Abstract
Embryologists working with livestock species were the pioneers in the field of reprogramming by somatic cell nuclear transfer (SCNT). Without the “Dolly experiment,” the field of cellular reprogramming would have been slow and induced plutipotent cells (iPSCs) would not have been conceived. The major drive of the work in mammalian cloning was the interest of the breeding industry to propagate superior genotypes. Soon it was realized that the properties of oocytes could be used also to clone endangered mammalian species or to reprogram the genomes of unrelated species through what is known as interspecies (i) SCNT, using easily available oocytes of livestock species. iSCNT for cloning animals works only for species that can interbreed, and experiments with taxonomically distant species have not been successful in obtaining live births or deriving embryonic stem cell (ESC) lines to be used for regenerative medicine. There are controversial reports in the literature, but in most cases these experiments have underlined some of the cellular and molecular mechanisms that are incomplete during cell nucleus reprogramming, including the failure to organize nucleoli, silence somatic cell genes, activate the embryonic genome, and resume mitochondrial replication and function, thus indicating nucleus–cytoplasmic incompatibility.
Introduction
The demonstration that the genome of a fully differentiated mammalian cell could be restored to full totipotency with the birth of Dolly (Wilmut et al., 1997) has provided a strong impetus to the area of cellular reprogramming. Following that milestone experiment, a number of mammals from different cell types have been cloned, demonstrating beyond any reasonable doubt that a fully differentiated mammalian genome could be reverted back to an embryonic state, albeit at a low efficiency, through the process later defined as somatic cell nuclear transfer (SCNT). The oocyte is responsible for the reprogramming; indeed, it is programmed to perform this function on the sperm chromatin soon after fertilization (Beaujean et al., 2004) and therefore contains all of the “magic” factors to do so. Some of these factors were later identified and used for cell reprogramming in vitro, leading to the development of induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka, 2006).
From the initial desire of livestock breeders to fill their herds with animals of superior genotype or to preserve endangered breeds and because of its rather low efficiency and peculiar failures, SCNT has become an important subject of investigation of basic biological mechanisms of genome (de)differentiation and a tool to generate specific embryonic stem cells (ESCs) to be used in regenerative medicine (Ogura et al., 2013). Nuclear transfer (nt) ntESCs have been generated in mice (Kishigami et al., 2006) and cattle (Lazzari et al., 2006; Wang et al., 2005) and proved to have a strong proliferation and differentiation potential equal to embryo-derived stem cells.
The availability of oocytes and the related technology of maturation in vitro and culture are central to cell reprogramming by nuclear transfer. Livestock species (mainly cattle and pigs) have been the main unlimited source of high-quality oocytes (Galli and Lazzari, 2008) for in vitro maturation and nuclear transfer experiments. The idea of using livestock or domestic species oocytes across other species has been conceived of since the early days of SCNT, as has the use of frog oocytes (Byrne et al., 2003). The events required for nuclear reprogramming are many and complex, and the assessment of their occurrence has a different stringency as development progresses from oocyte activation to full term development (Oback, 2009). Interspecies (i) SCNT is a way of generating autologous ESCs or cloning endangered or extinct animal species. It provides an extreme case of reprogramming failures from which much can be understood regarding the basic biological mechanisms underlying genome reprogramming.
This article reviews some aspects of iSCNT and outlines some of our work and that of others, examining the problem from an embryologist's perspective.
Achievements of iSCNT Embryo Development
Species that hybridize naturally are more likely to perform well in iSCNT experiments. This is understandable, because the natural production of living hybrid offspring shows that a certain nuclear–cytoplasmic compatibility exists between the two species (Mastromonaco et al., 2007). As a rule, iSCNT in mammals is more efficient when donor and recipient cells are from closely related species. Inter-subspecies SCNT has produced healthy offspring of Boar goat (Jian-Quan et al., 2007) and grey wolf (Kim et al., 2007). Inter-species SCNT embryos derived from mouflon (Ovis orientalis musimon) nucleus donor cells and sheep (Ovis aries) oocytes (Loi et al., 2001) can also develop to term. Wild cat (Felis silvestris lybica) (Gomez et al., 2004) and sand cat (Felis margarita) (Gomez et al., 2008) were produced using domestic cat (Felis catus) oocytes and coyote (Canis latrans) using dog (Canis lupus familiaris) oocytes (Hwang et al., 2012). In 2000, iSCNT embryos derived from gaur (Bos gaurus) adult nucleus donor cells and bovine (Bos taurus) oocytes were able to implant, and fetuses developed up to 200 days (Lanza et al., 2000). At last in 2012, a gaur–bovine offspring was born (Srirattana et al., 2012). Intergenus SCNT embryos derived from leopard cat (Prionailurus bengalensis) nucleus donor cells and domestic cat oocytes were able to implant and form fetuses (Yin et al., 2006).
On the other hand, many studies have reported production of iSCNT morulae and blastocysts when nucleus donor cells and recipient oocytes have had a very distant taxonomical relation, as in the case of interfamily bovine–pig (Dominko et al., 1999; Uhm et al., 2007), interorder cat– and panda–rabbit (Wen et al., 2005), camel– and Tibetan antelope–rabbit (Zhao et al., 2006), human–rabbit (Shi et al., 2008), dog–pig (Sugimura et al., 2009), tiger–pig (Hashem et al., 2007), rhesus monkey (Macaca mulata)–bovine (Kwon et al., 2011), human–bovine (Chang et al., 2003; Illmensee et al., 2006; Li et al., 2008), human–ovine (Hosseini et al., 2012), human–goat (Sha et al., 2009), mouse–pig (Jiang et al., 2011), or interclass chicken–rabbit (Liu et al., 2004) combinations. However, this approach remains ineffective and results are still not always reproducible.
Molecular Aspects of iSCNT Reprogramming
In the majority of the published work, only preimplantation development is described. Moreover, in only a few reports were hybrid embryos generated to address relevant biological questions, such as zygotic genome activation (ZGA) or mitochondrial/genomic DNA composition, to confirm the empirical nature of the experiments (Loi et al., 2011) published so far. However, the accumulated data on preimplantation embryo development of closely and distantly related species opens the wide perspective of deep investigation of cell/genomic organization of the process of early embryonic development as a whole.
Donor nucleus remodeling in recipient ooplasm upon nuclear trasfer
Now it is well understood that the first step of any iSCNT experiments, such us remodeling of donor nucleus in the recipient ooplasm, is very conserved among species and depends on the maternally inherited factors of oocyte cytoplasm. The normal pattern of nucleus remodeling was followed as previously reported in different studies on mammalian nuclear transfer and iSCNT embryos (Arat et al., 2003; Dominko et al., 1999; Lee et al., 2008; Tarkowski and Balakier, 1980; Uhm et al., 2007). Thus, the biochemical mechanism of this process is universal and it works also in intraclass nuclear transfer embryos derived from chicken blastodermal cells and rabbit oocytes (Liu et al., 2004).
Development before embryo genome activation
Nuclear transfer embryos, irrespective of donor nucleus origin, normally develop until the species-specific stage of maternal-to-embryonic transition (MET) that is determined by recipient oocyte (two cells in mouse, four to eight cells in pig, eight to 16 cells in ovine and bovine) and is under maternal control (Schultz, 1993). The preparation of successful embryo genome activation (EGA) fully depends on the ability of recipient oocyte to correctly block the donor cell DNA transcription and corresponding mRNAs translation. The use of metaphase II (MII) oocytes with high levels of maturation-promoting factor (MPF) leads to premature chromatin condensation (PCC), which guarantees the donor cell genome transcriptional silencing after nuclear transfer into the recipient ooplasm.
Silencing of donor nucleus transcription
The data on the absence of transcription of COL6A1 in iSCNT embryos (bovine–pig and pig–bovine) indicate that bovine cytoplasm can block de novo transcription of a fibroblast-specific gene irrespective of the species affiliation of the donor nucleus (Lagutina et al., 2010). This agrees with the results by Green et al. (2007) and Inoue et al. (2006) on the silencing of donor cell-specific genes in nuclear transfer embryos using muscle and hematopoietic cells as nucleus donors and with the statement by Vassena et al. (2007) that the donor genome is markedly silenced by the ooplasm at the one-cell stage of nuclear transfer embryo development. Conversely, the expression of avian feather KERATIN in chicken–rabbit intraclass nuclear transfer embryos (Liu et al., 2004) as early as in eight-cell embryos is an example of the inability of mammalian ooplasm to correctly reprogram an avian tissue-specific gene. The transcriptome analysis of eight- to 16-cell-stage rhesus monkey–bovine iSCNT embryos (Wang et al., 2011) using Affymetrix gene chips demonstrated that more than 7700 somatic genes were downregulated in iSCNT embryos. However, there was vast inability of recipient oocyte to silence about 860 rhesus monkey somatic genes, among which were Col1A1, Col3A1, and Col4A1 involved in the process of collagen production in fibroblasts. It should be mentioned that abnormal fibroblast-specific gene expression was also found, although to a lower extent in bovine SCNT embryos. These results suggest that neither iSCNT nor SCNT embryos can effectively silence the donor cell–specific genes, a phenomenon known as epigenetic memory (Ng and Gurdon, 2005), which can contribute to developmental failures (Wang et al., 2011). These data support the findings of Vassena et al. (2007), who used microarrays to analyze the transcriptome of mouse SCNT embryos at the time of EGA (two-cell stage) and found a large number of genes misexpressed. All of these findings demonstrate that recipient cytoplasm more likely cannot exactly reprogram donor nucleus.
Maternally inherited mRNA destruction
It is known that remnants of maternal RNA can detrimentally affect embryonic development after EGA (Paynton et al., 1988). This is why EGA and further embryo development require the degradation of maternal RNA (Alizadeh et al., 2005). Wang et al. (2011) monitored the degradation of the maternal RNA global profile and found broader maternal RNA degradation in SCNT embryos than in iSCNT embryos. Gdf9 was shown (Alizadeh et al., 2005) to be rapidly degraded along with c-mos and tissue plasminogen activator (tPA) soon after fertilization did not degrade in iSCNT embryos. As one of multiple reprogramming steps, faulty degradation of maternal RNA in iSCNT embryos could potentially be one of the causes of low reprogramming efficiency in iSCNT (Wang et al., 2011). The possibility of successful MET and EGA of SCNT and iSCNT embryos depends on the extent of damage that could produce this cocktail of mRNA and their protein products of improperly reprogrammed somatic genes and maternal non-degraded transcripts.
RNA polymerase II activity
In eukaryotes, RNA polymerase (Pol) II is responsible for transcription of mRNAs and of most of the small nuclear RNAs. RNA Pol II accumulation and activity through detection of polyadenylated (poly) mRNA accumulation in the nuclei was studied in bovine and porcine in vitro fertilization (IVF), nuclear transfer, and bovine–porcine iSCNT embryos as early as the two-cell stage (Lagutina et al., 2010). This confirms the data on low-grade transcription during the first three cell cycles in bovine embryos (Barnes and First, 1991; Hyttel et al., 1996; Memili and First, 1998; Plante et al., 1994; Svarcova et al., 2007; Viuff et al., 1996; Viuff et al., 1998) and extends our knowledge about porcine embryos that were considered transcriptionally inactive until late in the third cell cycle, i.e., at the four-cell stage (Freitag et al., 1991; Jarrell et al., 1991; Tomanek et al., 1989). However, during further in vitro culture, iSCNT embryos did not show the important increase in RNA Pol II activity observed in control embryos, which went through normal EGA.
Embryonic genome activation
The data on EGA in iSCNT embryos are very paradoxical. Porcine NANOG mRNA (Lagutina et al., 2010) was not detected even in the best (≥16-cell) pig–bovine iSCNT embryos at the time of porcine and bovine EGA, as previously reported for chimpanzee–bovine iSCNT embryos (Wang et al., 2009). The absence of NANOG gene expression could be a sign of inadequate reprogramming of OCT4 and SOX2, which have previously been described to drive pluripotent-specific expression of a number of genes, including NANOG (Rodda et al., 2005). Comparison of gene expression in eight- to 16-cell-stage human–bovine or human-rabbit embryos and human IVF or nuclear transfer embryos using single-embryo transcriptome profiling revealed general downregulation of human genes in the bovine and rabbit recipient cytoplasm (Chung et al., 2009). These data and the absence of eAp, Oct4, and e-Cad expression in MEF–bovine iSCNT embryos (Arat et al., 2003; Kim et al., 2004) suggests that bovine ooplasm does not reprogram donor nuclei properly from other species. However, there is a large set of data obtained during the last years demonstrating the different level of expression of embryonic genes in iSCNT embryos, suggesting partial EGA. Human embryonic genes OCT4, SOX2, NANOG, E-CADHERIN, as well as β-ACTIN were activated by enucleated bovine oocytes (Li et al., 2008). Real-time assessment of three developmentally important genes (Oct4, Sox2, and Nanog) indicated their upregulation in human–ovine iSCNT blastocysts (Hosseini et al., 2012) that were able to form blastocyst-derived outgrowths with alkaline phosphatase activity that was lost upon passage.
Partial EGA was found in chimpanzee–bovine iSCNT embryos at the eight-cell stage, as indicated by 5-bromouridine 5′-triphosphate (Br-UTP) incorporation and expression of chimpanzee embryonic genes. Oct4, Stella, Crabp1, CCNE2, CXCL6, PTGER4, H2AFZ, c-MYC, KLF4, and GAPDH transcripts were expressed, whereas Nanog, Glut1, DSC2, USF2, Adrbk1, and Lin28 failed to be activated (Wang et al., 2009). Another study (Wang et al., 2011) demonstrated activation of 2007 genes that were differentially expressed in the rhesus monkey–bovine iSCNT embryos, indicating active transcriptional activity and strongly suggesting that EGA was taking place in the iSCNT embryos.
However, activation of embryonic and pluripotency genes does not predict blastocyst development of iSCNT embryos; for example, crab-eating monkey (Maccaca fascicularis)–bovine NT embryos expressed OCT4 during early development but did not pass through the 16-cell stage of development (Lorthongpanich et al., 2008).
Structural Aspects of iSCNT Reprogramming
Origin and formation of the nucleolus
One of the important compartments of the cell nucleus is nucleolus. In 2008 Oguishi et al. demonstrated the maternal origin and inheritance of nucleoli experimentally (Ogushi et al., 2008). They proved that porcine and murine embryos failed to develop if the oocytes were enucleolated at the germinal vesicle stage and that fertilized/SCNT embryos restored their developmental ability after reinjection of isolated oocyte nucleoli at the MII stage. Nucleoli originating from fibroblast or embryonic stem cell nuclei were not able to substitute maternal nucleoli and support the development of NT embryos derived from enucleolated oocytes. The nature of these indispensable maternally inherited factors of nucleoli is unknown. This finding explained the formation of ruminant-type nucleolus precursor bodies (NPBs) in iSCNT embryos derived from porcine donor cells and ovine oocytes demonstrated earlier by means of transmission electron microscopy (Hamilton et al., 2004).
Nucleolus and nucleus compatibility in iSCNT embryos
In intraspecies SCNT embryos, there is full compatibility between the autologous cytoplasm, nucleus, and nucleoli. In contrast, iSCNT embryos are composed of maternally inherited cytoplasm, a xeno-nucleus, and NPBs originating from the oocyte. The highly conserved gene sequence between mammalian species does not guarantee interspecies compatibility of their products. Earlier studies had revealed that ribosomal (r) DNA transcription is species specific, requiring factors from either the same or very closely related species (Mishima et al., 1982). And human rDNA cannot be transcribed by mouse machinery and vice versa. Most of the factors, i.e., UBF, RNA Pol I, TIF-IA, and TIF-IC, are interchangeable between human and mouse, whereas TIF-IB/SL1 has been found to be the species-specific component in the preinitiation complex. It was shown (Heix et al., 1997) that the primary structure of human and mouse TATA box binding protein (TBP)-associated factors (TAFs) (proteins involved in formation of RNA Pol I complex) does not dramatically alter the network of protein–protein contacts responsible for assembly of the multimeric complex SL1/TIF-IB. The primate versus rodent promoter is likely to be the result of cumulative subtle differences between individual subunits that lead to species-specific properties of RNA Pol I transcription.
Activation of nucleoli formation and embryo development
It was found in Xenopus laevis that major embryonic genome activation initiated with activation of class II genes [messenger (m) RNA], followed by class III genes [transfer (t) RNA], and finally class I genes (rRNA) (Bjerregaard et al., 2007). In mammals, RNA Pol I as well as other key nucleolar proteins, upstream binding factor (UBF), and topoisomerase I, engaged in transcription of the rDNA, and fibrillarin (Svarcova et al., 2007), involved in early processing of rRNA, appeared in embryos at the time of EGA (Maddox-Hyttel et al., 2007; Svarcova et al., 2007). The impaired pre-rRNA transcription (Baran et al., 2003; Chen et al., 2008) or the inhibition of RNA Pol I transcription (Baran et al., 2003) led to fragmentation of nucleoli, apoptotic nuclei. and decreased cell proliferation prior to the morula stage. Similar phenotypes of preimplantation lethality before the blastocyst stage were observed in knockout mice or in knockdown embryos in the case of genes involved in ribosome biogenesis—Pescadillo (Lerch-Gaggl et al., 2002), fibrillarin (Newton et al., 2003), and Surf6 (Romanova et al., 2006). Thus, the inability of iSCNT embryos to correctly activate, transcribe, and translate genes involved in pre-rRNA synthesis at the time of EGA may cause the arrest of nucleoli formation and developmental block in embryos. The absence of mature nucleoli can indirectly indicate silencing or aberrant expression of genes encoding RNA Pol I, nucleolar proteins, and, as a result, rRNA genes.
Ability of oocytes of different species to support formation of nucleoli in xeno-nuclei
Bovine oocytes
Lagutina et al. (Lagutina et al., 2010; Lagutina et al., 2011) (Table 1 and Fig. 1) found differences in the capacities of bovine and porcine oocytes to support nucleoli formation in xeno-nuclei. Bovine oocytes were able to support nucleoli formation in the nuclei of only closely related species such as water buffalo (intergenus) and domestic sheep (inter-subfamily) that have very similar kinetics of embryo cleavage, time and stage of EGA, and likely a similar structure of NPBs. The existence of active RNA Pol I transcription of rDNA in buffalo–bovine iSCNT embryos was confirmed by actinomycin D test (Lagutina et al., 2011). Cattle–water buffalo hybrid IVF blastocysts serve as a confirmation of nuclear–cytoplasm compatibility between these species (Kochhar et al., 2002). Buffalo (Bubalus bubalis)–bovine (Bos indicus) iSCNT blastcysts were produced in several laboratories (Kitiyanant et al., 2001; Lu et al., 2005; Saikhun et al., 2002). Using trasnmission electron microscopy (TEM) (Song et al., 2009; Tao et al., 2008), normal functional changes in nucleoli during EGA were observed in goat–bovine iSCNT embryos. The data on development of ovine–bovine iSCNT embryos are very controversial. Hua et al. (2008) obtained almost 25% of blastocysts with 117±13 cells/blastocyst on day 6, whereas Lagutina et al. (2011) were not able to obtain morulae or blastocysts of such ovine–bovine iSCNT embryos. However, similar morphology of bovine, ovine, and buffalo nuclei and nucleoli in bovine cytoplasm was observed, which might indicate at least partial EGA. Hamilton et al. (2004) have shown that bovine–ovine iSCNT embryos were able to form ruminant-type NPBs as well as structures that appeared as fibrillar material surrounded by a rim of electron-dense granules, perhaps formerly of nucleolar origin. Taking in account similar kinetics of embryo cleavage, time and stage of EGA, likely a similar structure of NPBs in ovine and bovine embryos, and the formation of numerous small nucleoli in ovine–bovine iSCNT embryos into account, it is possible to suppose some nuclear–cytoplasmic compatibility that guarantees at least partial EGA, i.e., RNA Pol I function and rDNA transcription, in ovine–bovine iSCNT embryos.
Table 1.
iSCNT Embryos: Development and Nucleoli Formation
| |
|
|
|
|
|
D3b |
Advanced embryos n at the end of IVCd |
|---|---|---|---|---|---|---|---|
| Donor of nuclei | Donor of oocytes | Taxonomical relations | Embryos with nucleoli/embryos analyzeda | n | Cleaved % | n (%c) | (Day, n of nuclei) |
| Cattle | Cattle | Intraspecies | 20/20 | 45 | 96 | 26 (58) | 21 (D6, CM/BL) |
| Buffalo: | Intergenus | 24/24 | |||||
| 1) granulosa | 103 | 96 | 47 (46) | 10 (D6, 20–27 nuclei) | |||
| 2) AF | 133 | 85 | 64 (48) | 4 (D6, CM) | |||
| Sheep | Inter-subfamily | 26/26 | 247 | 99 | 220 (89) | 86 (D4, 12–16 cells) | |
| Mouse | Interorder | 0/30 | 62 | 98 | 43 (70) | 37 (D4, 12–16 cells) | |
| Cat | Interorder | 0/10 | 50 | 97 | 10 (20) | 10 (D4, 10–16 cells) | |
| Dog | Interorder | 0/31 | 50 | 97 | 37 (74) | 37 (D4, 10–16 cells) | |
| Rabbit | Interorder | 0/24 | 40 | ND | 24 (60) | 24 (D4, 10–16 cells) | |
| Horse | Interorder | 0/20 | 32 | 94 | 20 (63) | 20 (D4, 12–16 cells) | |
| Pig | Interfamily | 0/35 | 423 | 94 | 230 (54) | 230 (D6, 8–25 cells) | |
| Pig | Pig | Intraspecies | 18/18 | 30 | 85 | 22 (73) | 11 (D6, CM/BL) |
| Horse | Interorder | 21/43 | 79 | 81 | 41 (52) | 8 (D6, 6–7 nuclei) | |
| Rabbit | Interorder | 14/17 | 153 | 91 | 89 (58) | 22 (D6, 5–10 nuclei) | |
| Dog | Interorder | 6/21 | 70 | 96 | 58 (83) | 58 (D6, 4–6 cells) | |
| Cat | Interorder | 4/20 | 61 | 90 | 27 (44) | 27 (D4, 4–6 cells) | |
| Mouse | Interorder | 0/18 | 31 | ND | 18 (58) | 18 (D4, 4–6 cells) | |
| Sheep | Interfamily | 6/26 | 34 | 94 | 28 (82) | 28 (D4, 4–6 cells) | |
| Buffalo | Interfamily | 0/22 | 30 | ND | 22 (73) | 22 (D4, 4–6 cells) | |
| Cattle | Interfamily | 0/31 | 249 | 84 | 131 (53) | 131 (D7, 4–6 cells) | |
| Rabbit | Rabbit | Intraspecies | 26/26 | 96 | 83 | 44 (46) | 21 (D3, CM/BL) |
| Cattle | Interorder | 10/44 | 70 | 90 | 23 (33) | 23 (D3, 8–16 nuclei) | |
| Pig | Interorder | 0/26 | 111 | 70 | 10 (9) | 10 (D3, 8–9 nuclei) |
Bovine (≥8 cells) and porcine (≥4 cells) SCNT and iSCNT embryos were analyzed after 96 h of IVC. Rabbit SCNT and iSCNT embryos with ≥4 cells were analyzed after 72 h of IVC.
Advanced embryos D3—if cattle or rabbits are the oocyte donors, embryos with ≥8 blastomeres; if pigs are the oocyte donors, embryos with ≥4 blastomeres.
Percent from total number of reconstructed embryos
Development was estimated at the end of embryo culture as the number of cells or nuclei stained with Hoechst.
AF, adult fibroblasts; CM, compacting morula; BL, blastocyst; ND, not determined.
(Reprinted, with permission, from Lagutina el al., 2011.)
FIG. 1.
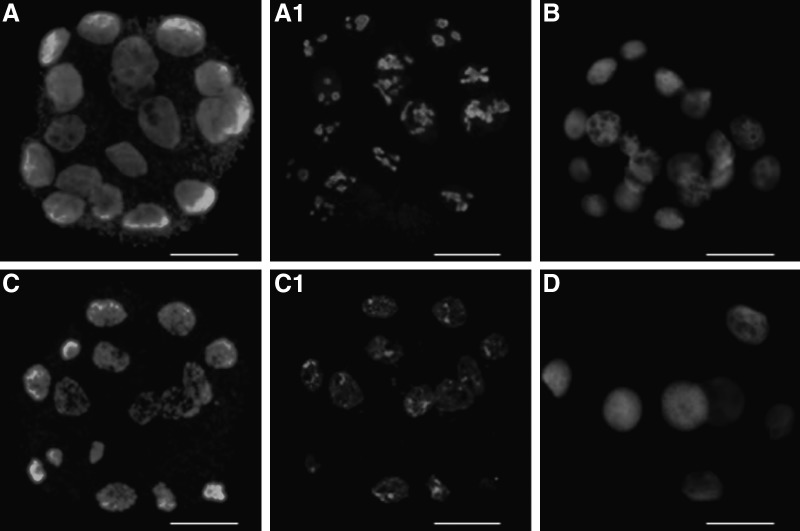
Bovine NT (A, A1, B, D) and pig–bovine iSCNT embryos (C, C1) labeled with anti-nucleolin (C23) antibody (A1, B, C1, D) and counterstained with DAPI for DNA staining (A, C) 96 h after activation. DAPI staining visualized homogeneously stained nuclei of NT embryos (A) and chromatin condensation in the nuclei of iSCNT embryos (C). C23 labeling revealed polygonal mature nucleoli in the nuclei of bovine NT embryos (A1), disappearance of functional nucleoli from the nuclei of bovine NT embryos after 3 h of treatment with 0.2 μg/mL AD (B); the lack of nucleoli formation in bovine NT embryos after 48 h treatment with 2 μg/mL AD (D); the absence of mature nucleoli in pig–bovine iSCNT embryos (C1). Scale bars, 100 μm. (Reprinted, with permission, from Lagutina et al. 2010.)
Lagutina et al. (2011) did not find signs of nucleoli formation in the nuclei of pig, horse, cat, dog, and rabbit cells transferred in bovine ooplasm. Using TEM, Song et al. (2009) observed only early compact nucleoli during EGA in monkey–bovine iSCNT embryos with large proportion of irregularly shaped NPBs. Interestingly, the absence of formation of active mature nucleoli in rhesus monkey–bovine iSCNT embryos (Song et al., 2009) and their inability to form blastocysts was associated with the presence of EGA-dependent nucleolar proteins, such as UBF and fibrillarin. However immunofluorescence confocal laser scanning microscopy results indicated that UBTF, fibrillarin, nucleophosmin, and nucleolin expression levels were significantly reduced in monkey–bovine iSCNT embryos compared to IVF and bovine SCNT embryos. Using TEM, Hamilton et al. showed that the ovine ooplasm directs initial nucleolar formation but is incompatible with the porcine nucleus for completing this event and forming fibrillar-granular nucleoli or restoring rRNA transcription (absence of 3H-uridine incorporation) (Hamilton et al., 2004). These authors demonstrated the absence of NUCLEOLIN-labeled nucleoli at 96 h in the most advanced eight- to 16-cell embryos.
Porcine oocytes
In contrast to bovine oocytes, porcine oocytes (Lagutina et al., 2011) are able to support nucleoli formation in nuclei of many species, including equine (Fig. 2), rabbit, canine, and feline that possess very close EGA at the four- to eight-cell stage (Grøndahl and Hyttel, 1996; Hoffert et al., 1997; Kanka, 2003; Maddox-Hyttel et al., 2005), and, interestingly, even in some ovine nuclei with EGA at the eight-cell stage (Crosby et al., 1988). The most intensively NUCLEOLIN-labeled nucleoli were found in equine and rabbit nuclei. The existence of active RNA Pol I transcription of rDNA in equine–porcine iSCNT embryos was confirmed by an actynomycin D test (Lagutina et al., 2011). None of these embryos with active RNA Pol I was able to overcome the developmental block at the four- to eight-cell stage, confirming the partial and aberrant character of EGA. However, development of canine–pig iSCNT blastocysts was reported in 2009 (Sugimura et al., 2009).
FIG. 2.
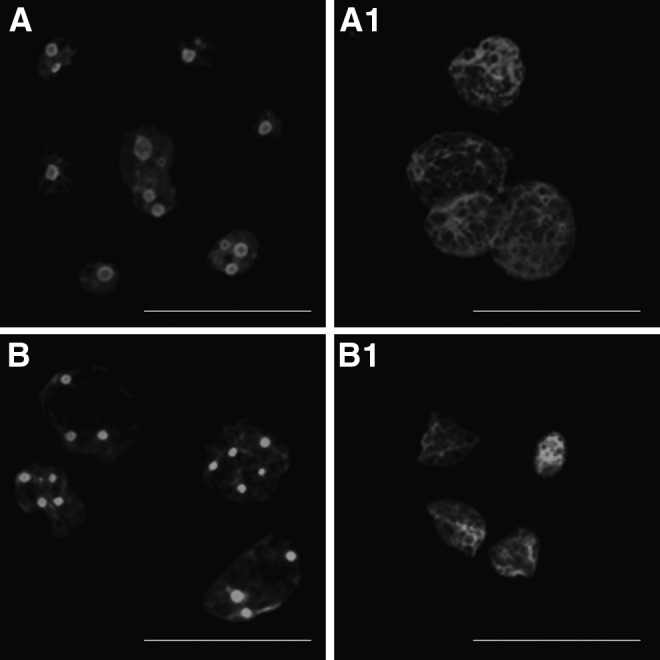
Porcine SCNT (A, A1) and equine–porcine iSCNT (B, B1) embryos 96 h after activation. NUCLEOLIN localization (C23) in mature nucleoli of porcine SCNT (A) and equine–porcine iSCNT (B) embryos, disappearance of mature nucleoli and dispersed nucleoplasmic NUCLEOLIN labeling in actinomycin D–treated porcine SCNT embryos (A1) and equine–porcine iSCNT (B1) embryos. Scale bar, 100 μm. (Reprinted, with permission, from Lagutina et al. 2011.)
Rabbit oocytes
Rabbit oocytes were found to be good recipients when shown to support preimplantation development of embryos derived from nuclei of several species, including bovine (Jiang et al., 2006), Capra ibex (Jiang et al., 2005), chicken (Liu et al., 2004), camel and Tibetan antelope (Zhao et al., 2006), macaca (Yang et al., 2003), cat and panda (Wen et al., 2005), human (Shi et al., 2008), and even chicken (Liu et al., 2004). However, microarray analysis failed to detect significant human genome reprogramming in human–rabbit iSCNT embryos (Chung et al., 2009).
The ability of rabbit oocytes to support nucleoli formation in bovine and porcine nuclei was evaluated by Lagutina et al. (2011). Anti-nucleolin staining of bovine–rabbit iSCNT embryos revealed numerous nucleoli of different sizes in almost half of embryos at the time of EGA determined by rabbit cytoplasm. Unexpectedly, in spite of mature nucleoli formation in rabbit nuclei transferred into pig oocytes, almost all nuclei of porcine–rabbit iSCNT embryos were blocked at the stage of mitotic chromosome condensation when the genome is silent without any sign of nucleoli formation, even in the most advanced embryos with eight to nine nuclei.
The simplest cause of nucleoli formation failure in iSCNT embryos may be a structural difference of promoter selectivity factors that play an important role in the formation of the RNA Pol I complex on the promoter. These promoter selectivity factors were found to be species specific in human (SL1) and mouse (TIF-IB) (Heix et al., 1997). These factors were not studied in other species. However the ability of Xenopus rRNA to be transcribed in mouse cell extract (Culotta et al., 1987) supposes that species divergence is not so large; nucleoli formation in donor nuclei of different unrelated species in rabbit and porcine cytoplasm also supports this.
Role of genome demethylation in genome reprogramming
Very little is known about the molecular mechanism of nuclear reprogramming. Simonsson and Gurdon (2004) analyzed the mechanism of activation of the stem cell marker gene oct4 by Xenopus oocytes using nuclear and DNA transfer from mammalian somatic cells. They found the demethylation of promoter DNA is a necessary step in the epigenetic reprogramming of somatic cell nuclei. The ooplasms from different species have different demethylation capacity (Beaujean et al., 2004; Chen et al., 2004; Chen et al., 2006). The demethylation of repetitive sequences of the donor genome is determined by the recipient ooplasm and not by intrinsic properties of the donor nucleus (Chen et al., 2004; Chen et al., 2006). It seems that the high demethylation capacity of porcine oocytes can explain their ability to support initiation of nucleoli formation in nuclei of several species. In contrast, it was shown that rabbit oocytes possess a much lower demethylation capacity than porcine oocytes (Shi et al., 2004). Together with hypermethylation of porcine fibroblast nuclei (Chen et al., 2006), this may be the reason for failure of nucleoli formation in porcine–rabbit iSCNT embryos. There are different methylation patterns of early rabbit and bovine SCNT/IVF embryos with equally high methylation levels of the paternal and maternal genomes from the zygote up to the 16-cell stage in rabbit IVF and SCNT embryos (Shi et al., 2004) in contrast to considerable demethylation in bovine embryos (Dean et al., 2001). The ability of oocyte to demethylate gene promoters has donor nucleus species-specific features (Wang et al., 2009). Although the trend of global demethylation seems to be similar in bovine SCNT and chimpanzee–bovine iSCNT embryos, bovine ooplasm could not recapitulate in chimpanzee nuclei the DNA demethylation events observed in the bovine SCNT embryos. These deficiencies could potentially significantly limit the transcription of chimpanzee-specific transcripts in the iSCNT embryos. This may be one of the reasons for nucleoli activation failure in bovine, buffalo, and mouse nuclei in porcine cytoplasm, as well as of nucleoli formation in bovine nuclei of bovine–rabbit iSCNT embryos.
Mitochondria-nuclear genome compatibility
Another most important component of cytoplasm is the mitochondrion, a maternally inherited organelle that possesses its own genome. From about 1500 genes regulating mitochondria, only 37 belong to mitochondrial genome whereas others are of nuclear origin.
Nuclear encoded mitochondrial mRNA around EGA
The detailed studies (May-Panloup et al. 2005; Mtango et al. 2008) of temporal expression of nuclear encoded mRNAs related to mitochondrial biogenesis revealed a persistent expression of maternally encoded mRNAs in combination with transcriptional activation and mRNA accumulation around the time of EGA. These findings confirm the observations of morphological changes in the shape and structure of oocyte mitochondria that associated with an increase of mitochondrial activity at the time of EGA.
Mitochondrial structure during early embryogenesis
Unlike their counterparts in differentiated cells, mitochondria in oocytes and newly fertilized eggs are structurally undeveloped and typically appear as small circular forms with an electron-dense matrix surrounded by truncated cristae that rarely penetrate or traverse the matrix. However, while structurally undeveloped, they are functional and active in adenosine triphosphate (ATP) generation by oxidative phosphorylation. During the cleavage and early blastocyst phases of preimplantation embryogenesis, mitochondria undergo changes in shape and structure that while species specific with respect to stage follow a similar pattern of transformation. At the time of EGA, mitochondrial geometry transitions from spherical to elliptical and the cristae become more numerous and able to traverse a matrix of lower electron density (Van Blerkom, 2009).
Mitochondria mass and activity around EGA
The changes in mitochondrial mass and activity were studied around EGA in bovine, porcine SCNT, and bovine–pig iSCNT embryos using JC-I staining. This method revealed that before EGA embryos possessed equal mitochondrial mass measured by J-monomers and low activity (accumulation of J-aggregates). However, at the initiation of EGA, there is a burst of JC-I accumulation (monomers and J-aggregates) in both intraspecies, bovine and porcine, SCNT embryos, whereas iSCNT embryos could be characterized by significantly lower JC-I accumulation, comparable to the pre-EGA stage (Lagutina et al., 2011).
Nuclear–mitochondrial incompatibility
The existence of nuclear–mitochondrial incompatibility between different species was demonstrated in cybrids of closely related primates (Barrientos et al., 2000; Kenyon and Moraes, 1997) and murids (Dey et al., 2000; McKenzie and Trounce, 2000). The mitochondrial protein synthesis in interspecies cybrids was unaffected whereas the activities of respiratory complexes I and IV were significantly reduced because of low steady-state levels of respective subunits, indicating problems in their assembly and existence of different compatibility of nuclear encoded proteins with their mitochondrial targets.
In iSCNT embryos, the function of recipient ooplasm inherited mitochondria depends on the donor nucleus genome of different species that could be in rather distant relationship (intergenus, interfamily, etc.). We investigated nuclear–mitochondria compatibility in iSCNT embryos derived from bovine and porcine oocytes and donor cells from different species using JC-1 labeling. The accumulation of J-monomeres that corresponds to mitochondrial mass was compared between iSCNT embryos with active nucleoli formation (Lagutina et al., 2011) and intraspecies SCNT embryos with the same ooplasm (bovine and porcine SCNT embryos) at the time of EGA (Table 2). We found that in bovine ooplasm buffalo and sheep nuclei (I. Lagutina, unpublished data) were able to support activation of mitochondrial mass accumulation at the same level as bovine ones. In the case of porcine ooplasm, mitochondria were activated neither by ovine, nor by horse or rabbit nuclei (I. Lagutina, unpublished data), demonstrating complete nuclear–mitochondria incompatibility in these iSCNT embryos in spite of activation of nucleoli formation.
Table 2.
Formation of Nucleoli and Mitochondrial Mass Growth around EGA in Different iSCNT Embryos
| Donor of nuclei | Oocyte | Taxonomical relations | Formation of nucleoli | Activation of mitochondria |
|---|---|---|---|---|
| Cattle | Cattle | Intraspecies | + | + |
| Buffalo | intergenus | + | + | |
| Sheep | Inter-subfamily | + | + | |
| Mouse | Interorder | − | NA | |
| Cat | Interorder | − | NA | |
| Dog | Interorder | − | NA | |
| Rabbit | Interorder | − | NA | |
| Horse | Interorder | − | NA | |
| Pig | Interfamily | − | − | |
| Pig | Pig | Intraspecies | + | + |
| Horse | Interorder | + | − | |
| Rabbit | Interorder | + | − | |
| Dog | Interorder | + | NA | |
| Cat | Interorder | + | NA | |
| Mouse | Interorder | − | NA | |
| Sheep | Interfamily | − | − | |
| Buffalo | Interfamily | − | NA | |
| Cattle | Interfamily | − | − | |
| Rabbit | Rabbit | Intraspecies | + | NA |
| Cattle | Interorder | + | NA | |
| Pig | Interorder | − | NA |
Effect of heteroplasmy on iSCNT embryo development
Nuclear–cytoplasmic incompatibility in iSCNT embryos was proved experimentally in goat–bovine (Sansinena, 2011) and pig–mouse (Amarnath et al., 2011) models. In contrast to injection of homologous ooplasm in bovine SCNT embryos that did not affect preimplantation embryo development, injection of goat ooplasm into goat–bovine iSCNT embryos led to a significant decrease of embryo cleavage of iSCNT embryos, demonstrating that heteroplasmy or mitochondrial incompatibilities may affect nuclear–ooplasmic events occurring at the time of EGA. Amarnath et al. (2011) constructed pig–mouse cytoplasmic hybrids by fusion of mouse zygotes with porcine cytoplasts of different volumes. The presence of pig cytoplasm significantly reduced the development of mouse zygotes to the blastocyst stage compared with control embryos and the extent of development failure positively correlated with the porcine ooplast volume. While mitochondrial DNA copy numbers remained relatively unchanged, expression of several important genes, namely Tfam, Polg, Polg2, Mfn2, Slc2a3 (Glut3), Slc2a1 (Glut1), Bcl2, Hspb1, Pou5f1 (Oct4), Nanog, Cdx2, Gata3, Tcfap2c, mt-Cox1, and mt-Cox2, was significantly reduced in cytoplasmic hybrids. These results demonstrate that the presence of even a small amount of porcine cytoplasm is detrimental to murine embryo development and suggest that a range of factors is likely to contribute to the failure of iSCNT embryos.
Conclusions
The most successful progress of iSCNT was achieved using donor cells and recipient ooplast of very closely related species. As the species divergence increases, the ability to sustain embryo development decreases to full incompatibility. The observed aberrant degradation of maternally inherited ooplasmic mRNA that should occur soon after oocyte activation and before EGA, the inability of maternally inherited factors to activate the embryonic genome, improper demethylation of the donor genome, and the nuclear–mitochondrial incompatibilities all contribute to the early death of iSCNT embryos. It has to be remembered that with same species SCNT embryos, although the preimplantation development is comparable to that of fertilized embryos, the development to term and into viable offspring is still low; not all of the reasons for this have been elucidated. Therefore, the iSCNT model, with its extreme molecular and structural failures, represents an important research tool that is advancing our knowledge in cellular and nuclear reprogramming but presents huge, albeit not insurmountable, challenges. In an interesting report, Jiang et al. (2011) improved the development of mouse–pig interspecies embryos by eliminating the mitochondria from the pig oocytes and injecting with the somatic cell nucleus mouse ESC extract that carried key pluripotent factors as well as mitochondria. Given the complexity of the events, it is clear that a variety of other factors are involved (Narbonne and Gurdon, 2012) and the number of potentially active reprogramming factors could be extremely vast (Awe and Byrne, 2013). The search for the most critical ones should be concentrated on those conserved across species.
Acknowledgments
This manuscript was prepared while funded by European Union grant FECUND no. 312097 and by Regione Lombardia grants InnovaB and Superpig. H.F. is supported from P302/11/P069 (Czech Science Foundation).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Alizadeh Z. Kageyama S. Aoki F. Degradation of maternal mRNA in mouse embryos: selective degradation of specific mRNAs after fertilization. Mol. Reprod. Dev. 2005;72:281–290. doi: 10.1002/mrd.20340. [DOI] [PubMed] [Google Scholar]
- Amarnath D. Choi I. Moawad A.R. Wakayama T. Campbell K.H. Nuclear-cytoplasmic incompatibility and inefficient development of pig-mouse cytoplasmic hybrid embryos. Reproduction. 2011;142:295–307. doi: 10.1530/REP-11-0044. [DOI] [PubMed] [Google Scholar]
- Arat S. Rzucidlo S.J. Stice S.L. Gene expression and in vitro development of inter-species nuclear transfer embryos. Mol. Reprod. Dev. 2003;66:334–342. doi: 10.1002/mrd.10362. [DOI] [PubMed] [Google Scholar]
- Awe J.P. Byrne J.A. Identifying candidate oocyte reprogramming factors using cross-species global transcriptional analysis. Cell. Reprogram. 2013;15:126–133. doi: 10.1089/cell.2012.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran V. Fabian D. Rehak P. Koppel J. Nucleolus in apoptosis-induced mouse preimplantation embryos. Zygote. 2003;11:271–283. doi: 10.1017/s0967199403002326. [DOI] [PubMed] [Google Scholar]
- Barnes F.L. First N.L. Embryonic transcription in in vitro cultured bovine embryos. Mol. Reprod. Dev. 1991;29:117–123. doi: 10.1002/mrd.1080290205. [DOI] [PubMed] [Google Scholar]
- Barrientos A. Muller S. Dey R. Wienberg J. Moraes C.T. Cytochrome c oxidase assembly in primates is sensitive to small evolutionary variations in amino acid sequence. Mol. Biol. Evol. 2000;17:1508–1519. doi: 10.1093/oxfordjournals.molbev.a026250. [DOI] [PubMed] [Google Scholar]
- Beaujean N. Taylor J.E. McGarry M. Gardner J.O. Wilmut I. Loi P. Ptak G. Galli C. Lazzari G. Bird A., et al. The effect of interspecific oocytes on demethylation of sperm DNA. Proc. Natl. Acad. Sci. USA. 2004;101:7636–7640. doi: 10.1073/pnas.0400730101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerregaard B. Pedersen H.G. Jakobsen A.S. Rickords L.F. Lai L. Cheong H.T. Samuel M. Prather R.S. Strejcek F. Rasmussen Z.R., et al. Activation of ribosomal RNA genes in porcine embryos produced in vitro or by somatic cell nuclear transfer. Mol. Reprod. Dev. 2007;74:35–41. doi: 10.1002/mrd.20594. [DOI] [PubMed] [Google Scholar]
- Byrne J.A. Simonsson S. Western P.S. Gurdon J. B. Nuclei of adult mammalian somatic cells are directly reprogrammed to oct-4 stem cell gene expression by amphibian oocytes. Curr. Biol. 2003;13:1206–1213. doi: 10.1016/s0960-9822(03)00462-7. [DOI] [PubMed] [Google Scholar]
- Chang K.H. Lim J.M. Kang S.K. Lee B.C. Moon S.Y. Hwang W.S. Blastocyst formation, karyotype, and mitochondrial DNA of interspecies embryos derived from nuclear transfer of human cord fibroblasts into enucleated bovine oocytes. Fertil. Steril. 2003;80:1380–1387. doi: 10.1016/j.fertnstert.2003.07.006. [DOI] [PubMed] [Google Scholar]
- Chen H. Li Z. Haruna K. Li Z. Li Z. Semba K. Araki M. Yamamura K. Araki K. Early pre-implantation lethality in mice carrying truncated mutation in the RNA polymerase 1–2 gene. Biochem. Biophys. Res. Commun. 2008;365:636–642. doi: 10.1016/j.bbrc.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Chen T. Zhang Y.L. Jiang Y. Liu J.H. Schatten H. Chen D.Y. Sun Q.Y. Interspecies nuclear transfer reveals that demethylation of specific repetitive sequences is determined by recipient ooplasm but not by donor intrinsic property in cloned embryos. Mol. Reprod. Dev. 2006;73:313–317. doi: 10.1002/mrd.20421. [DOI] [PubMed] [Google Scholar]
- Chen T. Zhang Y.L. Yang Liu S.Z. Schatten H. Chen D.Y. Sun Q.Y. The DNA methylation events in normal and cloned rabbit embryos. FEBS Lett. 2004;578:69–72. doi: 10.1016/j.febslet.2004.10.073. [DOI] [PubMed] [Google Scholar]
- Chung Y. Bishop C.E. Treff N.R. Walker S.J. Sandler V.M. Becker S. Klimanskaya I. Wun W. S. Dunn R. Hall R.M., et al. Reprogramming of human somatic cells using human and animal oocytes. Cloning Stem Cells. 2009;11:213–223. doi: 10.1089/clo.2009.0004. [DOI] [PubMed] [Google Scholar]
- Crosby I.M. Gandolfi F. Moor R.M. Control of protein synthesis during early cleavage of sheep embryos. J. Reprod. Fertil. 1988;82:769–775. doi: 10.1530/jrf.0.0820769. [DOI] [PubMed] [Google Scholar]
- Culotta V.C. Wilkinson J.K. Sollner-Webb B. Mouse and frog violate the paradigm of species-specific transcription of ribosomal RNA genes. Proc. Natl. Acad. Sci. USA. 1987;84:7498–7502. doi: 10.1073/pnas.84.21.7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean W. Santos F. Stojkovic M. Zakhartchenko V. Walter J. Wolf E. Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc. Natl. Acad. Sci. USA. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey R. Barrientos A. Moraes C. T. Functional constraints of nuclear-mitochondrial DNA interactions in xenomitochondrial rodent cell lines. J. Biol. Chem. 2000;275:31520–31527. doi: 10.1074/jbc.M004053200. [DOI] [PubMed] [Google Scholar]
- Dominko T. Mitalipova M. Haley B. Beyhan Z. Memili E. McKusick B. First N.L. Bovine oocyte cytoplasm supports development of embryos produced by nuclear transfer of somatic cell nuclei from various mammalian species. Biol. Reprod. 1999;60:1496–1502. doi: 10.1095/biolreprod60.6.1496. [DOI] [PubMed] [Google Scholar]
- Freitag M. Dopke H.H. Niemann H. Elsaesser F. 3H-uridine incorporation in early porcine embryos. Mol. Reprod. Dev. 1991;29:124–128. doi: 10.1002/mrd.1080290206. [DOI] [PubMed] [Google Scholar]
- Galli C. Lazzari G. The manipulation of gametes and embryos in farm animals. Reprod. Domest. Anim. 2008;43(Suppl 2):1–7. doi: 10.1111/j.1439-0531.2008.01136.x. [DOI] [PubMed] [Google Scholar]
- Gomez M.C. Pope C.E. Giraldo A. Lyons L.A. Harris R.F. King A.L. Cole A. Godke R.A. Dresser B.L. Birth of African Wildcat cloned kittens born from domestic cats. Cloning Stem Cells. 2004;6:247–258. doi: 10.1089/clo.2004.6.247. [DOI] [PubMed] [Google Scholar]
- Gomez M.C. Pope C.E. Kutner R.H. Ricks D.M. Lyons L.A. Ruhe M. Dumas C. Lyons J. Lopez M. Dresser B.L. Reiser J. Nuclear transfer of sand cat cells into enucleated domestic cat oocytes is affected by cryopreservation of donor cells. Cloning Stem Cells. 2008;10:469–483. doi: 10.1089/clo.2008.0021. [DOI] [PubMed] [Google Scholar]
- Green A.L. Wells D.N. Oback B. Cattle cloned from increasingly differentiated muscle cells. Biol. Reprod. 2007;77:395–406. doi: 10.1095/biolreprod.106.058164. [DOI] [PubMed] [Google Scholar]
- Grondahl C. Hyttel P. Nucleologenesis and ribonucleic acid synthesis in preimplantation equine embryos. Biol. Reprod. 1996;55:769–774. doi: 10.1095/biolreprod55.4.769. [DOI] [PubMed] [Google Scholar]
- Hamilton H.M. Peura T.T. Laurincik J. Walker S.K. Maddocks S. Maddox-Hyttel P. Ovine ooplasm directs initial nucleolar assembly in embryos cloned from ovine, bovine, and porcine cells. Mol. Reprod. Dev. 2004;69:117–125. doi: 10.1002/mrd.20160. [DOI] [PubMed] [Google Scholar]
- Hashem M.A. Bhandari D.P. Kang S.K. Lee B.C. Cell cycle analysis and interspecies nuclear transfer of in vitro cultured skin fibroblasts of the Siberian tiger (Panthera tigris Altaica) Mol. Reprod. Dev. 2007;74:403–411. doi: 10.1002/mrd.20528. [DOI] [PubMed] [Google Scholar]
- Heix J. Zomerdijk J.C. Ravanpay A. Tjian R. Grummt I. Cloning of murine RNA polymerase I-specific TAF factors: Conserved interactions between the subunits of the species-specific transcription initiation factor TIF-IB/SL1. Proc. Natl. Acad. Sci. USA. 1997;94:1733–1738. doi: 10.1073/pnas.94.5.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffert K.A. Anderson G.B. Wildt D.E. Roth T.L. Transition from maternal to embryonic control of development in IVM/IVF domestic cat embryos. Mol. Reprod. Dev. 1997;48:208–215. doi: 10.1002/(SICI)1098-2795(199710)48:2<208::AID-MRD8>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Hosseini S.M. Hajian M. Forouzanfar M. Moulavi F. Abedi P. Asgari V. Tanhaei S. Abbasi H. Jafarpour F. Ostadhosseini S., et al. Enucleated ovine oocyte supports human somatic cells reprogramming back to the embryonic stage. Cell. Reprogram. 2012;14:155–163. doi: 10.1089/cell.2011.0061. [DOI] [PubMed] [Google Scholar]
- Hua S. Zhang Y. Song K. Song J. Zhang Z. Zhang L. Zhang C. Cao J. Ma L. Development of bovine-ovine interspecies cloned embryos and mitochondria segregation in blastomeres during preimplantation. Anim. Reprod. Sci. 2008;105:245–257. doi: 10.1016/j.anireprosci.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Hwang I. Jeong Y.W. Kim J.J. Lee H.J. Kang M. Park K.B. Park J.H. Kim Y.W. Kim W.T. Shin T., et al. Successful cloning of coyotes through interspecies somatic cell nuclear transfer using domestic dog oocytes. Reprod. Fertil. Dev. Dec. 2012:7. doi: 10.1071/RD12256. [DOI] [PubMed] [Google Scholar]
- Hyttel P. Viuff D. Avery B. Laurincik J. Greve T. Transcription and cell cycle-dependent development of intranuclear bodies and granules in two-cell bovine embryos. J. Reprod. Fertil. 1996;108:263–270. doi: 10.1530/jrf.0.1080263. [DOI] [PubMed] [Google Scholar]
- Illmensee K. Levanduski M. Zavos P.M. Evaluation of the embryonic preimplantation potential of human adult somatic cells via an embryo interspecies bioassay using bovine oocytes. Fertil. Steril. 2006;85(Suppl 1):1248–1260. doi: 10.1016/j.fertnstert.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Inoue K. Ogonuki N. Miki H. Hirose M. Noda S. Kim J. M. Aoki F. Miyoshi H. Ogura A. Inefficient reprogramming of the hematopoietic stem cell genome following nuclear transfer. J. Cell. Sci. 2006;119:1985–1991. doi: 10.1242/jcs.02913. [DOI] [PubMed] [Google Scholar]
- Jarrell V.L. Day B.N. Prather R.S. The transition from maternal to zygotic control of development occurs during the 4-cell stage in the domestic pig, Sus scrofa: Quantitative and qualitative aspects of protein synthesis. Biol. Reprod. 1991;44:62–68. doi: 10.1095/biolreprod44.1.62. [DOI] [PubMed] [Google Scholar]
- Jian-Quan C. Juan C. Xu-Jun X. Guo-Hui L. Si-Guo L. Hong-Ying S. You-Bing W. Guo-Xiang C. Effect of cytoplast on the development of inter-subspecies nuclear transfer reconstructed goat embryo. Mol. Reprod. Dev. 2007;74:568–573. doi: 10.1002/mrd.20647. [DOI] [PubMed] [Google Scholar]
- Jiang Y. Chen T. Nan C.L. Ouyang Y.C. Sun Q.Y. Chen D.Y. In vitro culture and mtDNA fate of ibex-rabbit nuclear transfer embryos. Zygote. 2005;13:233–240. doi: 10.1017/s0967199405003254. [DOI] [PubMed] [Google Scholar]
- Jiang Y. Chen T. Wang K. Jiang M.X. Liu S.Z. Ouyang Y.C. Sun Q.Y. Chen D.Y. Different fates of donor mitochondrial DNA in bovine-rabbit and cloned bovine-rabbit reconstructed embryos during preimplantation development. Front. Biosci. 2006;11:1425–1432. doi: 10.2741/1893. [DOI] [PubMed] [Google Scholar]
- Jiang Y. Kelly R. Peters A. Fulka H. Dickinson A. Mitchell D.A. St John J.C. Interspecies somatic cell nuclear transfer is dependent on compatible mitochondrial DNA and reprogramming factors. PLoS One. 2011;6:e14805. doi: 10.1371/journal.pone.0014805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanka J. Gene expression and chromatin structure in the pre-implantation embryo. Theriogenology. 2003;59:3–19. doi: 10.1016/s0093-691x(02)01267-0. [DOI] [PubMed] [Google Scholar]
- Kenyon L. Moraes C.T. Expanding the functional human mitochondrial DNA database by the establishment of primate xenomitochondrial cybrids. Proc. Natl. Acad. Sci. USA. 1997;94:9131–9135. doi: 10.1073/pnas.94.17.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.K. Jang G. Oh H.J. Yuda F. Kim H.J. Hwang W.S. Hossein M.S. Kim J.J. Shin N.S. Kang S.K. Lee B.C. Endangered wolves cloned from adult somatic cells. Cloning Stem Cells. 2007;9:130–137. doi: 10.1089/clo.2006.0034. [DOI] [PubMed] [Google Scholar]
- Kim N.-H. Shin M.R. Park S.H. Bovine oocyte cytoplasm supports nuclear remodeling but not reprogramming of murine fibroblasts. Reprod. Reprod. Fertil. Dev. 2004;16:145. doi: 10.1002/mrd.20050. abstract. [DOI] [PubMed] [Google Scholar]
- Kishigami S. Wakayama S. van Thuan N. Wakayama T. Cloned mice and embryonic stem cell establishment from adult somatic cells. Hum. Cell. 2006;19:2–10. doi: 10.1111/j.1749-0774.2005.00001.x. [DOI] [PubMed] [Google Scholar]
- Kitiyanant Y. Saikhun J. Chaisalee B. White K.L. Pavasuthipaisit K. Somatic cell cloning in Buffalo (Bubalus bubalis): Effects of interspecies cytoplasmic recipients and activation procedures. Cloning Stem Cells. 2001;3:97–104. doi: 10.1089/153623001753205052. [DOI] [PubMed] [Google Scholar]
- Kochhar H.P. Rao K.B. Luciano A.M. Totey S.M. Gandolfi F. Basrur P.K. King W.A. In vitro production of cattle-water buffalo (Bos taurus–Bubalus bubalis) hybrid embryos. Zygote. 2002;10:155–162. doi: 10.1017/s0967199402002216. [DOI] [PubMed] [Google Scholar]
- Kwon D.K. Kang J.T. Park S.J. Gomez M.N. Kim S.J. Atikuzzaman M. Koo O.J. Jang G. Lee B.C. Blastocysts derived from adult fibroblasts of a rhesus monkey (Macaca mulatta) using interspecies somatic cell nuclear transfer. Zygote. 2011;19:199–204. doi: 10.1017/S0967199411000232. [DOI] [PubMed] [Google Scholar]
- Lagutina I. Fulka H. Brevini T. A. Antonini S. Brunetti D. Colleoni S. Gandolfi F. Lazzari G. Fulka J., Jr. Galli C. Development, embryonic genome activity and mitochondrial characteristics of bovine-pig inter-family nuclear transfer embryos. Reproduction. 2010;140:273–285. doi: 10.1530/REP-09-0578. [DOI] [PubMed] [Google Scholar]
- Lagutina I. Zakhartchenko V. Fulka H. Colleoni S. Wolf E. Fulka J., Jr. Lazzari G. Galli C. Formation of nucleoli in interspecies nuclear transfer embryos derived from bovine, porcine, and rabbit oocytes and nuclear donor cells of various species. Reproduction. 2011;141:453–465. doi: 10.1530/REP-10-0266. [DOI] [PubMed] [Google Scholar]
- Lanza R.P. Cibelli J.B. Diaz F. Moraes C.T. Farin P.W. Farin C.E. Hammer C.J. West M.D. Damiani P. Cloning of an endangered species (Bos gaurus) using interspecies nuclear transfer. Cloning. 2000;2:79–90. doi: 10.1089/152045500436104. [DOI] [PubMed] [Google Scholar]
- Lazzari G. Colleoni S. Giannelli S.G. Brunetti D. Colombo E. Lagutina I. Galli C. Broccoli V. Direct derivation of neural rosettes from cloned bovine blastocysts: A model of early neurulation events and neural crest specification in vitro. Stem Cells. 2006;24:2514–2521. doi: 10.1634/stemcells.2006-0149. [DOI] [PubMed] [Google Scholar]
- Lee E. Kim J.H. Park S.M. Jeong Y.I. Lee J.Y. Park S.W. Choi J. Kim H.S. Jeong Y.W. Kim S., et al. The analysis of chromatin remodeling and the staining for DNA methylation and histone acetylation do not provide definitive indicators of the developmental ability of inter-species cloned embryos. Anim. Reprod. Sci. 2008;105:438–450. doi: 10.1016/j.anireprosci.2007.12.021. [DOI] [PubMed] [Google Scholar]
- Lerch-Gaggl A. Haque J. Li J. Ning G. Traktman P. Duncan S.A. Pescadillo is essential for nucleolar assembly, ribosome biogenesis, and mammalian cell proliferation. J. Biol. Chem. 2002;277:45347–45355. doi: 10.1074/jbc.M208338200. [DOI] [PubMed] [Google Scholar]
- Li F. Cao H. Zhang Q. Li R. Chen X. Fang Z. Xue K. Chen da Y. Sheng H.Z. Activation of human embryonic gene expression in cytoplasmic hybrid embryos constructed between bovine oocytes and human fibroblasts. Cloning Stem Cells. 2008;10:297–305. doi: 10.1089/clo.2007.0084. [DOI] [PubMed] [Google Scholar]
- Liu S.Z. Zhou Z.M. Chen T. Zhang Y.L. Wen D.C. Kou Z.H. Li Z.D. Sun Q.Y. Chen D.Y. Blastocysts produced by nuclear transfer between chicken blastodermal cells and rabbit oocytes. Mol. Reprod. Dev. 2004;69:296–302. doi: 10.1002/mrd.20091. [DOI] [PubMed] [Google Scholar]
- Loi P. Ptak G. Barboni B. Fulka J., Jr. Cappai P. Clinton M. Genetic rescue of an endangered mammal by cross-species nuclear transfer using post-mortem somatic cells. Nat. Biotechnol. 2001;19:962–964. doi: 10.1038/nbt1001-962. [DOI] [PubMed] [Google Scholar]
- Loi P. Modlinski J.A. Ptak G. Interspecies somatic cell nuclear transfer: A salvage tool seeking first aid. Theriogenology. 2011;76:217–228. doi: 10.1016/j.theriogenology.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Lorthongpanich C. Laowtammathron C. Chan A. W. Ketudat-Cairns M. Parnpai R. Development of interspecies cloned monkey embryos reconstructed with bovine enucleated oocytes. J. Reprod. Dev. 2008;54:306–313. doi: 10.1262/jrd.20049. [DOI] [PubMed] [Google Scholar]
- Lu F. Shi D. Wei J. Yang S. Wei Y. Development of embryos reconstructed by interspecies nuclear transfer of adult fibroblasts between buffalo (Bubalus bubalis) and cattle (Bos indicus) Theriogenology. 2005;64:1309–1319. doi: 10.1016/j.theriogenology.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Maddox-Hyttel P. Bjerregaard B. Laurincik J. Meiosis and embryo technology: Renaissance of the nucleolus. Reprod. Fertil. Dev. 2005;17:3–14. doi: 10.1071/rd04108. [DOI] [PubMed] [Google Scholar]
- Maddox-Hyttel P. Svarcova O. Laurincik J. Ribosomal RNA and nucleolar proteins from the oocyte are to some degree used for embryonic nucleolar formation in cattle and pig. Theriogenology. 2007;68(Suppl 1):S63–S70. doi: 10.1016/j.theriogenology.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Mastromonaco G.F. Favetta L.A. Smith L.C. Filion F. King W.A. The influence of nuclear content on developmental competence of gaur×cattle hybrid in vitro fertilized and somatic cell nuclear transfer embryos. Biol. Reprod. 2007;76:514–523. doi: 10.1095/biolreprod.106.058040. [DOI] [PubMed] [Google Scholar]
- May-Panloup P. Vignon X. Chretien M.F. Heyman Y. Tamassia M. Malthiery Y. Reynier P. Increase of mitochondrial DNA content and transcripts in early bovine embryogenesis associated with upregulation of mtTFA and NRF1 transcription factors. Reprod. Biol. Endocrinol. 2005;3:65. doi: 10.1186/1477-7827-3-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie M. Trounce I. Expression of Rattus norvegicus mtDNA in Mus musculus cells results in multiple respiratory chain defects. J. Biol. Chem. 2000;275:31514–31519. doi: 10.1074/jbc.M004070200. [DOI] [PubMed] [Google Scholar]
- Memili E. First N.L. Developmental changes in RNA polymerase II in bovine oocytes, early embryos, and effect of alpha-amanitin on embryo development. Mol. Reprod. Dev. 1998;51:381–389. doi: 10.1002/(SICI)1098-2795(199812)51:4<381::AID-MRD4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Mishima Y. Financsek I. Kominami R. Muramatsu M. Fractionation and reconstitution of factors required for accurate transcription of mammalian ribosomal RNA genes: Identification of a species-dependent initiation factor. Nucleic Acids Res. 1982;10:6659–6670. doi: 10.1093/nar/10.21.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mtango N.R. Harvey A.J. Latham K.E. Brenner C.A. Molecular control of mitochondrial function in developing rhesus monkey oocytes and preimplantation-stage embryos. Reprod. Fertil. Dev. 2008;20:846–859. doi: 10.1071/rd08078. [DOI] [PubMed] [Google Scholar]
- Narbonne P. Gurdon J.B. Amphibian interorder nuclear transfer embryos reveal conserved embryonic gene transcription, but deficient DNA replication or chromosome segregation. Int. J. Dev. Biol. 2012;56:975–986. doi: 10.1387/ijdb.120150jg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K. Petfalski E. Tollervey D. Caceres J.F. Fibrillarin is essential for early development and required for accumulation of an intron-encoded small nucleolar RNA in the mouse. Mol. Cell. Biol. 2003;23:8519–8527. doi: 10.1128/MCB.23.23.8519-8527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R.K. Gurdon J.B. Maintenance of epigenetic memory in cloned embryos. Cell Cycle. 2005;4:760–763. doi: 10.4161/cc.4.6.1743. [DOI] [PubMed] [Google Scholar]
- Oback B. Cloning from stem cells: Different lineages, different species, same story. Reprod. Fertil. Dev. 2009;21:83–94. doi: 10.1071/rd08212. [DOI] [PubMed] [Google Scholar]
- Ogura A. Inoue K. Wakayama T. Recent advancements in cloning by somatic cell nuclear transfer. Phil. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20110329. doi: 10.1098/rstb.2011.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogushi S. Palmieri C. Fulka H. Saitou M. Miyano T. Fulka J., Jr. The maternal nucleolus is essential for early embryonic development in mammals. Science. 2008;319:613–616. doi: 10.1126/science.1151276. [DOI] [PubMed] [Google Scholar]
- Paynton B.V. Rempel R. Bachvarova R. Changes in state of adenylation and time course of degradation of maternal mRNAs during oocyte maturation and early embryonic development in the mouse. Dev. Biol. 1988;129:304–314. doi: 10.1016/0012-1606(88)90377-6. [DOI] [PubMed] [Google Scholar]
- Plante L. Plante C. Shepherd D.L. King W.A. Cleavage and 3H-uridine incorporation in bovine embryos of high in vitro developmental potential. Mol. Reprod. Dev. 1994;39:375–383. doi: 10.1002/mrd.1080390405. [DOI] [PubMed] [Google Scholar]
- Rodda D.J. Chew J.L. Lim L.H. Loh Y.H. Wang B. Ng H.H. Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- Romanova L.G. Anger M. Zatsepina O.V. Schultz R.M. Implication of nucleolar protein SURF6 in ribosome biogenesis and preimplantation mouse development. Biol. Reprod. 2006;75:690–696. doi: 10.1095/biolreprod.106.054072. [DOI] [PubMed] [Google Scholar]
- Saikhun J. Pavasuthipaisit K. Jaruansuwan M. Kitiyanant Y. Xenonuclear transplantation of buffalo (Bubalus bubalis) fetal and adult somatic cell nuclei into bovine (Bos indicus) oocyte cytoplasm and their subsequent development. Theriogenology. 2002;57:1829–1837. doi: 10.1016/s0093-691x(02)00667-2. [DOI] [PubMed] [Google Scholar]
- Sansinena M.J. Lynn J. Bondioli K.R. Denniston R.S. Godke R.A. Ooplasm transfer and interspecies somatic cell nuclear transfer: Heteroplasmy, pattern of mitochondrial migration, and effect on embryo development. Zygote. 2011;19:147–156. doi: 10.1017/S0967199410000419. [DOI] [PubMed] [Google Scholar]
- Schultz R.M. Regulation of zygotic gene activation in the mouse. Bioessays. 1993;15:531–538. doi: 10.1002/bies.950150806. [DOI] [PubMed] [Google Scholar]
- Sha H.Y. Chen J.Q. Chen J. Zhang P.Y. Wang P. Chen L.P. Cheng G.X. Zhu J.H. Fates of donor and recipient mitochondrial DNA during generation of interspecies SCNT-derived human ES-like cells. Cloning Stem Cells. 2009;11:497–507. doi: 10.1089/clo.2009.0021. [DOI] [PubMed] [Google Scholar]
- Shi L.H. Miao Y.L. Ouyang Y.C. Huang J.C. Lei Z.L. Yang J.W. Han Z.M. Song X.F. Sun Q.Y. Chen D.Y. Trichostatin A (TSA) improves the development of rabbit-rabbit intraspecies cloned embryos, but not rabbit-human interspecies cloned embryos. Dev. Dyn. 2008;237:640–648. doi: 10.1002/dvdy.21450. [DOI] [PubMed] [Google Scholar]
- Shi W. Dirim F. Wolf E. Zakhartchenko V. Haaf T. Methylation reprogramming and chromosomal aneuploidy in in vivo fertilized and cloned rabbit preimplantation embryos. Biol. Reprod. 2004;71:340–347. doi: 10.1095/biolreprod.103.024554. [DOI] [PubMed] [Google Scholar]
- Simonsson S. Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat. Cell Biol. 2004;6:984–990. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- Song B.S. Lee S.H. Kim S.U. Kim J.S. Park J.S. Kim C.H. Chang K.T. Han Y.M. Lee K.K. Lee D.S. Koo D.B. Nucleologenesis and embryonic genome activation are defective in interspecies cloned embryos between bovine ooplasm and rhesus monkey somatic cells. BMC Dev. Biol. 2009;9:44. doi: 10.1186/1471-213X-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srirattana K. Imsoonthornruksa S. Laowtammathron C. Sangmalee A. Tunwattana W. Thongprapai T. Chaimongkol C. Ketudat-Cairns M. Parnpai R. Full-term development of gaur-bovine interspecies somatic cell nuclear transfer embryos: Effect of trichostatin A treatment. Cell. Reprogram. 2012;14:248–257. doi: 10.1089/cell.2011.0099. [DOI] [PubMed] [Google Scholar]
- Sugimura S. Narita K. Yamashiro H. Sugawara A. Shoji T. Terashita Y. Nishimori K. Konno T. Yoshida M. Sato E. Interspecies somatic cell nucleus transfer with porcine oocytes as recipients: A novel bioassay system for assessing the competence of canine somatic cells to develop into embryos. Theriogenology. 2009;72:549–559. doi: 10.1016/j.theriogenology.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Svarcova O. Laurincik J. Avery B. Mlyncek M. Niemann H. Maddox-Hyttel P. Nucleolar development and allocation of key nucleolar proteins require de novo transcription in bovine embryos. Mol. Reprod. Dev. 2007;74:1428–1435. doi: 10.1002/mrd.20727. [DOI] [PubMed] [Google Scholar]
- Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tao Y. Cheng L. Zhang M. Li B. Ding J. Zhang Y. Fang F. Zhang X. Maddox-Hyttel P. Ultrastructural changes in goat interspecies and intraspecies reconstructed early embryos. Zygote. 2008;16:93–110. doi: 10.1017/S0967199407004492. [DOI] [PubMed] [Google Scholar]
- Tarkowski A.K. Balakier H. Nucleo-cytoplasmic interactions in cell hybrids between mouse oocytes, blastomeres and somatic cells. J. Embryol. Exp. Morphol. 1980;55:319–330. [PubMed] [Google Scholar]
- Tomanek M. Kopecny V. Kanka J. Genome reactivation in developing early pig embryos: an ultrastructural and autoradiographic analysis. Anat. Embryol. (Berl.) 1989;180:309–316. doi: 10.1007/BF00315889. [DOI] [PubMed] [Google Scholar]
- Uhm S.J. Gupta M.K. Kim T. Lee H.T. Expression of enhanced green fluorescent protein in porcine- and bovine-cloned embryos following interspecies somatic cell nuclear transfer of fibroblasts transfected by retrovirus vector. Mol. Reprod. Dev. 2007;74:1538–1547. doi: 10.1002/mrd.20755. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J. Mitochondria in early mammalian development. Semin. Cell Dev. Biol. 2009;20:354–364. doi: 10.1016/j.semcdb.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Vassena R. Han Z. Gao S. Baldwin D.A. Schultz R.M. Latham K.E. Tough beginnings: Alterations in the transcriptome of cloned embryos during the first two cell cycles. Dev. Biol. 2007;304:75–89. doi: 10.1016/j.ydbio.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viuff D. Avery B. Greve T. King W.A. Hyttel P. Transcriptional activity in in vitro produced bovine two- and four-cell embryos. Mol. Reprod. Dev. 1996;43:171–179. doi: 10.1002/(SICI)1098-2795(199602)43:2<171::AID-MRD6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Viuff D. Hyttel P. Avery B. Vajta G. Greve T. Callesen H. Thomsen P.D. Ribosomal ribonucleic acid is transcribed at the 4-cell stage in in vitro-produced bovine embryos. Biol. Reprod. 1998;59:626–631. doi: 10.1095/biolreprod59.3.626. [DOI] [PubMed] [Google Scholar]
- Wang K. Beyhan Z. Rodriguez R.M. Ross P.J. Iager A.E. Kaiser G.G. Chen Y. Cibelli J.B. Bovine ooplasm partially remodels primate somatic nuclei following somatic cell nuclear transfer. Cloning Stem Cells. 2009;11:187–202. doi: 10.1089/clo.2008.0061. [DOI] [PubMed] [Google Scholar]
- Wang K. Otu H.H. Chen Y. Lee Y. Latham K. Cibelli J.B. Reprogrammed transcriptome in rhesus-bovine interspecies somatic cell nuclear transfer embryos. PLoS One. 2011;6:e22197. doi: 10.1371/journal.pone.0022197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. Duan E. Sung L.Y. Jeong B.S. Yang X. Tian X.C. Generation and characterization of pluripotent stem cells from cloned bovine embryos. Biol. Reprod. 2005;73:149–155. doi: 10.1095/biolreprod.104.037150. [DOI] [PubMed] [Google Scholar]
- Wen D.C. Bi C.M. Xu Y. Yang C.X. Zhu Z.Y. Sun Q.Y. Chen D.Y. Hybrid embryos produced by transferring panda or cat somatic nuclei into rabbit MII oocytes can develop to blastocyst in vitro. J. Exp. Zoolog. A Comp. Exp. Biol. 2005;303:689–697. doi: 10.1002/jez.a.191. [DOI] [PubMed] [Google Scholar]
- Wilmut I. Schnieke A.E. McWhir J. Kind A.J. Campbell K.H. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Yang C.X. Han Z.M. Wen D.C. Sun Q.Y. Zhang K.Y. Zhang L.S. Wu Y.Q. Kou Z.H. Chen D.Y. In vitro development and mitochondrial fate of macaca-rabbit cloned embryos. Mol. Reprod. Dev. 2003;65:396–401. doi: 10.1002/mrd.10320. [DOI] [PubMed] [Google Scholar]
- Yin X.J. Lee Y.H. Jin J.Y. Kim N.H. Kong I.K. Nuclear and microtubule remodeling and in vitro development of nuclear transferred cat oocytes with skin fibroblasts of the domestic cat (Felis silvestris catus) and leopard cat (Prionailurus bengalensis) Anim. Reprod. Sci. 2006;95:307–315. doi: 10.1016/j.anireprosci.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Zhao Z.J. Ouyang Y.C. Nan C.L. Lei Z.L. Song X.F. Sun Q.Y. Chen D.Y. Rabbit oocyte cytoplasm supports development of nuclear transfer embryos derived from the somatic cells of the camel and Tibetan antelope. J. Reprod. Dev. 2006;52:449–459. doi: 10.1262/jrd.17095. [DOI] [PubMed] [Google Scholar]