Folliculin localizes to lysosomes and directly interacts with Rag GTPases, promoting mTORC1 activation, when amino acids are abundant.
Abstract
Birt-Hogg-Dubé syndrome, a human disease characterized by fibrofolliculomas (hair follicle tumors) as well as a strong predisposition toward the development of pneumothorax, pulmonary cysts, and renal carcinoma, arises from loss-of-function mutations in the folliculin (FLCN) gene. In this study, we show that FLCN regulates lysosome function by promoting the mTORC1-dependent phosphorylation and cytoplasmic sequestration of transcription factor EB (TFEB). Our results indicate that FLCN is specifically required for the amino acid–stimulated recruitment of mTORC1 to lysosomes by Rag GTPases. We further demonstrated that FLCN itself was selectively recruited to the surface of lysosomes after amino acid depletion and directly bound to RagA via its GTPase domain. FLCN-interacting protein 1 (FNIP1) promotes both the lysosome recruitment and Rag interactions of FLCN. These new findings define the lysosome as a site of action for FLCN and indicate a critical role for FLCN in the amino acid–dependent activation of mTOR via its direct interaction with the RagA/B GTPases.
Introduction
Birt-Hogg-Dubé syndrome arises from loss-of-function mutations in the folliculin (FLCN) gene and is characterized by a constellation of symptoms that includes the growth of benign hair follicle tumors (folliculomas), increased risk for the growth of pulmonary cysts, and pneumothorax, as well as an elevated incidence of renal carcinoma (Nickerson et al., 2002). The abnormal proliferation of select cell types in Birt-Hogg-Dubé syndrome raises questions about the molecular mechanisms whereby FLCN normally suppresses such pathology. Studies in human Birt-Hogg-Dubé syndrome patient cells and model organisms lacking FLCN have suggested roles for FLCN in multiple processes including mTOR signaling, transforming growth factor β (TGF-β) signaling, AMP-activated protein kinase (AMPK) signaling, JAK-STAT signaling, cell adhesion, membrane traffic, and cilia function (Roberg et al., 1997; Chan et al., 2000; Baba et al., 2006; Singh et al., 2006; van Slegtenhorst et al., 2007; Hartman et al., 2009; Hong et al., 2010b; Hudon et al., 2010; Cash et al., 2011; Nookala et al., 2012; Tee and Pause, 2012; Luijten et al., 2013). Although it is possible that the numerous changes arising in cells that lack FLCN reflect multiple sites of action for this protein, it is also possible that at least some of these phenotypes arise through indirect mechanisms. Addressing this issue requires a more mechanistic understanding of FLCN function, including the identification of its direct targets.
Of the large number of pathways that have been linked to FLCN, the FLCN–mTOR relationship stands out for having been observed across a very wide range of organisms, including fission yeast (van Slegtenhorst et al., 2007), budding yeast (Chan et al., 2000), fruit flies (Liu et al., 2013), mice (Hartman et al., 2009; Hudon et al., 2010), and humans (Baba et al., 2006; Takagi et al., 2008; Hartman et al., 2009). mTOR exerts its function as part of two distinct protein complexes known as mTORC1 and mTORC2 (Ma and Blenis, 2009; Dazert and Hall, 2011; Laplante and Sabatini, 2012). Based on pharmacological sensitivity, genetic interactions, and the specific substrates that show altered phosphorylation in cells lacking FLCN, it is specifically the mTORC1 complex that is linked to FLCN (Baba et al., 2006; van Slegtenhorst et al., 2007; Hartman et al., 2009).
mTORC1 integrates multiple inputs to regulate the balance between major anabolic and catabolic cellular processes (Ma and Blenis, 2009; Dazert and Hall, 2011; Laplante and Sabatini, 2012). mTORC1 is recruited to the cytoplasmic surface of lysosomes as an effector of Rag GTPase heterodimers (Sancak et al., 2008, 2010). This recruitment is sensitive to the guanine nucleotide bound state of the Rag GTPases, which is in turn controlled by amino acid availability (Kim et al., 2008; Sancak et al., 2008). Bringing mTOR to lysosomes is critical for the activation of its kinase activity by Rheb, a lysosome-enriched GTPase that is regulated by growth factor signaling and energy abundance (Inoki et al., 2003; Mihaylova and Shaw, 2011). Although previous studies of the relationship between FLCN and mTORC1 have detected changes in levels of mTORC1 substrate phosphorylation in cells lacking FLCN, no direct mechanistic connection between FLCN and the lysosome-localized mTORC1 or its regulators has been established to explain these changes.
It has also been shown that FLCN inhibits the nuclear accumulation of transcription factor E3 (TFE3) by promoting its phosphorylation (Hong et al., 2010a). This finding raises questions about the specific mechanism through which FLCN influences TFE3 phosphorylation and the identity of the specific kinase involved. TFE3 is a basic-helix-loop-helix transcription factor that is closely related to transcription factor EB (TFEB) and the microphthalmia transcription factor (MITF; Hemesath et al., 1994). Building on our previous observations that lysosome status plays a major role in regulating the subcellular localization of MITF, TFE3, and TFEB (MiT-TFE) through an mTORC1-dependent mechanism (Roczniak-Ferguson et al., 2012) as well as the established link between FLCN and mTOR signaling, we hypothesized that FLCN regulates MiT-TFE transcription factor nuclear localization through control of mTORC1 activity.
Following up on this hypothesis, we now show that FLCN regulates TFEB by promoting its phosphorylation on serine 211 (S211), a phosphorylation site we previously identified as a downstream target of mTORC1 that controls nuclear import of TFEB through promoting the interaction of TFEB with 14-3-3 proteins (Roczniak-Ferguson et al., 2012). TFEB has been described as a master regulator of lysosome biogenesis and function (Sardiello et al., 2009), and indeed the accumulation of nuclear TFEB in cells lacking FLCN results in TFEB-dependent changes in lysosome function. Following up on the previously established role for mTORC1 in the regulation of TFEB, we found that FLCN is required for efficient stimulation of mTORC1 activity by amino acids. Interestingly, FLCN itself is recruited to the cytoplasmic surface of lysosomes when cells are depleted of amino acids, and FLCN-interacting protein 1 (FNIP1) is required for this regulated localization. We directly connect FLCN to mTORC1 regulation at the lysosome with our observation that it selectively binds to the inactive forms of RagA and RagB (amino acid–regulated GTPases that recruit mTORC1 to lysosomes) and is critical for their activation. In summary, this study provides molecular insight into the actions of FLCN, the Birt-Hogg-Dubé syndrome tumor suppressor, and identifies FLCN as a critical component of the lysosome-localized machinery that transduces amino acid availability into the changes in Rag GTPase status that promote activation of mTORC1.
Results
FLCN regulates lysosome activity through control of TFEB localization
Although it was previously shown that the TFE3 transcription factor exhibited more nuclear localization and altered phosphorylation in Birt-Hogg Dubé syndrome renal carcinoma cells (Hong et al., 2010a), it was not known whether such observations reflected a general relationship between FLCN and this family of transcription factors, nor were the underlying mechanisms known. As a first step toward solving this problem, we examined the consequences of FLCN knockdown (KD) in HeLa cells stably expressing TFEB-GFP. The knockdown of FLCN elicited both an increase in the nuclear abundance of TFEB and a loss of the TFEB-positive cytoplasmic puncta that have previously been shown to reflect its localization to lysosomes (Fig. 1, A and B; Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2012). As the nuclear localization of TFEB is tightly controlled by mTORC1-dependent phosphorylation of TFEB on serine 211, which results in its interaction with 14-3-3 proteins (Martina et al., 2012; Roczniak-Ferguson et al., 2012), we next investigated the contribution of FLCN to regulation of TFEB-S211 phosphorylation and 14-3-3 interactions, and found that both were diminished after FLCN knockdown (Fig. 1 C). These observations suggested a role for FLCN in the regulation of mTORC1 and the possibility that enhanced nuclear localization of TFEB and/or related transcription factors contributes to the phenotype of cells lacking FLCN.
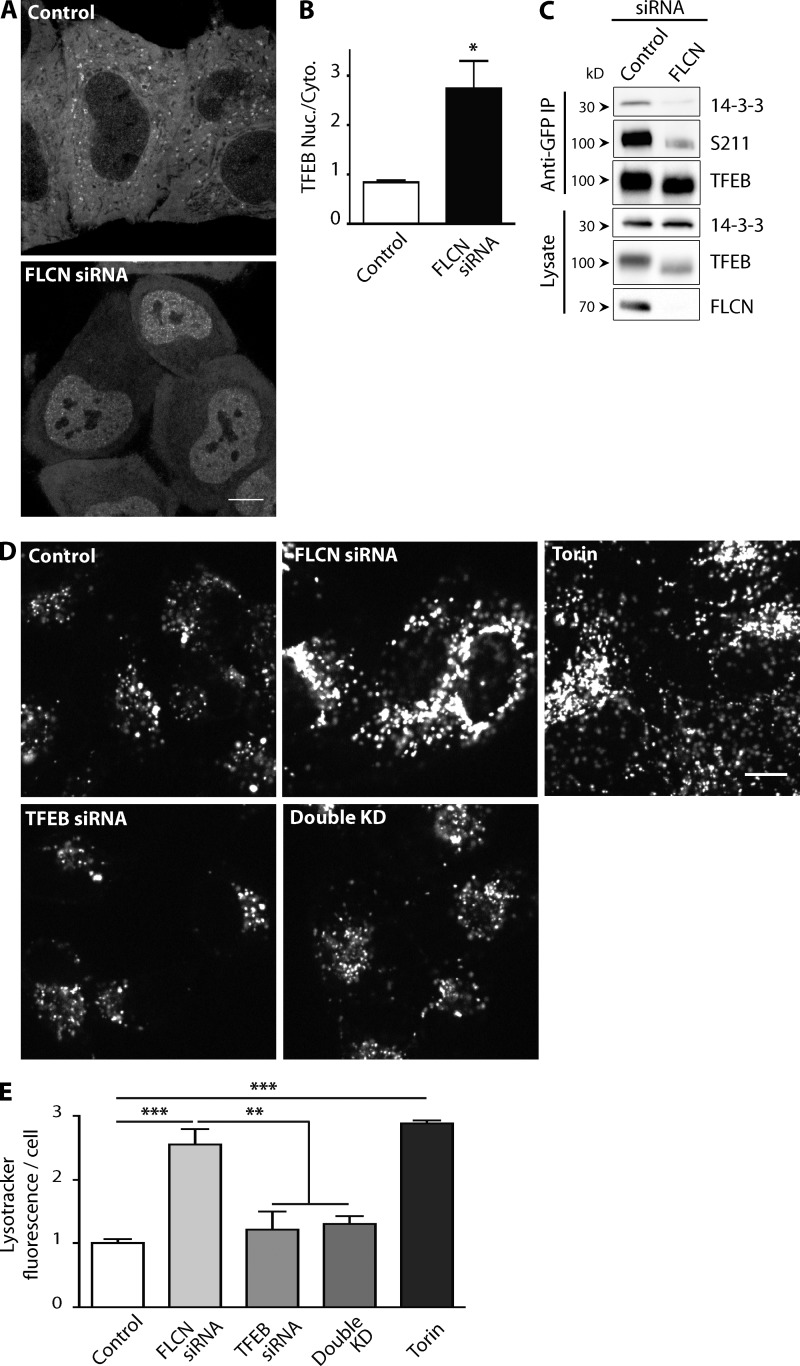
Figure 1.
FLCN controls TFEB-dependent regulation of lysosomes. (A) Spinning disk confocal imaging of TFEB-GFP localization ± FLCN knockdown. (B) Quantification of the effect of FLCN knockdown on the nuclear/cytoplasmic ratio of TFEB-GFP (n = 3 experiments, mean ± SEM; *, P < 0.05 by ANOVA with Bonferroni post-test). (C) Representative Western blots of anti-GFP immunoprecipitations from a stable TFEB-GFP HeLa cell line ± FLCN knockdown (n = 3). (D) Lysotracker labeling of HeLa cells transfected with the indicated siRNAs or after treatment with torin 1 (250 nM, 24 h). (E) Quantification of Lysotracker staining (n = 3 experiments, ANOVA with Bonferroni post-test, mean ± SEM; **, P < 0.01; ***, P < 0.001). Bars: (A and D) 10 µm. All images acquired by spinning disk confocal microscopy.
TFEB regulates the expression of multiple genes encoding lysosomal proteins and its robust overexpression was previously shown to increase the labeling of cells with Lysotracker, a fluorescent dye that selectively accumulates within the acidic lumen of lysosomes (Sardiello et al., 2009; Peña-Llopis et al., 2011). We thus investigated whether FLCN-dependent regulation of TFEB has lysosomal consequences and found that FLCN KD cells exhibit increased Lysotracker staining (Fig. 1, D and E). This effect requires TFEB, as it was abolished in cells where the expression of both FLCN and TFEB was suppressed (Fig. 1, D and E). Direct pharmacological inhibition of mTOR also elicited an increase in Lysotracker labeling of similar magnitude to that seen after FLCN KD (Fig. 1, D and E). These results showed that loss of FLCN affects lysosome function and support a model wherein such effects arise through mTORC1-dependent regulation of TFEB.
Loss of FLCN results in reduced mTORC1 activity
Building on our observation of diminished TFEB-S211 phosphorylation in FLCN knockdown cells (Fig. 1) and to more directly determine the contribution of FLCN to mTORC1 activity, we measured the phosphorylation of ribosomal protein S6 kinase (S6K1) on threonine 389 (T389), a well-characterized mTORC1 phosphorylation site (Ma and Blenis, 2009), and found that FLCN KD resulted in a significant reduction in its phosphorylation (Fig. 2, A and B). Additional analysis of ribosomal protein S6 phosphorylation (serine 235/236), a highly abundant target of S6K1, revealed a similar decrease (Fig. 2, A and C). As treatment with torin 1 (an ATP-competitive inhibitor of mTOR) completely abolished the phosphorylation of both S6K1 and S6 on these sites (Fig. 2, A and C; Fig. S1 B), the effects of FLCN knockdown on S6K1 and S6 phosphorylation must reflect a partial loss of mTOR activity. Surprisingly, phosphorylation of 4E-BP1 on mTORC1 target sites (threonines 37/46) was not affected by FLCN knockdown (Fig. S1 A). However, in accordance with a previous study (Thoreen et al., 2009) we also observed that inhibition of mTORC1-dependent phosphorylation of these sites was also less sensitive to direct pharmacological inhibition of mTOR (Fig. S1, B and C). Given our specific focus on the role of FLCN, we did not further seek to elucidate the mechanisms whereby TFEB and S6K1 are more sensitive than 4E-BP1 to inhibition of mTORC1 activity. The effects of FLCN knockdown on S6K, S6, and TFEB phosphorylation supported a role for FLCN in regulating steady-state levels of mTORC1 signaling, but raised further questions about the specific aspect of mTORC1 regulation that is influenced by FLCN.
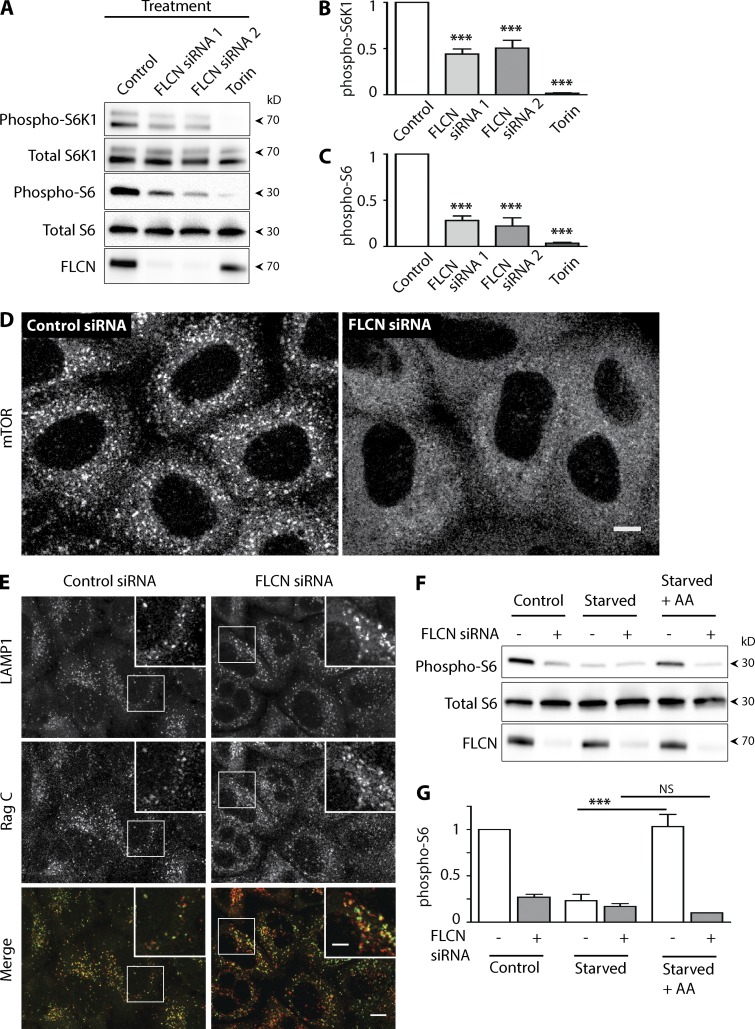
Figure 2.
FLCN regulates mTORC1 activity and localization. (A) Western blot analysis of the phosphorylation of reporters of mTORC1 activity ± FLCN siRNA. (B and C) Quantification of the effects of FLCN siRNA on S6K1 (T389) and S6 (S235/236) phosphorylation (mean ± SEM; ***, P < 0.001, ANOVA with Bonferroni post-test, n = 3–4 experiments). (D) Representative immunofluorescence images showing the localization of mTOR ± FLCN siRNA (line-scanning confocal imaging, n = 3; bar, 10 µm). (E) Representative immunofluorescence images showing the localization of LAMP1 and RagC in cells transfected with control vs. FLCN siRNA. Bars: (D and E) 10 µm; (inset) 5 µm. (F) Representative Western blots and quantification (G) showing the effects of FLCN knockdown on starvation (2 h in serum and amino acid–free RPMI) and amino acid–dependent stimulation (20 min after addition of 1× MEM amino acid supplement) of S6 phosphorylation (S235/236, mean ± SEM; ***, P < 0.001, ANOVA with Bonferroni post-test, n = 3 experiments). All experiments were performed in HeLa cells.
FLCN is required for the enrichment of mTOR on lysosomes
To begin to dissect the defect that underlies the reduced mTORC1 activity in FLCN-deficient cells, we investigated the localization of mTOR under such conditions. In support of a role for FLCN in promoting the lysosomal recruitment of mTOR, immunofluorescence staining revealed a dramatic reduction in the punctate, lysosomal (LAMP1-colocalized) staining pattern for mTOR in FLCN KD cells under basal growth conditions (Fig. 2 D and Fig. S2, A and B). As amino acid abundance plays a well-established role in regulating the Rag GTPase-mediated recruitment of mTORC1 to lysosomes (Inoki et al., 2012; Laplante and Sabatini, 2012), mTOR localization was further assessed under starved (2 h without serum or amino acids) and amino acid re-fed conditions. In cells transfected with a negative control siRNA, starvation resulted in diminished lysosome-localized mTOR staining and mTOR robustly returned to lysosomes after re-addition of amino acids to the culture media for 20 min (Fig. S2 A). Meanwhile, in FLCN-depleted cells, mTOR failed to be enriched on lysosomes regardless of amino acid availability (Fig. S2 B). Given the critical role for Rag GTPases in the recruitment of mTOR to lysosomes (Sancak et al., 2010), we investigated their localization in FLCN knockdown cells but did not detect any decrease in their lysosome enrichment (Fig. 2 E). The failure to recruit mTOR combined with the maintained presence of Rags on lysosomes in FLCN knockdown cells suggests that loss of FLCN results in a defect in the regulation of Rag activity rather than a problem with Rag abundance or localization.
FLCN is required for responsiveness of mTORC1 activity to amino acids
We next investigated the contribution of FLCN to the acute regulation of mTORC1 signaling by amino acids using phospho-S6 as our readout, due to its robust signal and established mTOR dependence in our cell culture model (Fig. 2, A and C; and Fig. S1 B). In these experiments, we observed that when cells were starved of serum and amino acids for 2 h, levels of S6 phosphorylation fell to a similar basal level in both control and FLCN siRNA-transfected cells (Fig. 2, F and G). Starting from this near equivalent minimal level of S6 phosphorylation, we next stimulated cells with amino acids for 20 min and found that although this completely restored S6 phosphorylation in the control siRNA-transfected cells, the FLCN KD cells failed to respond to this stimulus (Fig. 2, F and G). A similar insensitivity to amino acid stimulation was observed for mTORC1-dependent phosphorylation of TFEB-S211 (Fig. S2, C and D). These results demonstrate a critical role for FLCN in promoting the amino acid responsiveness of mTORC1 signaling.
FNIP1 promotes the recruitment of FLCN to lysosomes
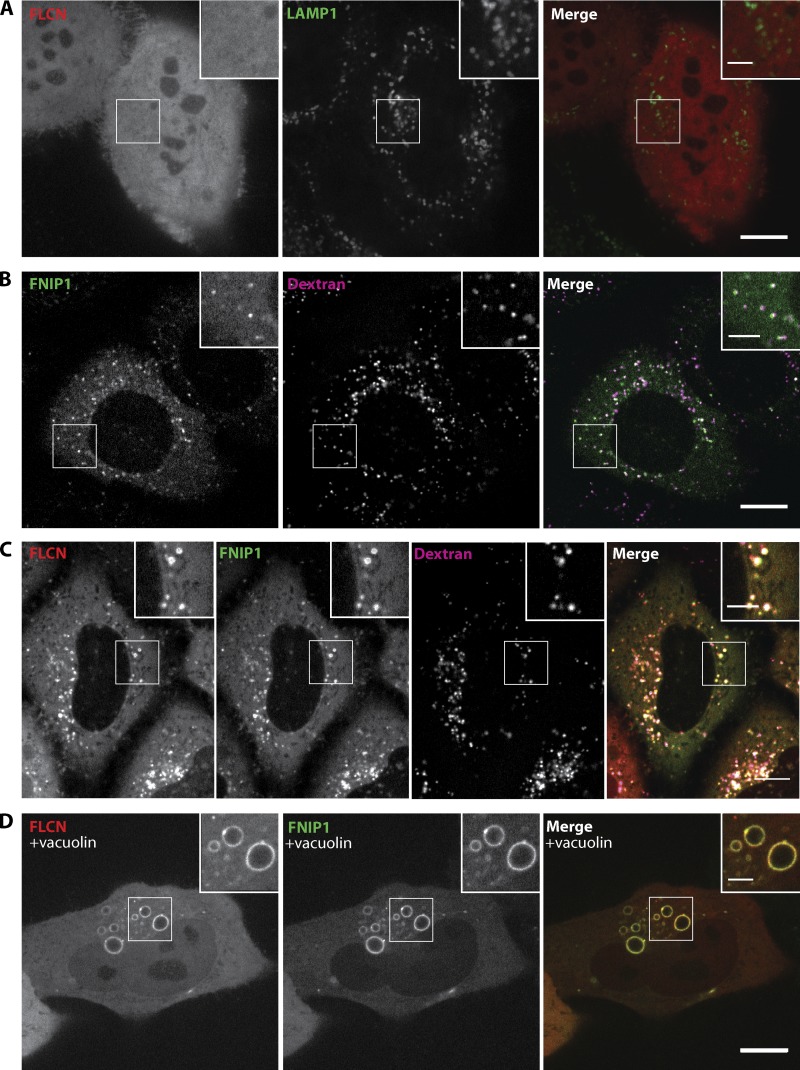
The contribution of FLCN to the recruitment of both TFEB and mTOR to lysosomes raised questions about the subcellular localization of FLCN itself. We observed a largely diffuse localization pattern in cells transfected with FLCN fused to either red or green fluorescent proteins (Fig. 3 A and Fig. S3 A, respectively). However, we wondered whether FLCN might function as part of a protein complex and that a critical component of this complex might be limiting for the proper subcellular targeting of FLCN in transfected cells. We predicted that FNIP1, the major binding partner of FLCN (Baba et al., 2006), might fulfill such a role. Indeed, consistent with a role for FNIP1 in the same pathway as FLCN, FNIP1 knockdown also evoked an increase in the nuclear localization of TFEB (Fig. S3, B–D) that paralleled our observations for FLCN knockdown (Fig. 1 A). Furthermore, an FNIP1-GFP fusion protein was enriched on dextran-labeled lysosomes (Fig. 3 B), and when cells were cotransfected with both FLCN-tdTomato and FNIP1-GFP, FLCN exhibited a punctate pattern that colocalized with both FNIP1 and lysosomes (Fig. 3 C). In cells where the size of endo-lysosomes was increased by treatment with vacuolin (Cerny et al., 2004), it was clear that FLCN and FNIP1 were selectively associated with the limiting membrane of lysosomes and did not fill their lumen (Fig. 3 D).
Figure 3.
FLCN and FNIP1 colocalize on lysosomes. (A) Localization of FLCN-tdTomato (red, diffuse) and LAMP1-mGFP (green, lysosomes). (B) FLCN-interacting protein 1 (FNIP1)-GFP (green) localizes to Alexa 647 dextran-positive (magenta) lysosomes. (C) Cells cotransfected with FLCN-tdTomato (red) and FNIP1-GFP (green) whose lysosomes were loaded with Alexa 647 dextran (magenta). (D) FLCN-tdTomato and FNIP1-GFP colocalization clearly occurs on the surface of the enlarged endosomes/lysosomes that form in vacuolin-treated (5 µM, 1 h) cells. All images were obtained by spinning disk confocal microscopy of live HeLa cells. Bars: (A–D) 10 µm; (insets) 5 µm.
Epitope tagging the endogenous FLCN protein via a CRISPR/Cas9 RNA guided genome editing strategy
The FNIP1-dependent recruitment of FLCN to lysosomes after their combined overexpression placed FLCN on the same organelle where mTORC1 activation takes place. To further investigate the specificity of this lysosomal enrichment, we next sought to assess the subcellular localization of the endogenous FLCN protein. Initial efforts with multiple anti-FLCN antibodies failed to yield a specific immunofluorescence signal, as assessed by siRNA-mediated knockdown experiments (unpublished data). We reasoned that the FLCN antibodies could have failed in these immunofluorescence experiments due to issues of epitope accessibility and/or antibody affinity. To circumvent these possible limitations, we used RNA-guided (CRISPR-Cas9) genome engineering (Chen et al., 2011; Mali et al., 2013) to insert a 3xHA epitope tag encoding sequence into the human FLCN gene (Fig. S4, A and B). Through this approach we generated a clonal HeLa cell line that expresses FLCN with a 3xHA tag at its N terminus (3xHA-FLCN) from the endogenous locus (Fig. 4 A and Fig. S4 C). In addition to insertion of the 3xHA tag at the site of Cas9-mediated DNA cleavage of the FLCN gene by homology-directed repair, cells can also repair such lesions by nonhomologous end joining, a lower fidelity process that can result in small insertions/deletions (“indels”). Analysis of the genomic PCR amplicons from the edited region of the FLCN gene revealed that the 3xHA tag had only been inserted into one FLCN allele by homologous recombination while errors in nonhomologous end joining introduced frameshift causing deletions of 1 bp and 16 bp respectively into the other two FLCN alleles of this near triploid cell line (Fig. S4 D). Thus, the 3xHA-FLCN completely replaces the untagged form of FLCN in this cell line.
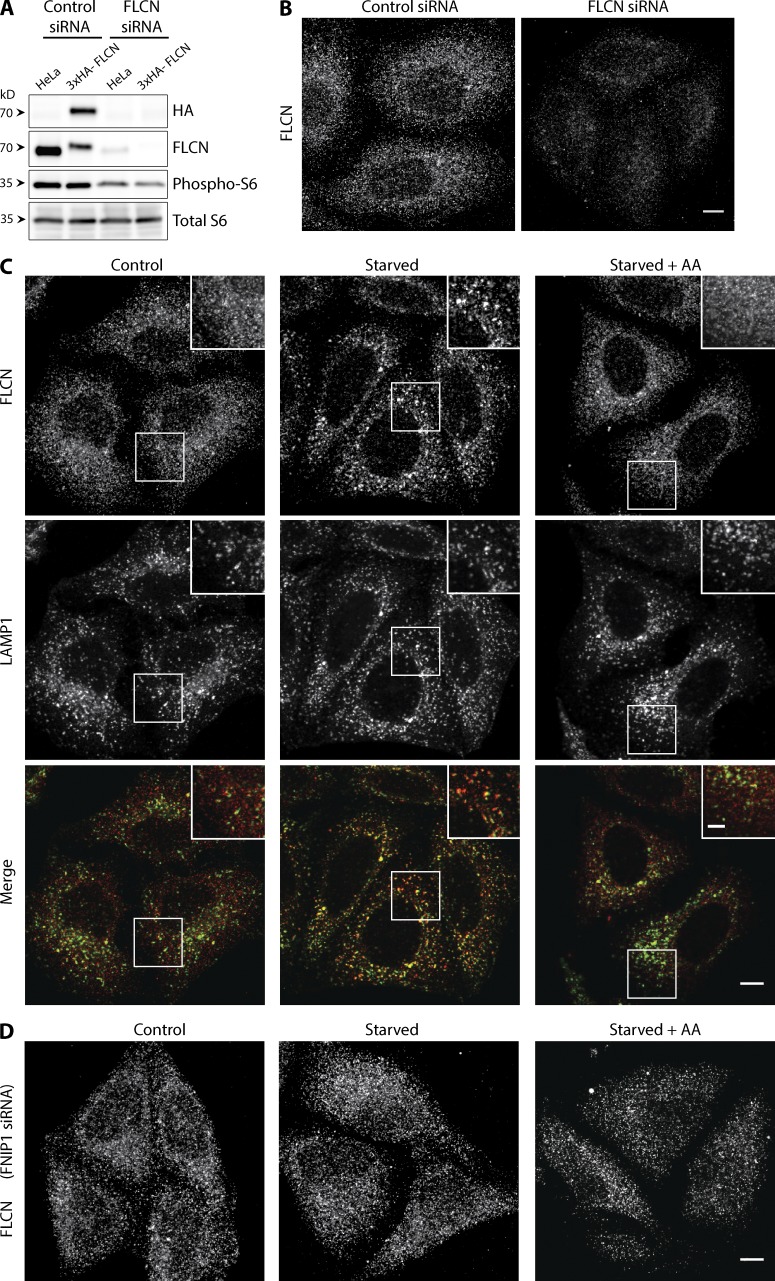
Figure 4.
Regulation of the lysosomal recruitment of FLCN by amino acid availability and FNIP1. (A) Western blot analysis of the 3xHA-tagged endogenous FLCN and phosphorylated S6 protein ± FLCN knockdown. (B) Representative immunofluorescence experiment showing the effect of FLCN knockdown on the levels of endogenous 3xHA-FLCN. (C) Representative images showing the effect of starvation (2 h serum and amino acid–free RPMI) and amino acid re-feeding (1× MEM amino acid solution, 15 min) on 3xHA-tagged endogenous FLCN and LAMP1 in control conditions or (D) after FNIP1 knockdown. All images obtained by line scanning confocal microscopy. Bars: (B–D) 10 µm; (inset) 5 µm. All experiments were performed in HeLa cells.
Exclusive expression of the 3xHA-FLCN protein was verified by immunoblots with anti-HA and FLCN antibodies that selectively detected a band in lysates from the 3xHA-FLCN cell line that migrates slightly higher than the untagged FLCN, and which is less abundant as expected based on its expression from just a single locus (Fig. 4 A). The identity of this band was further confirmed by the fact that it is effectively depleted after transfection of FLCN siRNA (Fig. 4 A). The functionality of the 3xHA-FLCN protein is supported by the normal levels of mTORC1 activity, as assessed by S6 phosphorylation in this cell line compared with the parental HeLa M cell line (Fig. 4 A), and the similar decrease in phospho-S6 after FLCN knockdown in both the parental and 3xHA-FLCN cell lines (Fig. 4 A). Having generated a cell line that exclusively expresses a functional 3xHA-tagged form of FLCN, we proceeded to take advantage of anti-HA antibodies to investigate the localization of FLCN.
Amino acid–dependent recruitment of FLCN to lysosomes
Surprisingly, given our observations of strong lysosome localization in cells overexpressing fluorescent protein–tagged versions of FLCN and FNIP1, the 3xHA-FLCN was not enriched on lysosomes under basal cell culture conditions; yet this signal was specific as it was depleted in cells transfected with FLCN siRNA (Fig. 4 B). As our previous experiments indicated a specific requirement for FLCN in the stimulation of mTORC1 activity by amino acids, we next examined the localization of FLCN across basal growth, starved, and re-fed conditions. These experiments revealed that 3xHA-FLCN is selectively recruited to lysosomes in response to starvation but rapidly disperses from them in response to amino acid resupplementation (Fig. 4 C). Thus, in contrast to the constitutive lysosomal localization after co-overexpression of both FLCN and FNIP1 (Fig. 3), the 3xHA-FLCN expressed from the endogenous locus exhibited a much more selective, starvation-dependent recruitment to lysosomes. Nonetheless, a role for FNIP1 in this starvation-stimulated recruitment of FLCN to lysosomes was still apparent, as knockdown of FNIP1 prevented the enrichment of 3xHA-FLCN on lysosomes under such conditions (Fig. 4 D).
FLCN interacts with Rag GTPases
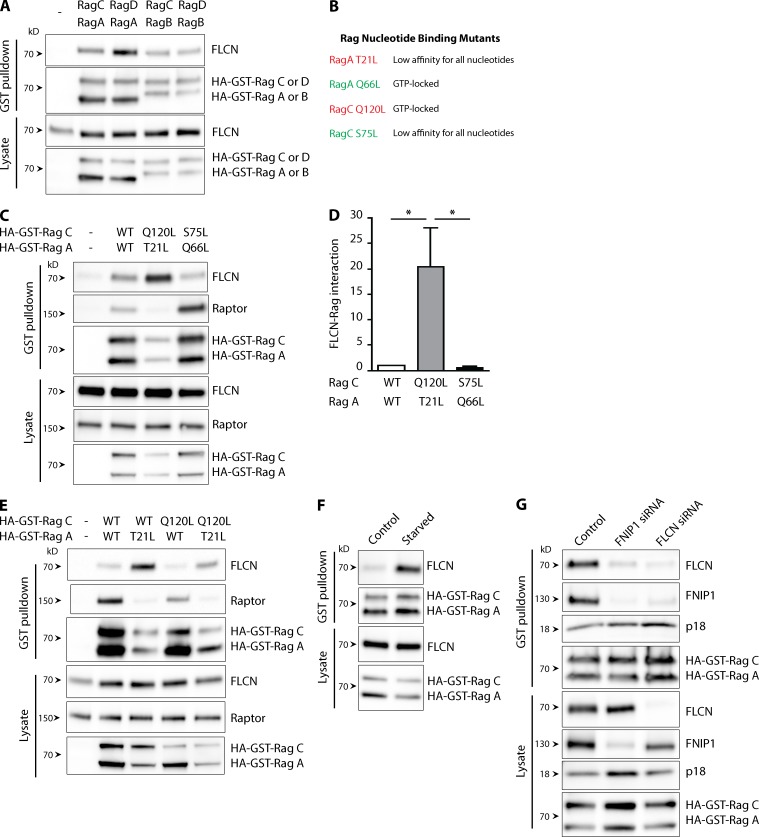
Our data show that FLCN dynamically localizes to the surface of lysosomes in a manner that depends on amino acid availability. Likewise, it is critical for both the amino acid–stimulated lysosome recruitment and activity of mTORC1 (Fig. 2), as well as the enrichment of TFEB on lysosomes (Fig. 1 A). Interestingly, both mTORC1 and TFEB represent the only currently known effectors of the amino acid–regulated and lysosome-localized Rag GTPases (Laplante and Sabatini, 2012; Martina and Puertollano, 2013). A role for FLCN in Rag GTPase regulation would also fit with recent structural and in vitro biochemical data that suggested a putative guanine nucleotide exchange factor (GEF) function for FLCN, but did not unambiguously identify the relevant GTPase target (Nookala et al., 2012). As a first step in testing the hypothesis that the Rag GTPases are the targets through which FLCN regulates mTORC1 and TFEB, we assayed for FLCN–Rag interactions and found that endogenously expressed FLCN co-purified with all four possible combinations of Rag GTPase heterodimers (Fig. 5 A) after their expression as HA-GST–tagged fusion proteins.
Figure 5.
FLCN selectively interacts with inactive Rag GTPases. (A) Representative immunoblots showing that endogenous FLCN co-purifies with all possible combinations of transfected HA-GST–tagged Rag GTPase heterodimers (n = 4). (B) Summary of the key properties of the Rag GTPase mutants used in our study. (C) Immunoblot analysis of GST pull-downs from cells transfected with either HA-GST tagged wild-type RagA+C or the indicated combinations of the constitutively active (RagA Q66L = RagAGTP + RagC S75L = RagCGDP/nucleotide free) and dominant-negative (RagA T21L = RagAGDP/nucleotide free + RagC Q120L = RagCGTP) mutants. (D) Quantification of the interaction between FLCN and the indicated Rags. The FLCN signal in each pull-down was normalized to the corresponding abundance of RagC in the same pull-down sample (mean ± SEM; *, P < 0.05, ANOVA with Bonferroni post-test, n = 4). (E) Immunoblot analysis of GST pull-downs from cells transfected with the indicated combinations of wild-type or dominant-negative recombinant Rag A and C. (F) Effect of starvation (2 h in Earle’s buffered saline solution) on the interaction between endogenous FLCN and wild-type HA-GST-RagA+C (n = 3). (G) Immunoblot analysis of GST pull-downs from cells transfected with wild-type HA-GST-RagAnucleotide-free+C along with control, FLCN, or FNIP1 siRNA. All experiments were performed in HeLa cells.
FLCN preferentially co-purifies with the inactive form of RagA/B
To determine the effect of Rag activation state on the binding between FLCN and Rags, we focused on the Rag A/C heterodimers and expressed combinations of Rag A and C mutants that lock these GTPases into active and inactive states (Sancak et al., 2008) and evaluated their ability to interact with endogenously expressed FLCN. Compared with wild-type Rags, FLCN bound most strongly to the RagAnucleotide-free-RagCGTP mutant heterodimer that was previously demonstrated to be inactive with respect to mTORC1 activation (Sancak et al., 2008). Consistent with expectations, raptor (an mTORC1 component and RagA/B effector) showed the opposite preference in its interactions with the mutant Rags (Fig. 5, B–D). Similar observations were made for the interactions between FLCN and equivalent RagB+D mutant heterodimers (Fig. S5 A). Through analysis of Rag heterodimers with single mutations in either the Rag A or Rag C subunits, it became clear that preventing the nucleotide loading of just RagA is sufficient for promoting maximal FLCN interactions (Fig. 5 E) under basal cell culture conditions.
FLCN–Rag interactions are enhanced in response to starvation
Amino acid availability is a physiological regulator of the loading of RagA/B with GTP such that when amino acids are scarce, RagA/B loading with GTP is diminished (Sancak et al., 2008). We next investigated the effect of cell starvation on the Rag–FLCN interaction and found that it was increased (Fig. 5 F). This starvation-induced stimulation of the FLCN–Rag interaction fits well with the preference of FLCN to bind RagA mutants that are defective in GTP binding (Fig. 5 C), as well as the selective enrichment of FLCN on lysosomes under such conditions (Fig. 4 C). However, the strong binding of FLCN to Rags in the absence of amino acids indicates that the simple binding of FLCN to Rags is not sufficient for their subsequent activation, and that an additional amino acid–dependent signal is required for this to occur.
FNIP1 is required for FLCN–Rag interactions
As FNIP1 is required for the recruitment of FLCN to lysosomes in response to starvation (Fig. 4 D), we hypothesized that FNIP1 would be required for the FLCN–Rag interactions that are also promoted by this same stimulus (Fig. 5 F). Indeed, co-purification of FLCN with RagA-C was greatly diminished in FNIP1 knockdown cells (Fig. 5 G). Interestingly, FNIP1 itself also co-purified with this RagA/C heterodimer in control cells, but not after FLCN knockdown (Fig. 5 G). This reciprocal requirement for both FLCN and FNIP1 to achieve Rag interactions is consistent with their interacting with the Rags as part of a complex wherein FNIP1 contributes to the lysosomal recruitment and/or Rag interactions of FLCN.
FLCN interacts directly with the GTPase domain of RagA
Purified recombinant proteins were used to directly assess the binding between FLCN and Rags A and C. We also included Rab35 in these binding assays, as FLCN was previously proposed to act as a Rab35 GEF (Nookala et al., 2012). In these experiments, which were performed in the absence of exogenous nucleotide, FLCN preferentially bound to RagA but not to either RagC or Rab35 (Fig. 6). Furthermore, the FLCN–RagA interaction was also observed with a truncated RagA mutant that comprised just the GTPase domain (amino acids 1–181) but which lacked the C-terminal roadblock domain (Fig. 6). These results indicate that FLCN can interact directly with RagA via its GTPase domain. As RagA and RagB share >99% amino acid identity (180 out of 181 amino acids are identical) within their GTPase domains, and FLCN co-purifies with RagB containing heterodimers from cells (Fig. 5 A and Fig. S5 A), FLCN is strongly predicted to also interact directly with the GTPase domain of RagB.
Figure 6.
FLCN binds directly to the GTPase domain of RagA. Left: in vitro binding assay where GST-FLCN was immobilized on GSH-beads and the binding of the indicated 6xHis-tagged-proteins (top) was detected by anti-6xHis immunoblotting. Equal loading of GST-FLCN was verified via anti-GST immunoblotting (bottom). Right: Ponceau S staining showing the migration of the various recombinant 6xHis-tagged proteins used in the in vitro binding assay.
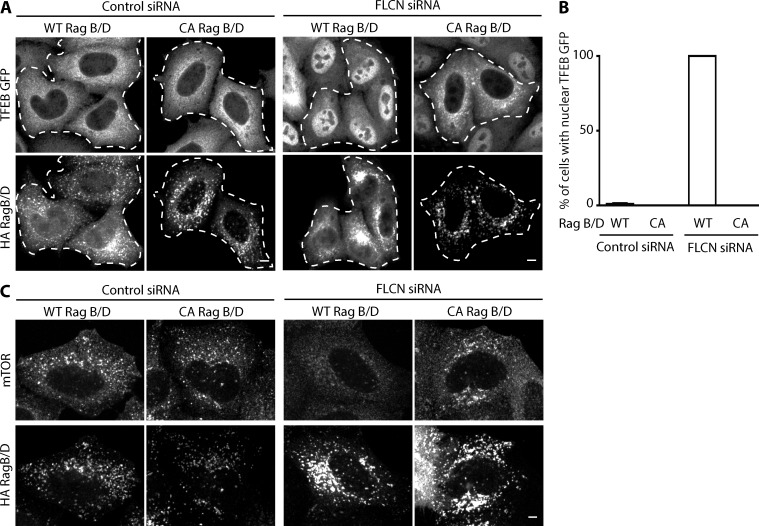
Constitutively active Rag GTPases rescue TFEB and mTOR localization in FLCN knockdown cells
If the specific lysosome-localized function for FLCN is to support the GTP loading of RagA/B, then the introduction of constitutively GTP-bound RagA/B mutants into FLCN-deficient cells should rescue the defects in the regulation of mTORC1 signaling from this organelle. We investigated this possibility using TFEB-GFP as well as endogenous mTOR localization as our readouts. Whereas overexpression of wild-type Rags (A+C and B+D heterodimers) had no detectable effect on the subcellular localization of TFEB under either control or FLCN knockdown conditions, the expression of constitutively active combinations of either RagB+D or RagA+C abolished the nuclear accumulation of TFEB in FLCN knockdown cells (Fig. 7, A and B; and Fig. S5 B). We also observed a rescue of mTOR localization to the lysosomes when FLCN knockdown cells were transfected with constitutively active RagB+D mutants (Fig. 7 C). The selective ability of constitutively active Rag GTPase mutants to rescue both TFEB and mTOR localization supports a model wherein FLCN promotes their activation (Fig. 8).
Figure 7.
Constitutively active Rag GTPase mutants rescue the effects of FLCN KD on TFEB and mTOR localization. (A) Immunofluorescence analysis of TFEB-GFP and HA-GST-Rag GTPase localization in control or FLCN KD cells cotransfected with either wild-type RagB+D or constitutively active (CA) Rag B+D (RagB Q99L + RagD S77L) heterodimers. Dashed lines highlight the Rag-transfected cells. (B) Quantification of the fraction of cells exhibiting predominantly nuclear (nuclear ≥ cytoplasmic signal intensity) TFEB-GFP localization after cotransfection of either control siRNA or FLCN knockdown cells with the indicated Rag GTPases (mean ± SEM, n = 3 experiments with >20 cells/condition/experiment). (C) Immunofluorescence analysis of mTOR localization in control or FLCN KD cells cotransfected with either wild-type RagB+D or CA RagB+D heterodimers. All images obtained by line scanning confocal imaging. Bars, 10 µm. All experiments were performed in HeLa cells.
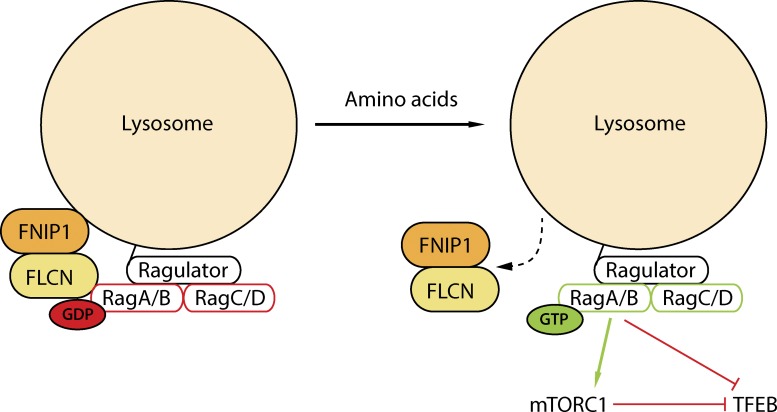
Figure 8.
Schematic diagram summarizing the proposed lysosome-localized role for FLCN and FNIP1 in the amino acid-dependent regulation of Rag GTPases, mTORC1 and TFEB.
Discussion
In this study, we identify the lysosome-localized Rag GTPases as targets of FLCN, the Birt-Hogg-Dubé syndrome tumor suppressor (Fig. 8). FLCN is selectively recruited to lysosomes (Fig. 4) and preferentially interacts with inactive RagA/B GTPases when amino acid levels are low (Fig. 5), but is required for their subsequent reactivation when amino acid levels are restored (Fig. 2). We also show that FLCN preferentially interacts with inactive RagA/B mutants (Fig. 5 C). In vitro binding assays with recombinant proteins demonstrate that such binding is direct and via the RagA/B GTPase domain (Fig. 6). FLCN-depleted cells fail to recruit either TFEB or mTORC1 (the only two known RagA/B effectors; Laplante and Sabatini, 2012; Martina and Puertollano, 2013) to lysosomes, and as a consequence further exhibit elevated nuclear levels of TFEB (Fig. 1, A and B), altered lysosome pH, and reduced mTORC1 activity (Fig. 1, D and E; Fig. 2, A–C, F, and G). FNIP1, the major previously characterized FLCN-binding protein, is critical for both the recruitment of FLCN to lysosomes (Figs. 3 and 4) and its interactions with Rag GTPases (Fig. 5 G). Collectively, these results: (1) define the lysosome as a major site of FLCN localization; (2) show that such FLCN localization is regulated by amino acid availability; (3) identify Rag GTPases as direct binding partners and lysosome-localized targets of FLCN function; and (4) establish a requirement for FLCN for the ability of cells to activate mTORC1 and sequester TFEB from the nucleus when amino acids are abundant.
Having now identified a direct physical and functional relationship between FLCN and RagA/B, it is possible to retrospectively appreciate numerous clues in studies of non-mammalian model organisms that are consistent with an evolutionarily conserved relationship between FLCN, responsiveness to changes in amino acid availability, and Rag GTPase regulation. For example, the major growth defects arising in Drosophila FLCN mutants were rescued by dietary amino acid supplementation (Liu et al., 2013). Even before the identification of FLCN mutations as the underlying cause of Birt-Hogg-Dubé syndrome (Nickerson et al., 2002), the budding yeast homologue of FLCN was first identified in a genetic screen designed to isolate genes that exhibited synthetic lethality with Sec13 mutations (Roberg et al., 1997) and named “lethal with Sec13 7” (LST7). Further analysis of Lst7 found that rather than being involved in COPII coat formation (the canonical site of action of Sec13), Lst7 along with Lst4, Lst8, and Sec13 played a distinct role in the nutrient-regulated trafficking of Gap1, an amino acid transporter (Roberg et al., 1997). A separate study from the same laboratory subsequently identified Gtr1 and 2 (the yeast Rag GTPase homologues) as important regulators of Gap1 trafficking (Gao and Kaiser, 2006). It has also been shown that Lst8 is an evolutionarily conserved TORC1 component (Loewith et al., 2002; Kim et al., 2003). More recently, Sec13 itself was found to have a vacuole/lysosome-localized, COPII-independent role in the regulation of TORC1 via Gtr/Rag GTPase regulation (Dokudovskaya et al., 2011; Bar-Peled et al., 2013; Panchaud et al., 2013). With respect to LST4, a very recent bioinformatic analysis predicted that despite minimal primary sequence similarity, this gene is most likely the yeast homologue of human FNIP1 (Levine et al., 2013). In agreement with these insights that arise from analyzing the context of the original identification of Lst7, a large-scale chemical genetic effort to annotate yeast gene function also independently linked Lst7 to Gtr1/2 (Rag GTPase homologues) and their regulators (Hillenmeyer et al., 2008).
Although our study as well as the abovementioned data derived from yeast support a role for FLCN as a positive regulator of mTORC1 activity via Rag GTPase regulation, it has been observed that some cysts and tumors arising from lack of FLCN in both humans and mice can actually have elevated mTORC1 activity (Chen et al., 2008; Hasumi et al., 2009; Nishii et al., 2013). It is not clear how absence of FLCN can have opposite effects on the mTORC1 pathway in different contexts. One possibility is that these opposing effects on the mTORC1 pathway reflect the existence of cell type–specific compensatory mechanisms that operate after the prolonged loss of FLCN. Our identification of direct contacts between FLCN, Rags, and lysosomes provides new targets that should contribute to the eventual resolution of this issue.
Even though amino acids have a well-established role in regulating mTORC1 activity via Rag GTPase regulation, the actual mechanism whereby intracellular amino acid abundance is sensed and transduced to the Rags has not been unambiguously established, and the abundance of proteins that have been very recently identified as contributing to this process make it clear that FLCN functions within the context of a much larger protein machinery that converges on mTORC1 regulation (Zoncu et al., 2011; Bar-Peled et al., 2012, 2013; Bonfils et al., 2012; Durán et al., 2012; Han et al., 2012; Kim et al., 2012; Ögmundsdóttir et al., 2012; Wauson et al., 2012; Panchaud et al., 2013). The integration of the roles of all these proteins into a coherent model represents an important challenge along the road to fully understanding how cells sense and respond to changing amino acid levels.
The role for amino acids in stimulating the conversion of RagA/B to their GTP-bound state suggests the need for an amino acid–regulated guanine nucleotide exchange factor (GEF) for these GTPases. The sole RagA/B GEF identified to date is the Ragulator complex that is also responsible for the recruitment of Rag heterodimers to the lysosomal membrane (Bar-Peled et al., 2012). Our observations that FLCN binds to GDP-bound/nucleotide-free RagA/B but is critical for the regulation of mTORC1 and TFEB, their two known effectors (Sancak et al., 2008; Martina and Puertollano, 2013), is consistent with a potential direct RagA/B GEF function for FLCN that could work in parallel with the previously described Ragulator pathway for RagA/B regulation. Interestingly, a crystal structure of the C terminus of FLCN combined with bioinformatic analysis of the full-length protein predicted that the FLCN protein contains a DENN (differentially expressed in normal and neoplastic cells) domain (Nookala et al., 2012). DENN domains are present in proteins that act as GEFs for members of the Rab family of GTPases (Allaire et al., 2010; Yoshimura et al., 2010; Wu et al., 2011), and the DENN1B–Rab35 co-crystal structure provided specific insight into the mechanisms of GEF action by DENN domains (Wu et al., 2011). FLCN was proposed to act as a GEF for Rab35 (Nookala et al., 2012), a protein implicated in the regulation of membrane trafficking early in the endocytic pathway (Marat et al., 2012). However, an in vivo role for FLCN as a Rab35 GEF was not established (Nookala et al., 2012), and the lysosomal localization, Rag GTPase interactions, and regulation of Rag effectors that we report here for FLCN suggests the alternative possibility of a physiologically relevant role for FLCN as a RagA/B GEF.
Although DENN domains are best characterized for their Rab GEF function, the identification of a partial DENN domain within the FLCN C terminus (Nookala et al., 2012) along with subsequent bioinformatic analyses (Zhang et al., 2012; Levine et al., 2013) have resulted in the prediction of multiple new putative DENN domain proteins whose precise functions remain uncharacterized. Surprisingly, FNIP1 is predicted to have a DENN domain by such studies (Zhang et al., 2012; Levine et al., 2013). Interestingly, similar to the FLCN–FNIP1 example of putative DENN domain proteins that dimerize with one another, Nprl2 and Nprl3 are two additional proteins with at least partial predicted homology to DENN domains (Zhang et al., 2012; Levine et al., 2013) that also heterodimerize (Neklesa and Davis, 2009). However, contrary to the expectations that DENN domain proteins should act as GEFs, the only known role for Nprl2/3 and their yeast homologues is to promote the activity of DEPDC5/Iml1, a RagA/B GAP (Bar-Peled et al., 2013; Panchaud et al., 2013). This example raises the possibility that FLCN–FNIP1 regulates RagA/B by a novel mechanism that is independent of a classical GEF function for their DENN domains. With this possibility in mind, it is also important to note that Lst7, the yeast FLCN homologue, is a much smaller protein that lacks the very region that was crystallized and found to have a DENN-like structure in human FLCN (Nookala et al., 2012) in spite of considerable evidence (discussed above) that supports an evolutionarily conserved function for Lst7 in Gtr1/2/Rag regulation. Thus, although our data are consistent with the possibility that FLCN acts via its DENN domain as a GEF for RagA/B, these additional considerations indicate that the actual situation is potentially more complex, and significant further experimentation will be required to address this topic.
From a technical perspective, it is noteworthy that although overexpressed FLCN and FNIP1 proteins provided a valuable clue about the ability of FLCN to localize to lysosomes (Fig. 3), we were only able to fully appreciate the amino acid–dependent regulation of FLCN localization upon detection of a functional epitope-tagged protein expressed from the endogenous locus (Fig. 4). Having shown the utility of this approach, we expect that the newly available CRISPR-Cas9/ssDNA oligonucleotide-based knock-in strategy that enables detection of proteins-of-interest with the robust 3xHA tag/antibody combination that we have used here will prove useful in future studies where reliable immunofluorescence detection is limited by either antibody availability, epitope accessibility, or target antigen abundance.
As elevated mTORC1 activity has been observed in many cancers (Laplante and Sabatini, 2012), it is perhaps puzzling that a tumor suppressor such as FLCN positively regulates mTORC1 activity. However, independent of any mutation affecting canonical components of the mTORC1 signaling pathway, the overexpression of MiT-TFE transcription factors due to chromosomal translocations is sufficient to cause renal carcinoma in humans (Haq and Fisher, 2011; Linehan, 2012). Thus, the strong nuclear localization of TFEB that we observed here in FLCN-deficient cells (Fig. 1, A and B), as well as the previous characterization of a role for FLCN as a negative regulator of the nuclear localization of TFE3 (Hong et al., 2010a; Betschinger et al., 2013), point to these transcription factors as being the candidate targets for the tumor suppressor function of FLCN in Birt-Hogg-Dubé syndrome. Although Rag GTPases were initially proposed to exert inhibitory effects on MiT-TFE transcription factors solely through their regulation of mTORC1 (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2012), it has since been discovered that MiT-TFE transcription factors are also direct RagA/B effectors (Martina and Puertollano, 2013). This interaction between RagA/BGTP and MiT-TFE proteins is critical for their recruitment to lysosomes and subsequent mTORC1-dependent phosphorylation. Thus, as negative regulation of MiT-TFE transcription factors depends on Rag GTPases for both their own recruitment as well as for mTORC1 recruitment and activation, even modest changes in Rag GTPase activation status will exert a stronger effect on MiT-TFE proteins than on other mTORC1 targets that lack this two-pronged dependence on the Rags.
In summary, our investigation of the relationship between FLCN, mTORC1, and MIT-TFE transcription factor regulation has provided evidence that FLCN functions at the lysosome where it interacts with both FNIP1 and the Rag GTPases, and is required for maximal amino acid–dependent stimulation of mTORC1 activity. Furthermore, we propose that it is through these combined effects on the Rag GTPases and mTORC1 that FLCN regulates the nuclear abundance of MiT-TFE transcription factors. Given the unique sensitivity of MiT-TFE transcription factors to alterations in Rag GTPase activity as well as the growing appreciation of additional roles for these proteins in the regulation of metabolism (O’Rourke and Ruvkun, 2013; Settembre et al., 2013) and stem cell pluripotency (Betschinger et al., 2013) along with their overexpression in multiple cancers (Haq and Fisher, 2011; Linehan, 2012; Vazquez et al., 2013), it is plausible that MiT-TFE dysregulation arising from reduced RagA/B activation contributes to Birt-Hogg-Dubé syndrome pathology. Building on our new findings, further investigation and eventual therapeutic manipulation of this signaling pathway could have an important impact on the treatment of Birt-Hogg-Dubé syndrome and other cancers that arise from alterations in MiT-TFE transcription factor activity.
Materials and methods
Cell culture and transfection
HeLa (American Type Culture Collection) or HeLa M subline (provided by P. De Camilli, Yale University, New Haven, CT) cells were grown in DMEM (+l-glutamine), 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin supplement (all from Invitrogen). Where indicated, cells were starved for 2 h by incubation in either amino acid–free RPMI (US Biologicals) or Earle’s buffered saline solution (Invitrogen). Amino acid stimulation was achieved with 1× MEM amino acid supplement (Invitrogen). As previously described (Roczniak-Ferguson et al., 2012), the TFEB-GFP stable HeLa cell line was generated by transfection of a pcDNA–human TFEB-GFP plasmid (provided by A. Ballabio, Telethon Institute of Genetics and Medicine, Naples, Italy) followed by selection with 500 µg/ml G418 (Invitrogen) and isolation of a clonal population of TFEB-GFP–positive cells. Transfections were performed with 4 µg of plasmid DNA, 12 µl of Fugene 6, and 800 µl of Opti-MEM (Invitrogen) per 100-mm dish of sub-confluent cells. For transfection of smaller or larger dishes, the volumes were scaled proportionally. Full-length mouse FLCN (BC015687.2) and FNIP1 (BC001956.1) sequences were PCR amplified from cDNAs purchased from Thermo Fisher Scientific, and cloned into pEGFP-N1 and ptdTomato-N1 plasmids (Takara Bio Inc.) as EcoRI+KpnI and XhoI+KpnI fragments, respectively. pRK5 plasmids encoding HA-GST–tagged human Rag GTPases and mutants thereof (RagA T21L, RagA Q66L, RagB T54L, RagB Q99L, RagC Q120L, RagC S75L, RagD S77L, and RagD Q121L) were obtained from D. Sabatini (Massachusetts Institute of Technology, Cambridge, MA) via Addgene (Sancak et al., 2008). LAMP1-mGFP (Falcón-Pérez et al., 2005) was provided by E. Dell’Angelica (UCLA, Los Angeles, CA). siRNA was transfected with the RNAiMAX transfection reagent (Invitrogen). siRNAs corresponding to the following target sequences were obtained from Thermo Fisher Scientific: TFEB (D-009798-03: 5′-AGACGAAGGUUCAACAUCA-3′); FLCN (D-009998-04: 5′-GAUAAAGAGACCUCCAUUA-3′); p18 (D-020556-04: 5′-CCAAGGAGACCGUGGGCUU-3′); and FNIP1 (D-032573-19: 5′-GGUAGUUGGCAAACGACAA-3′).
The following siRNAs were purchased from Integrated DNA Technologies: negative control (5′-CGUUAAUCGCGUAUAAUACGCGUAT-3′) and FLCN (5′-GGAUCUACCUCAUCAACUCCUGGCC-3′). Cells were analyzed 2 d after siRNA transfection.
RNA guided, Cas9-mediated engineering of the endogenous FLCN locus
Cas9 and guide RNA (gRNA) expression vectors (Mali et al., 2013) were obtained from Addgene. The following sequence: 5′-GGCACCATGAATGCCATCG-3′ (FLCN start codon underlined) was cloned into the gRNA expression vector in order to direct Cas9 nuclease activity toward the first coding exon of FLCN. To insert a 3xHA tag into this site via homologous recombination, we took advantage of a single-stranded DNA oligonucleotide-based genome editing strategy that was previously used in conjunction with zinc finger nucleases (Chen et al., 2011) and adapted it for the use of Cas9. To this end, an oligonucleotide was synthesized (IDT) that contained the 3xHA coding sequence followed by a GGGGS linker that was flanked on either side by ∼40 base pairs of sequence surrounding the FLCN start codon (Fig. S4). This sequence further included silent mutations to generate mismatches with the FLCN gRNA so as to prevent retargeting of the modified locus. 300 pmol of oligonucleotide was electroporated into one million HeLa M cells along with 2.5 µg each of the Cas9 and FLCN gRNA plasmids (Amaxa kit R, program A-24; Lonza). 1 wk after electroporation, cells were replated into 150-mm dishes at clonal density. Individual colonies were subsequently selected and expanded in 24-well dishes. After extraction of genomic DNA (QuickExtract DNA extraction solution; Epicentre), PCR (sense primer: 5′-GCTTTGTGGCTGTGGCTCTGTC-3′ and antisense primer: 5′-GACACTTGCCTCGCACATGTCC-3′) was performed using Phusion polymerase (New England Biolabs, Inc.) to amplify the genomic sequence surrounding the first FLCN coding exon to screen for clones that contained the 102 bp 3xHA tag + linker insertion (Fig. S4). To confirm the precise sequence of the targeted locus, PCR products were cloned into a plasmid (Zero Blunt TOPO PCR cloning kit; Invitrogen), transformed, and DNA from multiple colonies was directly sequenced.
Antibodies
The following primary antibodies were used in our experiments: anti–GFP-HRP (horseradish peroxidase; Miltenyi and Rockland Immunochemicals); pan–14-3-3 (Santa Cruz Biotechnology, Inc.); tubulin (Sigma-Aldrich); HA-HRP (Roche); GFP (Invitrogen); FNIP1 (Epitomics); and HA, 6xHis, LAMP1, mTOR, RagC, raptor, S6K1, phosphoT389–S6K1, S6, phospho–S6, FLCN, and p18/Lamtor1 (Cell Signaling Technology). Rabbit anti-GST antibody raised against full-length recombinant Schistosoma japonicum GST was provided by P. De Camilli (Yale University, New Haven, CT). The rabbit anti–phospho-TFEB serine 211 antibody was raised against the KVGVTSS-phosphoS-CPADLTQ peptide and the resulting serum was sequentially depleted and affinity purified with nonphosphorylated and phosphorylated versions of this peptide (Yenzym).
Expression and purification of recombinant proteins
To prepare recombinant proteins for in vitro binding assays, human Rag A and C were expressed individually from the pETDUET expression plasmid (EMD Millipore) to produce 6xHis-tagged proteins. Full-length mouse FLCN was cloned into the pGEX-6P-1 expression plasmid (GE Healthcare) for the production of a GST-FLCN fusion protein. The 6xHis-Rab35 construct was provided by K. Reinisch (Yale University, New Haven, CT; Wu et al., 2011). Plasmids were transformed into Escherichia coli BL21 (DE3) cells (Invitrogen). Cells were grown to an OD600 of 0.6 then shifted to 20°C and protein expression was induced by adding 0.5 mM IPTG and allowing the cultures to proceed overnight at 20°C.
For His-tagged proteins (full-length Rag A and C, GTPase domain of Rag A and C, and Rab35), cell pellets were resuspended in lysis buffer (20 mM Tris, pH 8.0, 300 mM NaCl, 15 mM imidazole, and 2 mM DTT) supplemented with protease inhibitor (Complete EDTA-free; Roche), lysed by sonication, and proteins were isolated on Ni-NTA columns (QIAGEN). Proteins were then eluted in 20 mM Tris, pH 8.0, 150 NaCl, 300 mM imidazole, and 2 mM DTT, and then extensively dialyzed against 20 mM Tris, pH 8.0, 150 mM NaCl, 20% glycerol, and 2 mM DTT.
For GST-FLCN, cells were resuspended in lysis buffer (50 mM Tris, pH 8.0, 0.5 mM MgCl2, 0.1% Triton X-100, and 1 mM DTT) supplemented with protease inhibitor, lysed by sonication, and proteins were isolated on glutathione Sepharose (GE Healthcare) columns. Proteins were then eluted in lysis buffer supplemented with 20 mM glutathione and extensively dialyzed against 20 mM Tris, pH 8.0, 150 mM NaCl, 20% glycerol, and 2 mM DTT.
In vitro binding assay
For the binding reactions, 2.5 µg of GST-FLCN was incubated for 1 h at 4°C with 25 µl of glutathione Sepharose slurry in 200 µl of binding buffer (40 mM Hepes, pH 7.4, 1% Triton X-100, 2.5 mM MgCl2, 2 mM DTT, and 1 mg/ml BSA). 10 µg of 6xHis-tagged protein (Rab35, full-length Rag A, full-length Rag C, GTPase domain of Rag A, or GTPase domain of Rag C) was added and the samples were rotated for 1 h at 4°C. The reactions were then washed three times with ice-cold binding buffer, followed by protein elution with 50 µl of Laemmli buffer.
Immunoprecipitations and immunoblotting
Cells were lysed in TBS + 1% Triton X-100 + protease and phosphatase inhibitor cocktails (Roche) and insoluble material was removed by centrifugation for 10 min at 20,000 g. Immunoprecipitations (TFEB-GFP) were performed on the resulting supernatant using anti-GFP agarose beads (Allele Biotechnology). HA-GST–tagged Rag proteins were purified from such extracts using glutathione Sepharose beads (GE Healthcare). Immunoblotting was performed using standard methods and the antibodies defined above. Chemiluminescent detection of HRP signals was performed via a Versadoc imaging station (Bio-Rad Laboratories).
Immunofluorescence and cell labeling
For standard immunofluorescence experiments, cells were grown on no. 1.0 glass coverslips and fixed with 4% paraformaldehyde/0.1 M sodium phosphate buffer (pH 7.2). Blocking was performed with 3% bovine serum albumin in PBS + 0.1% Triton X-100 or saponin. Primary and secondary antibody dilutions were performed in the same solution. Alexa 488– and Alexa 555–conjugated secondary antibodies were obtained from Invitrogen. Nuclei were stained with 1 µg/µl DAPI (Invitrogen) during one of the post-secondary antibody washes. Coverslips were finally mounted in ProLong Gold mounting medium (Invitrogen).
As we were unable to detect a signal above background using conventional indirect immunofluorescence for detection of 3xHA-FLCN expressed from its endogenous locus, we resorted to the use of a tertiary antibody in our staining protocol to increase detection sensitivity (Svensson, 1991). For this purpose, the mouse anti-HA primary antibody was followed by unlabeled goat anti–mouse IgG (Jackson ImmunoResearch Laboratories, Inc.) and then by Alexa 594–conjugated donkey anti–goat IgG (Invitrogen). Blocking and antibody incubations in these experiments were performed in PBS + 0.1% Triton X-100 + 5% normal donkey serum.
Lysosomes and nuclei were identified in live-cell imaging experiments with Lysotracker red DND-99 (50 nM for 10 min; Invitrogen) and Hoechst 33342 (1 µg/ml for 10 min; Cell Signaling Technology) staining, respectively, and the resulting spinning disk confocal images (20× or 40× objectives) were quantified using Cell Profiler to measure the average Lysotracker staining per cell. Dextran (10 µg/ml Alexa 647–Dextran, 10,000 MW; Invitrogen) labeling of lysosomes was performed via an overnight incubation followed by a 1-h washout before imaging.
Microscopy
Spinning disc confocal microscopy was performed using the UltraVIEW VoX system (PerkinElmer) including an inverted microscope (Ti-E Eclipse, Nikon; equipped with 60× CFI PlanApo VC, NA 1.4, oil immersion; 40× CFI Plan Apo, NA 1.0, oil immersion objective; and 20× CFI Plan Apo, NA 0.75, air objective) and a spinning disk confocal scan head (CSU-X1, Yokogawa) driven by Volocity (PerkinElmer) software. Images were acquired without binning with a 14-bit (1,000 × 1,000) EMCCD (Hamamatsu Photonics). Typical exposure times were 100–500 ms. Cells were imaged at room temperature (∼22°C). For live-cell imaging experiments the imaging medium consisted of 136 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1.3 mM MgCl2, and 10 mM Hepes, pH 7.4. Line-scanning confocal microscopy images (immunofluorescence samples) were acquired with a laser scanning confocal microscope (LSM 710; Carl Zeiss) using a 63× Plan Apo (NA 1.4) oil immersion objective and Efficient Navigation (ZEN) software (Carl Zeiss).
Image analysis
Post-acquisition image analysis was performed with ImageJ (National Institutes of Health) and Cell Profiler software (Carpenter et al., 2006). The absolute ratio of nuclear to cytoplasmic TFEB-GFP was quantified using Cell Profiler on images of paraformaldehyde-fixed cells acquired by spinning disk confocal microscopy using either the 20× or 40× objectives. Nuclei were identified by DAPI staining and nuclear edges were uniformly expanded to form a halo that defined the surrounding cytoplasmic compartment in each cell. The mean intensities of the nuclear and cytoplasmic regions were measured and used to calculate nuclear to cytoplasmic ratio. On average, greater than 200 cells per condition were analyzed per experiment.
TFEB nuclear localization in Figs. 7 and S5 was determined by visual inspection of photographs acquired by spinning disk confocal microscopy with a 20× CFI Plan Apo, NA 0.75 objective. Cells were visually scored as having nuclear localization if the nuclear levels of TFEB exceeded those in the cytoplasm. At least 20 cells were scored per condition per experiment.
Statistical analysis
Data were analyzed using Prism (GraphPad Software) and the tests specified in the figure legends. All error bars represent the SEM.
Online supplemental material
Fig. S1 provides a characterization of the sensitivity of mTORC1 target phosphorylation to torin 1 treatment. Fig. S2 demonstrates the effects of FLCN knockdown on both the lysosomal localization of mTOR and TFEB phosphorylation under basal, starved, and re-fed conditions. Fig. S3 shows the effect of FNIP1 knockdown on TFEB localization. Fig. S4 provides an overview of the strategy for introducing a 3xHA tag into the FLCN endogenous locus. Fig. S5 shows the effects of Rag B+D mutations on FLCN binding and the effect of Rag A+C mutations on TFEB localization ± FLCN knockdown. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201307084/DC1.
Supplementary Material
Acknowledgments
We thank Jennifer Ky, Sharon Qian, and Nicholas Roy for technical assistance and Xudong Wu, Karin Reinisch, Pietro De Camilli, and Michael Caplan for their scientific advice. We are grateful for the invaluable support of the Yale Program in Cellular Neuroscience, Neurodegeneration, and Repair imaging facility for enabling our microscopy experiments.
This research was supported by grants from the Ellison Medical Foundation and the National Institutes of Health (GM105718) to S.M. Ferguson. C.S. Petit was supported by an Anderson Fellowship from Yale University.
The authors have no conflicts of interest to declare.
Footnotes
Abbreviations used in this paper:
- CRISPR
- clustered regularly interspaced short palindromic repeats
- FLCN
- folliculin
- FNIP1
- folliculin-interacting protein 1
- GEF
- guanine nucleotide exchange factor
- KD
- knockdown
- mTOR
- mechanistic target of rapamycin
- mTORC1
- mechanistic target of rapamycin complex 1
- MITF
- microphthalmia transcription factor
- S6K1
- ribosomal protein S6 kinase
- TFEB
- transcription factor EB
- TFE3
- transcription factor E3
References
- Allaire P.D., Marat A.L., Dall’Armi C., Di Paolo G., McPherson P.S., Ritter B. 2010. The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol. Cell. 37:370–382 10.1016/j.molcel.2009.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M., Hong S.B., Sharma N., Warren M.B., Nickerson M.L., Iwamatsu A., Esposito D., Gillette W.K., Hopkins R.F., III, Hartley J.L., et al. 2006. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc. Natl. Acad. Sci. USA. 103:15552–15557 10.1073/pnas.0603781103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L., Schweitzer L.D., Zoncu R., Sabatini D.M. 2012. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 150:1196–1208 10.1016/j.cell.2012.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L., Chantranupong L., Cherniack A.D., Chen W.W., Ottina K.A., Grabiner B.C., Spear E.D., Carter S.L., Meyerson M., Sabatini D.M. 2013. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 340:1100–1106 10.1126/science.1232044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Nichols J., Dietmann S., Corrin P.D., Paddison P.J., Smith A. 2013. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 153:335–347 10.1016/j.cell.2013.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfils G., Jaquenoud M., Bontron S., Ostrowicz C., Ungermann C., De Virgilio C. 2012. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell. 46:105–110 10.1016/j.molcel.2012.02.009 [DOI] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash T.P., Gruber J.J., Hartman T.R., Henske E.P., Simon M.C. 2011. Loss of the Birt-Hogg-Dubé tumor suppressor results in apoptotic resistance due to aberrant TGFβ-mediated transcription. Oncogene. 30:2534–2546 10.1038/onc.2010.628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerny J., Feng Y., Yu A., Miyake K., Borgonovo B., Klumperman J., Meldolesi J., McNeil P.L., Kirchhausen T. 2004. The small chemical vacuolin-1 inhibits Ca(2+)-dependent lysosomal exocytosis but not cell resealing. EMBO Rep. 5:883–888 10.1038/sj.embor.7400243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T.F., Carvalho J., Riles L., Zheng X.F. 2000. A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR). Proc. Natl. Acad. Sci. USA. 97:13227–13232 10.1073/pnas.240444197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Pruett-Miller S.M., Huang Y., Gjoka M., Duda K., Taunton J., Collingwood T.N., Frodin M., Davis G.D. 2011. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods. 8:753–755 10.1038/nmeth.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Futami K., Petillo D., Peng J., Wang P., Knol J., Li Y., Khoo S.K., Huang D., Qian C.N., et al. 2008. Deficiency of FLCN in mouse kidney led to development of polycystic kidneys and renal neoplasia. PLoS ONE. 3:e3581 10.1371/journal.pone.0003581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dazert E., Hall M.N. 2011. mTOR signaling in disease. Curr. Opin. Cell Biol. 23:744–755 10.1016/j.ceb.2011.09.003 [DOI] [PubMed] [Google Scholar]
- Dokudovskaya S., Waharte F., Schlessinger A., Pieper U., Devos D.P., Cristea I.M., Williams R., Salamero J., Chait B.T., Sali A., et al. 2011. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol. Cell. Proteomics. 10:006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán R.V., Oppliger W., Robitaille A.M., Heiserich L., Skendaj R., Gottlieb E., Hall M.N. 2012. Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell. 47:349–358 10.1016/j.molcel.2012.05.043 [DOI] [PubMed] [Google Scholar]
- Falcón-Pérez J.M., Nazarian R., Sabatti C., Dell’Angelica E.C. 2005. Distribution and dynamics of Lamp1-containing endocytic organelles in fibroblasts deficient in BLOC-3. J. Cell Sci. 118:5243–5255 10.1242/jcs.02633 [DOI] [PubMed] [Google Scholar]
- Gao M., Kaiser C.A. 2006. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 8:657–667 10.1038/ncb1419 [DOI] [PubMed] [Google Scholar]
- Han J.M., Jeong S.J., Park M.C., Kim G., Kwon N.H., Kim H.K., Ha S.H., Ryu S.H., Kim S. 2012. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 149:410–424 10.1016/j.cell.2012.02.044 [DOI] [PubMed] [Google Scholar]
- Haq R., Fisher D.E. 2011. Biology and clinical relevance of the micropthalmia family of transcription factors in human cancer. J. Clin. Oncol. 29:3474–3482 10.1200/JCO.2010.32.6223 [DOI] [PubMed] [Google Scholar]
- Hartman T.R., Nicolas E., Klein-Szanto A., Al-Saleem T., Cash T.P., Simon M.C., Henske E.P. 2009. The role of the Birt-Hogg-Dubé protein in mTOR activation and renal tumorigenesis. Oncogene. 28:1594–1604 10.1038/onc.2009.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasumi Y., Baba M., Ajima R., Hasumi H., Valera V.A., Klein M.E., Haines D.C., Merino M.J., Hong S.B., Yamaguchi T.P., et al. 2009. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc. Natl. Acad. Sci. USA. 106:18722–18727 10.1073/pnas.0908853106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath T.J., Steingrímsson E., McGill G., Hansen M.J., Vaught J., Hodgkinson C.A., Arnheiter H., Copeland N.G., Jenkins N.A., Fisher D.E. 1994. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8:2770–2780 10.1101/gad.8.22.2770 [DOI] [PubMed] [Google Scholar]
- Hillenmeyer M.E., Fung E., Wildenhain J., Pierce S.E., Hoon S., Lee W., Proctor M., St Onge R.P., Tyers M., Koller D., et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 320:362–365 10.1126/science.1150021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.B., Oh H., Valera V.A., Baba M., Schmidt L.S., Linehan W.M. 2010a. Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS ONE. 5:e15793 10.1371/journal.pone.0015793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.B., Oh H., Valera V.A., Stull J., Ngo D.T., Baba M., Merino M.J., Linehan W.M., Schmidt L.S. 2010b. Tumor suppressor FLCN inhibits tumorigenesis of a FLCN-null renal cancer cell line and regulates expression of key molecules in TGF-beta signaling. Mol. Cancer. 9:160 10.1186/1476-4598-9-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudon V., Sabourin S., Dydensborg A.B., Kottis V., Ghazi A., Paquet M., Crosby K., Pomerleau V., Uetani N., Pause A. 2010. Renal tumour suppressor function of the Birt-Hogg-Dubé syndrome gene product folliculin. J. Med. Genet. 47:182–189 10.1136/jmg.2009.072009 [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Xu T., Guan K.L. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17:1829–1834 10.1101/gad.1110003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K., Kim J., Guan K.L. 2012. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu. Rev. Pharmacol. Toxicol. 52:381–400 10.1146/annurev-pharmtox-010611-134537 [DOI] [PubMed] [Google Scholar]
- Kim D.H., Sarbassov D.D., Ali S.M., Latek R.R., Guntur K.V., Erdjument-Bromage H., Tempst P., Sabatini D.M. 2003. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell. 11:895–904 10.1016/S1097-2765(03)00114-X [DOI] [PubMed] [Google Scholar]
- Kim E., Goraksha-Hicks P., Li L., Neufeld T.P., Guan K.L. 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10:935–945 10.1038/ncb1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.M., Stone M., Hwang T.H., Kim Y.G., Dunlevy J.R., Griffin T.J., Kim D.H. 2012. SH3BP4 is a negative regulator of amino acid-Rag GTPase-mTORC1 signaling. Mol. Cell. 46:833–846 10.1016/j.molcel.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M., Sabatini D.M. 2012. mTOR signaling in growth control and disease. Cell. 149:274–293 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T.P., Daniels R.D., Gatta A.T., Wong L.H., Hayes M.J. 2013. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 29:499–503 10.1093/bioinformatics/bts725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan W.M. 2012. Genetic basis of kidney cancer: role of genomics for the development of disease-based therapeutics. Genome Res. 22:2089–2100 10.1101/gr.131110.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Chen Z., Ma Y., Wu X., Jin Y., Hou S. 2013. Genetic characterization of the Drosophila birt-hogg-dubé syndrome gene. PLoS ONE. 8:e65869 10.1371/journal.pone.0065869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J.L., Bonenfant D., Oppliger W., Jenoe P., Hall M.N. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 10:457–468 10.1016/S1097-2765(02)00636-6 [DOI] [PubMed] [Google Scholar]
- Luijten M.N., Basten S.G., Claessens T., Vernooij M., Scott C.L., Janssen R., Easton J.A., Kamps M.A., Vreeburg M., Broers J.L., et al. 2013. Birt-Hogg-Dube syndrome is a novel ciliopathy. Hum. Mol. Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.M., Blenis J. 2009. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10:307–318 10.1038/nrm2672 [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. 2013. RNA-guided human genome engineering via Cas9. Science. 339:823–826 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat A.L., Ioannou M.S., McPherson P.S. 2012. Connecdenn 3/DENND1C binds actin linking Rab35 activation to the actin cytoskeleton. Mol. Biol. Cell. 23:163–175 10.1091/mbc.E11-05-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Puertollano R. 2013. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol. 200:475–491 10.1083/jcb.201209135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Chen Y., Gucek M., Puertollano R. 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 8:903–914 10.4161/auto.19653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova M.M., Shaw R.J. 2011. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13:1016–1023 10.1038/ncb2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa T.K., Davis R.W. 2009. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet. 5:e1000515 10.1371/journal.pgen.1000515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson M.L., Warren M.B., Toro J.R., Matrosova V., Glenn G., Turner M.L., Duray P., Merino M., Choyke P., Pavlovich C.P., et al. 2002. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dubé syndrome. Cancer Cell. 2:157–164 10.1016/S1535-6108(02)00104-6 [DOI] [PubMed] [Google Scholar]
- Nishii T., Tanabe M., Tanaka R., Matsuzawa T., Okudela K., Nozawa A., Nakatani Y., Furuya M. 2013. Unique mutation, accelerated mTOR signaling and angiogenesis in the pulmonary cysts of Birt-Hogg-Dubé syndrome. Pathol. Int. 63:45–55 10.1111/pin.12028 [DOI] [PubMed] [Google Scholar]
- Nookala R.K., Langemeyer L., Pacitto A., Ochoa-Montaño B., Donaldson J.C., Blaszczyk B.K., Chirgadze D.Y., Barr F.A., Bazan J.F., Blundell T.L. 2012. Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol. 2:120071 10.1098/rsob.120071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke E.J., Ruvkun G. 2013. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol. 15:668–676 10.1038/ncb2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ögmundsdóttir M.H., Heublein S., Kazi S., Reynolds B., Visvalingam S.M., Shaw M.K., Goberdhan D.C. 2012. Proton-assisted amino acid transporter PAT1 complexes with Rag GTPases and activates TORC1 on late endosomal and lysosomal membranes. PLoS ONE. 7:e36616 10.1371/journal.pone.0036616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchaud N., Péli-Gulli M.P., De Virgilio C. 2013. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 6:ra42 10.1126/scisignal.2004112 [DOI] [PubMed] [Google Scholar]
- Peña-Llopis S., Vega-Rubin-de-Celis S., Schwartz J.C., Wolff N.C., Tran T.A., Zou L., Xie X.J., Corey D.R., Brugarolas J. 2011. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 30:3242–3258 10.1038/emboj.2011.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberg K.J., Bickel S., Rowley N., Kaiser C.A. 1997. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7 and LST8. Genetics. 147:1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., Walther T.C., Ferguson S.M. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5:ra42 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., Sabatini D.M. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 320:1496–1501 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., Sabatini D.M. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141:290–303 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. 2009. A gene network regulating lysosomal biogenesis and function. Science. 325:473–477 [DOI] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., De Cegli R., Mansueto G., Saha P.K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T.J., et al. 2013. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 15:647–658 10.1038/ncb2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.R., Zhen W., Zheng Z., Wang H., Oh S.W., Liu W., Zbar B., Schmidt L.S., Hou S.X. 2006. The Drosophila homolog of the human tumor suppressor gene BHD interacts with the JAK-STAT and Dpp signaling pathways in regulating male germline stem cell maintenance. Oncogene. 25:5933–5941 10.1038/sj.onc.1209593 [DOI] [PubMed] [Google Scholar]
- Svensson M.A. 1991. Increased sensitivity of the indirect immunofluorescence method by use of a tertiary fluorochrome-labeled antibody. J. Histochem. Cytochem. 39:235–237 10.1177/39.2.1987267 [DOI] [PubMed] [Google Scholar]
- Takagi Y., Kobayashi T., Shiono M., Wang L., Piao X., Sun G., Zhang D., Abe M., Hagiwara Y., Takahashi K., Hino O. 2008. Interaction of folliculin (Birt-Hogg-Dubé gene product) with a novel Fnip1-like (FnipL/Fnip2) protein. Oncogene. 27:5339–5347 10.1038/onc.2008.261 [DOI] [PubMed] [Google Scholar]
- Tee A.R., Pause A. 2012. Birt-Hogg-Dubé: tumour suppressor function and signalling dynamics central to folliculin. Fam. Cancer. [DOI] [PubMed] [Google Scholar]
- Thoreen C.C., Kang S.A., Chang J.W., Liu Q., Zhang J., Gao Y., Reichling L.J., Sim T., Sabatini D.M., Gray N.S. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284:8023–8032 10.1074/jbc.M900301200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Slegtenhorst M., Khabibullin D., Hartman T.R., Nicolas E., Kruger W.D., Henske E.P. 2007. The Birt-Hogg-Dube and tuberous sclerosis complex homologs have opposing roles in amino acid homeostasis in Schizosaccharomyces pombe. J. Biol. Chem. 282:24583–24590 10.1074/jbc.M700857200 [DOI] [PubMed] [Google Scholar]
- Vazquez F., Lim J.H., Chim H., Bhalla K., Girnun G., Pierce K., Clish C.B., Granter S.R., Widlund H.R., Spiegelman B.M., Puigserver P. 2013. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 23:287–301 10.1016/j.ccr.2012.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauson E.M., Zaganjor E., Lee A.Y., Guerra M.L., Ghosh A.B., Bookout A.L., Chambers C.P., Jivan A., McGlynn K., Hutchison M.R., et al. 2012. The G protein-coupled taste receptor T1R1/T1R3 regulates mTORC1 and autophagy. Mol. Cell. 47:851–862 10.1016/j.molcel.2012.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Bradley M.J., Cai Y., Kümmel D., De La Cruz E.M., Barr F.A., Reinisch K.M. 2011. Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate. Proc. Natl. Acad. Sci. USA. 108:18672–18677 10.1073/pnas.1110415108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Gerondopoulos A., Linford A., Rigden D.J., Barr F.A. 2010. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191:367–381 10.1083/jcb.201008051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Iyer L.M., He F., Aravind L. 2012. Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front Genet. 3:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D.M. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 334:678–683 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.