A complex of KASH5 and Sun1 is required for meiotic homologous chromosome pairing through the coupling of telomere attachment sites to cytoplasmic dynein and microtubules.
Abstract
Chromosome pairing is an essential meiotic event that ensures faithful haploidization and recombination of the genome. Pairing of homologous chromosomes is facilitated by telomere-led chromosome movements and formation of a meiotic bouquet, where telomeres cluster to one pole of the nucleus. In metazoans, telomere clustering is dynein and microtubule dependent and requires Sun1, an inner nuclear membrane protein. Here we provide a functional analysis of KASH5, a mammalian dynein-binding protein of the outer nuclear membrane that forms a meiotic complex with Sun1. This protein is related to zebrafish futile cycle (Fue), a nuclear envelope (NE) constituent required for pronuclear migration. Mice deficient in this Fue homologue are infertile. Males display meiotic arrest in which pairing of homologous chromosomes fails. These findings demonstrate that telomere attachment to the NE is insufficient to promote pairing and that telomere attachment sites must be coupled to cytoplasmic dynein and the microtubule system to ensure meiotic progression.
Introduction
Eukaryote evolution has depended on sexual reproduction requiring both generation of haploid gametes and chromosome/genetic recombination. Haploidization and recombination, occurring during meiosis, are singularly dependent upon recognition and pairing of homologous chromosomes (Bhalla and Dernburg, 2008), a process in which the nuclear envelope (NE) plays a key role. In many organisms telomeres, or specialized pairing centers (PCs) in Caenorhabditis elegans, associate with the NE during meiotic prophase 1. Subsequent clustering of telomeres results in the formation of a “meiotic bouquet” (Scherthan, 2001). This transient chromosomal configuration, formation of which involves rapid cytoskeleton-driven chromosome movements, facilitates homologue recognition and pairing (Trelles-Sticken et al., 2005; Chikashige et al., 2007; Koszul et al., 2008). Thus, NE-linked chromosome dynamics are closely tied to meiotic progression (Conrad et al., 2008; Lee et al., 2012).
Architecture of the NE is conserved in all eukaryotes (Stewart et al., 2007; Wilson and Dawson, 2011). Its most prominent features are inner and outer nuclear membranes (INMs and ONMs) separated by a perinuclear space (PNS). This double-membrane structure raises the question of how telomeres are physically linked to the cytoskeleton during meiotic prophase, a period when the NE is still intact. A solution emerged from studies on interphase somatic cells which revealed that nuclear structures are mechanically coupled to elements of the cytoskeleton (Burke and Roux, 2009). The nature of this coupling is partly dependent on SUN (Sad1, Unc-84) domain proteins of the INM and KASH (Klarsicht, ANC-1, Syne Homology) domain proteins of the ONM (Starr and Han, 2003; Starr, 2009). KASH domain proteins play important roles in nuclear migration and positioning in a variety of cell and tissue types and interact with an assortment of cytoskeletal components (Mosley-Bishop et al., 1999; Starr and Han, 2002; Zhang et al., 2002; Zhen et al., 2002; Malone et al., 2003; Padmakumar et al., 2004; Wilhelmsen et al., 2005; Zhang et al., 2007; Roux et al., 2009; Horn et al., 2013).
The KASH domain is a C-terminal sequence of 50–60 amino acid residues (Starr and Han, 2002). It contains a single trans-membrane region followed by a short sequence of 40 residues or less that extends into the PNS. The KASH domain is both necessary and sufficient for targeting to the ONM, a function that is dependent upon the presence of one or more INM SUN proteins (Starr and Han, 2002).
Both mouse and human genomes encode at least six SUN proteins (Liu et al., 2007; Sohaskey et al., 2010). Of these, only two, SUN1 and SUN2, are widely expressed. Their N-terminal domains (∼200–400 residues) are exposed to the nucleoplasm and may interact with a variety of nuclear components (Hodzic et al., 2004; Crisp et al., 2006; Haque et al., 2006; Hasan et al., 2006), including nuclear lamins (Burke and Stewart, 2013). The C-terminal region of SUN1 and SUN2 extends into the PNS where a membrane-proximal sequence is predicted to form a coiled-coil (Hodzic et al., 2004; Crisp et al., 2006; Haque et al., 2006; Hasan et al., 2006; Liu et al., 2007) that terminates in the eponymous globular SUN domain. SUN proteins function as tethers for ONM KASH proteins (Padmakumar et al., 2005; Crisp et al., 2006; Haque et al., 2006; McGee et al., 2006). In this way, interacting pairs of SUN and KASH proteins form links in a molecular chain spanning both INM and ONM that physically couples nuclear components to the cytoskeleton. These SUN–KASH pairs are termed LINC complexes (linker of the nucleoskeleton and cytoskeleton; Crisp et al., 2006).
Different LINC isoforms have roles in nuclear movement and positioning in various cell types. These include nuclear repositioning during fibroblast migration (Luxton et al., 2010), cell cycle–dependent nuclear oscillation during neuronal differentiation (Zhang et al., 2009), and pronuclear migration in fertilized eggs (Malone et al., 2003; Lindeman and Pelegri, 2012). SUN and KASH proteins may also affect intranuclear organization. During meiosis LINC complexes play a crucial role in transmitting cytoskeletal forces across the NE to individual chromosomes (Chikashige et al., 2006; Ding et al., 2007; Chi et al., 2009; Sato et al., 2009), and so coordinate the complex chromosomal choreography that is a hallmark of meiotic prophase (Scherthan, 2001; Hiraoka and Dernburg, 2009).
Recent studies highlight roles played by LINC complexes in meiotic progression. In Schizosaccharomyces pombe, Kms1, an ONM KASH protein, is an adaptor for cytoplasmic dynein. Kms1 is tethered in the ONM by Sad1, an INM SUN protein (Miki et al., 2002, 2004). Sad1 aggregates in turn form NE attachment sites for meiotic telomeres. Association between Sad1 and telomeres is mediated by Taz1 and Rap1, a pair of telomeric proteins, as well as by meiosis-specific proteins, Bqt1-4 (Chikashige et al., 2006, 2009). Thus, Kms1-associated dynein is able to drive the clustering of telomeres that facilitates homologue pairing. Although budding yeast also uses LINC complexes to mediate bouquet formation (Jaspersen et al., 2002; Conrad et al., 2007, 2008), it is unusual in that the process is actin dependent rather than microtubule dependent (Trelles-Sticken et al., 2005; Koszul et al., 2008).
Meiosis in C. elegans conforms to the S. pombe paradigm, where pairing is facilitated by dynein-dependent bouquet formation. ZYG-12, an ONM KASH protein, is the adaptor for cytoplasmic dynein, whereas SUN-1/MTF-1 is the INM tether for ZYG-12. SUN-1/MTF-1 also defines attachment sites for chromosomal PCs. Association between SUN-1/MTF-1 and PCs is mediated by soluble chromosome- and PC-specific proteins (ZIM-1, -2, -3, and HIM-8; Phillips et al., 2005; Phillips and Dernburg, 2006; Penkner et al., 2007; Sato et al., 2009; Baudrimont et al., 2010) and regulated by phosphorylation (Penkner et al., 2009; Harper et al., 2011; Labella et al., 2011).
In mice, SUN1 is essential for gametogenesis (Ding et al., 2007; Chi et al., 2009). Both male and female mice deficient in SUN1 are infertile. Male infertility results from arrest of primary spermatocytes in meiotic prophase 1, and is associated with failure of telomeres to attach to the NE. As is the case in both fission yeast and C. elegans, telomere (or PC) association with the NE is a prerequisite for bouquet formation and efficient homologue pairing. Telomere clustering occurs at the nuclear pole closest to the centrosome (Scherthan, 2001; Harper et al., 2004). The prediction, therefore, is that this is likely to be a dynein-mediated process requiring a meiotic LINC complex coupling telomeres to the microtubule system. Although SUN1 clearly represents the INM portion of such a LINC complex, the identity of its ONM KASH domain portion has remained uncertain. Recently, Morimoto et al. (2012) described a new protein, KASH5, that is expressed in spermatocytes and which associates with both SUN1 and the dynactin complex (Morimoto et al., 2012). KASH5 is related to a zebrafish protein, futile cycle (Fue), that is required for pronuclear migration in early embryos (Lindeman and Pelegri, 2012). In this paper we demonstrate that KASH5 is an authentic LINC component and that KASH5 oligomers form ONM adaptors for cytoplasmic dynein. By deriving mice deficient in KASH5, we demonstrate that this protein is essential for male and female gametogenesis. Spermatocytes lacking KASH5 display a meiotic arrest in which there is little or no homologue pairing, although telomeres still associate with the NE. Derivation of these mice allows us for the first time to examine, in a mammalian system, the consequences of decoupling telomere attachment at the NE from telomere movement. These studies demonstrate an absolute requirement for cytoplasmic components, including dynein, for homologue pairing during meiotic prophase.
Results
C. elegans ZYG-12 is required for efficient homologue recognition and pairing (Penkner et al., 2009; Sato et al., 2009). ZYG-12 is an ONM adapter for cytoplasmic dynein and is connected in a meiotic LINC complex to chromosomal PCs via SUN-1/MTF-1 (Penkner et al., 2007). No genes encoding ZYG-12 orthologues are evident in mouse or human genomes. Consequently, a mammalian KASH protein linking meiotic telomeres to the microtubule system has, until recently, remained elusive. Certainly, of the known mammalian KASH proteins, Nesprins 1–4 and lymphocyte-restricted membrane protein (LRMP) can be ruled out individually as functional ZYG-12 homologues, because mice deficient in any one of these are fertile (Zhang et al., 2007, 2009; Horn et al., 2013; Ketema et al., 2013; unpublished data). Moreover, we have not detected any of these proteins in mouse spermatocytes (unpublished data).
A mammalian homologue of zebrafish Fue
Morimoto et al. (2012) reported the identification of an uncharacterized protein, CCDC155, in a yeast two-hybrid screen of a mouse testis library using the mouse cohesin protector protein, shugosin 2, as bait. Although this interaction is likely spurious, they concluded that CCDC155 was a new KASH protein and proposed renaming it KASH5 (Morimoto et al., 2012). They further showed that KASH5 was associated with SUN1 in spermatocytes, suggesting that it is a component of a meiotic LINC complex.
In complementary studies we detected CCDC155 (KASH5) by way of homology with other KASH proteins. We previously identified Nesprin 4 (NESP4) by homology with the NESP2 KASH domain (Roux et al., 2009). This approach also detected a fifth potential mammalian KASH protein, LRMP or JAW1 (Fig. 1, A and B; Behrens et al., 1994). LRMP was initially described in preB cells and its function remains largely unknown.
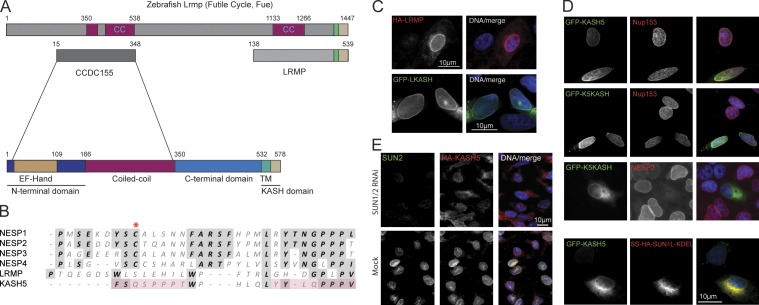
Figure 1.
Identification of a new KASH protein, KASH5. (A) A BLASTP search with mouse LRMP identified its zebrafish homologue, futile cycle (Fue). Fue contains an N-terminal extension sharing an ∼343-residue region of similarity with a mouse CCDC155. CCDC155 features a central coiled-coil flanked by an N-terminal domain, containing an EF hand–like sequence, and a C-terminal region terminating in a KASH domain. In all subsequent panels CCDC155 is referred to as KASH5. (B) Alignment of the luminal regions of KASH domains from mouse Nesprins 1–4 (NESP1, 2, 3, and 4), LRMP, and KASH5. A conserved cysteine (asterisk, absent from LRMP and KASH5) may form a disulphide bond with a SUN domain cysteine. Pink shading highlights a possible duplication within KASH5. (C) Mouse LRMP behaves as an authentic KASH protein. HA-tagged LRMP (HA-LRMP) localizes to the NE in HeLa cells, as does GFP fused to the LRMP KASH domain (GFP-LKASH). (D) KASH5 is a bona fide KASH protein. GFP-tagged KASH5 (GFP-KASH5) localizes to the NE of HeLa cells (top row). The NE is detected using a monoclonal antibody (SA1) against Nup153, a nuclear pore complex protein. The KASH5 KASH domain alone targets GFP (GFP-K5KASH) to the NE (second row). Overexpression of GFP-K5KASH displaces endogenous Nesprin 2 (NESP2) from the NE (third row). Co-expression of the dominant-negative SUN1 mutant (SS-HA-SUN1L-KDEL) blocks localization of GFP-KASH5 to the NE (bottom row). (E) HA-KASH5 localization to the NE of HEK293 cells is lost upon co-depletion of SUN1 and SUN2 by RNA interference. In C and D, nuclei are co-stained with DAPI.
LRMP, which associates with the inositol trisphosphate receptor (Shindo et al., 2010), accumulates at the nuclear periphery when ectopically expressed (Fig. 1, A and C). This localization is conferred entirely by its C-terminal KASH sequence and is eliminated by coexpression of a dominant-negative SUN1 mutant (SS-HA-SUN1L-KDEL; Fig. S1 A; Crisp et al., 2006), but not by coexpression of wild-type SUN1 or SUN2 (Fig. S1 A). Mammalian LRMP therefore behaves as an authentic KASH protein. A BLASTP search with mouse LRMP detected a zebrafish homologue (CAI20727), Fue (Fig. 1 A; Dekens et al., 2003; Lindeman and Pelegri, 2012). Fue binds dynein and is essential for pronuclear migration in fertilized eggs. This protein features an enlarged N terminus that is absent from both human and mouse LRMP. A second BLASTP search using the entire Fue/Lrmp sequence detected both mouse (NP_958762) and human (NP_653289) CCDC155/KASH5. The two proteins share 54% (mouse) and 57% (human) sequence similarity over 331 and 327 residues, respectively within the unique Fue/Lrmp N-terminal extension. In silico analysis of mouse CCDC155/KASH5 (64.4 kD, 62.7 kD in humans) revealed a degenerate C-terminal KASH domain (Fig. 1 B). CCDC155/KASH5 features a central sequence of ∼190 residues predicted to form a coiled-coil (Fig. 1 A). Upstream of this is an N-terminal domain of 166 residues containing a calmodulin-like EF hand sequence between residues 18 and 109 (Kretsinger and Barry, 1975). The region of similarity with zebrafish Fue/Lrmp lies between residues 15 and 348 (344 in humans) encompassing the CCDC155/KASH5 N-terminal domain and most of the coiled-coil.
KASH5 is a dynein-binding protein of the ONM
If KASH5 is a bona fide KASH domain protein it should localize to the ONM in a SUN- and KASH-dependent fashion. To test this, HA- or GFP-tagged versions of KASH5 were expressed in both HeLa and HEK293 cells. GFP-tagged KASH5 concentrated at the NE (Fig. 1 D, top), a localization that was abolished by coexpression of SS-HA-SUN1L-KDEL (Fig. 1 D, bottom). Similarly, the KASH5 KASH domain alone (K5KASH) was sufficient to target GFP to the NE (Fig. 1 D, second row). Moreover, this same GFP fusion protein efficiently displaced endogenous NESP2 from the NE (Fig. 1 D, third row). Conversely, NE localization of HA-KASH5 stably expressed in HEK293 cells was abolished after depletion of SUN1 and SUN2 by RNA interference (Fig. 1 E). Differential permeabilization of HEK293 cells using digitonin versus Triton X-100 revealed that GFP-tagged KASH5 localizes at least in part to the cytoplasmic face of the NE, indicating that it is an ONM component (Fig. S1 B; Adam et al., 1990). Together these data demonstrate that KASH5 behaves as an authentic KASH protein.
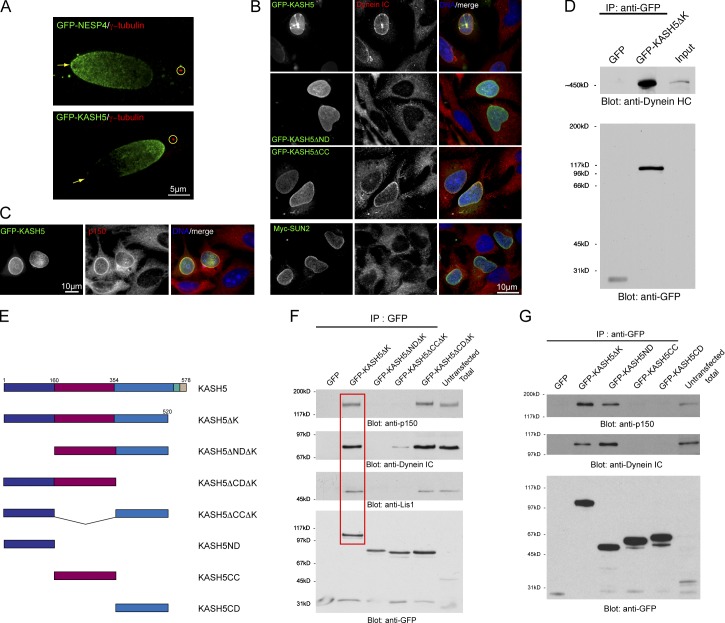
When expressed in HeLa cells, we noted that in ∼10% of nuclei KASH5 displayed an asymmetric distribution in the NE, concentrating at the nuclear pole proximal to the centrosome (Fig. 2 A). This is exactly the reverse of what we previously reported for NESP4, a kinesin-1–binding protein (Roux et al., 2009). This distribution of NESP4 (Fig. 2 A) is both microtubule dependent and kinesin-1 dependent. Because kinesin-1 moves toward the plus end of microtubules, these observations prompted us to test the idea that KASH5 might be a partner for a minus end–directed motor protein such as cytoplasmic dynein. Indeed, expression of GFP-KASH5, but not a Myc-tagged SUN2 control, resulted in the recruitment of dynein intermediate chain (dynein IC) to the NE (Fig. 2 B). Similarly, p150Glued, a subunit of dynactin (Schroer, 2004), a dynein regulatory complex, was also recruited to the NE by GFP-KASH5 (Fig. 2 C). We then performed a series of coimmunoprecipitations (coIPs) using a soluble form of GFP-tagged KASH5 (KASH5ΔK) lacking the C-terminal KASH domain. Western blot analysis of anti-GFP IPs revealed the presence of dynein heavy chain with GFP-KASH5ΔK, but not with GFP alone (Fig. 2 D). These data imply that KASH5 is an adaptor for cytoplasmic dynein. Additional coIP experiments revealed associations with dynein IC and p150Glued as well as with LIS1, another dynein regulator (Fig. 2 F; Vallee and Tsai, 2006). We next prepared a series of KASH5 deletion mutants (Fig. 2 E). Dynein subunits were only observed in coIPs with those forms of KASH5 retaining the N-terminal domain (residues 1–166; Fig. 2, F and G). Indeed, the N-terminal domain alone is sufficient to confer dynein binding (Fig. 2 G). Correspondingly, immunofluorescence microscopy revealed that KASH5 lacking its N-terminal domain (KASH5ΔND) was unable to recruit dynein IC to the NE (Fig. 2 B, second row). In contrast, deletion of the coiled-coil domain (KASH5ΔCC) had no such effect on dynein IC recruitment (Fig. 2 B, third row). In our coIP experiments, KASH5ΔCCΔK association with dynein IC (Fig. 2 F) is reduced. This discrepancy between coIP and immunofluorescence data may reflect reduced avidity of KASH5ΔCCΔK for dynein. The reason being is that, as will be described below, the coiled-coil domain is required for KASH5 oligomerization.
Figure 2.
KASH5 is a dynein-binding protein. (A) GFP-tagged Nesprin 4 (GFP-NESP4) expressed in HeLa cells concentrates at the nuclear pole (arrows) distal to the centrosome (anti–γ-tubulin, circled). GFP-KASH5 concentrates at the pole proximal to the centrosome. (B) GFP-KASH5 (top row) recruits dynein intermediate chain, (dynein IC) to the NE of HeLa cells. Myc-tagged SUN2 (Myc-SUN2) has no such effect (bottom row). GFP-tagged KASH5 lacking its N-terminal domain (GFP-KASH5ΔND) is unable to recruit dynein IC to the NE. Deletion of the coiled-coil domain (GFP-KASH5ΔCC) has little effect on dynein IC recruitment. (C) GFP-KASH5 recruits dynactin p150Glued (p150) to the NE. (D) Immunoprecipitation (IP) reveals that a soluble GFP-tagged form of KASH5 lacking its KASH domain, GFP-KASH5ΔK, forms a complex containing dynein heavy chain (Dynein HC). (E) A series of KASH5 deletion mutants. (F) Co-IPs using GFP-tagged KASH5 deletion constructs (E) reveal that GFP-KASH5ΔK associates with dynein IC, p150glued, and LIS1 (red box). These interactions are lost upon deletion of the N-terminal domain (GFP-Kask5ΔNDΔK). In the absence of the coiled-coil domain (GFP-KASH5ΔCCΔK), while LIS1 and p150Glued binding is lost, interaction with dynein is maintained, albeit at a lower level. Deletion of the C-terminal domain (GFP-KASH5ΔCDΔK) has no effect on these interactions. (G) Co-IPs using individual KASH5 domains. The N-terminal domain alone (GFP-KASH5ND) associates with the dynein motor complex. The coiled-coil (GFP-KASH5CC) and C-terminal (GFP-Kask5CD) domains show no such interaction.
Self-association of KASH5
The presence of a predicted coiled-coil suggests that KASH5 might self-associate. To explore this, we used our series of differentially tagged deletion mutants (Fig. 2 E), which we coexpressed with versions of KASH5 containing an intact cytoplasmic domain. Full-length KASH5 recruits the soluble cytoplasmic domain of KASH5 (KASH5ΔK) to the NE (Fig. S2 A). Corresponding coIP studies with HA- and GFP-tagged versions of KASH5 provided corroborating evidence that KASH5 self-associates (Fig. S2 C). A similar result was observed with the coiled-coil domain alone fused to GFP, which is also recruited to the NE by full-length KASH5 (Fig. S2 B). Complementary coIP analyses confirmed that KASH5 self-association was mediated by the central coiled-coil domain (Fig. S2 D).
Kash5 is expressed in spermatocytes
Immunofluorescence microscopy and RT-PCR (Fig. 3, A and B) revealed expression of Kash5 in adult testes, in agreement with Morimoto et al. (2012). Kash5 transcripts were also identified in bone marrow, albeit at a lower level (Fig. 3 B), as well as in fetal liver (unpublished data). In the testis, Kash5-expressing cells were located in a distinct band at the periphery of the seminiferous tubules (Fig. 3 A). These cells were positive for SCP3, an axial element protein of synaptonemal complexes (SCs; Fig. 3 A; Fraune et al., 2012), indicating that they are spermatocytes in meiotic prophase 1. Round and elongated spermatids, which are localized closer to the centers of tubules, display KASH5 aggregates at one nuclear pole. In mature sperm cells the protein is undetectable.
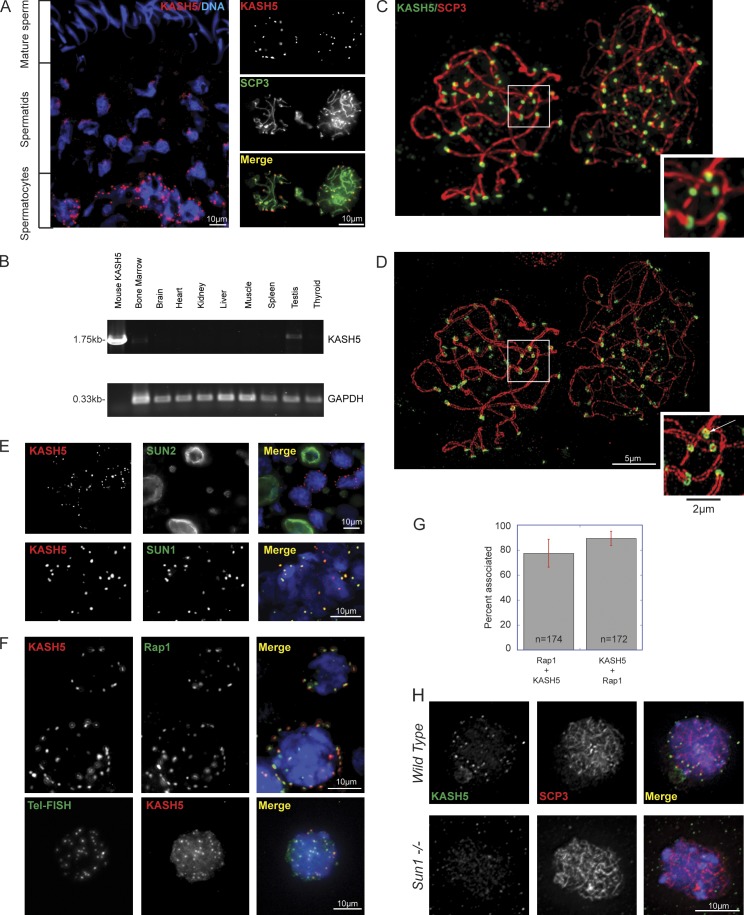
Figure 3.
Kash5 is expressed in the testis. (A) A testis section labeled with anti-KASH5 reveals a punctate distribution in spermatocyte NEs. These are localized toward the periphery of seminiferous tubules (bottom of image). Double labeling with an antibody against SCP3 confirms that KASH5-positive cells are spermatocytes. Spermatids feature polar KASH5 aggregates. In mature sperm KASH5 is undetectable. Nuclei are visualized with DAPI. (B) Reverse-transcription PCR from adult mouse tissues. KASH5 is expressed in testis and to a lesser extent in bone marrow. GAPDH is the loading control. (C) Spermatocyte spreads, labeled with anti-KASH5 and anti-SCP3 antibodies, imaged by conventional deconvolution microscopy. KASH5 localizes to the tips of SCP3-positive SCs, where it appears as a single patch (inset). (D) Structured illumination microscopy (SIM) of the same field (C) reveals both single (right-hand cell in leptotene/zygotene) and double (left-hand cell in pachytene) SCP3-positive strands. SIM reveals KASH5 rings at the tips of SCP3 axial strands (inset). Where paired axial strands are evident KASH5 displays “figure eight”–like distributions (inset, arrow). (E) KASH5 colocalizes with SUN1 (bottom panels), but not SUN2 (top panels) in mouse spermatocyte spreads. KASH5 and SUN2 are clearly expressed in different cell types. (F) KASH5 colocalizes with Rap1, a telomere protein, in spermatocyte spreads. A similar result is observed when telomeres are visualized by fluorescence in situ hybridization (Tel-FISH). (G) Quantitation of Rap1 and KASH5 colocalization. 77.4 ± 3.7% of Rap1 foci have associated KASH5, whereas 89.4 ± 1.7% of KASH5 foci have associated Rap1. Bars indicate standard error. (H) KASH5 localization is dependent on SUN1. In wild-type spermatocytes KASH5 localizes to the NE at telomere-associated foci. This distribution is lost in Sun−/− spermatocytes. SCP3 labeling confirms that the spermatocytes are in similar meiotic phases (leptotene/zygotene).
KASH5 localizes to the spermatocyte NE in multiple foci. This contrasts with the more uniform distribution of KASH5 in HeLa and HEK293 NEs. KASH5 foci, which first appear in leptotene and become increasingly pronounced as meiotic prophase 1 progresses, precisely colocalize with the tips of SCs (Fig. 3, C and D; and Fig. S3). Using conventional wide-field microscopy combined with deconvolution, little substructure could be discerned in either the SCs or in their associated KASH5 foci (Fig. 3, C and inset). However, the same fields imaged using structured illumination microscopy (SIM; Schermelleh et al., 2008), with its twofold improvement in resolution, revealed paired SCP3-positive axial elements within each SC (Fig. 3 D and Fig. S3). In this way, SIM can provide a simple yet robust criterion for assessing homologue pairing and synapsis. SIM also revealed that KASH5 itself appears to be arranged in ring-like assemblies at the tip of each axial element. During homologue pairing, individual KASH5 rings combine to form “figure eight”–like structures (Fig. 3 D inset, arrow).
A meiotic LINC complex
Analyses of testis cryosections and spermatocyte spreads revealed that KASH5 and SUN1 precisely colocalized in the spermatocyte NEs (Fig. 3 E). SUN2, in contrast, was not detected in spermatocytes but was present at the NEs of Sertoli cells (Fig. 3 E). SUN1 is required for telomere anchoring at the NE (Ding et al., 2007). It follows therefore that KASH5 should also associate with NE telomere attachment sites. Indeed, such an association is implied in the SCP3-KASH5 double label data (Fig. 3, A, C, and D; and Fig. S3). This was confirmed in further experiments using antibodies against Rap1, a telomere protein (Fig. 3, F and G), as well as by fluorescence in situ hybridization using a telomere-specific oligonucleotide probe (Tel-FISH; Fig. 3 F). Quantitative analysis of Rap1-KASH5 colocalization (Fig. 3 G) indicated that 77.4 ± 3.7 (SE)% of Rap1 foci were associated with KASH5 while 89.4 ± 1.7 (SE)% of KASH5 foci were associated with Rap1.
Colocalization of SUN1 and KASH5 in spermatocytes suggests that these two proteins function together as a meiotic LINC complex. Accordingly, spermatocytes from Sun1-null mice display a complete absence of NE-associated KASH5 foci (Fig. 3 H). Evidently, SUN1 is required for the appropriate localization of KASH5, confirming the earlier report of Morimoto et al. (2012).
Infertility in Kash5-null mice
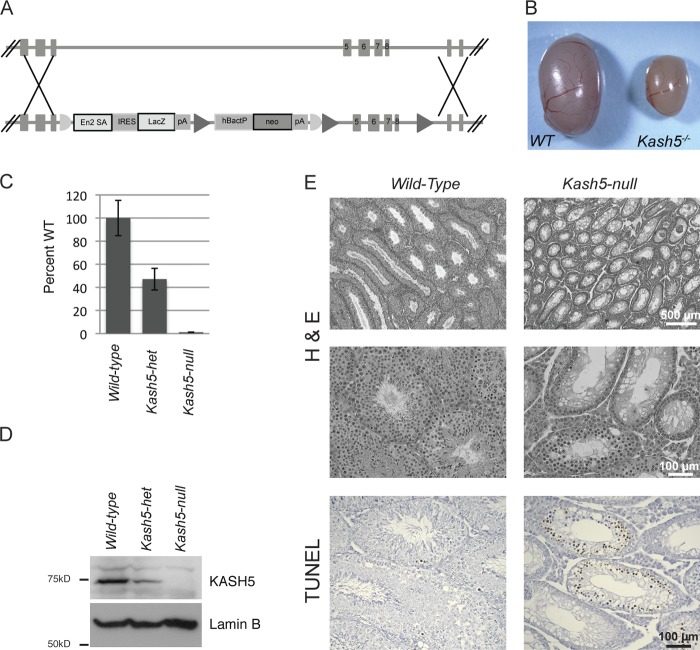
To further explore meiotic LINC complex function we derived mice deficient in KASH5. Targeting of Kash5 eliminated exons 5–8, resulting in a frame shift and appearance of a stop codon 36 bp downstream of the 3′ end of exon 4. Homozygous-null mice appeared overtly normal. They were born with the expected Mendelian frequency, indicating no lethal developmental defects. Although KASH5 transcripts could be detected in bone marrow, homozygous-null mice displayed no obvious hematological pathologies. Although lifespan of these animals was unaffected by Kash5 deficiency, mating with wild-type partners revealed that Kash5-null mice, both male and female, were infertile. Macroscopically, testes from homozygous-null males were only ∼25% the size of those derived from wild-type siblings (Fig. 4 B). Quantitative RT-PCR and Western blot analyses (Fig. 4, C and D) revealed a complete absence of both KASH5 mRNA and protein from the Kash5-null animals. Testis samples from heterozygous animals displayed an intermediate (roughly 50%) level of KASH5 mRNA and protein.
Figure 4.
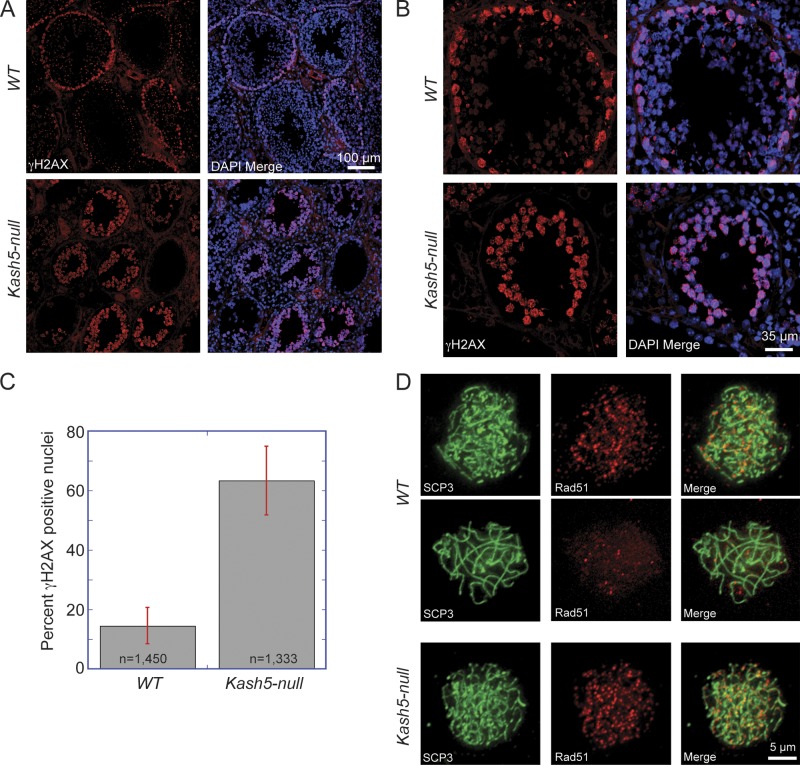
Derivation of Kash5-null mice. (A) Vector and targeting strategy for generating Kash5-null mice. (B) Testes from Kash5-null animals are only ∼25% of the size of those from wild-type littermates. (C) Quantitative PCR of whole testis lysates from littermate males. In Kash5-null mice Kash5 mRNA was undetectable. In Kash5 heterozygotes Kash5 mRNA was present at ∼50% of the level observed in wild-type mice. (D) Immunoblot analysis of whole testis lysates from littermate males. In the Kash5-null animals, no KASH5 was detectable. In Kash5 heterozygotes, roughly half the level of KASH5 was present compared with that seen in wild-type mice. (E) Hematoxylin and eosin (H&E) stained testis sections from wild-type and Kash5-null littermates. The overall structure of Kash5-null testes is preserved with identifiable, albeit smaller, seminiferous tubules. At higher magnification (center panels) it is apparent that Kash5-null seminiferous tubules lack sperm. Wild-type testes have occasional TUNEL-positive nuclei (bottom panels). In Kash5-null testes, widespread TUNEL staining is seen in ∼30% of tubule cross sections.
Histological analyses revealed normal tissue organization in Kash5-null testes (Fig. 4 E). However, the seminiferous tubules were narrower and devoid of both elongated spermatids and mature sperm. Indeed many tubules (∼30%) featured only a single layer of cells, likely including Sertoli cells, at the tubule periphery. TUNEL analysis, detecting nicked DNA, highlighted a further difference between wild-type and Kash5-null testes (Fig. 4 E). Although cross sections of tubules in wild-type samples displayed only a scattering of TUNEL-positive cells, in Kash5-null testis sections ∼30% of the tubules contained numerous cells that were TUNEL positive. Because the formation of DNA double-strand breaks (DSBs) is an essential feature of meiosis, it is possible that accumulation of TUNEL-positive cells in the Kash5-null testis samples reflected early meiotic arrest. However, apoptotic or necrotic cell death might also contribute to the TUNEL signal.
Like their male siblings, female Kash5-null mice were infertile. Ovaries from such animals were barely visible after necropsy. Histological analyses revealed the presence of little more than stromal tissue (Fig. S4). We were unable to identify any follicles in these samples.
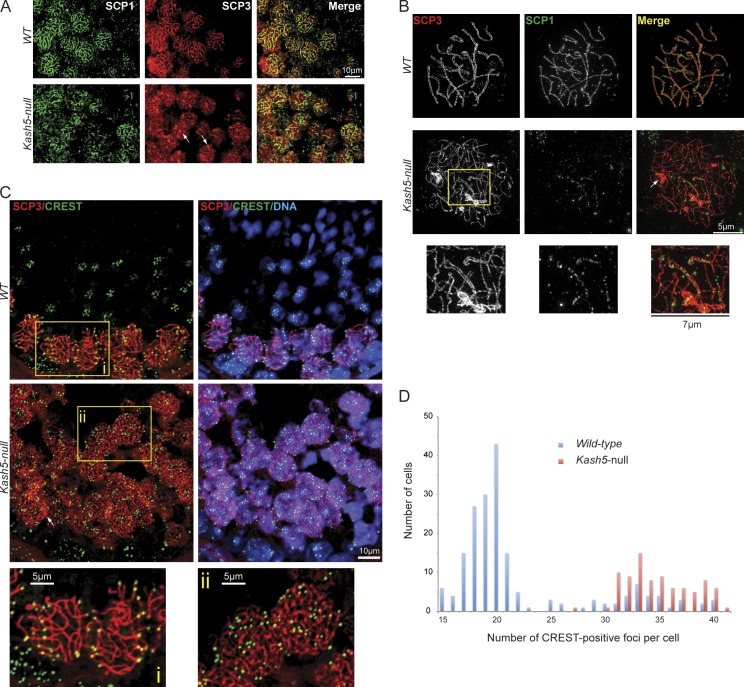
KASH5 deficiency prevents synapsis
Labeling of testis sections and spermatocyte spreads with antibodies against SCP3 and SCP1, a SC transverse filament protein, revealed an apparent absence of pachytene spermatocytes in Kash5-null samples (Fig. 5 A). However, numerous leptotene/zygotene spermatocytes were evident, indicating that loss of KASH5 is associated with a prepachytene arrest. We subsequently used SIM to examine homologue pairing and synapsis. In conventional wide-field microscopy, synaptonemal complexes appear as a single thread-like structure in which SCP3 and SCP1 labeling are coincident. SIM, however, revealed paired threads of SCP3 corresponding to the SC axial elements flanking SCP1-containing transverse filaments. In many cases the axial elements coiled around each other in an extended double helix (Fig. 5 B and Fig. S5). Examination of Kash5-null spermatocytes using SIM yielded only a few examples of seemingly partially paired homologues (Fig. 5 B, boxed area) with telomeres in precise alignment. We could also find occasional examples of what may be nonhomologous pairing where telomeres were clearly misaligned (see Fig. 7 B, arrowhead). We never observed fully synapsed homologues in Kash5-null spermatocytes. This contrasts with Sun1-null mice, where complete pairing of some homologues was documented (Ding et al., 2007).
Figure 5.
Homologue pairing is defective in Kash5-null spermatocytes. (A) SCP1 and SCP3 distribution in testis sections from wild-type (WT) and Kash5-null mice. Wild-type testes show consistent colocalization of SCP1 and SCP3 where most SCP3-positive strands are also SCP1 positive (i.e., the cells are in pachytene). In Kash5-null testes, SCP1 vs. SCP3 colocalization is more variable. SCP3 tends to form aggregates in Kash5-null spermatocytes (arrows). These can also be seen in B and C (arrows). (B) Spermatocyte spreads from wild-type (WT) and Kash5-null littermates. SCP3 and SCP1 were localized using SIM. In wild-type pachytene spermatocytes sister chromosomes are paired as indicated by both SCP3 and SCP1 labeling. SIM reveals strings of SCP1 foci lying between easily resolvable SCP3 axial strands. In Kash5-null spermatocytes SCP3 appears mainly in single axial strands. Only limited regions of paired SCP3 strands are seen (inset) that are weakly positive for SCP1. (C) Testis sections labeled with an anti-centromere CREST auto-antibody and anti-SCP3. In wild-type (WT) sections spermatocytes positive for both CREST and SCP3 are restricted to the periphery of the seminiferous tubules. In Kash5-null specimens CREST- and SCP3-positive cells persist into the center of the tubules. Higher magnification (i and ii, bottom panels) reveals that the number of CREST-positive foci in Kash5-null spermatocytes (34.80 ± 0.33 SEM) (ii) is noticeably greater than that in wild-type cells (21.96 ± 0.45) (i). This difference is statistically significant (P < 0.0001). (D) SCP3-positive cells in testis sections (C) were also plotted according to the numbers of CREST-positive foci, providing a graphical comparison of wild-type and Kash5-null spermatocytes. The displayed data are from a single representative experiment out of two repeats. For the experiment shown, n = 273.
Figure 7.
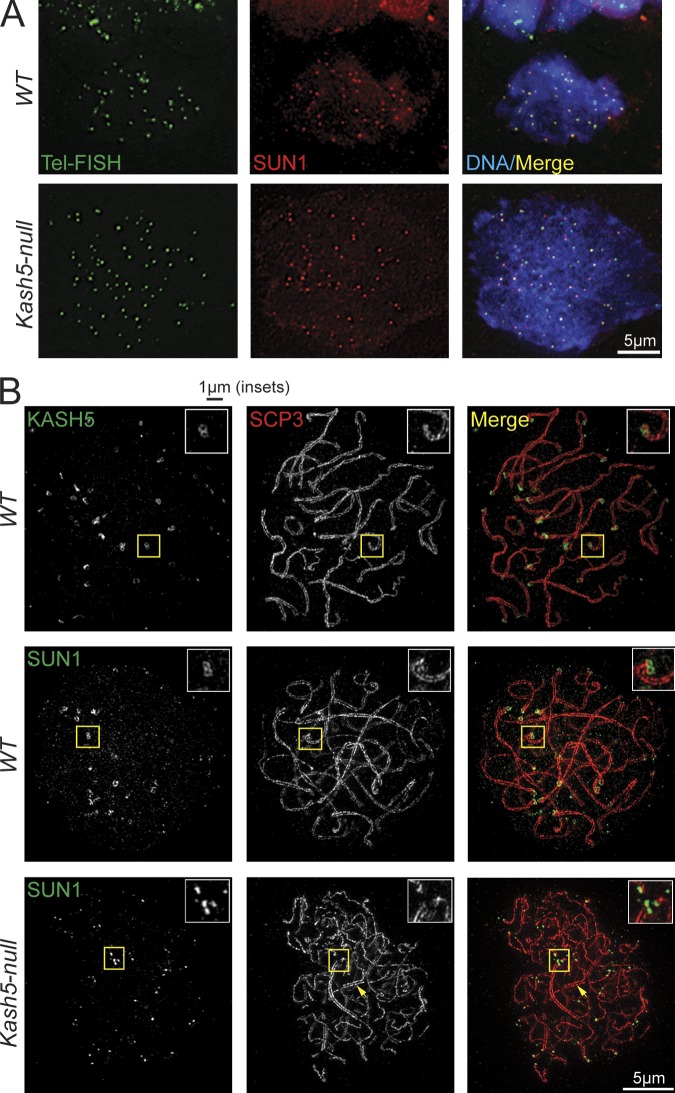
Telomeres associate with SUN1 in wild-type and Kash5-null spermatocytes. (A) Spermatocyte spreads processed for telomere fluorescence in situ hybridization (Tel-FISH) and immunostained with anti-SUN1. In wild-type (WT) specimens 96.8 ± 0.6% of telomeres are associated with SUN1 foci. In Kash5-null spermatocytes the figure is lower at 55.8 ± 2.0%. Nevertheless, telomeres can still associate with SUN1 in the absence of KASH5. (B) Spermatocyte spreads visualized by SIM. SUN1 exhibits the same ring-like structure as KASH5 at the tips of SCP3-positive axial strands in WT spermatocytes (insets). In Kash5-null spermatocytes, SUN1 also localizes to the tips of SCP3-positive strands. However, the ring-like organization is replaced by seemingly unstructured foci (insets).
That homologue pairing was defective in Kash5-null mice, and was reinforced by labeling sections with a CREST anti-centromere antiserum (Fig. 5 C). In wild-type samples, CREST- and SCP3-positive cells were concentrated toward the periphery of seminiferous tubules, the central regions being occupied by spermatids and mature sperm. By contrast, in Kash5-null samples CREST- and SCP3-positive cells persisted in the central regions of the tubule. Significantly, wild-type spermatocytes contained fewer CREST foci than their Kash5-null counterparts (Fig. 5 D), consistent with reduced pairing in the latter (mice have 20 chromosome pairs).
DSBs are never resolved in Kash5-null spermatocytes
DSBs are a key feature of meiosis, and while required for recombination, are not a prerequisite for pairing (Boateng et al., 2013). DSBs are normally resolved by pachytene. To determine the fate of such breaks in Kash5-null mice we labeled testis sections with an antibody against phosphorylated histone H2AX (γ-H2AX), a marker for DNA breakage sites (Fig. 6, A–C; Dickey et al., 2009). We also used an antibody against Rad51, a protein required for DSB repair by homologous recombination (Fig. 6 D; San Filippo et al., 2008). In wild-type sections, γ-H2AX–positive spermatocytes were restricted to a single layer of cells at the periphery of seminiferous tubules, with many tubule sections containing no γ-H2AX–positive cells (Fig. 6, A and B). In Kash5-null sections large numbers of γ-H2AX–positive cells were evident, which filled entire tubules (Fig. 6, A and B). Quantitative analyses of cells localized within the lumen of seminiferous tubules revealed a preponderance of γ-H2AX–positive nuclei in the Kash5-null samples (63.3 ± 11.5% vs. 14.5 ± 6.1%; Fig. 6 C). A similar situation was seen with Rad51 in spermatocyte spreads. In wild-type specimens only the occasional SCP3-positive cell was also positive for Rad51. In contrast, in Kash5-null specimens virtually all SCP3-positive cells contained Rad51 foci (Fig. 6 D). The conclusion is that Kash5-null spermatocytes accumulate DSBs that are never resolved.
Figure 6.
DSBs persist in Kash5-null testes. (A) Low and (B) high magnification views of testis sections from wild-type (WT) and Kash5-null animals, showing the distribution of DSBs revealed by γ-H2AX labeling. In WT testis sections, a ring of γ-H2AX–positive nuclei can sometimes be seen at the periphery of seminiferous tubules. The DSBs are always resolved, with a concomitant decline in γ-H2AX levels, as spermatocytes mature and progress spatially toward the center of the tubule. Kash5-null testes display persistent γ-H2AX–positive cells that accumulate toward the center of the tubule. (C) Percentage of cells in wild-type (WT, 14.5 ± 6.1%) vs. Kash5-null (63.3 ± 11.5%) seminiferous tubules with γ-H2AX–positive nuclei. Bars indicate standard deviation. (D) Spermatocyte spreads immunostained with anti-SCP3 and anti-Rad51 antibodies. In spreads from WT animals only the occasional cell is positive for both proteins (top row). Distribution of SCP3 indicates that these are leptotene/zygotene cells. The bulk of cells positive for SCP3, typically in pachytene, have resolved the majority of the Rad51 foci (middle row). The preponderance of SCP3-positive Kash5-null spermatocytes that never reach pachytene remain Rad51-positive (bottom row).
KASH5 recruits dynein to NE–telomere attachment sites
Earlier studies showed that SUN1 is essential for telomere attachment to the NE (Ding et al., 2007). Tel-FISH double-label experiments reveal that telomere association with SUN1 foci is partially conserved in Kash5-null spermatocytes (Fig. 7 A). Although 96.8 ± 0.6% of telomeres were associated with SUN1 foci in wild-type spermatocytes, the corresponding figure in Kash5-null cells was 55.8 ± 2.0%. What is not clear is whether this ∼40% reduction represents a failure of some telomeres to attach to the NE or whether it is due to an increased rate of detachment, possibly associated with early stages of apoptosis. SIM also revealed some subtle differences in the telomere attachment sites themselves in wild-type versus Kash5-null spermatocytes. Using conventional microscopy, both SUN1 and KASH5 appeared to concentrate in single foci colocalizing with telomeres. SIM, however, revealed that SUN1 (Fig. 7 B), like KASH5 (Figs. 3 D, 7 B, and S3), was organized in ring-like assemblies. Before homologue pairing, single rings were observed. After pairing, the single telomere-associated rings combined to form “figure eight” structures (Fig. 7 B). In Kash5-null spermatocytes, the ring-like distribution of SUN1 was no longer evident. Instead, SUN1 appeared in single foci lacking discernible substructure (Fig. 7 B). One interpretation is that not only does SUN1 function as a transluminal tether for KASH5, but KASH5 itself may contribute to the organization of SUN1 within the INM.
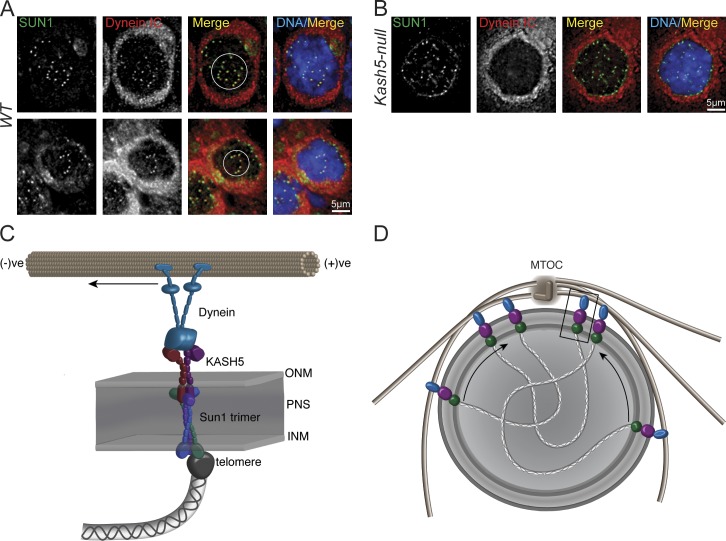
Because KASH5 is a dynein-binding protein, we would anticipate that cytoplasmic dynein should be recruited to SUN1-positive telomere attachment sites at the NE. As revealed in Fig. 8 A, NE-associated dynein IC does colocalize with SUN1 in wild-type spermatocytes. The cells displayed in this figure were in zygotene where partial clustering of the SUN1 foci (circled) was observed. In Kash5-null spermatocytes no such colocalization was detected (Fig. 7 B). Furthermore, we never observed clustered SUN1 foci in these cells. KASH5 is therefore essential for the recruitment of dynein to telomere attachment sites at the NE during meiosis. This confirms and extends the observations of Morimoto et al. (2012), who documented colocalization of p150Glued with SUN1/KASH5 LINC complexes in wild-type spermatocytes.
Figure 8.
KASH5 recruits dynein to NE telomere attachment sites. (A) SUN1 foci and dynein intermediate chain (Dynein IC) colocalize in WT spermatocyte spreads. (B) In Kash5-null spermatocytes, SUN1 foci have no associated dynein. In both A and B nuclei were co-stained with DAPI. Circles enclose clusters of SUN1 foci. (C) Model of a meiotic LINC complex. KASH5 and SUN1 link telomere ends to dynein motor proteins. For simplicity, only a single SUN1 trimer tethering a single KASH5 dimer is shown. In reality several KASH5 oligomers might cross-link multiple SUN1 trimers, creating larger LINC complex clusters. (D) Recruitment of dynein (blue) to LINC complex clusters by KASH5 (purple) should drive the entire assemblage, including telomeres, toward the minus end of microtubules focused at the microtubule organizing center (MTOC). In this way, SUN1, KASH5, and dynein would mediate meiotic bouquet formation.
Our results indicate that KASH5 is a LINC complex component and ONM adaptor for cytoplasmic dynein. KASH5 is an important determinant of telomere dynamics and is essential for successful completion of meiosis. In the absence of KASH5, spermatocytes arrest at the leptotene/zygotene stage of meiosis with the majority of chromosomes never forming homologous pairs. Ultimately, these arrested spermatocytes, which maintain numerous DSBs, are eliminated. It is likely that KASH5 plays a similar essential role in oogenesis.
Discussion
We have extensively characterized a new mammalian KASH protein, KASH5, which is expressed largely in meiotic cells. KASH5 is a dynein-binding protein tethered in the ONM via interactions with SUN1, an INM resident (Fig. 8 C). Together, SUN1 and KASH5 comprise a meiotic LINC complex. A recurring theme of meiosis is association of telomeres (or PCs) with the nuclear face of the NE during prophase 1 (Scherthan, 2001; Hiraoka and Dernburg, 2009). This association is mechanistically linked to both rapid telomere and nuclear movements and transient telomere clustering that facilitate homologue pairing (Fig. 8 D). In mammals, SUN1 foci define attachment sites for telomeres at the NE in meiotic cells (Chi et al., 2009; Ding et al., 2007). In both spermatocytes and oocytes derived from Sun1-deficient mice, telomeres fail to attach to the NE. Although some homologue pairing still occurs (Ding et al., 2007), the process is inefficient and culminates in meiotic arrest.
In C. elegans, SUN-1/MTF-1 aggregates form attachment sites at the NE for chromosome-specific PCs (Penkner et al., 2007). These attachment sites are micron-scale features, significantly larger than the SUN1 foci seen in mouse spermatocytes. SUN-1/MTF-1 as a LINC complex component also behaves as a tether for ZYG-12 in the ONM (Minn et al., 2009). ZYG-12, by virtue of its function as an adaptor for cytoplasmic dynein, serves to couple SUN-1/MTF-1–associated PCs to the microtubule system (Penkner et al., 2009; Sato et al., 2009).
The zygotene stage of meiosis is characterized by microtubule-dependent clustering of PCs at one pole of the nucleus. The establishment of this “bouquet” configuration facilitates search and recognition between homologues. In the absence of dynein, PC movement is curtailed and homologue pairing and synapsis are abolished (Sato et al., 2009). Significantly, elimination of SUN-1/MTF-1 partially overcomes the meiotic block imposed by dynein loss. However, in this situation the frequency of promiscuous nonhomologous pairing is elevated. It is possible that, in the absence of SUN-1/MTF-1, chromosomes are no longer physically constrained by their PCs at the NE, and may be free to sample the entire nuclear volume by diffusion. Although highly inefficient, this would allow some pairing to occur. These findings also suggest that ZYG-12 and dynein may together have a role in quality control or licensing of the pairing process (Sato et al., 2009).
As a mammalian ONM dynein-binding protein, KASH5 displays the hallmarks of a functional ZYG-12 homologue. In mouse spermatocytes, SUN1 defines NE attachment sites for telomeres. KASH5 also localizes to these sites as does both dynactin and dynein. Because the localization of KASH5 is SUN1 dependent, the implication is that SUN1 and KASH5 together comprise a meiotic LINC complex that couples telomeres to microtubules. Mice deficient in KASH5 are infertile. During spermatogenesis, loss of KASH5 results in the accumulation of spermatocytes arrested in leptotene/zygotene of meiotic prophase 1. Predictably, spermatocytes deficient in KASH5 fail to recruit cytoplasmic dynein to NE-associated telomere attachment sites.
SIM is a valuable tool in our studies of spermatogenesis. The twofold improvement in resolution over conventional wide-field microscopy allowed us to directly assess homologue pairing and synapsis by following the alignment of SC axial elements. In Kash5-null mice, only limited homologous and possibly nonhomologous pairing occurs. SIM has also provided images of telomere-associated SUN1/KASH5 LINC complex clusters, suggesting that these are organized into ring-like structures. Surprisingly, in the absence of KASH5 the distribution of SUN1 appears to undergo a subtle change from ring-like to focal. However, SUN1 can still remain telomere associated in Kash5-null spermatocytes. Thus, KASH5 may help define the distribution of LINC complexes at the nano-scale level. There is, however, an important caveat here that brings to the forefront potential labeling artifacts that may only become apparent at the higher resolutions achieved with SIM. It is possible that SUN1/KASH5 LINC complexes are indeed organized in homogenous patches, but that the ring pattern may arise due to antigen masking or reduced antibody access to the center of the patch. This would not be evident by conventional microscopy. Elimination of KASH5 might simply improve access of antibodies to centrally located SUN1. Notwithstanding these qualifications, our view is that the SIM images do represent an authentic portrayal of LINC complex distribution. Significantly, the ring structures are observed with two different antibodies against two separate LINC components (SUN1 and KASH5) and recognize epitopes in two separate compartments, the PNS (SUN1) and the cytoplasm (KASH5). Because loss of KASH5 is associated with an ∼40% reduction in the number of NE-associated telomeres, this might also imply a change in the functional organization of SUN1 clusters.
The effects of KASH5 on telomere anchoring and the nano-scale distribution of SUN1 could be a reflection of KASH5 oligomerization. SUN2, and most likely SUN1, exist as coiled-coil trimers (Sosa et al., 2012; Zhou et al., 2012). KASH domain binding occurs at the interface between adjacent SUN domains (Sosa et al., 2012). Hence, each SUN trimer has the capacity to bind three KASH domains. Accordingly, interaction between KASH5 oligomers and SUN1 trimers could stabilize extensive arrays of SUN1 within the INM. This may be manifest as increased avidity for telomeres or telomere-associated proteins. Conversely, loss of KASH5 would lead to reduced avidity and concomitant increase in the number of free telomeres.
There are striking parallels in the mechanics of meiosis between C. elegans and mice. In worms, deletion of SUN-1/MTF-1 can partially overcome prophase arrest linked to dynein mutations (Sato et al., 2009). In mice, although loss of SUN1 does result in a prophase arrest, some homologue pairing and synapsis may occur (Ding et al., 2007). In contrast, deletion of KASH5 (which disrupts the link between telomeres and cytoplasmic dynein) has a more profound effect with complete homologue pairing never observed. This may be due to constraints on chromosome movement imposed by the maintenance of at least partial telomere attachment at the NE. The suggestion is that co-deletion of SUN1 would have a mild relaxation effect on KASH5-linked meiotic arrest. These results also leave open the possibility that KASH5 could possess a meiotic “licensing” function as suggested for ZYG-12 (or more accurately, its association with dynein) in C. elegans (Sato et al., 2009).
We originally identified KASH5 by virtue of its homology with zebrafish Fue/Lrmp. This protein encodes both KASH5- and LRMP-specific functions (Lindeman and Pelegri, 2012). Terrestrial vertebrates, on the other hand, all possess separate Kash5 and Lrmp genes. It is possible that during vertebrate evolution an ancestral fish fue/lrmp gene underwent a duplication followed by elimination of 5′ versus 3′ exons.
Fue/Lrmp is required for pronuclear migration in zebrafish zygotes (Lindeman and Pelegri, 2012). This activity is dependent upon association of Fue/Lrmp with cytoplasmic dynein and suggests that KASH5 might have a similar role in mammals. Indeed, ZYG-12, a functional homologue of KASH5, is essential for pronuclear migration in C. elegans embryos. We can already rule out LRMP as a mediator of pronuclear migration because Lrmp-null mice are fully fertile (unpublished data). In mouse zygotes the female pro-nucleus migrates toward its male counterpart along microtubules that are focused at the single “male” centrosome. This migration is likely dynein mediated. We propose that KASH5 may function as an NE adaptor for dynein in the female pronucleus. Future studies, including the derivation of conditional mouse strains, will be designed to test this hypothesis and to further explore the function of KASH5 in the female germline.
Materials and methods
Plasmids
Mouse KASH5 (CCDC155) cDNA (clone ID: 30008752) was obtained from Thermo Fisher Scientific and amplified by using primers 5′-ACCCTCGAGGACCTGCCGGAGGGCCAGGCTGGT-3′ and 5′-CTACTTAAGTCACACTGGTGGTGGCGGCTGTAGGTAGTA-3′. The PCR product was digested with XhoI and AflII and fused to the 3′ end of an HA tag sequence within pcDNA3.1(−). Alternatively, it was fused to the 3′ end of a GFP cDNA within pcDNA 4T/O. GFP-K5KASH, GFP-KASH5ΔNΔK, GFP-KASH5ΔCCΔK, GFP-KASH5ΔCDΔK, GFP-KASH5ND, GFP-KASH5CC, and GFP-KASH5CD were generated in GFP-pcDNA 4T/O using similar PCR-based cloning techniques. Human LRMP cDNA (clone ID: 8992133) was obtained from Thermo Fisher Scientific. The N-terminally epitope tagged LRMP expression vector was generated by using XhoI and AflII flanked primers 5′-AGCTCGAGAATGATGACCCAAGTATGGAA-3′ and 5′-TGCTTAAGTCACACTGGTGGTGGCCCATTG-3′. Myc-SUN2 and HA-SUN2 were constructed in pcDNA3.1(−). In both cases the tag sequence was placed at the 5′ end of the coding sequence to generate an N-terminally tagged protein. The junction between the tag sequence and cDNA consisted of a single XhoI site exactly spanning the pair of codons for leucine and glutamic acid. GFP-NESP4 was constructed in pEGFP-C1, which yields full-length NESP4 fused to the C terminus of EGFP. The dominant-negative SUN1 mutant ss-HA-SUN1L-KDEL was constructed in pcDNA3.1(−) (Crisp et al., 2006). Each of these plasmids was used as described previously (Crisp et al., 2006; Liu et al., 2007; Roux et al., 2009).
Cell lines and transfection
HeLa and HEK293 cells were maintained in 6.0% CO2 at 37°C in DMEM supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 2 mM l-glutamine. To generate cells stably expressing GFP-KASH5, HEK293 cells were transfected with GFP-KASH5 pcDNA 4T/O plasmids and selected with Zeocin (Life Technologies) as recommended by the manufacturer. To generate cells stably expressing HA-KASH5, HA-KASH5pcDNA3.1(−), HEK293 cells were transfected with the appropriate plasmids and selected with G418 (600 µg/ml) before subcloning. The moderate expression of GFP-KASH5 or HA-KASH5 in individual surviving clones was verified by fluorescence microscopy. Transfections were performed using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions.
Short inhibitory RNA (siRNA) methods
To deplete SUN1 and SUN2, SmartPool ON-TARGETplus human siRNAs targeting to the ORF of human SUN1 and SUN2 were purchased from Thermo Fisher Scientific. Each siRNA was dissolved in RNase/DNase-free water (Promega) to make 20 µM stock and stored in aliquots at −20°C before use. Both siRNAs were transfected into HEK293 cells stably expressing HA-KASH5 at a final concentration of 60 nM using Lipofectamine 2000. After 48–72 h of siRNA exposure, the depletion of SUN1 and SUN2 was confirmed by immunofluorescence (Harborth et al., 2001).
Generation of knockout mouse
Ccdc155 (Kash5) targeted ES cell clones (EPD0094_2_G08) were obtained from the Knockout Mouse project (KOMP) Repository, expanded in our laboratory and injected into blastocysts. Chimeric mice were obtained and mated to wild-type females to obtain founder animals. To remove loxP-flanked sequences, Kash5 mutant mice carrying the conditional allele (Fig. 4 A) were crossed with C57/B6 animals harboring a transgene consisting of a Cre-recombinase cDNA under the control of the Zp3 promoter (Zp3-Cre, a gift from B. Knowles, Institute of Medical Biology, A*STAR, Singapore; de Vries et al., 2000). These Kash5 deleted animals were bred to remove Cre and were maintained as heterozygotes.
RT-PCR
Total RNA was isolated from mouse tissues by using Trizol (Life Technologies) and following chloroform extraction and ethanol precipitation. 1 µg of total RNA was reverse-transcribed with Superscript II First-Strand Synthesis kit (Life Technologies) using oligo-dT primers as recommended by the manufacturer. Subsequent PCR was performed with the cloning primers for full-length mouse KASH5. For the GAPDH control the following primer pair was used: 5′-CTGCACCACCAACTGCTTAG-3′ and 5′-CCTGCTTCACCACCTTCTTG-3′.
QPCR
Testes from wild-type, heterozygous, and Kash5-null littermates were weighed and snap frozen in liquid nitrogen and homogenized using lysing matrix D in an MP FastPrep-24 tissue homogenizer in weight-adjusted volumes of RNABle (Eurobio), followed by chloroform extraction and ethanol precipitation. Samples were further purified by using the RNeasy Mini kit (QIAGEN). cDNA was generated using Thermoscript reverse transcription (Invitrogen), and qRT-PCR was performed on a 7500 Fast Real-Time PCR system using Fast SYBR Green master mix (Applied Biosystems).
Immunoprecipitation
HEK293 cells transiently expressing GFP-KASH5ΔK and/or its deletion mutants were mechanically lysed in 1 ml of hypotonic lysis buffer (50 mM Tris, pH 7.5, 10 mM NaCl, 2.5 mM MgCl2, 1 mM DTT, 0.01% digitonin, and 1× proteinase inhibitor [Thermo Fisher Scientific]). For the self-association study, HA- and GFP-KASH5ΔK or its deletion mutants were coexpressed in HEK293 cells and pulled down with rabbit anti-GFP (or HA) antibodies after lysis in an isotonic buffer containing nonionic detergent (50 mM Tris, pH 7.5, 150 mM NaCl, 2.5 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, and 1× proteinase inhibitor). Lysates were passed through a 21-gauge needle ten times and centrifuged at 16,000 g for 10 min at 4°C. The supernatants were rotated for 4 h at 4°C with protein A–Sepharose beads (Sigma-Aldrich) coupled to rabbit anti-HA or anti-GFP antibodies. After incubation with the beads, samples were thoroughly washed five times with lysis buffers and then twice with a final wash buffer (50 mM Tris, pH 7.5, and 50 mM NaCl). Proteins eluted from the bead by adding SDS-PAGE sample buffer followed by boiling at 98°C for 5 min were analyzed by SDS-PAGE and immunoblotting.
Immunoblot analysis
Protein samples, both cell lysates, and immunoprecipitates were fractionated on polyacrylamide gels and transferred to nitrocellulose membrane (Bio-Rad Laboratories) by using semi-dry transfer unit (Bio-Rad Laboratories). The membrane was then immersed in blocking buffer (10% adult bovine serum and 0.2% Triton X-100) for 30 min and then incubated with appropriate primary antibody overnight at 4°C. After washes with blocking buffer, blots were incubated with HRP-conjugated secondary antibody (Life Technologies) for 1 h, washed again with blocking buffer, and visualized by using ECL. For the immunoprecipitation studies, HRP-conjugated native secondary antibodies (Sigma-Aldrich) were used. Testes from wild-type, heterozygous, and Kash5-null littermates were weighed and placed in PBS to remove the tunica to release the seminiferous tubules. Samples were dissociated using two forceps in a small volume of PBS and placed into 15-ml Falcon tubes on ice, with volume brought to 10 ml. When all samples were dissociated, the suspensions were centrifuged for 5 min at 300 RCF to pellet the cells. Cells were resuspended in SDS-PAGE loading buffer with volumes corresponding to testes weights. Samples were boiled for 5 min and resolved on 10% polyacrylamide gels, transferred using the semi-dry transfer system, blocked with 10% nonfat dry milk in TBS-T, and incubated with primary antibody overnight at 4°C. After washes with TBS-T, blots were incubated with HRP-conjugated secondary antibody (GE Healthcare) for 1 h, washed in TBS-T, and visualized using Luminata Forte Western HRP Substrate (EMD Millipore).
Antibodies
Antibodies used were as follows: mouse monoclonal anti–γ-tubulin (GTU-88,T6557; Sigma-Aldrich), anti-dynein intermediate chain (74–1, MMS400R; Covance), anti-dynein IC1/2, cytosolic antibody (74-1; Santa Cruz Biotechnology, Inc.), p150 (610474; BD), anti-synaptonemal complex protein 3 (SCP3) (D-1; Santa Cruz Biotechnology, Inc.), anti–phospho-Histone H2A.X (Ser139) antibody, clone JBW301 (EMD Millipore), rabbit polyclonal anti-Rad51 antibody (ab63801; Abcam), anti-Rap1 (ab11191; Abcam), anti-SUN1 (HPA008346; Sigma-Aldrich), anti-SUN2 (HPA001209; Sigma-Aldrich), anti-dynein heavy chain (sc9115; Santa Cruz Biotechnology, Inc.), anti-Lis1 (ab2607; Abcam), anti-synaptonemal complex 1 (SCP1) (ab15090; Abcam), and anti-lamin B1 (ab16048; Abcam). The rabbit antibody against the N terminus (aa 9–24) of human KASH5 was raised by Yenzym antibodies, LLC. The mouse monoclonal anti-Nup153 (SA1) raised against the Nup153 FG repeat domain (Bodoor et al., 1999), and a mouse monoclonal anti-nesprin2 antibody (MANNES2A-11A3), a gift from G. Morris (Wolfson Centre for Inherited Neuromuscular Disease, RJAH Hospital, Oswestry, England, UK) were used as described previously (Crisp et al., 2006; Roux et al., 2009). The monoclonal antibodies 9E10 and 12CA5 against the myc, HA, and GFP tags were obtained from the American Type Culture Collection, Covance, and Abcam, respectively. A human CREST autoantibody was a gift from J.B. Rattner (University of Calgary, Calgary, Alberta, Canada). Rabbit anti-SUN1 is a gift from Y.-H. Chi (Institute of Cellular and System Medicine, Taipei, Taiwan). Secondary antibodies were as follows. Alexa Fluor (Invitrogen): donkey/goat anti–mouse 568, donkey/goat anti–rabbit 488, and goat anti–rabbit 350.
3D structured illumination microscopy (3D-SIM)
A microscope (DeltaVision OMX v4; Applied Precision) equipped with 405-, 488-, and 568-nm lasers for excitation and the BGR filter drawer (emission wavelengths 436/31 for DAPI, 528/48 for Alexa Fluor 488, and 609/37 for Alexa Fluor 568) was used for acquisition of 3D-SIM images. An Plan Apochromat 100x/1.4 PSF oil immersion objective lens (Olympus) was used with liquid-cooled EM-CCD cameras (Evolve; Photometrics) for each channel. 15 images per section per channel were acquired (made up of three rotations and five phase movements of the diffraction grating) at a z-spacing of 0.125 µm as described previously (Gustafsson et al., 2008; Schermelleh et al., 2008). Structured illumination reconstruction and alignment was completed using the SoftWorX (Applied Precision) program.
Wide-field fluorescence and deconvolution
A microscope (DeltaVision CORE; Applied Precision) equipped with a xenon light source and bandpass filters was used for acquisition of wide-field fluorescence images. Either a Plan Apochromat 40×/1.35 NA (Olympus) or a Plan Apochromat 60x/1.4 NA oil immersion objective lens (Olympus) was used with a CCD camera (no binning; CoolSNAP HQ, Photometrics). The z-spacing was fixed at 0.2 µm. Deconvolution was then completed using the SoftWorX program (Applied Precision) with figure preparation in Fiji (Schindelin et al., 2012) and Adobe Photoshop and Illustrator.
Confocal microscopy
A confocal microscope (LSM510; Carl Zeiss) equipped with 405-, 488-, and 561-nm lasers for excitation and bandpass emission filters was used for acquisition of confocal images and z-stacks. A Plan Neofluor 40×/1.3 NA oil immersion objective lens (Carl Zeiss) was used. The confocal pinhole was set to 1 Airy unit for the green channel and other channels adjusted to the same optical slice thickness. For z-stacks the z-spacing was fixed at 0.48 µm.
Figure preparation for all of the microscopy techniques was performed with Fiji (Schindelin et al., 2012) and both Adobe Photoshop and Adobe Illustrator.
Fluorescence in situ hybridization (FISH)
Spermatocyte spreads were prepared as above. For telomere FISH and immunofluorescence labeling on the same sample, a modified FISH protocol was used (Bastos et al., 1996). Samples were brought to room temperature and fixed in 4% PFA for 10 min. This was followed by a 10-min incubation with 0.2% Triton X-100 PBS, a 1-h block in 10% normal goat serum (Abcam), and a 1-h incubation with primary antibody at room temperature. Samples were washed in 0.2% Triton X-100 PBS three times. At this point a telomere PNA FISH kit (Dako) was used to label the telomeres. The kit protocol was adhered to with the following changes: The 10-min pretreatment step was reduced to 5 min. After the last wash (5 min at 65°C) samples were put in PBS, fixed again in 4% PFA for 2 min, blocked in 10% normal goat serum for 30 min, and incubated with secondary antibody for 1 h at room temperature. Samples were washed 1 time in Triton X-100 PBS followed by two washes in PBS, mounted using ProLong Gold Antifade reagent (Invitrogen), and visualized using the inverted microscope system (DeltaVision CORE; Applied Precision).
Immunostaining methods
For HeLa and HEK293 cells stably or transiently expressing KASH5 and its deletion mutants, fixation was performed in 3% formaldehyde for 10 min. This was followed by permeabilization using 0.4% Triton X-100 for 15 min. For experiments involving selective permeabilization, HEK293 cells stably expressing GFP-KASH5 were first fixed in 3% PFA and permeabilized in 0.001% digitonin in PBS on ice for 15 min. For the dynein intermediate chain and p150Glued immunostaining, samples were fixed with methanol for 4 min at −20°C. After fixation and permeabilization, samples were labeled with appropriate primary and secondary antibodies for 20 min at room temperature. Hoechst dye or DAPI was used to visualize DNA. Samples were washed two times with 0.2% Triton X-100 after each antibody labeling step. Images were recorded using a fluorescence microscope (model DMRB; Leica) equipped with a CoolSNAP HQ (Photometrics) CCD camera linked to a Macintosh G4 computer running IPLab Spectrum software (Scanalytics). Alternatively, cells were imaged using the DeltaVision CORE system. For the testis sections or spermatocyte spreads, specimens were brought to room temperature and fixed for 10 min in 4% PFA in PBS. Samples were washed in PBS and permeabilized for 10 min in 0.2% Triton X-100 PBS followed by a brief wash in PBS and blocked in blocking buffer (10% NGS in 0.2% Triton X-100 PBS) for 1 h. Samples were rinsed briefly in PBS and incubated with primary antibody diluted in blocking buffer in a moist chamber at 4°C overnight. Samples were washed in PBS in a Coplin jar, which was immersed in a beaker with running water for 10 min. Samples were incubated with secondary antibody diluted in blocking buffer for 1 h at room temperature and then subjected to the same wash regimen. Excess water was removed and samples were mounted using ProLong Gold Antifade reagent (Invitrogen) and visualized on the confocal microscope (LSM510; Carl Zeiss) or the DeltaVision CORE microscope as detailed above. For 3D-SIM, samples were fixed again for 10 min in 4% PFA in PBS after the final wash, washed in PBS, and mounted using VectaShield mounting medium (H-1000; Vector Laboratories).
Spermatocyte spreads
Kash5-null and wild-type littermate male mice were sacrificed by CO2 intoxication. Testes were removed and placed in PBS. The tunica was removed and seminiferous tubules were washed in PBS and then transferred to a small aliquot of fresh PBS. The seminiferous tubules were dissociated by mechanical disruption using a pair of forceps, and the tissue suspension was passed through a 100-µm cell strainer (BD) to remove undissociated tubules and large pieces of tissue. Thoroughly cleaned Polysine slides (Thermo Fisher Scientific) were dipped in 1% PFA and 0.15% Triton X-100/PBS. Excess liquid was allowed to drip off briefly and a few drops of testis suspension were added to each slide. They were incubated in a moist chamber for 2 h, washed for 3 min in 0.1% Triton X-100/PBS, and allowed to air dry. Slides were stored at −80°C. To detect dynein intermediate chain and p150Glued at the NE, free spermatocytes, after seminiferous tubule dissociation, were attached to coverslips, which had been pretreated with poly-l-lysine (Sigma-Aldrich), by centrifugation at 37°C for 10 min at 1,000 g.
Tissue sections
Kash5-null and wild-type littermate mice were sacrificed as described above; testes and other tissues were removed and briefly placed in 2-methy butane. These specimens were transferred in Tissue-Tek OCT (Sakura Finetek, USA) and frozen in liquid nitrogen. 12-µm sections were cut on a cryostat and placed on Polysine slides (Thermo Fisher Scientific) and stored at −80°C.
Online supplemental material
Fig. S1 shows that LRMP behaves as an authentic KASH domain protein and that KASH5 localizes to the outer nuclear membrane. Fig. S2 provides evidence that KASH5 self-associates and that this is mediated by the central coiled-coil domain. Fig. S3 compares conventional wide-field and structured illumination microscopy to follow meiotic progression in wild-type spermatocytes. Fig. S4 presents the histology of wild-type versus Kash5-null mouse ovaries. Fig. S5 shows synaptonemal complexes visualized using structured illumination microscopy. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201304004/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201304044.dv.
Supplementary Material
Acknowledgments
This work was funded by the Singapore Biomedical Research Council and the Singapore Agency for Science Technology and Research, A*STAR (B. Burke and C.L. Stewart), as well as by a research grant from the National Institutes of Health (B. Burke).
Footnotes
Abbreviations used in this paper:
- DSB
- double-strand break
- Fue
- futile cycle
- INM
- inner nuclear membrane
- IP
- immunoprecipitation
- KASH
- klarsicht, ANC-1, syne homology
- LINC
- linker of the nucleoskeleton and cytoskeleton
- LRMP
- lymphocyte-restricted membrane protein
- NE
- nuclear envelope
- ONM
- outer nuclear membrane
- PC
- pairing center
- PNS
- perinuclear space
- SC
- synaptonemal complex
- SIM
- structured illumination microscopy
- SUN
- Sad1, Unc-84
References
- Adam S.A., Marr R.S., Gerace L. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807–816 10.1083/jcb.111.3.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R., Lin A., Enarson M., Burke B. 1996. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 134:1141–1156 10.1083/jcb.134.5.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudrimont A., Penkner A., Woglar A., Machacek T., Wegrostek C., Gloggnitzer J., Fridkin A., Klein F., Gruenbaum Y., Pasierbek P., Jantsch V. 2010. Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet. 6:e1001219 10.1371/journal.pgen.1001219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens T.W., Jagadeesh J., Scherle P., Kearns G., Yewdell J., Staudt L.M. 1994. Jaw1, A lymphoid-restricted membrane protein localized to the endoplasmic reticulum. J. Immunol. 153:682–690 [PubMed] [Google Scholar]
- Bhalla N., Dernburg A.F. 2008. Prelude to a division. Annu. Rev. Cell Dev. Biol. 24:397–424 10.1146/annurev.cellbio.23.090506.123245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boateng K.A., Bellani M.A., Gregoretti I.V., Pratto F., Camerini-Otero R.D. 2013. Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev. Cell. 24:196–205 10.1016/j.devcel.2012.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodoor K., Shaikh S., Salina D., Raharjo W.H., Bastos R., Lohka M., Burke B. 1999. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112:2253–2264 [DOI] [PubMed] [Google Scholar]
- Burke B., Roux K.J. 2009. Nuclei take a position: managing nuclear location. Dev. Cell. 17:587–597 10.1016/j.devcel.2009.10.018 [DOI] [PubMed] [Google Scholar]
- Burke B., Stewart C.L. 2013. The nuclear lamins: flexibility in function. Nat. Rev. Mol. Cell Biol. 14:13–24 10.1038/nrm3488 [DOI] [PubMed] [Google Scholar]
- Chi Y.H., Cheng L.I., Myers T., Ward J.M., Williams E., Su Q., Faucette L., Wang J.Y., Jeang K.T. 2009. Requirement for Sun1 in the expression of meiotic reproductive genes and piRNA. Development. 136:965–973 10.1242/dev.029868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y., Tsutsumi C., Yamane M., Okamasa K., Haraguchi T., Hiraoka Y. 2006. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 125:59–69 10.1016/j.cell.2006.01.048 [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Haraguchi T., Hiraoka Y. 2007. Another way to move chromosomes. Chromosoma. 116:497–505 10.1007/s00412-007-0114-8 [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Yamane M., Okamasa K., Tsutsumi C., Kojidani T., Sato M., Haraguchi T., Hiraoka Y. 2009. Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J. Cell Biol. 187:413–427 10.1083/jcb.200902122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M.N., Lee C.Y., Wilkerson J.L., Dresser M.E. 2007. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 104:8863–8868 10.1073/pnas.0606165104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M.N., Lee C.Y., Chao G., Shinohara M., Kosaka H., Shinohara A., Conchello J.A., Dresser M.E. 2008. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 133:1175–1187 10.1016/j.cell.2008.04.047 [DOI] [PubMed] [Google Scholar]
- Crisp M., Liu Q., Roux K., Rattner J.B., Shanahan C., Burke B., Stahl P.D., Hodzic D. 2006. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172:41–53 10.1083/jcb.200509124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries W.N., Binns L.T., Fancher K.S., Dean J., Moore R., Kemler R., Knowles B.B. 2000. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 26:110–112 [DOI] [PubMed] [Google Scholar]
- Dekens M.P., Pelegri F.J., Maischein H.M., Nüsslein-Volhard C. 2003. The maternal-effect gene futile cycle is essential for pronuclear congression and mitotic spindle assembly in the zebrafish zygote. Development. 130:3907–3916 10.1242/dev.00606 [DOI] [PubMed] [Google Scholar]
- Dickey J.S., Redon C.E., Nakamura A.J., Baird B.J., Sedelnikova O.A., Bonner W.M. 2009. H2AX: functional roles and potential applications. Chromosoma. 118:683–692 10.1007/s00412-009-0234-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Xu R., Yu J., Xu T., Zhuang Y., Han M. 2007. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell. 12:863–872 10.1016/j.devcel.2007.03.018 [DOI] [PubMed] [Google Scholar]
- Fraune J., Schramm S., Alsheimer M., Benavente R. 2012. The mammalian synaptonemal complex: protein components, assembly and role in meiotic recombination. Exp. Cell Res. 318:1340–1346 10.1016/j.yexcr.2012.02.018 [DOI] [PubMed] [Google Scholar]
- Gustafsson M.G., Shao L., Carlton P.M., Wang C.J., Golubovskaya I.N., Cande W.Z., Agard D.A., Sedat J.W. 2008. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94:4957–4970 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F., Lloyd D.J., Smallwood D.T., Dent C.L., Shanahan C.M., Fry A.M., Trembath R.C., Shackleton S. 2006. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell. Biol. 26:3738–3751 10.1128/MCB.26.10.3738-3751.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J., Elbashir S.M., Bechert K., Tuschl T., Weber K. 2001. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114:4557–4565 [DOI] [PubMed] [Google Scholar]
- Harper L., Golubovskaya I., Cande W.Z. 2004. A bouquet of chromosomes. J. Cell Sci. 117:4025–4032 10.1242/jcs.01363 [DOI] [PubMed] [Google Scholar]
- Harper N.C., Rillo R., Jover-Gil S., Assaf Z.J., Bhalla N., Dernburg A.F. 2011. Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev. Cell. 21:934–947 10.1016/j.devcel.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S., Güttinger S., Mühlhäusser P., Anderegg F., Bürgler S., Kutay U. 2006. Nuclear envelope localization of human UNC84A does not require nuclear lamins. FEBS Lett. 580:1263–1268 10.1016/j.febslet.2006.01.039 [DOI] [PubMed] [Google Scholar]
- Hiraoka Y., Dernburg A.F. 2009. The SUN rises on meiotic chromosome dynamics. Dev. Cell. 17:598–605 10.1016/j.devcel.2009.10.014 [DOI] [PubMed] [Google Scholar]
- Hodzic D.M., Yeater D.B., Bengtsson L., Otto H., Stahl P.D. 2004. Sun2 is a novel mammalian inner nuclear membrane protein. J. Biol. Chem. 279:25805–25812 10.1074/jbc.M313157200 [DOI] [PubMed] [Google Scholar]
- Horn H.F., Brownstein Z., Lenz D.R., Shivatzki S., Dror A.A., Dagan-Rosenfeld O., Friedman L.M., Roux K.J., Kozlov S., Jeang K.T., et al. 2013. The LINC complex is essential for hearing. J. Clin. Invest. 123:740–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Giddings T.H., Jr, Winey M. 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 159:945–956 10.1083/jcb.200208169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketema M., Kreft M., Secades P., Janssen H., Sonnenberg A. 2013. Nesprin-3 connects plectin and vimentin to the nuclear envelope of Sertoli cells but is not required for Sertoli cell function in spermatogenesis. Mol. Biol. Cell. 24:2454–2466 10.1091/mbc.E13-02-0100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Kim K.P., Prentiss M., Kleckner N., Kameoka S. 2008. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 133:1188–1201 10.1016/j.cell.2008.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsinger R.H., Barry C.D. 1975. The predicted structure of the calcium-binding component of troponin. Biochim. Biophys. Acta. 405:40–52 10.1016/0005-2795(75)90312-8 [DOI] [PubMed] [Google Scholar]
- Labella S., Woglar A., Jantsch V., Zetka M. 2011. Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev. Cell. 21:948–958 10.1016/j.devcel.2011.07.011 [DOI] [PubMed] [Google Scholar]
- Lee C.Y., Conrad M.N., Dresser M.E. 2012. Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet. 8:e1002730 10.1371/journal.pgen.1002730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman R.E., Pelegri F. 2012. Localized products of futile cycle/lrmp promote centrosome-nucleus attachment in the zebrafish zygote. Curr. Biol. 22:843–851 10.1016/j.cub.2012.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Pante N., Misteli T., Elsagga M., Crisp M., Hodzic D., Burke B., Roux K.J. 2007. Functional association of Sun1 with nuclear pore complexes. J. Cell Biol. 178:785–798 10.1083/jcb.200704108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G.W., Gomes E.R., Folker E.S., Vintinner E., Gundersen G.G. 2010. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 329:956–959 10.1126/science.1189072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.J., Misner L., Le Bot N., Tsai M.C., Campbell J.M., Ahringer J., White J.G. 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 115:825–836 10.1016/S0092-8674(03)00985-1 [DOI] [PubMed] [Google Scholar]
- McGee M.D., Rillo R., Anderson A.S., Starr D.A. 2006. UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol. Biol. Cell. 17:1790–1801 10.1091/mbc.E05-09-0894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki F., Okazaki K., Shimanuki M., Yamamoto A., Hiraoka Y., Niwa O. 2002. The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol. Biol. Cell. 13:930–946 10.1091/mbc.01-11-0543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki F., Kurabayashi A., Tange Y., Okazaki K., Shimanuki M., Niwa O. 2004. Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol. Genet. Genomics. 270:449–461 10.1007/s00438-003-0938-8 [DOI] [PubMed] [Google Scholar]
- Minn I.L., Rolls M.M., Hanna-Rose W., Malone C.J. 2009. SUN-1 and ZYG-12, mediators of centrosome-nucleus attachment, are a functional SUN/KASH pair in Caenorhabditis elegans. Mol. Biol. Cell. 20:4586–4595 10.1091/mbc.E08-10-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto A., Shibuya H., Zhu X., Kim J., Ishiguro K., Han M., Watanabe Y. 2012. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J. Cell Biol. 198:165–172 10.1083/jcb.201204085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley-Bishop K.L., Li Q., Patterson L., Fischer J.A. 1999. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr. Biol. 9:1211–1220 10.1016/S0960-9822(99)80501-6 [DOI] [PubMed] [Google Scholar]
- Padmakumar V.C., Abraham S., Braune S., Noegel A.A., Tunggal B., Karakesisoglou I., Korenbaum E. 2004. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp. Cell Res. 295:330–339 10.1016/j.yexcr.2004.01.014 [DOI] [PubMed] [Google Scholar]
- Padmakumar V.C., Libotte T., Lu W., Zaim H., Abraham S., Noegel A.A., Gotzmann J., Foisner R., Karakesisoglou I. 2005. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 118:3419–3430 10.1242/jcs.02471 [DOI] [PubMed] [Google Scholar]
- Penkner A., Tang L., Novatchkova M., Ladurner M., Fridkin A., Gruenbaum Y., Schweizer D., Loidl J., Jantsch V. 2007. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell. 12:873–885 10.1016/j.devcel.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Penkner A.M., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T., Woglar A., Csaszar E., Pasierbek P., Ammerer G., Gruenbaum Y., Jantsch V. 2009. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 139:920–933 10.1016/j.cell.2009.10.045 [DOI] [PubMed] [Google Scholar]
- Phillips C.M., Dernburg A.F. 2006. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell. 11:817–829 10.1016/j.devcel.2006.09.020 [DOI] [PubMed] [Google Scholar]
- Phillips C.M., Wong C., Bhalla N., Carlton P.M., Weiser P., Meneely P.M., Dernburg A.F. 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 123:1051–1063 10.1016/j.cell.2005.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Crisp M.L., Liu Q., Kim D., Kozlov S., Stewart C.L., Burke B. 2009. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. USA. 106:2194–2199 10.1073/pnas.0808602106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J., Sung P., Klein H. 2008. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77:229–257 10.1146/annurev.biochem.77.061306.125255 [DOI] [PubMed] [Google Scholar]
- Sato A., Isaac B., Phillips C.M., Rillo R., Carlton P.M., Wynne D.J., Kasad R.A., Dernburg A.F. 2009. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 139:907–919 10.1016/j.cell.2009.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L., Carlton P.M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M.C., Agard D.A., Gustafsson M.G., et al. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 320:1332–1336 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H. 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2:621–627 10.1038/35085086 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer T.A. 2004. Dynactin. Annu. Rev. Cell Dev. Biol. 20:759–779 10.1146/annurev.cellbio.20.012103.094623 [DOI] [PubMed] [Google Scholar]
- Shindo Y., Kim M.R., Miura H., Yuuki T., Kanda T., Hino A., Kusakabe Y. 2010. Lrmp/Jaw1 is expressed in sweet, bitter, and umami receptor-expressing cells. Chem. Senses. 35:171–177 10.1093/chemse/bjp097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohaskey M.L., Jiang Y., Zhao J.J., Mohr A., Roemer F., Harland R.M. 2010. Osteopotentia regulates osteoblast maturation, bone formation, and skeletal integrity in mice. J. Cell Biol. 189:511–525 10.1083/jcb.201003006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa B.A., Rothballer A., Kutay U., Schwartz T.U. 2012. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 149:1035–1047 10.1016/j.cell.2012.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A. 2009. A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 122:577–586 10.1242/jcs.037622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Han M. 2002. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 298:406–409 10.1126/science.1075119 [DOI] [PubMed] [Google Scholar]
- Starr D.A., Han M. 2003. ANChors away: an actin based mechanism of nuclear positioning. J. Cell Sci. 116:211–216 10.1242/jcs.00248 [DOI] [PubMed] [Google Scholar]
- Stewart C.L., Roux K.J., Burke B. 2007. Blurring the boundary: the nuclear envelope extends its reach. Science. 318:1408–1412 10.1126/science.1142034 [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken E., Adelfalk C., Loidl J., Scherthan H. 2005. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J. Cell Biol. 170:213–223 10.1083/jcb.200501042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee R.B., Tsai J.W. 2006. The cellular roles of the lissencephaly gene LIS1, and what they tell us about brain development. Genes Dev. 20:1384–1393 10.1101/gad.1417206 [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K., Litjens S.H., Kuikman I., Tshimbalanga N., Janssen H., van den Bout I., Raymond K., Sonnenberg A. 2005. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 171:799–810 10.1083/jcb.200506083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K.L., Dawson S.C. 2011. Evolution: functional evolution of nuclear structure. J. Cell Biol. 195:171–181 10.1083/jcb.201103171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ragnauth C., Greener M.J., Shanahan C.M., Roberts R.G. 2002. The nesprins are giant actin-binding proteins, orthologous to Drosophila melanogaster muscle protein MSP-300. Genomics. 80:473–481 10.1006/geno.2002.6859 [DOI] [PubMed] [Google Scholar]
- Zhang X., Xu R., Zhu B., Yang X., Ding X., Duan S., Xu T., Zhuang Y., Han M. 2007. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 134:901–908 10.1242/dev.02783 [DOI] [PubMed] [Google Scholar]
- Zhang X., Lei K., Yuan X., Wu X., Zhuang Y., Xu T., Xu R., Han M. 2009. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 64:173–187 10.1016/j.neuron.2009.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y.Y., Libotte T., Munck M., Noegel A.A., Korenbaum E. 2002. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 115:3207–3222 [DOI] [PubMed] [Google Scholar]
- Zhou Z., Du X., Cai Z., Song X., Zhang H., Mizuno T., Suzuki E., Yee M.R., Berezov A., Murali R., et al. 2012. Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J. Biol. Chem. 287:5317–5326 10.1074/jbc.M111.304543 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.