Abstract
Insufficient pharmacokinetic properties and poor cellular uptake are the main hurdles for successful therapeutic development of oligonucleotide agents. The covalent attachment of various ligands designed to influence the biodistribution and cellular uptake or for targeting specific tissues is an attractive possibility to advance therapeutic applications and to expand development options. In contrast to advanced formulations, which often consist of multiple reagents and are sensitive to a variety of preparation conditions, oligonucleotide conjugates are defined molecules, enabling structure-based analytics and quality control techniques. This review gives an overview of current developments of oligonucleotide conjugates for therapeutic applications. Attached ligands comprise peptides, proteins, carbohydrates, aptamers and small molecules, including cholesterol, tocopherol and folic acid. Important linkage types and conjugation methods are summarized. The distinct ligands directly influence biochemical parameters, uptake machanisms and pharmacokinetic properties.
Although oligonucleotides have been regarded as a new class of drugs for more than three decades, their therapeutic applications have so far failed to fully live up to the expectations [1,2]. Today, there are many different subclasses, which are mainly divided by different mechanisms of biological actions, and include antisense [3], splice-switching oligonucleotides [4,5], siRNA [6], miRNA [7], aptamers [8] and immunostimulatory oligonucleotides [9]. Structural differences are minimal, and consequently all nucleic acid-based drugs generally suffer from poor pharmacokinetics [10], the lack of cell membrane permeation [11], and often insufficient stability and sometimes relevant off-target effects [12]. Those properties are generally caused by their hydrophilicity, multiple anionic charges and ready interaction with degrading enzymes. Decades of therapeutic development have brought advances in enzymatic stability through chemical derivatization [3], and a multitude of distinct liposomal or polymeric delivery systems for improving their pharmacokinetic behavior [13]. Until 2012, one antisense agent (fomivirsen) and one aptamer (pegaptanib) had won market approval. Both are restricted to local application, circumventing the major problems. Mipomersen (Kynamro® [CA, USA]), an antisense agent targeted at apoB-100 for treatment of homozygous familial hypercholesterolemia [14-16], has been approved by the US FDA in January 2013, and could mark a major breakthrough for this type of biological therapeutic. Mipomersen, like other phosphorothioate drugs, accumulates in the liver, the site of apoB production. Clinical safety profiling showed a risk of hepatotoxic effects, including elevation of liver enzymes alanine aminotransferase and aspartate aminotransferase, and hepatic steatosis [17]. As a consequence, the FDA is requiring a risk evaluation and mitigation strategy program for mipomersen, including an enhanced pharmacovigilance program and a long-term registry of patients to determine the long-term safety.
Contrastingly, the European Medicines Agency has rejected the submission for market approval due to safety concerns. In Europe, the marketing application included a broader patient population, encompassing not only homozygous, but also severe heterozygous familial hyper cholesterolemia. However, mipomersen and other front-running oligonucleotide development candidates target the liver (Table 1), the organ most easily accessible after parenteral application. Other approaches for oligonucleotides intended for action in other tissues were ultimately unsuccessful in clinical evaluation. Prime examples are oblimersen and aprinocarsen, for which comprehensive Phase III trials were conducted, but ended with disappointing outcomes [18].
Table 1.
Clinical status of selected antisense and siRNA agents.
| Compound | Application route | Formulation/modification | Target | Indication | Company | Clinical status |
|---|---|---|---|---|---|---|
| siRNA oligonucleotides | ||||||
| Bevasiranib | Intravitreal | - | VEGF | AMD | Opko Health Inc. | Phase III, terminated |
| AGN-745 (siRNA027) | Intravitreal | - | VEGF | AMD | Allergan/Sirna | Phase II, terminated |
| SYL040012 | Ophthalmic drops | - | β2 adrenoreceptor | Glaucoma | Sylentis | Phase II |
| ALN-RSV01 | Inhalation | - | RSV nucleocapsid gene | RSV infection after lung transplantation |
Alnylam Pharmaceuticals |
Phase II |
| RXI109† | Intradermal | Modified siRNA (phosphorothioate, lipophilic ligands)† |
Connective tissue growth factor |
Dermal scarring after surgery |
RXi Pharmaceuticals | Phase I |
| QPI-1002 | Intravenous | Modified siRNA (alternating 2′-O-Me) |
p53 | Delayed graft function and acute kidney injury |
Quark Pharmaceuticals | Phase II |
| CALAA-01 | Intravenous | RONDEL™ (cyclodextrin polymer, PEG and transferrin ligands) |
M subunit of ribonucleotide reductase |
Solid tumors | Arrowhead Research Corporation |
Phase I |
| ALN-TTR02 | Intravenous | SNALP | TTR | TTR-mediated amyloidosis | Alnylam Pharmaceuticals |
Phase II |
| ALN-TTRsc† | Intravenous | GalNAc conjugate† | TTR | TTR-mediated amyloidosis | Alnylam Pharmaceuticals |
Phase I |
| ARC-520† | Intravenous | Dynamic polyconjugate† | Coagulation factor 7 (F7) | Hepatitis B virus infection | Arrowhead Research Corporation |
Phase I (mid-2013) |
| Antisense oligonucleotides | ||||||
| Mipomersen | Subcutaneous | PS-2′-MOE gapmer ODN | apo-B-100 | Homozygous familial hypercholesterolemia |
ISIS/Genzyme | Approved in USA |
| Alicaforsen | Intravenous, enema | PS-ODN | ICAM-1 | Pouchitis, ulcerative colitis | ISIS/Atlantic | Phase III (orphan drug status) |
| Drisapersen (PR0051, GSK-2402968) |
Intramuscular | PS-2′-O-Me ASO | Dystrophin (splice correction) |
Duchenne muscular dystrophy |
Prosensa/GSK | Phase III |
| Custirsen (OGX-011) |
Intravenous | PS-2′-MOE gapmer ODN | Clusterin | Prostate cancer, lung cancer |
ISIS/OncogeneX | Phase III |
| Miravirsen | Intravenous, subcutaneous |
PS-LNA gapmer ODN | miR-122 (Hepatitis C) |
Hepatitis C virus infection | Santaris Pharma | Phase II |
| Eteplirsen (AVI-4658) |
Intravenous | PMO ASO | Dystrophin (splice correction) |
Duchenne muscular dystrophy |
Sarepta Therapeutics |
Phase II |
| ISIS-TTR02 | Intravenous | PS-2′-MOE gapmer ODN, liposomal formulation |
TTR | TTR-mediated amyloidosis | ISIS | Phase II |
| Cenersen (EL-625) | Intravenous | PS-ODN | p53 | Acute myeloid leukemia | Eleos, Inc. | Phase II |
| GTI-2040 | Intravenous | PS-ODN | RRM2 (ribonucleotide reductase M2) |
Acute myeloid leukemia | Lorus Therapeutics | Phase II |
| Imetelstat (GRN163L)† |
Intravenous | N3′–P5′ thio- phosphoramidate, palmitoyl conjugate† |
Telomerase | Essential thrombocythemia, multiple myeloma, solid tumors |
Geron | Phase II |
| ISIS-STAT3RX, ISIS-481464 |
Intravenous | PS-cEt-BNA gapmer ODN | STAT 3 | Solid tumors, lymphoma | ISIS | Phase II |
| Liposomal Grb-2 (BP-100–1.01) |
Intravenous | P-ethoxy ODN liposomal formulation |
L-Grb-2 | Leukemia | Bio-Path Holdings, Inc. |
Phase I |
Oligonucleotide conjugates.
AMD: Age-related macular degeneration; ASO: Antisense oligonucleotide; cEt-BNA: Constrained ethyl bicyclic nucleic acid; GalNAc: N-acetyl galactosamine; LNA: Locked nucleic acid; MOE: Methyl oxethyl; ODN: Oligodeoxynucleotide; PMO: Phosphorodiamidate morpholino oligomer; PS: phosphorothioate; RSV: Respiratory syncytial virus; SNALP: Stable nucleic acid lipid particles.
Drisapersen is a promising representative of splice-correcting oligonucleotides. Recently, encouraging clinical results have been reported against Duchenne muscular dystrophy (DMD) by inducing the expression of a truncated, but functional splicing pattern of dystrophin, mutations of which cause the disease [19]. Similar to mipomersen, it exhibits a full phosphorothioate backbone, and 2′-O-methylated nucleosides. In contrast to antisense agents, RNase H activation is unwanted for splice correction, and consequently all of drisapersen’s nucleosides are methylated. Eteplirsen is a similar agent, also targeted at dystrophin, but featuring a different chemical modification; it is a phosphorodiamidate morpholino oligomer (PMO) [20]. A Phase II trial reported significant clinical benefit for DMD patients, and biochemical analysis proved the expression of a functional dystrophin protein in skeletal muscles [20,21]. Both agents take advantage of increased accumulation in muscular tissue due to tissue damage caused by the disease. Eteplirsen is rapidly eliminated by renal filtration because PMOs lack substantial binding to plasma proteins. As a result, high doses are necessary, and splice correction was not detected in all muscle tissues in animal experiments and clinical trials [20]. Dystrophin expression in cardiac muscle fibers is still a major hurdle.
These examples illustrate that despite some progress, the challenges of therapeutic application of nucleic acid-derived compounds are not yet solved. Delivery to tissues and organs other than the liver has not been achieved in a satisfactory manner. Toxicology issues, which are linked to the chemical modifications, are not sufficiently understood and even after market approval a major cause for concern. In some cases, oligonucleotides are rapidly eliminated by renal filtration, demanding high doses. For full realization of the enormous potential of oligonucleotide therapeutics, the pharmacokinetic properties need to be improved. Besides structural optimization and the development of delivery systems, the chemical coupling of ligands that modulate the pharmacokinetic behavior or target specific receptors is an attractive strategy for improving oligonucleotide therapy.
Over the last decades, the chemical derivatization of antisense and siRNA oligonucleotides has seemingly been carried out exhaustively [3,22-24]. It is probable that thousands of different modifications have been prepared and many of those evaluated for their therapeutic use. The outcome of these efforts suggest that structural optimization is a viable option to improve stability, but is, on its own, not sufficient to achieve proper bioavailability. Careful structural evolution has resulted in second generation antisense molecules, consisting of terminal 2′-O-methyl (2′-O-Me) or 2′-O-methoxyethyl (2′-O-MOE) nucleosides and a full phosphorothioate backbone. The backbone modification seems to be essential for triggering plasma protein binding and at least some extent of cellular uptake, however, the mechanism is not yet fully understood [11,25]. While 2′-alkylation supports resistance against enzymatic degradation, an internal stretch of deoxynucleosides is essential for activation of RNase H. This nuclease is vital for degradation of the corresponding mRNA and a catalytic antisense effect.
There are also high hopes for RNA interference by siRNAs. Compared with antisense agents, they offer improved effects through activation of the RNA-induced silencing complex. On the downside, this mechanism tolerates a significantly lower number of structural modifications. Advanced delivery systems, such as lipid particles, are essential for adequate half-lives and effective cell membrane permeation [26-30]. For achieving sufficient stability, a limited number of 2′-O-Me nucleosides are usually distributed within the sequence, but no backbone modifications are employed. Therapeutic development of siRNA is still in its infancy and major Phase III clinical trials are set to begin within the next few years (Table 1).
In addition, locked nucleic acids (LNAs) derivatives have been successful primarily in molecular biology techniques through their increased selective binding affinity to matching nucleic acids. Consequently, LNA modifications enable shorter nucleic acid sequences for effective hybridization [31]. This modification does not significantly influence the pharmacokinetic properties, and higher binding affinity is of minor importance for several therapeutic applications, including antisense and siRNA [31]. The use of LNAs is particularly promising for those clinical applications in which only short oligonucleotides sequences are used. Among those, miRNA antagonists are particularly attractive, because they are designed to bind to a short RNA, which leaves little space for sequence optimization. One promising substance is miravirsen, targeted at a short RNA, miRNA-122, which is currently being tested for the treatment of hepatitis C [32].
Currently, many research programs focus on the generation of delivery systems, primarily for siRNA [28]. The nucleic acid cargo can be covalently attached to a carrier, or loaded, usually by charge complexation or absorption, to supramolecular delivery devices. The latter includes lipid particles, multi-componental liposomes, cationic polymers and nanoparticulate systems of different classes. Without a doubt, the pharmacokinetic properties can be positively influenced by the use of these systems, but the successful therapeutic application in the clinical setting remains to be proven. In particular the challenges include reliable and reproducible constitution of the systems, which often consist of up to six different components, as well as immunogenicity, adequate stability with sufficient release and problems associated with large-scale manufacturing.
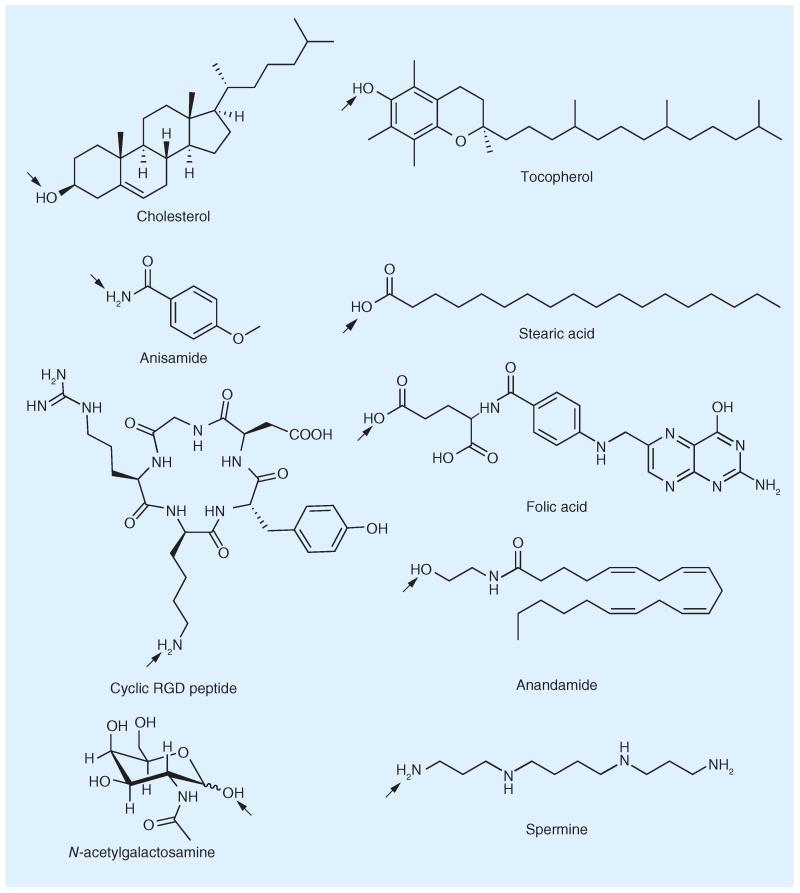
Conjugations of uptake-enhancing or targeting ligands to oligonucleotides offer the advantage of generating a defined molecule, which, in turn, enables traditional pharmaceutical quality assessment. Scientists are widely using oligonucleotide–dye conjugates as probes for qPCR [33], for nucleic acid sequencing and hybridization, as well as in microscopy applications such as fluorescence in situ hybridization and fluorescence resonance energy transfer [34]. A number of diverse molecules have been attached to therapeutic oligonucleotides in order to improve their delivery, biodistribution and cellular uptake [35,36]. Most conjugation strategies aim to simply improve biochemical properties and cellular uptake characteristics, but some attempts to combine tissue or tumor-targeting functionalities with oligonucleotide effector molecules have also been described. Among the ligands primarily designed to increase the extent of uptake into the cytosol are the lipophilic compounds cholesterol and tocopherol (Figure 1), cell-penetrating peptides (CPPs) such as penetratin, and polyamines such as spermine. In terms of achieving tissue-specific accumulation, proteins for antigen-specific binding, as well as small-molecule-based targeting moieties such as the carbohydrate N-acetyl galactosamine (GalNAc), have been employed as oligonucleotide ligands.
Figure 1. Selection of ligands for oligonucleotides for influencing pharmacokinetic behavior, membrane permeation or receptor-specific cellular uptake.
Arrows indicate attachment sites.
Conjugates with uptake-enhancing ligands
For therapeutic applications, conjugates of antisense and siRNA oligonucleotides have primarily been investigated, as both need to reach intracellular sites of action. Stability and biodistribution of siRNA oligonucleotides can be easily improved by the attachment of small lipophilic molecules. Cholesterol was tethered to siRNA for one of the first reports of silencing of an endogenous gene in vivo under physiological conditions with a normal pressure injection in mice [37]. Cholesterol was attached to the controlled pore glass solid support prior to oligonucleotide synthesis, and an aminocaproic acid pyrrolidine phosphate linker was used between ligand and siRNA. Cholesterol–siRNA conjugates resulted in an mRNA reduction of the targeted apoB of around 50%, while unconjugated siRNA had no effect. Similar results were reported for the lipid docosanyl and stearoyl ligands [38]. In vivo mRNA downregulation efficiency was found to be dependent on fatty acid length. Shorter compounds (<C18) were unable to reduce apoB mRNA levels, but stearoyl (C18) and docosanyl (C22) resulted in 30 and 50% reductions, respectively. Circulation time is increased through association of the conjugates with lipoproteins. Uptake in liver, but also in the kidney and adrenal glands is dependent on binding to lipoproteins and mediated by lipoprotein receptors [38]. Repeated applications of high doses were necessary for target downregulation, which limits the application, and the conjugation of cholesterol markedly increases accumulation in the liver. Thus, this strategy is all but confined to targeting hepatic diseases. Cholesterol–siRNA can be employed for noncovalent association to polymers, demonstrated by in vivo gene silencing in combination with a targeted engineered polymer [39]. Later developments focused on the improvement of lipid particles [26,27,40,41], which have similar characteristics, but are more effective and allows easy structural optimization for increased stability and effect.
Imetelstat is a N3′-P5′ thio-phosphoramidate oligonucleotide conjugated with a palmitic acid ligand tethered to the 5′-phosphate via an ethylene glycol linker [42]. The 13-mer oligonucleotide is not an antisense agent, but directly binds to the active site of the human telomerase enzyme, and the lipid conjugate is currently in Phase I/II clinical evaluation as an anticancer agent [43]. The chemical oligonucleotide modification induces sufficient stability, and the lipid ligand markedly increases cellular uptake and bioavailability. In cell lines, IC50 values of the palmitoyl conjugates were five- to ten-times lower than those of the parent oligonucleotides. In a mouse xenograft tumor model, the lipid conjugate demonstrated significantly more uptake in the tumor and the liver, and inhibited telomerase activity by more than 50%, while the unconjugated siRNA was ineffective [42].
α-tocopherol (vitamin E) was attached to an apoB1-targeted siRNA at the 5′-position via a phosphate linkage [44]. Gene silencing in the liver was achieved at lower concentrations as needed for cholesterol conjugates, but uptake is similarly mainly limited to the liver. In vitro uptake relied on the presence of serum, indicating an interaction with tocopherol-binding proteins with subsequent uptake into the cytosol. Pre-absorption of the tocopherol conjugate to HDL allowed efficient gene silencing in neurons after intracerebroventricular infusion [45]. The LDL receptor was identified as an essential factor for successful uptake, which was absent for unconjugated siRNA.
Because the high anionic charge density is a major factor for the poor pharmacokinetics of oligonucleotides, complexation or covalent attachment of cationic molecules has been extensively performed and evaluated. In particular, basic peptides have been tethered to oligonucleotides in a large variety of ways. Several, usually highly cationic, peptides have the ability to cross cellular membranes [46]. Those peptides are known as CPPs or protein transduction domains [47,48]. Although the exact mechanism of uptake is not completely understood, it has been established that CPPs interact with several anionic cell-surface molecules. Proteoglycans such as syndecans are negatively charged anchoring groups on cellular membranes, and CPPs, including their cargo, are subsequently internalized into the endosome [49]. Depending on poorly characterized properties including size and structure of the internalized complex, the effectiveness of cytosolic delivery is variable [36]. Peptide–oligonucleotide conjugates have been investigated extensively [50], and a considerable number of different possibilities of chemical conjugation have been developed. Overall, uncharged oligonucleotides seem to give better results in conjugates with CPPs than their charged counterparts. Interaction of lysine and arginine side chains with phosphates or phosphorothioates lead to aggregation into larger particles, which, in turn, seems to hinder effective cytoplasmatic delivery. Initially, CPP-mediated uptake was believed to be a nonendocytotic mechanism, but recent evidence strongly points to cellular uptake via clathrin-mediated endocytosis [51]. The observed higher efficiency of CPP-mediated transfection is probably attributable to a higher rate of endosomal escape. While a considerably high number of different CPP–oligonucleotide conjugates have demonstrated successful uptake in vitro and in vivo, no clinical trials have been undertaken so far.
Morpholino oligonucleotides, which particularly suffer from poor cellular uptake, have been conjugated to arginine-rich CPPs [52-54]. Lacking any charged backbone functionalities, they especially profit from CPP conjugation. Preclinical data reports that by conjugation of arginine-rich peptides to the parent PMO, significantly lower doses are sufficient for exon skipping in mouse models of DMD [54]. Dystrophin expression was found in all tested muscle tissue, including cardiac muscle [55]. Conjugations increased cellular uptake, but the majority of the dose ends up in liver and kidney. Attempts to increase muscle selectivity by fusing a muscle-specific peptide selected by phage display illustrated lower effects, but combining this peptide with a basic peptide for enhancing cellular uptake resulted in an enhanced effect [56]. However, little tissue selectivity was achieved [54]. These data indicate that conjugations of short basic peptides may be a viable option for overcoming some of the limitations of uncharged PMOs. In a comparison of different CPPs conjugated to either the 3′- or the 5′-end of PMOs in Ebola virus-infected mice, a correlation of virus replication inhibition with a number of arginines in the peptide ligand was found. Efficient replication inhibition was observed with a peptide containing eight arginine residues [57], while PMOs lacking any positive charge had no effect at all.
Similar to basic peptides, polyamines can be used to alleviate the negative consequences of the anionic charges of oligonucleotides. Fusing serveral spermine molecules to an oligonucleotide results in increased binding affinity to its target counter strand, mediated by a reduction of electrostatic repulsion between the two strands [58,59]. No data on biological activity of these conjugates have been reported. In a similar approach, an attachment of polyamines at the 2′-position of antisense oligonucleotides demonstrated a dependency on binding affinity and in vitro gene silencing effect on the length of the polyamine, and, thus, the number of cationic charges [60]. Short polyamines all but abolished the gene-silencing activity, but conjugates with spermidine and an artificial pentaamine ligand had a higher effect on the gene target than the unmodified antisense agent. Shorter ligands hinder the hybridization to the mRNA due to steric effects, but with longer polyamine chains, the charge interaction prevails. In both approaches, an aggregation towards nanoparticulate structures can be assumed, which can be speculated to increase cellular uptake.
Pegaptanib is one of only three oligonucleotide drugs having been approved for therapy. It is a PEGylated aptamer targeted at VEGF and applied locally for treatment of age-dependent macular degeneration [61]. The branched, 40 kDa PEG chain attached at the 5′-end prolongs half-life and increases retention in the eye [62]. In vitro, a conjugate of PEG 5000 and an anti-VEGF siRNA was transfected into cultured cells using polyethylene imine, resulting in reduction of VEGF protein release [63]. The PEGylation slightly prolonged serum stability, however there is no indication of uptake enhancement in the absence of a transfection agent.
Conjugates with ligands for receptor-specific targeting
Antibodies, antibody fragments [64,65] and alternative binding protein scaffolds [66] promise antigen-dependent and cell-type specific delivery of drug cargoes. This is certainly an attractive option for antisense and particularly siRNA agents, which rely on delivery enhancers. While a number of approaches using complexes of oligonucleotides and antibody derivatives [30,67-69], biotin–streptavidin coupling [70] and antibodies fused to liposomes [71,72] have been reported, little research has been undertaken on direct covalent conjugation of protein binders to oligonucleotides. Site-specific derivatization of antibodies is a challenging task because of the protein structure. Linkers for oligonucleotide attachment can be tethered at lysine-amines [73] or cysteine-thiols [74], usually rendering mixtures of multiple-loaded antibodies, but not defined chemical structures. Together with high costs, these challenges and concerns of immunogenicity have limited the approach [75,76]. Alternative protein scaffolds generally offer easier derivatization [77-79], but their use for oligonucleotide targeting remains to be evaluated.
A recent example of antibody–oligonucleotide conjugate employed an α-CD19 monoclonal antibody and a phosphorothioate antisense oligonucleotide targeted at the fusion gene E2A-PBX1 [80]. α-CD19 is a B-lineage-specific surface receptor and a biomarker for acute lymphoblastic leukemia (ALL), and the E2A-PBX1 fusion gene is a transcriptional activator, and the result of a common chromosomal translocation in ALL. A disulfide linker was used, and a 15-fold surplus of antisense oligonucleotide was added to the antibody, but only a fraction successfully reacted [80]. A high extent of specific in vitro and in vivo silencing of the fusion gene was reported, with the pharmacological effect depending on both active antibody binding and the functional antisense sequence. A near complete reduction of clonogenic activity of ALL cell lines as well as significant increase in the survival of mice with xenograft tumors was found.
Small-molecule ligands offer a much easier chemical conjugation methodology. Among the receptor-binding molecules that have been used for shuttling covalently attached oligonucleotide cargo into cells, folate [29,81] and anisamide [82] have been used most extensively. Folic acid (vitamin B9) binds with high affinity to the folate-receptor protein and triggers cellular uptake via an endosomal pathway. The presence of the folate receptor on many cancer types has prompted the use for targeted therapy [83]. Unlike the use of folate on liposomes or polyplexes, tethering folate to siRNA has resulted in specific uptake, but not silencing of reporter genes [81,84]. It was concluded that the conjugates end up in endosomes with insufficient escape to the cytosol for gene silencing. Complexing the constructs with a polycationic transfection enhancer resulted in effective, and targeted downregulation of the reporter [81]. The gene-silencing effect was dependent on the presence of the folate, as unconjugated siRNA absorbed to the transfection enhancer was inactive.
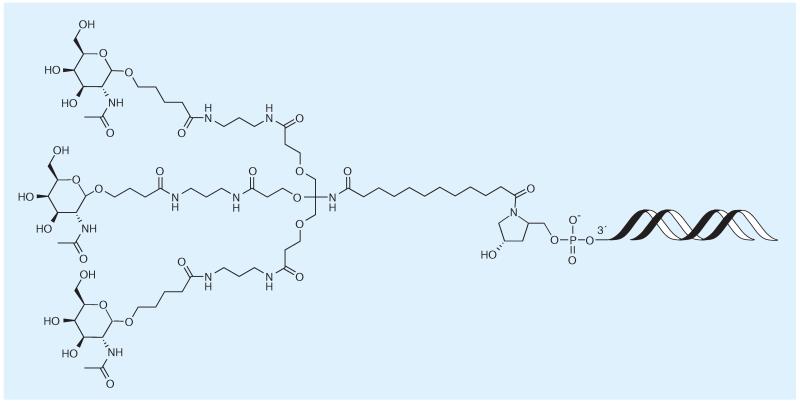
The monosaccharide GalNAc is able to induce targeting to hepatocytes by binding to the asialoglycoprotein receptor (Figure 2) [85]. It has been used both as a targeting functionality on lipid particles and as a component in siRNA conjugates [86]. Alnylam Inc. (MA,USA) is advancing its GalNAc conjugates for subcutaneous injection towards clinical trials. For increasing binding avidity through multivalency, three GalNAc molecules are attached via a tridentate linker to the 3′-terminus of the sense strand of an siRNA. Animal experiments demonstrated encouraging efficacy data after subcutaneous injection of ALN–TTRsc, targeted at transthyretin for the treatment of amyloidosis [201]. In a similar drug candidate, a GalNAc conjugate is tested against hemophilia A and B by siRNA-mediated silencing of antithrombin III, which is an important regulatory protein for maintaining hemostasis. No comprehensive report on preclinical data has been published, but preliminary results have been presented at several scientific meetings and on the company’s homepage [201].
Figure 2. N-acetyl galactosamine–siRNA oligonucleotide conjugates.
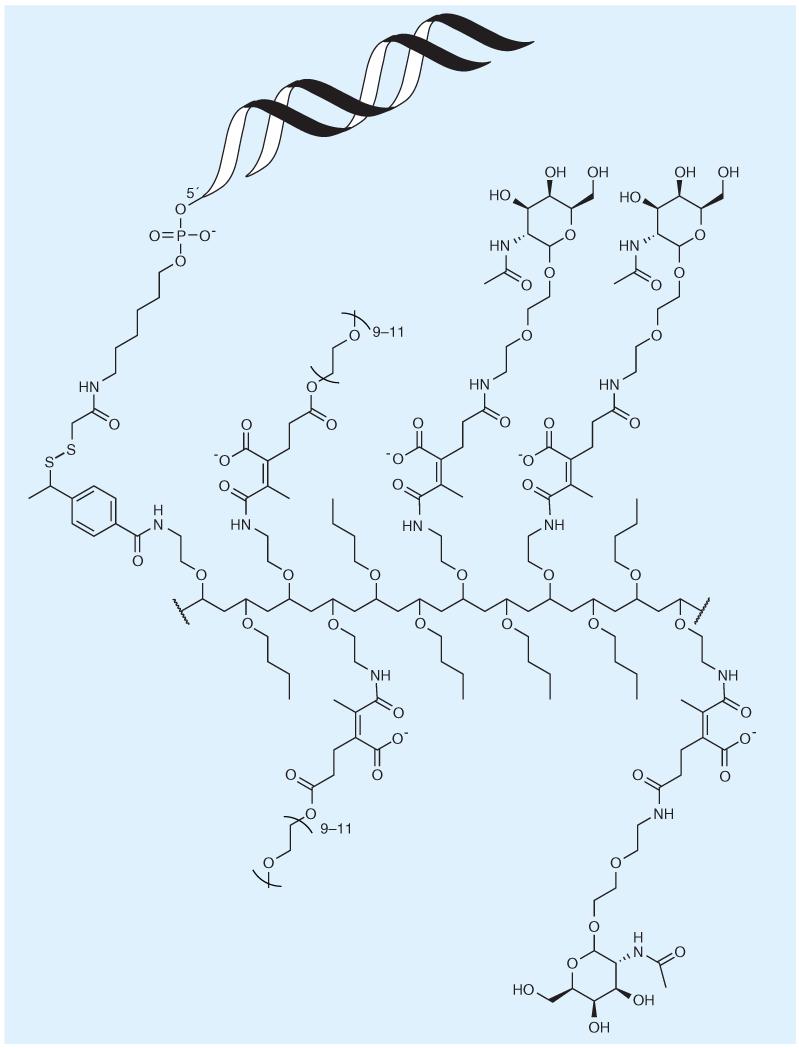
Arrowhead Research Coperation (CA, USA) is developing dynamic polyconjugates for siRNA delivery (Figure 3). Herein, GalNAc is also used for hepatocyte targeting via the asialoglycoprotein receptor. Instead of directly attaching GalNAc to the siRNA, the cargo is tethered to the polybutyl aminovinyl ether polymer via a disulfide linkage [87]. The endosomolytic polymer is consequently masked by attachment of PEG chains and GalNAc with a dialkylmaleic acid linker [88]. After successful cellular uptake, the delivery system is designed to be cleaved into the single components, because the disulfide and acid-labile dialkylmaleic acid tether are unstable in the reducing and acidic endosomal environment [29]. The siRNA is either directly linked to the polymer via a disulfide tether [88], or a cholesterol–siRNA conjugate is co-injected with the dynamic polyconjugate [89]. Clinical testing of a corresponding agent, ARC-520, is scheduled to begin in 2013.
Figure 3. Dynamic polyconjugates with covalent attachment of siRNA.
Anisamide is a high-affinity ligand for sigma receptors, and has been used in targeted delivery approaches on lipid drug and siRNA nano carriers [90], as well as a ligand covalently attached to a splice switching oligonucleotide [82]. Both uptake and splice switching was increased compared with an unconjugated oligonucleotides. A trivalent construct demonstrated an increase in uptake more than twofold compared with the unconjugated oligonucleotides, while a monovalent compound had only approximately 1.5-fold higher intracellular accumulations. In terms of splice switching, the trivalent conjugate was three- to four-times more effective than the naked siRNA and the monovalent conjugate, which resulted in no increase compared with the siRNA [82].
The RGD tripeptide is an attractive, small peptide for integrin targeting [91]. Integrins are cellsurface receptors with high expression in various cancer types like breast cancers, melanoma and glioblastoma [92]. Cyclic RGD peptides (cRGD) are ligands for αvβ3 integrin. Bivalent attachment of cRGD via a lysine to a splice-switching oligonucleotide [93] or siRNA [94] markedly increased uptake. Interestingly, when comparing bi-, tri-, and tetra-valent cRGD–siRNA conjugates, the uptake efficiencies of fluorescently tagged oligonucleotides were very similar, but gene silencing effect was correlated to the number of cRGD peptides [94]. Obviously, effects after the initial uptake are responsible for the divergent pharmacological results, however the detailed mechanisms of uptake and trafficking are not yet completely understood.
Another recent example of a ligand introduced for receptor-mediated endocytosis involves anand amide, designed for binding to the can-nabinoid receptor [95]. In a leukemia cell line reporter gene system, the anandamide conjugate was more potent than isosequential cholesterol and folate conjugates [96]. High concentrations of conjugates of up to 1 μM were necessary to achieve up to 50% reduction of the reporter gene.
The cyclic insulin-like growth factor receptorbinding peptide D-(CSLC) was tethered to an siRNA to enable specific cellular uptake [97]. Relatively high doses were necessary for efficient target downregulation in the breast cancer cell line MCF-7, and the uptake and gene silencing efficiency of approximately 50% was very similar to that of cholesterol–siRNA conjugates.
The gastrin-releasing peptide receptor is a G-protein coupled receptor that regulates a number of functions in the gastrointestinal system. It is up regulated in many human cancers, and the 14 amino acid-peptide bombesin is a specific ligand. Maleimide–thiol coupling of a splice-switching oligonucleotide to bombesin improved cellular uptake in gastrin-releasing peptide receptor-expressing cells approximately twofold compared with unconjugated siRNA [98]. The splice-switching effect was successfully competed with free bombesin. Compared with lipofectamine transfection, uptake efficiency and pharmacological response were rather low. The maximal biological effect was detected after 3 days, indicating slow intracellular trafficking.
Aptamers can be engineered to target virtually any protein. Aptamers binding to extracellular proteins are an obvious choice for the use as the cell-recognition proportion of conjugates with antisense or siRNA agents. Aptamers are typically 20–60 base long oligonucleotides and mediate selective ligand binding through their characteristic 3D shape [99]. Aptamers against specific antigens can be developed by selection and evolution of a library by the systematic evolution of ligands by exponential enrichment process. In addition, aptamers specific for cell-types, without prior identification of a molecular target, can be engineered using cell-systematic evolution of ligands by exponential enrichment [100]. Aptamer-siRNA or aptamer–antisense chimera, that is oligonucleotides with an aptamer and an antisense or siRNA part, can easily be prepared by chemical synthesis of long oligonucleotides [101,102]. Other options for aptamermediated delivery are post-synthetic linkages, which afford endosomal cleavage by using appropriate tethers, and noncovalent linkage by partial sequence overlaps [103].
Conjugation reactions & linkage types
There are several positional options for ligand attachment to oligonucleotides, which can have an influence on the biophysical properties (Figure 4). For antisense agents, tethering ligands to the 3′- or 5′-hydroxyl terminal groups is the most popular method, because little inference with base pairing, even when sterically large ligands are used, can be expected. Furthermore, these positions are technically easy to access, and even enable online conjugation methodologies that prepare the final bioconjugates on a single solid support [104]. Similarly, aptamer chimeras are synthesized using solid-phase chemistry for long-chain oligonucleotides [102]. In the case of double-stranded siRNA, the sense strand is typically preferred for ligand tethering over the antisense strand, because, although the duplex is recruited in the RNA-induced silencing complex, only the antisense strand is needed for mRNA hybridization and cleavage [105]. Consequently, less influence on RNAi activity is expected for sense-modified siRNAs.
Figure 4. Selection of ligand attachment sites in oligonucleotides.
Most linkers can be used at the 3′- and 5′-ends, which are technically the easiest to access, followed by 2′-hydroxy groups and nucleobases.
Small-molecule ligands, such as cholesterol, tocopherol, GalNAc and fatty acids, have been directly added to controlled pore glass resins for use in solid-phase oligonucleotides synthesis. By appropriate design of the structure, including protecting groups, DMT-containing linker for nucleotide coupling, and a cleavable site, such resins are available for standard oligonucleotides synthesis [104]. This elegant approach avoids the need for separate ligand coupling and elaborate purification of the conjugate from a mixture of educts.
The generation of peptide–oligonucleotide conjugates is possible via post-synthetic coupling, that is separate assembly of peptide and oligonucleotides using their respective automated solid-phase syntheses, and stepwise (online) solid-phase assembly in succession on the same solid support. While the latter holds obvious advantages in terms of purification, the noncompatible standard syntheses of the two components have so far limited the development of general and feasible procedures [106]. Postsynthetic protein or peptide coupling yields strongly depend on structure and solubility of the protein or peptide. Highly basic compounds typically interact with anionic oligonucleotides, impeding efficient conjugation. Workup and purification of the final conjugate can be tedious.
The 2′-O-hydroxy function of RNA–oligonucleotides offers the possibility to attach one or multiple ligands at internal positions [23,24,107,108]. Since groups attached there are localized in the minor groove of a duplex, interference of counter-strand hybridization is limited. Except for a change towards the A-form of nucleic acids of DNA-based oligonucleotides, the impact on helical structure can expected to be negligible. The attractiveness of 2′-O-conjugations is adequately reflected in the importance of 2′-modification for structural stability of antisense and siRNA oligonucleotides, including 2′-O-Me, 2′-O-MOE and 2′-fluor chemistries [1].
Other options for ligand attachment include the nucleobases [96], especially the 5-C-atom of pyrimidine bases cytosine, thymine, uracil, the 8-C of adenine, and the exocyclic amino group (2-N) of guanine, as well as the linker tethering at the backbone phosphate. Using these anchoring groups will generally result in a more profound impact on base pairing. By incorporation at a terminal position within the oligonucleotide sequence, this detrimental effect can be minimized [96]. Ligand attachment to nucleobases can either be afforded by using prederivatized nucleoside building blocks [104], or by incorporating functional linkers for post-synthetic reactions.
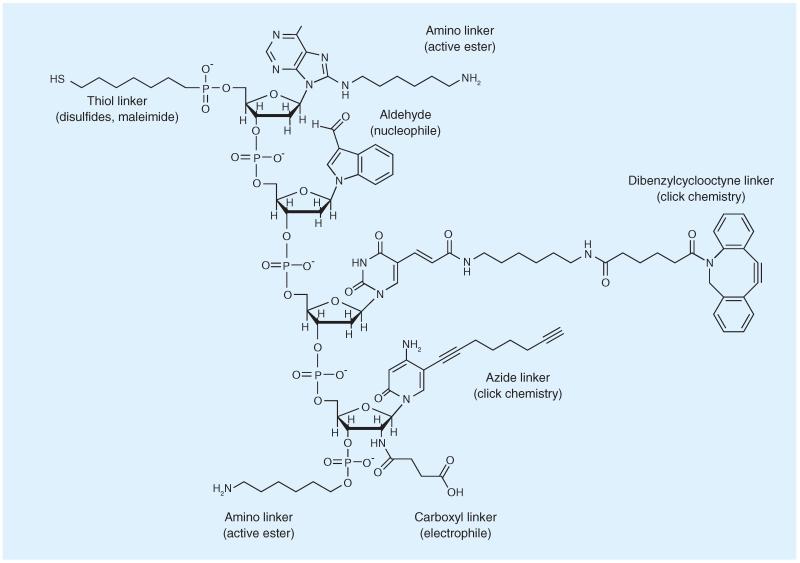
The employed linker holds great importance for biophysical properties and can play a role in the pharmacokinetic behavior of the conjugate. A large number of different ligation methods have been used for oligonucleotide conjugations [109]. Among the most important ones are disulfides, amide bonds and click chemistry (Figure 5). Disulfides offer linkages that are sufficiently stable for characterization, storage and handling, but are designed to be cleaved in the reductive environment in endosomes after successful cellular uptake [110]. The generation of disulfide linkages can be achieved by reaction of two free thiol groups – necessitating the removal of homodimeric byproducts – or by prior activation of one of the reacting thiols. Disulfide bridges are particularly attractive for peptide–oligonucleotide conjugates, for which a cysteine amino acid can be used as a handle. Thiol linkers are commercially available for attachment to oligonucleotides at the 5′-end during solid-phase synthesis, but can also be attached to the more widely used amino linkers. A common example is the use of thiol-amine hetero-bifunctional crosslinkers such as succinimidyl 3-(2-pyridyldithio)propionate [80].
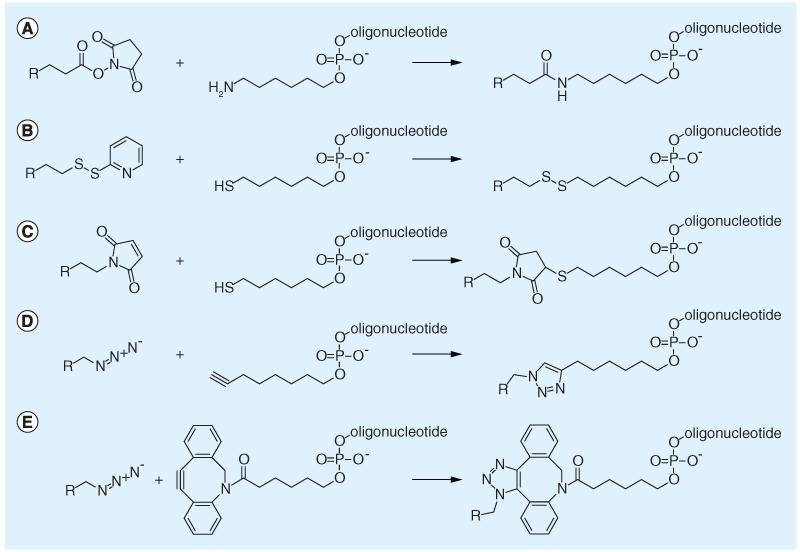
Figure 5. Ligand attachments to oligonucleotides.
(A) Amide coupling via active ester method. (B) Disulfide linkages with pyridyldithiol activated ligand. (C) Thiol-maleimide coupling resulting in stable covalent linkages. (D) Copper-catalyzed click chemistry by coupling an azide-modified linker to an alkyne-oligonucleotide. (E) Copper-free click chemistry using dibenzo-cyclooctyne-azide reaction, proceeding in aqueous buffers without catalysts.
Thiol functions can also be used for preparing maleimide-type linkages. This reaction, widely applied for labeling proteins, proceeds without a catalyst in aqueous buffers, and yields covalent stabile linkages [98].
Due to the highly specific, efficient and fast reaction time, click chemistry is an attractive methodology for the generation of bioconjugates. Azide–alkyne additions with or without copper catalysts can be carried out in aqueous buffers at room temperature [111]. Copper-based catalysts are difficult to remove after the reaction and due to their cytotoxicity are prone to influence biological assays. Copper-free click chemistry is available by using cyclooctyne-based educts, which react without catalysts with azides. Linkers for introducing an alkyne- or dibenzo-cyclooctyne group during oligonucleotide synthesis are commercially available, and many ligands can be easily derivatized for click chemistry reactions [96]. Azides and alkynes can be attached to terminal hydroxyl groups with phosphoramidite reagents or by coupling to amine linkers. Alkyne handles tethered to C5 of uridine can be employed for click reactions at the nucleobase. Procedures for introducing azido or propargyl anchors into proteins have been established [112], and can be achieved by metabolically engineering an unnatural amino acid, such as azidohomoalanine or homopropargylglycine into the protein sequence.
The linker length can be crucial for successful attachment of larger molecules to oligonucleotides. To avoid a negative steric impact, longer linkers often need to be used. Because alkyl chains are exceptionally lipophilic, poorly soluble in aqueous buffers and have a tendency for aggregation, PEG linkers are preferred for tethers exceeding about eight to ten atoms [113]. In addition to playing a role during conjugate preparations, linker structure and length can also influence the pharmacokinetic behavior.
Pharmacokinetics of oligonucleotides conjugates
Short and rigid linkers can impede efficient ligand binding to the receptor, especially in multivalent approaches. Biochemical data suggest that six to ten atom spacers are sufficient to allow binding of small-molecule ligands to several receptors on the cell surface [82,85]. Long PEG tethers not only improve the solubility and reaction yields, but can also increase the hydrodynamic size, in turn decreasing renal filtration rates, and shielding the conjugate from degrading enzymes and plasma–protein interactions [114].
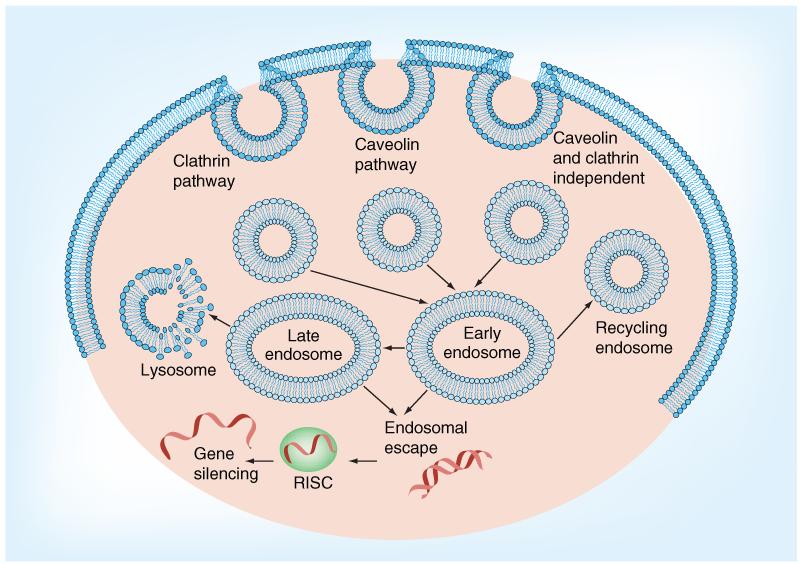
There is a general consensus that the bottle neck of cellular uptake of oligonucleotides is not the association to membrane components and initial uptake into endosomes, but rather the transition from those endomembrane compartments to the cytosol. The uptake mechanisms are of enormous complexity and we are only at the beginning of dissecting the distinct pathways (Figure 6) [36,115]. Most receptor-mediated uptake events including transferrin-, LDL-, and many G-protein coupled receptor-mediated endocytoses function via the clathrin-coated pit pathway [116]. Clathrin forms triskeletons of heterodimers, and assembles in a lattice-like structure. After receptor binding, the membrane is coated with clathrin and associated adaptor proteins followed by invagination and subsequent decoating of the vesicle [115]. Clathrin-coated pits are of between 100 and 200 nm in size. Another important pathway is caveolar-dependent uptake, often associated with protein toxins. Caveolar structures are generally smaller, below 100 nm, than other vesicles. For both clathrin- and caveolin-mediated uptake, dynamin is a key protein for scission of the newly formed vesicles. Several other clathrin- and caveolin-independent processes have been described recently, some of which seem to be independent of dynamin [117,118]. In addition, uptake can take place in the form of macropino-cytosis, and phagocytosis, restricted to certain cell types such as macrophages, granulocytes and Kupffer cells.
Figure 6. Uptake routes of oligonucleotides conjugates.
The different endocytotic vesicles all converge at the formation of early endosomes, which are transported along the cytoskeleton to the target organelle and are eventually fused with membranes to deliver the contents or mature into lysosomes, ending in degradation of the cargo.
Free unmodified oligonucleotides are not readily capable of passing cellular membranes. One of the most widely used antisense modification, phosphorothioates, induce cellular uptake through a slow and ineffective process [10,11,25]. The precise mechanisms is equally unclear as is the proposed existence of an oligonucleotides receptor [119]. By conjugating receptor ligands, the uptake process clearly shifts toward receptormediated endocytosis [120]. Consequently, uptake efficiency is mainly dependent on the choice and properties of the ligand, but also has to eventually rival unspecific uptake induced by the phosphorothioate backbone. Cholesterol, transferrin and anisamide, among others, bind to their respective receptor before being internalized by receptor-mediated endocytosis. For efficient binding, high affinity to the respective receptor is desirable. Multivalency is an easy strategy for improving affinity or exceeding a certain threshold. In that regard, multivalent linkers can be attached to the 5′- or 3′-end of oligonucleotides during solid-phase synthesis.
The relevance of the distinct uptake mechanisms for oligonucleotide delivery and the eventual possibilities for guidance uptake towards one or several of those are yet not well understood. There seem to be productive and unproductive pathways [120,121], that is, some result in a pharmacodynamic effect while others do not, despite cellular uptake, because of degradation in lysosomes. The exact mechanism of uptake is probably dependent on ligand, targeted receptor and tissue, and potentially on the chemical structure of the oligonucleotides. Some cargoes change the type of uptake dependent on the concentration. For example, EGF is internalized via clathrin-dependent endocytosis at low concentrations, but by a clathrin-independent mechanism in higher concentrations [122].
Most of the different endocytosis pathways seem to converge at the stage of early endosomes [36]. Subsequently, intracellular trafficking involves the formation of late and recycling endosomes, lysosomes, as well as the endoplasmatic reticulum and the golgi apparatus. After pinching off from the membrane and uncoating, the vesicle eventually wanders along actin- and tubulin-based cytoskeletal structures to its destination compartment, where it delivers its content by membrane fusion. These processes are regulated by Rab proteins [36]. Several reports have indicated enhanced activity of conjugates over naked oligonucleotides, despite only minor changes in cellular accumulation being found using fluorescently labeled cargoes [93,98,120]. These data indicate a principal dependence of the intracellular trafficking and ultimately the pharmacological result on the initial uptake mechanism. However, the underlying biological basics are not sufficiently understood for rational direction towards a productive uptake.
Mechanisms of transition from the initially formed vesicles (endosomal escape) to the cytosol or target compartment are generally inefficient. Strategies for enhancing endosomal escape include basic functionalities, which are supposed to lead to endosome destabilization by osmotic swelling through protonation in the acidic milieu (the proton sponge effect), and fusogenic peptides, which undergo conformational changes inducing fusion to the endosomal membrane [122]. The addition of respective components, either by chemical conjugation or complexation, is an approach for improving uptake efficiency of oligonucleotides conjugates. Amine-rich polymers or peptides induce the proton sponge effect by becoming protonated in the acidic endosomal environment. In addition to protons, chloride anions enter the endosome to maintain a neutral charge, followed by a volume increase caused by osmotic effects. Finally, the endosome ruptures, setting free its contents into the cytosol.
Release from bulky carriers is often necessary for endosomal escape and, particularly for siRNA agents, for induction of the biological effect through the cytosolic effector molecules [88,123]. Click chemistry and the formation of amide bonds, for example through the active ester method, yield covalent conjugates that will not be readily cleaved. For many small ligands, such as short peptides, cholesterol or defined PEG chains, this is not an impediment. The corresponding conjugates have been demonstrated to be active as antisense, splice-switching, or siRNA agents [82,94,96,98]. In covalent conjugates using larger ligands for which carrier release is beneficial, the use of disulfide linkages is thus particularly attractive, because of the reducing environment in the endosome [80,81,110]. Despite the oxidative environment, the presence of cysteine and glutathione in systemic circulation account for a certain extent of disulfide cleavage after prolonged circulation time. Consequently, a more stable linkage is preferred in cases where no dissociation from the carrier is necessary after successful intracellular uptake. Other approaches include the incorporation of linkages that are cleaved under mild acidic conditions, or peptide motifs such as furin, which are substrates for endosomal proteases [124]. Dynamic carrier systems are designed to be structurally modified into endosomolytic agents. Triggered by the acidic and reducing endosomal environment, ligands and masking groups are cleaved to produce polycationic polymers, which cause endosomal disruption. While this approach has resulted in increased gene silencing effects in model systems, the clinical benefit remains to be proven.
Contrasting delivery strategies in which the oligonucleotides are packaged into liposomes or nanoparticles, chemical conjugation alone does generally not protect from degradation by nucleases. Consequently, suitable chemical modifications need to be employed to ensure sufficient stability in biological fluids. For single-stranded antisense agents, the phosphorothioate backbone modification confers adequate stability, but may, in turn, mediate unspecific (plasma) protein binding, and result in a competing cell membrane permeation mechanism. Off-target effects have been demonstrated for phosphorothioates, undermining their therapeutic value [10,12,125]. Other possibilities to for shielding from degradation include 2′-modifications (2′-O-Me, 2′-O-MOE or LNAs). Antisense oligonucleotides with phosphodiester backbones and 2′-modifications seem to be the optimal choice for ligand conjugations. In the case of siRNA, the molecular effectors demonstrate lower tolerance for chemical modifications, aside from 2′-methylation, 2′-fluoridation and isolated phosphorothioate bonds. Respective compounds are usually used for the preparation of siRNA conjugates.
For adequate bioavailability, biologics need to be above the renal glomerular filtration limit to prevent rapid elimination from the bloodstream. In the absence of plasma protein binding, oligonucleotides, with a size of approximately <50 kDa, are rapidly lost in the urine [8,10]. Conjugates with small-molecule ligands usually fall below this limit. An increase in apparent mass can be achieved with PEGylation, or by using modifications that mediate plasma protein binding.
In conclusion, tethering ligands to antisense or siRNA oligonucleotides is a viable way to alter their biophysical properties and result in increased membrane permeation or specif receptor-mediated uptake. Compared with the more widely used encapsulation into liposomal or nanoparticulate delivery systems, they offer the advantage of being distinct molecules with defined structures, enabling analytical tech niques and standard methods of quality assurance. Additionally, problems associated with dispersion of particle size and composition, as well as batch variations are eliminated. However, despite recent progress, the extent of uptake mediated by the distinct ligand conjugation generally seems to be inferior to what has been achieved with lipid nanoparticles [126,127]. Further development and optimization are needed for ligand-specific oligonucleotides uptake to be applied in the therapeutic setting.
Future perspective
After several years of setbacks to gene-silencing technology, the prospect for therapeutic gene silencing mediated by oligonucleotides is currently looking brighter. Stimulated by the approval of mipomersen, and encouraging clinical results from splice-switching agents eteplirsen and drisapersen against DMD, as well as the preclinical advancement of siRNA agents, the enthusiasm for this class of biological has increased. The gained experience now seems to allow a fair assessment of the potential therapeutic value, and has highlighted the necessity for careful selection of molecular target and disease. The main challenges are identified and include improvement of pharmacokinetic behavior and increase of cellular uptake. Bioconjugates of oligonucleotides with ligands for enhancing uptake or for specific receptor binding are valuable means for tackling those challenges. The next few years will bring further preclinical and clinical evaluation of these compounds, finally leading to a clearer picture of their therapeutic potential. In particular, the GalNAc conjugate ALN–TTRsc is expected to be the first siRNA bioconjugate to be evaluated clinically for efficacy, pharmacokinetics and toxicity. If successful, its subcutaneous application could be an advantage over intravenous injections of stable nucleic acid lipid particle delivered siRNA and naked phosphorothioate antisense oligonucleotides. It seems unlikely that an effective oligonucleotide bioconjugate will reach the market in less than 5 years from now, and further clinical development will depend on the rivalry with other delivery systems, especially lipid nanoparticles and related advanced nanoparticle formulations.
For more efficient targeting beyond the liver, superior binding ligands will have to be developed. The current preclinical approaches primarily rely on small molecules and peptides. Ligands for specific high-affinity binding, such as antibody derivatives and alternative protein scaffolds, could enable tissue-selective delivery. A better understanding of uptake mechanisms is required to facilitate distinction between functional and nonfunctional uptake pathways and consequently design delivery systems with preference for effective intracytoplasmatic oligonucleotide delivery.
Executive summary.
Oligonucleotide conjugates for improving cellular uptake and specific targeting
-
■
Small-molecule ligands and short peptides are technically easy to attach to oligonucleotides, while the attachment of large proteins is more demanding.
-
■
Specific receptor binding is possible by folic acid, GalNAc, RGD peptide and antibody derivatives.
-
■
GalNAc conjugates show promise for subcutaneous application, and increase bioavailability in hepatocytes by binding to the asialoglycoprotein receptor.
Conjugation reactions and linkage types
-
■
Labile linkers, such as disulfide or those cleavable in weak acids, release the oligonucleotides cargo from their carrier after uptake in endosomes.
-
■
A variety of conjugation sites are available in oligonucleotides. The 3′- and 5′-hydroxy groups are preferred sites because of easy chemical conjugation, and minimal interference with oligonucleotides base pairing.
Pharmacokinetics
-
■
Reaching a minimal hydrodynamic size prevents rapid elimination. Choosing larger molecules or adding PEG chains can avoid renal filtration.
-
■
The relevance of different uptake mechanisms – clathrin- and caveolin-dependent, as well as clathrin- and caveolin-independent pathways – are not completely understood and not all are productive.
-
■
Endosomal escape is often the limiting factor for a successful gene silencing effect. Strategies to enhance functional uptake include cleavage in the endosome, and addition of endosomolytic agents.
Glossary
- Aptamers
Nucleic acids that bind specifically to molecular targets including proteins, other nucleic acids and small molecules. Aptamers are selected from libraries through repeat rounds of selection called systematic evolution of ligands by exponential enrichment.
- PEGylation
Attachment of PEG chains. A versatile technique to increase the molecular mass and hydrodynamic size of a drug or antibody. It reduces immunogenicity (often referred to as ‘stealth’ properties), prolongs circulation time by lowering renal filtration rates, and the amphiphilicity increases solubility of hydrophobic substances.
- Protein scaffolds
Polypeptide sequences of a structured core with variable domains, which account for modulation of specific binding to antigens. Alternative (non-IgG) scaffolds are attractive developments for scientific, diagnostic and therapeutic applications, and include anticalins, avimers, DARPins and monobodies (adnectins). Some of those scaffolds are extremely stable, and offer easy and site-specific derivatization opportunities.
- Dynamic polyconjugates
Macromolecules that undergo structural modification after application. Typically, disulfide and acid-labile linkages are cleaved after cellular uptake in the endosome, releasing cationic groups, which result in endosomal destabilization.
- Splice-switching oligonucleotide
Designed for inducing functional protein expression by influencing aberrant splicing. They block their corresponding sequence in pre-mRNA, and thus redirect mRNA splicing. In a therapeutic strategy, the splicing of dystrophin, which is expressed in a non-functional manner in Duchenne muscular dystrophy, is reconstituted to produce a truncated, but functional protein.
- Solid-phase synthesis
Technology for chemical synthesis of short nucleic acid sequences. The process has been highly optimized over several decades and is carried out in automated synthesizers on controlled pore glass resin, which facilitates reagent removal and purification.
Footnotes
Financial & competing interests disclosure
The work of the author is supported by the Austrian Science Fund (FWF): I519-B11 and the Johanna Mahlke Obermann foundation. The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 2.Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem. Biol. 2012;19(1):60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamamoto T, Nakatani M, Narukawa K, Obika S. Antisense drug discovery and development. Future Med. Chem. 2011;3(3):339–365. doi: 10.4155/fmc.11.2. [DOI] [PubMed] [Google Scholar]
- 4.Opar A. Exon-skipping drug pulls ahead in muscular dystrophy field. Nat. Med. 2012;18(9):1314. doi: 10.1038/nm0912-1314. [DOI] [PubMed] [Google Scholar]
- 5.Bauman J, Jearawiriyapaisarn N, Kole R. Therapeutic potential of splice-switching oligonucleotides. Oligonucleotides. 2009;19(1):1–13. doi: 10.1089/oli.2008.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Rao DD, Senzer N, Nemunaitis J. RNA interference and cancer therapy. Pharm. Res. 2011;28(12):2983–2995. doi: 10.1007/s11095-011-0604-5. [DOI] [PubMed] [Google Scholar]
- 7.Yeung ML, Jeang KT. MicroRNAs and cancer therapeutics. Pharm. Res. 2011;28(12):3043–3049. doi: 10.1007/s11095-011-0526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9(7):537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanagata N. Structure-dependent immunostimulatory effect of CpG oligodeoxynucleotides and their delivery system. Int. J. Nanomed. 2012;7:2181–2195. doi: 10.2147/IJN.S30197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dirin M, Winkler J. Influence of diverse chemical modifications on the ADME characteristics and toxicology of antisense oligonucleotides. Expert Opin. Biol. Ther. 2013;13(6):875–888. doi: 10.1517/14712598.2013.774366. [DOI] [PubMed] [Google Scholar]
- 11.Koller E, Vincent TM, Chappell A, De S, Manoharan M, Bennett CF. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39(11):4795–4807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stessl M, Noe CR, Winkler J. Off-target effects and safety aspects of phosphorothioate oligonucleotides. In: Erdmann VA, Barciszewski J, editors. From Nucleic Acids Sequences to Molecular Medicine, RNA Technologies. Springer Verlag; Berlin, Germany: 2012. pp. 67–83. [Google Scholar]
- 13.Gomes-da-Silva LC, Fonseca NA, Moura V, Pedroso de, Lima MC, Simões S, Moreira JN. Lipid-based nanoparticles for siRNA delivery in cancer therapy: paradigms and challenges. Acc. Chem. Res. 2012;45(7):1163–1171. doi: 10.1021/ar300048p. [DOI] [PubMed] [Google Scholar]
- 14.McGowan MP, Tardif J-C, Ceska R, et al. Randomized, placebo-controlled trial of mipomersen in patients with severe hypercholesterolemia receiving maximally tolerated lipid-lowering therapy. PLoS ONE. 2012;7(11):e49006. doi: 10.1371/journal.pone.0049006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelsinger C, Steinhagen-Thiessen E, Kassner U. Therapeutic potential of mipomersen in the management of familial hypercholesterolaemia. Drugs. 2012;72(11):1445–1455. doi: 10.2165/11635060-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 16.Parhofer KG. Mipomersen: evidence-based review of its potential in the treatment of homozygous and severe heterozygous familial hypercholesterolemia. Core Evid. 2012;7:29–38. doi: 10.2147/CE.S25239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visser ME, Wagener G, Baker BF, et al. Mipomersen, an apolipoprotein B synthesis inhibitor, lowers low-density lipoprotein cholesterol in high-risk statin-intolerant patients: a randomized, double-blind, placebo-controlled trial. Eur. Heart J. 2012;33(9):1142–1149. doi: 10.1093/eurheartj/ehs023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bedikian AY, Lebbé C, Robert C, et al. Survival in a Phase III, randomized, double-blind study of dacarbazine with or without oblimersen (Bcl-2 antisense) in patients with advanced melanoma and low-normal serum lactate dehydrogenase (LDH; AGENDA) J. Clin. Oncol. 2011;29(15 Suppl.):8531. [Google Scholar]
- 19■■.Goemans NM, Tulinius M, van den Akker JT, et al. Systemic administration of PRO051 in Duchenne’s muscular dystrophy. N. Engl. J. Med. 2011;364(16):1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]; Clinical results demonstrate dystrophin restoration and improvement of muscular function in Duchenne muscular dystrophy with drisapersen (PRO051).
- 20.Cirak S, Arechavala-Gomeza V, Guglieri M, et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, Phase 2, dose-escalation study. Lancet. 2011;378(9791):595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21■■.Cirak S, Feng L, Anthony K, et al. Restoration of the dystrophin-associated glycoprotein complex after exon skipping therapy in Duchenne muscular dystrophy. Mol. Ther. 2012;20(2):462–467. doi: 10.1038/mt.2011.248. [DOI] [PMC free article] [PubMed] [Google Scholar]; Eteplirsen, a phosphordiamidate morpholino oligomer, is well tolerated in Duchenne muscular dystrophy patients and induces dystrophin expression.
- 22.Noe CR, Winkler J, Urban E, Gilbert M, Haberhauer G, Brunar H. Zwitterionic oligonucleotides: a study on binding properties of 2′-O-aminohexyl modifications. Nucleosides Nucleotides Nucleic Acids. 2005;24(8):1167–1185. doi: 10.1081/NCN-200067400. [DOI] [PubMed] [Google Scholar]
- 23.Winkler J, Urban E, Noe CR. Oligonucleotides conjugated to short lysine chains. Bioconjug. Chem. 2005;16(4):1038–1044. doi: 10.1021/bc049729d. [DOI] [PubMed] [Google Scholar]
- 24.Winkler J, Gilbert M, Kocourková A, Stessl M, Noe CR. 2′-O-Lysylaminohexyl oligonucleotides: modifications for antisense and siRNA. ChemMedChem. 2008;3(1):102–110. doi: 10.1002/cmdc.200700169. [DOI] [PubMed] [Google Scholar]
- 25.Stein CA, Hansen JB, Lai J, et al. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2010;38(1):e3. doi: 10.1093/nar/gkp841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semple SC, Akinc A, Chen J, et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28(2):172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 27.Jayaraman M, Ansell SM, Mui BL, et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. Engl. 2011;51(34):8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Shum K-T, Burnett J, Rossi J. Nanoparticle-based delivery of RNAi therapeutics: progress and challenges. Pharmaceuticals. 2013;6(1):85–107. doi: 10.3390/ph6010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner E. Polymers for siRNA delivery: inspired by viruses to be targeted, dynamic, and precise. Acc. Chem. Res. 2011;45(7):1005–1013. doi: 10.1021/ar2002232. [DOI] [PubMed] [Google Scholar]
- 30.Winkler J. Nanomedicines based on recombinant fusion proteins for targeting therapeutic siRNA oligonucleotides. Ther. Deliv. 2011;2(7):891–905. doi: 10.4155/tde.11.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Straarup EM, Fisker N, Hedtjärn M, et al. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 2010;38(20):7100–7111. doi: 10.1093/nar/gkq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindow M, Kauppinen S. Discovering the first microRNA-targeted drug. J. Cell Biol. 2012;199(3):407–412. doi: 10.1083/jcb.201208082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44(5):619–626. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 34.Amann R, Fuchs BM. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 2008;6(5):339–348. doi: 10.1038/nrmicro1888. [DOI] [PubMed] [Google Scholar]
- 35.Juliano RL, Ming X, Nakagawa O. The chemistry and biology of oligonucleotide conjugates. Acc. Chem. Res. 2012;45(7):1067–1076. doi: 10.1021/ar2002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36■.Juliano RL, Ming X, Nakagawa O. Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjug. Chem. 2012;23(2):147–157. doi: 10.1021/bc200377d. [DOI] [PMC free article] [PubMed] [Google Scholar]; Describes the current knowledge of oligonucleotides uptake mechanisms depending on distinct chemical modifications and delivery systems.
- 37.Soutschek J, Akinc A, Bramlage B, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 38.Wolfrum C, Shi S, Jayaprakash KN, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007;25(10):1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 39.Wong SC, Klein JJ, Hamilton HL, et al. Co-injection of a targeted, reversibly masked endosomolytic polymer dramatically improves the efficacy of cholesterol-conjugated small interfering RNAs in vivo. Nucleic Acids Therap. 2012;22(6):380–390. doi: 10.1089/nat.2012.0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmermann TS, Lee AC, Akinc A, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441(7089):111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 41.Akinc A, Zumbuehl A, Goldberg M, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 2008;26(5):561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herbert B-S, Gellert GC, Hochreiter A, et al. Lipid modification of GRN163, an N3′→P5′ thio-phosphoramidate oligonucleotide, enhances the potency of telomerase inhibition. Oncogene. 2005;24(33):5262–5268. doi: 10.1038/sj.onc.1208760. [DOI] [PubMed] [Google Scholar]
- 43.Gryaznov SM. Oligonucleotide N3′→P5′ phosphoramidates and thio-phoshoramidates as potential therapeutic agents. Chem. Biodivers. 2010;7(3):477–493. doi: 10.1002/cbdv.200900187. [DOI] [PubMed] [Google Scholar]
- 44.Nishina K, Unno T, Uno Y, et al. Efficient in vivo delivery of siRNA to the liver by conjugation of alpha-tocopherol. Mol. Ther. 2008;16(4):734–740. doi: 10.1038/mt.2008.14. [DOI] [PubMed] [Google Scholar]
- 45.Uno Y, Piao W, Miyata K, Nishina K, Mizusawa H, Yokota T. High-density lipoprotein facilitates in vivo delivery of α-tocopherol-conjugated short-interfering RNA to the brain. Hum. Gene Ther. 2010;22(6):711–719. doi: 10.1089/hum.2010.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Said Hassane F, Saleh AF, Abes R, Gait MJ, Lebleu B. Cell penetrating peptides: overview and applications to the delivery of oligonucleotides. Cell. Mol. Life Sci. 2010;67(5):715–726. doi: 10.1007/s00018-009-0186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Berg A, Dowdy SF. Protein transduction domain delivery of therapeutic macromolecules. Curr. Opin. Biotechnol. 2011;22(6):888–893. doi: 10.1016/j.copbio.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 48.Eguchi A, Dowdy SF. siRNA delivery using peptide transduction domains. Trends Pharmacol. Sci. 2009;30(7):341–345. doi: 10.1016/j.tips.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 49.Gump JM, June RK, Dowdy SF. Revised role of glycosaminoglycans in TAT protein transduction domain-mediated cellular transduction. J. Biol. Chem. 2010;285(2):1500–1507. doi: 10.1074/jbc.M109.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindberg S, Copolovici DM, Langel Ü . Therapeutic delivery opportunities, obstacles and applications for cell-penetrating peptides. Ther. Deliv. 2011;2(1):71–82. doi: 10.4155/tde.10.78. [DOI] [PubMed] [Google Scholar]
- 51.Ivanova GD, Arzumanov A, Abes R, et al. Improved cell-penetrating peptide-PNA conjugates for splicing redirection in HeLa cells and exon skipping in mdx mouse muscle. Nucleic Acids Res. 2008;36(20):6418–6428. doi: 10.1093/nar/gkn671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abes R, Arzumanov AA, Moulton HM, et al. Cell-penetrating-peptide-based delivery of oligonucleotides: an overview. Biochem. Soc. Trans. 2007;35(Pt 4):775–779. doi: 10.1042/BST0350775. [DOI] [PubMed] [Google Scholar]
- 53.Moulton JD, Jiang S. Gene knockdowns in adult animals: PPMOs and vivo-morpholinos. Molecules. 2009;14(3):1304–1323. doi: 10.3390/molecules14031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moulton HM, Moulton JD. Morpholinos and their peptide conjugates: therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim. Biophys. Acta. 2010;1798(12):2296–2303. doi: 10.1016/j.bbamem.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 55.Yin H, Moulton HM, Seow Y, et al. Cell-penetrating peptide-conjugated antisense oligonucleotides restore systemic muscle and cardiac dystrophin expression and function. Hum. Mol. Genet. 2008;17(24):3909–3918. doi: 10.1093/hmg/ddn293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yin H, Moulton HM, Betts C, et al. A fusion peptide directs enhanced systemic dystrophin exon skipping and functional restoration in dystrophin-deficient mdx mice. Hum. Mol. Genet. 2009;18(22):4405–4414. doi: 10.1093/hmg/ddp395. [DOI] [PubMed] [Google Scholar]
- 57.Swenson DL, Warfield KL, Warren TK, et al. Chemical modifications of antisense morpholino oligomers enhance their efficacy against Ebola virus infection. Antimicrob. Agents Chemother. 2009;53(5):2089–2099. doi: 10.1128/AAC.00936-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pons B, Kotera M, Zuber G, Behr J-P. Online synthesis of diblock cationic oligonucleotides for enhanced hybridization to their complementary sequence. ChemBioChem. 2006;7(8):1173–1176. doi: 10.1002/cbic.200600178. [DOI] [PubMed] [Google Scholar]
- 59.Noir R, Kotera M, Pons B, Remy JS, Behr JP. Oligonucleotide-oligospermine conjugates (zip nucleic acids): a convenient means of finely tuning hybridization temperatures. J. Am. Chem. Soc. 2008;130(40):13500–13505. doi: 10.1021/ja804727a. [DOI] [PubMed] [Google Scholar]
- 60.Winkler J, Saadat K, Díaz Gavilán M, Urban E, Noe CR. Oligonucleotide polyamine conjugates: influence of length and position of 2′-attached polyamines on duplex stability and antisense effect. Eur. J. Med. Chem. 2009;44(2):670–677. doi: 10.1016/j.ejmech.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 61.Ng EWM, Shima DT, Calias P, Cunningham ET, Jr, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5(2):123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 62.Rosina C, Bottoni F, Staurenghi G. Clinical experience with pegaptanib sodium. Clin. Ophthalmol. 2008;2(3):485–488. doi: 10.2147/opth.s3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. PEG conjugated VEGF siRNA for anti-angiogenic gene therapy. J. Control. Release. 2006;116(2):123–129. doi: 10.1016/j.jconrel.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 64.Caravella J, Lugovskoy A. Design of next-generation protein therapeutics. Curr. Opin. Chem. Biol. 2010;14(4):520–528. doi: 10.1016/j.cbpa.2010.06.175. [DOI] [PubMed] [Google Scholar]
- 65.Cuesta ÁM, Sainz-Pastor N, Bonet J, Oliva B, Álvarez-Vallina L. Multivalent antibodies: when design surpasses evolution. Trends Biotechnol. 2010;28(7):355–362. doi: 10.1016/j.tibtech.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 66.Boersma YL, Plückthun A. DARPins and other repeat protein scaffolds: advances in engineering and applications. Curr. Opin. Biotechnol. 2011;22(6):849–857. doi: 10.1016/j.copbio.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 67.Song E, Zhu P, Lee SK, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 2005;23(6):709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 68.Yao YD, Sun TM, Huang SY, et al. Targeted delivery of PLK1-siRNA by ScFv suppresses Her2+ breast cancer growth and metastasis. Sci. Transl Med. 2012;4(130):130ra148. doi: 10.1126/scitranslmed.3003601. [DOI] [PubMed] [Google Scholar]
- 69.Winkler J, Martin-Killias P, Pluckthun A, Zangemeister-Wittke U. EpCAM-targeted delivery of nanocomplexed siRNA to tumor cells with designed ankyrin repeat proteins. Mol. Cancer Ther. 2009;8(9):2674–2683. doi: 10.1158/1535-7163.MCT-09-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xia C-F, Boado RJ, Pardridge WM. Antibody-mediated targeting of siRNA via the human insulin receptor using avidin–biotin technology. Mol. Pharm. 2008;6(3):747–751. doi: 10.1021/mp800194y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manjappa AS, Chaudhari KR, Venkataraju MP, et al. Antibody derivatization and conjugation strategies: application in preparation of stealth immunoliposome to target chemotherapeutics to tumor. J. Control. Release. 2011;150(1):2–22. doi: 10.1016/j.jconrel.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 72.Sawant R, Torchilin V. Challenges in development of targeted liposomal therapeutics. AAPS J. 2012;14(2):303–315. doi: 10.1208/s12248-012-9330-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He J, Liu G, Dou S, Gupta S, Rusckowski M, Hnatowich D. An improved method for covalently conjugating morpholino oligomers to antitumor antibodies. Bioconjug. Chem. 2007;18(3):983–988. doi: 10.1021/bc060208v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y, Cheng D, Liu X, et al. Comparing the intracellular fate of components within a noncovalent streptavidin nanoparticle with covalent conjugation. Nucl. Med. Biol. 2012;39(1):101–107. doi: 10.1016/j.nucmedbio.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 75.Ikeda Y, Taira K. Ligand-targeted delivery of therapeutic siRNA. Pharm. Res. 2006;23(8):1631–1640. doi: 10.1007/s11095-006-9001-x. [DOI] [PubMed] [Google Scholar]
- 76.Yu B, Zhao X, Lee LJ, Lee R. Targeted delivery systems for oligonucleotide therapeutics. AAPS J. 2009;11(1):195–203. doi: 10.1208/s12248-009-9096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wurch T, Pierré A, Depil S. Novel protein scaffolds as emerging therapeutic proteins: from discovery to clinical proof-of-concept. Trends Biotechnol. 2012;30(11):575–582. doi: 10.1016/j.tibtech.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 78.Tamaskovic R, Simon M, Stefan N, Schwill M, Plückthun A. Designed ankyrin repeat proteins (DARPins): from research to therapy. In: Wittrup KD, Gregory LV, editors. Methods Enzymol. Academic Press; MA, USA: 2012. pp. 101–134. [DOI] [PubMed] [Google Scholar]
- 79.Simon M, Zangemeister-Wittke U, Plückthun A. Facile double-functionalization of designed ankyrin repeat proteins using click and thiol chemistries. Bioconjug. Chem. 2012;23(2):279–286. doi: 10.1021/bc200591x. [DOI] [PubMed] [Google Scholar]
- 80.Uckun FM, Qazi S, Dibirdik I, Myers DE. Rational design of an immunoconjugate for selective knock-down of leukemia-specific E2A-PBX1 fusion gene expression in human Pre-B leukemia. Integr. Biol. 2013;5(1):122–132. doi: 10.1039/c2ib20114c. [DOI] [PubMed] [Google Scholar]
- 81.Dohmen C, Frohlich T, Lachelt U, et al. Defined folate–PEG–siRNA conjugates for receptor-specific gene silencing. Mol. Ther. Nucleic Acids. 2012;1:e7. doi: 10.1038/mtna.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakagawa O, Ming X, Huang L, Juliano RL. Targeted intracellular delivery of antisense oligonucleotides via conjugation with small-molecule ligands. J. Am. Chem. Soc. 2010;132(26):8848–8849. doi: 10.1021/ja102635c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Low PS, Henne WA, Doorneweerd DD. Discovery and development of folic-acid-based receptor targeting for imaging and therapy of cancer and inflammatory diseases. Acc. Chem. Res. 2007;41(1):120–129. doi: 10.1021/ar7000815. [DOI] [PubMed] [Google Scholar]
- 84.Thomas M, Kularatne SA, Qi L, et al. Ligand-targeted delivery of small interfering RNAs to malignant cells and tissues. Ann. NY Acad. Sci. 2009;1175:32–39. doi: 10.1111/j.1749-6632.2009.04977.x. [DOI] [PubMed] [Google Scholar]
- 85.Khorev O, Stokmaier D, Schwardt O, Cutting B, Ernst B. Trivalent, Gal/GalNAc-containing ligands designed for the asialoglycoprotein receptor. Bioorg. Med. Chem. 2008;16(9):5216–5231. doi: 10.1016/j.bmc.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 86.Akinc A, Querbes W, De S, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18(7):1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wakefield DH, Klein JJ, Wolff JA, Rozema DB. Membrane activity and transfection ability of amphipathic polycations as a function of alkyl group size. Bioconjug. Chem. 2005;16(5):1204–1208. doi: 10.1021/bc050067h. [DOI] [PubMed] [Google Scholar]
- 88.Rozema DB, Lewis DL, Wakefield DH, et al. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl Acad. Sci. USA. 2007;104(32):12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89■.Wooddell CI, Rozema DB, Hossbach M, et al. Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Mol. Ther. 2013;21(5):973–985. doi: 10.1038/mt.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]; Co-injection of cholesterol-siRNA and dynamic polymers with targeting and PEG ligands is efficient in mice against hepatitis B virus.
- 90.Li SD, Chono S, Huang L. Efficient oncogene silencing and metastasis inhibition via systemic delivery of siRNA. Mol. Ther. 2008;16(5):942–946. doi: 10.1038/mt.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Waite CL, Roth CM. PAMAM-RGD conjugates enhance siRNA delivery through a multicellular spheroid model of malignant glioma. Bioconjug. Chem. 2009;20(10):1908–1916. doi: 10.1021/bc900228m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dunehoo AL, Anderson M, Majumdar S, Kobayashi N, Berkland C, Siahaan TJ. Cell adhesion molecules for targeted drug delivery. J. Pharm. Sci. 2006;95(9):1856–1872. doi: 10.1002/jps.20676. [DOI] [PubMed] [Google Scholar]
- 93.Alam MR, Dixit V, Kang H, et al. Intracellular delivery of an anionic antisense oligonucleotide via receptor-mediated endocytosis. Nucleic Acids Res. 2008;36(8):2764–2776. doi: 10.1093/nar/gkn115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94■.Alam MR, Ming X, Fisher M, et al. Multivalent cyclic RGD conjugates for targeted delivery of small interfering RNA. Bioconjug. Chem. 2011;22(8):1673–1681. doi: 10.1021/bc200235q. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gene silencing effect of RGD conjugates is dependent on valency, and not correlated with uptake efficiency.
- 95.Glaser ST, Kaczocha M, Deutsch DG. Anandamide transport: a critical review. Life Sci. 2005;77(14):1584–1604. doi: 10.1016/j.lfs.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 96.Willibald J, Harder J, Sparrer K, Conzelmann K-K, Carell T. Click-modified anandamide siRNA enables delivery and gene silencing in neuronal and immune cells. J. Am. Chem. Soc. 2012;134(30):12330–12333. doi: 10.1021/ja303251f. [DOI] [PubMed] [Google Scholar]
- 97.Cesarone G, Edupuganti OP, Chen CP, Wickstrom E. Insulin receptor substrate 1 knockdown in human MCF7 ER+ breast cancer cells by nuclease-resistant IRS1 siRNA conjugated to a disulfide-bridged D-peptide analogue of insulin-like growth factor 1. Bioconjug. Chem. 2007;18(6):1831–1840. doi: 10.1021/bc070135v. [DOI] [PubMed] [Google Scholar]
- 98.Ming X, Alam MR, Fisher M, Yan Y, Chen X, Juliano RL. Intracellular delivery of an antisense oligonucleotide via endocytosis of a G protein-coupled receptor. Nucleic Acids Res. 2010;38(19):6567–6576. doi: 10.1093/nar/gkq534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mayer G. The chemical biology of aptamers. Angew. Chem. Int. Ed. Engl. 2009;48(15):2672–2689. doi: 10.1002/anie.200804643. [DOI] [PubMed] [Google Scholar]
- 100.Sefah K, Shangguan D, Xiong X, O’Donoghue MB, Tan W. Development of DNA aptamers using Cell-SELEX. Nat. Protocols. 2010;5(6):1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 101.McNamara JO, Andrechek ER, Wang Y, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24(8):1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 102.Dassie JP, Liu XY, Thomas GS, et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009;27(9):839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou J, Rossi J. Aptamer-targeted cell-specific RNA interference. Silence. 2010;1(1):4. doi: 10.1186/1758-907X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104■.Lonnberg H. Solid-phase synthesis of oligonucleotide conjugates useful for delivery and targeting of potential nucleic acid therapeutics. Bioconjug. Chem. 2009;20(6):1065–1094. doi: 10.1021/bc800406a. [DOI] [PubMed] [Google Scholar]; Comprehensive review of methods for online synthesis of oligonucleotides conjugates.
- 105.Rao M, Sockanathan S. Molecular mechanisms of RNAi: implications for development and disease. Birth Defects Res. C Embryo Today. 2005;75(1):28–42. doi: 10.1002/bdrc.20030. [DOI] [PubMed] [Google Scholar]
- 106.Lu K, Duan Q-P, Ma L, Zhao D-X. Chemical strategies for the synthesis of peptide–oligonucleotide conjugates. Bioconjug. Chem. 2009;21(2):187–202. doi: 10.1021/bc900158s. [DOI] [PubMed] [Google Scholar]
- 107.Winkler J, Urban E, Losert D, Wacheck V, Pehamberger H, Noe CR. A novel concept for ligand attachment to oligonucleotides via a 2′-succinyl linker. Nucleic Acids Res. 2004;32(2):710–718. doi: 10.1093/nar/gkh229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Winkler J, Giessrigl B, Novak C, Urban E, Noe CR. 2′-O-lysylaminohexyladenosine modified oligonucleotides. Monatsh. Chem. 2010;141(7):809–815. [Google Scholar]
- 109.Singh Y, Murat P, Defrancq E. Recent developments in oligonucleotide conjugation. Chem. Soc. Rev. 2010;39(6):2054–2070. doi: 10.1039/b911431a. [DOI] [PubMed] [Google Scholar]
- 110.Saito G, Swanson JA, Lee K-D. Drug delivery strategy utilizing conjugation via reversible disulfide linkages: role and site of cellular reducing activities. Adv. Drug Deliv. Rev. 2003;55(2):199–215. doi: 10.1016/s0169-409x(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 111.El-Sagheer AH, Brown T. Click chemistry with DNA. Chem. Soc. Rev. 2010;39(4):1388–1405. doi: 10.1039/b901971p. [DOI] [PubMed] [Google Scholar]
- 112.Dieterich DC, Hodas JJL, Gouzer G, et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat. Neurosci. 2010;13(7):897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jeffery DA, Bogyo M. Chemical proteomics and its application to drug discovery. Curr. Opin. Biotechnol. 2003;14(1):87–95. doi: 10.1016/s0958-1669(02)00010-1. [DOI] [PubMed] [Google Scholar]
- 114.Harris JM, Chess RB. Effect of pegylation on pharmaceuticals. Nat. Rev. Drug Discov. 2003;2(3):214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 115.Kumari S, Mg S, Mayor S. Endocytosis unplugged: multiple ways to enter the cell. Cell Res. 2010;20(3):256–275. doi: 10.1038/cr.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 117.Sandvig K, Pust S, Skotland T, van Deurs B. Clathrin-independent endocytosis: mechanisms and function. Curr. Opin. Cell Biol. 2011;23(4):413–420. doi: 10.1016/j.ceb.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 118.Mayor S, Pagano RE. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 2007;8(8):603–612. doi: 10.1038/nrm2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hanss B, Leal-Pinto E, Bruggeman LA, Copeland TD, Klotman PE. Identification and characterization of a cell membrane nucleic acid channel. Proc. Natl Acad. Sci USA. 1998;95(4):1921–1926. doi: 10.1073/pnas.95.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Alam MR, Ming X, Dixit V, Fisher M, Chen X, Juliano RL. The biological effect of an antisense oligonucleotide depends on its route of endocytosis and trafficking. Oligonucleotides. 2010;20(2):103–109. doi: 10.1089/oli.2009.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Geary RS, Wancewicz E, Matson J, et al. Effect of dose and plasma concentration on liver uptake and pharmacologic activity of a 2′-methoxyethyl modified chimeric antisense oligonucleotide targeting PTEN. Biochem. Pharmacol. 2009;78(3):284–291. doi: 10.1016/j.bcp.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 122.Sigismund S, Woelk T, Puri C, et al. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl Acad. Sci. USA. 2005;102(8):2760–2765. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J. Control. Release. 2011;151(3):220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 124.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005;7(5):657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 125.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J. Cell Sci. 2010;123(8):1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 126.Richards RM, Lowy DR, Schiller JT, Day PM. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc. Natl Acad. Sci. USA. 2006;103(5):1522–1527. doi: 10.1073/pnas.0508815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Winkler J, Stessl M, Amartey J, Noe CR. Off-target effects related to the phosphorothioate modification of nucleic acids. ChemMedChem. 2010;5(8):1344–1352. doi: 10.1002/cmdc.201000156. [DOI] [PubMed] [Google Scholar]
Website
- 201.Alnylam Pharmaceuticals. www.alnylam.com.