Single-cell analyses in budding yeast reveal that neighboring replicons are assembled stochastically and stay associated to maintain stable replication factories.
Abstract
Inside the nucleus, DNA replication is organized at discrete sites called replication factories, consisting of DNA polymerases and other replication proteins. Replication factories play important roles in coordinating replication and in responding to replication stress. However, it remains unknown how replicons are organized for processing at each replication factory. Here we address this question using budding yeast. We analyze how individual replicons dynamically organized a replication factory using live-cell imaging and investigate how replication factories were structured using super-resolution microscopy. Surprisingly, we show that the grouping of replicons within factories is highly variable from cell to cell. Once associated, however, replicons stay together relatively stably to maintain replication factories. We derive a coherent genome-wide mathematical model showing how neighboring replicons became associated stochastically to form replication factories, which was validated by independent microscopy-based analyses. This study not only reveals the fundamental principles promoting replication factory organization in budding yeast, but also provides insight into general mechanisms by which chromosomes organize sub-nuclear structures.
Introduction
DNA replication initiates at replication origins and proceeds as replication forks move along parental DNA. At each replication fork, DNA polymerases and accessory proteins, such as proliferating cell nuclear antigen (PCNA), form a large complex (called the replisome) that conducts de novo DNA synthesis. Current evidence suggests that in eukaryotes, the two sister replisomes initiated from each origin stay associated (Falaschi, 2000; Kitamura et al., 2006; Ligasová et al., 2009) and normally coordinate replication of the entire replicon (Conti et al., 2007; Natsume and Tanaka, 2010). Nevertheless, if one fork stalls, for example due to DNA damage, the other fork can still continue DNA synthesis (Doksani et al., 2009; Yardimci et al., 2010). In eukaryotes, multiple pairs of sister replisomes are grouped together at discrete sites called replication factories (Berezney et al., 2000; Kitamura et al., 2006; Gillespie and Blow, 2010). Upon replication stress or DNA damage, a replication factory defines an important boundary, inside of which dormant origins can initiate replication and thus evade checkpoint-dependent inhibition (Dimitrova and Gilbert, 2000; Ge and Blow, 2010; Thomson et al., 2010). Moreover, replication factories may facilitate efficient DNA synthesis by creating high concentrations of replication proteins (Natsume and Tanaka, 2010).
However, in spite of their functional importance, principles that underpin the organization of replication factories remain elusive. There has been a long-standing debate about whether replication factories are organized by deterministic constraints or by stochastic processes. Several reports have suggested that the organization is facilitated by particular regulatory factors (Yan et al., 1998) or post-translational modifications (Rossi et al., 1999). Nevertheless, these observations still leave open whether or not replicons are grouped together stochastically.
Results and discussion
Live-cell microscopy reveals that neighboring replicons often form replication factories but their grouping is highly variable from cell to cell
To investigate the organization of replication factories, we analyzed the behavior of replicons using live-cell microscopy in the budding yeast Saccharomyces cerevisiae. We chose a region on chromosome VII with four adjacent replicons (Fig. 1 A). We selected one locus on each replicon such that all four loci show the same average replication timing (Yabuki et al., 2002). We integrated tetO and lacO arrays at paired loci in three different yeast strains (strains #1–3; Fig. 1 A, bottom). These arrays bound TetR and LacI proteins, fused with cyan and green fluorescent proteins (CFP, GFP), respectively, and were thus visualized as small fluorescent dots. The fluorescent dots increased in intensity upon DNA replication as the number of arrays was doubled, thus defining their replication timing by microscopy (Kitamura et al., 2006). Fluorescent dots did not increase their intensity if DNA replication was inhibited.
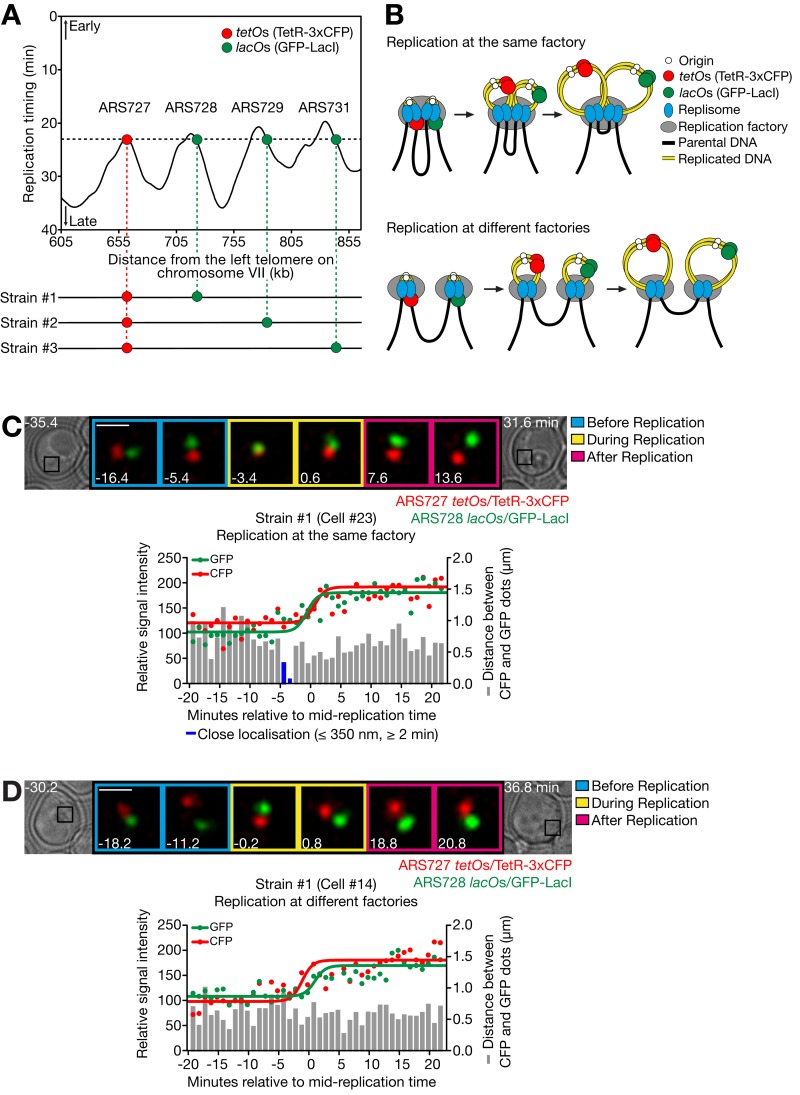
Figure 1.
Live-cell imaging reveals assembly of replicons at replication factories. (A) Three yeast strains were constructed to analyze assembly of three different replicon pairs at an individual factory. In strains #1 (T7278), #2 (T9184), and #3 (T7277) with TetR-3xCFP and GFP-LacI, the indicated two chromosomal loci were marked by integration of tetO×224 and lacO×256. The replication timing profile (top) was obtained from Yabuki et al. (2002); 0 min represents the time of release from α-factor arrest. (B) Models for replication of two replicons at the same factory and at different factories. (C and D) Representative examples of strain #1 showing replication of CFP and GFP dots at the same factory (C) and at different factories (D). Cells were released from α-factor treatment. CFP and GFP images were acquired every 1 min for 65 min. The intensity of CFP/GFP dots and their distance were measured in 23 cells of strain #1 in single experiments. The change in intensity of each dot was fitted by a sigmoidal dose–response curve (see Materials and methods; see R2, representing fitness, below) in which a midpoint in the increase of its intensity was defined as its replication time. Then 0 min was defined as the mid-replication time of two dots. R2 = 0.70 (C, CFP), 0.81 (C, GFP), 0.78 (D, CFP), and 0.74 (D, GFP). Bars, 1 µm.
It is known that there is cell-to-cell variation in the time that any particular origin initiates (Bechhoefer and Rhind, 2012), so that the two marked loci in each strain did not always replicate with similar timing (Fig. S1 A). To analyze how replicons are gathered into factories, we focused on the cells in which the two marked loci replicated with similar timing (difference <3 min). We then determined whether the two loci replicated in close spatial proximity (≤350 nm apart for ≥2 min), meaning that they replicated in the same factory (Fig. 1, B [top] and C). Alternatively, they could replicate in different factories (Fig. 1, B [bottom] and D). The same proximity threshold was previously used to define the association of sister replisomes (Kitamura et al., 2006) and was independently validated as described below.
Using this protocol, the two marked loci in strain #1 that are 70 kb apart (the distance from mid-tetO to mid-lacO) replicated in the same factory in 43% of cells (10/23) and in different factories in 57% of cells (13/23; Fig. 2 A), suggesting that grouping of replicons within factories can vary from cell to cell. In contrast, in strains #2 (loci 131 kb apart) and #3 (loci 192 kb apart), the two marked loci replicated in the same factory less frequently (Fig. 2 A and Fig. S1, B–D): 11% (2/19) and 5% (1/19), respectively (P = 0.037 and 0.0059, respectively, compared with strain #1). Thus, replicons close along a chromosome were often processed for replication in the same factory, but replicons farther apart replicated more frequently in different factories.
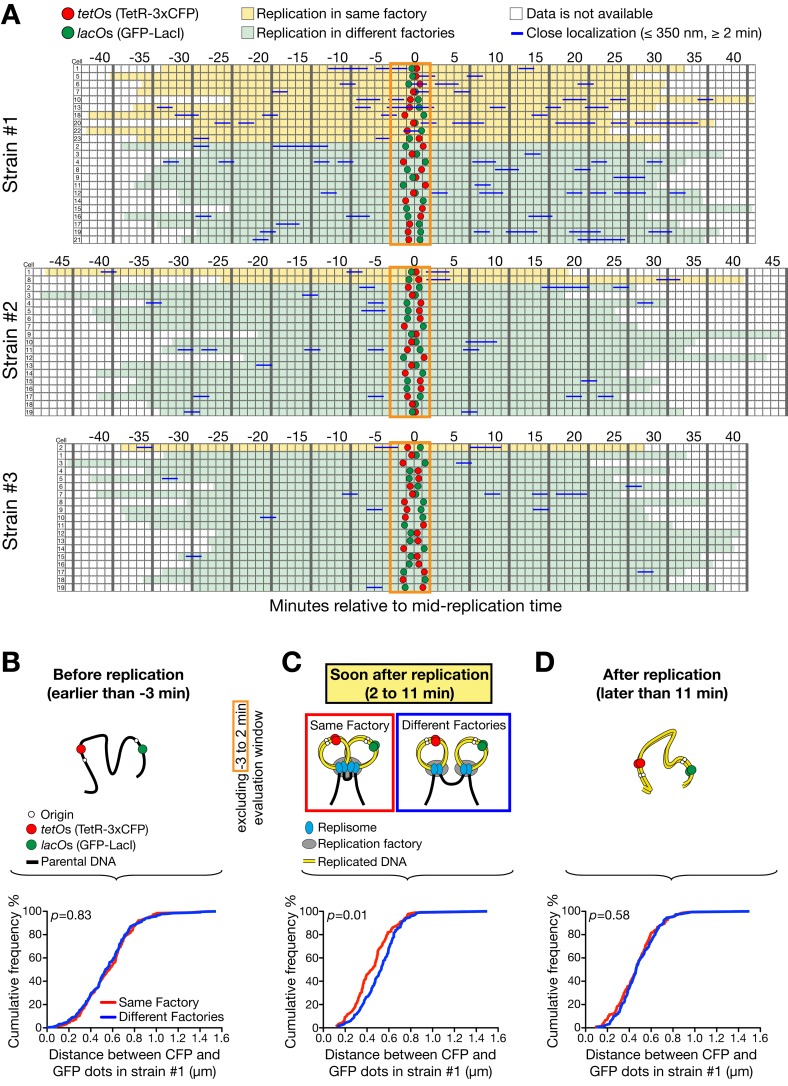
Figure 2.
Assembly of replicons for a replication factory varies from cell to cell and closer ones show more frequent assembly. (A) In each strain of #1–3 (see Fig. 1 A), 19–23 cells were analyzed as in Fig. 1, C and D, and categorized based on their two fluorescent dots being replicated at the same (yellow) or different factories (light green). 0 min was defined as in Fig. 1, C and D. Replication at the same factory was evaluated by close proximity (≤350 nm for 2 min or longer) of the dots within the time window from −3 min to +2 min (orange rectangle). (B–D) The distance between the two dots was measured at each time point in 23 cells of strain #1, as shown in A. Its distribution was then plotted as cumulative frequency, separately for replication at the same (red line) and different factories (blue line), in three time windows; i.e., (B) before replication, (C) soon after replication (excluding the time window −3 min to +2 min for the evaluation; see A), and (D) ≥11 min after replication. n (number of time points) = 190 (B, same), 254 (B, different), 87 (C, same), 117 (C, different), 140 (D, same), 182 (D, different).
We next addressed if the two marked loci being close together before replication leads to a higher probability of them replicating in the same factory. We determined the distance between the loci in strain #1 before replication, and showed that its distribution had no correlation with whether the loci were replicated in the same or different factories (Fig. 2 B and Fig. S2 A). In contrast, when we measured the distance between loci soon after replication (until +11 min when the fork that passed through the tetO array is expected to have terminated; Fig. 1 A), loci that replicated in the same factory tended to remain closer than loci that replicated in different factories (Fig. 2 C; refer to a control in Fig. S2 B). If the same measurement is made >11 min after replication, the difference in the distance between loci replicating in the same and different factories was again lost (Fig. 2 D). This suggests that the factory configuration persists for a few minutes after loci have replicated, but is lost within 11 min (perhaps when replication of a replicon is completed). This is consistent with a relatively high rate of diffusion of chromosomal loci within the nucleus (see Materials and methods, Mathematical modeling ii). Moreover, the difference between the same versus different factory groups (Fig. 2 C) validates the original loci-proximity threshold to estimate replication in the same factory, as an inaccurate threshold would not create such a difference.
Super-resolution microscopy reveals the distribution of replicons within individual replication factories
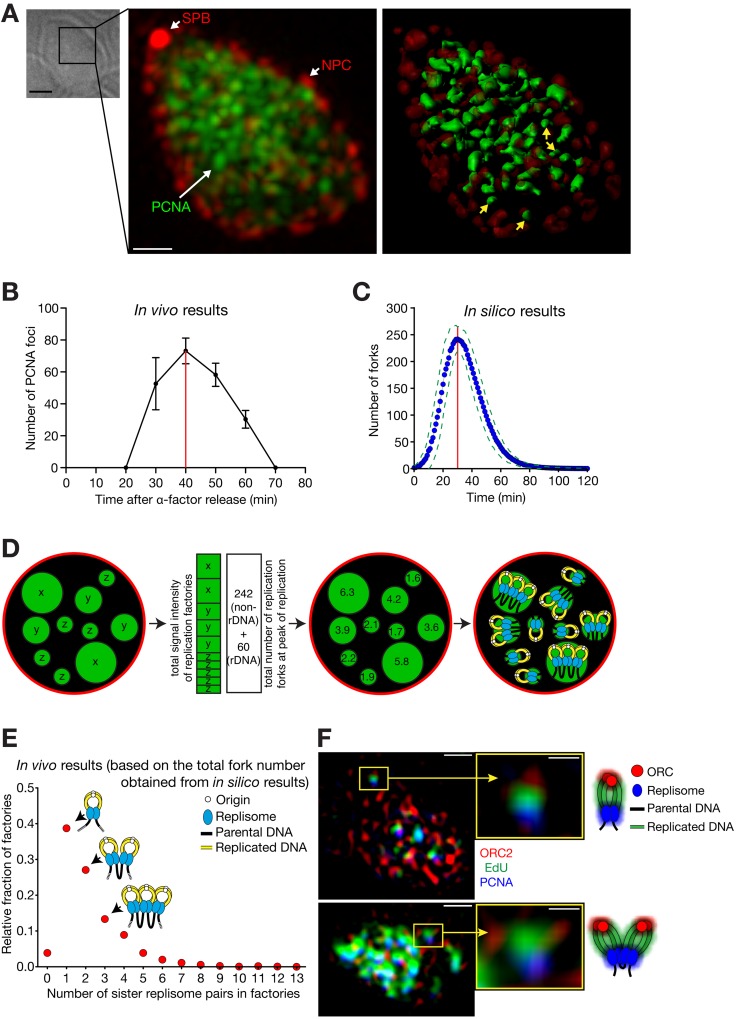
Using live-cell imaging, we previously visualized replication factories as bright globular foci of GFP-PCNA, which showed dynamic changes in shape and location during S phase (Kitamura et al., 2006). To provide a better understanding of replication factory organization, we observed GFP-PCNA by super-resolution structured-illumination microscopy (Schermelleh et al., 2008). Cells were fixed and analyzed at different times after release from α-factor treatment (time 0), allowing us to monitor factory number and fluorescence intensity at different times. This showed that the number of replication factories increased to a peak value of 73 ± 8 (mean ± SD) in mid-S phase (Fig. 3, A and B).
Figure 3.
Super-resolution microscopy reveals the organization of replication factories. (A and B) Replication factories observed by super-resolution microscopy. Cells (T8375) with GFP-POL30 (PCNA), SPC42-mCherry (a component of spindle pole body, SPB), and NIC96-mCherry (a component of the nuclear pore complex, NPC) were released from α-factor treatment (defined as 0 min). (A) A bright-field image, a fluorescence image (GFP, green; mCherry, red), and its 3D rendering in a representative cell, which was fixed at 40 min. Yellow arrows on the 3D rendering image indicate representative replication factories that are estimated to contain a single replisome pair (see D). Bars: (bright-field image) 2 µm; (fluorescence image) 0.5 µm. (B) The number of PCNA foci (mean ± SD) within the nucleus along the time course. Red line indicates the peak of DNA replication. (C) The number of replication forks along the time course (0 min; DNA replication initiated in 50% cells), based on the in silico analysis. The blue dots represent the mean fork numbers with 1-min intervals (green broken lines: ± SD). The number of forks reached the maximum (242 ± 24, mean ± SD) at 30 min (red line). The data did not include the forks along the rDNA region. (D) Schematic diagrams explaining how the number of replisomes (at replication forks) was estimated at each factory. This number was binned to even integers as sister replisomes associate (right; Kitamura et al., 2006). (E) Relative fractions of replication factories containing each number of sister replisome pairs. The intensities of individual replication factories were analyzed at the peak of replication (40 min in B) as explained in D, collectively in 23 cells. (F) Representative super-resolution fluorescence images, with schematic diagrams of interpretation. Cells (T8659) with TagBFP-POL30 (PCNA) and ORC2-mCherry were treated as in A. At 45 min, cells were labeled with the thymidine analogue EdU for 12 min, followed by fixation. Bars: (left) 0.5 µm; (right) 0.15 µm.
We next evaluated the number of replisome pairs present in each replication factory. Published replication profiles showing the replication timing of the whole genome (Raghuraman et al., 2001; Yabuki et al., 2002) are an average from a large number of cells and do not accurately represent replication in individual cells. For example, the number of “origin” peaks found in an averaged profile is usually larger than the number of replication initiation events in individual cells because replication origins initiate with <100% efficiency (de Moura et al., 2010). To estimate the total number of forks, we simulated the replication in individual cells, using a technique originally established for chromosome VI (de Moura et al., 2010) and subsequently extended to the whole genome (unpublished data), excluding the ribosomal DNA (rDNA) region. Using this simulation, we estimated that 242 ± 24 (mean ± SD) replication forks were present at the peak of DNA replication (Fig. 3 C). Using published data (Linskens and Huberman, 1988; Pasero et al., 2002), we estimated that 60 replication forks were present, on average, in the rDNA region. We estimated that on average, 302 forks (242 + 60) are found in the whole nucleus at the peak of DNA replication. We then assigned these 302 replication forks to each of the replication factories, assuming that the integrated GFP-PCNA signal in each factory is proportional to the number of forks it contains (Fig. 3 D). In this way, we were able to estimate the number of replisome pairs (at sister replication forks) present in each replication factory (Fig. 3 E). Intriguingly, our result suggests that many replication factories (39%) consist of just one pair of sister replisomes, as was previously predicted (Berezney et al., 2000). Factories with more replisome pairs are less abundant, with only 17% of factories having ≥4 replisome pairs.
If the majority of replication factories comprise one or two pairs of sister replisomes, as shown in Fig. 3 E, we may be able to visualize their association with nascent DNA using super-resolution microscopy. To observe both factories and replication origins, we fused PCNA and origin recognition complex subunit 2 (Orc2) with different fluorescent proteins (Fig. 3 F; blue and red, respectively). We also visualized nascent DNA by a 12-min pulse of 5-ethynyl-2′-deoxyuridine (EdU, green). To simplify our interpretation, we focused on factories that were well separated from others and were closely associated with Orc2 and EdU. We observed a high proportion of cases (22/40; P = 0.0037) where the labels were present in the order PCNA–EdU–Orc2 (Fig. 3 F, top). This may represent a replicon emerging from a factory during replication, with nascent DNA lying behind the factory, and the origin (marked by Orc2) behind the nascent DNA. In addition, we found several examples that were consistent with two replicons coming out of a single factory (Fig. 3 F, bottom). We also studied the direction in which replicated DNA came out of a factory, and found it to be random relative to the factory–nucleus center axis (Fig. S3 A).
Genome-wide stochastic assembly of replicons recapitulates the organization of replication factories within the whole nucleus
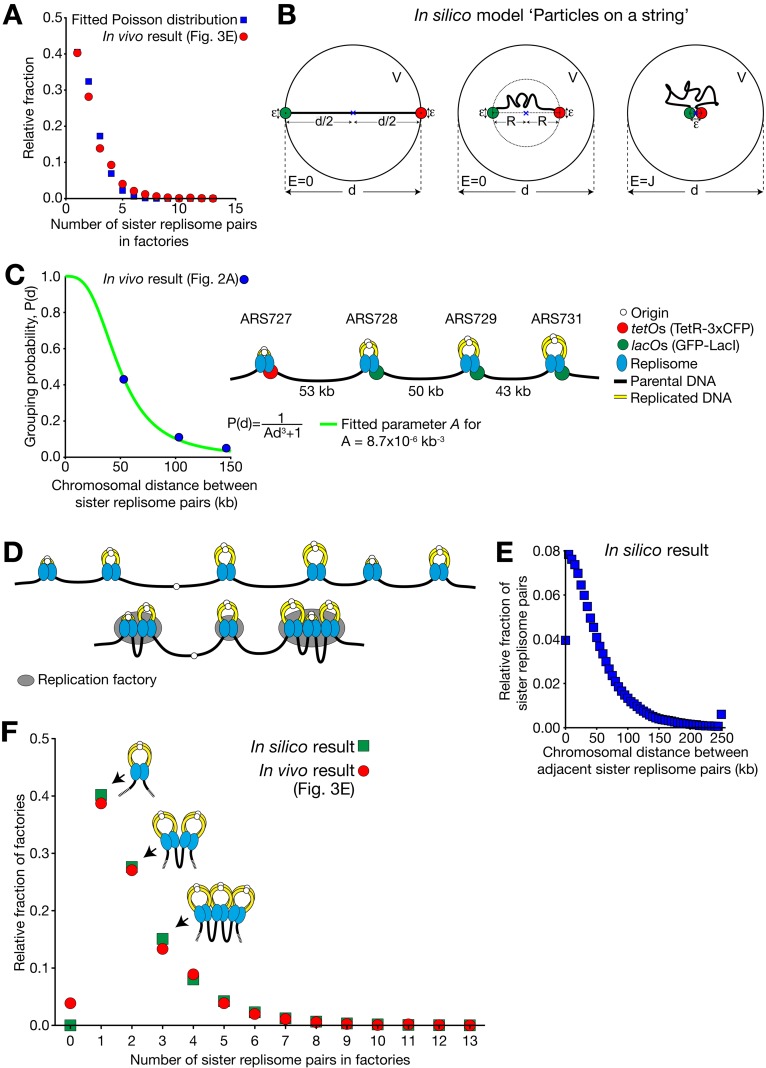
How are replisome pairs assembled into replication factories? As shown in Fig. 2 A, this assembly differed from cell to cell. Given this, the simplest hypothesis is that factory assembly is stochastic. If replisome pairs are dispersed in the nucleus and they randomly assemble into a factory, the number of replisome pairs per factory should follow a Poisson distribution (Motulsky, 2010). Consistent with this, our estimate of the number of replisomes in each factory (Fig. 3 E) approximately matched a Poisson distribution (Fig. 4 A).
Figure 4.
Stochastic assembly of sister replisome pairs recapitulates their observed distribution at individual replication factories. (A) Distribution of the number of sister replisome pairs per replication factory (Fig. 3 E) approximately fits a Poisson distribution (λ = 1.60, R2 = 0.98). (B) Schematic diagram of “Particles on a string” model. See detail in Materials and methods, Mathematical modeling i. (C) We fitted the parameter A in P(d) = 1/(Ad3 + 1), using the replicon assembly frequency result, obtained in strain #1 and #2 (Fig. 2 A; 23 and 19 cells were analyzed, respectively). Fig. S3 B explains how the chromosomal distance between sister replisome pairs was obtained in strain #1, #2, and #3. (D) Schematic diagram showing the example of positions of sister replisome pairs along a chromosome (top), and their stochastic assembly (bottom) based on the grouping probability shown in C. (E) Distribution of the distance (replicated DNA is not counted, as it is looped out; see C) between neighboring sister replisome pairs along a chromosome, obtained from the simulation. Relative fractions of the pairs at the indicated distance (each 5-kb window) were obtained from one million simulations, at the peak of DNA replication (at 30 min in Fig. 3 C). Fractions for >250 kb are shown together at 250 kb. (F) Distributions of the number of sister replisome pairs per replication factory: comparison between in vivo data (obtained in Fig. 3 E) and in silico data (as shown in B–E). Pearson correlation coefficient r = 0.9953. P < 0.0001.
Prompted by this finding, we developed a simple mathematical model for factory formation (Fig. 4 B; see Materials and methods, Mathematical modeling i and ii) where two adjacent pairs of sister replisomes, connected by a chromosome region (length d), are represented by two particles on a string. The two particles diffusing rapidly within the nucleus can aggregate with a binding energy J if they come within distance ε from each other. Using basic statistical physics, we derived the probability P(d) of the two particles being associated as:
where A is a constant depending on J, ε, and temperature. We derived the parameter A as 8.7 × 10−6 kb−3 by fitting this equation (Fig. 4 C and Fig. S3 B) to the results of replicon assembly frequency, obtained in Fig. 2 A. The model fitted the data well (R2 = 0.99; Fig. 4 C). If the diameter of a sister replisome pair is 90 nm (i.e., ε = 90 nm; Baddeley et al., 2010), J is about −12.5 kJ/mol (−5.1 kBT) at ambient temperature, which is an energy involved in a typical weak protein–protein interaction (Baxter et al., 1998; Rippe, 2007).
Next, we extended this model to the whole genome by combining it with the mathematical simulation of the replication profiles in individual cells (de Moura et al., 2010; unpublished data). Note that all parameters in this simulation were determined from the known profiles of DNA replication and we did not tune any of the parameters. For each cell in one million simulations we acquired a “snapshot” of the replication fork positions on the genome at the peak of DNA replication (Fig. 4 D, top; see Materials and methods, Mathematical modeling iii). The collection of these snapshots gave the distribution of the distance between neighboring sister fork pairs, as shown in Fig. 4 E. We then used our equation for P(d) as described above (Fig. 4 C) to determine the probability of fork pairs aggregating into a factory given their distance d along a chromosome (Fig. 4 D, bottom). This gave an “in silico” distribution of numbers of sister fork pairs per replication factory (Fig. 4 F, green squares). Remarkably, the in silico result was very similar to the microscopy observations (Fig. 4 F, red circles). Thus, from the frequency that adjacent sister replisome pairs associate with one another (Fig. 2 A) we were able to accurately recapitulate the genome-wide replicon distribution in replication factories (Fig. 3 E) by assuming stochastic assembly of replicons.
Implications of this study
Our results suggest that it is mainly neighboring replicons on a chromosome that are brought together in factories (Figs. 2 A and 4 C). This conclusion is widely anticipated (Jackson and Pombo, 1998; Berezney et al., 2000; Gillespie and Blow, 2010) and is consistent with the observed clustering of active replicons on DNA fibers (Tuduri et al., 2010) and the high rate of association of neighboring DNA sequences observed in chromosome conformation capture assays (Duan et al., 2010).
Previous data suggested that sister replisomes stay associated with each other during replication (Kitamura et al., 2006). Do the sister replisome pairs present in the same factory remain continuously associated with each other, or do they often dissociate? We found that the physical distance between two marked loci tends to remain shorter after they were replicated in the same factory than after being replicated in different factories (Fig. 2 C). This suggests that replicons are stably associated for a significant period once they are brought together. Furthermore, the association between replicons fitted very well a stochastic thermodynamic process (Fig. 4, C and F) in which their association represents a low energy state. Thus, the association between replicons is expected to be relatively stable.
Our results suggest that a replication factory is mainly organized by associated neighboring replicons on the same chromosome. However, there may be other types of replicon association that contributes to factory formation, although they are unlikely to constitute a major population. For example, as centromeres cluster at a spindle pole in budding yeast (Duan et al., 2010), replicons at centromeres on different chromosomes may be associated to organize a factory (Natsume et al., 2013). Moreover, a DNA binding protein that recognizes a common DNA motif may bring together a subgroup of replicons that are distant on the same chromosome or present on different chromosomes (Knott et al., 2012).
It is intriguing to consider whether replication factories are organized in mammalian cells in similar or different ways compared with budding yeast. Some mammalian factories formed in late S phase are as large as the whole yeast nucleus (see Fig. 3 in Natsume and Tanaka, 2010). A chromosome region, replicated at such a large mammalian factory, represents a chromosome territory—a stable unit maintained until the next cycle (Jackson and Pombo, 1998). It is unlikely that a chromosome region replicated at a yeast factory represents such a stable unit because the motion of a chromosome locus (on a chromosome arm) covers a large proportion of the yeast nuclear volume (see Materials and methods, Mathematical modeling ii). Nonetheless, a mammalian chromosome territory is composed of smaller units called topologically associating domains (TADs), inside of which chromosomes show more dynamic behaviors (Gibcus and Dekker, 2013). Stochastic assembly of yeast replicons forming replication factories may correspond to the way that chromosomal DNA associates within a mammalian TAD. Indeed, mammalian cells also have small factories, some of which contain only 1–3 replicons (Berezney et al., 2000), as we found in yeast. Moreover, the large factories seen in mammalian cells may actually be composed of several small ones (Leonhardt et al., 2000; Baddeley et al., 2010). Such small factories of mammalian cells may be organized in the same way as we found in yeast cells.
A stochastic assembly mechanism may provide robustness to factory organization. It is relatively easy to establish—all that is required is that some replisome components have an affinity for another replisome component. In a deterministic assembly scheme, failure to incorporate one component might cause failure of the entire factory network, whereas in a stochastic scheme, each individual interaction is independent of the status of the others. This could be important in vertebrate cells in responding to replication stress when a replication factory defines the boundary, inside of which dormant origins can initiate and complete replication for the region between two stalled replication forks (Ge and Blow, 2010; Thomson et al., 2010).
In addition to organizing DNA replication, replication factories (foci) are likely to represent a fundamental feature of chromosome organization (Jackson and Pombo, 1998; Berezney et al., 2000). Using budding yeast as a model organism, we have found that the replicons making up individual replication factories are highly variable from cell to cell. Our results suggest that neighboring replicons are assembled stochastically and stay associated together to maintain replication factories relatively stably. Our study gives an important insight not only to the organization of DNA replication within the nucleus, but also to general mechanisms by which chromosomes organize sub-nuclear structures such as transcription factories and repair foci (Lisby and Rothstein, 2004; Sutherland and Bickmore, 2009).
Materials and methods
Yeast genetics and molecular biology
All S. cerevisiae strains used in this study were MATa haploid strains with the W303 genetic background (trp1-1, leu2-3,112, ura3-1, his3-11,15, ade2-1, can1-100). The methods in yeast genetics, α-factor treatment for cell synchronization (Amberg et al., 2005; Kitamura et al., 2006), the TetR-GFP/tet and GFP-LacI/lac operator system (Straight et al., 1996; Michaelis et al., 1997), and the construct of TetR-3xCFP (Bressan et al., 2004) were as described previously. Cells were cultured in YPA medium containing 2% glucose at 25°C unless otherwise stated.
tetOx224 (11.2 kb) and lacOx256 (10.1 kb) were integrated by a two-step “pop-in and pop-out” method (Struhl, 1983), at the chromosomal loci shown in Fig. 1 A, as follows. tetOx224 was integrated to chromosome VII at 660847 (base pairs from the left telomere) within the replicon including autonomously replicating sequence, ARS727 (the ARS727 replicon), in strains #1, #2, and #3. lacOx256 was integrated to chromosome VII at 720007 within the ARS728 replicon, at 781554 within the ARS729 replicon and at 842709 within the ARS731 replicon in strains #1, #2, and #3, respectively. To make constructs for integration, DNA fragment spanning ∼500 bp upstream (toward the left telomere) and ∼1 kb downstream (toward the right telomere) from integration sites were amplified by PCR and cloned into the pRS306 plasmids containing URA3 as an auxotroph marker. The operator arrays were then inserted between these upstream and downstream genomic DNA fragments. The plasmids were subsequently cut within the upstream fragment and inserted into each locus. The cells with the plasmid backbone popped out were selected on 5-fluoroorotic acid–containing media (Amberg et al., 2005). Plasmid integrations and subsequent loss of plasmid backbones were confirmed by PCR amplifications of relevant regions.
When the N terminus of PCNA (POL30) was tagged with a fluorescent protein at its original locus, the growth of haploid cells was severely retarded (Kitamura et al., 2006). We therefore integrated a single copy of PCNA, tagged at its N terminus, into an auxotroph marker TRP while PCNA is intact at its original locus. It has been shown in fission yeast that fluorescently tagged PCNA behaves similarly to untagged PCNA (Meister et al., 2007). PCNA and its promoter were first cloned into pRS404 (pT1046) and then DNA fragments of yEGFP (from pKT128; Sheff and Thorn, 2004) and TagBFP (from pTagBFP-C; Evrogen) were inserted between the promoter and the coding region to construct pT1056 and pT1448, respectively. SPC42 and NIC96 were tagged at their C termini at the original gene loci with a one-step PCR method (Knop et al., 1999), using 4xmCherry cassette from pT909 as a PCR template. The plasmid pT909 was constructed by multiplying the mCherry gene in pKS391 (Snaith et al., 2005). Strains with the tagged genes grew normally at temperatures used in this study.
To facilitate EdU incorporation (Salic and Mitchison, 2008), five copies of the herpes simplex thymidine kinase gene were expressed from GPD1 promoters (Dahmann et al., 1995), and the human equilibrative nucleoside transporter 1 (ENT1) gene (with codon usage optimized for yeast) was expressed from ADH1 promoter (a gift from K. Shirahige, University of Tokyo, Tokyo, Japan).
Live-cell imaging
The procedures for time-lapse fluorescence microscopy were as described previously (Kitamura et al., 2006; Tanaka et al., 2010) unless otherwise stated. During image acquisition, cells were suspended in synthetic complete/YPA (3:1 ratio) medium containing 2% glucose at 25°C (ambient temperature). For image acquisition, we used a microscope (DeltaVision RT; Applied Precision), UPlanSApo 100×/NA 1.40 objective lens (Olympus), a CCD camera (CoolSnap HQ; Photometrics), and SoftWoRx software (Applied Precision). CFP and GFP signals were discriminated with the 89006 ET filter set (Chroma Technology Corp.). We acquired nine z-sections (0.7 µm apart), which were subsequently analyzed with SoftWoRx, Volocity (PerkinElmer), and Image-Pro Plus (Media Cybernetics) software. For two-dimensional presentation in figures, z-sections were deconvoluted with SoftWoRx and projected to two-dimensional images. For quantitative analyses, images before deconvolution were used.
Super-resolution structured illumination microscopy
Cells were fixed with 2% paraformaldehyde for 30 min and washed with PBS. When cells were pulse-labeled with 1 mM EdU (for 12 min; Fig. 3 F), they were further processed for the Click-iT EdU reaction as described in the protocol C10337 (Invitrogen), followed by a PBS wash. The images were acquired with structured illumination microscopy (Schermelleh et al., 2008) as described previously (Hattersley et al., 2011). In brief, we used the OMX system (version 3; Applied Precision) equipped with 405-, 488-, and 593-nm solid-state lasers. To acquire images, we used a 512 × 512 electron-multiplying charge-coupled device camera (Cascade II; Photometrics) and a UPlanSApochromat 100×/1.4 NA lens. A coherent scrambled laser through a diffraction grating was used to generate the structured illumination. We used SoftWorx to process raw images for reconstruction to reveal structures in three dimensions (Gustafsson et al., 2008).
Image and data analyses
The grouping of replicons into the same factory or different factories was evaluated based on the distance between two dots (CFP and GFP signals) during replication. For an accurate evaluation, we analyzed individual cells where the two dots had always been on focal planes within the acquired z-stack during observation. We manually tracked each signal (both CFP and GFP) in individual cells and chose the brightest four pixels (2 × 2) after the z-stack had been projected to a two-dimensional image at each time point. The signal intensities of the selected four pixels along the z-stack were summed using Volocity, after background subtraction. The background was defined as the most frequently observed signal level among individual pixels in images. The intensities of CFP and GFP dots were normalized in relevant graphs (Fig. 1, C and D; Fig. S1, B–D) so that their intensities were in the range of 80–120 before replication and their regression curves (see below) did not overlap before or after replication (for clear presentation). The nonlinear regression curve following the increase in CFP and GFP signal intensities was drawn with Prism 5 (GraphPad Software), fitting to curves
where a, b, and LogEC50 (mid-replication time of each dot) were derived constants. The calculated LogEC50 from the nonlinear regression curve allowed us to estimate the mid-replication times at a sub-minute level (to the first decimal place). The average replication time of CFP and GFP dots were defined as 0 min. Cells, in which the difference between CFP and GFP mid-replication times was equal to, or less than, 3 min were further analyzed; the centroid coordinates of signals from CFP and GFP dots were obtained at each time point using Image-Pro Plus and the distances between the two dots were measured in three dimensions (Fig. 1, C and D; Fig. S1, B–D). Cells were evaluated for replication at the same factory based on the close localization of dots (≤350 nm, ≥2 min) within a −3 to +2 min time window (Fig. 2 A, orange rectangle).
When the distances between the two dots were analyzed before (Fig. 2 B) and after replication (Fig. 2 C), the data within the −3 to +2 min window were not included; inclusion of such data would have given a bias based on the classification itself between the same vs. different factories. Moreover, in some cells, close localization continued beyond this evaluation time window; such data points were also excluded from the distance analyses in Fig. 2, B and C, for the same reason. In Fig. 2 C, we aimed to analyze the distances between CFP and GFP dots while a replication fork actively underwent replication along the right side of the ARS727 replicon in the majority of cells. We estimated that such a situation continued, on average, until +11 min for the following reasons: (1) we assumed that, on average, a half of the tetO array completed replication at 0 min; then it took ∼4 min to replicate the other half, given that the fork speed is 1.5 kb/min (Sekedat et al., 2010); (2) it took ∼9 min for a replication fork to move from the right end of the tetO array to the lowest point between the ARS727 and ARS728 replicons in the replication profile (see Fig. 1 A); and (3) we did not include the last 2 min because another fork from ARS728 might move in to the relevant region in some cells in the population.
To define the contours of replication factories in Fig. 3, A and D, surface rendering was applied to the GFP-Pol30 (PCNA) signal using Imaris software (Bitplane). The objects (factories), which were less than 15 voxels (∼0.002 µm3) in size, were excluded from the analyses to remove the background. The total signal intensity of all factories was calculated in individual cells (n = 23). The average total signal intensity measured in the population was divided by an average number of forks (302) to obtain the average signal intensity for a single replication fork. Using this value, we estimated the numbers of forks at individual factories (Fig. 3 D). These numbers were then binned to even numbers (e.g., 3–5 binned to 4), as sister replisomes (therefore sister forks) are associated (Kitamura et al., 2006); thus, the numbers of sister replisome pairs were estimated at individual factories (Fig. 3 E). In rare cases (3%), the signal intensity of a factory was less than the average intensity of a single fork; such a factory was considered to contain zero sister replisome pairs (Fig. 3 E).
The general expression for the Poisson distribution that k events may occur is
where k = {0,1,2,3,…} and λ is the mean of distribution. We revised the expression considering that there is no factory without a replication fork. We calculated the probability for an event of count k to occur under the condition that k = 0 was unobservable, as the following:
The distribution of the number of sister replisome pairs per replication factory (Fig. 3 E) was normalized by the sum of the observable counts (for k = 1,2,3,…) and then fitted by this zero-truncated Poisson distribution P(X = k|X > 0) (Fig. 4 A). The fitted value of λ was 1.60.
Statistical analyses
Statistical analyses were performed with Prism Software (GraphPad) by choosing Fisher’s exact test (Fig. 2 A), unpaired t test (Fig. S2 A), chi-square test (Fig. 3 F), and Pearson correlation (Fig. 4 F), or with R (http://www.r-project.org) by choosing Kolmogorov-Smirnov test (Fig. 2, B–D; Fig. S2 B; and Fig. S3 Aii). The null hypotheses in these tests (except for Fig. 4 F) were that the samples were collected randomly and independently, from the same population. For P value in Fig. 4 F, we calculated the chance that random sampling from two groups with no correlation would give such correlation coefficient. All P values were two-tailed, and the null hypotheses were reasonably discarded when P values were <0.05. To represent goodness in fitting, R2 was calculated with Prism software (GraphPad) as coefficient of determination (Fig. 1, C and D; Fig. S1, B–D; Fig. 4, A and C).
Mathematical modeling
(i) “Particles on a string” model.
We use an analogy of two particles tethered by a string of length d to describe two sister replisome pairs that are apart from each other at a chromosomal distance d (Fig. 4 B). Each sister replisome pair is thereby a particle fixed to the end of the string. Each particle is considered to be a sphere with diameter ε. We assume the string has no stiffness, given that the persistence length of yeast chromatin is short (2.5 kb; Dekker et al., 2002) relative to the distance between replication origins and between marked chromosome loci analyzed here. The particles perform a random walk within a sphere of radius d/2 in three dimensions (illustrated in two dimensions, for simplicity, in Fig. 4 B). If both particles come within interaction radius (i.e., the distance between their centers is ε or less), they associate. We fix the coordinate system at the center of mass (midpoint) of the two particles. The system can be in two conditions. First, when particles are separated; the energy of the system is then E = 0 (Fig. 4 B, left and middle). Second, when particles are in close proximity and become associated; the energy of the system in this state is E = J, where J is a binding energy (Fig. 4 B, right). Therefore, J is negative, meaning that the particles’ interaction is attractive.
Our aim is to estimate the probability of finding the system in each condition—particles separated or particles associated—depending on the string length between them. The probability that the two particles meet and associate with each other when the system is in thermodynamic equilibrium is
| (1) |
where na and ns are the normalized numbers of states in which particles are associated and separated, respectively. Ba and Bs represent corresponding Boltzmann factors (weighing factors). Each Boltzmann factor,
depends on temperature, T, and energy, E, of the system. kB is the Boltzmann constant.
The normalized number of states is derived as follows. As the reference frame is centered at the midpoint, states corresponding to the particles separated by a distance R lie on a spherical shell of radius R/2. Particles are considered to associate once the distance between their centers is less than ε, i.e., they are within a sphere of radius ε/2 around the origin. The volume of this sphere is
We normalize the number states to the total volume
The normalized number of states in which the two particles associate is then given by
| (2) |
The energy of the system at this association state is minimized, thus Ea = J, with the corresponding Boltzmann factor
The normalized number of states in which particles are not associated is
| (3) |
for a small interaction radius (ε << d). The energy of the system when the particles are apart is Es = 0, and this Boltzmann factor is Bs = 1.
The association probability from equation (1) is then
| (4) |
where
is a constant with a constant temperature. Equation (4) describes the probability of the two particles, separated by a distance ≤d, being associated in an equilibrium system. We can estimate the parameter A from the grouping frequencies of replicons in strain #1 and #2. The function Pa(d) (shown as P(d) in other parts for simplicity) fits the data well (Fig. 2 A), with the best-fitting A = 8.7 x 10−6 kb−3 (R2 = 0.99, Fig. 4 C). The binding energy of two sister replisome pairs is J = kBT ln(Aε3) = −5.1 kBT = −12.5 kJ/mol, for the best-fitting A, ε = 90 nm and T = 298.2 K. Here we estimate the diameter of a single sister replisome pair from the minimum size of a replication factory of ∼90 nm (Baddeley et al., 2010). We apply the chromatin packaging ratio of 10 nm/kb, based on the ratio of measured spatial distances over the chromosomal distances between two fluorescently labeled chromosomal loci, which matches a reported value (Dekker et al., 2002). The calculated binding energy of sister replisome pairs (−12.5 kJ/mol) is in the range of a typical weak protein–protein interaction (Baxter et al., 1998; Rippe, 2007). It is also in agreement with the estimated energy for the association of DNA polymerases bound on two replication origins (Marenduzzo et al., 2006).
(ii) Diffusion time-scale.
Because replication factories are formed by assembly of replisomes that are actively involved in replication (Kitamura et al., 2006), it is thought that sister replisome pairs can be associated with each other only after replication is initiated at replication origins. Next, we addressed whether diffusion of replisome pairs, assumed in the model above, is rapid enough for two replisome pairs to meet with each other after replication initiation at origins but before fluorescently labeled loci are replicated.
For this, we needed to estimate the diffusion coefficient D of a sister replisome pair. We estimated D based on the diffusion coefficient of a marked locus around its replication (Fig. 1 A). To measure this, we synchronized cells with α-factor and then released them into S phase. We started acquiring images of the spindle pole body (SPC42-4mCherry) and a fluorescently labeled locus (on ARS728 replicon) 35 min after α-factor release. Images were acquired every 7.5 s for 8 min. This time window covered the average replication time of this locus (41 min after α-factor release). Because the spindle pole body (SPB) is embedded in the nuclear envelope and less mobile than a chromosome locus, it was used as a control point for analyzing the position of the fluorescently labeled locus. We calculated the mean squared displacement (MSD) between SPB and the locus as a function of time. Diffusion coefficient was obtained from MSD (in four cells) as described previously (Marshall et al., 1997) and we found D ≈ 0.2 µm2/min.
To estimate diffusion time-scale, we can compare diffusion size-scale with the distance between the two particles. The typical time for one particle to diffuse through a distance L is (Sneppen and Zocchi, 2005)
| (5) |
Because we have two particles diffusing, this is equivalent to one particle diffusing with 2D. Hence, we double the diffusion coefficient in equation (5), D2 = 2D. For the maximum distance between the origins in strain #2, L ≈ 1.3 µm, corresponding to chromosomal distance of 129 kb. With D ≈ 0.2 µm2/min, the diffusion time-scale is tL ≈ 2 min. In strain #2 it takes 4–6 min after replication initiation at ARS727 and ARS729 until tetO/lacO dots are replicated (i.e., until replication forks reach the middle of tetO/lacO arrays, which are 10–11 kb in length). We therefore conclude that diffusion is rapid enough for two relevant replisome pairs to come together before replication of the dots.
Because the chromosomal distance between the relevant replication origins in strain #3 (ARS727 and 731) is larger (186 kb) than those in strains #1 and #2, diffusion may not be rapid enough for two replisome pairs (generated at ARS727 and 731) to meet before the replication of two fluorescently labeled loci in strain #3. For this reason, the grouping probability of the replicon pair in strain #3 was not considered in fitting the parameter A (Fig. 4 C). Nonetheless, after A was fitted using the other data points, the model was in good agreement with the data point from strain #3 (Fig. 4 C).
(iii) In silico distribution of sister replisome pairs in replication factories.
With the “particles on a string” model (Mathematical modeling i), we could estimate grouping probabilities of neighboring sister replisome pairs as a function of chromosomal distance between them. We then aimed to extend our analysis of the replicon assembly from the region on chromosome VII to the whole genome context in budding yeast where the positions of origins have been well defined by various studies (Nieduszynski et al., 2007). For this, we required information about locations of sister replisome pairs along all chromosomes, during replication in individual cells. How can we obtain such information?
The replication timing profile of the yeast genome, which was obtained from population-based studies (Raghuraman et al., 2001; Yabuki et al., 2002), helped our understanding of the global temporal organization of replication. However, these data provide only the average replication timing in the relevant cell population, whereas replication timing is actually different from cell to cell. For instance, the same replication origin does not fire at the same timing in different cells. Intriguingly, if replication timing is obtained in individual cells, the average timing from such dataset could recapitulate replication timing obtained from a population-based analysis (Czajkowsky et al., 2008). Importantly, the converse approach is also possible. For example, we can set the parameters that determine the behaviors of replication origins, such as (1) the fraction of cells where an origin was competent to fire, (2) mean activation time of an origin, and (3) distribution of the origin activation time (a standard deviation, assuming the Gaussian distribution). Then, these parameters can be determined so that they recapitulate the replication profile obtained from a population-based analysis (de Moura et al., 2010). In this approach (de Moura et al., 2010), it was assumed that the speed of replication fork movement along a chromosome was reasonably constant (1.5 kb/min; Friedman et al., 1997; Sekedat et al., 2010). Such a mathematical approach was applied first to recapitulate replication timing profile of chromosome VI (de Moura et al., 2010) and then further extended to recapitulate replication timing of the whole genome (Retkute et al., 2012). Once the above parameters were determined, we could simulate replication progression in individual cells by assigning timing of replication initiation at origins, randomly but still based on the relevant parameters.
In the current study, we used this mathematical approach on two occasions. First, using this approach, we obtained the distribution of the fork numbers on the whole genome in individual cells over the duration of DNA replication, as shown in Fig. 3 C. The average number of forks at the peak of replication (when the total fork number became maximum) was subsequently used to estimate in vivo distribution of sister replisome pairs in replication factories (Fig. 3, D and E). Note that, in Fig. 3 C, we aimed to estimate how the number of forks in individual cells changes during S phase. For accurate estimation, we needed to consider temporal variation in S phase entry among cell population after α-factor treatment and subsequent release. We estimated that cells entered S phase with 4-min variation (in standard deviation) based on the following result: Globular signals of PCNA represented replication factories and its first appearance could define S phase onset (Kitamura et al., 2006). We investigated this timing in individual cells (n = 56) after α-factor treatment and subsequent release, and found that timing of S phase onset varies with 4-min standard deviation. The standard deviation was reproducible in repeated experiments.
Second, we intended to obtain in silico distribution of sister replisome pairs in replication factories. For this, we ran the simulation in one million cells and took snapshots of replisome positions on chromosomes at the peak of replication (Fig. 4, D and E) after cells had entered S phase with 4-min variation (see above). Based on these snapshots, we determined whether adjacent sister replisome pairs were grouped in the same factory or not, based on the chromosomal distance between them and corresponding probability of grouping (Fig. 4 C and Mathematical modeling i), as follows. Let us designate each sister replisome pair along a chromosome as A, B, C, D, … etc. in the order. To determine whether A and B are grouped to the same factory, we drew a uniformly distributed random number between 0 and 1 (1, but not 0, is included), which was then tested against the distance-dependent value P(d) of those two pairs. If the random number was below P(d), the pairs were assumed to be part of the same factory. Next, we examined the association of pairs B and C in the same way. If A and B were in the same factory and if B and C were in the same factory, then we concluded that A, B, and C are grouped together in the same factory. We performed this pairwise clustering of adjacent sister replisome pairs into factories in the rightward direction along each chromosome. Nonetheless, we confirmed that clustering in the left direction gave a very similar result (Fig. S3 C).
In this study, we assumed that sister replisomes were always associated with each other during replication of a relevant replicon. This assumption was based on our previous result that sister replisomes were associated in vivo in most of the cells (Kitamura et al., 2006). We also assumed that, when two replisome pairs had encountered upon completion of DNA between them, one sister replisome in each pair disappeared, leaving the remaining two replisomes (which originally belonged to two different pairs) associated and allowing the new pair to undergo replication. This assumption is consistent with a low energy state of associated replisome pairs; i.e., once two pairs become associated, we can expect that they stay associated for a while (see Mathematical modeling i). Nonetheless, in the above mathematical simulation, we observed a low number of cases where one replisome was present without its sister. This happened when one replisome completed replication at the end of a chromosome (which is linear) while its sister was still engaged in replication. This led to generation of a small number of replication factories containing odd numbers of replisomes (Fig. S3 C). However, for a direct comparison of the distribution of sister replisome pairs in factories obtained from in vivo and in silico data, we partitioned factories with odd numbers of forks (replisomes) proportionally to the nearby categories with even numbers of forks (e.g., factories with three forks were recategorized to those with two and four forks proportionally to their factory numbers).
In the above mathematical modeling, we assumed that replisome pairs A and C could only associate when both A/B and B/C associate. In other words, we considered association between immediate neighbors but not between others. It was actually not simple to consider direct association between A and C because we needed to consider the presence and absence of A/C association separately depending on whether A/B and B/C association was present or not. In practice, it was impossible to compute all possible cases for all possible associations. Nonetheless, our approach is justified only when A/C association is relatively low compared with A/B and B/C association. We tested this in a simplified case where the chromosomal distances between A and B and between B and C are both d. The ratio of the A/C association probability to the A/B and B/C association probability was calculated as follows.
| (6) |
When d = 36 kb (the median chromosomal distance between two neighboring replisome pairs; see Fig. 4 E) and A = 8.7 × 10−6 kb−3, this ratio was 0.17. Thus, it was ∼6 times more likely to observe A/B and B/C associations than A/C association.
Online supplemental material
Fig. S1 and S2 provide results supplemental to Figs. 1 and 2, respectively. Fig. S3 A provides results supplemental to Fig. 3 F and analyses the direction in which replicated DNA was reeled out of a replication factory. Fig. S3 B provides results supplemental to Fig. 4 C. Fig. S3 C gives supplemental information to Mathematical modeling iii in Materials and methods. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201306143/DC1.
Supplementary Material
Acknowledgments
We thank all laboratory members, especially E. Kitamura, for discussions and technical advices; L. Clayton for reading the manuscript; G. Barton for supervising the Data Analysis Group; E. King, M. Posch, and S. Swift for help with microscopy; J.E. Haber, K. Nasmyth, K.E. Sawin, A.F. Straight, K. Shirahige, R. Tsien, and EUROSCARF for reagents; and K. Kitada for yeast replication timing profile.
This work was supported by the Wellcome Trust (080737, 081918, 083524, 096535, and 097945), CRUK (A6996 and A7399), BBSRC (BB/E023754/1 and BB/G001596/1), and SULSA. N. Saner was supported by a Wellcome Trust studentship and ORSAS. C.A. Nieduszynski is a BBSRC David Phillips Fellow. T.U. Tanaka is a Wellcome Trust Principal Research Fellow.
Footnotes
Abbreviations used in this paper:
- ARS
- autonomously replicating sequence
- EdU
- 5-ethynyl-2′-deoxyuridine
- Orc2
- origin recognition complex, subunit 2
- PCNA
- proliferating cell nuclear antigen
- rDNA
- ribosomal DNA
References
- Amberg D.C., Burke D.J., Strathern J.N. 2005. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 230 pp [Google Scholar]
- Baddeley D., Chagin V.O., Schermelleh L., Martin S., Pombo A., Carlton P.M., Gahl A., Domaing P., Birk U., Leonhardt H., et al. 2010. Measurement of replication structures at the nanometer scale using super-resolution light microscopy. Nucleic Acids Res. 38:e8 10.1093/nar/gkp901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter C.A., Murray C.W., Clark D.E., Westhead D.R., Eldridge M.D. 1998. Flexible docking using Tabu search and an empirical estimate of binding affinity. Proteins. 33:367–382 [DOI] [PubMed] [Google Scholar]
- Bechhoefer J., Rhind N. 2012. Replication timing and its emergence from stochastic processes. Trends Genet. 28:374–381 10.1016/j.tig.2012.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R., Dubey D.D., Huberman J.A. 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 108:471–484 10.1007/s004120050399 [DOI] [PubMed] [Google Scholar]
- Bressan D.A., Vazquez J., Haber J.E. 2004. Mating type-dependent constraints on the mobility of the left arm of yeast chromosome III. J. Cell Biol. 164:361–371 10.1083/jcb.200311063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti C., Saccà B., Herrick J., Lalou C., Pommier Y., Bensimon A. 2007. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol. Biol. Cell. 18:3059–3067 10.1091/mbc.E06-08-0689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowsky D.M., Liu J., Hamlin J.L., Shao Z. 2008. DNA combing reveals intrinsic temporal disorder in the replication of yeast chromosome VI. J. Mol. Biol. 375:12–19 10.1016/j.jmb.2007.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmann C., Diffley J.F., Nasmyth K.A. 1995. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol. 5:1257–1269 10.1016/S0960-9822(95)00252-1 [DOI] [PubMed] [Google Scholar]
- de Moura A.P., Retkute R., Hawkins M., Nieduszynski C.A. 2010. Mathematical modelling of whole chromosome replication. Nucleic Acids Res. 38:5623–5633 10.1093/nar/gkq343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. 2002. Capturing chromosome conformation. Science. 295:1306–1311 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- Dimitrova D.S., Gilbert D.M. 2000. Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat. Cell Biol. 2:686–694 10.1038/35036309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y., Bermejo R., Fiorani S., Haber J.E., Foiani M. 2009. Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell. 137:247–258 10.1016/j.cell.2009.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y.J., Lee C., Shendure J., Fields S., Blau C.A., Noble W.S. 2010. A three-dimensional model of the yeast genome. Nature. 465:363–367 10.1038/nature08973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaschi A. 2000. Eukaryotic DNA replication: a model for a fixed double replisome. Trends Genet. 16:88–92 10.1016/S0168-9525(99)01917-4 [DOI] [PubMed] [Google Scholar]
- Friedman K.L., Brewer B.J., Fangman W.L. 1997. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells. 2:667–678 10.1046/j.1365-2443.1997.1520350.x [DOI] [PubMed] [Google Scholar]
- Ge X.Q., Blow J.J. 2010. Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J. Cell Biol. 191:1285–1297 10.1083/jcb.201007074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus J.H., Dekker J. 2013. The hierarchy of the 3D genome. Mol. Cell. 49:773–782 10.1016/j.molcel.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie P.J., Blow J.J. 2010. Clusters, factories and domains: The complex structure of S-phase comes into focus. Cell Cycle. 9:3218–3226 10.4161/cc.9.16.12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M.G., Shao L., Carlton P.M., Wang C.J., Golubovskaya I.N., Cande W.Z., Agard D.A., Sedat J.W. 2008. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94:4957–4970 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley N., Shen L., Jaffray E.G., Hay R.T. 2011. The SUMO protease SENP6 is a direct regulator of PML nuclear bodies. Mol. Biol. Cell. 22:78–90 10.1091/mbc.E10-06-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A., Pombo A. 1998. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140:1285–1295 10.1083/jcb.140.6.1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E., Blow J.J., Tanaka T.U. 2006. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 125:1297–1308 10.1016/j.cell.2006.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 15(10B):963–972 [DOI] [PubMed] [Google Scholar]
- Knott S.R., Peace J.M., Ostrow A.Z., Gan Y., Rex A.E., Viggiani C.J., Tavaré S., Aparicio O.M. 2012. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell. 148:99–111 10.1016/j.cell.2011.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H., Rahn H.P., Weinzierl P., Sporbert A., Cremer T., Zink D., Cardoso M.C. 2000. Dynamics of DNA replication factories in living cells. J. Cell Biol. 149:271–280 10.1083/jcb.149.2.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligasová A., Raška I., Koberna K. 2009. Organization of human replicon: singles or zipping couples? J. Struct. Biol. 165:204–213 10.1016/j.jsb.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M.H., Huberman J.A. 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:4927–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Rothstein R. 2004. DNA damage checkpoint and repair centers. Curr. Opin. Cell Biol. 16:328–334 10.1016/j.ceb.2004.03.011 [DOI] [PubMed] [Google Scholar]
- Marenduzzo D., Micheletti C., Cook P.R. 2006. Entropy-driven genome organization. Biophys. J. 90:3712–3721 10.1529/biophysj.105.077685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W.F., Straight A., Marko J.F., Swedlow J., Dernburg A., Belmont A., Murray A.W., Agard D.A., Sedat J.W. 1997. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 7:930–939 10.1016/S0960-9822(06)00412-X [DOI] [PubMed] [Google Scholar]
- Meister P., Taddei A., Ponti A., Baldacci G., Gasser S.M. 2007. Replication foci dynamics: replication patterns are modulated by S-phase checkpoint kinases in fission yeast. EMBO J. 26:1315–1326 10.1038/sj.emboj.7601538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 91:35–45 10.1016/S0092-8674(01)80007-6 [DOI] [PubMed] [Google Scholar]
- Motulsky H. 2010. Poisson distribution. In Intuitive Biostatistics. Oxford University Press, New York: 47–49 [Google Scholar]
- Natsume T., Tanaka T.U. 2010. Spatial regulation and organization of DNA replication within the nucleus. Chromosome Res. 18:7–17 10.1007/s10577-009-9088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T., Müller C.A., Katou Y., Retkute R., Gierliński M., Araki H., Blow J.J., Shirahige K., Nieduszynski C.A., Tanaka T.U. 2013. Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment. Mol. Cell. 50:661–674 10.1016/j.molcel.2013.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski C.A., Hiraga S., Ak P., Benham C.J., Donaldson A.D. 2007. OriDB: a DNA replication origin database. Nucleic Acids Res. 35(Database issue):D40–D46 10.1093/nar/gkl758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P., Bensimon A., Schwob E. 2002. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev. 16:2479–2484 10.1101/gad.232902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman M.K., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L. 2001. Replication dynamics of the yeast genome. Science. 294:115–121 10.1126/science.294.5540.115 [DOI] [PubMed] [Google Scholar]
- Retkute R., Nieduszynski C.A., de Moura A. 2012. Mathematical modeling of genome replication. Phys. Rev. E. Stat. Nonlin. Soft Matter Phys. 86:031916 10.1103/PhysRevE.86.031916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K. 2007. Dynamic organization of the cell nucleus. Curr. Opin. Genet. Dev. 17:373–380 10.1016/j.gde.2007.08.007 [DOI] [PubMed] [Google Scholar]
- Rossi R., Villa A., Negri C., Scovassi I., Ciarrocchi G., Biamonti G., Montecucco A. 1999. The replication factory targeting sequence/PCNA-binding site is required in G(1) to control the phosphorylation status of DNA ligase I. EMBO J. 18:5745–5754 10.1093/emboj/18.20.5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A., Mitchison T.J. 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA. 105:2415–2420 10.1073/pnas.0712168105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L., Carlton P.M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M.C., Agard D.A., Gustafsson M.G., et al. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 320:1332–1336 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekedat M.D., Fenyö D., Rogers R.S., Tackett A.J., Aitchison J.D., Chait B.T. 2010. GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome. Mol. Syst. Biol. 6:353 10.1038/msb.2010.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff M.A., Thorn K.S. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Snaith H.A., Samejima I., Sawin K.E. 2005. Multistep and multimode cortical anchoring of tea1p at cell tips in fission yeast. EMBO J. 24:3690–3699 10.1038/sj.emboj.7600838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneppen K., Zocchi G. 2005. Timescales for target location in a cell. In Physics in Molecular Biology. Cambridge University Press, New York: 146–208 [Google Scholar]
- Straight A.F., Belmont A.S., Robinett C.C., Murray A.W. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599–1608 10.1016/S0960-9822(02)70783-5 [DOI] [PubMed] [Google Scholar]
- Struhl K. 1983. The new yeast genetics. Nature. 305:391–397 10.1038/305391a0 [DOI] [PubMed] [Google Scholar]
- Sutherland H., Bickmore W.A. 2009. Transcription factories: gene expression in unions? Nat. Rev. Genet. 10:457–466 10.1038/nrg2592 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Kitamura E., Tanaka T.U. 2010. Live-cell analysis of kinetochore-microtubule interaction in budding yeast. Methods. 51:206–213 10.1016/j.ymeth.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson A.M., Gillespie P.J., Blow J.J. 2010. Replication factory activation can be decoupled from the replication timing program by modulating Cdk levels. J. Cell Biol. 188:209–221 10.1083/jcb.200911037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuduri S., Tourrière H., Pasero P. 2010. Defining replication origin efficiency using DNA fiber assays. Chromosome Res. 18:91–102 10.1007/s10577-009-9098-y [DOI] [PubMed] [Google Scholar]
- Yabuki N., Terashima H., Kitada K. 2002. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 7:781–789 10.1046/j.1365-2443.2002.00559.x [DOI] [PubMed] [Google Scholar]
- Yan H., Chen C.Y., Kobayashi R., Newport J. 1998. Replication focus-forming activity 1 and the Werner syndrome gene product. Nat. Genet. 19:375–378 10.1038/1263 [DOI] [PubMed] [Google Scholar]
- Yardimci H., Loveland A.B., Habuchi S., van Oijen A.M., Walter J.C. 2010. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol. Cell. 40:834–840 10.1016/j.molcel.2010.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.