Abstract
Retinoic acid (RA) is present at sites of neurogenesis in both the embryonic and adult brain. While it is widely accepted that RA signaling is involved in the regulation of neural stem cell differentiation, little is known about vitamin A utilization and biosynthesis of active retinoids in the neurogenic niches, or about the details of retinoid metabolism in neural stem cells and differentiating progenies. Here we provide data on retinoid responsiveness and RA production of distinct neural stem cell/neural progenitor populations. In addition, we demonstrate differentiation-related changes in the expression of genes encoding proteins of the retinoid machinery, including components responsible for uptake (Stra6) and storage (Lrat) of vitamin A, transport of retinoids (Rbp4, CrbpI, CrabpI-II), synthesis (Rdh10, Raldh1-4), degradation of RA (Cyp26a1-c1) and RA signaling (Rarα,β,γ, Rxrα,β,γ). We show that both early embryonic neuroectodermal (NE-4C) stem cells and late embryonic or adult derived radial glia like progenitors (RGl cells) are capable to produce bioactive retinoids but respond differently to retinoid signals. However, while neuronal differentiation of RGl cells can not be induced by RA, neuron formation by NE-4C cells is initiated by both RA and RA-precursors (retinol or retinyl acetate). The data indicate that endogenous RA production, at least in some neural stem cell populations, may result in autocrine regulation of neuronal differentiation.
Introduction
All-trans retinoic acid (RA), a derivative of vitamin A, is one of the most powerful morphogens governing the development of various tissues and organs, including the central nervous system. RA has been reported to regulate neuron formation both in the embryonic, as well as in the postnatal/adult brain [1–4] and has been widely used as a potent inducer of neuronal differentiation in various multipotent cell populations (embryonic carcinoma cells, embryonic and neural stem cells, induced pluripotent stem cells), in vitro.
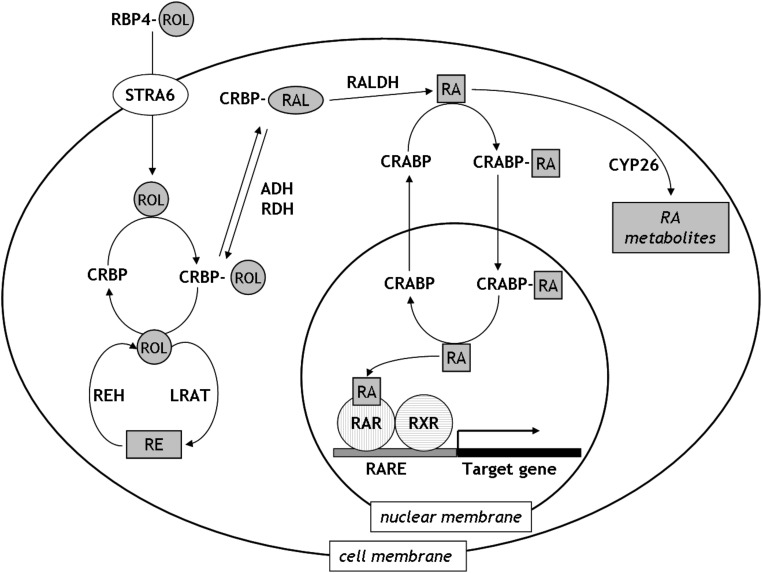
The major molecular mechanisms that control RA availability and signaling were thoroughly characterized over the last decades [5–9]. In brief, extracellular retinol (ROL) is delivered to the tissues via ROL binding protein 4 (RBP4), which associates with the transthyretin carrier in the circulation (Fig. 1). ROL can be taken up by cells via facilitated transport upon RBP4-ROL binding to the STRA6 transporter [10]. Once within the cell, ROL can either be stored after conversion to retinyl esters (RE) by lecithin retinol acyltransferase (LRAT, [11]) or it can be directed towards RA synthesis. In the latter case, ROL is reversibly converted to retinaldehyde (RAL) by ROL/alcohol dehydrogenases (RDHs/ADHs), among which RDH10 seems to play a primary role [12,13]. RAL is irreversibly oxidized to all-trans RA by RALDH1-3 retinaldehyde dehydrogenases, while RALDH4 was shown to be involved in the biosynthesis of 9-cis RA [14]. Oxidation of RA into polar metabolites is mediated by CYP26 hydroxylases, a family of cytochrome P450 enzymes. RA exerts most of its effects through activation of nuclear retinoid receptors, the heterodimers of the RA receptor (RAR) and the retinoid X receptor (RXR) [8]. All-trans RA activates RARs and 9-cis RA binds to both RARs and RXRs [15]. The activated nuclear receptor dimers attach to RA response elements (RAREs) in the promoter regions of target genes [8,15,16]. Because of the multiplicity of responsive genes and interacting transcription factors, the actual actions of RA highly depend on the type, as well as the physiological and developmental stage of the target cells.
FIG. 1.
Retinoid metabolism. In the circulation, ROL is bound to RBP4. It can be taken up by cells by facilitated transport via STRA6 receptors. Within the cell, ROL is carried by CRBPs and can be stored in the form of REs or converted to RA in a two step process. RA is transported to the nucleus upon binding to CRABPs and acts through the activation of RA receptor heterodimers. Excess RA can be metabolized by the CYP26 enzymes. RE, retinyl ester; RAL, retinaldehyde; ROL, retinol; RA, retinoic acid; RBP4, retinol binding protein 4; STRA6, receptor for the ROL/RBP4 complex; CRBP, cellular ROL binding protein; CRABP, cellular RA binding protein; ADH, alcohol dehydrogenase; RDH, ROL dehydrogenase; RALDH, retinaldehyde dehydrogenase; REH, retinyl ester hydrolase; LRAT, lecithin ROL acyltransferase; RAR, RA receptor; RXR, retinoid X receptor; CYP26, a family of cytochrome P450 (CYP) enzymes.
During development, RA plays important regulatory roles in the formation of the neural tube and regional patterning of the future hindbrain and spinal cord [1]. Morphogenic roles of retinoids were also demonstrated in the developing forebrain [17–19] and the presence and production of RA was described in a subdivision of the developing rostro-ventral telencephalon, the lateral ganglionic eminence [19–21]. RA-responsive cells persist in the main neural stem cell niches of the postnatal rodent brain, including the subventricular zone (SVZ) adjacent to the lateral ventricles and the dentate gyrus of the hippocampal formation [2–4,22–24]. Depletion of RA in adult mice was shown to result in significantly decreased neuronal differentiation within the dentate gyrus [25,26] and RA synthesis seems to regulate proliferation and gene transcription of at least a subset of neural stem cells in the SVZ [27,28]. Most likely, RA signaling can determine the fate of resident neural stem cells throughout life. However, despite the accumulating knowledge on RA's action, the contribution of intracerebral production and distribution of retinoids to the genesis and renewal of neural tissue still needs to be specified.
The aim of the present work was to investigate the retinoid sensitivity of distinct neural stem/progenitor populations and to map the expression of retinoid metabolism related genes during their in vitro neuronal differentiation. For this purpose, neural stem cells cloned from early embryonic (E9) mouse neuroectoderm (NE-4C cells; [29–34]) and stem/progenitor cells isolated from late embryonic (E14.5) or adult (P62) neurogenic zones (RGl cells; [35]) were used. The results demonstrated that the investigated stem/progenitor populations showed significant differences in many aspects of both RA metabolism and retinoid responsiveness. Each neural stem cell population generated bioactive retinoids capable of activating the RARE-lacZ reporter transgene from precursor (RE, ROL) molecules, while the initiation of neuronal differentiation via RA signaling was restricted to NE-4C neural stem cells.
Materials and Methods
NE-4C neuroectodermal stem cells
NE-4C cells (ATCC no.: CRL-2925; [32]) derived from the anterior brain vesicles of p53 deficient 9 day old mouse embryos were maintained in serum containing medium, composed of minimum essential medium (MEM; Sigma-Aldrich) supplemented with 5% fetal calf serum (FCS; Invitrogen-Gibco), 4 mM glutamine (Sigma-Aldrich) and 40 μg/mL gentamycin (Chinoin), at 37°C temperature with 5% CO2. For maintenance, subconfluent cultures were regularly split by trypsinization (0.05 w/v% trypsin in PBS) into poly-L-lysine-coated Petri dishes. Neural differentiation was induced either by addition of RA/ROL/RE (see Results) or by changing the maintenance medium to defined medium (DM), formulated as MEM/F12 [1:1] supplemented with insulin–transferrin–selenite cocktail (Invitrogen-Gibco).
Radial glia-like cells
Radial glia like (RGl) cells were isolated from late embryonic/fetal (E14,5–16) or adult (P50-180) mouse brains via selective attachment to adhesive surfaces covered with AK-cyclo[RGDfC] peptides [36], as described previously [35]. The RGl clones used in the presented experiments were prepared either from late embryonic (E14.5) forebrains (RGl-1 and A2 clones) or from the SVZ (SVZ_M clone) or hippocampus (HC_A clone) of adult (P62) mouse brain [35].
The cells were maintained in DM composed of DMEM/F12 [1:1] (Sigma-Aldrich), 1% (v/v) B27 (Invitrogen-Gibco) and 20 ng/mL epidermal growth factor (EGF) (Peprotech) (EGF+). According to the aim of the experiment, B27 supplement without (DM) or with retinyl acetate (DM+RE) [37,38] was used. For large-scale neuron-production, EGF was withdrawn (EGF−) from the media of confluent cultures of RGl cells.
Treatment with retinoids and RAR antagonist
Neural stem cells were treated for varying periods with various concentrations of retinoids according to the actual experimental design. NE-4C cells collected for RT-PCR or real-time PCR were treated with 100 nM all-trans RA (Sigma-Aldrich) for an initial 48 h. In other experiments retinoids in 100 nM–10 μM concentrations were applied for 48 h or on every 2nd day. The pan RAR antagonist AGN193109 (Allergan, Inc.) was applied daily, at a final concentration of 100 nM [24]. In experiments using retinoids, the cultures and samples were protected from light.
RA reporter bioassay
The level of biologically active retinoids was determined by RA reporter cells as described previously [24]. In brief, F9 embryonic carcinoma cells, carrying a RARE-lacZ construct [39] were maintained in DMEM containing 10% FCS, in the presence of 400 μg/mL G418 (Sigma-Aldrich). F9 cells, plated 24 h before the onset of the experiments in 50,000 cell/cm2 density, were treated with media conditioned by stem cells for 24 h. In other experiments, F9 cells were seeded on top of confluent layers of neural stem cells to detect the actual retinoid production. After 18 h, the cultures were homogenized and the β-galactosidase activity of the homogenates was determined by the chromogenic substrate ortho-nitrophenyl-β-D-galactopyranoside (Sigma-Aldrich). The optical density was measured at a wavelength of 420 nm. The data were related to optical densities of retinoid-free cultures of the reporter cells (100%) and averages and standard deviations of relative values (as percentages) were calculated from 6–8 assays. Student's t-test was used to calculate the P-values.
In experiments with NE-4C cells, the culture media were changed to defined media either lacking retinoids (DM) or supplemented with 1 μM ROL, RE or RA for the initial 48 h of differentiation. After induction, the cultures were washed several times with retinoid free DM and were subjected to the F9 reporter assay on the 8th day. In agreement with published data [40] and our tests (not shown), RA used in the 48 h induction period was successfully removed by the media changes and/or degraded within no more than 4 days.
RGl cells were maintained in DM supplemented either with retinoid free or retinyl acetate supplemented B27 for 2 weeks before the F9 bioassays. Then the cells were differentiated by EGF removal and conditioned media were collected on the 7th day of induction.
Immunocytochemical staining and evaluation of neuron number
Cultures were fixed with 4% paraformaldehyde in PBS at room temperature for 20 min. Before staining, the cells were treated with 0.1% Triton X-100 in PBS for 5 min. After permeabilization, nonspecific antibody binding was blocked by incubating with 5% FCS in PBS (PBS-FCS) for 1.5 h at room temperature. Primary antibodies were diluted in PBS-FCS and were used at 4°C, overnight. Monoclonal antibodies against neuron-specific β-III tubulin or GFAP (Sigma-Aldrich) were diluted in 1:2,000. For fluorescent detection, Alexa-488 and Alexa-594 conjugated secondary antibodies (Invitrogen) were used at 1:1,000 dilutions, for 60 min at room temperature. Independent experimental series and subsequent immunostainings were performed at least four times. The analysis of the images (15 image/culture, 10×objective, Zeiss Axiovert 200M microscope) was performed by using AxioVision 4.8 and ImageJ softwares. As an alternative approach, the number of neurons (β-III tubulin positive cells) and the total number of DAPI stained nuclei were manually counted on 15 images taken from each of four parallel cultures grown in the presence or absence of retinoids or RAR antagonist for 4 days. The percentages of neurons were calculated for each culture, and were averaged among identical treatment-groups. Averages and standard deviations were related to those obtained from retinoid free, control cultures. Student's t-test was used to calculate the P-values.
Microarray
Samples were collected from noninduced or RA-induced NE-4C cells being at the 1st, 3rd, 6th and 10th day of differentiation. The cells were grown in the standard MEM medium complemented with 5% FCS. The RNA integrity was checked with Agilent 2100 Bioanalyzer. The generation of cDNA, production of labeled cRNA, and hybridization to Agilent G4120A Mouse Development Microarrays were performed according to standard protocols provided by the manufacturer at the Microarray Core Facility at the Department of Genetics, Cell- and Immunobiology at Semmelweis University (Budapest, Hungary). All materials were purchased from Agilent (Agilent Technologies) except RNA isolation kit (Qiagen).
RT-PCR analysis
Total RNA was isolated from cells with Tri Reagent (Sigma-Aldrich) according to the manufacturer's protocol. DNA contamination was eliminated by DNase-I (Fermentas) treatment and RNA fraction was dissolved in RNase/DNase free water at a concentration of 1 mg/mL. Reverse transcription reactions were undertaken from 3 μg total RNA using First Strand cDNA Synthesis Kit (Fermentas). The quantity of the cDNA product was determined by PCR (Hotstart Taq PCR Kit; Qiagen) using the hypoxanthine phosphoribosyltransferase 1 (Hprt1) coding gene. For each primer pair (see table below), the PCR conditions (temperatures, cycle numbers and MgCl2 concentration) have been optimized. The PCR products were analysed by electrophoresis on agarose gel with ethidium bromide, and were visualized by UV trans-illumination.
The presented RT-PCR results are representatives of several independent experiments. In NE-4C cells five completely different series, while in the case of RGl cells six independent (three embryonic and three adult derived) clones were investigated. For negative control (−) water, for positive controls total brain tissue of newborn mice (Lrat, Stra6, Math2, Ngn2, Oct4, Gfap), cultured astrocytes (Raldh1, Raldh2, RARβ, Cyp26a1) or adult liver tissue (all other genes) were used. As molecular weight marker (M), GeneRuler™ DNA Ladder Mix (#SM0331, Fermentas) was used.
| Gene | Sense and antisense primers |
|---|---|
| CrabpI | 5′acgccatgctgaggaaggtgg3′ and 5′cctgcatttgcgtccgtccac3′ |
| CrabpII | 5′tgatgaggaagatcgctgtg3′ and 5′ttccactctcccatttcacc3′ |
| CrbpI | 5′ccaaaatgcctgtggactt3′ and 5′cttgcagatcacaccctcag3′ |
| Cyp26a1 | 5′gaggggagagaggctggata3′ and 5′cagtggggcttgtcttcatt3′ |
| Cyp26b1 | 5′ttctctctgccagtggacct3′ and 5′cagcacagcagggtgtttta3′ |
| Cyp26c1 | 5′ttggtacagggctctcgttt3′ and 5′atcacctggctctgctgtct3′ |
| Gfap | 5′gactatcgccgccaactgc3′ and 5′cgtccttgtgctcctgcttc3′ |
| Hprt1 | 5′cacaggactagaacacctgc3′ and 5′gctggtgaaaaggacctct3′ |
| Lrat | 5′ctgaccaatgacaaggaacgcactc3′ and 5′ctaatcccaagacagccgaagcaagac3′ |
| Math2 | 5′tgagaatggcttgtccagaagg3′ and 5′tggtagggtgggtagaatgtgg3′ |
| Ngn2 | 5′aagaggactatggcgtgtgg3′ and 5′atgaagcaatcctccctcct3′ |
| Oct4 | 5′ggcgttctctttggaaaggtgttc3′ and 5′ctcgaaccacatccttctct3′ |
| Rdh10 | 5′ acttgtgacgtggggaagag3′ and 5′ caaggtaagggcaaaccaga3′ |
| Raldh1 | 5′gccagcagagcaaactcct3′ and 5′tcgctcaacactccttttca3′ |
| Raldh2 | 5′acatcgatttgcagggagtc3′ and 5′gtccaagtcagcatctgcaa3′ |
| Raldh3 | 5′cgaagagtgcgaaccagtta3′ and 5′cttggtgaacttgacctcca3′ |
| Raldh4 | 5′ggaacgttctctcgtcctga3′ and 5′gggacacctgctttatccaa3′ |
| Rarα | 5′gcgagctcattgagaaggtt3′ and 5′gtgtcttgctcaggcgtgta3′ |
| Rarβ | 5′ttcaaagcaggaatgcacag3′ and 5′gctgggtcgtcgttttctaa3′ |
| Rarγ | 5′ctgcaagggcttcttcagac3′ and 5′ctggcagagtgagggaaaag3′ |
| Rbp4 | 5′ccgagtcaaggagaacttcg3′ and 5′attggggtcacgagaaaaca3′ |
| Rxrα | 5′ctcctatcagcaccctgagc3′ and 5′accccatagtgtttgcctga3′ |
| Rxrβ | 5′tgggggtgagaaaagagatg3′ and 5′gagcgacactgtggagttga3′ |
| Rxrγ | 5′tgtgtacagctgtgaaggttgc3′ and 5′tctgagaagtgggggatgc3′ |
| Stra6 | 5′gcctccctgcctctacttct3′ and 5′gccagcaggtaggagacatc3′ |
RNA isolation for real-time PCR
Total RNA was isolated using RNAzol RT solution (Molecular Research Center Inch.) according to the manufacturer's protocol. Although intron-spanning primer pairs (see table below) were used in the Real-Time PCR experiments, genomic DNA contamination was regularly checked by Real-Time PCR using ACTB (beta-actin) primers on 100 ng total RNA templates. The RNA samples were considered to be free of genomic DNA contamination, when CT-s was higher, than cycle 35. To remove DNA contamination, recombinant DNase treatment was carried out using rDNase set (Macherey-Nagel) then the samples were subsequently cleaned and concentrated using NucleoSpin RNA Clean-up XS kit (Macherey-Nagel) according to the manufacturer's protocol. The quantity and quality of the RNA were checked by spectrophotometry and gel electrophoresis.
| Target | Primer sequence | CC | Amplicon (nt) |
|---|---|---|---|
| Hprt1 | tcctcctcagaccgctttt | 900 nM | 90 |
| cctggttcatcatcgctaatc | 900 nM | ||
| Gfap | cgccacctacaggaaattg | 300 nM | 60 |
| gtctgtacaggaatggtgatgc | 900 nM | ||
| Oct4 | gaggctacagggacacctttc | 300 nM | 71 |
| gtgccaaagtggggacct | 300 nM | ||
| Ngn2 | acatctggagccgcgtag | 900 nM | 79 |
| cagcagcatcagtacctcctc | 900 nM | ||
| Math2 | cgacactcagcctgaaaaga | 900 nM | 155 |
| caaactttctgcacatctggg | 900 nM | ||
| Stra6 | cgaggaacctctaggagctg | 900 nM | 60 |
| agctggcaaaggggaatc | 300 nM | ||
| Cyp26a1 | ccggcttcaggctacaga | 900 nM | 125 |
| ggagctctgttgacgattgtt | 900 nM | ||
| Aldh1a1 | ctcctctcacggctcttca | 300 nM | 71 |
| aatgtttaccacgccaggag | 900 nM | ||
| Aldh1a2 | catggtatcctccgcaatg | 900 nM | 65 |
| gcgcatttaaggcattgtaac | 300 nM | ||
| Aldh1a3 | aacctggacaaagcactgaag | 900 nM | 74 |
| aatgcattgtagcagttgatcc | 300 nM | ||
| Lrat | tatggctctcggatcagtcc | 300 nM | 102 |
| taatcccaagacagccgaag | 900 nM |
Reverse transcription for real-time PCR
Briefly 2 μg total RNA, 200 ng Random Primers (Invitrogen, Life Technologies), and 1 μL dNTP (10 mM each) were combined and incubated for 5 min at 65°C. After 2 min chilling on ice, 200 U SuperScript III Reverse Transcriptase (Invitrogen, Life Technologies), 40 U RNaseOUT Recombinant Ribonuclease Inhibitor (Invitrogen, Life Technologies), and 5 mM dithiothreitol were added to the mixture in 20 μL reaction volume. The reaction mixture was incubated at 25°C for 10 min to anneal the random hexamers, at 50°C for 5 h to transcribe cDNA and at 70°C for 15 min to inactivate the enzymes.
Relative quantitative real-time PCR assay
1 μL cDNA was used for quantifying mRNA together with 300 or 900 nM intron-spanning primers and 10 μL 2×Power Syber Green PCR Master Mix (Applied Biosystems). Each reaction was run in triplicate on differentiating NE-4C (DIV2, 4, 6, 10, 14), embryonic RGl or adult RGl (DIV4, DIV8) samples, as well as on nondifferentiated NE-4C or RGl (DIV0) samples. Amplification was performed in StepOnePlus (Applied Biosystems) equipment.
Real-time PCR data evaluation
Data were normalized to hypoxanthine phosphoribosyltransferase 1 (Hprt1) gene expression and evaluated with StepOne Software v2.1 (Applied Biosystems) utilizing the Relative Quantitative Real-Time PCR assay evaluation with comparative CT (ΔΔCT) method. The Relative Quantity values were analyzed with one-way analysis of variance together with the two-sided Dunnett post-hoc test to evaluate whether difference in mRNA levels of the given target between a sample and the reference of means is significant. The statistics were carried out using the SPSS for Windows 9.0 software. On the diagrams gene expression data of differentiating and undifferentiated stem cells are compared. All data are presented as Means±SD of the biological parallels and P-values below 0.1 are shown. Detailed statistical analysis is shown in Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/scd
Results
Retinoid-reponsiveness and the expression of retinoid metabolism (Fig. 1) related genes were investigated in different neural stem cell populations cloned from embryonic or adult brain tissues (Table 1) and in their progenies at various stages of in vitro induced neuronal differentiation.
Table 1.
Characteristics of NE-4C and Radial Glia Like (RGl) Neural Stem Cell Clones
| |
|
Neural stem/progenitor clones |
||
|---|---|---|---|---|
| Characteristics | NE-4C cells | Radial glia like cells (RGl) | ||
| Origin | embryonic (E9) anterior brain vesicles | embryonic (E14,5) forebrain | adult (P62) SVZ, HC | |
| Principle of isolation | continuous proliferation in serum+medium | selective adhesion to and serum-free propagation on AK-c(RGDfC) peptide | ||
| Cell cycle time | 16 h | ∼20 h | 22–24 h | |
| Cytochemical markers | nestin | + | + | + |
| RC2 | − | + | + | |
| GFAP | − | − | + | |
| βIII-tub | − | − | − | |
| Gene expression (based on RT-PCR data) | Oct4 | + | − | − |
| Nanog | + | − | − | |
| Sox2 | + | + | + | |
| Pax6 | − | + | + | |
| Blbp | + | + | + | |
| GLAST | + | + | + | |
| Ngn2 | − | + | + | |
| Mash1 | − | + | + | |
| Math2 | − | − | − | |
| Induction of neural commitment/differentiated progenies | retinoic acid or serum withdrawal (neurons and astrocytes)* | EGF withdrawal (neurons); serum supplementation (astrocytes); PDGF+FGF+forskolin/T3+AA (oligodendrocytes) | ||
Undifferentiated NE-4C cells display many embryonic stem cell features (Oct4, Nanog) with complete inactivity of tissue-specifying bHLH (Ngn2, Mash1, Math2) or radial glia specifying (Pax6, RC2) markers [32,57,58]. Their neural commitment is indicated by the expression of Sox2 [59] and by the fact that they could develop exclusively to neural cell types [60]. The lack of Oct4 and Nanog and the presence of RGl (RC2, GLAST) immunoreactivities together with the expression of proneural bHLH (Ngn and Mash1) genes indicate that noninduced RGl cells represent a more advanced progenitor state than NE-4C cells [35].
Differentiation of NE-4C cells towards the oligodendroglial lineage was observed upon implantation into the lesioned cerebral cortex [61].
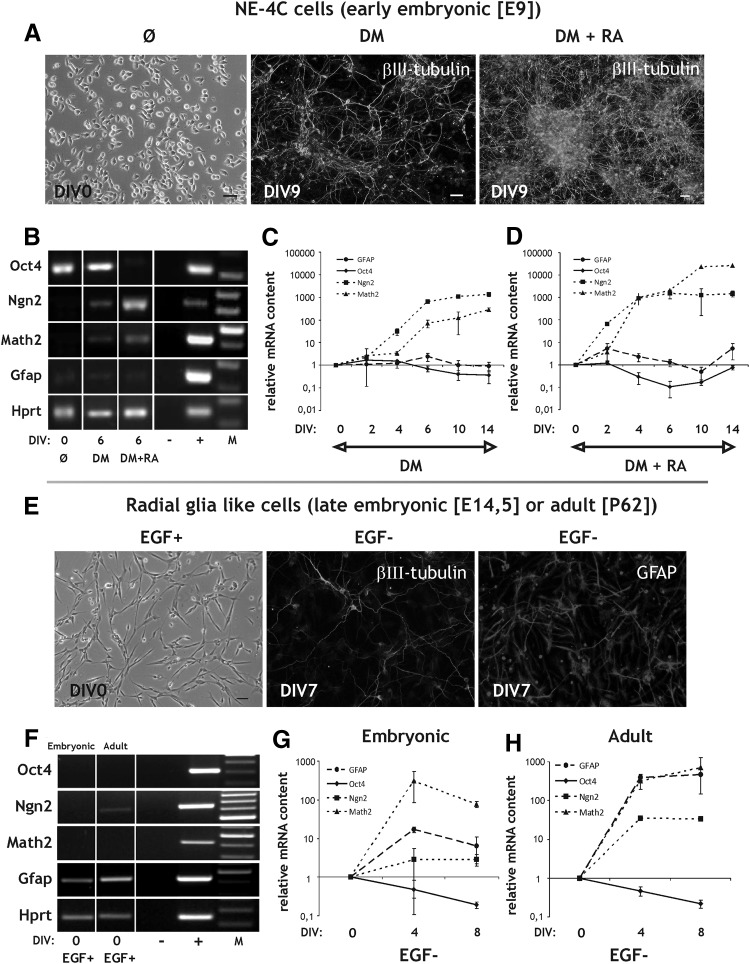
NE-4C neuroectodermal stem cells (Fig. 2A) were derived from the anterior brain vesicles of E9 mouse embryos [32]. These cells display many characteristics of radial neurepithelial cells, residents of the neural plate and early neural tube, including Oct4 and Sox2 expression, the lack of Pax6 or proneuronal/neurogenic (Ngn2/Math2) gene transcription, and the lack of RC2 or GFAP protein production (Fig. 2B and Table 1). If induced by all-trans RA, NE-4C cells develop into neurons and glial cells through well characterized stages of differentiation, including transient states of radial glia like development characterized with Pax6 and Emx2 expression and RC2-immunopositivity [29,33,41,42]. Mature neurons with synapsin expression and specific machinery for neurotransmitter production, release and uptake develop by the 9–10th day after RA induction [31].
FIG. 2.
Characteristics of NE-4C neural stem cells and radial glia like (RGl) cells. (A) Phase-contrast view of noninduced NE-4C cells (Ø) and β−III tubulin immunocytochemical staining of NE-4C derived neurons on the 7th day of induction (DIV7) (bars: 50 μm). Neuronal differentiation of NE-4C cells was initiated either by serum removal (DM: defined medium) or by RA treatment (DM+RA). (B) RT-PCR of the expression of some development-related (Oct4 stem cell-specific, Ngn2 proneural, Math2 neurogenic/neuronal and Gfap astrocyte-specific) genes in nondifferentiated (Ø) or differentiated cells. (C, D) Relative expression of the same genes during differentiation (DIV2–DIV14) compared to noninduced cells (DIV0). (E) The characteristic elongated morphology of noninduced (EGF+) adult-derived RGl cells and immunofluorescent stainings of their neuronal and astroglial derivatives on the 7th day of induction (DIV7) (bars: 20 μm). Differentiation of RGl cells was induced by EGF withdrawal (EGF−). (F) RT-PCR demonstration of transcription of Oct4, Ngn2, Math2 and Gfap genes by embryonic or adult-derived RGl cells. (G, H) Relative expression of the same genes during differentiation (EGF−; DIV4 and DIV8) compared to noninduced cells (DIV0). DIV indicates in vitro days after the onset of induction. “−”and “+” represent the negative (water) and positive (total brain) controls of the RT-PCR reactions. “M” indicates molecular weight marker. All real-time PCR data are means±SD from triplicate data. EGF, epidermal growth factor; RT, reverse transcription.
Smaller-scale neuronal differentiation of NE-4C cells was observed in the absence of exogenous RA too, in response to growth factor/serum withdrawal. Under such conditions, however, lesser neurons were formed as it was indicated by the markedly lower percentage of βIII-tubulin immunreactive cells (5.5%±1% neurons in serum withdrawal induced cultures versus 25.9%±1.5% in RA treated cultures) at DIV4. The difference in neuronal density was more evident by DIV9 (Fig. 2A) and was further supported by RT-PCR and real-time PCR data on Ngn2 and Math2 expression in a longer (14 day) time scale (Fig. 2B–D; Supplementary Fig. S1). While transcription of both Ngn2 and Math2 was upregulated upon serum withdrawal, Ngn2 expression increased with a delay and Math2 expression remained below the levels observed in RA-induced cultures. Without RA, transcription of the stem cell specific Oct4 was preserved in the cultures during the entire course of differentiation showing only a minor decline, after DIV6 (Fig. 2C). During RA-induced differentiation Oct4 expression dropped considerably by DIV6 (Fig. 2B, D) followed by a later rise back to the basal levels by DIV14. The data are in accord with our previous findings [34] on the persistence of noncommitted stem cells in differentiating cultures.
Upon RA-priming, NE-4C stem cells generate neurons and the gliogenesis is blocked for a 10–12 day period [29]. By the end of the 2nd week of RA-induction, neurogenesis ceases and astrocytes appear. Without RA-priming, however, GFAP-immunpositive astrocytes are not generated at all [29]. The earlier observations together with the recent data on Gfap expression (Fig. 2C, D) indicate that neuronal differentiation dominates over gliogenesis in the investigated period of NE-4C cell development.
RGl neural stem/progenitor clones were isolated by their preferential adhesion to a synthetic polypeptide AK-c(RGDfC) [36] and by propagation on peptide-coated surfaces under serum-free conditions with EGF supplementation [35]. Regardless of the age (eg, E14.5 or P62) and the region of the source brain tissue, peptide-adhering cells displayed elongated morphology (Fig. 2E), RC2 immunoreactivity and expressed Sox2 and Pax6 mRNAs, but did not transcribe the early pluripotency genes Oct4 and Nanog [35] (Fig. 2F). They gave rise to Ngn2 and Math2 mRNAs (Fig. 2G, H) and βIII-tubulin (Fig. 2E) expressing neurons upon induction by EGF withdrawal, in the absence of exogenous RA ([35]). Gfap mRNA was present in both embryonic and adult derived RGls cells (Fig. 2F) but GFAP protein could be demonstrated only in adult derived cultures [35]. Upon EGF withdrawal, the increase of Gfap level was significantly higher in adult RGl cells than in embryonic ones (Fig. 2G, H).
RA-production by NE-4C neural stem cells and RGl progenitors
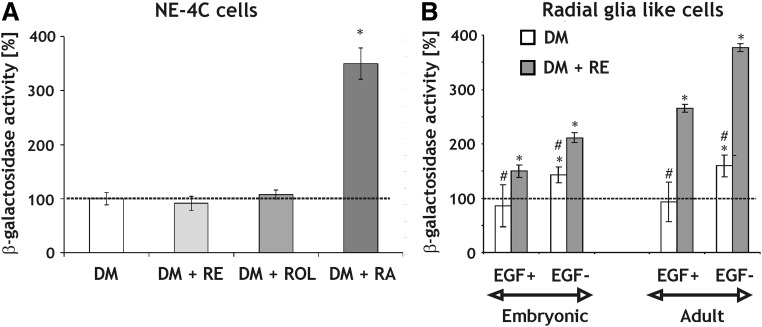
As we reported earlier [24], nondifferentiated NE-4C cells did not produce enough retinoids to activate RA-reporter F9 cells. However, if neuronal differentiation was induced by an initial 48 h treatment with RA, the cells generated bioactive retinoids 6 days later, as it was indicated by the RA-reporter assay (Fig. 3A). RA production, on the other hand, was not detected if neural differentiation was initiated by serum withdrawal without RA-treatment.
FIG. 3.
Retinoid production by NE-4C neural stem cells and RGl cells assayed by measuring β-galactosidase activity in F9 RA-reporter cells. (A) NE-4C cells were kept either in DM or treated with 1 μM RE, ROL or RA for 48 h (DIV0-2), then washed thoroughly and kept in retinoid-free medium for 6 more days (DIV2-8). The RA-reporter cell assay was carried out on DIV8 incubating the differentiated NE-4C cells in cocultures with the reporter cells. The method allowed direct, cell to cell activation of the transgene. *P<0.01. (B) β-galactosidase response of F9 RA-reporter cells to conditioned media obtained either from nondifferentiated (EGF+) or differentiated (EGF−) embryonic or adult RGl cells grown in the absence (DM) or presence (DM+RE) of retinyl acetate for at least 2 weeks before, as well as during the assay. The optical density (OD) values of the assays (A, B) were related to the β-galactosidase activity (100%; indicated by the dashed line) of control F9 cells. *P<0.01: significant difference from F9 cells; #P<0.05: significant difference from cells grown in DM+RE.
In contrast to NE-4C cells, nondifferentiated (EGF+) RGl cells produced sufficient retinoids to be detected by F9 RA-reporter cells (Fig. 3B). Cells differentiated upon EGF removal (EGF−, DIV7) produced even higher amounts of RA, both in embryonic and adult-derived cultures. The potential for retinoid production, however, seemed to depend on RE supply in the culture medium.
The different RA production and RA-sensitivity of NE-4C and RGl cells suggested major differences in their retinoid machinery. To explore this, we investigated the transcription of genes encoding enzymes, transporters and storage proteins involved in RA metabolism in both noninduced neural stem cells and in their differentiating progenies (Table 2 and see details below).
Table 2.
Expression of Components of RA Metabolism in NE-4C and RGl Neural Stem Cells
| |
|
|
|
|
RGl cells |
|||
|---|---|---|---|---|---|---|---|---|
| |
|
NE-4C cells |
nondifferentiated |
differentiated |
||||
| |
|
nondifferentiated |
differentiated |
EGF+ |
EGF− |
|||
| Function | mRNA | serum + | serum withdrawal | retinoic acid | e | a | e | a |
| Uptake and storage | Rbp4 | + | + | + | + | + | + | + |
| Stra6 | + | ↑ | ↑ | − | − | ↓ | ↑ | |
| Lrat | + | + | ↓ | − | − | ↓ | ↑ | |
| RA synthesis | Rdh10 | + | ↑ | ↑ | + | + | + | + |
| Raldh1 | − | ↑ | ↑ | + | + | ↓ | ↑ | |
| Raldh2 | − | ↓ ↑ | ↑ | − | − | ↓ | ↑ | |
| Raldh3 | + | ↓ ↑ | ↑ | + | + | ↓ | ↓ | |
| Raldh4 | + | + | + | − | − | − | − | |
| Intracellular transport | CrbpI | + | + | ↑ | + | + | ↓ | + |
| CrabpI | − | − | ↑ | + | +/− | + | +/− | |
| CrabpII | + | + | ↑ | + | + | + | + | |
| RA catabolism | Cyp26a1 | − | ↑ | ↑ | − | − | − | ↑ |
| Cyp26b1 | + | + | ↑ | − | − | +/− | − | |
| Cyp26c1 | − | − | ↑ | − | − | − | − | |
| RA receptors | Rarα | + | + | + | + | + | + | + |
| Rarβ | − | ↑ | ↑ | + | + | + | + | |
| Rarγ | + | + | + | + | + | + | + | |
| Rxrα | + | + | + | + | + | + | + | |
| Rxrβ | + | + | + | + | + | + | + | |
| Rxrγ | + | + | + | − | − | − | − | |
“+” and “−” indicate the presence or absence of the transcripts; the arrows indicate up- or downregulation of gene expression upon differentiation, not specifying the magnitude of growth or decrease. The table summarizes the results of real time PCR and RT-PCR data obtained from several experimental series (see Materials and Methods), represented in Figs. 4–8.
e, embryonic; a, adult.
Expression of elements of the retinoid machinery in NE-4C cells and RGl cells and in their differentiating progenies
Uptake and storage of vitamin A
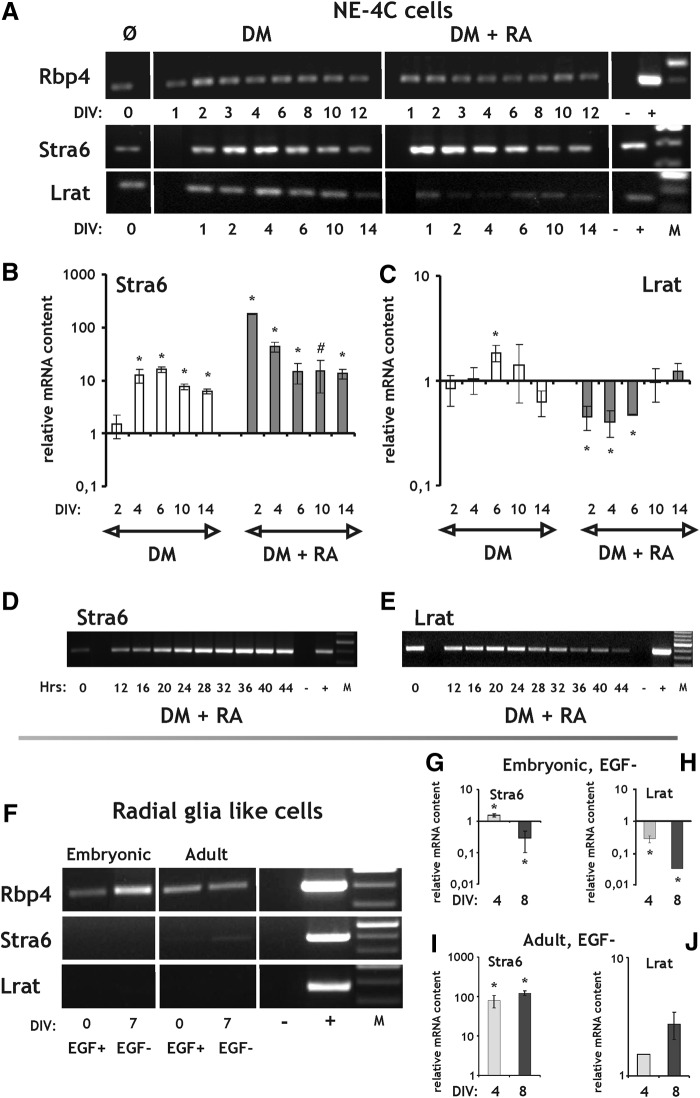
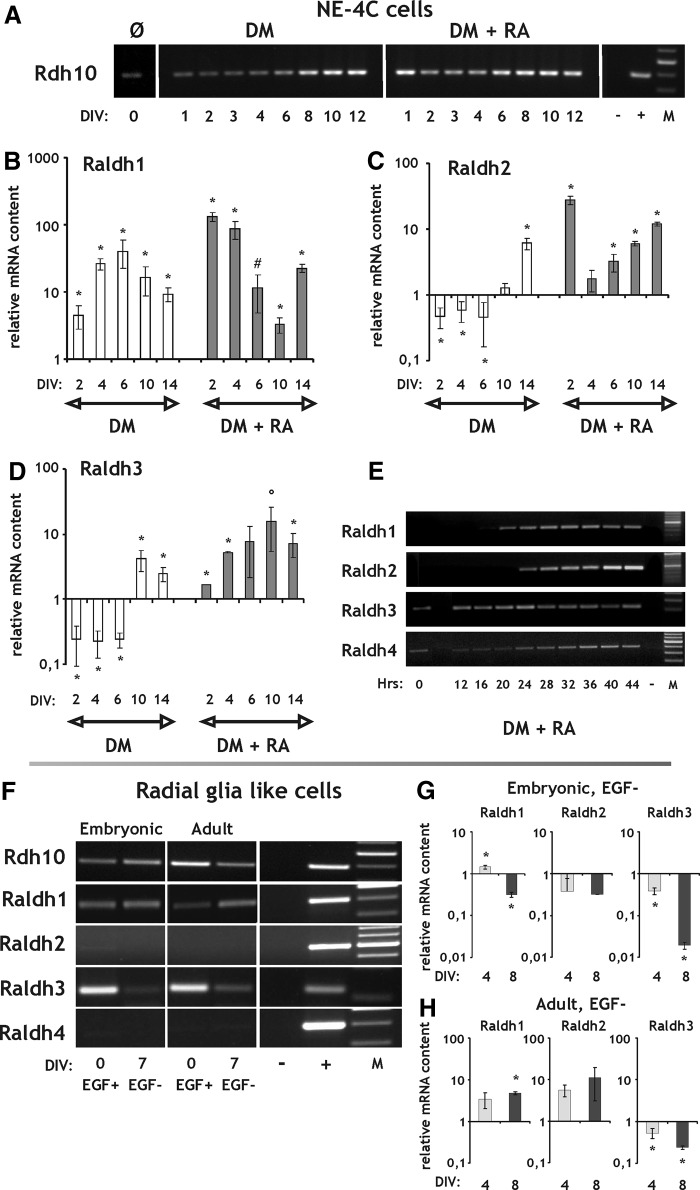
The mRNA of ROL binding protein (Rbp4), the major extracellular carrier of ROL [43] was present both in NE-4C and RGl cells, regardless of the stage of differentiation (Fig. 4A, F). The transcript of the RBP4 receptor Stra6 [10] was present in nondifferentiated NE-4C neural stem cells (Fig. 4A). Its expression increased significantly in response to RA treatment by DIV1-2 (Fig. 4A, B, D), and decreased thereafter, but remained elevated in comparison to noninduced cells, even 10 days after the removal of RA (Fig. 4B). Smaller, but significant increase was observed in Stra6 expression during serum-deprivation induced differentiation too, but with a few day delay (Fig. 4A, B). The Stra6 transcript was absent from or was expressed at a very low level in RGl cells (Fig. 4F) and was upregulated appreciably only in differentiating adult RGl clones (Fig. 4G, I).
FIG. 4.
RT and real-time PCR analysis of elements of the retinoid machinery involved in binding, uptake and storage of vitamin A during differentiation of NE-4C neural stem cells and RGl cells. (A) Rbp4, Stra6, and Lrat expression in NE-4C cells demonstrated by RT-PCR. Neuronal differentiation of the NE-4C cells was initiated either by serum removal (DM) or by RA treatment (DM+RA). (B, C) Real-time PCR analysis of Stra6 and Lrat expression during a 2-week period in NE-4C cells. (D, E) RT-PCR of Stra6 and Lrat in the first 44 h after the onset of induction of NE-4C cells with RA. (F–J) RT-PCR (F) and real-time PCR (G–J) analysis of Rbp4, Stra6 and Lrat expression in nondifferentiated (EGF+) and differenting (EGF−) embryonic (G, H) and adult (I, J) RGl cells. DIV and Hrs indicate in vitro days and hours, respectively, after the onset of induction. “−”and “+” represent the negative (water) and positive (brain, liver) controls of the RT-PCR reactions. “M” indicates molecular weight marker. All real-time PCR data are means±SD from triplicate data. *P<0.05; #P=0.064.
Lrat, the major enzyme responsible for the esterification and storage of ROL, was transcribed by NE-4C neural stem cells at all stages of differentiation (Fig. 4A). In the course of NE-4C cell differentiation its expression showed a minor and transient decrease in RA-induced cultures, while only a minor and transient elevation was observed under serum free conditions (Fig. 4A, C, E). Lrat expression in RGl cells was around the detection limit and showed only a small decrease in embryonic and a small increase in adult RGl cells upon differentiation (Fig. 4F, H, J).
RA synthesis
RA synthesis depends on the presence of ROL/alcohol (RDHs/ADHs) and RALDHs (Fig. 1) catalyzing the first and second steps of ROL↔RAL→RA conversion, respectively. ROL dehydrogenase 10 (Rdh10), a prominent member of the Rdh family [12,13], was present at the mRNA level in both noninduced and differentiated neural stem cells regardless of their origin (Fig. 5A, F). Its expression increased upon induction in NE-4C cell cultures (Fig. 5A). In noncommitted NE-4C cells, only Raldh3 and Raldh4 mRNAs were detected, as it was shown by RT-PCR (Fig. 5E; [24]). During differentiation, Raldh1 displayed the highest elevations in expression among Raldhs as it is indicated by the real-time PCR data (Fig. 5B–D). Raldh1 and also Raldh2 messages were rapidly upregulated (within 24–28 h) upon RA treatment followed by a transient decline by DIV6 or DIV4, respectively (Fig. 5B, C, E). Raldh3 did not respond to RA treatment with an immediate upregulation but it was slightly increased during differentiation (Fig. 5D), while Raldh4 mRNA content was slightly elevated during RA induced differentiation according to the RT-PCR data (Fig. 5E). Under serum free conditions, Raldh1 levels increased considerably, with a peak at DIV6 (Fig. 5B), while Raldh2 and Raldh3 levels decreased slightly during the 1st week of differentiation and increased thereafter (Fig. 5C, D).
FIG. 5.
RT and real-time PCR analysis of genes involved in the synthesis of RA during differentiation of NE-4C neural stem cells and RGl cells. (A) Expression of Rdh10 demonstrated by RT-PCR both in nondifferentiated (Ø) and differentiating NE-4C cells. Differentiation was induced either by serum withdrawal (DM) or RA treatment (DM+RA). (B–E) Expression of Raldh1-4 in differentiating NE-4C cells demonstrated by real-time PCR (B–D) in the first 2 weeks and by RT-PCR assays (E) in the first 44 h after the onset of induction. (F–H) RT-PCR (F) and real-time PCR (G, H) analysis of Rdh10 and Raldh1-4 expression in nondifferentiated (EGF+) and differentiating (EGF−) embryonic (G) and adult (H) RGl cells. DIV and Hrs indicate in vitro days and hours, respectively, after the onset of induction. “−” and “+” represent the negative (water) and positive (liver, glia) controls of the RT-PCR reactions. “M” indicates molecular weight marker. All real-time PCR data are means±SD from triplicate data. *P<0.05; #P=0.058; °P=0.071.
Raldh1 and Raldh3 mRNAs were present in nondifferentiated RGl cells, while Raldh2 was absent or expressed at a very low level (Fig. 5F). Real-time PCR data indicated a slight increase in Raldh1 and Raldh2 expression in the adult-derived RGl cells, while both Raldh1 and Raldh2 levels dropped by DIV8 in the differentiating embryonic RGl cells (Fig. 5G, H). A significant downregulation of Raldh3 upon EGF withdrawal was detected in all investigated RGl cell clones (Fig. 5F–H) and Raldh4 was not found in RGl cells by RT-PCR (Fig. 5F).
The expression patterns of Raldhs were in accordance with our earlier [24,29] and recent findings that noncommitted and differentiating neural stem cells possess the capability to produce RA.
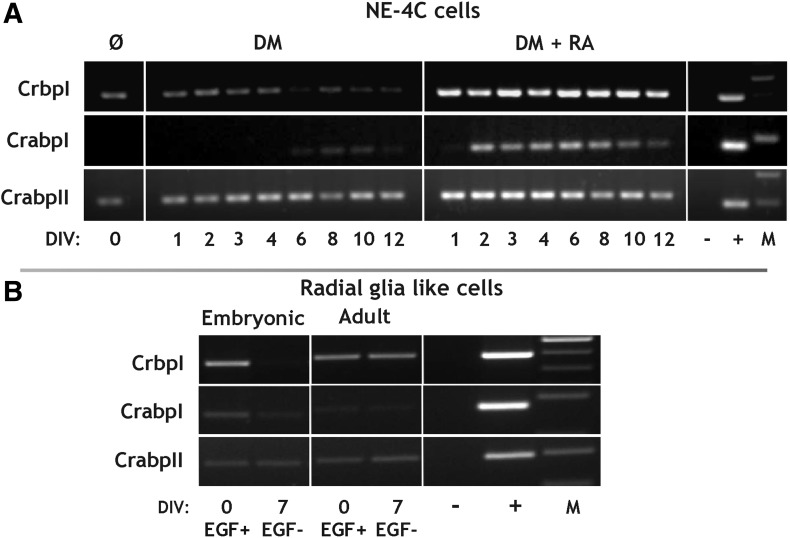
Intracellular ROL and RA binding proteins
Cellular ROL binding protein I (CrbpI) mRNA levels increased in NE-4C neural stem cells after RA exposure, with no apparent changes in later stages of differentiation. CrbpI level was, however, not elevated in cultures induced by serum deprivation (Fig. 6A). CrbpI mRNA was present in undifferentiated RGl cells (Fig. 6B) too. With the advancement of neural differentiation, the expression decreased in RGl cells of embryonic origin, while no marked changes were observed in adult derived cells.
FIG. 6.
RT-PCR analysis of genes involved in the intracellular binding of ROL and RA in NE-4C neural stem cells and RGl cells. (A) RT-PCR analysis of CrbpI and CrabpI-II in nondifferentiated (Ø) and differentiating NE-4C cells. Differentiation was induced either by serum withdrawal (DM) or RA treatment (DM+RA). (B) RT-PCR analysis of CrbpI and CrabpI-II expression in nondifferentiated (EGF+) and differentiating (EGF−) RGl cells. DIV indicates in vitro days after the onset of induction. “−” and “+” represent the negative (water) and positive (liver) controls of the RT-PCR reactions. “M” indicates molecular weight marker.
In noncommitted NE-4C cells, CrabpI mRNA was not expressed, while CrabpII mRNA was present. The mRNA levels of both binding proteins increased after RA treatment with CrabpI responding more slowly to RA priming and declining markedly after DIV8 (Fig. 6A). Both CrabpI and II mRNAs were expressed in all RGl clones at a relatively low level (Fig. 6B).
RA catabolism
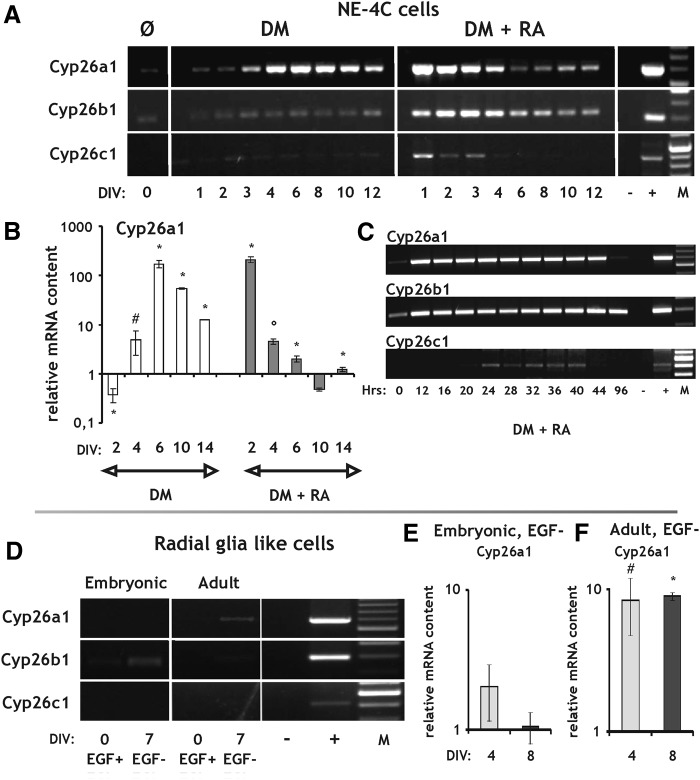
Members of the cytochrome P450 family 26 (Cyp26) enzymes are responsible for metabolizing RA (Fig. 1). RA treatment of NE-4C cells resulted in a marked upregulation of Cyp26a1 and Cyp26b1 mRNAs within 12 h and a moderate increase of Cyp26c1 within 24 h after RA exposure (Fig. 7A, C). The initial elevations in all Cyp26 mRNA expressions, however, were transient and after varying periods turned to a decline. A transient but significant rise of Cyp26a1 mRNA was observed in NE-4C cells differentiating without exogenous RA too, with a peak at DIV6 (Fig. 7A, B). The latter finding may suggest that these cells produced sufficient amounts of RA to activate the transcription of the direct RA-responsive Cyp26a1 gene. Alternatively, upregulation of the Cyp26a1 enzyme mRNA might be explained by turning on some unidentified, RA-independent and/or developmental stage-related pathways.
FIG. 7.
RT and real-time PCR analysis of genes involved in the catabolism of RA in NE-4C neural stem cells and RGl cells. (A–C) Expression of Cyp26a1, Cyp26b1, and Cyp26c1 enzyme mRNAs demonstrated by RT-PCR (A, C) and real-time PCR (B) both in nondifferentiated (Ø) and differentiated NE-4C cells. Differentiation of the cells was induced either by serum withdrawal (DM) or RA treatment (DM+RA). (D–F) RT-PCR (D) and real-time PCR (E, F) analysis of Cyp26a1-c1 enzyme expression in nondifferentiated (EGF+) and differentiated (EGF−) RGl cells. DIV and Hrs indicate in vitro days and hours, respectively, after the onset of induction. “−” and “+” represent the negative (water) and positive (liver, glia) controls of the RT-PCR reactions. “M” indicates molecular weight marker. All real-time PCR data are means±SD from triplicate data. *P<0.05; #P=0.06; °P=0.097.
In embryonic RGl cells a low level Cyp26b1 expression was observed, while none of the Cyp26 enzymes could be detected in adult derived RGl clones by RT-PCR (Fig. 7D). By real-time PCR, low level activation of Cyp26a1 was revealed during the differentiation of adult-derived RGl cells (Fig. 7E, F).
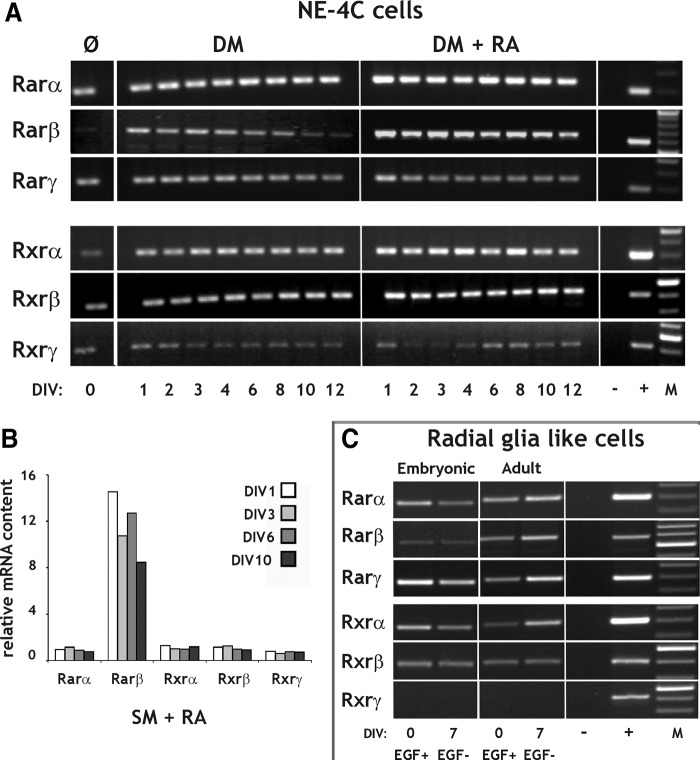
RA receptors
Multiple RAR and RXR subunits were expressed by both NE-4C and RGl cells (Fig. 8A, C). Rarβ mRNA level, however, was extremely low in nondifferentiated NE-4C neural stem cells (Fig. 8A) and Rxrγ mRNA was missing from all RGl cell populations (Fig. 8C). EGF withdrawal-induced differentiation of the RGl cells was not accompanied by consistent changes in the expression of retinoid receptors, when several clones were investigated (from which Fig. 8C represents only one embryonic and one adult clone). In NE-4C cells, however, the Rarβ message increased rapidly (within 12 h, data not shown) in response to induction regardless whether it was initiated with RA or serum deprivation, as it was shown both by RT-PCR (Fig. 8A) and microarray data (Fig. 8B). Rarβ expression declined considerably in later stages of serum withdrawal (Fig. 8A) and slightly during RA induced (Fig. 8A, B) differentiation.
FIG. 8.
RT-PCR and microarray analysis of RARs and RXRs in NE-4C neural stem cells and RGl cells. (A) Expression of Rar and Rxr mRNAs were demonstrated both in nondifferentiated (Ø) and differentiating NE-4C cells by RT-PCR. (B) Microarray data show the expression of retinoid receptor subunits in differentiating cells compared to undifferentiated ones. Differentiation was induced either by serum withdrawal (DM; A) or RA treatment either in DM (DM+RA; A) or in serum containing medium (SM+RA; B). (C) RT-PCR analysis of Rar and Rxr receptor expression in nondifferentiated (EGF+) and differentiating (EGF−) embryonic and adult-derived RGl cells. DIV and Hrs indicate in vitro days and hours, respectively, after the onset of induction. “−” and “+” represent the negative (water) and positive (liver, glia) controls of the RT-PCR reactions. “M” indicates molecular weight marker.
Responsiveness of neural stem cells to RA-precursor molecules, ROL and retinyl acetate
ROL induces robust neuronal differentiation in NE-4C neural stem cells
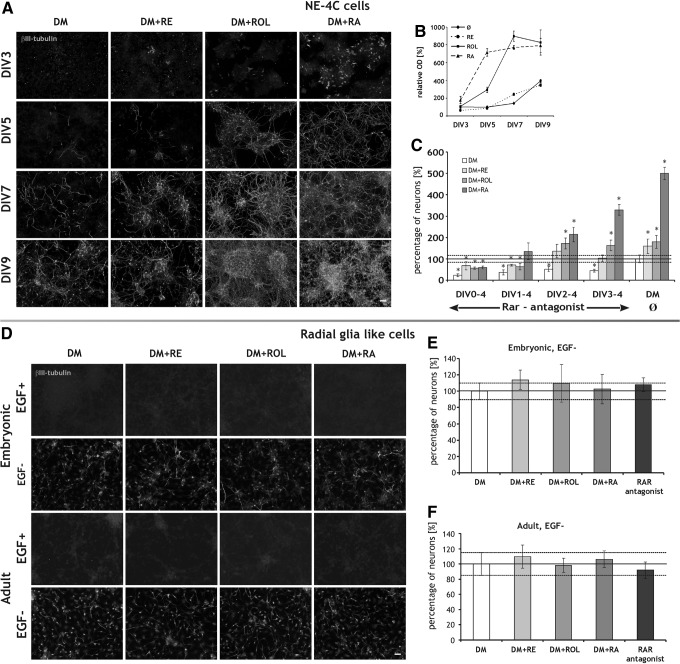
As genes encoding the components of the uptake and storage of vitamin A and the key enzymes of RA synthesis were active in noninduced NE-4C cells, we raised the question whether RA can be replaced by RA precursors in inducing neural differentiation. ROL or retinyl-acetate (RE) was added to the cells and neuron formation was followed by immunocytochemical staining for neuron-specific β-III tubulin on the 3rd, 5th, 7th, and 9th days after retinoid addition (Fig. 9A, B). β-III tubulin immunoreactive neurons appeared in RA-primed cultures as early as the 3rd day of induction. Large scale neuron formation in ROL treated cultures was observed from the 5th day on. In RE treated or retinoid free cultures massive neuron formation was detected not sooner than the 7–9th days (Fig. 9A), but the neuron density did not reach the levels observed in RA or ROL treated cultures (Fig. 9B). Counting β-III tubulin immunopositive neurons revealed that all investigated retinoids significantly elevated the neuron number if applied for 4 days. Compared to retinoid free control cultures (100%±19%), the percentage of neurons was 159%±24% in RE, 179%±29% in ROL and 499%±29% in RA treated cultures (Fig. 9C). Neuron forming effects of all retinoids were completely prevented if the pan RAR antagonist AGN193109 was included in the culture media during the 1st day of induction demonstrating that the induction was mediated through nuclear retinoid receptors regardless if it was initiated by RA, ROL, or RE. The antagonist was effective in the initial 48 h period of retinoid application, but did not arrest neuron formation if applied later (Fig. 9C).
FIG. 9.
Diverse responses of NE-4C neural stem cells and RGl cells to retinoids. (A) Neuron-specific β-III tubulin immunoreactive neurons in cultures of NE-4C cells grown for 3, 5, 7 or 9 days (DIV) either in retinoid free DM or in media supplemented with 1 μM RE, ROL or RA. (B) Densitometric analysis of fluorescence in cultures (n=4) represented in (A). The data were normalized to the average OD of cultures grown in retinoid free DM for 3 days (100%). (C) Percentage of neurons in cultures induced by 1 μM RE, ROL or 10 nM RA and grown in the presence or absence of 100 nM pan RAR antagonist AGN193109 for various periods (DIVstarting-ending). The percentages of neurons were determined in β-III tubulin immunostained preparations (n=4 for each) fixed on the 4th day. The data were related to the percentage of neurons in cultures grown in retinoid and antagonist free DM (100%, DM Ø). DIV indicates in vitro days after the onset of induction. *P<0.05. (D) β-III tubulin immunoreactive neurons in cultures of embryonic and adult-derived RGl cells grown (EGF+) or differentiated (EGF−) in the absence or presence of different retinoid (1 μM RE, ROL, RA) supplements for 4 days. (E, F) Percentage of neurons in cultures grown in 1 μM RE, ROL, RA, or 100 nM RAR antagonist containing media for 4 days. The percentages of neurons were determined in β-III tubulin immunostained preparations (n=4 for each) fixed on the 4th day. Data obtained from control (retinoid and antagonist free) cultures were set to 100%. Scale bars: 100 μm.
The schedule of differentiation was monitored also by real time PCR and RT-PCR analysis of differentiation marker gene expression (Supplementary Fig. S1). Real-time PCR data showed an order of DM+RA>DM+ROL>DM+RE>DM in both the magnitude and timing of the activation of proneural Ngn2 and neurogenic/neuronal Math2 genes (Supplementary Fig. S1). The statistical analysis of the data revealed, that RE induced cells express significantly more Ngn2 (P<0.005) at DIV2 and Math2 mRNAs (P<0.005) at DIV4 than cells differentiating in retinoid free DM, indicating that RE, probably through conversion to RA, had an influence on gene expression too, even if the increase in neuron number in RE containing medium lagged behind that of ROL. The induction of neuronal differentiation by different retinoids was concentration-dependent. RA initiated neuronal differentiation effectively in a concentration as low as 1 pM when applied under serum free conditions [24], while ROL and RE induced neuron formation in 100 nM and 1 μM doses, respectively (Supplementary Fig. S2). Higher concentrations resulted in formation of more compact aggregates with higher neuron density (Supplementary Fig. S2A).
Application of RA precursors evoked changes in the expression of genes of the retinoid machinery too. Raldh1 and Stra6 mRNAs were significantly upregulated in the presence of ROL (Supplementary Fig. S1). The expression of Cyp26 enzymes, known to be directly regulated by RA [44], was increased by both ROL and RE. In RE containing medium the peak of Cyp26a1 mRNA expression was at DIV6, while in ROL or RA around DIV4 and DIV2, respectively (Supplementary Fig. S1; Fig. 7A, C). Rarβ, another direct RA-response gene [45,46] showed similar quick activation in response to ROL, RE (Supplementary Fig. S1) and RA (Fig. 8A).
While the cellular responses to ROL (and RE) indicated an effective conversion of the precursor molecules to active retinoids, significant retinoid-activity in the fluid environment of RE or ROL treated NE-4C cells could not be detected by the F9 RARE-lacZ reporter cell line (Fig. 3A). These data, however, did not exclude the possibility that ROL- or RE-induced NE-4C cells produced small amounts of RA sufficient to initiate neuronal differentiation but this amount of RA, if dissolved in the large volume of culture medium, is below the detection limit (1 nM, [24]) of the reporter assay. This assumption was supported by the fact that the RAR-antagonist AGN193109 could effectively block the ROL-induced differentiation even in a concentration as low as 100 pM (Supplementary Fig. S2B).
RGl cells do not differentiate in response to retinoids
To investigate the influence of retinoids on neuronal differentiation of RGl cells, we treated the cultures with 1 μM RE, ROL, or RA. In the presence of EGF, neuronal differentiation could not to be initiated by any of the retinoids as it was demonstrated by the complete lack of β-III tubulin expression (Fig. 9D). Besides, in RGl cell cultures differentiating in response to EGF withdrawal, the percentage of neurons was not altered by either retinoid or RAR antagonist treatment (Fig. 9D–F). Meantime, significant retinoid production by both embryonic and adult derived RGl cells was detected with F9 RA-reporter cells (Fig. 3B).
In experiments presented in Fig. 9D–F, RGl cells were maintained in retinoid-free medium for at least 2 weeks before addition of exogenous retinoids. Retinoid depletion, however, did not affect the neurogenic capacity of the cells. These findings were further supported by experiments, when RGl cells isolated from adult brains (P153, SVZ) were prepared and maintained in DM lacking retinoids for more than 2 month. The neurogenic capacity of these cells was maintained under such conditions too (data not shown). These observations together with the inability of the pan RAR antagonist AGN193109 to prevent neuronal differentiation (Fig. 9E, F) demonstrated that endogenous RA synthesis did not contribute to enhanced neurogenesis in RGl cells.
Discussion
A number of recent investigations have demonstrated that RA-responsive cells are present not only in the developing rodent brain but they also persist in the neural stem cell niches in the postnatal CNS [2,3,22–24]. Retinoids are known to regulate many aspects of neural cell development, including regional and phenotype-determination [47], gliogenesis [29], neurite growth [48], and synaptic functions [49]. Discussing the influence of retinoids on fine-tuning of neural cell type specification is beyond the scope of this article. The presented data, however, clearly indicate that distinct neural stem cell/progenitor populations respond to retinoids in diverse ways. The major similarities and differences between the investigated NE-4C and RGl stem cell populations in the context of their developmental potential, retinoid responsivity and metabolism are summarized in Tables 1–3.
Table 3.
Major Similarities and Differences Between NE-4C and RGl Cells in the Context of Their Retinoid Responsivity, Production and Metabolism
| Similarities | |
|---|---|
| i | Capacity for RA synthesis from RA precursors (ROL, RE) indicated either by direct (RA-reporter assay) or indirect (neuronal differentiation in ROL or RE containing medium; activation of direct RA-responsive genes) evidences (Fig. 3, Fig. 9, Supplementary Fig. S1) |
| ii | Expression of ROL/RA binding (Rbp4, CrbpI, CrabpII) and RA synthesizing protein (Rdh10 and Raldh) mRNAs (Fig. 4, Fig. 5, Fig. 6) |
| iii | The lack of Raldh2 expression in non-committed cells (Fig. 5) |
| iv | Expression of a broad panel of RA receptor mRNAs (Fig. 8) |
| Differences | |
|---|---|
| i | Higher RA production by RGl cells (Fig. 3) |
| ii | Retinoids induce extensive neuronal differentiation of NE-4C neural stem cells but not of RGl progenitors (Fig. 9, Supplementary Fig. S1) |
| iii | Pan-RAR antagonist prevents neuronal differentiation of NE-4C neural stem cells but not of RGl progenitors (Fig. 9) |
| iv | Lack or very low level expression of enzymes responsible for vitamin A uptake (Stra6) and storage (Lrat) in RGl cells (Fig. 4) |
| v | Lack or low level expression of enzymes responsible for RA catabolism (Cyp26) in RGl cells (Fig. 7) |
| vi | Fewer retinoid pathway related genes are induced during differentiation of RGl cells (7/20) compared to NE-4C cells (13/20)* (Figs. 4–8) |
Statements are based on detailed data on RA responsiveness, RA production, and retinoid machinery of NE-4C and RGl neural stem/progenitor cells.
7/20 and 13/20: the number of genes showing alteration in expression upon differentiation/total number of genes investigated.
RE, retinyl ester; ROL, retinol; RA, retinoic acid; RAR, RA receptor.
In NE-4C cells, despite of hardly measurable levels of RA production, ROL and RE initiated differentiation and activated RA-responsive (Cyp26a1, Rarβ [44–46]) genes. Delays in neuron formation and gene activation suggested a rather slow accumulation of active retinoids upon RE/ROL-induction. Even low (picomolar) levels of RA, however, can initiate neural development of NE-4C cells [24], similarly to ES cells [47]. The lack of catabolising Cyp26 enzymes might contribute to high RA-responsiveness of noninduced NE-4C cells. During differentiation, Cyp26 enzymes are upregulated, but while balancing the intrinsic RA level, they may generate 4-oxo- and 4-hydroxy-RA compounds, known to contribute to the differentiation of ES [50] and neural progenitor [51] cells.
In contrast to NE-4C stem cells, RGl cells produced significant amounts of RA, but displayed RA-independent differentiation program. The rate of neuronal differentiation of these cells was not affected either by the application of retinoids or RAR-antagonist. Also, RGl cells, which were prepared and maintained for several weeks in retinoid free conditions, could undergo neuronal differentiation upon EGF removal. The RA-production by RGl cells, however, seemed to depend on exogenous retinoid resources, which may be explained by the lack of Lrat expression resulting in a missing capacity to transform retinoids to the stored RE form. The lack or very low level expression of the ROL transporter Stra6 mRNA in RGl cells suggested the involvement of alternative retinoid uptake mechanisms [52,53]. Interestingly, mRNA for the extracellular ROL carrier (Rbp4) was present in both NE-4C and RGl cells regardless of the stage of their development. Rbp4 expression may contribute to the regulation of vitamin A availability in the proximate environment of neural stem/progenitor cells [27,28] or developing/mature neurons [43,54].
Both retinoid sensitivity and production by differentiating neural stem cells highly depend on the cellular composition of the cultures. In NE-4C cells, glial differentiation was negligible in the investigated period [29]. Accordingly, the presented data reflect mainly the features of neuronal progenies of NE-4C cells. However, in RGl cells, the contribution of glial cells to the observed changes in both RA production and the retinoid machinery has to be taken into account. In mammals, many adult neural stem cells belong to GFAP-positive populations [55,56], and our adult-derived RGl clones also show GFAP-immunoreactivity [35]. Infinite serum-free, EGF-dependent proliferation, spindle-shaped morphology and ready neuron formation upon EGF withdrawal distinguishes RGl cells from mature astrocytes. GFAP-positive, flattened astrocytes; however, appear in differentiated cultures of both embryonic and adult RGl cells [35]. Their contribution to the retinoid metabolism can hardly be assessed. Differences in Gfap expression, in the level of RA production together with opposite changes in Stra6, Lrat, Raldh1 and Raldh2 expressions show a nonidentical cellular composition of differentiating cultures of embryonic and adult-derived RGl cells.
Our results presented here outline distinct features of retinoid metabolism in different neural stem cell populations. The highly retinoid-sensitive NE-4C neural stem cells can run the complete retinoid machinery and seem to regulate their own differentiation via autocrine or cell-autonomous RA signalling. The high sensitivity and low endogenous RA-levels, however, imply that small disturbances in extracellular retinoid supply or intrinsic RA metabolism can modulate cell-fate determining processes. As a contrast, the high RA-production of RGl cells is disconnected from immediate cell-fate decisions. The role and developmental significance of active retinoids generated by these cells are to be determined.
Supplementary Material
Acknowledgments
This work was supported by Hungarian National Science Foundation (OTKA; grant no.: K-68939, K-82090, K-106191), by National Institution for Science and Development (NKTH; grant: Bio_Surf) and by Richter Gedeon Pharmaceutical, Inc. (grant no.: IPI/266/2011).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Rhinn M. Dolle P. Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 2.Mey J. McCaffery P. Retinoic acid signaling in the nervous system of adult vertebrates. Neuroscientist. 2004;10:409–421. doi: 10.1177/1073858404263520. [DOI] [PubMed] [Google Scholar]
- 3.Lane MA. Bailey SJ. Role of retinoid signalling in the adult brain. Prog Neurobiol. 2005;75:275–293. doi: 10.1016/j.pneurobio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Goodman T. Crandall JE. Nanescu SE. Quadro L. Shearer K. Ross A. McCaffery P. Patterning of retinoic acid signaling and cell proliferation in the hippocampus. Hippocampus. 2012;22:2171–2183. doi: 10.1002/hipo.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 6.Blomhoff R. Blomhoff HK. Overview of retinoid metabolism and function. J Neurobiol. 2006;66:606–630. doi: 10.1002/neu.20242. [DOI] [PubMed] [Google Scholar]
- 7.Gudas LJ. Wagner JA. Retinoids regulate stem cell differentiation. J Cell Physiol. 2011;226:322–330. doi: 10.1002/jcp.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samarut E. Rochette-Egly C. Nuclear retinoic acid receptors: conductors of the retinoic acid symphony during development. Mol Cell Endocrinol. 2012;348:348–360. doi: 10.1016/j.mce.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 9.Napoli JL. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim Biophys Acta. 2012;1821:152–167. doi: 10.1016/j.bbalip.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawaguchi R. Yu J. Honda J. Hu J. Whitelegge J. Ping P. Wiita P. Bok D. Sun H. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- 11.Amengual J. Golczak M. Palczewski K. von Lintig J. Lecithin:retinol acyl transferase is critical for cellular uptake of vitamin A from serum retinol binding protein. J Biol Chem. 2012;287:24216–24227. doi: 10.1074/jbc.M112.353979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farjo KM. Moiseyev G. Nikolaeva O. Sandell LL. Trainor PA. Ma JX. RDH10 is the primary enzyme responsible for the first step of embryonic Vitamin A metabolism and retinoic acid synthesis. Dev Biol. 2011;357:347–355. doi: 10.1016/j.ydbio.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar S. Sandell LL. Trainor PA. Koentgen F. Duester G. Alcohol and aldehyde dehydrogenases: retinoid metabolic effects in mouse knockout models. Biochim Biophys Acta. 2012;1821:198–205. doi: 10.1016/j.bbalip.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin M. Zhang M. Abraham M. Smith SM. Napoli JL. Mouse retinal dehydrogenase 4 (RALDH4), molecular cloning, cellular expression, and activity in 9-cis-retinoic acid biosynthesis in intact cells. J Biol Chem. 2003;278:9856–9861. doi: 10.1074/jbc.M211417200. [DOI] [PubMed] [Google Scholar]
- 15.Bastien J. Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Gillespie RF. Gudas LJ. Retinoid regulated association of transcriptional co-regulators and the polycomb group protein SUZ12 with the retinoic acid response elements of Hoxa1, RARbeta(2), and Cyp26A1 in F9 embryonal carcinoma cells. J Mol Biol. 2007;372:298–316. doi: 10.1016/j.jmb.2007.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halilagic A. Ribes V. Ghyselinck NB. Zile MH. Dolle P. Studer M. Retinoids control anterior and dorsal properties in the developing forebrain. Dev Biol. 2007;303:362–375. doi: 10.1016/j.ydbio.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 18.Li H. Wagner E. McCaffery P. Smith D. Andreadis A. Drager UC. A retinoic acid synthesizing enzyme in ventral retina and telencephalon of the embryonic mouse. Mech Dev. 2000;95:283–289. doi: 10.1016/s0925-4773(00)00352-x. [DOI] [PubMed] [Google Scholar]
- 19.Smith D. Wagner E. Koul O. McCaffery P. Drager UC. Retinoic acid synthesis for the developing telencephalon. Cereb Cortex. 2001;11:894–905. doi: 10.1093/cercor/11.10.894. [DOI] [PubMed] [Google Scholar]
- 20.Chatzi C. Brade T. Duester G. Retinoic acid functions as a key GABAergic differentiation signal in the basal ganglia. PLoS Biol. 2011;9:e1000609. doi: 10.1371/journal.pbio.1000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toresson H. Mata de Urquiza A. Fagerstrom C. Perlmann T. Campbell K. Retinoids are produced by glia in the lateral ganglionic eminence and regulate striatal neuron differentiation. Development. 1999;126:1317–1326. doi: 10.1242/dev.126.6.1317. [DOI] [PubMed] [Google Scholar]
- 22.Wagner E. Luo T. Drager UC. Retinoic acid synthesis in the postnatal mouse brain marks distinct developmental stages and functional systems. Cereb Cortex. 2002;12:1244–1253. doi: 10.1093/cercor/12.12.1244. [DOI] [PubMed] [Google Scholar]
- 23.Wagner E. Luo T. Sakai Y. Parada LF. Drager UC. Retinoic acid delineates the topography of neuronal plasticity in postnatal cerebral cortex. Eur J Neurosci. 2006;24:329–340. doi: 10.1111/j.1460-9568.2006.04934.x. [DOI] [PubMed] [Google Scholar]
- 24.Kornyei Z. Gocza E. Ruhl R. Orsolits B. Voros E. Szabo B. Vagovits B. Madarasz E. Astroglia-derived retinoic acid is a key factor in glia-induced neurogenesis. FASEB J. 2007;21:2496–2509. doi: 10.1096/fj.06-7756com. [DOI] [PubMed] [Google Scholar]
- 25.Bonnet E. Touyarot K. Alfos S. Pallet V. Higueret P. Abrous DN. Retinoic acid restores adult hippocampal neurogenesis and reverses spatial memory deficit in vitamin A deprived rats. PLoS One. 2008;3:e3487. doi: 10.1371/journal.pone.0003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs S. Lie DC. DeCicco KL. Shi Y. DeLuca LM. Gage FH. Evans RM. Retinoic acid is required early during adult neurogenesis in the dentate gyrus. Proc Natl Acad Sci U S A. 2006;103:3902–3907. doi: 10.1073/pnas.0511294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haskell GT. LaMantia AS. Retinoic acid signaling identifies a distinct precursor population in the developing and adult forebrain. J Neurosci. 2005;25:7636–7647. doi: 10.1523/JNEUROSCI.0485-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang TW. Zhang H. Parent JM. Retinoic acid regulates postnatal neurogenesis in the murine subventricular zone-olfactory bulb pathway. Development. 2005;132:2721–2732. doi: 10.1242/dev.01867. [DOI] [PubMed] [Google Scholar]
- 29.Hadinger N. Varga BV. Berzsenyi S. Kornyei Z. Madarasz E. Herberth B. Astroglia genesis in vitro: distinct effects of retinoic acid in different phases of neural stem cell differentiation. Int J Dev Neurosci. 2009;27:365–375. doi: 10.1016/j.ijdevneu.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Herberth B. Pataki A. Jelitai M. Schlett K. Deak F. Spat A. Madarasz E. Changes of KCl sensitivity of proliferating neural progenitors during in vitro neurogenesis. J Neurosci Res. 2002;67:574–582. doi: 10.1002/jnr.10155. [DOI] [PubMed] [Google Scholar]
- 31.Jelitai M. Schlett K. Varju P. Eisel U. Madarasz E. Regulated appearance of nMDA receptor subunits and channel functions during in vitro neuronal differentiation. J Neurobiol. 2002;51:54–65. doi: 10.1002/neu.10049. [DOI] [PubMed] [Google Scholar]
- 32.Schlett K. Madarasz E. Retinoic acid induced neural differentiation in a neuroectodermal cell line immortalized by p53 deficiency. J Neurosci Res. 1997;47:405–415. [PubMed] [Google Scholar]
- 33.Tarnok K. Pataki A. Kovacs J. Schlett K. Madarasz E. Stage-dependent effects of cell-to-cell connections on in vitro induced neurogenesis. Eur J Cell Biol. 2002;81:403–412. doi: 10.1078/0171-9335-00262. [DOI] [PubMed] [Google Scholar]
- 34.Varga BV. Hadinger N. Gocza E. Dulberg V. Demeter K. Madarasz E. Herberth B. Generation of diverse neuronal subtypes in cloned populations of stem-like cells. BMC Dev Biol. 2008;8:89. doi: 10.1186/1471-213X-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marko K. Kohidi T. Hadinger N. Jelitai M. Mezo G. Madarasz E. Isolation of radial glia-like neural stem cells from fetal and adult mouse forebrain via selective adhesion to a novel adhesive peptide-conjugate. PLoS One. 2011;6:e28538. doi: 10.1371/journal.pone.0028538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marko K. Ligeti M. Mezo G. Mihala N. Kutnyanszky E. Kiss E. Hudecz F. Madarasz E. A novel synthetic peptide polymer with cyclic RGD motifs supports serum-free attachment of anchorage-dependent cells. Bioconjug Chem. 2008;19:1757–1766. doi: 10.1021/bc8000134. [DOI] [PubMed] [Google Scholar]
- 37.Brewer GJ. Torricelli JR. Isolation and culture of adult neurons and neurospheres. Nat Protoc. 2007;2:1490–1498. doi: 10.1038/nprot.2007.207. [DOI] [PubMed] [Google Scholar]
- 38.Brewer GJ. Torricelli JR. Evege EK. Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- 39.Sonneveld E. van den Brink CE. van der Leede BJ. Maden M. van der Saag PT. Embryonal carcinoma cell lines stably transfected with mRARbeta2-lacZ: sensitive system for measuring levels of active retinoids. Exp Cell Res. 1999;250:284–297. doi: 10.1006/excr.1999.4513. [DOI] [PubMed] [Google Scholar]
- 40.Sharow KA. Temkin B. Asson-Batres MA. Retinoic acid stability in stem cell cultures. Int J Dev Biol. 2012;56:273–278. doi: 10.1387/ijdb.113378ks. [DOI] [PubMed] [Google Scholar]
- 41.Jelitai M. Anderova M. Chvatal A. Madarasz E. Electrophysiological characterization of neural stem/progenitor cells during in vitro differentiation: study with an immortalized neuroectodermal cell line. J Neurosci Res. 2007;85:1606–1617. doi: 10.1002/jnr.21282. [DOI] [PubMed] [Google Scholar]
- 42.Schlett K. Herberth B. Madarasz E. In vitro pattern formation during neurogenesis in neuroectodermal progenitor cells immortalized by p53-deficiency. Int J Dev Neurosci. 1997;15:795–804. doi: 10.1016/s0736-5748(97)00015-4. [DOI] [PubMed] [Google Scholar]
- 43.Komatsu Y. Watakabe A. Hashikawa T. Tochitani S. Yamamori T. Retinol-binding protein gene is highly expressed in higher-order association areas of the primate neocortex. Cereb Cortex. 2005;15:96–108. doi: 10.1093/cercor/bhh112. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y. Zolfaghari R. Ross AC. Multiple retinoic acid response elements cooperate to enhance the inducibility of CYP26A1 gene expression in liver. Gene. 2010;464:32–43. doi: 10.1016/j.gene.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balmer JE. Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res. 2002;43:1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- 46.de The H. Marchio A. Tiollais P. Dejean A. Differential expression and ligand regulation of the retinoic acid receptor alpha and beta genes. EMBO J. 1989;8:429–433. doi: 10.1002/j.1460-2075.1989.tb03394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim M. Habiba A. Doherty JM. Mills JC. Mercer RW. Huettner JE. Regulation of mouse embryonic stem cell neural differentiation by retinoic acid. Dev Biol. 2009;328:456–471. doi: 10.1016/j.ydbio.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clagett-Dame M. McNeill EM. Muley PD. Role of all-trans retinoic acid in neurite outgrowth and axonal elongation. J Neurobiol. 2006;66:739–756. doi: 10.1002/neu.20241. [DOI] [PubMed] [Google Scholar]
- 49.Aoto J. Nam CI. Poon MM. Ting P. Chen L. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron. 2008;60:308–320. doi: 10.1016/j.neuron.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langton S. Gudas LJ. CYP26A1 knockout embryonic stem cells exhibit reduced differentiation and growth arrest in response to retinoic acid. Dev Biol. 2008;315:331–354. doi: 10.1016/j.ydbio.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 51.Goncalves MB. Agudo M. Connor S. McMahon S. Minger SL. Maden M. Corcoran JP. Sequential RARbeta and alpha signalling in vivo can induce adult forebrain neural progenitor cells to differentiate into neurons through Shh and FGF signalling pathways. Dev Biol. 2009;326:305–313. doi: 10.1016/j.ydbio.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 52.McCarthy RA. Argraves WS. Megalin and the neurodevelopmental biology of sonic hedgehog and retinol. J Cell Sci. 2003;116:955–960. doi: 10.1242/jcs.00313. [DOI] [PubMed] [Google Scholar]
- 53.Wicher G. Larsson M. Rask L. Aldskogius H. Low-density lipoprotein receptor-related protein (LRP)-2/megalin is transiently expressed in a subpopulation of neural progenitors in the embryonic mouse spinal cord. J Comp Neurol. 2005;492:123–131. doi: 10.1002/cne.20673. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y. Reese DH. The retinol signaling pathway in mouse pluripotent P19 cells. J Cell Biochem. 2011;112:2865–2872. doi: 10.1002/jcb.23200. [DOI] [PubMed] [Google Scholar]
- 55.Doetsch F. Garcia-Verdugo JM. Alvarez-Buylla A. Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J Neurosci. 1997;17:5046–5061. doi: 10.1523/JNEUROSCI.17-13-05046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kriegstein A. Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cavaleri F. Scholer HR. Nanog: a new recruit to the embryonic stem cell orchestra. Cell. 2003;113:551–552. doi: 10.1016/s0092-8674(03)00394-5. [DOI] [PubMed] [Google Scholar]
- 58.Rossant J. Stem cells from the Mammalian blastocyst. Stem Cells. 2001;19:477–482. doi: 10.1634/stemcells.19-6-477. [DOI] [PubMed] [Google Scholar]
- 59.Chambers I. The molecular basis of pluripotency in mouse embryonic stem cells. Cloning Stem Cells. 2004;6:386–391. doi: 10.1089/clo.2004.6.386. [DOI] [PubMed] [Google Scholar]
- 60.Herberth B. Minko K. Csillag A. Jaffredo T. Madarasz E. SCL, GATA-2 and Lmo2 expression in neurogenesis. Int J Dev Neurosci. 2005;23:449–463. doi: 10.1016/j.ijdevneu.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 61.Anderova M. Kubinova S. Jelitai M. Neprasova H. Glogarova K. Prajerova I. Urdzikova L. Chvatal A. Sykova E. Transplantation of embryonic neuroectodermal progenitor cells into the site of a photochemical lesion: immunohistochemical and electrophysiological analysis. J Neurobiol. 2006;66:1084–1100. doi: 10.1002/neu.20278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.