Live cell imaging, high-resolution microscopy, and computational modeling show that dynamic formin-filamin-actin asters self-organize into an actomyosin contractile network that may maintain mechanical coherence of cytoplasm.
Abstract
A ctin filaments, with the aid of multiple accessory proteins, self-assemble into a variety of network patterns. We studied the organization and dynamics of the actin network in nonadhesive regions of cells bridging fibronectin-coated adhesive strips. The network was formed by actin nodes associated with and linked by myosin II and containing the formin disheveled-associated activator of morphogenesis 1 (DAAM1) and the cross-linker filamin A (FlnA). After Latrunculin A (LatA) addition, actin nodes appeared to be more prominent and demonstrated drift-diffusion motion. Superresolution microscopy revealed that, in untreated cells, DAAM1 formed patches with a similar spatial arrangement to the actin nodes. Node movement (diffusion coefficient and velocity) in LatA-treated cells was dependent on the level and activity of myosin IIA, DAAM1, and FlnA. Based on our results, we developed a computational model of the dynamic formin-filamin-actin asters that can self-organize into a contractile actomyosin network. We suggest that such networks are critical for connecting distant parts of the cell to maintain the mechanical coherence of the cytoplasm.
Introduction
Cellular processes in eukaryotic cells depend on the architecture of the actin cytoskeleton (Mogilner and Keren, 2009; Pollard, 2010). Numerous specialized structures formed by actin filaments, such as the dense filament network that fills lamellipodia, actin bundles in microvilli, filopodia, stress fibers, and cytokinetic rings have been relatively well described. Some of these structures contain myosins and are contractile. In addition to these specialized and highly ordered actin filament arrays, less well-defined networks also exist adjacent to the plasma membrane or distributed throughout the bulk of the cytoplasm, as documented by numerous electron microscopy studies (see, for example Schliwa, 1982; Svitkina et al., 1984, 1997; Medalia et al., 2002). A more recent study, which used superresolution optical microscopy techniques, revealed that in cultured cells, two layers of actin networks, each with distinct densities and structural organizations, exist in sheet-like cell protrusions (Xu et al., 2012). Contractile cellular actin networks appear to be important in the maintenance of cell shape and coherence of the cytoplasm (Cai and Sheetz, 2009; Rossier et al., 2010). However, the organization and dynamics of these networks are still poorly understood.
One way to understand the mechanical and dynamic characteristics of an actomyosin network is to use purified actin, myosin II, and some associated proteins to build up the actomyosin network in vitro. Such studies showed that pure actomyosin gels are unstable and undergo “super-precipitation.” However, gels containing actin, myosin II, and cross-linking proteins such as filamin (Koenderink et al., 2009), fascin (Gordon et al., 2012), or even artificial cross-linkers such as streptavidin, which bridges biotinylated actin filaments (Mizuno et al., 2007; Soares e Silva et al., 2011), demonstrated apparent self-organization into a system of dynamic actin nodes that coalesce due to myosin II activity. In each case however, these networks were only transiently maintained, resulting in collapse of the gel.
Interestingly, structures resembling these myosin-containing actin nodes have been observed in some in vivo systems. For example, during the formation of the contractile ring in dividing cells (Wu et al., 2006; Werner et al., 2007; Laporte et al., 2011), during the establishment and maintenance of anterior-posterior polarity in the Caenorhabditis elegans zygote (Munro et al., 2004), in the course of “punctuated actin contractions” of embryonic mesenchymal cells in the Xenopus laevis mesoderm (Kim and Davidson, 2011), and finally in apical actomyosin networks that are dynamically coupled to adherens junctions of epithelial cells in Drosophila melanogaster and C. elegans embryos (Martin et al., 2009; Rauzi et al., 2010; Roh-Johnson et al., 2012). Wound closure in Xenopus oocytes is also accompanied by the formation of multiple myosin-containing actin nodes that are connected by thin actin filaments at the wound border (Mandato and Bement, 2001). Interestingly, in at least some of these in vivo systems, formin family proteins (formins), which are potent activators of actin polymerization (Chesarone et al., 2010), were found to be involved in the organization of these multinodal networks (Wu et al., 2006; Werner et al., 2007; Laporte et al., 2011).
Treatment of cells with small doses of drugs that can interfere with actin assembly, such as the actin monomer sequestering drug Latrunculin A (LatA) or the actin polymerization inhibitor cytochalasin D, revealed multiple nodes of actin filaments scattered over the entire cell area (Schliwa, 1982; Verkhovsky et al., 1997; Rossier et al., 2010). Because very similar patterns are also observed in untreated cells of various types (Werner et al., 2007; Roh-Johnson et al., 2012; Xu et al., 2012, etc.), it is probable that LatA treatment reveals preexisting multinodal structures rather than creating them de novo. This highlights the potential for such drug treatments as tools to investigate the organization and dynamics of cellular actin networks in more detail.
Using this approach we have developed an experimental system in which the formation and dynamics of multinodal actin arrays can be reproducibly observed. We have obtained quantitative data characterizing actin node movement and growth following their fusion with each other. We have also investigated the role of major types of molecular building blocks in this process, including motors, actin nucleators, and actin filament cross-linkers. We found that the myosin IIA motor, the cross-linking protein filamin A (FlnA), and the formin family actin filament nucleator and elongator, disheveled-associated activator of morphogenesis 1 (DAAM1), were strongly involved in the organization and dynamics of the multinodal cellular actin network.
Moreover, we propose a simple model in which actin nodes, represented as asters, are generated via formin-driven actin polymerization, stabilized by cross-linker proteins, and connected with bipolar myosin minifilaments (or antiparallel dimers). Such modeling faithfully reproduced the experimentally observed behavior of actin and myosin structures in the evolution of the multinodal actin network, particularly the relocation of myosin from the position in between actin asters to the center of the asters. This model also showed that continual filament growth from asters, and the random character of aster movement, prevents the connected actin network from collapsing, while maintaining potential contraction ability. Thus, formin- and filamin-dependent actin filament asters that are dynamically connected by myosin II form a network that supports cytoplasm coherence.
Results
LatA treatment reveals numerous actin nodes in nonadherent regions of the cell
Mouse embryonic fibroblasts (MEFs) or HeLa JW cells transfected with Lifeact-Ruby were plated on fibronectin-patterned surfaces comprised of 2–4-µm-wide adhesive strips and 5–12-µm-wide nonadhesive gaps (Fig. 1 A). Consistent with previous studies (Rossier et al., 2010), attachment of cells to this pattern lead to the formation of broad cytoplasmic bridges that connected the adhesive strips. These areas, marked green in Fig. 1 A, did not have focal adhesions or lamellipodia. Thus, they provided a suitable system for the visualization of the other cellular actin networks.
Figure 1.
Actin node formation triggered by LatA-mediated F-actin depolymerization. (A) Schematic diagram of the experimental setup. Transfected cells were placed on fibronectin-patterned glass coverslips. The “zone of observation” (green) represents the area of the cell observed in all experiments and corresponds to the actin network that spanned the nonadhesive areas. (B) Representative images of LatA treatment results. HeLa JW cells transfected with Lifeact-Ruby were treated with 800 nM LatA. The yellow dotted lines mark the adhesive areas patterned with fibronectin. In phase 1, stress fibers shortened and eventually disappeared (red arrows). Actin nodes appeared in phase 2. (C) Actin node aggregation. In phase 2, actin nodes merged and became bigger. In the event where two actin nodes merge together (yellow arrows), only one track is continued. At time 1,060 s, the red track ends and the green track continues, while at time 1,088 s, the blue track ends and the green track continues. (D) The histogram was obtained from the measurements of 10 s displacement in one of the control samples that contained 100-s long trajectories of ∼100 nodes. Distribution analysis of the angle (θ) between direction of movement (yellow arrow) of a given node (red dot) at a given moment, and the neighboring nodes within the distance of R. Blue region: R < 2 µm. Pink region: 2 µm ≤ R < 4 µm. Orange region: 4 µm ≤ R < 6 µm. The histograms plot the distribution of θ in the respected color-coded regions. In the range of 0–4 µm, θ has a biased distribution toward small angles. When the distance is >4 µm, the distribution of θ is uniform. (E) STORM images of Alexa Fluor 647–phalloidin staining of actin structures before (left) and after treatment with 200 nM LatA for 20 min (middle and right). The image on the right represents the same field as the middle panel visualized by TIRF. Blue arrowheads indicate some of the actin nodes. The boxed regions are enlarged below. Bars: (A) 20 µm; (B) 10 µm; (C) 2 µm; (D) 5 µm; (E, top) 5 µm; (E, bottom) 1 µm.
Treatment of cells with LatA at concentrations ranging from 200 nM to 800 nM resulted in a biphasic alteration of the actin pattern. During “phase 1,” retraction and the subsequent disappearance of actin stress fibers was observed, while in “phase 2,” the appearance of numerous micron-sized actin nodes occurred (Fig. 1 B and Video 1). The appearance of nodes usually preceded the complete disappearance of stress fibers (Fig. S1 A); however, a brief period was occasionally observed whereby both stress fibers and actin nodes were absent (for example in Fig. 1 B, at 240 s, in the central region of the cell). No significant alteration to the actin pattern was observed at LatA concentrations <200 nM. Increasing the concentration of LatA from 200 to 800 nM accelerated the onset of phase 2 (Fig. S1 A), but did not alter the node density or size (∼10 nodes per 100 µm2, ∼1 µm2 in size; Fig. S1 B). Cells treated with 200 nM of LatA maintained a continuous cytoskeleton network that filled the bridges between adhesive strips for up to 3 h. However, after a 30–40-min treatment with 800 nM of LatA, or 40–60 min with 400 nM of LatA, cell rounding was observed. LatA effects were reversible and the actin organization returned to a normal state within an hour of washing off the LatA (unpublished data). The appearance of actin nodes upon LatA treatment was a general observation seen in all cell types studied (MEFs, human HeLa JW cells, and a human lung cancer cell line H460). The drug dose-dependence of the time course varied in different cell types (unpublished data).
The actin nodes observed after LatA treatment were motile. To determine whether their movement can be characterized as an undirected random walk (pure diffusion) or a biased random walk (drift-diffusion), the mean square displacement (MSD) of the nodes at different time intervals Δt was examined for best fit to either model (Qian et al., 1991). Pure diffusion predicts linear MSD growth with time; drift-diffusion predicts a quadratic growth. To determine whether there was a statistically significant evidence for drift, an F-test (Kutner et al., 2004), which compares the two-parameter drift-diffusion model (MSD(Δt) = 4D · Δt + (v · Δt)2) with the one-parameter pure-diffusion model (MSD(Δt) = 4D · Δt) was applied. Data with Δt > 100 s were discarded because of paucity. Using a cut-off of P = 0.05, we rejected the pure-diffusion model for the eight control datasets collected from independent experiments (summarized in Table 1). Thus, the actin nodes in LatA-treated cells demonstrated a biased random walk, or drift-diffusion motion.
Table 1.
MSD fitting of the diffusion-drift model for control and perturbed cells
| Related figures | Samples | na | D | v | Type of motion |
| ×10−3 µm2/s | ×10−3 µm/s | ||||
| Fig. 3 C (MEF) | Control | 8 | 17.9 ± 13.0 | 10.8 ± 6.50 | Diffusion-drift |
| LatA-blebbistatin | 4 | 8.31 ± 5.46 | N/A | Diffusion only | |
| Myosin IIA knockdown | 1 | 7.43 | N/A | Diffusion only | |
| Blebbistatin − LatA | 4 | 5.67 ± 4.19 | N/A | Diffusion only | |
| Fig. 5 D (HeLa JW) | Control | 3 | 5.17 ± 3.95 | 6.40 ± 1.18 | Diffusion-drift |
| Non-targeting control siRNA | 1 | 5.21 | 4.87 | Diffusion-drift | |
| DAAM1 siRNA | 4 | 2.66 ± 2.04 | N/A | Diffusion only | |
| SMIFH2 | 7 | 1.27 ± 0.53 | N/A | Diffusion only | |
| DAAM1 siRNA + DAAM1 | 4 | 7.27 ± 0.69 | 9.26 ± 2.81 | Diffusion-drift | |
| Fig. 6 D (MEF) | Control | 8 | 17.9 ± 13.0 | 10.8 ± 6.50 | Diffusion-drift |
| Control + FlnAb | 3 | N/A | N/A | N/A | |
| FlnA (−/−) | 5 | 16.8 ± 14.7 | N/A | Diffusion only | |
| FlnA (−/−) + FlnA | 7 | 15.5 ± 10.9 | 11.4 ± 3.5 | Diffusion-drift |
N/A, not applicable.
Number of independent experimental samples used for the tracking and calculation.
The experimental data obtained from this perturbation does not fit either drift-diffusion or simple diffusion processes.
We next analyzed the direction by which actin nodes moved relative to their neighboring nodes. The data were generated from one control sample that contained 100-s trajectories of ∼100 nodes. The histogram of the angle (θ) between the direction of movement of an actin node at a given moment and the vectors connecting this node with the neighboring nodes in a few distance ranges were plotted in Fig. 1 D. There is a strong bias for nodes to move toward the neighboring node when the distance from the node center to the neighboring node is <2 µm. This bias still exists when the distance between neighboring nodes is 2–4 µm. When the distance is >4 µm, the distribution of θ is uniform. This indicates that actin nodes attract each other strongly when they are within a 2-µm distance. The interaction diminished at 2–4 µm distance and disappeared when the distance is >4 µm.
Stochastic optical reconstruction microscopy (STORM) was used to obtain images of untreated cells stained with Alexa Fluor 647 phalloidin, which selectively labels F-actin. This revealed a sparse nodal network located between actin stress fibers, where the distance between each node was ∼1–2 µm (Fig. 1 E, left). After LatA treatment, stress fibers were not observed; however, the actin nodal network remained spatially unchanged despite showing enlarged actin nodes (Fig. 1 E, middle). Examining the same specimens using total internal reflection fluorescence (TIRF) microscopy (Fig. 1 E, right) revealed images that were indistinguishable from those obtained using Lifeact to label actin in live cells (Fig. 1 B). These observations indicated that the nodal organization of the actin network existed in the cell before LatA treatment.
Identification of actin-binding proteins associated with the actin nodes
To determine which proteins might be important in the organization of actin multinodal arrays, we analyzed the distribution and dynamics of several major actin-associated proteins relative to the actin nodes. Among those tested were actin cross-linking proteins (FlnA, α-actinin, and β-spectrin), formins (mDia1 and DAAM1), and the major actin-based motor, myosin II (detected by labeling myosin regulatory light chain [MRLC]). Some of these proteins (α-actinin, β-spectrin, and mDia1) did not show any clear association with the actin nodes (Fig. S2). However, three other candidate proteins—myosin II, DAAM1, and FlnA—appeared to be closely associated with actin nodes that emerged after LatA treatment.
Localization and dynamics of myosin foci in the multinodal actin arrays
To follow myosin II dynamics in live cells, we used a GFP-MRLC construct that can label both myosin IIA and IIB. Here, we refer to the MRLC-positive patches as “foci” to distinguish them from the actin-positive patches that we call “nodes.” Myosin foci were usually smaller in size and higher in contrast than actin nodes, but their movement and fusion was similar as revealed by a kymograph (see Fig. 5 A and Fig. S1, C–E) and MSD analysis (see Fig. 5, B and C). Treatment of cells with LatA revealed some overlap between myosin foci and actin nodes (Fig. 2 A). However in most cases, when the actin nodes were small (<1 µm2), myosin foci localized in between neighboring actin nodes aligned with thin actin filaments (Fig. 2 B, 0 s). Larger actin nodes (>1 µm2), which appeared at later times after LatA addition, usually overlapped with the myosin foci (Fig. 2 B, 60 s).
Figure 5.
Perturbation of formins activity altered the actin-myosin node dynamics. (A) 3D kymographs of actin node and myosin foci formation and aggregation. (A, left) 800 nM LatA. (A, right) 20 µM pretreatment of SMIFH2 for 24 h and then 800 nM LatA. Bars, 7 µm. (B and C) MSD analysis from imaging data shown in A, for one single experiment in each case. Data obtained from the experiments are plotted as individual dots. (D) MSD of the control and perturbed samples from HeLa JW cells (summarized in Table 1; error bars indicate SD). (E and F) Comparisons of MSD data from experiment results (red for MSD of actin nodes in the control sample, green for actin nodes in the SMIFH2 treated sample), drift-diffusion model fitting (solid lines), and pure diffusion model fitting (dashed lines). (G) RNAi silencing of mouse DAAM1 was confirmed by immunoblotting.
Figure 2.

Localization and dynamics of actin nodes and myosin II. (A) The dynamic reorganization of F-actin and myosin II was visualized using Lifeact-Ruby and EGFP-tagged MRLC (EGFP-MRLC). MEFs were plated on fibronectin-patterned glass coverslips and then treated with 200 nM LatA. The cell area was marked as the yellow contour. Regions marked in white boxes are magnified in B and show the dynamic reorganization of actin and MRLC. Actin nodes and MRLC first exhibited an alternating pattern (0–20 s, yellow arrows), but the degree of colocalization increases with time. (C) The 3D kymograph of time-lapsed images in B. As multiple actin node fusion and fission events occurred, zigzag patterns appeared in the 3D kymograph. Bars: (A) 10 µm; (B) 2 µm; (C) 5 µm.
Observations of actin and myosin dynamics in LatA-treated cells, as well as kymograph analysis of such dynamics (Fig. 2, B and C; Video 2; and Fig. S1, F and G), revealed that, on average, the degree of overlap between actin and myosin increased as small actin nodes fused to form into bigger nodes (Fig. 2 C). Occasionally, the fission of large nodes was accompanied by an apparent decrease in the overlap of actin and myosin distribution (Video 2 and Fig. S1, F and G).
Movement and fusion of actin nodes depends on myosin II activity
Inhibition of myosin II activity inhibited the movement of actin nodes and myosin foci, and reduced the frequency of fusion events. Time-lapse imaging and kymograph analysis showed that movement and fusion of actin nodes, which were revealed after LatA addition, were significantly suppressed by the addition of 25 µM blebbistatin (Fig. 3, A and B; and Video 3). MSD analysis showed that the character of node movement changed considerably after the addition of blebbistatin. In blebbistatin-treated cells, MSD grew linearly with time so that the node motion follows the pure diffusion model (Fig. 3 C and Table 1). Pretreatment of cells with blebbistatin before LatA addition did not prevent the appearance of actin nodes, but again significantly inhibited their movement and made it purely diffusional (Fig. 3 C).
Figure 3.

Perturbation of myosin altered the actin node dynamics. (A) Actin node movement is reduced by myosin II inhibition. Cells were treated with 200 nM LatA (0 s) until the cell entered phase 2. When active aggregation of actin nodes occurred, 25 µM blebbistatin was added at 35 min. (B) The 3D kymograph of Lifeact-Ruby was created from the marked area (yellow box, 5 × 5 µm2) in A. Initial LatA treatment triggered the appearance and merging of actin nodes. Subsequent blebbistatin treatment decelerated the lateral movement of actin nodes and prevented further aggregation events. (C) MSD of the control and perturbed samples from MEFs (summarized in Table 1, error bars indicate SD). (D) Actin nodes formed in myosin IIA knockdown cells but did not actively move compared with the control. The last frame is the merged images of nodes at 0 s (in red) and 90 s (in green). Node position was strongly altered in control cells but not in myosin IIA knockdown cells. (E) Histogram of the actin node size detected in control and myosin IIA knockdown cells. Total actin nodes n = 2 × 104 for two experiments in control cells and n = 1.6 × 104 for two experiments in myosin IIA knockdown cells. (F) Knockdown of mouse myosin IIA by siRNA was confirmed by Western blotting. Bars: (A) 10 µm; (B) 5 µm; (D) 5 µm.
We have also confirmed the blebbistatin data using myosin IIA knockdown cells. Consistent with the blebbistatin treatment, myosin IIA knockdown cells (Fig. 3 F) showed reduced actin node movement in phase 2 of LatA treatment (Fig. 3, C and D; and Video 4). The nodes still emerged in LatA-treated myosin IIA knockdown cells and their size distribution did not differ significantly from those in control cells (Fig. 3 E). However, the nodes moved slower in the knockdown cells, and the MSD curves characterizing their movement grew with time at the same rate as in control cells treated with blebbistatin (Fig. 3 C).
Localization of formin DAAM1 patches in multinodal actin arrays
The formin DAAM1 has been shown to localize to a subset of stress fibers (Ang et al., 2010). We identified an apparent colocalization of DAAM1 with the actin structures in both untreated and LatA-treated cells. By immunostaining for endogenous DAAM1, and by examining the expression of GFP-DAAM1 in cells, we show that DAAM1 was not only localized to a subset of stress fibers, but also demonstrated patchy distributions outside the stress fibers (Fig. 4 A and Video 1). Moreover, DAAM1 colocalized with actin nodes in LatA-treated cells (Fig. 4 B, Fig. S2 B, and Video 1). To investigate the role of formins in the organization and dynamics of actin nodes, we used a small molecule inhibitor of formin FH2 domains, SMIFH2 (Rizvi et al., 2009). Cells treated with 20 µM of SMIFH2 showed less stress fiber formation, but remaining stress fibers still contained DAAM1 (unpublished data). Pre-incubation with SMIF2 did not prevent the appearance of actin nodes in LatA-treated cells; however, in this case colocalization of DAAM1 patches and actin nodes was absent (Fig. 4, C and D).
Figure 4.
Localization of actin nodes and DAAM1 patches. (A) In untreated cells, DAAM1 antibody and phalloidin staining showed patchy distribution of DAAM1, which colocalized along the actin filamentous network. Boxed regions are enlarged on the right. (B) In live cells, and after 20 min of 800 nM LatA treatment, EGFP-DAAM1 localized with actin nodes. B1 and B2, enlarged views of the boxed regions. (C) In cells pretreated with 20 µM of SMIFH2 for 4 h then 800 nM LatA for 20 min, DAAM1 patches either did not form (C1) or did not colocalize with actin nodes (C2). (D) Colocalization analysis showed that the SMIFH2-LatA–treated samples show significantly less colocalization of EGFP-DAAM1 to actin compared with samples that only received LatA treatment. (N, number of cells for each sample). Error bars indicate SD. (E) Localization of DAAM1 on F-actin by superresolution microscopy. Left: F-actin staining of the entire cell, visualized by Alexa Fluor 633–phalloidin staining. The boxed region is enlarged: PALM image of photoactivatable PA-TagRFP-DAAM1 (middle) and overlaid with the F-actin network (right). Bars: (A–C) 5 µm; (B1, B2, C1, and C2) 2 µm; (E, left) 10 µm; (middle and right) 2 µm.
A closer inspection of DAAM1 distribution using superresolution photoactivated localization microscopy (PALM) with TIRF illumination (PALM-TIRF) confirmed that DAAM1-positive patches are present in cells before LatA treatment (Fig. 4 E). Analysis of these images using radially averaged autocorrelation measurements (Schmid, 2011) enabled the estimation of the mean patch diameter as well as the distance between patches as 0.55 µm and 1.10 µm, respectively. The estimated distance between DAAM1 patches is consistent with the distance between actin nodes in untreated cells (Fig. 1 E, left).
Inhibition of formins or knockdown of DAAM1 reduce the dynamics of the actin nodes
HeLa JW cells were preincubated with 20–40 µM of SMIFH2 for 24 h and then treated with LatA. Actin nodes as well as myosin foci were observed after LatA treatment in both control and formin inhibited cells (Fig. 5 A and Video 5). We analyzed the kymographs characterizing the motion of actin nodes and myosin foci in control and SMIFH2-treated cells. In SMIFH2-treated cells, actin nodes and myosin foci moved slower and fused less frequently (if at all) than in control cells (Fig. 5 A). The MSD time dependence curves for actin nodes (Fig. 5, B and D) and myosin foci (Fig. 5 C) also showed that inhibition of formins led to a shift from drift-diffusion to pure diffusion mode with a very low diffusion coefficient (Table 1).
Since DAAM1, but not mDia1, was shown to localize to the actin nodes, we examined the effect of DAAM1 knockdown on actin node dynamics (Fig. 5, D and E). Two independent siRNA sequences, si2318 and si2832 (Ang et al., 2010), were used for the silencing of endogenous DAAM1. The level of DAAM1 in the siRNA-transfected cells was analyzed using immunoblotting (Fig. 5 E and Fig. S3 C), immunofluorescence (Fig. S3 A), and RT-PCR (Fig. S3 B), and was found to be at least fivefold lower than in control cells. At this reduced level, DAAM1 still demonstrated patchy distribution, as revealed by immunostaining (Fig. S3 A). The MSD analysis demonstrated that DAAM1 depletion, similar to SMIFH2 treatment, inhibited the movement of actin nodes, even though the effect of SMIFH2 was somewhat stronger (Fig. 5 D and Table 1). We were able to rescue the phenotype in DAAM1 siRNA-treated cells by expressing full-length DAAM1-GFP. Expression of exogenous DAAM1-GFP in siRNA-treated cells restored the motility of actin nodes back to the control level (Fig. 5 D and Table 1). Altogether these results indicated that formin- (and, in particular DAAM1)-driven actin polymerization played an important role in the dynamics of actin nodes.
The actin cross-linker FlnA affects the structure and movement of actin nodes
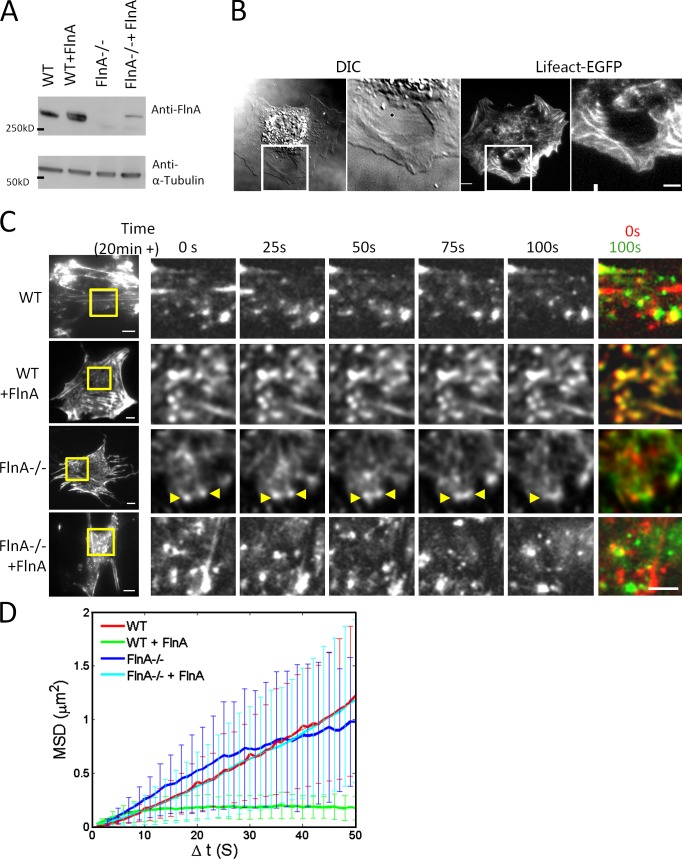
FlnA, a major actin cross-linker, was found to be localized to the actin nodes as revealed by observations of GFP-FlnA in live cells (Fig. S2 A). In all experiments with GFP-FlnA and Lifeact-Ruby expression, localization of FlnA to the actin nodes was clearly seen. To investigate its role in actin node dynamics, we compared the effect of LatA treatment in four cell types: control wild-type MEFs, FlnA knockout cells, and FlnA (−/−) MEFs (Lynch et al., 2011), as well as in control and FlnA (−/−) MEFs transiently transfected with GFP-FlnA. Immunoblot analysis revealed that transfection with GFP-FlnA leads to overexpression of FlnA in control MEFs and can partially restore the level of FlnA in FlnA (−/−) knockout cells (Fig. 6 A). The morphological characteristics of FlnA knockout cells have been described previously (Lynch et al., 2011); in brief, the cells were less spread and have less pronounced actin bundles than their control counterparts (unpublished data). The changes in shape and actin cytoskeleton organization of FlnA (−/−) cells upon LatA treatment were more dramatic as compared with those of control cells. The integrity of the actin network was drastically perturbed, and big holes in the cytoplasm were transiently formed and resealed (Fig. 6 B and Video 6). Large-scale “pulsations” (contraction and relaxation with 1–2 min periodicity) of the actin network in the central region of the cell were often observed (Video 6).
Figure 6.
FlnA levels affect actin node appearance and movement. (A) Immunoblot analysis showing the expression level of FlnA in samples corresponding to the images in C and D. (B) A FlnA (−/−) cell shows a hole in the cytoplasm (white box) during 400 nM LatA treatment as revealed in both differential interference contrast (DIC) and Lifeact-EGFP images. (C) The shape and movement of actin nodes are affected by the level of FlnA. Initial actin node formation was triggered by the addition of 400–800 nM LatA. Actin nodes moved in wild-type samples and FlnA (−/−) rescued samples, but were frozen in FlnA overexpressed cells. FlnA (−/−) cells demonstrated exaggerated dynamics of actin nodes. The boxed regions are enlarged on the right. The yellow arrows showed the merging of one pair of such nodes. The rightmost column shows merged images of actin nodes at 0 s (red) and 100 s (green). (D) MSD of the control and perturbed samples from MEFs or FlnA (−/−) MEFs corresponding to the images in C (summarized in Table 1; error bars indicate SD). Bars: (B, whole cell image) 10 µm; (enlarged image) 5 µm; (C, first column) 10 µm; (C, enlarged images) 5 µm.
When comparing actin organization in LatA-treated wild-type cells with that of FlnA (−/−) cells at higher magnification, we found that FlnA (−/−) cells do not display typical multinodal actin arrays characteristic of control cells. The actin structures observed in these cells were larger in size, and had irregular shapes and diffuse outlines that were difficult to distinguish (Fig. 6 C). Quantification of MSD revealed that these structures were moving faster than the typical actin nodes in wild-type cells for Δt < 35 s, and their motion fitted the pure diffusion mode (Fig. 6 D and Table 1). These fuzzy nodes rapidly aggregated and disassociated in a random manner so that after 50 s only a few original structures were left and further MSD calculations were unreliable. In contrast, the actin multinodal arrays were well developed in the LatA-treated FlnA overexpressing cells (Fig. 6 C); however, the displacement of these nodes was significantly reduced (Fig. 6, C and D). In this case, the motion of the nodes fitted neither pure diffusion nor drift-diffusion modes, but was instead constrained in space. Finally, expression of the exogenous GFP-FlnA in FlnA (−/−) cells rescued the control phenotype. In such cells, the actin nodes were normal in size and shape and moved in a drift-diffusion mode with a velocity comparable to that of control nodes (Fig. 6, C and D; Table 1; and Video 7). Thus, filamin-dependent cross-linking was required for the maintenance and stabilization of the multinodal actomyosin network.
Modeling of the cellular actin network
To better understand how the cytoskeletal network formed and moved, we developed a model of basic cytoskeletal network dynamics. This model assumes that actin filament polymerization occurs by growth from formin aggregates, that filamin cross-links filaments, and that myosin II–driven filament sliding occurs. Previous models (Ziebert and Zimmermann, 2005; Goswami et al., 2008; Gordon et al., 2012) proposed that actomyosin asters self-organize when multivalent myosin clusters slide along actin filaments and subsequently bring their barbed ends together.
Instability in the myosin-sorted actin asters could be due to two reasons: (1) strong contractions could tear actin filaments from the focused asters and destroy them; or (2) formins at the barbed ends of the actin filaments could continue to elongate the filaments, essentially causing the barbed ends to move out from the center of the asters and carry the formins away. However, the repetitively formed, actively moved actin nodes observed after LatA treatment suggest that these nodes are relatively stable; an exception being in the case of filamin depletion.
Thus, our model postulates that formins form multimeric complexes (Copeland et al., 2004; Madrid et al., 2005), bind to each other either directly, or through accessory proteins, and nucleate growing actin filaments that point away from the aggregates, thereby making an aster. Additionally, filamin stabilizes aster focal points by cross-linking actin filaments in the regions of highest filament density, which is near each aster’s focal point. We then assume that myosin filaments slide to the aster focal point, and subsequently “slide off,” or dissociate, from the barbed ends. In the regions where the pointed ends of one aster overlap with antiparallel filaments from neighboring asters, myosin filaments pull the actin filaments together, thereby pulling the asters toward each other. Because actin filaments may radiate in all directions from an aster, contraction would create tension in the actin network. The effect of LatA would be to decrease the number and mean length of the actin filaments within the actin mesh into which the asters are embedded; as a result, inter-aster interactions become more pronounced and eventually create contractile instability across the entire system. Filamin depletion would reduce the strength of the cross-links needed to slow down aster movement and support tension in the network.
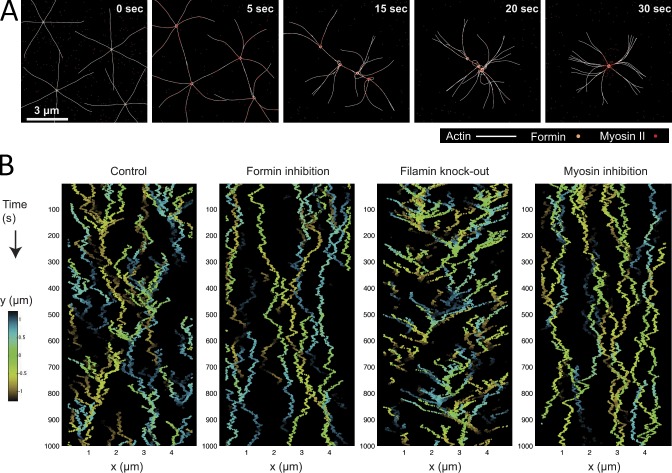
We implemented this model computationally on three scales: First, to address microscopic aster dynamics, we used the Cytosim software (Nedelec and Foethke, 2007) to simulate myosin contraction in regions with overlapping antiparallel filaments from neighboring asters. The results (Fig. 7 A, Fig. S4, and Video 8) showed that the density of myosin molecules increased both in regions where antiparallel filaments from neighboring asters overlapped, and in the immediate vicinity of the asters. This was in agreement with the experimental data (Fig. S4 C). We believe this can be explained by the movement of myosin in regions between asters where antiparallel filaments overlap. In this case half of myosin heads attempt to move toward one aster, while the other half move toward a neighboring cluster. This would result in the contraction and convergence of asters. Furthermore, although myosin filaments do not move and accumulate in regions of overlap, other myosin filaments, which attach to single actin filaments, glide to each aster focal point and subsequently “slide off” or dissociate from the barbed ends. Because of myosin convergence at the focal points, and its transient localization close to them, myosin density is high in these regions.
Figure 7.
Modeling of the cellular actin network. (A) A simulation of myosin II–powered merging of formin-actin nodes. With a simulation condition of myosin abundance and filamin absence, the formin-actin nodes aggregate rapidly. White, actin; green, formin clusters; red, myosin II. (B) A kymograph generated by the computer simulations of aster trajectories. Trajectories of the asters are color-coded so that cold/hot colors correspond to asters that are behind/in front of the visible plane. Frequent aster merging was consistently observed in the wild-type control, whereas this process is abolished when cells had either formin or myosin inhibited. In the case of filamin inhibition, the merging takes less time and the asters are more dynamic. See Materials and methods for detailed parameter settings in the simulation.
As a result of myosin-mediated contraction, asters were seen, in the simulation, to move toward each other and gradually collapse to a single location (Fig. 7 A, 30 s). Such phenomena can only be observed experimentally after treatment with a relatively high concentration of LatA, as at low LatA levels, asters collapsed and disassociated repetitively. By quantitatively analyzing the tracking data of the asters from our experiments, we developed a second, mesoscopic, computational model (see Fig. S5 and description of the computational model in the Materials and methods). The histogram in Fig. S5 E is generated by simulation of the number of nodes for the time period similar to that in the experimental setup shown in Fig. 1 D. This model was calibrated using MSD measurements of the asters in time (Fig. S5 B). After calibration, the predicted kymograph of the asters (Fig. 7 B; by changing the strength of aster interactions corresponding to the experimental perturbations) appeared very similar to those of experimental images.
After calibration, the mesoscopic model correctly predicted several quantitative characteristics of aster movement (Fig. S5, C–F), thereby reinforcing the finding that the model assumptions were correct. Furthermore, the mesoscopic and computational models, as well as a third, analytical macroscopic model that is described in the Materials and methods, predict that there is a randomness involved in aster movements where any given aster can be pulled in various directions, both random and toward multiple neighboring asters, over random time intervals. The pulling toward neighboring asters originates with myosin-powered contraction of the inter-aster antiparallel actin filaments while pulling in random directions could be due to interactions of aster filaments with myosin clusters as well as filaments of the nonaster actin mesh. The macroscopic model showed that these random displacements cannot by themselves prevent network collapse, and this result reflects conclusions drawn from another cytoskeletal system (Ojkic and Vavylonis, 2010). Together with the continuous merging and formation of asters these random movements prevent the network collapse; keeping the network in a connected and contractile state. Thus, formation of nascent asters in random locations (importantly, with the given observed rates) is a factor that ensures the network of asters will not collapse.
These models also helped to explain other observations made during our experiments. Formin inhibition would result in fewer, shorter filaments, and that would cause weaker contraction (Fig. 7 B). Myosin inhibition slowed aster movement because weaker contractile force generates slower aster drift (Fig. 7 B). Overexpression of filamin should have caused an effective increase in cross-linking and resistance to the contraction. Probably not all filamin localizes to aster foci; and some would cross-link overlapping actin filaments from neighboring asters, causing fewer, slower movements. Filamin depletion, however, should cause lower effective resistance and greater contraction between the neighboring asters (Fig. 7 B). Note that in the case of filamin depletion, our microscopic model indicated that greater contractions would result in the collapse of the asters and the loss of network continuity (Fig. 7 B), as observed. These results agree with a recent experimental and theoretical study into the roles of myosin and cross-linkers in contractile ring self-organization in yeast (Laporte et al., 2012).
Discussion
Treatment of cells with LatA enhances the appearance of a dynamic multinodal array of actin. These nodes contain the actin cross-linker FlnA and actin nucleator formin DAAM1. Myosin II foci appear either at the center of the nodes or in between them. These actin nodes move and fuse rapidly, and, occasionally, split. Movement of the nodes is biased toward neighboring nodes, indicating that nodes may attract each other when the distance between them is <4 µm. According to the percolation theory (Quintanilla et al., 2000; Mertens and Moore, 2012), this provides evidence that at a given actin node density (1–2 µm distance between the nodes), the network of nodes is interconnected throughout the cell. Filamin is required for the stability of the actin nodes, while myosin II and formin are required for their rapid movement and fusion. The fact that numerous nodes can be visualized by TIRF microscopy suggests that they are concentrated near the ventral plasma membrane. However, using confocal microscopy, we also observed nodes that were located >300 nm away from the plasma membrane and could therefore hardly be considered to be elements of the cortical actin layer. In addition, the nodes never colocalized with the typical components of the cortical layer, such as spectrin.
Several pieces of evidence indicate that this multinodal actin array was present in the cells before LatA treatment. We demonstrated that in untreated cells, both endogenous DAAM1, visualized by confocal microscopy after immunofluorescence staining, and transfected PA-TagRFP-DAAM1, visualized by PALM microscopy, were located not only at the stress fibers, as shown previously (Ang et al., 2010), but also in numerous patches that showed a similar spatial arrangement to the actin nodes observed in the initial stage of LatA treatment. Moreover, STORM images of endogenous F-actin in untreated cells revealed tiny nodes with a spatial arrangement similar to DAAM1 nodes. These resembled the more prominent nodes found in the cells treated with LatA. These results are consistent with recently published STORM images that revealed multiple aster-like structures in the ventral layer of the actin filament network in cultured cells (Xu et al., 2012). Thus, actin nodes that appear after LatA treatment could be enhanced elements of a preexisting actin network. LatA treatment makes the actin nodes more prominent, most probably by reducing the density of actin filaments not associated with the nodes and, in addition, increasing the density of the filaments associated with the nodes through the activation of formin-mediated actin polymerization, as was previously demonstrated (Higashida et al., 2008).
We introduced a multimodal actin network model that incorporated actin filaments, an actin cross-linker protein, formin, and myosin II. Formulation of the model represents our working hypothesis of the organization of a generic contractile network. According to our model, actin asters are generated by formin driven actin polymerization (we assume that formin molecules form oligomeric complexes as shown in Copeland et al. [2004] and Madrid et al. [2005]), and these asters are stabilized by cross-linker proteins. Both formins and cross-linkers are localized at the center of the asters. Furthermore, the actin filaments from neighboring asters connect to each other via the association of peripheral minus ends of actin filaments with bipolar myosin filaments. These bipolar myosin minifilaments cross-link and contract actin filaments originating from different asters, then slide along them, and transiently accumulate in the centers of fused asters before being released.
The conclusions from this model are in good agreement with the experimental results. The simulations based on the model faithfully reproduce the movement of the actin nodes in various situations (including after the inhibition or depletion of formin, filamin, and myosin), and predict the relocation of myosin from a position in between actin asters to the center of the asters over time. Altogether, the modeling results are in agreement with the notion that a multinodal actin network exists in the cell before LatA treatment. At the same time, the model predicts that a LatA-induced decrease in the number of actin filaments may partially destabilize the multinodal network to subsequently accelerate node movement and fusion events.
A dynamic mode of aster self-organization may provide the optimal structure for mechanical connectivity and contractility associated with dynamic actomyosin networks. Indeed, the level of connectivity in an actin network will be substantially greater when filaments are organized into asters rather than in random arrangements. Such a nodal network can rapidly reorganize upon perturbation to change the mechanical properties of the cell. Thus, cells can rapidly adapt their mechanical properties to chemical perturbations, as well as to mechanics of the extracellular matrix or externally applied forces, as both polymerization of actin by formins, and myosin contractility, are force-dependent
Furthermore, the optimal organization for potential contractility appears to involve asters with actin pointed ends at the periphery and myosin concentrating in between the asters. Myosin action, if unrestrained by the juxtaposed actin mesh, would be streamlined into purely contractile movements, leading to the convergence of the aster focal points. This was recently demonstrated in vitro with preorganized large actin-myosin asters (Thoresen et al., 2011). Cells could control this effect by regulating the actin mesh density. A randomly arranged and cross-linked actin mesh can also contract, but there is no obvious way to reform the mesh once it has contracted (Dasanayake et al., 2011). A potentially analogous actomyosin aster-mediated network contraction was recently proven to underlie yeast cytokinesis (Vavylonis et al., 2008). Last, but not least, a similar actomyosin aster-mediated network contraction is a crucial component of some developmental processes (Munro et al., 2004; Werner et al., 2007; Roh-Johnson et al., 2012). Thus, we suggest that the basic process by which dynamic contractile networks form in cells (Cai et al., 2010; Rossier et al., 2010) relies upon the basic elements of asters; including formins catalyzing actin polymerization, cross-linkers stabilizing the asters, and bipolar myosin filaments to cross-link asters as well as generate the force needed for the dynamics.
Materials and methods
Fabrication of the fibronectin pattern
The procedure for the fabrication of the fibronectin pattern was adapted from Carpi et al. (2011). Here, clean No. 1.0 glass coverslips (VWR VistaVision) were incubated in 0.1 mg/ml PLL(20)-g[3.5]-PEG(2) (SuSoS) as a passivation layer for 1 h, washed with deionized (DI) water, and then dried in a pressurized stream of nitrogen gas. A quartz photomask was generated in-house with line array patterns alternating between 2–4-µm UV-transparent lines and 5–12-µm UV-opaque gaps. PLL-g-PEG–coated glass coverslips were spatially patterned by exposing deep UV (185–254 nm wavelength, 5 min) through the photomask in the contact-printing mode, washing with DI water, and then drying in a pressurized stream of nitrogen gas. PLL-g-PEG–coated glass coverslips with 2–4-µm line areas removed by UV treatment were incubated in 10 µg/ml fibronectin (Sigma-Aldrich) for 1 h, and washed in 1× PBS. Fibronectin selectively bound to areas without PLL-g-PEG and filled the 2–4-µm lines to create adhesive strips for cell attachment.
Cell culture, plasmids, and transfection
Immortalized MEFs (Giannone et al., 2007), FlnA (−/−) MEFs (Lynch et al., 2011), and HeLa JW cells (Paran et al., 2006) were maintained in DMEM medium (Gibco) supplemented with 10% FBS (Gibco), 1% l-glutamine, and 100 IU/mg penicillin-streptomycin (Invitrogen) at 37°C and 5% CO2. Human large cell lung cancer cell line H460 (ATCC HTB-177, purchased from American Type Culture Collection) were maintained in ATCC-formulated RPMI-1640 medium (catalog No. 30-2001).
EGFP-Lifeact (Riedl et al., 2008) and Lifeact-Ruby were gifts from R. Wedlich-Söldner (Institute of Cell Dynamics and Imaging, University of Münster, Münster, Germany). EGFP-MRLC, mRFP-MRLC, mRFP–β-actin, EGFP–β-actin, EGFP–α-actinin, EGFP–β-spectrin, and EGFP-FlnA were from our previous work (Giannone et al., 2004, 2007; Rossier et al., 2010). Full-length GFP-DAAM1 was a gift from E. Manser (Institute of Molecular and Cell Biology, A*STAR Singapore, Singapore, Ang et al., 2010). PATag-RFP was provided by V. Verkhusha (Gruss-Lipper Biophotonics Center, Albert Einstein College of Medicine, Bronx, NY; Subach et al., 2010). The PATag-RFP construct containing full-length DAAM1 was generated by subcloning DAAM1 from the GFP-DAAM1 plasmid. Transient transfection of plasmids was performed through electroporation using a Neon Transfection System (Invitrogen) 24 h before imaging.
Transfection of siRNA
Cells were transfected with 120 pmol of DAAM1 siRNA using Lipofectamine RNAiMAX (Invitrogen) and incubated for 48 h before imaging experiments. In all cases, transfection of plasmid was done 24 h after the initial siRNA transfection. The forward strands of the DAAM1 siRNA were si2318 = 5′-GGUUGCAAUCGCUGUACUU-3′ and si2832 = 5′-CCUUCUAGCAGAAGCUAAA-3′ corresponding to the nucleotide position. The nontargeting control siRNA sequence used was 5′-GCUGUCACAGGGGAGUUUACG-3′ (Ang et al., 2010). A Cy5 tag at the 5′ end of the sense strand was added to check the transfection efficiency.
Chemicals, antibodies, and immunoblotting
All reagents used in this study are chemical grade and were purchased as indicated. LatA (L5163), Blebbistatin (B0560), and Cytochalasin D (C8273) were purchased from Sigma-Aldrich. The small molecule formin inhibitor, SMIFH2 (catalogue No. 5992446) was purchased from ChemBridge Corporation. All chemicals were dissolved in DMSO and then use at the indicated concentrations.
Affinity-purified rabbit polyclonal antibody against human DAAM1 was provided by E. Manser (Ang et al., 2010). Rabbit anti-Filamin A antibody (SAB4500951) and mouse monoclonal anti–α-tubulin (T6199) were purchased from Sigma-Aldrich. Non-muscle myosin heavy chain II-A polyclonal antibody (PRB-440P) and nonmuscle myosin heavy chain II-B polyclonal antibody (PRB-445P) were purchased from Covance. Alexa Fluor–conjugated secondary antibodies, Alexa Fluor 647–phalloidin (A22287), and Alexa Fluor 633–phalloidin (A22284) were purchased from Invitrogen.
Transfected cells were lysed in RIPA buffer and cell extracts were then subjected to SDS-PAGE. Proteins were transferred to PVDF membranes at 100 V for 2 h and blocked for 1 h with 10% low-fat milk before the addition of primary antibodies. Primary antibodies were either added for 2 h at room temperature or overnight at 4°C. After washes, the membrane was incubated with HRP-conjugated secondary antibodies for 1 h. Bound antibodies were detected by Immun-Star HRP Chemiluminescent reagent (Bio-Rad Laboratories). All antibodies were diluted in 3% BSA in PBS.
Immunofluorescence
Cells were fixed and simultaneously permeabilized at 37°C in a mixture of 3% paraformaldehyde, 0.2% glutaraldehyde, and 0.25% Triton X-100 (Sigma-Aldrich) in PBS for 15 min, then washed twice in PBS for 10 min each time. Before blocking and antibody staining, cells were treated with 10 mg/ml sodium borohydride in cytoskeleton buffer (10 mM MES, 150 mM NaCl, 5 mM EGTA, 5 mM MgCl2, and 5 mM glucose, pH 6.1) for 15 min on ice. 3% of BSA was used to block overnight before PALM imaging. Fixed cells were incubated with primary antibodies, washed, and then followed by Alexa Fluor–conjugated secondary antibodies.
STORM sample preparation
Cells were cultured on cover glasses coated with gold nanoparticles patterned with fibronectin lines. Gold nanoparticles of 80–100 nm were sparsely absorbed (∼2,000 per mm2) to the coverglass surface and immobilized by 30–50 nm of sputtered SiO2 (Kanchanawong et al., 2010). Cells were fixed by first using 0.3% glutaraldehyde mixed with 0.25% Triton X-100 in PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH 6.9) for 2 min and then in 2% glutaraldehyde for 10 min. After rinsing three times with PHEM buffer, the cells were treated with freshly prepared 0.1% NaBH4 for 10 min. The cells were washed with PHEM three times. After washing, cells were incubated with 0.5 µM Alexa Fluor 647–phalloidin in 4°C overnight. Before imaging, the imaging buffer containing glucose-oxidate/catalase oxygen scavenger system was freshly prepared as follows: 0.1 M cysteamine (30070; Sigma-Aldrich), 0.25 M glucose (Fluka G0350500; Sigma-Aldrich), 613 µM Trolox (238813; Sigma-Aldrich), 1 g/liter glucose oxidase (G2133; Sigma-Aldrich), and 0.4 mg/liter catalase (C9322; Sigma-Aldrich). The samples were sealed quickly after mixing the enzyme.
Microscope image acquisition
Transfected cells were fully spread on fibronectin-patterned cover glass for 2–4 h, and then loaded into a sealed live-cell imaging chamber (37°C, 5% CO2) for imaging in DMEM. Time-lapse TIRF images were acquired every 1–5 s using an inverted TIRF microscope (IX81; Olympus) with a 100× (NA 1.49) oil UApo N TIRF objective lens (Olympus), equipped with dual Photometrics Evolve512 EM charge-coupled device (CCD) cameras for simultaneously acquiring GFP and RFP channels. Image acquisition was controlled by MetaMorph software (Molecular Devices). Time-lapse confocal images were acquired every 4–10 s using a confocal microscope (LSM 710 ConfocorIII; Carl Zeiss) with an Plan-Apochromat 100× oil objective lens (NA 1.46) or C-Apochromat 63× water objective lens (NA 1.2). Adjustments to brightness and contrast were performed with FIJI software. Images were acquired using ZEN microscope software.
PALM and STORM images were taken at room temperature. A superresolution microscope (Elyra; Carl Zeiss) with an EM CCD camera (iXon 897; Andor Technology) was used for PALM imaging via TIRF illumination, using a Plan-Apochromat 100× oil objective lens (NA 1.46; Carl Zeiss). Images then reconstructed using QuickPALM in FIJI (Henriques et al., 2010). A Nikon N-STORM system equipped with an EM CCD camera (DU897; Andor Technology) was used for STORM imaging (CFI Plan-Apochromat TIRF 100× NA 1.49 objective lens). 20,000–60,000 images were acquired at 33 ms per frame then reconstructed using the PeakSelector software developed in IDL (ITT Visual Information Solutions; Betzig et al., 2006; Shtengel et al., 2009; Kanchanawong et al., 2010). The images were rendered using the position and localization uncertainty of the identified peaks from single molecule localization analysis. Each localization point was represented by a normalized two-dimensional Gaussian, where width was proportional to localization uncertainty.
Image analysis
An ImageJ plugin of Intensity Correlation Analysis (Li et al., 2004) was used for colocalization analysis. A time series of stack images of 100 slides at 20 × 20 µm2 was first subjected to background subtraction and then analyzed. Each stack was selected from the nonadhesive region of one cell from one experiment. Radially averaged autocorrelation was performed using an ImageJ Macro of the same name provided from the ImageJ wiki (Schmid, 2011).
Particle tracking and MSD calculation
The 3D kymograph and particle tracking was done using Imaris 7.3 (Bitplane AG). Particle identification was first performed to obtain node statistics such as the spatial and temporal positions (x, y, t), node fluorescence intensity, node size, and density. Particle tracking was performed using an autoregressive motion algorithm. This algorithm models the motion of each identified object as an autoregressive process of order 1. A merging event of two particles was identified as the disappearance of one of the two merging particles. Tracking data from Imaris with position (x, y) and time (t) information was further processed using Matlab (R2010a; MathWorks). To adapt to different acquisition rates (0.1–1 Hz), the trajectories were first interpolated into 1 Hz. Instead of presenting subtrajectories one by one, the N trajectories detected within one sample were averaged to avoid bias in selecting trajectories. For N trajectories each with mn time points identified (track length = mn s), the total number of displacement for interval Δt = 1, 2, … s will be
The MSD is then calculated as
As ND decreases exponentially when Δt increases, MSD becomes noisy with small ND and large Δt. Therefore, all the MSD calculated with ND < 100 were discarded. The maximum of Δt was chosen to be 100 s for pure diffusion or drift-diffusion model fitting using ordinary least square fitting using
for pure diffusion and
for drift-diffusion.
Statistical test
An F-test for nested models (Kutner et al., 2004) was used for testing the statistical significance of drift in addition to diffusion of the actin node movement, instead of the pure diffusion model. The best fit for both models was first computed and then the F-statistics established. If the p-value was <0.05, the pure diffusion model was rejected and diffusion-drift was favored.
The mean values and SD (mean ± SD) of diffusion and drift coefficients were reported from the fits to the MSD data from all samples pooled together. The SDs were calculated as follows: first, polynomial fits are made for each cell separately, and respective diffusion and drift coefficients are calculated for each cell. After this, the SDs are calculated to account for variance between the cells. Note that although the calculated coefficient means for all cells would be different from those reported from the MSD data fits from all cells pooled together, the difference is negligible. The SDs are not reported for n = 1 cases.
Simulations
Both an agent-based stochastic modeling approach and analysis of partial differential equations were used to simulate the self-organization of the actin network and aster dynamics.
Tracking data used to calibrate the model.
We used the quantitative data from one control sample containing ∼100 aster trajectories to establish four characteristics of the aster movements. First, we computed the MSD of asters that have a small parabolic component in addition to the straight line (Fig. S5 B, experimental data), which means that the asters both drift and diffuse (the F-test confirmed that the drift part is statistically significant). Second, we measured the mean aster displacement over 10-s intervals (Fig. S5 C, left bar), which was ∼0.1 µm. Third, we measured the consecutive displacement vectors of the asters over 10-s intervals and found the average dot products of these displacements normalized by dividing by the MSDs (Fig. S5 D, left bar). The average normalized dot product, ∼0.2, is nonzero, indicating that over the short time intervals there is a nonrandom drift component of aster movement, in addition to the random walk. Fourthly, by taking the direction of the 10-s displacement of the given aster angle as equal to zero, we established the angular distributions of asters around (≤4 µm) any given aster. After averaging such angular distributions, it was clear (Fig. S5 E) that there was a higher density of neighboring asters in the direction of movement of any given aster. Qualitatively, the data from all samples exhibited the directional basis presented in Figs. 1 D and S5 E. This observation explains the origin of the effective drift apparent in the MSD: the neighboring asters effectively attract each other. The observation that the aster MSD increased with time dropped drastically after myosin inhibition (Fig. 3 C), which suggests that the asters effectively attract each other through the actin-myosin contraction. Furthermore, the significant variance of aster displacement, together with direct observations of aster tracks, show that there is a significant random element to aster movement. This randomness is likely caused by the random time of interaction between aster pairs, the randomness of the subset of asters with which a given aster interacts at any given time, and the interactions between asters with the nonaster actin mesh. The tracking data indicates that most of the asters disappeared by merging, and the lifetime of an aster ranges from 10 to 500 s. Also, nascent asters appeared at apparently random locations, balancing the number of disappearing asters.
Discrete stochastic computational model.
We incorporated these data into a discrete stochastic computational model, in which the asters interacted randomly with subsets of other neighboring asters within a 4-µm radius (which was achieved by a probability, exponentially decreasing with distance, to interact, with characteristic length in the exponential function on the order of 2.5 µm), converge in small increments according to these interactions and made random displacements. The random displacements amplitude was proportional to the local number of neighboring asters. More specifically, a few hundred nodes moved in the simulations during each time step by (1) making a diffusion-like random displacement, the magnitude of which is proportional to the average diffusion coefficient and the average local node density (in the ∼4-µm vicinity of the given node); and (2) by directed displacement, which is determined by weighted vector sum of pairwise attractions to the neighboring nodes. This directed displacement is characterized by the effective drift rate. Thus, at a given average node density, which is achieved at known rates of fusion and emergence of the nodes, and at the measured radius of interactions, two parameters—effective diffusion and drift coefficients—fully characterize node movement. We constrained these two coefficients by fitting the observed MSD as a function of time (Fig. S5 B, red solid line), as well as the average 10-s displacement (Fig. S5 C, right bar). Effectively, the asters diffused with the effective diffusion coefficient on the order of 0.001–0.01 µm2/s and drifted up the gradient of aster density with an mean drift rate of 0.001–0.01 µm/s. Additional model parameters were: a threshold distance of 0.1 µm below which the neighboring nodes merged; the rate of random node emergence was chosen to maintain the observed node density. The simulations confirmed that with these parameters, the average lifetime of the node was 200–300 s. After that, without additional parameter fitting, the model correctly predicted: (1) average dot products of consecutive displacements (Fig. S5 D, right bar) and (2) directionality of displacement up the density gradient (Fig. S5 F). At these parameters, the aster population in the simulations remained dynamic but relatively uniform in space (Fig. S5 A). This illustrated that random displacement and random emergence of asters prevent network collapse. Fig. S5 A shows that at any given moment the number of myosin-mediated interactions between neighboring asters is such that the overlapping interacting actin-myosin pairs interconnect the whole network. To simulate the formin inhibition case, we decreased the effective actin filament length to 40% of that in the control case. To simulate the filamin depletion case, we decreased the resistance to myosin pulling twice, and to simulate the myosin inhibition case, we decreased the myosin strength threefold compared with the control. Simulations of this stochastic computational model also produced the kymographs shown in Fig. 7 B. The simulations of the discrete stochastic model were done using Matlab. The M-file used in the simulations of the discrete stochastic computational model is available as a Supplemental text file.
Cytosim model.
To simulate aster merging, we used the freely available Cytosim software. In these simulations, we used actin bending rigidity 0.05 pN × µm2, aster’s effective viscous drag coefficient 10 pN × s/µm, 11 myosin molecules per square micrometer, myosin-actin on-rate 100 s−1, and off-rate 0.02 s−1, myosin free velocity 0.1 µm/s, and stall force 6 pN. In addition, in the analytical model, we used the measured rate by which asters merge on the order of 0.01/s.
Macroscopic mathematical model.
The following continuous model can be investigated analytically and helps to understand the conditions for the stability of the aster network. In the continuous deterministic approximation, the asters can be described by the density (number of asters per unit area). The asters undergo effective density-dependent diffusion (they displace randomly only when there are other asters in the local vicinity), and their effective diffusion coefficient, D = αA, is proportional to the local aster density. The observations show that the asters drift up the gradient of the aster density, so their instantaneous velocity can be approximated by the expression . The rate of merging is proportional to the square of the aster density, assuming mass action kinetics of the merging, so the resulting equation governing this density has the form:
where S is the rate of nascent asters emergence and m is a constant coefficient. Substitution of the expressions for drift and diffusion gives us:
or
The uniform constant density is the steady-state. We scale the density by its steady-state value: and rescale time and spatial coordinate as follows: , where L is a characteristic size of the cell domain where the asters are observed. Then, the rescaled nondimensional equation has the form:
where
To investigate the stability, we introduce the small deviation of the density from the steady-state: and obtain the linearized equation:
Substituting the form , we obtain the dispersion relation:
According to parameters of the discrete stochastic model at which we achieved a good fit with our data, β/α ≈ 4: we found good fit at D = 0.001 – 0.01 µm2/s and V = 0.001 – 0.01 µm/s, so D/V = 1 µm. However, D/V = α/(β/l), where l = ∼4 µm is the characteristic range of aster interactions that is the spatial scale for the density gradient in the expression for the velocity. Thus, β/α ≈ 4 µm/1 µm ≈ 4. Parameter ε is defined as the diffusion coefficient of a characteristic aster multiplied by the mean time of the aster existence before its merger with another aster and divided by the size of the domain where asters exist,
Thus, λ ≈ 0.03 q2 − 2. The collapse takes place for q > 8, which means that the characteristic spatial scale on which the asters would collapse onto each other is L/q ≈ 10 µm/8 ≈ 1.25 µm, but this is the average distance between the neighboring asters, so there will be no macroscopic collapse of the aster network.
Online supplemental material
Fig. S1 shows that LatA concentrations affected the duration of phase 1 but not the node size or density. The figure is a montage from Video 2 showing examples of fission and fusion of actomyosin nodes after LatA treatment. Fig. S2 shows the distribution and the dynamics of the major actin-associated proteins with the multi-nodal actin array. Fig. S3 shows additional data on DAAM1 knockdown (related to Fig. 5). Fig. S4 shows the simulation of myosin-powered collapse of the initially square-lattice-like network of formin-actin asters (related to Fig. 7). Fig. S5 is a snapshot of the discrete stochastic model simulation and comparison of experimental data and model simulation. Video 1 shows actin and DAAM1 dynamics in a HeLa JW cell after 800 nM LatA treatment (related to Fig. 4 B). Video 2 shows a typical response of MEF cell to mild LatA treatment (related to Fig. 2). Video 3 shows active aster movement appearing after LatA addition was stopped by blebbistatin treatment (related to Fig. 3, A and B). Video 4 compares actin node movement in control cells and myosin IIA knockdown cells (related to Fig. 3 D). Video 5 compares actin node movement with and without formin inhibition by SMIFH2 (related to Fig. 5 A). Video 6 compares actin node movement in different FlnA perturbations (related to Fig. 6 C). Video 7 shows the response to LatA treatment in FlnA (−/−) cell (related to Fig. 6 B). Video 8 is the simulation of myosin-powered merging of a few formin-actin asters (related to Fig. 7 A). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201210123/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201210123.dv.
Supplementary Material
Acknowledgments
The authors acknowledge helpful discussions with Dr. Edward Manser and Drs. Naila Alieva and Ronen Zaidel-Bar (Mechanobiology Institute [MBI] Singapore). We thank Dr. Edward Manser and Dr. Roland Wedlich-Soldner for kind gifts of plasmid and antibodies, and Dr. Pakorn (Tony) Kanchanawong (MBI Singapore) for guidance of STORM imaging. We are grateful to Steven Wolf (MBI Singapore) for expert help in preparing this article for publication. All microscopy was carried out at the microscopy core facility, MBI, National University of Singapore.
This research was supported by the National Research Foundation (NRF), Singapore, and Ministry of Education (MOE), Singapore, and funding to the MBI, National University of Singapore. J. Allard and A. Mogilner were supported by the National Science Foundation grant DMS-1118206 and National Institutes of Health grant GM068952. A.D. Bershadsky holds the Joseph Moss Professorial Chair in Biomedical Research at the Weizmann Institute and is a visiting professor at the National University of Singapore, and acknowledges support from Israel Science Foundation (grant No. 956/10), and a Lord David Alliance Manchester-Weizmann grant.
Footnotes
Abbreviations used in this paper:
- DAAM1
- disheveled-associated activator of morphogenesis 1
- FlnA
- filamin A
- LatA
- Latrunculin A
- MEF
- mouse embryonic fibroblast
- MRLC
- myosin regulatory light chain
- MSD
- mean square displacement
- PALM
- photoactivated localization microscopy
- STORM
- stochastic optical reconstruction microscopy
- TIRF
- total internal reflection fluorescence
References
- Ang S.F., Zhao Z.S., Lim L., Manser E. 2010. DAAM1 is a formin required for centrosome re-orientation during cell migration. PLoS ONE. 5:e13064 10.1371/journal.pone.0013064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S., Davidson M.W., Lippincott-Schwartz J., Hess H.F. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 313:1642–1645 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- Cai Y., Sheetz M.P. 2009. Force propagation across cells: mechanical coherence of dynamic cytoskeletons. Curr. Opin. Cell Biol. 21:47–50 10.1016/j.ceb.2009.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Rossier O., Gauthier N.C., Biais N., Fardin M.-A., Zhang X., Miller L.W., Ladoux B., Cornish V.W., Sheetz M.P. 2010. Cytoskeletal coherence requires myosin-IIA contractility. J. Cell Sci. 123:413–423 10.1242/jcs.058297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpi N., Piel M., Azioune A., Fink J. 2011. Micropatterning on glass with deep UV. Nature Protocol Exchange. 10.1038/protex.2011.238 [DOI] [Google Scholar]
- Chesarone M.A., DuPage A.G., Goode B.L. 2010. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 11:62–74 10.1038/nrm2816 [DOI] [PubMed] [Google Scholar]
- Copeland J.W., Copeland S.J., Treisman R. 2004. Homo-oligomerization is essential for F-actin assembly by the formin family FH2 domain. J. Biol. Chem. 279:50250–50256 10.1074/jbc.M404429200 [DOI] [PubMed] [Google Scholar]
- Dasanayake N.L., Michalski P.J., Carlsson A.E. 2011. General mechanism of actomyosin contractility. Phys. Rev. Lett. 107:118101 10.1103/PhysRevLett.107.118101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G., Dubin-Thaler B.J., Döbereiner H.-G., Kieffer N., Bresnick A.R., Sheetz M.P. 2004. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 116:431–443 10.1016/S0092-8674(04)00058-3 [DOI] [PubMed] [Google Scholar]
- Giannone G., Dubin-Thaler B.J., Rossier O., Cai Y., Chaga O., Jiang G., Beaver W., Döbereiner H.-G., Freund Y., Borisy G., Sheetz M.P. 2007. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 128:561–575 10.1016/j.cell.2006.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D., Bernheim-Groswasser A., Keasar C., Farago O. 2012. Hierarchical self-organization of cytoskeletal active networks. Phys. Biol. 9:026005 10.1088/1478-3975/9/2/026005 [DOI] [PubMed] [Google Scholar]
- Goswami D., Gowrishankar K., Bilgrami S., Ghosh S., Raghupathy R., Chadda R., Vishwakarma R., Rao M., Mayor S. 2008. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell. 135:1085–1097 10.1016/j.cell.2008.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R., Lelek M., Fornasiero E.F., Valtorta F., Zimmer C., Mhlanga M.M. 2010. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat. Methods. 7:339–340 10.1038/nmeth0510-339 [DOI] [PubMed] [Google Scholar]
- Higashida C., Suetsugu S., Tsuji T., Monypenny J., Narumiya S., Watanabe N. 2008. G-actin regulates rapid induction of actin nucleation by mDia1 to restore cellular actin polymers. J. Cell Sci. 121:3403–3412 10.1242/jcs.030940 [DOI] [PubMed] [Google Scholar]
- Kanchanawong P., Shtengel G., Pasapera A.M., Ramko E.B., Davidson M.W., Hess H.F., Waterman C.M. 2010. Nanoscale architecture of integrin-based cell adhesions. Nature. 468:580–584 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.Y., Davidson L.A. 2011. Punctuated actin contractions during convergent extension and their permissive regulation by the non-canonical Wnt-signaling pathway. J. Cell Sci. 124:635–646 10.1242/jcs.067579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenderink G.H., Dogic Z., Nakamura F., Bendix P.M., MacKintosh F.C., Hartwig J.H., Stossel T.P., Weitz D.A. 2009. An active biopolymer network controlled by molecular motors. Proc. Natl. Acad. Sci. USA. 106:15192–15197 10.1073/pnas.0903974106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutner M.H., Nachtsheim C.J., Neter J., Li W. 2004. Applied linear statistic models. McGraw-Hill/Irwin, Boston 1,396 pp [Google Scholar]
- Laporte D., Coffman V.C., Lee I.J., Wu J.Q. 2011. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J. Cell Biol. 192:1005–1021 10.1083/jcb.201008171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Ojkic N., Vavylonis D., Wu J.Q. 2012. α-Actinin and fimbrin cooperate with myosin II to organize actomyosin bundles during contractile-ring assembly. Mol. Biol. Cell. 23:3094–3110 10.1091/mbc.E12-02-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Lau A., Morris T.J., Guo L., Fordyce C.B., Stanley E.F. 2004. A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J. Neurosci. 24:4070–4081 10.1523/JNEUROSCI.0346-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch C.D., Gauthier N.C., Biais N., Lazar A.M., Roca-Cusachs P., Yu C.H., Sheetz M.P. 2011. Filamin depletion blocks endoplasmic spreading and destabilizes force-bearing adhesions. Mol. Biol. Cell. 22:1263–1273 10.1091/mbc.E10-08-0661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid R., Gasteier J.E., Bouchet J., Schröder S., Geyer M., Benichou S., Fackler O.T. 2005. Oligomerization of the diaphanous-related formin FHOD1 requires a coiled-coil motif critical for its cytoskeletal and transcriptional activities. FEBS Lett. 579:441–448 10.1016/j.febslet.2004.12.009 [DOI] [PubMed] [Google Scholar]
- Mandato C.A., Bement W.M. 2001. Contraction and polymerization cooperate to assemble and close actomyosin rings around Xenopus oocyte wounds. J. Cell Biol. 154:785–797 10.1083/jcb.200103105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.C., Kaschube M., Wieschaus E.F. 2009. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 457:495–499 10.1038/nature07522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medalia O., Weber I., Frangakis A.S., Nicastro D., Gerisch G., Baumeister W. 2002. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 298:1209–1213 10.1126/science.1076184 [DOI] [PubMed] [Google Scholar]
- Mertens S., Moore C. 2012. Continuum percolation thresholds in two dimensions. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 86:061109 10.1103/PhysRevE.86.061109 [DOI] [PubMed] [Google Scholar]
- Mizuno D., Tardin C., Schmidt C.F., Mackintosh F.C. 2007. Nonequilibrium mechanics of active cytoskeletal networks. Science. 315:370–373 10.1126/science.1134404 [DOI] [PubMed] [Google Scholar]
- Mogilner A., Keren K. 2009. The shape of motile cells. Curr. Biol. 19:R762–R771 10.1016/j.cub.2009.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E., Nance J., Priess J.R. 2004. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell. 7:413–424 10.1016/j.devcel.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Nedelec F., Foethke D. 2007. Collective Langevin dynamics of flexible cytoskeletal fibers. New J. Phys. 9:427 10.1088/1367-2630/9/11/427 [DOI] [Google Scholar]
- Ojkic N., Vavylonis D. 2010. Kinetics of myosin node aggregation into a contractile ring. Phys. Rev. Lett. 105:048102 10.1103/PhysRevLett.105.048102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paran Y., Lavelin I., Naffar-Abu-Amara S., Winograd-Katz S., Liron Y., Geiger B., Kam Z. 2006. Development and application of automatic high-resolution light microscopy for cell-based screens. Methods Enzymol. 414:228–247 10.1016/S0076-6879(06)14013-6 [DOI] [PubMed] [Google Scholar]
- Pollard T.D. 2010. Mechanics of cytokinesis in eukaryotes. Curr. Opin. Cell Biol. 22:50–56 10.1016/j.ceb.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Sheetz M.P., Elson E.L. 1991. Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys. J. 60:910–921 10.1016/S0006-3495(91)82125-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla J., Torquato S., Ziff R.M. 2000. Efficient measurement of the percolation threshold for fully penetrable discs. J. Phys. A Math. Gen. 33:L399–L407 10.1088/0305-4470/33/42/104 [DOI] [Google Scholar]
- Rauzi M., Lenne P.F., Lecuit T. 2010. Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature. 468:1110–1114 10.1038/nature09566 [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi S.A., Neidt E.M., Cui J., Feiger Z., Skau C.T., Gardel M.L., Kozmin S.A., Kovar D.R. 2009. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16:1158–1168 10.1016/j.chembiol.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh-Johnson M., Shemer G., Higgins C.D., McClellan J.H., Werts A.D., Tulu U.S., Gao L., Betzig E., Kiehart D.P., Goldstein B. 2012. Triggering a cell shape change by exploiting preexisting actomyosin contractions. Science. 335:1232–1235 10.1126/science.1217869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossier O.M., Gauthier N., Biais N., Vonnegut W., Fardin M.A., Avigan P., Heller E.R., Mathur A., Ghassemi S., Koeckert M.S., et al. 2010. Force generated by actomyosin contraction builds bridges between adhesive contacts. EMBO J. 29:1055–1068 10.1038/emboj.2010.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M. 1982. Action of cytochalasin D on cytoskeletal networks. J. Cell Biol. 92:79–91 10.1083/jcb.92.1.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M. 2011. Radially Averaged Autocorrelation. http://imagejdocu.tudor.lu/doku.php?id=macro:radially_averaged_autocorrelation (accessed September 27, 2011)
- Shtengel G., Galbraith J.A., Galbraith C.G., Lippincott-Schwartz J., Gillette J.M., Manley S., Sougrat R., Waterman C.M., Kanchanawong P., Davidson M.W., et al. 2009. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc. Natl. Acad. Sci. USA. 106:3125–3130 10.1073/pnas.0813131106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares e Silva M., Depken M., Stuhrmann B., Korsten M., MacKintosh F.C., Koenderink G.H. 2011. Active multistage coarsening of actin networks driven by myosin motors. Proc. Natl. Acad. Sci. USA. 108:9408–9413 10.1073/pnas.1016616108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach F.V., Patterson G.H., Renz M., Lippincott-Schwartz J., Verkhusha V.V. 2010. Bright monomeric photoactivatable red fluorescent protein for two-color super-resolution sptPALM of live cells. J. Am. Chem. Soc. 132:6481–6491 10.1021/ja100906g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T.M., Shevelev A.A., Bershadsky A.D., Gelfand V.I. 1984. Cytoskeleton of mouse embryo fibroblasts. Electron microscopy of platinum replicas. Eur. J. Cell Biol. 34:64–74 [PubMed] [Google Scholar]
- Svitkina T.M., Verkhovsky A.B., McQuade K.M., Borisy G.G. 1997. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 139:397–415 10.1083/jcb.139.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen T., Lenz M., Gardel M.L. 2011. Reconstitution of contractile actomyosin bundles. Biophys. J. 100:2698–2705 10.1016/j.bpj.2011.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D., Wu J.-Q., Hao S., O’Shaughnessy B., Pollard T.D. 2008. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 319:97–100 10.1126/science.1151086 [DOI] [PubMed] [Google Scholar]
- Verkhovsky A.B., Svitkina T.M., Borisy G.G. 1997. Polarity sorting of actin filaments in cytochalasin-treated fibroblasts. J. Cell Sci. 110:1693–1704 [DOI] [PubMed] [Google Scholar]
- Werner M., Munro E., Glotzer M. 2007. Astral signals spatially bias cortical myosin recruitment to break symmetry and promote cytokinesis. Curr. Biol. 17:1286–1297 10.1016/j.cub.2007.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.Q., Sirotkin V., Kovar D.R., Lord M., Beltzner C.C., Kuhn J.R., Pollard T.D. 2006. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 174:391–402 10.1083/jcb.200602032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Babcock H.P., Zhuang X. 2012. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat. Methods. 9:185–188 10.1038/nmeth.1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebert F., Zimmermann W. 2005. Nonlinear competition between asters and stripes in filament-motor systems. Eur. Phys. J. E Soft Matter. 18:41–54 10.1140/epje/i2005-10029-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.