PLP levels are regulated by Centrobin and act to suppress mother centrosome maturation by blocking the localization of Polo kinase.
Abstract
Centrosomes determine the mitotic axis of asymmetrically dividing stem cells. Several studies have shown that the centrosomes of the Drosophila melanogaster central brain neural stem cells are themselves asymmetric, organizing varying levels of pericentriolar material and microtubules. This asymmetry produces one active and one inactive centrosome during interphase. We identify pericentrin-like protein (PLP) as a negative regulator of centrosome maturation and activity. We show that PLP is enriched on the inactive interphase centrosome, where it blocks recruitment of the master regulator of centrosome maturation, Polo kinase. Furthermore, we find that ectopic Centrobin expression influenced PLP levels on the basal centrosome, suggesting it may normally function to regulate PLP. Finally, we conclude that, although asymmetric centrosome maturation is not required for asymmetric cell division, it is required for proper centrosome segregation to the two daughter cells.
Introduction
Drosophila melanogaster neural stem cells, or neuroblasts (NBs), undergo invariant asymmetric cell divisions (ACDs). NBs orient their mitotic spindle along a fixed polarity axis and divide to produce a ganglion mother cell and a self-renewing NB (Kraut et al., 1996). Misalignment of the mitotic spindle leads to detrimental symmetric divisions that produce tumors (Cabernard and Doe, 2009).
Efficient orientation of the NB mitotic spindle is mediated by centrosomes, which comprise a pair of centrioles surrounded by pericentriolar material (PCM) that includes factors needed for their microtubule (MT)-organizing center (MTOC) activity, such as γ-tubulin (γ-Tub). The interphase centrosome must duplicate once in S phase to produce a mother and a daughter centrosome, which then segregate to distal sides of the cell and mature, where the amount of PCM and MTOC activity peak as cells enter mitosis (Khodjakov and Rieder, 1999). Each mitotic division asymmetrically partitions the apical (daughter) centrosome to the NB and the basal (mother) centrosome to the ganglion mother cell (Conduit and Raff, 2010; Januschke et al., 2011).
To ensure the faithful pattern of centrosome inheritance, NBs use an asymmetric maturation cycle in which the daughter centrosome remains active and immobilized at the apical side, whereas the mother is transiently inactivated and traverses the cell to a distant basal site (Rebollo et al., 2007; Rusan and Peifer, 2007). Once this centriole pair is positioned at the basal cortex, it matures and contributes to spindle formation. Recent work indicates that this centrosome asymmetry is Centrobin (Cnb) dependent but dispensable for ACD (Januschke et al., 2013). Therefore, the significance of this mechanism is little understood.
Here, we demonstrate that the Pericentrin (PCNT)-like protein (PLP) is required to suppress mother centrosome maturation by blocking the localization of the mitotic kinase, Polo. Our data also indicate that the asymmetric centrosome maturation cycle is required for efficient segregation of stem cell centrosomes.
Results and discussion
PLP is enriched on the inactive basal centrosome during interphase
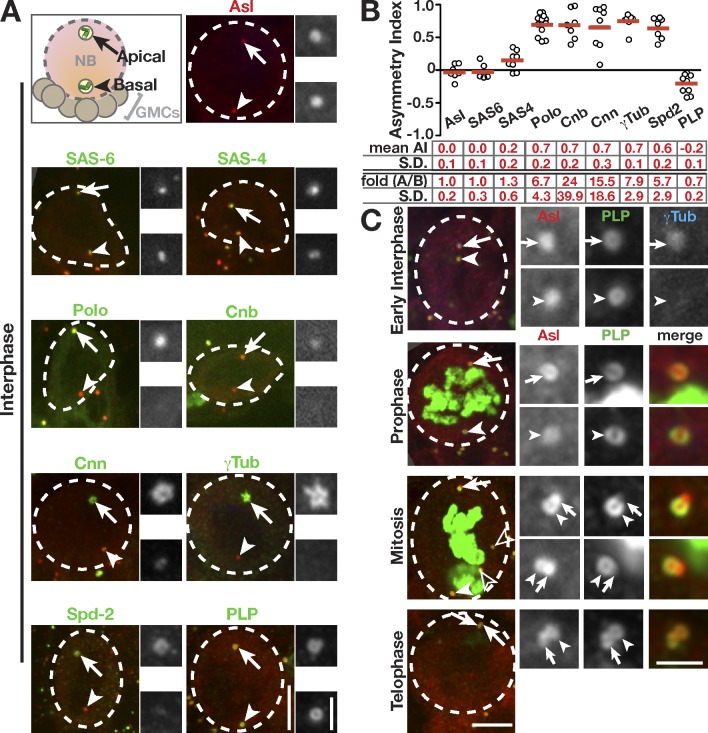
To identify factors that regulate asymmetric maturation of NB centrosomes, we assayed the interphase distribution of centrosome proteins using an asymmetry index (AI; see Materials and methods). We visualized the localization of the centriolar proteins Asterless (Asl; Varmark et al., 2007) and SAS6 (Rodrigues-Martins et al., 2007). Both Asl and SAS6 are equally present on the apical and basal centrioles (AI = 0; Fig. 1, A and B). We next examined the distribution of SAS4, which has been described both as a centriole protein and centriole–PCM scaffold (Dzhindzhev et al., 2010; Gopalakrishnan et al., 2011). SAS4 shows a bias for the apical centrosome (Fig. 1, A and B), suggesting that SAS4 is not strictly a centriole protein. Finally, we examined the distribution of several PCM proteins. As previously described, Polo, Cnb, Centrosomin, and γ-Tub (Fig. 1, A and B) preferentially associate with the apical centrosome (Rusan and Peifer, 2007; Conduit and Raff, 2010; Januschke et al., 2011). Spd2 (Spindle defective 2), which is less characterized (Giansanti et al., 2008), also localizes to the apical centrosome (Fig. 1, A and B). In contrast to all other proteins examined, PLP is selectively enriched on the basal, inactive centrosome (Fig. 1, A and B). Previous work in NBs indicates that PLP functions during mitosis to organize PCM (Martinez-Campos et al., 2004). However, given its unique distribution, we hypothesized that PLP may also contribute to asymmetric centrosome activity in interphase.
Figure 1.
PLP is enriched on the mother centrosome in interphase. The indicated proteins (green) were detected by immunofluorescence or direct fluorescence in interphase NBs (dashed circles) counterstained for Asl to localize apical/daughter (arrows) and basal/mother (arrowheads) centrosomes. The adjacent boxes (grayscale) are magnifications of the indicated proteins showing the apical (top) and basal (bottom) centrioles. (A) Projections of NBs. Cnn, Centrosomin. (B) Quantification of AI. Each data point from the graph represents one measurement from a single cell; data were collected from five to nine NBs from three to six brains. The means ± SD are shown by the red bars and numbers below the graph. Absolute fold enrichment (A/B) is also indicated. See Materials and methods for details. (C) Projections of WT NBs showing PLP distribution on centrioles. Mitotic cells are marked by pH3 (green), and open arrowheads indicate centrosomes from other cells. GMCs, ganglion mother cells. Bars: (main images) 5 µm; (insets) 1.5 µm.
Levels of PLP inversely correlate with centrosome activity
PLP was identified as the orthologue to mammalian PCNT based on the conservation of its PCNT/AKAP-450 centrosome targeting (PACT) domain (Kawaguchi and Zheng, 2004; Martinez-Campos et al., 2004). Moreover, work reveals that both PLP and PCNT share the same radial configuration that facilitates PCM scaffolding (Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012).
To understand how PLP might regulate centrosome maturation in NBs, we examined its endogenous distribution. Analysis of early interphase NBs stained for PLP and γ-Tub revealed a reciprocal relationship between PCM and PLP staining intensity (Fig. 1, A and C, early interphase). These data suggest that the enriched, and perhaps more compact, PLP on the basal centrosome does not support PCM recruitment or maintenance. By prophase, PLP levels are more uniform between the two centrosomes (Fig. 1 C), which can now both support γ-Tub recruitment. Interestingly, a drop in fluorescence on the basal centrosome is coincident with the transition of PLP from a ring to a horseshoe shape, which becomes more prominent in mitosis (Fig. 1 C). This is consistent with data from cultured Drosophila cells that note a horseshoe configuration during procentriole formation (Mennella et al., 2012). It is interesting to speculate that a ring of PLP acts to prevent both procentriole nucleation and the maturation of the basal centrosome. By telophase, PLP moves onto the daughter centrioles (Fig. 1 C), and the cycle repeats.
Our data show a negative correlation between PLP and γ-Tub intensity in interphase NBs, which implicates PLP as a negative regulator of centrosome maturation. In mitosis, PLP may transition to promote γ-Tub localization as previously shown (Martinez-Campos et al., 2004). In agreement with this model, recent work indicates that NB centrosome maturation is differentially regulated in interphase versus mitosis (Januschke et al., 2013).
Loss of PLP causes precocious activation of the mother centrosome
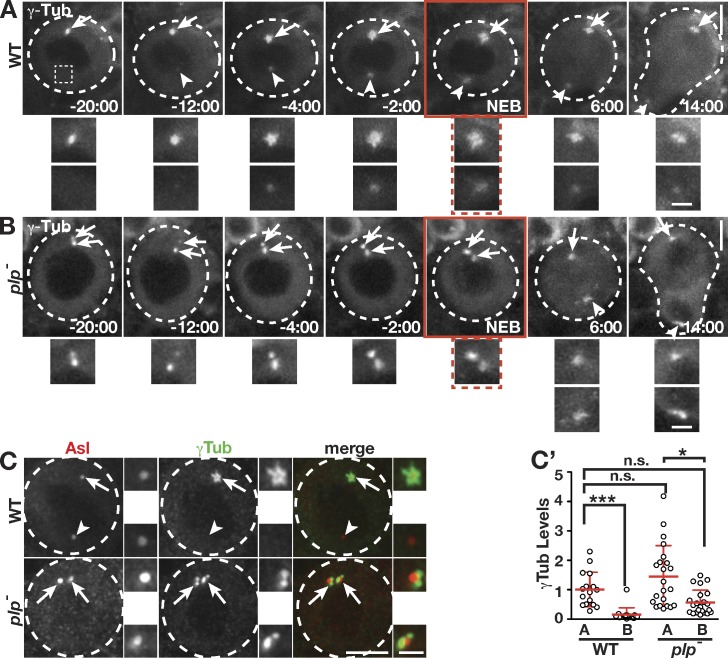
Given the inverse relationship between the levels of γ-Tub and PLP on interphase centrosomes, we hypothesized that reducing the amount of PLP in NBs might cause a concomitant increase in γ-Tub. To test this, we assayed the distribution of γ-Tub in live NBs. In control cells, a single apical centrosome associates with γ-Tub during interphase (Fig. 2 A, −20 min relative to nuclear envelope breakdown [NEB]; and Video 1). In contrast, visualization of γ-Tub in plp− (plp2172/Df) NBs revealed two active centrosomes in ∼40% (13/33) of interphase NBs (Fig. 2 B, −20 min; and Video 2). Normally, γ-Tub levels on both centrosomes rise during centrosome maturation (−12 min) and peak in prophase (−2 min; Fig. 2 A and Fig. S1, A and A′). Quantification indicates γ-Tub is enriched on apical centrosomes in excess of fourfold in interphase (−20 min) to a low of 1.5-fold in prophase (Fig. 2 A and Fig. S1 B), indicating centrosomes are most asymmetric during interphase. In contrast, both interphase and mitotic plp− NBs average 1.5-fold more γ-Tub on apical centrosomes (Fig. 2 B and Fig. S1 B). Thus, both interphase plp− centrosomes resemble control mitotic centrosomes. To quantify the frequency at which loss of PLP results in this phenotype, we assayed the localization of endogenous γ-Tub in fixed samples and found that ∼40% (48/128) of plp− interphase NBs exhibit two active centrosomes (Fig. 2 C). Quantification confirms that both centrosomes of plp− NBs recruit γ-Tub at levels that are statistically similar to the apical, active centrosome of wild-type (WT) cells (Fig. 2 C′). Although centrosome asymmetry is highly reduced in interphase plp− NBs, the apical centrosome still shows twofold more γ-Tub (Fig. 2 C′), similar to the 1.5-fold enrichment found in live cells (Fig. S1 B). We conclude that PLP normally regulates interphase centrosomes by preventing the localization of γ-Tub. Moreover, PLP plays a greater role on the basal/mother centrosome, where it is enriched.
Figure 2.
PLP prevents the maturation of the mother centrosome. (A and B) Live GFP–γ-Tub in WT (A) and plp− (B) NBs. Apical (arrows) and basal (arrowheads) centrosomes are enlarged and shown below. A basal location from −20 min WT is enlarged to indicate no detectable γ-Tub (dotted box). Times are minutes and seconds relative to NEB (red boxes). (C) Projections of interphase NBs showing anti–γ-Tub. (C′) Quantification is of γ-Tub levels at interphase apical (A) and basal (B) centrosomes from single NBs (WT: n = 16, 5 brains; plp−: n = 22, 6 brains). Data shown are from a single representative experiment from four repeats. See Materials and methods for statistical analysis. Data are means ± SD. *, P < 0.01. The adjacent boxes (grayscale) are magnifications of the indicated proteins showing the apical (top) and basal (bottom) centrioles. Dashed circles, NBs. Bars: (main images) 5 µm; (insets) 1.5 µm.
PLP blocks centrosome maturation by preventing Polo localization
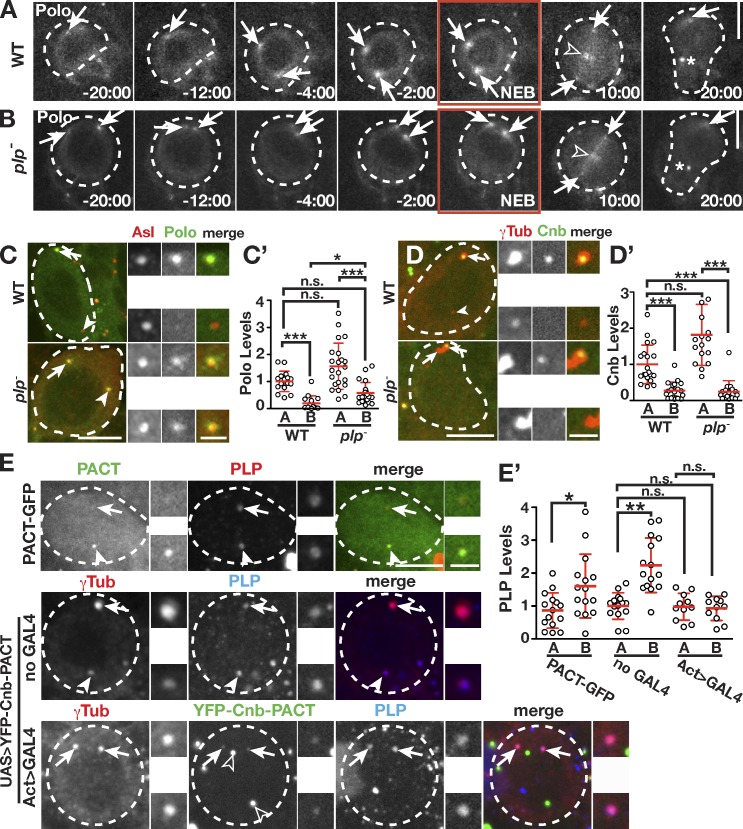
To elucidate the molecular basis of the precocious mother centrosome activation in plp− mutants, we turned to Polo, a critical centrosome maturation factor (Sunkel and Glover, 1988). In NBs, Polo localizes to apical centrosomes during interphase, where it promotes centrosome activity (Fig. 1 A). Polo is only detected at both apical and basal centrosomes during prophase (−4 min), when the centrosomes undergo mitotic maturation (Fig. 3 A). Therefore, we hypothesized that PLP functions to keep basal centrosomes inactive by preventing Polo localization. To test this, we followed Polo in cycling plp− NBs (Fig. 3 B). Strikingly, we observe interphase (−20 min) plp− NBs that display two Polo-positive centrosomes. For detailed analysis, we assayed Polo localization to centrosomes in fixed NBs and found that 34% (22/65 interphase NBs) of plp− NBs localize Polo to both centrosomes (Fig. 3 C). Direct comparison of the basal centrosomes shows a significant enrichment of Polo on plp− centrosomes versus WT (Fig. 3 C′). Therefore, PLP functions to prevent localization of Polo to centrosomes. These data support our observations that PLP is normally required to act as a brake to selectively block Polo localization to, and maturation of, interphase centrosomes.
Figure 3.
PLP prevents the localization of Polo to the mother centrosome. (A and B) Live Polo-GFP in WT (A) and plp− (B) NBs. Polo also localizes to kinetochores (open arrowheads) and the midbody ring (asterisks). Times are minutes and seconds relative to NEB (red boxes). (C–E) Projections of NBs showing Polo-GFP (C), Cnb-GFP (D), and PLP (E). Open arrowheads in E show nonspecific YFP signals. Quantification in NBs of Polo (C′) from WT (n = 16, 5 brains) and plp− (n = 22, 5 brains), Cnb (D′) from WT (n = 20, 6 brains) and plp− (n = 17, 7 brains), and PLP (E′) levels from PACT-GFP (n = 16, 7 brains), UAS>YFP-Cnb-PACT/+ (no GAL4; noninduced; n = 15, 6 brains), and UAS>YFP-Cnb-PACT/Act>GAL4 (induced; n = 11, 4 brains). Data shown are from a single representative experiment from two repeats. See Materials and methods for statistical analysis. Only plp− cells containing two γ-Tub–positive centrosomes were selected for quantification of Cnb in D′. The α-PLP antibody is against the N terminus and does not detect the PACT constructs. In all panels, apical (arrows) and basal (arrowheads) centrosomes are indicated. Data are means ± SD. *, P < 0.01; **, P < 0.001; ***, P < 0.0001. The adjacent boxes (grayscale) are magnifications of the indicated proteins showing the apical (top) and basal (bottom) centrioles. Dashed circles, NBs. A, apical; B, basal. Bars: (main images) 5 µm; (insets) 1.5 µm.
Cnb can regulate PLP levels
Recent work suggests that the daughter centriole–specific protein, Cnb, is both necessary and sufficient to promote interphase centrosome activity (Januschke et al., 2013). One model that emerges from these data is that Cnb recruits Polo to the interphase daughter centriole. In combination with our data, this predicts that PLP might function to prohibit the localization of Cnb on the mother centriole, which would then prevent Polo localization. To test this possibility, we analyzed plp− NBs expressing GFP-Cnb. The distribution of Cnb on mother versus daughter centrioles shows Cnb is restricted to a single centrosome in both control (28/28) and plp− (26/26) interphase NBs (Fig. 3 D). Even in plp− NBs that have two active interphase centrosomes, we detected no change in the level of GFP-Cnb that localizes to basal centrioles (Fig. 3 D′). Therefore, PLP does not function upstream of Cnb to regulate Polo. However, recent data show that ectopic targeting of Cnb using Cnb-PACT is sufficient to activate the mother centrosome during interphase (Januschke, et al., 2013). One possibility is that this activation is achieved because Cnb-PACT–expressing NBs exhibit reduced PLP at the basal centrosome. Indeed, in contrast to control NBs expressing GFP-PACT or noninduced Cnb-PACT, the induced expression of Cnb-PACT does reduce and symmetrize PLP localization (Fig. 3, E and E′). Although these data indicate that ectopic Cnb can regulate PLP function, the distribution of PLP (basal enriched) and Cnb (apical enriched) observed in WT NBs suggests that asymmetric centrosome maturation is established via mechanisms that combine both positive regulation on the daughter centrosome by Cnb and negative regulation on the mother centrosome by PLP.
Our data predict that overexpression of PLP would inactivate the apical centrosome. To test this, we attempted to overexpress PLP using four isoforms (PF, PE, PD, and PC; see Materials and methods), none of which recapitulated endogenous PLP localization. Therefore, this experiment was inconclusive. Further understanding of PLP isoforms and detailed genetic analysis are required to clarify the multiple mechanisms (Cnb and PLP) that instruct interphase centrosome maturation.
PLP is required for centrosome separation and the initial mitotic spindle alignment
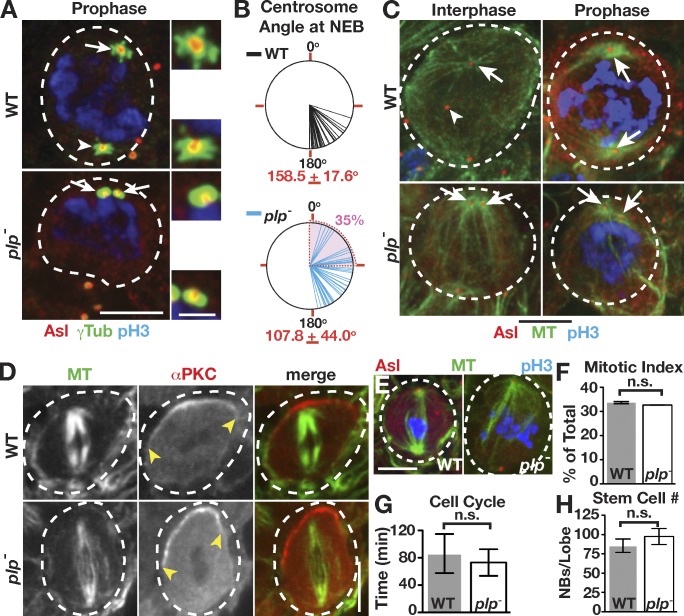
The presence of two active interphase centrosomes in plp− NBs allowed us to test the importance of centrosome asymmetry observed in normal interphase NBs. In WT cells, centrosomes are well separated in prophase (Fig. 4 A). In contrast, plp− NBs often fail to separate their centrosomes by the same time point (Fig. 4 A). Quantification of the degree of centrosome separation (θ) in live cells indicates that by NEB, the centrosomes of plp− NBs average a 108° separation, significantly lower than the 159° separation in WT cells (Fig. 4 B). Furthermore, θ < 90° at NEB was observed in 35% (16/46) of plp− NBs, which strongly correlates with the frequency at which we observe NBs with two active centrosomes (40%). We favor a hypothesis in which the two active, apical centrosomes present in plp− cells fail to segregate as a result of physical association of both centrosomes to the apical cortex via astral MTs. In support of this hypothesis, both plp− centrosomes maintain robust MTOC activity in interphase and mitosis (Fig. 4 C). These data suggest that inactivation of the basal centrosome in normal cells is an early requisite for the alignment of the mitotic spindle. However, all plp− NBs we imaged corrected this centrosome separation defect immediately after NEB (Fig. 2 B). Thus, the mitotic centrosome separation machinery can robustly overcome early centrosome positioning defects.
Figure 4.
PLP is required for centrosome inheritance but not ACD. (A) Projections of prophase WT and plp− NBs. Active (arrows) and inactive (arrowheads) centrosomes are indicated. The adjacent boxes are magnifications of the indicated proteins showing the apical (top) and basal (bottom) centrioles. (B) Angle of centrosome separation at NEB from live γ-Tub analysis of WT and plp− NBs: WT (n = 42) and plp− (n = 46). The mean angle of centrosome separation is significantly different: P < 0.0001, two-tailed Student’s t test. The means ± SD are indicated in red, and the shaded area corresponds to <90°. (C) Interphase and mitotic MTOCs from WT and plp− NBs. (D) WT and plp− NBs showed no difference in proper spindle alignment along the apical (α-PKC, yellow arrowheads) and basal axis. (E) Some plp− mitotic spindles are abnormal compared with WT. (F–H) Mitotic index (WT: n = 38/169; plp−: n = 32/150; F), cell cycle length (WT: n = 26; plp−: n = 39; G), and NB number from anterior and posterior optic lobe (stained for Mira; H) are all unaffected and displayed as means ± SD. Dashed circles, NBs. Bars: (main images) 5 µm; (insets) 1.5 µm.
Asymmetric centrosome activity is required for proper centrosome inheritance
One long-standing hypothesis posits that NBs must have one active and one inactive centrosome for proper ACD and fate determinant segregation. However, plp− NBs show a normal distribution of cortical polarity proteins and metaphase spindle alignment (Fig. 4 D). Likewise, the mitotic-like plp− centrosomes do not alter the interphase distribution of cortical polarity cues (Fig. S1, C and C′). Despite previously noted (Martinez-Campos et al., 2004) aberrant spindles (Fig. 4 E), we found no difference in either the frequency or duration of mitosis (Fig. 4, F and G). Finally, the standard measure of defects in ACD is to count the number of NBs and assay for brain tumor formation (Castellanos et al., 2008). Our analysis revealed no difference in the number of NBs or brain size in mutants (Fig. 4 H and Fig. S2 A). Therefore, in agreement with a recent study (Januschke et al., 2013), we find that the asymmetry of centrosome activity in interphase appears dispensable for NB ACD.
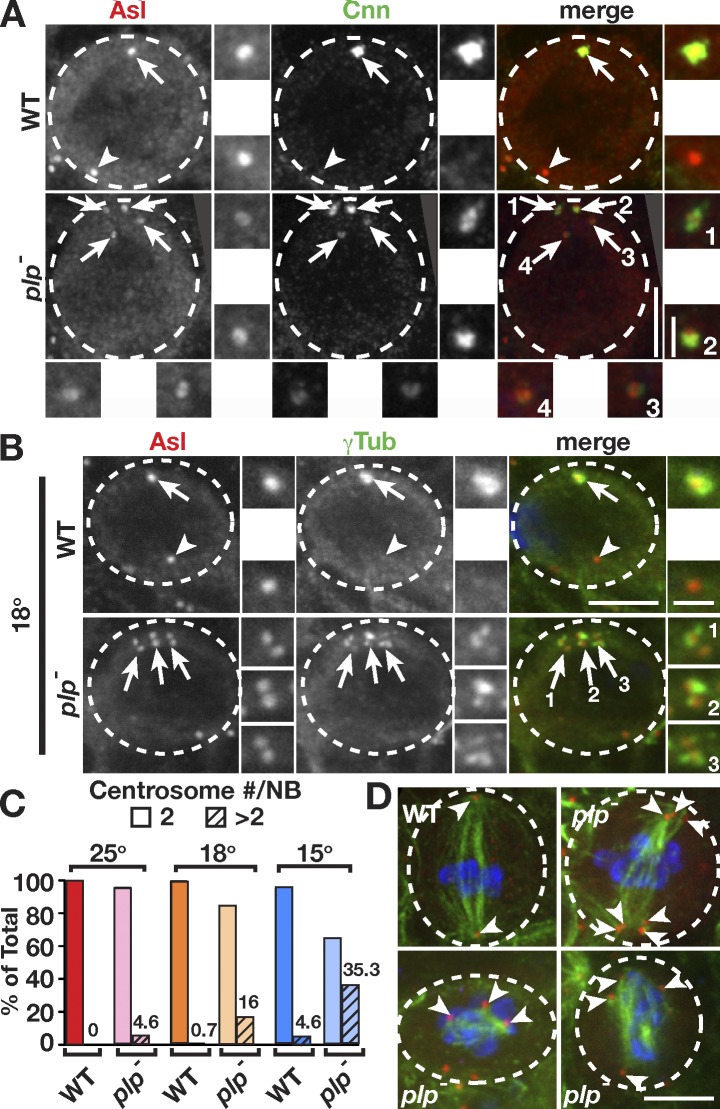
To detect subtle abnormalities in plp− NBs, we analyzed a very large number of fixed brains. Our analysis revealed some plp− NBs (4.6%; 15/323 NBs), but no controls, inherit too many centrosomes (Fig. 5 A), some with >10 centrosomes. It is likely that active centrosomes from earlier cell cycles remained associated at the apical pole and were segregated into the NB. These observations suggest that the asymmetric centrosome maturation cycle evolved as a mechanism to ensure proper spindle formation and the faithful inheritance of centrosomes.
Figure 5.
plp− mutant cells contain supernumerary centrosomes. (A and B) WT and plp− larvae were grown from first to third instar at 25°C (A) or 18°C (B). plp− NBs contain supernumerary centrosomes (arrows). The adjacent boxes (grayscale) are magnifications of the indicated proteins showing the apical (top) and basal (bottom) centrioles. Cnn, Centrosomin. (C) Quantification of supernumerary centrosomes was determined by counting the number of NBs containing more than two Asl-positive centrosomes divided by the total number of NBs. Frequency is indicated on each column, and means ± SD percentages were as follows: 25°C (WT: n = 0/57, 7 brains, 0%; plp−: n = 15/323, 24 brains, 4.6 ± 0.9%), 18°C (WT: n = 1/148, 9 brains, 0.7 ± 0.2%; plp−: n = 30/187, 9 brains, 16 ± 1.3%), and 15°C (WT: n = 6/109, 8 brains, 4.6 ± 0.6%; plp−: n = 36/102, 5 brains, 35 ± 1.4%). (D) Mitotic spindles from one WT and three plp− cold-treated NBs; green (MTs), red (Asl), and blue (pH3). Arrowheads label spindle poles. Dashed circles, NBs. Bars: (main images) 5 µm; (insets) 1.5 µm.
To assay whether loss of PLP sensitizes NBs to mitotic aberrations, we reared first-instar larvae under cold conditions. We reasoned that lower temperatures would slow the dynamics of MTs and their associated motors, both of which are required to drive centrosome separation. We found no difference between control larvae grown at 25°C versus 18°C (Fig. 5, B and C). In contrast, supernumerary centrosomes were markedly increased to 16% in plp− NBs from larvae grown at 18°C, and this phenotype was exacerbated to 35% at 15°C (Fig. 5, B and C; and Fig. S2 B). We note that although some control NBs do fail to properly segregate their centrosomes at 15°C (4.6%), the defect is more prevalent in plp− mutants. To test whether this level of centrosome amplification can induce brain tumors (Basto et al., 2008), we counted NBs from cold-reared plp− larvae. We did not see a significant increase in NBs (Fig. S2 C), despite the presence of many active centrosomes in mitotic cells (Fig. S2 B), suggesting that our levels of centrosome amplification are insufficient to drive tumorigenesis or that cold temperature may be refractory to NB proliferation. Nonetheless, we find an increase in the severity of mitotic spindle defects in plp− NBs from cold-reared larvae, including the formation of multipolar spindles (Fig. 5 D). We predict that these multipolar spindles generally permit normal ACD because of centrosome clustering forces (Basto et al., 2008) but may gradually contribute to chromosomal instability and tumor formation over time.
Models
We present a model in which PLP functions as a negative regulator of centrosome maturation in interphase NBs by preventing the localization of Polo to the basal centrosome (Fig. S3 A). Loss of PLP leads to the activation of both centrosomes in interphase and greatly reduces their mobility, resulting in the atypical apical positioning and inheritance of both centrosomes (Fig. S3 B). Our temperature shift experiments illustrate that plp− mutants are sensitized to increased centrosome segregation and mitotic spindle defects (Fig. S3 B). We conclude that inhibition of basal/mother centrosome maturation during interphase is critical for proper centrosome partitioning and mitotic spindle organization in neural stem cells.
It is unknown how PLP functions as a negative regulator, but one model may involve an indirect role for PLP. For example, PLP might normally promote early centriole separation to ensure efficient movement of the mother centriole away from the apical cortex to allow for its inactivation. We performed two experiments to test this model. First, we analyzed the timing of centriole separation in cells exiting mitosis in WT and plp− NBs (Fig. S2 D) and found that centriole disengagement (WT = 10 ± 5 vs. plp− = 7 ± 6, min ± SD relative to anaphase onset) and the earliest signs of independent centriole movement (WT = 16 ± 6 vs. plp− = 15 ± 8) are unaffected (see Materials and methods). Therefore, centrosomes remain apically positioned in early interphase in both WT and plp− NBs. Next, we tested whether apical positioning of centrioles is sufficient for their activation. We analyzed early interphase WT NBs that contained two apical centrioles (<30° separation) and found no correlation between centrosome position and activity (Fig. S2, E and E′). Collectively, these observations argue that PLP does not influence early centrosome separation and that centrosome location is not critical for mother centrosome inactivation.
Instead, we favor the alternative hypothesis that PLP directly regulates centrosome activity, which, in turn, influences centrosome position. One possible model is that PLP acts on the centrosome by physically shielding the docking of promaturation factors until mitotic entry. A biochemical modification and/or conformational change would then allow PLP to transition to a positive regulator in mitosis that scaffolds other PCM factors. Another possible direct function for PLP might be to differentially modulate the behavior of proteins at the apical and basal centrosomes. It will be critical to examine how the dynamics of other centrosome proteins are affected by the loss of PLP. Given the importance of interphase centrosome asymmetry in stem cell division, we believe that an in depth understanding of the interplay between PCM proteins in both interphase and mitosis is a critical area of future research.
Materials and methods
Fly stocks
The following mutant strains and transgenic lines were used: plp2172 (Spradling et al., 1999), Ppolo-GFP-Polo expressing Polo under the endogenous promoter (Moutinho-Santos et al., 1999), Pubi-GFP-SAS4 expressing SAS4 ubiquitously (Dix and Raff, 2007), Df(3L)BrdR15, which is a deficiency that uncovers plp and Pubi-GFP-PACT expressing PACT ubiquitously (gift from J. Raff, University of Oxford, Oxford, England, UK; Martinez-Campos, et al., 2004), upstream activation sequence (UAS)–YFP-Cnb-PACT that only expresses Cnb-PACT when coexpressed with GAL4 (gift from C. Gonzalez, Institute for Research in Biomedicine, Barcelona, Spain; Januschke, et al., 2013), Psqh-GFP-Moesin that expresses Moesin under the spaghetti squash promoter (Kiehart et al., 2000), and PAct5C-GAL4 for ubiquitous GAL4 expression under the Actin5C promoter (Bloomington Drosophila Stock Center). y−w− flies were controls for immunofluorescence experiments, unless otherwise noted. plp− mutants are plp2172/Df(3L)BrdR15 trans-heterozygotes; plp2172 is a strong hypomorph induced by P element insertion.
Construction of transgenic animals
All transgenic animals were generated by BestGene, Inc. Ubiquitin-GFP–γ-Tub23C and Ubiquitin-GFP-Cnb animals were generated by cloning cDNA of each gene into the pUGW destination vector using the Gateway system (Invitrogen). γ-Tub23C was PCR amplified from EST clone LD40196 using the primers 5′-CACCATGCCAAGTGAAATAATTACTTTGCAGCTTG-3′ and 5′-GGAACCGGCGCTGGTCACAGATCGAC-3′. Cnb was PCR amplified from EST clone SD06673 using the primers 5′-CACCATGAGTGATACCGATACGGACG-3′ and 5′-GCACTTCCAAGGTGGAGGCTTACTG-3′. GFP-SAS6 animals were generated by recombineering as previously described (Venken et al., 2008). PBac-GFP-SAS6 was generated from the bacterial artificial chromosome clone CH322-126P4 (BACPAC Resources Center, Children’s Hospital Oakland Research Institute) using the following primers to amplify the N-terminal GFP-Kanamycin cassette from the N-EGFP (N-terminal EGFP) template vector: 5′-ACCAATTTTAAAATTTTAAAAATTAAGGTTTCTCACGAGCCTGATCTATCATGGTGAGCAAGGGCGAGGAG-3′ and 5′-TTGCCATAGTCCATTTTGGCGGAGTAGCTATCCTCGCTCCCTGGAGGCCAACTAGTGGATCCCCTCGAGGGAC-3′. This modified bacterial artificial chromosome was directionally integrated into the genome using the PhiC31 system and the attP-3BVK00002 stock (stock 9723; Bloomington Drosophila Stock Center). The plp gene is predicted to encode 12 protein isoforms, which are annotated in Flybase as Px, in which P denotes a protein and x represents a unique isoform named in the order of discovery. PLP isoforms PC, PD, PE, and PF failed to recapitulate expression of endogenous PLP protein in NBs.
Live-cell imaging
Whole brains from crawling third-instar larvae were explanted into Schneider’s Drosophila medium (Invitrogen) supplemented with Antibiotic-Antimycotic (Gibco; Invitrogen), dissected from peripheral tissues, and mounted into a drop of the same medium on a 50-mm gas-permeable lumox dish (Sarstedt). Media were surrounded by Halocarbon oil 700 (Sigma-Aldrich) to support a 22 × 22–mm glass coverslip (#1.5; Thermo Fisher Scientific). The anterior-ventral NBs were imaged using a 40×, 1.3 NA oil immersion objective in a stage incubator (Binomic System BC-110 Controller; 20/20 Technologies) heated to 25°C on a microscope (Eclipse Ti; Nikon) fitted with a spinning-disk confocal head (CSU-22; Yokagawa Corporation of America), a cooled charge-coupled device camera (Clara; Andor Technology), and the Perfect Focus System (Nikon), all run by an automated controller (MAC 6000; Ludl Electronic Products) using MetaMorph software (Molecular Devices). Laser excitation was supplied by a laser merge module equipped with 491-, 561-, and 642-nm solid-state lasers (VisiTech International). Images were collected at 1-µm z intervals over a 10–15-µm volume at 120-s time intervals. Images were assembled using Photoshop (Adobe) and QuickTime Player 7 software (Apple).
Immunofluorescence
Whole brains were fixed in 9% formaldehyde in PBSTx (PBS and 0.3% Triton X-100) for 15 min, washed 3 × 15 min in PBSTx, blocked for 1 h in PBT (PBS, 0.1% Tween 20, and 1% BSA), and then incubated in primary antibody in PBT plus 4% normal goat serum overnight at 4°C. After several washes, samples were further blocked in modified PBT (PBS, 0.1% Tween 20, 2% BSA, and 4% normal goat serum) and incubated for 2 h at room temperature with a secondary antibody. Samples were finally washed 3 × 15 min in PBST (PBS and 0.1% Tween 20) and mounted in mounting medium (Aqua-Poly/Mount; Polysciences, Inc.). Samples were imaged as described in the Live-cell imaging section, using a 100×, 1.49 NA oil immersion objective unless otherwise noted, and images were collected at 0.25-µm z intervals. Maximum intensity projections were generated with ImageJ software (National Institutes of Health). Primary antibodies: rabbit anti–α-PKC C20 (1:1,000; Santa Cruz Biotechnology, Inc.), mouse anti–-γ-Tub ascites GTU-88 (1:300; Sigma-Aldrich), rabbit antiphospho–Histone H3 Ser 10 (pH3; 1:1,000; EMD Millipore), mouse anti–β-Tubulin E7 (1:250; Drosophila Studies Hybridoma Bank), rabbit anti-Bazooka (made against aa 1–311, used at 1:2,000; gift from T. Harris, University of Toronto, Toronto, Ontario, Canada; McKinley et al., 2012), rat anti-Mira (made against the SPPQKQVLKARNI peptide; used at 1:1,000; gift from C. Doe, University of Oregon, Eugene, OR; Peng et al., 2000), rabbit anti-Centrosomin (made against aa 271–1,034; used at 1:2,000; gift from T. Megraw, Florida State University; Tallahassee, FL; Heuer et al., 1995), rabbit anti-Spd2 (made against aa 915–1,146; used at 1:3,000; gift from M. Gatti, Insituto Pasteur; Rome, Italy; Giansanti et al., 2008), rabbit anti-PLP (made against aa 8–351; used at 1:5,000; Rogers et al., 2008), and guinea pig anti-Asl GP1 (made against full-length Asl; used at 1:3,000; gift from G. Rogers, University of Arizona Cancer Center, Tucson, AZ). Secondary antibodies were as follows: Alexa Fluor 488, 568, or 647 (1:500; Molecular Probes).
Manipulation of temperature
Unless otherwise noted, all experiments were performed at 23–25°C. For cold sensitization experiments, bottles seeded with first-instar larvae were shifted to either an 18°C incubator (Precision Large-Capacity 30MR; Thermo Fisher Scientific) or a 15°C incubator (MIR-145 refrigerated incubator; Sanyo). Larvae were left to develop until third-instar crawling stage, when they were dissected for analysis.
Image analysis
ImageJ was used for all image analysis. Several criteria were used to select S/G2-stage NBs: duplicated and nearly complete separation of centrosomes, absence of pH3 staining, and an enlarged nucleus. Apical versus basal centrosomes were identified based on their relative position in the cell. For plp− mutants with centrosome segregation defects, we assigned the apical designation to that centrosome that organized the most PCM or associated with GFP-Cnb. Centrosome fluorescence (CF) was calculated from maximum intensity projections by taking the difference between the integrated densities of a centrosome and an equal area of proximal intracellular space. AIs were calculated from stage S/G2 NBs to compare the asymmetric localization of proteins at apical versus basal centrosomes (Hotz et al., 2012). AI is defined by the equation (A − B)/(A + B), in which A represents the CF of an apical centrosome and B represents the CF of the basal centrosome from the same cell. Positive values indicate apical centrosome enrichment, whereas negative values indicate enrichment on basal centrosomes. The fold enrichment of centrosome proteins represents the quotient of protein localization to apical versus basal centrosomes and is defined by the equation A/B. The angle of centrosome separation (θ) was measured from live NBs by tracing the angle formed from the nucleus center to each of the two centrosomes. To calculate the mitotic index, we counted the number of pH3-labeled NBs and divided by the total number of NBs.
Statistical analysis
Data were plotted, and statistical analyses were performed using Excel (Microsoft) and Prism (GraphPad Software). For Fig. 2 C′ and Fig. 3 (C′, D′, and E′), data are normalized to mean (red bars) of WT apical measurements and plotted as means ± SD, in which P < 0.0001 (***), P < 0.001 (**), P < 0.01 (*), and not significant were determined by analysis of variance followed by Tukey’s post test. Except in Fig. 3 E′, significance of PLP levels in PACT-GFP NBs was determined by Student’s two-tailed t test; P < 0.01 (*).
Online supplemental material
Fig. S1 shows that PLP restricts maturation of the interphase mother centrosome. Fig. S2 shows cold sensitization of NBs. Fig. S3 shows a proposed model of the establishment of interphase NB centrosome asymmetry. Video 1 shows live Imaging of GFP–γ-Tub23C in a control NB. Video 2 shows live Imaging of GFP–γ-Tub23C in a plp− NB. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201303141/DC1.
Supplementary Material
Acknowledgments
We are grateful to C. Fagerstrom for assistance with molecular biology and B. Galletta for contributing to the live γ-Tub23C data. We thank the Bloomington Drosophila Stock Center, C. Doe, M. Gatti, C. Gonzalez, T. Megraw, J. Raff, and G. Rogers for reagents. We thank J. Smyth, B. Galletta, and C. Shebelut for their comments and suggestions.
This work was supported by the division of intramural research at the National Institutes of Health/National Heart, Lung, and Blood Institute (1ZIAHL006126) and a Lenfant Biomedical Postdoctoral Fellowship awarded to D.A. Lerit.
Footnotes
Abbreviations used in this paper:
- ACD
- asymmetric cell division
- AI
- asymmetry index
- Asl
- Asterless
- Cnb
- Centrobin
- γ-Tub
- γ-tubulin
- MT
- microtubule
- MTOC
- MT-organizing center
- NEB
- nuclear envelope breakdown
- NB
- neuroblast
- PACT
- PCNT/AKAP-450 centrosome targeting
- PCM
- pericentriolar material
- PCNT
- Pericentrin
- PLP
- PCNT-like protein
- UAS
- upstream activation sequence
- WT
- wild type
References
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., Raff J.W. 2008. Centrosome amplification can initiate tumorigenesis in flies. Cell. 133:1032–1042 10.1016/j.cell.2008.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabernard C., Doe C.Q. 2009. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev. Cell. 17:134–141 10.1016/j.devcel.2009.06.009 [DOI] [PubMed] [Google Scholar]
- Castellanos E., Dominguez P., Gonzalez C. 2008. Centrosome dysfunction in Drosophila neural stem cells causes tumors that are not due to genome instability. Curr. Biol. 18:1209–1214 10.1016/j.cub.2008.07.029 [DOI] [PubMed] [Google Scholar]
- Conduit P.T., Raff J.W. 2010. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr. Biol. 20:2187–2192 10.1016/j.cub.2010.11.055 [DOI] [PubMed] [Google Scholar]
- Dix C.I., Raff J.W. 2007. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 17:1759–1764 10.1016/j.cub.2007.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., Glover D.M. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature. 467:714–718 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Fu J., Glover D.M. 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2:120104 10.1098/rsob.120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M.G., Bucciarelli E., Bonaccorsi S., Gatti M. 2008. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 18:303–309 10.1016/j.cub.2008.01.058 [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan J., Mennella V., Blachon S., Zhai B., Smith A.H., Megraw T.L., Nicastro D., Gygi S.P., Agard D.A., Avidor-Reiss T. 2011. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat Commun. 2:359 10.1038/ncomms1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer J.G., Li K., Kaufman T.C. 1995. The Drosophila homeotic target gene centrosomin (cnn) encodes a novel centrosomal protein with leucine zippers and maps to a genomic region required for midgut morphogenesis. Development. 121:3861–3876 [DOI] [PubMed] [Google Scholar]
- Hotz M., Leisner C., Chen D., Manatschal C., Wegleiter T., Ouellet J., Lindstrom D., Gottschling D.E., Vogel J., Barral Y. 2012. Spindle pole bodies exploit the mitotic exit network in metaphase to drive their age-dependent segregation. Cell. 148:958–972 10.1016/j.cell.2012.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Llamazares S., Reina J., Gonzalez C. 2011. Drosophila neuroblasts retain the daughter centrosome. Nat. Commun. 2:243 10.1038/ncomms1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Reina J., Llamazares S., Bertran T., Rossi F., Roig J., Gonzalez C. 2013. Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nat. Cell Biol. 15:241–248 10.1038/ncb2671 [DOI] [PubMed] [Google Scholar]
- Kawaguchi S., Zheng Y. 2004. Characterization of a Drosophila centrosome protein CP309 that shares homology with Kendrin and CG-NAP. Mol. Biol. Cell. 15:37–45 10.1091/mbc.E03-03-0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C.L. 1999. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 146:585–596 10.1083/jcb.146.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart D.P., Galbraith C.G., Edwards K.A., Rickoll W.L., Montague R.A. 2000. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J. Cell Biol. 149:471–490 10.1083/jcb.149.2.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut R., Chia W., Jan L.Y., Jan Y.N., Knoblich J.A. 1996. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 383:50–55 10.1038/383050a0 [DOI] [PubMed] [Google Scholar]
- Lawo S., Hasegan M., Gupta G.D., Pelletier L. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14:1148–1158 10.1038/ncb2591 [DOI] [PubMed] [Google Scholar]
- Martinez-Campos M., Basto R., Baker J., Kernan M., Raff J.W. 2004. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165:673–683 10.1083/jcb.200402130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley R.F., Yu C.G., Harris T.J. 2012. Assembly of Bazooka polarity landmarks through a multifaceted membrane-association mechanism. J. Cell Sci. 125:1177–1190 10.1242/jcs.091884 [DOI] [PubMed] [Google Scholar]
- Mennella V., Keszthelyi B., McDonald K.L., Chhun B., Kan F., Rogers G.C., Huang B., Agard D.A. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14:1159–1168 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho-Santos T., Sampaio P., Amorim I., Costa M., Sunkel C.E. 1999. In vivo localisation of the mitotic POLO kinase shows a highly dynamic association with the mitotic apparatus during early embryogenesis in Drosophila. Biol. Cell. 91:585–596 [PubMed] [Google Scholar]
- Peng C.Y., Manning L., Albertson R., Doe C.Q. 2000. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature. 408:596–600 10.1038/35046094 [DOI] [PubMed] [Google Scholar]
- Rebollo E., Sampaio P., Januschke J., Llamazares S., Varmark H., González C. 2007. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell. 12:467–474 10.1016/j.devcel.2007.01.021 [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Bettencourt-Dias M., Riparbelli M., Ferreira C., Ferreira I., Callaini G., Glover D.M. 2007. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 17:1465–1472 10.1016/j.cub.2007.07.034 [DOI] [PubMed] [Google Scholar]
- Rogers G.C., Rusan N.M., Peifer M., Rogers S.L. 2008. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol. Biol. Cell. 19:3163–3178 10.1091/mbc.E07-10-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan N.M., Peifer M. 2007. A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 177:13–20 10.1083/jcb.200612140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A.C., Stern D., Beaton A., Rhem E.J., Laverty T., Mozden N., Misra S., Rubin G.M. 1999. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 153:135–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel C.E., Glover D.M. 1988. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 89:25–38 [DOI] [PubMed] [Google Scholar]
- Varmark H., Llamazares S., Rebollo E., Lange B., Reina J., Schwarz H., Gonzalez C. 2007. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 17:1735–1745 10.1016/j.cub.2007.09.031 [DOI] [PubMed] [Google Scholar]
- Venken K.J., Kasprowicz J., Kuenen S., Yan J., Hassan B.A., Verstreken P. 2008. Recombineering-mediated tagging of Drosophila genomic constructs for in vivo localization and acute protein inactivation. Nucleic Acids Res. 36:e114 10.1093/nar/gkn486 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.