Abstract
Under conditions of reduced tissue oxygenation, hypoxia-inducible factor (HIF) controls many processes, including angiogenesis and cellular metabolism, and also influences cell proliferation and survival decisions. HIF is centrally involved in tumour growth in inherited diseases that give rise to renal cell carcinoma (RCC), such as Von Hippel–Lindau syndrome and tuberous sclerosis complex. In this study, we examined whether HIF is involved in tumour formation of RCC in Birt–Hogg–Dubé syndrome. For this, we analysed a Birt–Hogg–Dubé patient-derived renal tumour cell line (UOK257) that is devoid of the Birt–Hogg–Dubé protein (BHD) and observed high levels of HIF activity. Knockdown of BHD expression also caused a threefold activation of HIF, which was not as a consequence of more HIF1α or HIF2α protein. Transcription of HIF target genes VEGF, BNIP3 and CCND1 was also increased. We found nuclear localization of HIF1α and increased expression of VEGF, BNIP3 and GLUT1 in a chromophobe carcinoma from a Birt–Hogg–Dubé patient. Our data also reveal that UOK257 cells have high lactate dehydrogenase, pyruvate kinase and 3-hydroxyacyl-CoA dehydrogenase activity. We observed increased expression of pyruvate dehydrogenase kinase 1 (a HIF gene target), which in turn leads to increased phosphorylation and inhibition of pyruvate dehydrogenase. Together with increased protein levels of GLUT1, our data reveal that UOK257 cells favour glycolytic rather than lipid metabolism (a cancer phenomenon termed the ‘Warburg effect’). UOK257 cells also possessed a higher expression level of the l-lactate influx monocarboxylate transporter 1 and consequently utilized l-lactate as a metabolic fuel. As a result of their higher dependency on glycolysis, we were able to selectively inhibit the growth of these UOK257 cells by treatment with 2-deoxyglucose. This work suggests that targeting glycolytic metabolism may be used therapeutically to treat Birt–Hogg–Dubé-associated renal lesions.
Keywords: Birt–Hogg–Dubé, HIF, folliculin, warburg effect
Introduction
Birt–Hogg–Dubé syndrome is an autosomal dominant disorder characterized by skin fibrofolliculomas, lung cyst, spontaneous pneumothorax and renal cancer and is caused by inactivating mutations of BHD (Baba et al., 2006). The Birt–Hogg–Dubé protein (BHD) (sometimes called folliculin) seems to function as a classical ‘Knudson’ tumour suppressor, as loss of heterozygosity of the BHD locus is a frequent event in renal cell carcinoma (RCC) (Khoo et al., 2002). There is a 15–30% occurrence of RCC within Birt–Hogg–Dubé patients, which includes oncocytoma, chromophobe, papillary, clear cell and oncocytic hybrid types (Woodward et al., 2008). This varying spread of histological types of RCC within Birt–Hogg–Dubé patients might be explained by different genetic backgrounds or ‘secondary hits’ of additional genes that have not yet been determined. Although it is considered that BHD represses cell growth, the role that BHD has in cancer progression and/or initiation is currently unresolved.
Other inherited genetic disorders that give rise to RCC include Von Hippel–Lindau (VHL) (Hemminki et al., 2002) and hereditary leiomyomatosis renal cell carcinoma (HLRCC) (Kiuru and Launonen, 2004). Higher levels of hypoxia-inducible factor (HIF)-mediated signalling is a shared feature attributed to tumour growth within these inherited disorders (Kaelin, 2005; Isaacs et al., 2005). HIF1α and HIF2α are transcription factors that interact with HIF-β (also known as aryl hydrocarbon receptor nuclear translocator) and, as heterodimers, enhance expression of over 70 genes containing hypoxia-response elements that are involved in angiogenesis, erythropoiesis, glucose metabolism, cell survival and metastasis (Semenza, 2004). HIF-α proteins are rapidly degraded in an oxygen-dependent manner through hydroxylation of two proline residues within their oxygen-dependent degradation domain catalysed through HIF prolyl hydroxylases (Semenza, 2004). Proline hydroxylation leads to rapid degradation of HIF-α proteins, which is dependent on the VHL tumour suppressor protein. VHL is a component of the E3 ubiquitin ligase complex that targets proline-hydroxylated HIF-α proteins for degradation through the proteosome. Consequently, low oxygen or loss of VHL function leads to stabilization of HIF-α proteins and to an upregulation of HIF-mediated gene expression (Semenza, 2004). VHL patients develop clear-cell RCC, which is caused by loss of function of the tumour suppressor protein VHL (Hemminki et al., 2002). HLRCC is caused through fumarate hydratase (FH) gene mutations that consequently lead to an accumulation of fumarate, inhibition of HIF prolyl hydroxylases and enhanced stability of HIF-α proteins (Sudarshan et al., 2009). HLRCC patients are predisposed to type 1 papillary renal carcinoma.
Recent studies have also implicated HIF as a factor involved in tumour growth in tuberous sclerosis complex (TSC) (Liu et al., 2003). TSC has overlapping clinical features to those of Birt–Hogg–Dubé (Slegtenhorst et al., 2007) and arises through mutations of either TSC1 or TSC2 that function as a heterodimer to repress mammalian target of rapamycin (mTOR). mTOR is a positive regulator of HIF in an apparently multifaceted manner through levels of gene expression, translation, protein stability and activity (see review Dunlop and Tee, 2009). Previously, we showed that HIF transcriptional activity within cells devoid of either VHL or TSC2 was robustly impaired when the respective functional VHL or TSC2 proteins were reexpressed (Land and Tee, 2007). Given that HIF seems to be centrally involved in the pathology associated with inherited diseases such as VHL, HLRCC and TSC, we wanted to examine whether cells derived from a kidney tumour devoid of BHD had elevated levels of HIF-mediated gene expression that would lead to a cancer phenotype. In this study, we analysed HIF-mediated signalling events within a Birt–Hogg–Dubé patient renal tumour cell line, UOK257. These UOK257 cells lacking functional BHD (referred to as BHD−) display the same morphological and ultrastructural features that were present within the tumour from which they were derived (Yang et al., 2007). In addition, we analysed the expression of HIF targets in a chromophobe renal carcinoma from a patient with proven Birt–Hogg–Dubé syndrome.
Results
Cells lacking BHD have elevated levels of HIF1α- and HIF2α-driven gene expression
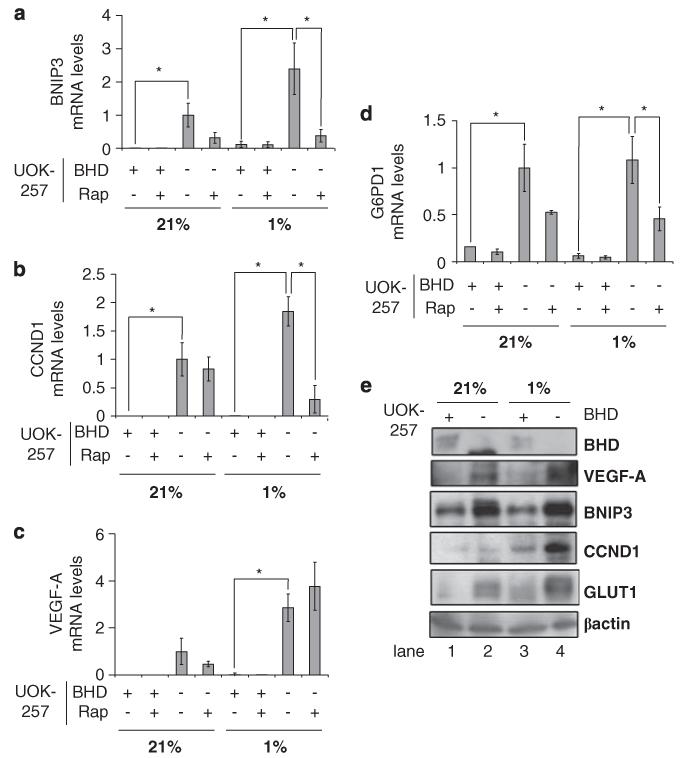
To examine whether the transcriptional activity of HIF was upregulated within BHD− cells, we compared the levels of HIF1α and HIF2α gene expression within BHD− cells with a stably generated UOK257-2 cell line that reexpresses wild-type BHD (Baba et al., 2006) (now referred to as BHD+). For this, we analysed the mRNA levels of genes that are transcribed by HIF1α (BNIP3 (Figure 1a); Guo et al., 2001), HIF2α (CCND1 (Figure 1b); Maxwell, 2005) or both (VEGF-A (Figure 1c); Maxwell, 2005). Loss of BHD markedly increases the transcription of mRNAs that are regulated by both HIF1α and HIF2α under conditions of hypoxia. Furthermore, there is an elevated level of HIF1α- and HIF2α-mediated gene expression under normoxic conditions. Treatment of BHD− cells with the specific mTOR inhibitor, rapamycin, inhibited the expression of BNIP3 (Figure 1a) and CCND1 (Figure 1b) under hypoxic conditions, suggesting that mTOR has a role in hypoxia-mediated signalling in BHD− cells. We did not observe any significant change in the levels of VEGF-A mRNA on rapamycin treatment in BHD− cells (Figure 1c). Of interest, the mRNA levels of BNIP3 (Figure 1a), CCND1 (Figure 1b) and VEGF-A (Figure 1c) were very low in BHD+ cells under both normoxic and hypoxic conditions.
Figure 1.
BHD negatively regulates HIF-induced gene expression during hypoxia. The mRNA levels of (a) BNIP3, (b) CCND1, (c) VEGF-A and (d) G6PD1 were compared in BHD+ (UOK257-2) and BHD− (UOK257) cells treated overnight with 50 nM rapamycin under normoxia (21%) and hypoxia (1%), where indicated, in this study by quantitative reverse-transcriptase PCR. mRNA levels were standardized against β-Actin and fold activation was compared with that in BHD− cells under normoxia. n = 3. *P<0.05 when comparing BHD+ and BHD− cells under hypoxia and normoxia and rapamycin treatment. (e) Western blot analysis was carried out on cell lysates prepared from BHD+ and BHD− cells under normoxia (21%) and hypoxia (1%), where indicated. Protein levels of BHD, VEGF-A, BNIP3, CCND1, GLUT1 and β-actin (as loading control) were determined.
One of the ways by which cellular redox homeostasis can be maintained during hypoxia is through HIF-dependent glucose-6-phosphate dehydrogenase 1 (G6PD1) gene expression (Gao et al., 2004). G6PD1 is the rate-limiting enzyme within the pentose phosphate pathway (also referred to as the hexose-monophosphate shunt) that is induced on oxygen stress. The pentose phosphate pathway maintains the level of coenzyme nicotinamide adenine dinucleotide phosphate, the primary reducing agent within mammalian cells, and in doing so helps maintain cellular redox homeostasis (Gao et al., 2004). Of interest, we observe a higher level of G6PD1 mRNA within BHD− cells when compared with BHD+ cells (Figure 1d). The mRNA levels of G6PD1 under normoxic conditions were not further enhanced when BHD− cells were cultured in low oxygen, suggesting that G6PD1 is maximally transcribed under normoxic conditions.
The protein levels of HIFα transcriptional targets VEGF-A, BNIP3, CCND1 and GLUT1 were compared between BHD− and BHD+ cells through western blotting (Figure 1e). We observed an increase in the protein levels of VEGF, BNIP3, CCND1 and GLUT1 in BHD− cells (lane 4) compared with BHD+ cells (lane 3) under hypoxic conditions. Even under normoxia, there is an increase in the levels of BNIP3, VEGF-A and GLUT1 in BHD− cells (lane 2 compared with lane 1). These results show that the levels of HIF1α- and HIF2α-regulated proteins are increased in this Birt–Hogg–Dubé patient renal tumour cell line.
Loss of BHD in UOK257 does not affect the mRNA or protein levels of HIF1α
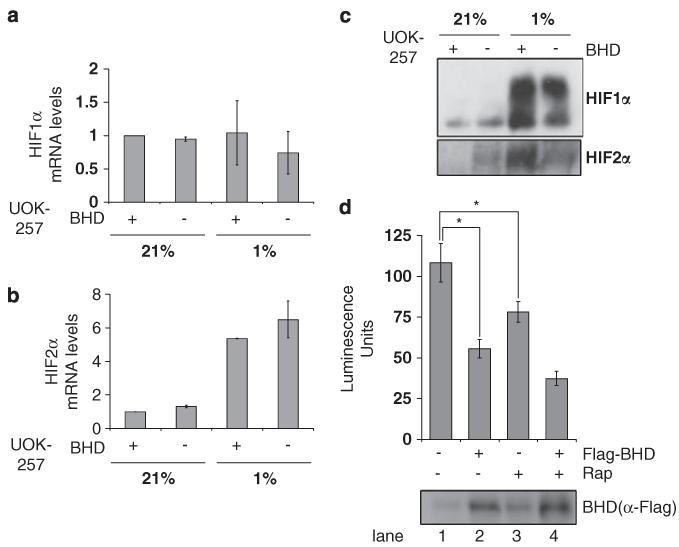
To determine whether BHD regulates HIF1α and HIF2α at the level of its gene expression, we analysed the mRNA levels of HIF1α (Figure 2a) and HIF2α (Figure 2b) within UOK257 cells. Our data showed no significant differences in the levels of HIF1α mRNA between BHD− and BHD+ cells under both normoxic and hypoxic conditions (Figure 2a). Our data also reveal that there was a significant increase in HIF2α mRNA levels under hypoxic compared with normoxic conditions; however, this increase did not appear to be regulated by BHD (Figure 2b). Similarly, Li et al. observed an increase of HIF2α mRNA, but not of HIF1α mRNA, in glioma stem cells after hypoxia (Li et al., 2009).
Figure 2.
Increased transcriptional activity of HIF1α and HIF2α in BHD− cells. The mRNA of (a) HIF1α and (b) HIF2α was compared in BHD+ (UOK257-2) and BHD− (UOK257) cells after 18 h of normoxia (21%) or hypoxia (1%), where indicated, by reverse transcriptase PCR. mRNA levels were standardized against β-actin and fold activation was compared with that in BHD+ cells under normoxia. n = 3. (c) The protein levels of both HIF1α and HIF2α were analysed by western blot analysis (d) BHD− cells transiently transfected with an HIF-inducible luciferase reporter and either empty pRK7 or Flag-BHD vector and then treated with 50nm rapamycin ‘Rap’ (where indicated) were maintained at 1% O2 for 18 h. Lysates were analysed for luciferase fluorescence to determine the transcriptional activity of HIF. n = 3. *P<0.05. Protein levels of Flag-tagged BHD were determined with anti-Flag antibodies.
We next analysed the protein level of HIF1α and HIF2α under the same conditions (Figure 2c). Hypoxia caused an accumulation of HIF1α in both BHD− and BHD+ cell lines and to a similar level. We also observed an increase in HIF2α protein upon hypoxia in BHD− cells, but the intensity of the HIF2α protein band was much weaker than that of the HIF2α protein band in BHD+ cells after hypoxia. These experiments suggest that the enhanced HIF-mediated gene expression that we observed in these BHD-null (UOK257) cells is not as a consequence of increased HIF mRNA or protein levels upon hypoxia. To examine whether the transcriptional activity of HIF was altered in these cells, we used a HIF-induced luciferase reporter construct (Figure 2d). In this assay, we observed a high level of HIF activity in BHD− cells (lane 1, Figure 2d), which was dramatically repressed in the rescue experiment after BHD was reexpressed (lane 2, Figure 2d). Of interest, treatment with rapamycin was not as potent as reexpression of BHD to repress HIF activity. Collectively, these data suggest that overexpression of BHD does not affect HIF1α protein levels through gene expression or protein stability but rather that BHD impairs the transcriptional activity of HIF1α.
BHD knockdown by short hairpin (sh)RNA enhances HIF activity during hypoxia
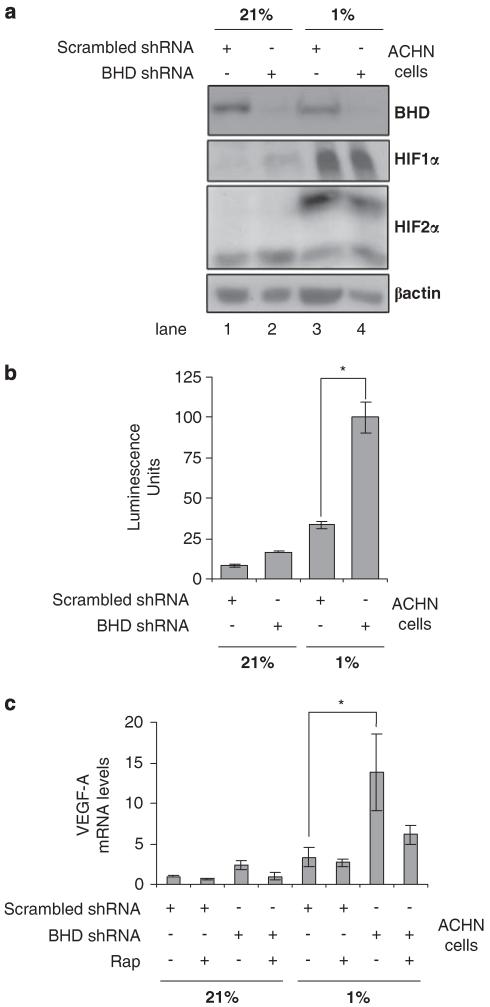
To examine whether stable knockdown of BHD influences HIF activity, we used a adenocarcinoma human (ACHN) cell line. ACHN cells after selection with scrambled shRNA (termed ‘ACHN/scrambled-shRNA’) still expressed endogenous BHD and were used as a control. These ACHN/scrambled-shRNA cells were analysed alongside ACHN cells that had BHD stably knocked down with BHD shRNA (termed ‘ACHN/BHD-shRNA’). We analysed the total protein levels of HIF1α and HIF2α during normoxia (21% O2) and hypoxia (1% O2) within these cells (Figure 3a). We found that the accumulation of HIF1α and HIF2α protein on hypoxia was not enhanced on knockdown of BHD. Interestingly, even with similar levels of HIF1α and HIF2α protein in both ACHN/scrambled-shRNA and ACHN/BHD-shRNA cell lines after hypoxia, the activity of HIF was threefold higher in ACHN cells lacking BHD (Figure 3b). Consistent with higher levels of HIF activity in cells lacking BHD, ACHN/BHD-shRNA also had higher levels of VEGF-A mRNA, an HIF-gene target (Figure 3c). These data support our previous findings in the UOK257 cell lines, wherein loss of BHD enhances the activity of HIF during hypoxia.
Figure 3.
Knockdown of BHD expression increases HIF activity under hypoxia in human kidney ACHN cells. (a) The protein levels of HIF1α and HIF2α were compared in ACHN cells, stably transfected with either BHD shRNA or scrambled shRNA, after 18 h of normoxia (21%) or hypoxia (1%) by western blot. β-actin serves as a loading control and endogenous BHD expression was determined to verify efficient BHD knockdown. (b) ACHN cells, stably transfected with either BHD shRNA or scrambled shRNA were transiently transfected with an HIF-inducible luciferase reporter and then maintained at either 21 or 1% O2 for 18 h. Lysates were analysed for luciferase fluorescence to determine the transcriptional activity of HIF. n = 3. *P<0.05. (c) The mRNA levels of VEGF-A were determined from ACHN/scrambled-shRNA and ACHN/BHD-shRNA cells that were treated overnight with 50 nm rapamycin under normoxia (21%) and hypoxia (1%), where indicated, by quantitative reverse-transcriptase PCR. mRNA levels were standardized against β-actin. Fold activation was compared with that in ACHN/BHD-shRNA cells under normoxia. n = 3. *P<0.05 when comparing ACHN cells lines under hypoxia.
HIF gene expression is upregulated in a Birt–Hogg–Dubé tumour
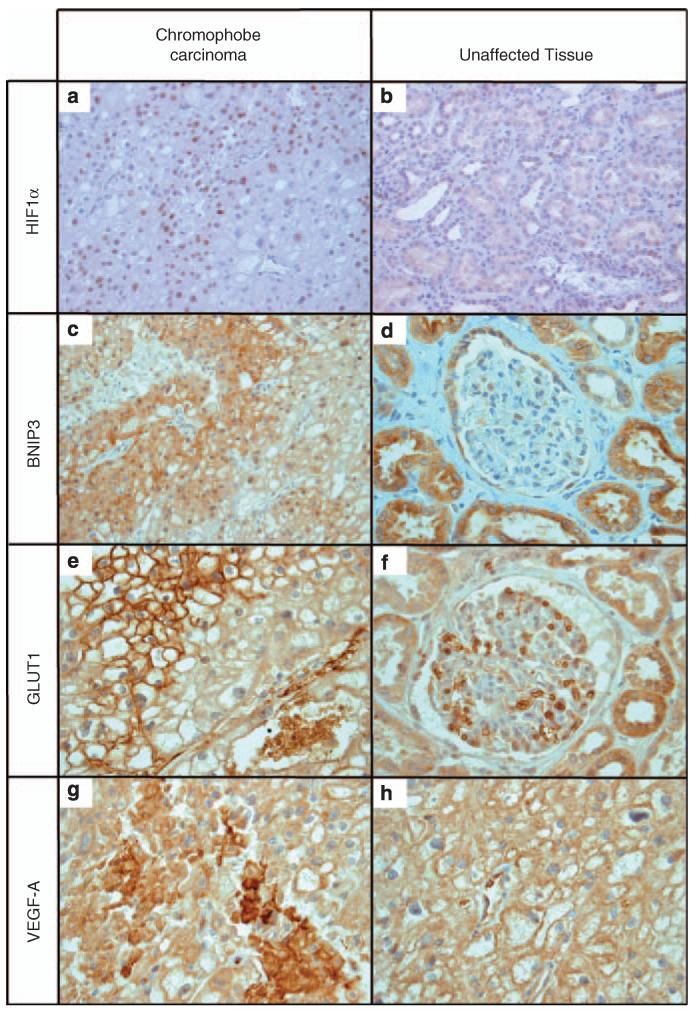
Immunohistochemical examination of a chromophobe renal carcinoma from a patient with BHD supports the in vitro data. Using antibodies against HIF1α, BNIP3, GLUT1 and VEGF-A (Figure 4), we observed staining patterns within the carcinoma that differed from those in unaffected parts of the kidney. Positive nuclear staining of HIF1α is evident within the cells of the chromophobe renal carcinoma showing a hypoxic environment (Figure 4a) and the intensity of HIF1α staining is higher when compared with unaffected tissue (compare Figures 4a and b). For BNIP3, staining of tumour cells was prominent but uneven, being more pronounced near the cell membrane (Figure 4c). This deviates from the pattern observed in unaffected tissue, in which diffuse staining is observed in tubules (Figure 4d). Tumour cells showed intense GLUT1 staining, again concentrated near the membrane (Figure 4e), whereas unaffected tubules displayed weaker and more diffuse staining (Figure 4f). Similarly, tumour cells demonstrated intense, uneven but diffuse VEGF-A staining (Figure 4g). Unaffected tissue did not stain appreciably above background levels (Figure 4h).
Figure 4.
Increased levels of HIF target proteins. Paraffin-embedded samples were obtained from a chromophobe renal carcinoma from a patient with Birt–Hogg–Dubé. There is strong and specific staining with antibodies directed against HIF1α (a, b) and the HIF targets BNIP3 (c, d), GLUT1 (e, f) and VEGF-A (g, h). Magnification is ×400 for all panels, apart from the HIF1α panels that are at ×200. Note the different staining patterns for BNIP3 and GLUT1 in chromophobe carcinoma compared with unaffected tissue. For VEGF, staining is much more intense in the tumour than in normal tissue but the intracellular distribution seems to be more diffuse. (e, f) Erythrocytes are clearly stained, serving as a positive internal control. In panels (g) and (h) vascular endothelium does the same for VEGF.
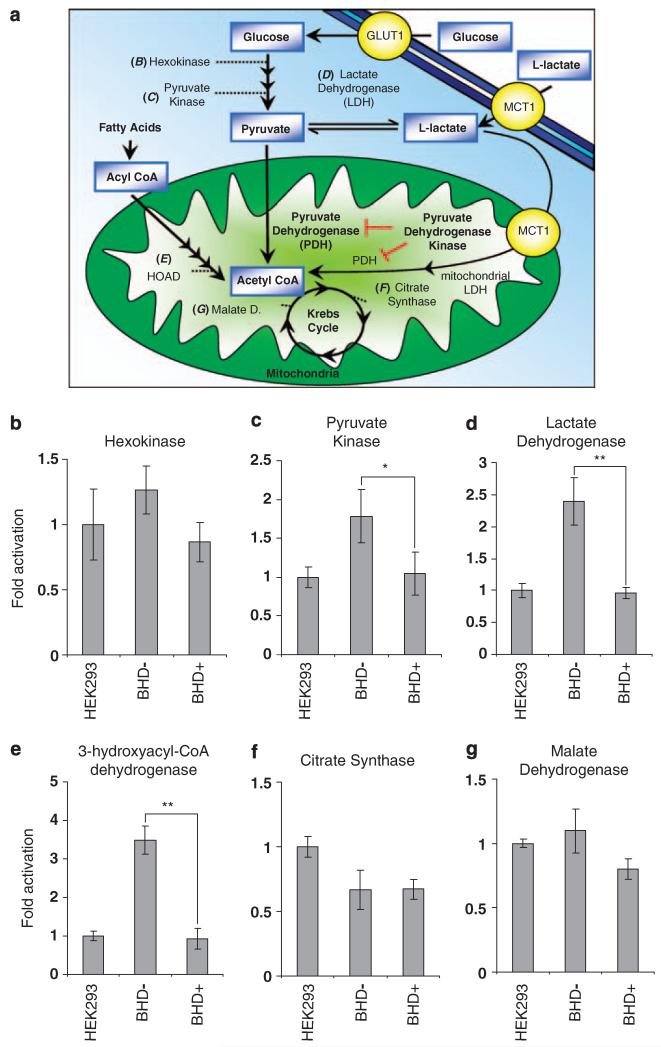
Analysis of metabolic enzyme activities within UOK257 cells is suggestive of an altered metabolic phenotype that reflects the ‘Warburg effect’
We hypothesized that the high level of HIF-mediated gene expression within the UOK257 cell line would alter their metabolism. To explore this notion, we analysed the activity of a series of metabolic enzymes involved in glucose metabolism, fatty acid oxidation and the Krebs cycle (also known as the tricarboxylic acid cycle). The enzymes examined are depicted in Figure 5a and were analysed from cells cultured in normoxic conditions. We did not observe any significant differences in hexokinase activity between HEK293 cells (the cell line that we used to standardize these metabolic assays) and the BHD− and BHD+ cell lines (Figure 5b). Hexokinase regulates the first step of glycolysis (phosphorylation of glucose to form glucose-6-phosphate), which is necessary to maintain a positive influx of glucose though glucose transporters such as GLUT1 (Robey and Hay, 2005). We observed a marked increase in the activity of pyruvate kinase (Figure 5c) and lactate dehydrogenase (LDH) (Figure 5d) in cells devoid of BHD. These higher levels of pyruvate kinase and LDH activity within BHD− cells would encourage the production of l-lactate. Indeed, the glycolytic enzyme, LDH-A, is known to be induced by oxygen stress (Firth et al., 1994). We also saw a significant increase of 3-hydroxyacyl-CoA dehydrogenase activity, which suggests that fatty acid oxidation might be also upregulated in BHD− cells (Figure 5e). Analysis of two Krebs cycle enzymes, citrate synthase (Figure 5f) and malate dehydrogenase (Figure 5g), showed no marked difference of activity between BHD− and BHD+ cells, and their activity was comparable to that of HEK293 cells. The ‘Warburg effect’ is a term used to describe a phenomenon in which cancerous cells convert glucose to l-lactate rather than use oxidative phosphorylation even when oxygen levels are adequate (Warburg, 1956). The data presented here suggest that BHD− cells possess a metabolic profile that parallels the observation that Otto Warburg made of cancer cells.
Figure 5.
Increased enzyme activity levels of pyruvate kinase and lactate dehydrogenase in BHD− cells. (a) Diagram of glucose and fatty acid metabolism depicting aerobic and anaerobic respiration. Enzyme activity assays were conducted to compare activity levels of (b) hexokinase, (c) pyruvate kinase, (d) lactate dehydrogenase, (e) 3-hydroxyacyl-CoA dehydrogenase, (f) citrate synthase, (g) malate dehydrogenase in BHD− (UOK257), BHD+ (UOK257-2) and HEK-293 cells. n = 3. *P<0.01; **P<0.05 when comparing enzyme activity in BHD+ and BHD− cells.
Aerobic glycolysis is favoured within UOK257 cells lacking BHD
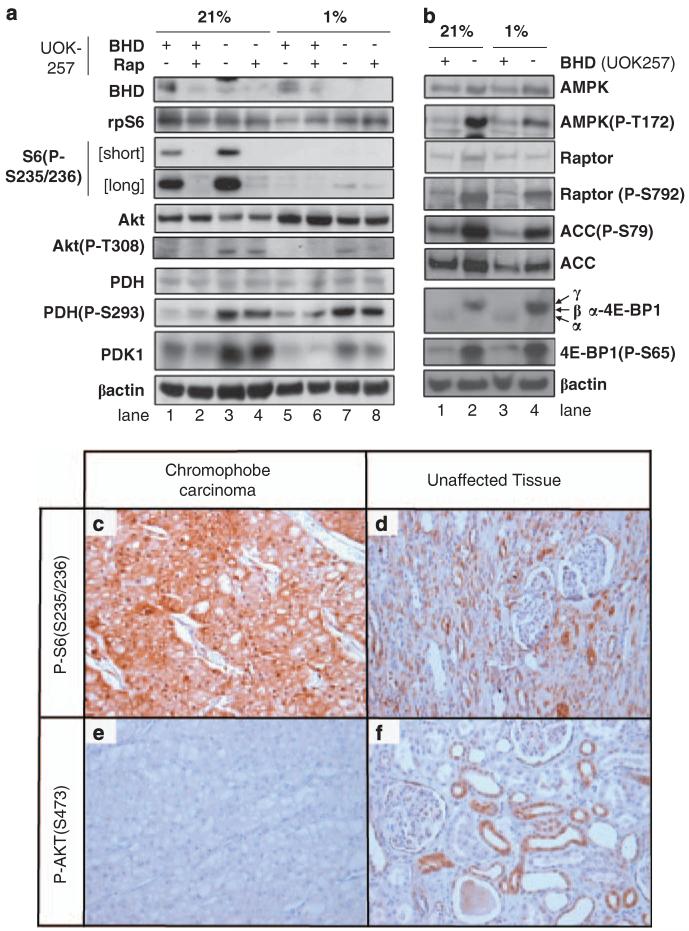
Pyruvate lies at the intersection of two glycolytic pathways: as a substrate for LDH enabling l-lactate production or as a substrate of the pyruvate dehydrogenase (PDH) reaction to generate the Krebs cycle entry molecule, acetyl CoA (see Figure 5a). Under low oxygen, PDH is typically inhibited through phosphorylation by pyruvate dehydrogenase kinase 1 (PDK1). As a consequence of PDH phosphorylation, pyruvate is preferentially converted to l-lactate by LDH (Wigfield et al., 2008) and oxidative phosphorylation is reduced, or entry of acetyl-CoA from fatty acid oxidation is preferred. To examine whether the activity of PDH was affected in BHD− cells, we analysed the protein expression of PDK1 and levels of PDH phosphorylation (Figure 6a). Of interest, we observed a robust increase in the levels of PDK1 in BHD− cells, when compared with BHD+ cells under conditions of both normoxia (Figure 6a, compare lanes 1 and 3) and hypoxia (Figure 6a, compare lanes 5 and 7). β-actin and PDH total protein expression levels are shown as protein loading controls. In accordance with the higher levels of PDK1 protein levels, PDH phosphorylation was markedly enhanced in BHD− cells (Figure 6a, lanes 3 and 7), indicating that PDH is inhibited in these cells. We also analysed the phosphorylation of both ribosomal protein S6 (rpS6) and Akt (also known as protein kinase B). rpS6 is routinely used to determine relative levels of signal transduction through the mTOR/70 kDa ribosomal protein S6 kinase 1 signalling pathway. As previously reported, the level of rpS6 phosphorylation is elevated in BHD− cells (Figure 6a, lane 3), indicating that they have a higher basal level of mTOR signalling (Hartman et al., 2009). Analysis of rpS6 phosphorylation revealed that rapamycin effectively repressed mTOR signalling within these BHD− cells (Figure 6a, lane 4). Under hypoxia, which is known to potently repress mTOR through multiple negative feedback loops (Liu et al., 2003), we were only able to detect a modest level of rpS6 phosphorylation within BHD− cells (Figure 6a, lane 7). We also observed an elevated level of Thr308 phosphorylation of Akt, which suggested that phosphoinositide-dependent kinase-1 activity within these BHD− cells is also elevated (Figure 6a, lanes 3 and 7). Our data reveal that there is an increase in the activity of LDH (Figure 5d) and in the phosphorylation of PDH within BHD− cells (Figure 6a), which implies that pyruvate might be converted more readily to l-lactate. If this is the case, the energy demands of the cell would have to be provided by either anaerobic glycolysis or by a shift towards fat-derived acetyl-CoA.
Figure 6.
Inactivation of PDH and activation of mTOR and AMPK in BHD− cells. (a) Western blot analyses were carried out on cell lysates obtained from BHD+ (UOK257-2) and BHD− (UOK257) cells after 18 h of normoxia (21%) or hypoxia (1%), with and without 50 nm rapamycin, where indicated. Protein levels of BHD, rpS6, phosphorylated rpS6 at Ser235 and Ser236, Akt, phosphorylated Akt at Thr308, PDH, phosphorylated PDH at Ser293, PDK1 and β-actin (used as a loading control) were determined. (b) Western blot analyses were carried out on cell lysates obtained from BHD+ (UOK257-2) and BHD− (UOK257) cells after 18 h of normoxia (21%) or hypoxia (1%), where indicated. Protein levels of AMPKα, phosphorylated AMPKα at Thr172, raptor, phosphorylated raptor at Ser792, acetyl-CoA carboxylase (ACC), phosphorylated ACC at Ser79, 4E-BP1, phosphorylated 4E-BP1 at Ser65 and β-actin (used as a loading control) were determined. Chromophobe renal carcinoma paraffin-embedded samples from a Birt–Hogg–Dubé patient show a strong and specific staining with phosphorylated rpS6 (c, d) but not Akt (e, f) antibodies. Magnification is ×200 for all panels.
BHD is considered a tumour suppressor, but it is currently unclear how BHD functions to repress cell growth. Folliculin-interacting protein 1 was found to interact with BHD and this protein complex is phosphorylated in an mTOR and adenosine monophosphate-dependent protein kinase (AMPK)-dependent manner (Baba et al., 2006). This evidence implies that BHD is a downstream signalling component of both mTOR and AMPK. Therefore, we examined AMPK signalling within these BHD− cells and observed a higher level of AMPK phosphorylation (Thr172 phosphorylation of the α-subunit, Figure 6b, lanes 2 and 4). Thr172, within the active loop of the catalytic-α subunit of AMPK, is indicative of its activity. To verify that AMPK activity was heightened in UOK257 cells, we analysed the phosphorylation status of two AMPK substrates, raptor and acetyl-CoA carboxylase. Indeed, we observed phosphorylation of both raptor and acetyl-CoA carboxylase at Ser792 and Ser79, respectively (Figure 6b, lanes 2 and 4), which supports our finding that AMPK activity is basally high in UOK257 cells lacking BHD. To examine mTOR signalling in more depth in these UOK257 cells, we analysed eukaryotic initation factor 4E-binding protein 1 (4E-BP1) phosphorylation, which is a well-characterized substrate of mTOR. 4E-BP1 resolves as three bands on SDS–PAGE, where the upper band represents the hyperphosphorylated ‘γ-isoform’ and the lowest band is the least phosphorylated ‘α-isoform’. We observe a higher level of expression of 4E-BP1 within UOK257 cells lacking BHD. Interestingly, 4E-BP1 is hyperphosphorylated in these BHD− cells. For instance, the majority of 4E-BP1 resolved as the γ-isoform band, and we observe an increase in the phosphorylation of 4E-BP1 on Ser65 (Figure 6b, lanes 2 and 4). The higher level of rpS6 and 4E-BP1 phosphorylation in UOK257 cells suggests that mTOR signalling is upregulated in these cells lacking BHD. We next examined mTOR activity in a chromophobe renal carcinoma from a patient with Birt–Hogg–Dubé by immunohistochemistry using antibodies against phosphorylated rpS6 (Figure 6c) and compared this with unaffected kidney cells (Figure 6d). Tumour cells showed intense rpS6 staining, showing that mTOR signalling is upregulated. Of interest, the level of Akt phosphorylation at Ser473 was lower in tumour cells compared with that in unaffected kidney tissue and suggests that Akt signalling is downregulated in the chromophobe renal carcinoma (Figures 6e and f).
UOK257 cells use l-lactate as a metabolic fuel
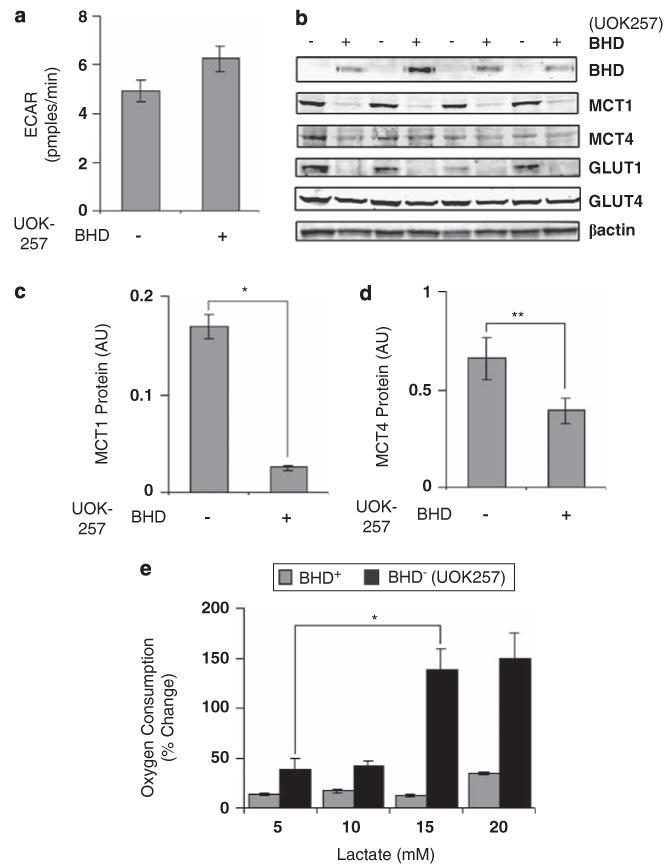
To test whether anaerobic glycolysis is increased in BHD− cells, we next sought to determine whether these cells produced more l-lactate through facilitated diffusion. Surprisingly, the extracellular acidification rate, a measure of glycolytic rate and l-lactate production, was not different between cells (Figure 7a). Of interest, BHD− cells had significantly more of the monocarboxylate transporter 1 (MCT1) associated with l-lactate influx and only marginally more of the efflux transporter MCT4 (Figures 7b, c and d). Consistent with these findings, BHD− cells were able to uptake and oxidize l-lactate better, when exogenous l-lactate was added to the medium (Figure 7e). These data suggest that the metabolic shift is not towards anaerobic glycolysis, as lactate can be taken up and oxidized.
Figure 7.
BHD− cells utilize l-lactate as a metabolic fuel. (a) The extracellular acidification rate (ECAR) was determined in BHD+ (UOK257-2) and BHD− (UOK257) cells under normoxia. (b) Western blot analyses were performed to determine protein levels of BHD, MCT1, MCT4, GLUT1 and GLUT4 from lysates prepared from BHD− and BHD+ grown in normoxia. Lysates from four different experiments were analysed. β-actin was used as a loading control. Densitometry analyses were also performed on (c) MCT1 and (d) MCT4 for the purpose of comparison. n = 4. *P<0.05; **P<0.01 relative to BHD+ and BHD− cells. (e) Oxygen consumption was also measured in BHD+ and BHD− cells treated with 5, 10, 15 and 20 mm l-lactate. n = 3. *P<0.05 relative to oxygen consumption within BHD− cells at 5 mm and 15 mm l-lactate.
2-deoxyglucose selectively prevents proliferation of BHD− cells
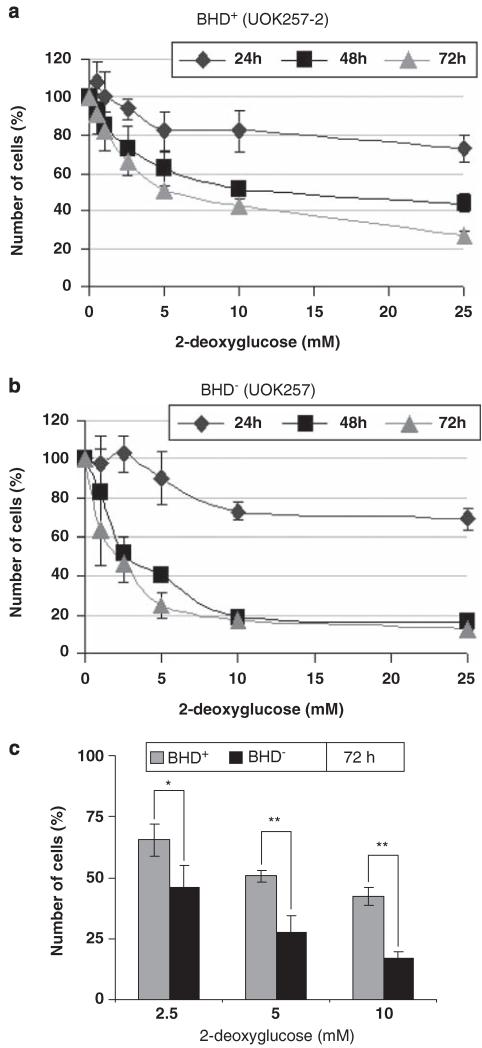
Our pyruvate kinase, GLUT1 and G6PD1 data suggest that the UOK257 renal-derived cell line from the Birt–Hogg–Dubé patient has adapted its metabolism to possibly use fatty acid-derived acetyl-CoA or to increase the production and utilization of the glycolytic intermediate l-lactate. To determine which hypothesis was more likely, we tested the dependency of BHD− cells on glucose by competitively blocking glycolysis using 2-deoxyglucose, and examined cell proliferation (Figures 8a and b). Consistent with a glycolytic requirement, BHD− cells were more sensitive to 2-deoxyglucose (IC50 at 48 h: 2.5 mm) than were BHD+ cells (IC50 at 48 h: 10 mm) and showed a significant reduction in cell proliferation after 72 h of treatment (Figure 8c). This suggests that BHD− cells have a higher dependence on glucose as a fuel. This inability to shift fuel oxidation to match availability is known as a loss of ‘metabolic flexibility’ (Galgani et al., 2008).
Figure 8.
Proliferation of BHD− cells is selectively inhibited with 2-deoxyglucose. (a) BHD+ (UOK257-2) and (b) BHD− (UOK257) were treated with 0, 1.25, 2.5, 5, 10 and 25 mm 2-deoxyglucose. The percentage of the original number of cells present in each cell line was then determined after 24, 48 and 72 h. n = 3. (c)The percentage of the number of BHD+ and BHD− cells after 72 h of treatment with 2.5, 5 and 10 mm 2-deoxyglucose from a and b was directly compared. n = 3. *P<0.05; **P<0.01.
Discussion
By altering their metabolism to favour aerobic glycolysis over lipid oxidation, cancerous cells meet their bioenergetic and biosynthetic demands and thus elicit a proliferative advantage (reviewed in Van der Heiden et al., 2009). This phenomenon was first described in 1924 by Otto Warburg, in whose study proliferating tumour cells consumed glucose at a high rate and released l-lactate and not CO2 (Warburg et al., 1924; Warburg, 1956). On first impression, it would seem more advantageous for cancer cells in an adequate oxygen environment to generate as much ATP as possible. Therefore, the Warburg effect might initially appear counterintuitive, as aerobic glycolysis generates less ATP than mitochondrial oxidative phosphorylation. However, the Warburg effect does incur some advantages to highly proliferating cells. For instance, glucose more readily enters the pentose phosphate pathway that leads to the generation of nicotinamide adenine dinucleotide phosphate, ribose-5-phosphate and erythrose-4-phosphate, which are used in the synthesis of fatty acids, nucleotides and aromatic amino acids, respectively. Such biosynthetic precursors are essential for rapidly proliferating cells (Chen et al., 2007).
Although the ‘Warburg effect’ is not apparent in all cancerous cells, these UOK257 cells emulate Warburg’s observation in that they seem to utilize glucose rather than fatty acids for generating ATP. The higher expression levels of the HIF gene target, GLUT1 (Figures 1e, 3c and 6b), would enhance glucose uptake. Inhibition of PDH through phosphorylation by PDK1 (Figure 6a) would block the entry of pyruvate into the Krebs cycle, whereas the elevated level of LDH activity within the cells (Figure 5d) would increase l-lactate production (Wigfield et al., 2008). Although secretion of l-lactate is a marker of aerobic or anaerobic glycolysis, we did not observe any noticeable difference in the extracellular acidification rate (Figure 7a) or an increase of l-lactate secretion in UOK257 cells under normoxia (data not shown). The lack of l-lactate accumulation within culture media could be due to an increase in MCT1 expression (Figures 7b and c), which is known to regulate l-lactate uptake (Garcia, 1995), as well as favoured entry of glucose into the pentose phosphate pathway (Figure 1d). Supporting a higher uptake of l-lactate by MCT1, we do observe a high capacity for these UOK257 cells to oxidize l-lactate (Figure 7e). Recent studies revealed that l-lactate can function as a major source of fuel in adequately oxygenated tumour cells (Sonveaux et al., 2008). l-lactate produced by the cells within the hypoxic core of a tumour is consequently taken up and oxidized by the surrounding oxygenated tumour cells. Utilization of l-lactate within the periphery of tumours as the main metabolic fuel might increase the amount of glucose that reaches and feeds the hypoxic tumour core. Such a mechanism would provide a better environment for tumour growth and expansion.
In the context of other inherited genetic disorders that give rise to RCC, HIF has a pivotal role in tumour progression in VHL (Kaelin, 2005), HLRCC (Isaacs et al., 2005) and TSC (Liu et al., 2003). Our data suggest that aberrant activation of HIF has a role in cancer progression of renal tumours in Birt–Hogg–Dubé patients. It might be argued that our data may be particular to UOK257 cells, which are derived from a clear-cell carcinoma and have chromosomal aberrations. However, our immunohistochemical data argue against this notion and suggest that our findings are more broadly applicable. In a chromophobe carcinoma from a patient with Birt–Hogg–Dubé syndrome, we find robustly increased levels of HIF targets BNIP3, GLUT1 and VEGF-A, and nuclear localization of HIF1α (Figure 4), paralleling our findings in UOK257 cells, in which we observe a higher activity of HIF. Therefore, we think that our findings can be generalized to multiple Birt–Hogg–Dubé tumour types. It is of interest to note the pronounced membranous localization of the GLUT1 staining (Figure 4e), which likely reflects the cells’ increased glycolytic potential. Of note, the expression pattern of BNIP3 has changed in the tumour compared with unaffected tissue (compare Figures 4c and d). BNIP3 does have a transmembrane domain and has a proapoptotic function. In the latter context, BNIP3 translocates to mitochondria (Van de Velde et al., 2000). However, BNIP3 has emerging functions in regulating autophagy and may therefore have protective functions during hypoxia. The apparent shift in staining intensity from cytoplasm to the cell membrane is a quite intriguing observation in this respect and might be consistent with BNIP3 fulfilling a protective rather than apoptotic function in the chromophobe carcinoma.
It is well appreciated that HIF1α and HIF2α are critical factors in promoting tumour progression by driving gene expression that leads to cellular changes in energy metabolism, cell survival, angiogenesis, glucose transport and metastasis (Semenza, 2004). When BHD expression is restored in UOK257 cells, we observe a robust inhibition of HIF-mediated gene expression (Figure 1) and HIF activity (Figure 2C), implying that BHD negatively regulates HIF. The high level of HIF-mediated gene expression observed in UOK257 cells is partially accounted for by the absence of BHD. This notion is supported by BHD knockdown experiments in ACHN cells, in which HIF activity was significantly enhanced upon loss of BHD expression (Figure 3). Collectively, our data suggest that BHD does not modulate HIF1α protein levels through gene expression or protein stability, but rather BHD regulates (directly or indirectly) the ability of HIF1α to function as a transcription factor. Given that BHD is implicated in downstream signalling from AMPK (Baba et al., 2006), a known modulator of HIF and energy metabolism (Treins et al., 2006), it is plausible that loss of BHD might dysregulate signalling through AMPK and consequently activate HIF. There are different mechanisms of dysregulated HIF signalling between inherited genetic disorders such as VHL, HLRCC and TSC. Enhanced stability of HIF-α proteins is the main contributing factor of cancer progression in both VHL (Bratslavsky et al., 2007) and HLRCC (Isaacs et al., 2005), whereas an upregulation of mTOR signalling is the main driving influence of HIF in TSC. BHD has also been implicated in the regulation of Akt and mTOR (Baba et al., 2006; Hasumi et al., 2009), which are signalling pathways known to potently activate HIF (Hudson et al., 2002). It is possible that dysregulated signalling through AMPK removes a negative regulator of mTOR signalling (Inoki et al., 2006 and Gwinn et al., 2008). It is of interest that the phosphorylation of the AMPK site within raptor, Ser792, is basally high in these UOK257 cells, which is a known signalling event that inhibits mTORC1 (Gwinn et al., 2008). However, mTORC1 signalling appears to still be active within these cells, as observed by an increase in rpS6 and 4E-BP1 phosphorylation levels. In particular, 4E-BP1 is maximally phosphorylated (Figure 6b). At first glance, high levels of mTORC1 substrate phosphorylation in cells, when mTORC1 activity should be repressed by AMPK-mediated phosphorylation of raptor, present a cell signalling paradox. One likely explanation is that BHD could be involved in the regulation of phosphatase complexes. Serine/threonine protein phosphatase 2A is known to be involved in mTORC1 signalling. It is possible that BHD could be involved in the regulation of phosphatase complexes, such as protein phosphatase 2A. The higher level of mTORC1 signalling within UOK257 cells (Figure 6) likely promotes the activity of HIF. In line with this notion, inhibition of mTOR with rapamycin treatment was able to impair hypoxia-driven gene expression of BNIP3 (Figure 1a), CCND1 (Figure 1b) and G6PD1 (Figure 1d) within these UOK257 cells. It is interesting that Akt phosphorylation was downregulated in the Birt–Hogg–Dubé patient’s chromophobe carcinoma (Figure 6e), and that parallels findings in which high levels of mTOR signalling due to loss of function of either TSC1 or TSC2 cause attenuation of Akt signalling (Harrington et al., 2004; Shah and Hunter, 2006). This is likely due to a higher level of 70 kDa ribosomal protein S6 kinase 1 activity in the tumour tissue, as indicated by the high levels of rpS6 phosphorylation (Figure 6c), which represses PI3K signalling through a negative feedback loop involving 70 kDa ribosomal protein S6 kinase 1-mediated phosphorylation of the insulin receptor substrates (Harrington et al., 2004).
Our study reveals that UOK257 cells have a higher dependency on glucose metabolism. This higher glucose dependency is likely caused by increased HIF activity and by an alteration of their metabolism. By targeting glycolysis with 2-deoxyglucose, which cannot be fully metabolized and thereby competitively blocks glycolysis, we were able to selectively inhibit the growth of UOK257 cells (Figure 8). This dependence on a single fuel for metabolic production of ATP is known as a loss of ‘metabolic flexibility.’ A loss of metabolic flexibility is a common feature of insulin resistance. In the case of insulin resistance, cells are unable to increase lipid metabolism in response to increased lipid supply. In BHD− cells, there is a similar inability to increase lipid metabolism when glycolytic production of l-lactate is lost. Therefore, impairing glycolytic production of l-lactate may be a feasible strategy for the treatment of BHD-associated lesions in renal carcinomas. A number of inhibitors of glucose metabolism are currently in preclinical or clinical development (Pelicano et al., 2006) and we suggest that these might be of use for the treatment of (unresectable or metastasized) renal cancer in Birt–Hogg–Dubé syndrome. Although our data show that HIF has mediated metabolic changes in the UOK257 cells, the exact mechanism by which BHD influences HIF signalling is unclear. Evidently, further studies to determine the cellular signalling mechanics of how BHD functions to regulate cell growth are still required.
Materials and methods
Antibodies and other biochemicals
Anti-BHD antibodies were kindly provided by Dr Arnim Pause, McGill University, Canada). Anti-PDK1, anti-β-actin, anti-BNIP3, anti-GLUT1 and anti-VEGF-A antibodies were bought from Abcam (Cambridge, UK). Anti-phospho-rpS6 (Ser235/236), anti-rpS6, anti-phospho-Akt (Thr308, Ser476), anti-Akt antibodies, antiacetyl choline carboxylase and antiphospho acetyl choline carboxylase (Ser79), anti-raptor and anti-phospho raptor (Ser792), anti-4E-BP1 and anti-phospho 4E-BP1 (Ser65), as well as anti-AMPKα and phospho-AMPKα (Thr172) antibodies, were purchased from Cell Signalling Technology (Danvers, MA, USA). Anti-phospho-PDH (Thr306, Ser473), anti-PDH, anti-GLUT4, anti-MCT1 and anti-MCT4 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). HIF1α and HIF2α antibodies for western blotting were from Novartis (Surrey, UK) and HIF1α and BNIP3 antibodies for immunohistochemistry were from New England Biolabs (Herts, UK) and Sigma-Aldrich Company Ltd. (Dorset, UK), respectively. Flag (clone: M2) and CCND1 antibodies and antibodies specifically used for immunohistochemistry (anti-BNIP3) and all other reagents used if not otherwise stated were obtained from Sigma-Aldrich. Rapamycin was purchased from Calbiochem/Merck (Beeston, Nottingham, UK). The N-terminal Flag-tagged BHD vector was a kind gift from Dr Laura S Schmitt (Bethesda, MD, USA, described in Baba et al. (2006).
Cell culture
UOK257 and UOK257-2 cells that were kind gifts from Dr Laura S Schmitt (Bethesda, MD, USA) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% (v/v) fetal calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco, Paisley, UK). UOK257-2 and UOK257 cells were incubated at either 21 or 1% oxygen, either with or without 50 nm rapamycin treatment. After 18 h, these cells were harvested. Stable clones of ACHN cells (purchased from ATCC No. CRL-1611) expressing scrambled or BHD-shRNA were generated using Mission shRNA constructs (purchased from Sigma-Aldrich Company Ltd.).
mRNA extraction and reverse transcription
Cells were first washed in phosphate-buffered saline, then lysed from 6 cm plates using 0.5 ml RNAprotect Cell Reagent (Qiagen, West Sussex, UK). RNA was extracted using a RNeasy Plus mini kit. Cell lysates were homogenized using Qiashredder tubes (purchased from Qiagen) during the RNA extraction procedure as described in the manufacturer’s protocol.
Quantitative real-time PCR
Total RNA from each sample (1 μg) was transcribed into complementary DNA using a Quantitect reverse transcription kit (Qiagen) in a thermal cycler (Applied Biosystems, Carlsbad, CA, USA). The sequences of the VEGF-A I-II primers used were forward 5′-CTGCTGTCTTGGGTGCATTG-3′ and reverse 5′-TTCACAATTTGTTGTGCTGTAG-3′, as described in Roland et al. (2000). All other primer sets were purchased from Qiagen, who have the right to withhold primer sequence information. Quantitative real-time PCR reactions were conducted in 96-well plates using appropriate primer assays and Sybr Green PCR Master mix (Qiagen). Assays were performed as follows: initial denaturation step (95 °C, 15 min), 40 cycles of denaturation (94 °C, 15 s), annealing step (55 °C, 30 s) and extension step (72 °C, 40 s). The amplification products were quantified during the extension step in the fortieth cycle. The results were then determined using the ddCT (delta-delta-Ct) method, and normalized first to β-actin. A dissociation step was performed, which verified that only one PCR product was produced with each primer set and shows their specificity. The correct size of PCR products was also verified by resolution on a 2% polyacrylamide gel. The expected size of the amplified products was approximately 70 bp for VEGF-A, 104 bp for β-actin (Catalogue number QT01680476), 73 bp for BNIP3 (Catalogue number QT00024178), 96bp for CCND1 (Catalogue number QT00495285), 91bp for G6PD1 (Catalogue number QT00071596) and 104 bp for HIF1α (Catalogue number QT00083664). The information given above is in accordance with the minimum information for publication of quantitative real-time PCR data as described in Bustin et al. (2009).
Western blotting
Cells were washed in phosphate-buffered saline and then lysed from the 6 cm plates using lysis buffer (20 mm Tris, 135 mm NaCl, 5% (v/v) glycerol, 50 mm NaF and 0.1% (v/v) triton X-100, pH 7.5 supplemented with complete mini protease inhibitor cocktail (Roche Diagnostics Ltd. Burgess Hill, UK) and 1 mm dithiothreitol (DTT)) at 4 °C. Following centrifugation at 13 000 r.p.m. for 8 min at 4 °C, the samples were prepared in ×4 NuPAGE LDS sample buffer (Invitrogen, Paisley, UK) with 25 mm DTT and boiled at 70 °C for 10 min. For preparation of HIF1α samples, cells were lysed directly into 62.5 mm Tris–HCl (pH 6.8), 2% (w/v) sodium dodecyl sulphate, 10% (v/v) glycerol, 50 mm DTT and 0.1 (w/v) bromophenol blue, followed by pulse sonication, and then subsequently boiled at 95 °C for 5 min. Samples were then separated by electrophoresis using the NuPAGE gel system (Invitrogen). Proteins were transferred to polyvinylidene difluoride membranes (Millipore, Watford, UK), blocked in 5% (w/v) dry milk powder/Tris-buffered saline 0.1% (v/v) tween, then probed using the required primary antibody and horseradish peroxidase-conjugated secondary antibody in Tris-buffered saline tween. Proteins were visualized using Enhanced Chemiluminescent solution and Hyperfilm (both from GE Healthcare, Buckinghamshire, UK). All western blots shown are representative of at least three independent experiments.
Luciferase assay
UOK257 cells were transfected with either pRK7 empty vector or Flag-tagged BHD with the firefly luciferase reporter pGL2-TK-HRE plasmid (a gift from G Melillo (National Cancer Institute at Frederick, Maryland, USA) using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s protocol. The pGL2-TK-HRE plasmid was generated by subcloning three hypoxia-response elements (5′-GTG ACTACGTGCTGCCTAG-3′) from the inducible nitricoxide synthase promoter into the promoter region of the pGL2-TK vector as previously described (Rapisarda et al, 2002). The medium was changed on cells 4 h post transfection. After 24 h post transfection, the cells were washed in phosphate-buffered saline and then lysed in lysis buffer (20 mm Tris, 135 mm NaCl, 5% (v/v) glycerol, 50 mm NaF and 0.1% (v/v) triton X-100, pH 7.5, supplemented with complete mini-protease inhibitor cocktail (Roche Diagnostics Ltd) and 1 mm DTT) at 4 °C. Following centrifugation at 13 000 r.p.m. for 8 min at 4 °C, 20 μl of each sample was injected with 50 μl luciferase reagent (20 mm Hepes (pH 7.7), 5 mm MgSO4, 1 mm d-luciferin and 2 mm ATP). A volume of 20 μg of protein lysate was analysed for luciferase levels using a luminometer 2 s and 10 s after initial injection.
Enzyme assays
The enzyme assay protocol in this study was carried out as described in Suarez et al. (1986), in which the cells were lysed directly into homogenization buffer (20 mm Hepes (pH 7.4), 2 mm EDTA, 0.1% (v/v) triton X-100, and 10 mm DTT). Lysates were pulse sonicated and centrifugated at 13 000 r.p.m. for 8 min at 4 °C. Protein levels were quantified using the Bradford assay. For hexokinase, pyruvate kinase and LDH enzyme assays, the samples were assayed in 50 mm imidazole-HCl (pH 7.4) under the following conditions: hexokinase: 5 mm glucose (omitted for control), 1 mm ATP, 5 mm MgCl2, 5 mm DTT, 0.5 mm nicotinamide adenine dinucleotide phosphate and excess G6PD1; pyruvate kinase: 5 mm phospho(enol)pyruvate (omitted for control), 5 mm adenosine diphosphate, 0.15 mm nicotinamide adenine dinucleotide (NADH), 10 mm MgCl2, 100 mm KCl, 0.02 mm fructose-1,6-bisphosphate, 5 mm DTT, excess LDH; LDH assay: 4 mm pyruvate (omitted for control), 0.15 mm NADH, 5 mm DTT. For the citrate synthase, malate dehydrogenase and 3-hydroxyacyl-CoA dehydrogenase enzyme assays, samples were assayed in 50 mm Tris–Cl (pH 8.0). For citrate synthase, 0.5 mm oxaloacetate (omitted for control), 0.3 mm acetyl CoA and 0.1 mm 5,5′-dithiobis(2-nitrobenzoic acid); for malate dehydrogenase, 50 mm imidazole-HCl (pH 7.4), 10 mm oxaloacetate (omitted for control), 0.15 mm NADH and 5 mm DTT; and for 3-hydroxyacyl-CoA dehydrogenase, 50 mm imidazole-HCl (pH 7.4), 0.1 mm acetoacetyl CoA (omitted for control), 0.15 mm NADH, 1 mm EDTA and 5 mm DTT were used. Enzyme activity (U/mg/P; U = 1 umol substrate converted to product per minute) was determined from the maximum rate of change in absorbance at 340 nm (NADH-linked assays) or 412 nm (DNTB (5,5′-dithiobis-(nitrobenzoic acid))-linked assays) at 37 °C using a Beckman Coulter DU650 spectrophotometer (Beckman Coulter (UK) Ltd., High Wycombe, UK). Results were normalized to the respective enzyme activity measured in parallel in HEK293 cells.
Immunohistochemistry
For BNIP-3, GLUT-1, VEGF-1A and phospho-rpS6 (Ser235/236), immunohistochemical staining was performed on paraffin-embedded samples of a chromophobe renal cell carcinoma (samples obtained during total nephrectomy). After deparaffination with xylene and rehydration, sections were incubated in 0.3% (w/v) hydrogen peroxide (H2O2) diluted in methanol (30 min) to inactivate endogenous peroxidases. Antigen retrieval was carried out by microwave treatment using citrate buffer (pH 6) for 10 min (90 W). Sections were then incubated for 60 min at room temperature with the primary antibody, and diluted in 3% (w/v) bovine serum albumin. After washing the samples in phosphate-buffered saline, antibody visualization was performed with Power-Vision+ (ImmunoVision Technology, Brisbane, CA, USA) using a horseradish peroxidase-conjugated secondary antibody that was incubated for 30 min, apart from phospho-rpS6 (Ser235/236), which was diluted in antibody diluent (Dako, Heverlee, Belgium) and incubated at 4 °C overnight. EnVision (Peroxidase/3,3-diaminobenzidine (DAB) detection systems, Dako) was used for secondary detection in phospho-rpS6. 3,3-diaminobenzidine was applied at all sections for 10 min to demonstrate the antigenic sites. Tissue was then counterstained with haematoxylin, dehydrated and coverslipped. For phospho-Akt(Ser473), sections were deparaffinized, rehydrated and boiled in citrate buffer (pH 6) for 20 min (microwave 90 W). Subsequently, endogenous peroxidase was inactivated in 3% (w/v) hydrogen peroxide (H2O2) diluted in methanol for 10 min. Primary antibody was diluted in 1% bovine serum albumin in Tris-buffered saline tween-20 and incubated overnight at 4 °C. For the remaining steps, the protocol was used as described above. HIF1α staining was performed using the Dako EnVison Flex kit (Dako). Briefly, sections were pretreated using the EnVision FLEX Target Retrieval Solution, of high pH (Dako), in a pretreatment module. After blocking endogenous peroxidase, sections were washed and incubated with the HIF1α antibody for 1h at room temperature. To improve antibody visualization, sections were washed and incubated with linker for 20 min. For secondary detection, EnVison was applied for 20 min and slides were subsequently stained with 3,3-diaminobenzidine for 10 min. Sections were counterstained with Gill II haematoxylin, dehydrated and coverslipped.
Cellular respiration
Cells were seeded in XF 24-well cell culture microplates (Seahorse Bioscience, Fisher Scientific, Vastra Frolunda, Sweden) at 2.0–3.0 × 104 cells per well (0.32 cm2) in 500 μl growth medium and incubated at 37 °C/5% CO2 for 24 h. The medium was changed before the cell respiration assay. Following a 30 min stabilization period, 75 μl of sodium lactate (Sigma, UK) was injected into each cell chamber to elicit a 5 mm concentration. This procedure was repeated on three more occasions resulting in a final concentration of 20 mm. A total of five measurements were made throughout each injection phase, with data being reported as the mean value for each phase. Following the protocol, cells from each well were lysed and determined for protein content as previously described (Deldicque et al., 2007).
2-deoxyglucose proliferation assay
Cells were counted and 5 × 103 cells were plated per well of a black 96-well plate (one plate for each time point). In total, 0.5, 1, 2.5, 5, 10, 25 mm 2-deoxyglucose were added 4 h after cells were plated and the plates were incubated at 37 °C for 24, 48 or 72 h. At the required time point, medium was removed from the cells and the plates were frozen at −80 °C. Following thawing, 200 μl Cyquant mix, prepared as described in the CyQUANT Cell Proliferation Assay (Invitrogen) manufacturer’s handbook, was added to each well. Fluorescence was then measured using a fluorometer at 485 and 520 nm.
Acknowledgements
This work was supported by The Association for International Cancer Research Career Development Fellowship (No 06-914/915) (to A Tee). The Myrovlytis Trust supports the labs of A Tee, M v Steensel and A Pause and Partnership for Cures supports A Tee and Mv Steensel. Funding was also provided by the Wellcome Trust (No 088032/Z/08/Z) (to S Land) and the Dutch Cancer Society (grants UM2009-4352 to M v Steensel and UM2010-4609 to T Brinkhuizen). M Kamps, A van Marion and V Winnepenninckx (all of the MUMC) provided valuable assistance with immunohistochemistry. Funding was also provided by the Engineering and Physical Sciences Research Council (EP/E008925/1) and the Biotechnology and Biological Sciences Research Council (BB/F002084/1) (to K Baar). We also thank Wales Gene Park.
Abbreviations
- AMPK
Adenosine monophosphate-dependent protein kinase
- BHD
Birt–Hogg–Dubé protein
- DTT
dithiothreitol
- G6PD1
glucose-6-phosphate dehydrogenase 1
- HIF
hypoxia-inducible factor
- HLRCC
hereditary leiomyomatosis-renal cell carcinoma
- LDH
lactate dehydrogenase
- mTOR
mammalian target of rapamycin
- MCT
monocarboxylate transporter
- PDH
pyruvate dehydrogenase
- PDK1
pyruvate dehydrogenase kinase 1
- RCC
renal cell carcinoma
- rpS6
ribosomal protein S6
- TSC
tuberous sclerosis complex
- VHL
Von Hippel-Lindau
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Baba M, Hong S-B, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signalling. P Natl Acad Sci USA. 2006;103:15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratslavsky G, Sudarshan S, Neckers L, Linehan WM. Pseudohypoxic pathways in renal cell carcinoma. Clin Cancer Res. 2007;13:4667–4671. doi: 10.1158/1078-0432.CCR-06-2510. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Chen Z, Lu W, Garcia-Prieto C, Huang P. The Warburg effect and its cancer therapeutic implications. J Bioenerg Biomembr. 2007;39:267–274. doi: 10.1007/s10863-007-9086-x. [DOI] [PubMed] [Google Scholar]
- Deldicque L, Theisen D, Bertrand L, Hespel P, Hue L, Francaux M. Creatine enhances differentiation of myogenic C2C12 cells by activating both p38 and Akt/PKB pathways. Am J Physiol. 2007;293:1263–1271. doi: 10.1152/ajpcell.00162.2007. [DOI] [PubMed] [Google Scholar]
- Dunlop EA, Tee AR. Mammalian target of rapamycin complex 1: signalling inputs, substrates and feedback mechanisms. Cell Signal. 2009;21:827–835. doi: 10.1016/j.cellsig.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Firth JD, Ebert BL, Pugh CW, Ratcliffe PJ. Oxygen-regulated control elements in the phosphoglycerate kinase 1 and lactate dehydrogenase A genes: similarities with the erythropoietin 3′ enhancer. Proc Natl Acad Sci USA. 1994;91:6496–6500. doi: 10.1073/pnas.91.14.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgani JE, Moro C, Ravussin E. Metabolic flexibility and insulin resistance. Am J Physiol. 2008;295:1009–1017. doi: 10.1152/ajpendo.90558.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Mejias R, Echevarria M, López-Barneo J. Induction of the glucose-6-phosphate dehydrogenase gene expression by chronic hypoxia in PC12 cells. FEBS Lett. 2004;569:256–260. doi: 10.1016/j.febslet.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Garcia CK, Brown MS, Pathak RK, Goldstein JL. cDNA cloning of MCT2, a second monocarboxylate transporter expressed in different cells than MCT1. J Biol Chem. 1995;270:1843–1849. doi: 10.1074/jbc.270.4.1843. [DOI] [PubMed] [Google Scholar]
- Guo K, Searfoss G, Krolikowski D, Pagnoni M, Franks C, Clark K, et al. Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Differ. 2001;8:367–376. doi: 10.1038/sj.cdd.4400810. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova NM, Mery A, Vasquez DS, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell Biol. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, et al. The TSC1-2 tumour suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman TR, Nicolas E, Klein-Szanto A, Al-Saleem T, Cash TP, Simon MC, et al. The role of the Birt–Hogg–Dubé protein in mTOR activation and renal tumourigenesis. Oncogene. 2009;28:1594–1604. doi: 10.1038/onc.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasumi Y, Baba M, Ailma R, Linehan W. Homozygous loss of BHD causes early embryonic lethality and kidney tumour development with activation of mTORC1 and mTORC2. Proc Natl Acad Sci USA. 2009;106:18722–18727. doi: 10.1073/pnas.0908853106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminki K, Jiang Y, Ma X, Yang K, Egevand L, Lindbland P. Molecular epidemiology of VHL gene mutations in renal cell carcinoma patients: relation to dietary and other factors. Carcinogenesis. 2002;23:809–815. doi: 10.1093/carcin/23.5.809. [DOI] [PubMed] [Google Scholar]
- Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, et al. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol. 2002;22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, et al. TSC2 integrates wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung Y-L, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Kaelin WG. The von Hippel–Lindau protein, HIF hydroxylation, and oxygen sensing. Biochem Bioph Res Commun. 2005;338:627–638. doi: 10.1016/j.bbrc.2005.08.165. [DOI] [PubMed] [Google Scholar]
- Khoo SK, Giraud S, Kahnoski K, Chen J, Motorna O, Nickolov R, et al. Clinical and genetic studies of Birt-Hogg-Dubé syndrome. Med Genet. 2002;39:906–912. doi: 10.1136/jmg.39.12.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuru M, Launonen V. Hereditary leiomyomatosis and renal cell cancer (HLRCC) Curr Mol Med. 2004;4:869–875. doi: 10.2174/1566524043359638. [DOI] [PubMed] [Google Scholar]
- Land S, Tee A. Hypoxia-inducible factor 1alpha is regulated by the mammalian target of rapamycin (mTOR) via an mTOR signalling motif. J Biol Chem. 2007;282:20534–20543. doi: 10.1074/jbc.M611782200. [DOI] [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, et al. Hypoxia-inducible factors regulate tumourigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MY, Poellinger L, Walker CL. Up-regulation of hypoxia-inducible factor 2 in renal cell carcinoma associated with loss of Tsc-2 tumour suppressor gene. Cancer Res. 2003;63:2675–2680. [PubMed] [Google Scholar]
- Maxwell PH. The HIF pathway in cancer. Semin Cell Dev Biol. 2005;16:523–530. doi: 10.1016/j.semcdb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Pelicano H, Martin DS, Xu R-H, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- Rapisarda A, Uranchimeg B, Scudiero DA, Selby M, Sausville EA, Shoemaker RH, et al. Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway. Cancer Res. 2002;62:4316–4324. [PubMed] [Google Scholar]
- Robey RB, Hay N. Akt, hexokinase, mTOR: targeting cellular energy metabolism for cancer therapy. Drug Discov Today. 2005;2:239–246. [Google Scholar]
- Roland I, Minet E, Ernest I, Pascal T, Michel G, Remacle J, et al. Identification of hypoxia-responsive messengers expressed in human microvascular endothelial cells using differential display RT-PCR. Eur J Biochem. 2000;267:3567–3574. doi: 10.1046/j.1432-1327.2000.01385.x. [DOI] [PubMed] [Google Scholar]
- Semenza G. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology. 2004;19:176–182. doi: 10.1152/physiol.00001.2004. [DOI] [PubMed] [Google Scholar]
- Shah OJ, Hunter T. Turnover of the active fraction of IRS1 involves raptor-mTOR- and S6K1-dependent serine phosphorylation in cell culture models of tuberous sclerosis. Mol Cell Biol. 2006;26:6425–6434. doi: 10.1128/MCB.01254-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slegtenhorst MV, Khabibullin D, Hartman TR, Nicolas E, Kruger WD, Henske EP. The Birt-Hogg-Dube and tuberous sclerosis complex homologs have opposing roles in amino acid homeostasis in schizosaccharomyces pombe. J Biol Chem. 2007;282:24583–24590. doi: 10.1074/jbc.M700857200. [DOI] [PubMed] [Google Scholar]
- Sonveaux P, Végran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumour cells in mice. J Clin Invest. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez RK, Brown GS, Hochachka PW. Metabolic sources of energy for hummingbird flight. Am J Physiol. 1986;251:537–542. doi: 10.1152/ajpregu.1986.251.3.R537. [DOI] [PubMed] [Google Scholar]
- Sudarshan S, Sourbier C, Kong H-S, Block K, Romero VV, Yang Y, et al. Fumarate hydratase deficiency in renal cancer induces glycolytic addiction and HIF-1 stabilisation by glucose-dependent generation of reactive oxygen species. Mol Cell Biol. 2009;29:4080–4090. doi: 10.1128/MCB.00483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treins C, Murdaca J, Obberghen EV, Giorgetti-Peraldi S. AMPK activation inhibits the expression of HIF-1a induced by insulin and IGF-1. Biochem Bioph Res Commun. 2006;342:1197–1202. doi: 10.1016/j.bbrc.2006.02.088. [DOI] [PubMed] [Google Scholar]
- Van der Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde C, Cizeau J, Dubik D, Alimonti J, Brown T, Israels S, et al. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol Cell Biol. 2000;20:5454–5468. doi: 10.1128/mcb.20.15.5454-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. The origin of cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Warburg O, Posener K, Negelein E. On the metabolism of tumours. Biochem Z. 1924;152:319–344. [Google Scholar]
- Wigfield SM, Winter SC, Giatromanolaki A, Taylor J, Koukourakis ML, Harris AL. PDK-1 regulates lactate production in hypoxia and is associated with poor prognosis in head and neck squamous cancer. Brit J Cancer. 2008;98:1975–1984. doi: 10.1038/sj.bjc.6604356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward ER, Ricketts C, Killick P, Gad S, Morris MR, Kavalier F, et al. Familial non-VHL clear cell (conventional) renal cell carcinoma: clinical features, segregation analysis, and mutation analysis of FLCN. Clin Cancer Res. 2008;14:5925–5930. doi: 10.1158/1078-0432.CCR-08-0608. [DOI] [PubMed] [Google Scholar]
- Yang Y, Padilla-Nash HM, Vira MA, Abu-Asab MS, Val D, Worrell R, et al. The UOK 257 cell line: a novel model for studies of the human Birt–Hogg–Dubé gene pathway. Cancer Genet Cytogen. 2007;96:336–340. doi: 10.1016/j.cancergencyto.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]