-wide epigenetic alterations in cloned bovine fetuses have been previously analyzed by detecting the total 5mC content (Curchoe et al., 2009). Using this approach in the above study, the genomes of the cloned bovines showed no significant differences among the adult clones, lactating clones, or similarly aged lactating cows (Curchoe et al., 2009). Differential methylation modifications of satellite genes have also been studied in different embryonic manipulation processes. These studies revealed that the methylation percentages were 9% of in vitro–fertilized embryos, 3% of parthenogenesis-generated embryos, and 5% of in vivo–fertilized embryos (Kang et al., 2005). Moreover, the artificially generated blastocysts exhibited hypomethylation of satellite genes when compared with their donor cell (72%) (Kang et al., 2001). Global DNA methylation and histone acetylation both showed aberrant patterns in cloned buffalo embryos (Suteevun et al., 2006). However, the maintenance of methylation was observed in the Satellite I locus during the preimplantation development of normal bovine embryos (Kang et al., 2005). We suggest that during developmental stages in SCNT clones or normal individuals, the reprogramming process may act to maintain the genome-wide methylation system. VNTR and Art2 satellites in cloned bovine fetuses showed higher methylation levels than in in vitro–fertilized fetuses (Kang et al., 2001). In this study, we assessed the genome-wide methylation patterns of Satellite I, Satellite II, Art2, and VNTR, and our data showed that there were fewer methylation changes in these satellite loci (Fig. 4) compared with the imprinted genes in cloned bovine genomes.
In previous studies, major defects in the cloned animals were usually accompanied by a certain amount of abnormal gene expression (Humpherys et al., 2001; Long and Cai, 2007; Oishi et al., 2006; Schrader et al., 2003). The gene clusters that have been shown to be misregulated as a result of cloning include imprinted genes (Long and Cai, 2007), X-linked genes (Xue et al., 2002), apoptosis genes (Nakagawa and Yamaguchi, 2005), and other developmentally related genes. Furthermore, the expression of histone acetylation modifiers and transcription factors are also correlated with DNA methylation (Jones and Takai, 2001; Morgan et al., 2005). In this study, we identified abnormal methylation patterns for IGF2, XIST, and H19 in various tissues of cloned bovines. Additionally, we detailed the altered expression of these genes in the very short-lived NT-5 individual (Table S1).
For further confirmation of the correlation between DNA methylation and mRNA expression, we treated bovine ear fibroblast primary cultured cells with different doses of the demethylation agent, 5-Aza-dC, according to the protocol described in a previous study (Magdinier et al., 2000). We also demonstrated that the mRNA expression levels of H19 and XIST, but not IGF2, increased significantly in a 5-Aza-dC dose-dependent manner (Fig. 5). The increasing mRNA expression levels of H19 and XIST genes correlated with a decrease of DNA methylation in their putative DMRs (Table 3). These results indicate that H19 and XIST gene expression may be directly affected by the DNA methylation of their putative DMR. The disparity of imprinting pattern and its mRNA expression between naturally fertilized newborns and the cloned animals is likely to affect by the improper activity of the demethylation machinery in SCNT. This idea is supported by an increasing number of studies that show improvement in development of SCNT embryos through treatment with chromatin-modifying compounds, such as 5-Aza-dC or deacetylase inhibitors, that coordinate epigenetic reprogramming or chromatin remodeling (Peat and Reik, 2012).
In conclusion, we found that the frequent appearance of aberrant methylation profiles was presented in the imprinted genes, IGF2, XIST, and H19, in various tissues from five short-lived cloned bovines (NT-1 to NT-5), but not from a little longer life span NT-6 cloned bovine. The phenomenon showed that the putative DMRs of imprinted genes are more vulnerable than genome-wide satellite loci. Aberrant DNA methylation of imprinted genes increased from abnormal reprogramming caused by SCNT. The cloned bovines exhibited a number of organ defects and abnormal gene expressions. SCNT techniques require optimization to replicate the normal methylation status of DMRs of imprinted genes from embryos to adults. A better understanding of the aberrant methylation that occurs in the genomes of cloned animals may provide insight into improving the efficiency of SCNT and the subsequent health of cloned animals.
Supplementary Material
Acknowledgments
The authors would like to thank Prof. Jiung-Wang Liao for his help with the pathology analysis and our colleagues (Drs. Yu-Tang Tung, Cheng-Wei Lai, and Tung-Chou Tsai) in the Molecular Embryology & DNA Methylation Laboratory for their help with discussions and technical issues. This research was supported by grants NSC-98-2313-B-005-012 and NSC-100-2313-B-005-028-MY3 from the National Science Council, and the Ministry of Education, Taiwan, under the Aiming for Top University (ATU) plan.
Author Disclosure Statement
The authors declare that no competing interests exist.
References
- Cezar G.G. Bartolomei M.S. Forsberg E.J., et al. Genome-wide epigenetic alterations in cloned bovine fetuses. Biol. Reprod. 2003;68:1009–1014. doi: 10.1095/biolreprod.102.010181. [DOI] [PubMed] [Google Scholar]
- Chen C.M. Chen H.L. Hsiau T.H., et al. Methylation target array for rapid analysis of CpG island hypermethylation in multiple tissue genomes. Am. J. Pathol. 2003;163:37–45. doi: 10.1016/S0002-9440(10)63628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.M. Shyu M.P. Au L.C., et al. Analysis of deletion of the integrated human papillomavirus 16 sequence in cervical cancer: A rapid multiplex polymerase chain reaction approach. J. Med. Virol. 1994;44:206–211. doi: 10.1002/jmv.1890440216. [DOI] [PubMed] [Google Scholar]
- Chen H.L. Huang J.Y. Chu T.W., et al. Expression of VP1 protein in the milk of transgenic mice: a potential oral vaccine protects against enterovirus 71 infection. Vaccine. 2008a;26:2882–2889. doi: 10.1016/j.vaccine.2008.03.041. [DOI] [PubMed] [Google Scholar]
- Chen Y.J. Wu C.Y. Chang C.C., et al. Nuclear Kruppel-like factor 4 expression is associated with human skin squamous cell carcinoma progression and metastasis. Cancer Biol. Ther. 2008b;7:777–782. doi: 10.4161/cbt.7.5.5768. [DOI] [PubMed] [Google Scholar]
- Curchoe C.L. Zhang S. Yang L., et al. Hypomethylation trends in the intergenic region of the imprinted IGF2 and H19 genes in cloned cattle. Anim. Reprod. Sci. 2009;116:213–225. doi: 10.1016/j.anireprosci.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Dindot S.V. Farin P.W. Farin C.E., et al. Epigenetic and genomic imprinting analysis in nuclear transfer derived Bos gaurus/Bos taurus hybrid fetuses. Biol. Reprod. 2004;71:470–478. doi: 10.1095/biolreprod.103.025775. [DOI] [PubMed] [Google Scholar]
- Farin C.E. Farin P.W. Piedrahita J.A. Development of fetuses from in vitro-produced and cloned bovine embryos. J. Anim. Sci. 2004;82:E53–E62. doi: 10.2527/2004.8213_supplE53x. [DOI] [PubMed] [Google Scholar]
- Gabory A. Ripoche M.A. Le Digarcher A., et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- Gao W.L. Li D. Xiao Z.X., et al. Detection of global DNA methylation and paternally imprinted H19 gene methylation in preeclamptic placentas. Hypertens. Res. 2011;34:655–661. doi: 10.1038/hr.2011.9. [DOI] [PubMed] [Google Scholar]
- Gebert C. Wrenzycki C. Herrmann D., et al. The bovine IGF2 gene is differentially methylated in oocyte and sperm DNA. Genomics. 2006;88:222–229. doi: 10.1016/j.ygeno.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Hendrich B.D. Plenge R.M. Willard H.F. Identification and characterization of the human XIST gene promoter: Implications for models of X chromosome inactivation. Nucleic Acids Res. 1997;25:2661–2671. doi: 10.1093/nar/25.13.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins M.P. Moore G.E. Genomic imprinting in fetal growth and development. Expert. Rev. Mol. Med. 2002;4:1–19. doi: 10.1017/S146239940200457X. [DOI] [PubMed] [Google Scholar]
- Humpherys D. Eggan K. Akutsu H., et al. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- Hung C.M. Yeh C.C. Chen H.L., et al. Porcine lactoferrin administration enhances peripheral lymphocyte proliferation and assists infectious bursal disease vaccination in native chickens. Vaccine. 2010;28:2895–2902. doi: 10.1016/j.vaccine.2010.01.066. [DOI] [PubMed] [Google Scholar]
- Jones P.A. Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Kang Y.K. Koo D.B. Park J.S., et al. Aberrant methylation of donor genome in cloned bovine embryos. Nat. Genet. 2001;28:173–177. doi: 10.1038/88903. [DOI] [PubMed] [Google Scholar]
- Kang Y.K. Lee H.J. Shim J.J., et al. Varied patterns of DNA methylation change between different satellite regions in bovine preimplantation development. Mol. Reprod. Dev. 2005;71:29–35. doi: 10.1002/mrd.20249. [DOI] [PubMed] [Google Scholar]
- Li S. Li Y. Du W., et al. Aberrant gene expression in organs of bovine clones that die within two days after birth. Biol. Reprod. 2005;72:258–265. doi: 10.1095/biolreprod.104.029462. [DOI] [PubMed] [Google Scholar]
- Lin L. Li Q. Zhang L., et al. Aberrant epigenetic changes and gene expression in cloned cattle dying around birth. BMC Dev. Biol. 2008;8:14. doi: 10.1186/1471-213X-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.H. Yin S. Xiong B., et al. Aberrant DNA methylation imprints in aborted bovine clones. Mol. Reprod. Dev. 2008;75:598–607. doi: 10.1002/mrd.20803. [DOI] [PubMed] [Google Scholar]
- Long J.E. Cai X. Igf-2r expression regulated by epigenetic modification and the locus of gene imprinting disrupted in cloned cattle. Gene. 2007;388:125–134. doi: 10.1016/j.gene.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Magdinier F. Billard L.M. Wittman G., et al. Regional methylation of the 5′ end CpG island of BRCA1 is associated with reduced gene expression in human somatic cells. FASEB J. 2000;14:1585–1594. doi: 10.1096/fj.14.11.1585. [DOI] [PubMed] [Google Scholar]
- Mohana Kumar B. Jin H.F. Kim J.G., et al. DNA methylation levels in porcine fetal fibroblasts induced by an inhibitor of methylation, 5-azacytidine. Cell Tissue Res. 2006;325:445–454. doi: 10.1007/s00441-006-0201-9. [DOI] [PubMed] [Google Scholar]
- Morgan H.D. Santos F. Green K., et al. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005;1:R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Nakagawa T. Yamaguchi M. Overexpression of regucalcin suppresses apoptotic cell death in cloned normal rat kidney proximal tubular epithelial NRK52E cells: Change in apoptosis-related gene expression. J. Cell. Biochem. 2005;96:1274–1285. doi: 10.1002/jcb.20617. [DOI] [PubMed] [Google Scholar]
- Oishi M. Gohma H. Hashizume K., et al. Early embryonic death-associated changes in genome-wide gene expression profiles in the fetal placenta of the cow carrying somatic nuclear-derived cloned embryo. Mol. Reprod. Dev. 2006;73:404–409. doi: 10.1002/mrd.20345. [DOI] [PubMed] [Google Scholar]
- Peat J.R. Reik W. Incomplete methylation reprogramming in SCNT embryos. Nat. Genet. 2012;44:965–966. doi: 10.1038/ng.2393. [DOI] [PubMed] [Google Scholar]
- Sasaki H. Ishihara K. Kato R. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J. Biochem. 2000;127:711–715. doi: 10.1093/oxfordjournals.jbchem.a022661. [DOI] [PubMed] [Google Scholar]
- Schrader A.D. Iqbal M.J. Jones K.L. Gene expression in cloned bovine fetal liver. Cloning Stem Cells. 2003;5:63–69. doi: 10.1089/153623003321512175. [DOI] [PubMed] [Google Scholar]
- Shen C.J. Cheng W.T. Wu S.C., et al. Differential differences in methylation status of putative imprinted genes among cloned swine genomes. PLoS One. 2012;7:e32812. doi: 10.1371/journal.pone.0032812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadafora C. Geraci G. The subunit structure of sea urchin sperm chromatin: A kinetic approach. FEBS Lett. 1975;57:79–82. doi: 10.1016/0014-5793(75)80156-6. [DOI] [PubMed] [Google Scholar]
- Surani M.A. Barton S.C. Norris M.L. Nuclear transplantation in the mouse: heritable differences between parental genomes after activation of the embryonic genome. Cell. 1986;45:127–136. doi: 10.1016/0092-8674(86)90544-1. [DOI] [PubMed] [Google Scholar]
- Suteevun T. Parnpai R. Smith S.L., et al. Epigenetic characteristics of cloned and in vitro-fertilized swamp buffalo (Bubalus bubalis) embryos. J. Anim. Sci. 2006;84:2065–2071. doi: 10.2527/jas.2005-695. [DOI] [PubMed] [Google Scholar]
- Tsai T.C. Lin W. Yang S.H., et al. Granzyme G is expressed in the two-cell stage mouse embryo and is required for the maternal-zygotic transition. BMC Dev. Biol. 2010;10:88. doi: 10.1186/1471-213X-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung Y.T. Chen H.L. Lai C.W., et al. Curcumin reduces pulmonary tumorigenesis in vascular endothelial growth factor (VEGF)-overexpressing transgenic mice. Mol. Nutr. Food Res. 2011;55:1036–1043. doi: 10.1002/mnfr.201000654. [DOI] [PubMed] [Google Scholar]
- Wang Y. Su J. Wang L., et al. The effects of 5-aza-2′-deoxycytidine and trichostatin A on gene expression and DNA methylation status in cloned bovine blastocysts. Cell. Reprogram. 2011;13:297–306. doi: 10.1089/cell.2010.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weksberg R. Shen D.R. Fei Y.L., et al. Disruption of insulin-like growth factor 2 imprinting in Beckwith-Wiedemann syndrome. Nat. Genet. 1993;5:143–150. doi: 10.1038/ng1093-143. [DOI] [PubMed] [Google Scholar]
- Wilmut I. Schnieke A.E. McWhir J., et al. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. Matzke M.A. Epigenetics: Regulation through repression. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- Xue F. Tian X.C. Du F., et al. Aberrant patterns of X chromosome inactivation in bovine clones. Nat. Genet. 2002;31:216–220. doi: 10.1038/ng900. [DOI] [PubMed] [Google Scholar]
- Yang H. Chen C.M. Yan P., et al. The androgen receptor gene is preferentially hypermethylated in follicular non-Hodgkin's lymphomas. Clin. Cancer Res. 2003;9:4034–4042. [PubMed] [Google Scholar]
- Yang L. Chavatte-Palmer P. Kubota C., et al. Expression of imprinted genes is aberrant in deceased newborn cloned calves and relatively normal in surviving adult clones. Mol. Reprod. Dev. 2005;71:431–438. doi: 10.1002/mrd.20311. [DOI] [PubMed] [Google Scholar]
- Yen C.C. Lin C.Y. Chong K.Y., et al. Lactoferrin as a natural regimen for selective decontamination of the digestive tract: recombinant porcine lactoferrin expressed in the milk of transgenic mice protects neonates from pathogenic challenge in the gastrointestinal tract. J. Infect. Dis. 2009;199:590–598. doi: 10.1086/596212. [DOI] [PubMed] [Google Scholar]
- Young L.E. Fairburn H.R. Improving the safety of embryo technologies: possible role of genomic imprinting. Theriogenology. 2000;53:627–648. doi: 10.1016/s0093-691x(99)00263-0. [DOI] [PubMed] [Google Scholar]
- Zhang S. Kubota C. Yang L., et al. Genomic imprinting of H19 in naturally reproduced and cloned cattle. Biol. Reprod. 2004;71:1540–1544. doi: 10.1095/biolreprod.104.031807. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Tycko B. Monoallelic expression of the human H19 gene. Nat. Genet. 1992;1:40–44. doi: 10.1038/ng0492-40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.